Abstract
The genotype Blastocystis hominis is highly polymorphic. Therefore, a genetic marker would be a powerful tool for the identification or classification of B. hominis subtypes and could be used as a means to resolve the transmission route or origin of the parasite. To this end, 32 B. hominis isolates were collected from patients and/or staff members of two long-term health care facilities (facilities A and B), and these organisms were subjected to genotype analysis based on diagnostic PCR primers and restriction fragment length polymorphism (RFLP) of small subunit rRNA gene (rDNA). Based on PCR amplification using diagnostic primers which were developed from randomly amplified polymorphic DNA analysis of known strains of B. hominis, the 32 isolates of B. hominis were classified into three different subtypes. Thirty isolates, including twenty-four that were isolated from patients and a staff member, from facility A and all isolates isolated from six patients from facility B showed the same genotype. Two of six patients of facility B had been transferred from facility A, and these two patients also had the same-genotype B. hominis that corresponded to 24 isolates from facility A. This genotype strain may have been transmitted by these two patients from facility A to facility B, suggesting human-to-human transmission. In contrast, 2 of 26 isolates from facility A showed distinct genotypes, suggesting that the colonization by these two isolates is attributable to another infectious route. These different subtypes were subjected to RFLP analysis, and the RFLP profiles were correlated with the results obtained by diagnostic PCR primers. This study presents the first molecular evidence of possible human-to-human B. hominis infection between and/or among two small communities.
Since Blastocystis hominis was originally found in stool samples of humans as a harmless yeast in 1912 (3), the organism was left unstudied for more than a half-century (1, 10, 16). Although many recent studies characterize this organism as an intestinal parasite, there were many conflicting reports on its pathogenicity as a causative agent of diarrhea (1, 10). Current thought describes B. hominis as organisms that have been isolated from humans. However, many B. hominis-like organisms have been isolated from a wide range of animals, such as nonhuman primates, as well as rodents, birds, amphibians, reptiles, and insects (1, 14, 15). Although most of these organisms were indistinguishable from B. hominis based on light and electron microscopy, genetic diversity among B. hominis strains has been demonstrated (2, 6, 12). Therefore, it is important to develop a precise method for identifying or classifying B. hominis.
Recently, molecular biological approaches were attempted to subcategorize B. hominis strains based on amplification with the diagnostic PCR primers or on the analysis of restriction fragment length polymorphism (RFLP) of the small subunit (SSU) rDNA (4, 13). These studies successfully classified subtypes or subgroups among B. hominis strains and also revealed the presence of zoonotic strains. Use of SSU rRNA gene (rDNA)-RFLP was recently termed as riboprinting, and groups with the same riboprint pattern were designated ribodemes (4). Since the transmission route of B. hominis was not exclusively identified (10), molecular epidemiology patterns in local communities will be important in elucidating the transmission route or origins of infection and the usefulness of the diagnostic PCR primers. However, most epidemiological studies report the prevalence of B. hominis infection among communities and estimate the pathogenicity symptomatically (10). Currently, no systematic genetic study to compare B. hominis populations based on their genetic markers have been reported. This study was undertaken to examine genotypes among B. hominis strains isolated from two long-term health care facilities by using the PCR diagnostic primers and riboprinting. Using the combined methods, B. hominis strains were subclassified. The results revealed genetic diversity among B. hominis strains and allowed for the proposal of human-to-human transmission of B. hominis infection between two small communities.
MATERIALS AND METHODS
Sources and culture of B. hominis.
A total of 26 isolates were used from patients and two staff members of health care facility A in Osaka city in 1996. Since B. hominis infection was confirmed in 29 patients and 2 staff members of facility A by fecal examination, these isolates were designated HJ96A-1 to A-29 and as HJAS-1 and HJAS-2 since they were isolated from patients and staff members, respectively. However, several strains were uncultivatable; therefore, only 24 strains isolated from patients and 2 strains isolated from staff members of facility A were used in this study. Six isolates were isolated from patients of another facility, facility B, in Osaka City at the same time. These six isolates were designated HJ96B-1 to B-6. A total of 32 isolates were successfully cultured in diphasic agar slant medium or whole-egg slant medium at 37°C as described previously (11).
Control subtypes of B. hominis organisms were used in this study, namely, Nand II and HE87-1 strains were used as subtype 1, B strain was used as subtype-2, and HV93-13 strain was used as another subtype (13).
Genomic DNA preparation.
Genomic DNA of B. hominis was extracted by using DNAzol reagent (Gibco BRL/Life Technologies, Inc., Grand Island, N.Y.) according the manufacturer's protocol.
PCR amplification.
The PCR conditions for random amplified polymorphic DNA (RAPD) and conventional PCR with diagnostic primers were as described previously (12, 13). The PCR products and a size marker of a 100-bp ladder (Pharmacia Biotech, Uppsala, Sweden) were electrophoresed in 1.5% agarose gels and Tris-borate buffer using a Mupid gel electrophoresis (Advance).
Development of diagnostic primers.
Diagnostic PCR primer sets SB82, SB83, and SB155 were as described previously (13) (Table 1). The diagnostic PCR primer sets SB227, SB228, SB229, and SB332 were developed from unique bands of RAPD fragments amplified with B. hominis DNA by using arbitrary 10-base primer PCR as described previously (13). Briefly, sample DNA fragments of RAPD products were separated from agarose gel using an Ultrafree-C3 HV unit (Nihon Milipore, Ltd.) and amplified again with the same RAPD-PCR conditions. DNA fragments were purified by using a Geneclean II Kit (Bio 101, Inc., Buena Vista, Calif.) and then ligated into a pGEM-T plasmid vector by using the pGEM-T vector system (Promega Corp., Madison, Wis.). The recombinant plasmids were introduced into competent cells of Escherichia coli JM109. The plasmid DNA was isolated from E. coli by using a FlexiPrep Kit (Pharmacia Biotech) and subjected to Taq cycle sequencing reactions with a Dye Primer Cycle Sequencing Kit (Perkin-Elmer Co., Newark, N.J.) using −21M13 forward and M13 reverse primers. The products were sequenced using an automatic sequencer (model 373A; Applied Biosystems). Based on the DNA sequences, the two primer sets SB227 and SB228 were designed from the sequence of HV93-13 strain by using Oligo 4 · 04 software (Table 1). These two diagnostic PCR primers produced 526 and 473 bp, respectively. Two different primer sets, SB229 and SB332, were designed from the sequence data of HJ96A-26 and HJ96AS-1 strains, respectively, isolated from facility A. These PCR primers yielded 631- and 338-bp products, respectively (Table 1). The GenBank accession numbers of sequence data used for primer development are summarized in Table 1.
TABLE 1.
Diagnostic primer sets used in this study
| Primer set | Product size (bp) | Sequence of forward (F) and reverse (R) primers (5′ to 3′) | GenBank accession no. |
|---|---|---|---|
| SB82a | 462 | F:TCTTGCTTCATCGGAGTC | AF166085 |
| R:CCTTCTCGCAGTTCTTTATC | |||
| SB83a | 351 | F:GAAGGACTCTCTGACGATGA | AF166086 |
| R:GTCCAAATGAAAGGCAGC | |||
| SB155a | 650 | F:ATCAGCCTACAATCTCCTC | AF166087 |
| R:ATCGCCACTTCTCCAAT | |||
| SB227 | 526 | F:TAGGATTTGGTGTTTGGAGA | AF166088 |
| R:TTAGAAGTGAAGGAGATGGAAG | |||
| SB228 | 473 | F:GACTCCAGAAACTCGCAGAC | AF166089 |
| R:TCTTGTTTCCCCAGTTATCC | |||
| SB229 | 631 | F:CACTGTGTCGTCATTGTTTTG | AF166090 |
| R:AGGGCTGCATAATAGAGTGG | |||
| SB332 | 338 | F:GCATCCAGACTACTATCAACATT | AF166091 |
| R:CCATTTTCAGACAACCACTTA |
These primers were as reported previously (11).
RFLP analysis of SSU rDNA.
Two restriction enzymes, Hinf I and RsaI, had been reported to successfully classify seven patterns of RFLP of amplified SSU rDNA among 30 B. hominis strains (4). Therefore, these two enzymes were used for analysis of RFLP of SSU rDNA in this study. The SSU rDNAs were amplified by using the forward primer SR1F (5′-GCTTATCTGGTTGATCCTGCCAGTAGT-3′) and the reverse primer SR1R (5′-TGATCCTTCCGCAGGTTCACCTA-3′), which were designed to prime conserved region of SSU rRNA sequence obtained from three strains: B. hominis Nand II (GenBank U51151) (9), Blastocystis sp. (GenBank U51152) (9), and B. hominis HE87-1 (GenBank AB023578) (T. Hashimoto, unpublished data). These primer pairs produced an approximately 1,780-bp product. The PCR amplification was performed with 35 cycles of 94°C for 40 s, 57°C for 60 s, and 72°C for 2 min, after an initial denaturation at 94°C for 3 min. The PCR products were purified with a Geneclean II Kit (Bio 101). Then the purified DNA were digested with HinfI or RsaI in a reaction mixture of 2 μl of 10 × buffer, 0.2 μl of bovine serum albumin solution (10 mg/ml), 0.5 μl (5 U) of restriction endonuclease (Promega Corp.), 8 μl of DNA solution, and 9.3 μl of distilled water to a final volume of 20 μl at 37°C for 3 h. The digested products were electrophoresed with size marker of a 100-bp ladder (New England BioLabs, Inc.) in 2.0% agarose gels and Tris-borate buffer. Gels were stained with ethidium bromide, and the approximate sizes of the band profiles were estimated from the photographs.
RESULTS
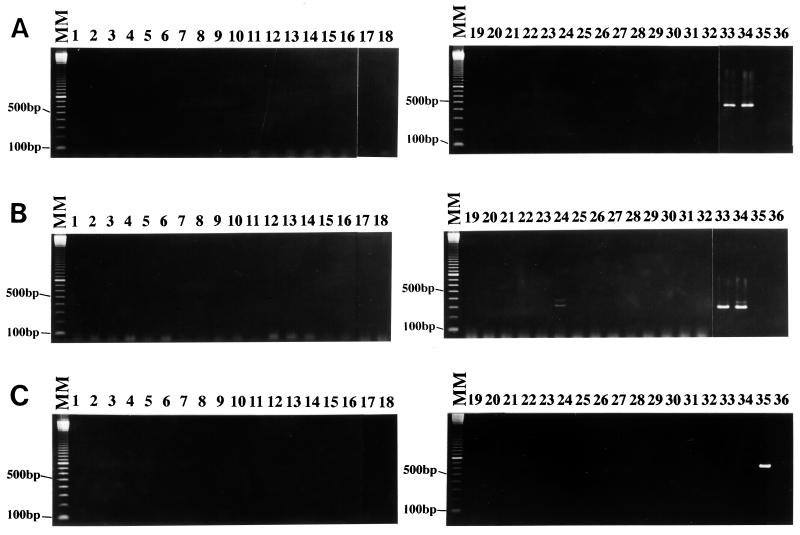
The diagnostic PCR primer sets SB82 and SB83 had been designed from sequence data of unique bands of RAPD-PCR products of the reference strain of B. hominis Nand II strain (13). Since Nand II, HE87-1, and CK86-1 strains were amplified by these primers, these three strains had been subclassified as subtype 1 (13). Genomic DNA isolated from samples of the patients and/or staff members of two health care facilities was subjected to amplification with the diagnostic primers SB82 and SB83. All 32 isolates were negative with the primer set SB82 (Fig. 1A), while only HJ96A-29 isolate produced two faint bands, of approximately 350 and 400 bp (Fig. 1B, lane 24). Since SB83 primer amplified 351 bp with Nand II strain (13), the former band was the target size. These results indicate the partial genomic homology between Nand II and HJ96A-29 isolate. Therefore, this isolate was subclassified as a variant of subtype 1 (Table 2).
FIG. 1.
The specificity of diagnostic primer sets SB82 (A), SB83 (B), and SB155 (C) with 32 isolates from patients and staff members of two facilities (lanes 1 to 32) and four control strains (lanes 33 to 36) of B. hominis. Strains Nand II (lane 33) and HE87-1 (lane 34) were amplified with primer sets SB82 (462 bp) and SB83 (351 bp), while HJ96A-29 (lane 24) showed two minor bands (350 and 400 bp) with primer set SB83. With primer set SB155, strain B (lane 35) was only amplified and showed a single band of 650 bp. Lanes: MM, molecular marker of a 100-bp ladder; 1, HJ96A-1; 2, HJ96A-2; 3, HJ96A-3; 4, HJ96A-4; 5, HJ96A-5; 6, HJ96A-6; 7, HJ96A-7; 8, HJ96A-10; 9, HJ96A-13; 10, HJ96A-14; 11, HJ96A-15; 12, HJ96A-16; 13, HJ96A-17; 14, HJ96A-18; 15, HJ96A-19; 16, HJ96A-20; 17, HJ96A-22; 18, HJ96A-23; 19, HJ96A-24; 20, HJ96A-25; 21, HJ96A-26; 22, HJ96A-27; 23, HJ96A-28; 24, HJ96A-29; 25, HJ96AS-1; 26, HJ96AS-2; 27, HJ96B-1;28, HJ96B-2; 29, HJ96B-3; 30, HJ96B-4; 31, HJ96B-5; 32, HJ96B-6; 33, Nand II; 34, HE87-1; 35, B; 36, HV93-13.
TABLE 2.
Classification of subtypes of B. hominis strains based on the diagnostic PCR primers
| Group | Strain(s) | Primer set(s) |
|---|---|---|
| Subtype 1a | Nand II, HE87-1, CK86-1, HJ96A-29b | SB82, SB83 |
| Subtype 2a | B, C, E, G, H | SB155 |
| Subtype 3 | HV93-13, HJ96A-1, HJ96A-2, HJ96A-3, HJ96A-4, HJ96A-5, HJ96A-6, HJ96A-7, HJ96A-10, HJ96A-13, HJ96A-14, HJ96A-15, HJ96A-16, HJ96A-17, HJ96A-18, HJ96A-19, HJ96A-20, HJ96A-22, HJ96A-23, HJ96A-24, HJ96A-25, HJ96A-26, HJ96A-27, HJ96A-28, HJ96AS-2, HJ96B-1, HJ96B-2, HJ96B-3, HJ96B-4, HJ96B-5, HJ96B-6 | SB227, SB228, SB229 |
| Subtype 4 | HJ96AS-1 | SB332 |
These subtypes were as reported previously (11).
Since HJ96A-29 strain was only amplified by using primer SB83, it was classified as a variant of subtype 1.
The diagnostic PCR primer set SB155 had been developed from B. hominis B strain, and this primer had amplified 650-bp products of five B. hominis strains isolated in Singapore; these five strains had been designated as B. hominis subtype 2 (13). When all 32 isolates isolated from both facilities A and B were screened with the diagnostic primer set SB155, no strain was amplified except the positive control (Fig. 1C).
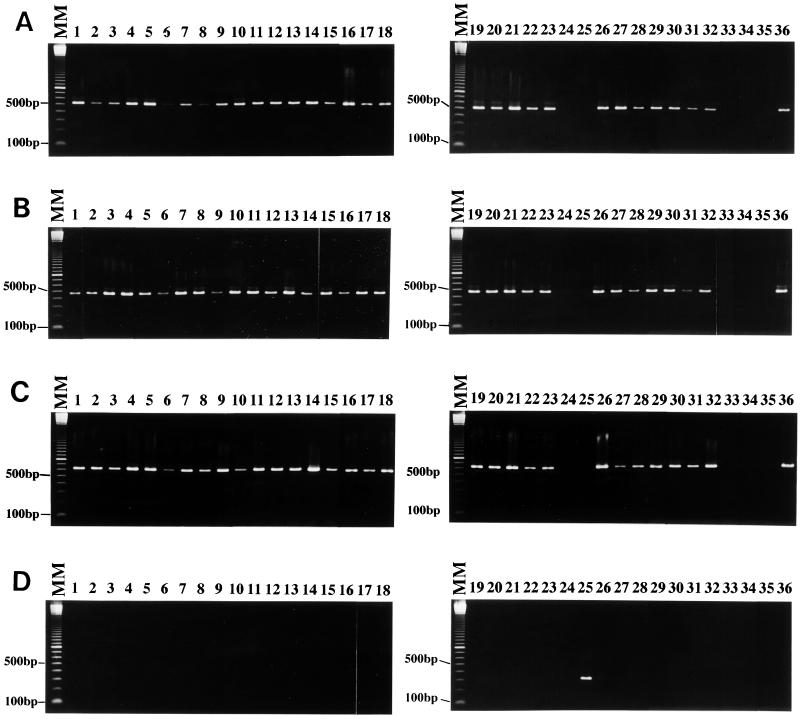
Two diagnostic PCR primer sets, SB227 and SB228, were developed from the HV93-13 strain. This strain had been confirmed as another subtype (13), but no diagnostic primer was available at this time. These two primer sets successfully amplified 24 isolates of facility A and all 6 isolates of facility B (Fig. 2A and B). On the other hand, HJ96A-29 (lane 24) and HJ96AS-1 (lane 25) isolates of facility A were not amplified by primer sets SB227 and SB228, and also the other subtype Nand II (lane 33), HE87-1 (lane 34), and B (lane 35) strains were not amplified.
FIG. 2.
The specificity of diagnostic primer sets SB227 (A), SB228 (B), SB229 (C), and SB332 (D). The molecular size marker and sample for each lane were as described in the legend of Fig. 1. Of 32 isolates from patients and staff members of two facilities, 30 isolates (lanes 1 to 23 and 26 to 32) and strain HV93-13 (lane 36) showed a single band with primer sets SB227 (526 bp), SB228 (473 bp), and SB229 (631 bp). Isolates HJ96A-29 (lane 24) and HJ96AS-1 (lane 25) and strains Nand II (lane 33), HE87-1 (lane 34), and B (lane 35) were not amplified with these three primer sets. HJ96AS-1 was only amplified with primer set SB332 (panel D, lane 25) and showed a single band of 338 bp.
In order to demonstrate genotype homology between HV93-13 strain and all 30 isolates amplified by both SB227 and SB228, another diagnostic primer set SB229 was developed from the HJ96A-26 isolate of facility A. As expected, the SB229 primer set amplified all 30 isolates (HJ96A-1, HJ96A-2, HJ96A-3, HJ96A-4, HJ96A-5, HJ96A-6, HJ96A-7, HJ96A-10, HJ96A-13, HJ96A-14, HJ96A-15, HJ96A-16, HJ96A-17, HJ96A-18, HJ96A-19, HJ96A-20, HJ96A-22, HJ96A-23, HJ96A-24, HJ96A-25, HJ96A-26, HJ96A-27, HJ96A-28, HJ96AS-2, HJ96B-1, HJ96B-2, HJ96B-3, HJ96B-4, HJ96B-5, and HJ96B-6) and HV93-13 strain (Fig. 2C). Since HJ96AS-1 isolate was not amplified with any diagnostic PCR primer sets, an additional new diagnostic primer, SB332, was developed from this isolate. Primer SB332 amplified only the HJ96AS-1 isolate (Fig. 2D, lane 25). These primers also did not amplify the Nand II, HE87-1, B, and HV93-13 strains (lanes 33 to 36). Based on the amplification of the diagnostic PCR primers, 32 isolates isolated from two facilities were classified into three groups of variants of subtypes 1, 3, and 4 (Table 2). No isolates were classified as subtype 2. Based on the diagnostic PCR primer sets, 32 isolates obtained from two long-term health care facilities were classified into three groups of subtypes (Table 3).
TABLE 3.
Subtype classification among 32 strains isolated from two long-term health care facilities based on the diagnostic PCR primer sets
| Group | No. of strains (%)
|
Total (%) | |
|---|---|---|---|
| Facility A | Facility B | ||
| Subtype 1 | 1 (3.8) | 0 | 1 (3.1) |
| Subtype 2 | 0 | 0 | 0 |
| Subtype 3 | 24 (92.3) | 6 (100) | 30 (93.8) |
| Subtype 4 | 1 (3.8) | 0 | 1 (3.1) |
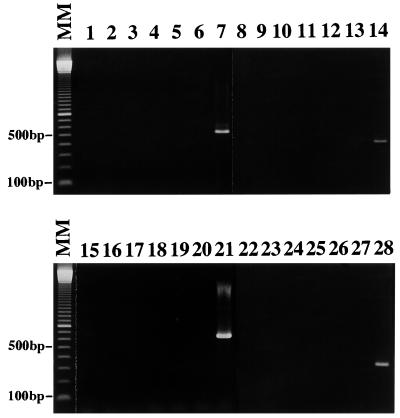
The specificity of the newly developed diagnostic primer sets SB227, SB228, SB229, and SB332 were tested by using some intestinal parasites and a yeast, as described previously (13). No PCR products were observed in any of these genomic DNAs isolated from Entamoeba histolytica, Entamoeba moshkovskii, Giardia intestinalis, Cryptosporidium muris, C. parvum, and Saccharomyces cerevisiae, except for the positive controls (Fig. 3). A GenBank search using these four sequences yielded no significant homology to the functional genes.
FIG. 3.
The specificity of four diagnostic primers was tested against other intestinal protozoa and a yeast. Lanes 1 to 7 were amplified by using primer sets SB227, lanes 8 to 14 were amplified with SB228, lanes 15 to 21 were amplified with SB229, and lanes 22 to 28 were amplified with SB332. Only positive controls (lanes 7, 14, 21, and 28) showed a single band. Lanes: MM, molecular marker of a 100-bp ladder; 1, 8, 15, and 22, E. histolytica; 2, 9, 16, and 23, E. moshkovskii; 3, 10, 17, and 24, G. intestinalis; 4, 11, 18, and 25, C. muris; 5, 12, 19, and 26, C. parvum; 6, 13, 20, and 27, S. cerevisiae; 7 and 14, HV93-13; 21, HV96A-26; 28, HJ96AS-1.
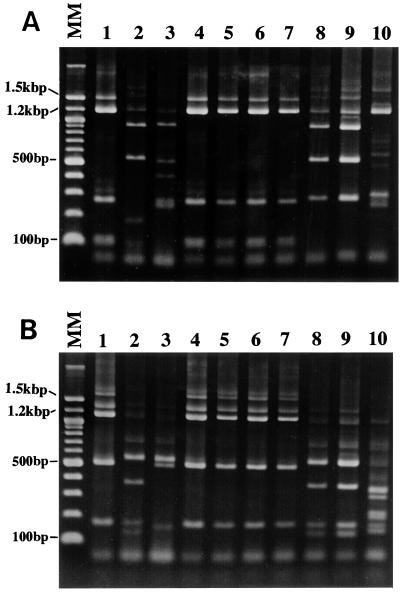
A total of 10 Blastocystis isolates were picked from different subtypes to analyze the RFLP pattern when digested with HinfI and RsaI restriction enzymes. Strains HJ96A-26, HJ96A-29, and HJ96AS-1 were selected from facility A because two of the three strains were the sources of diagnostic primer sets SB229 and SB332, and these three strains were classified as subtype 3, a variant of subtype 1, and subtype 4, respectively. For the evaluation of human-to-human transmission, three HJ96B-1, HJ96B-4, and HJ96B-6 strains were selected from facility B because the latter two strains were isolated from patients who had been transferred from facility A. In addition, all 6 strains isolated from facility B were classified as subtype 3 and were identical to 24 strains from facility A. For the controls, four strains, Nand II, HE87-1, B, and HV93-13, were used for the evaluation of the RFLP pattern. The SSU rRNA of the two former strains had been sequenced (GenBank U51151 and AB023578) (9) and corresponded to subtype 1 based on the primer classification (13). The latter B and HV93-13 strains were sources of the diagnostic PCR primer sets SB155, SB227, and SB228 and were classified as subtype 2 (13) and subtype 3, respectively. These 10 strains were compared by RFLP profile after digestion with HinfI and RsaI enzymes.
HinfI restriction enzyme produced five kinds of patterns (Fig. 4A), while the RsaI restriction product showed four kinds of patterns (Fig. 4B). The RFLP pattern of the HE87-1 strain was identical to that of the Nand II strain with both HinfI and RsaI enzymes, while the pattern of HJ96A-29 strain was different from the Nand II and HE87-1 strains with the HinfI enzyme. Since the Nand II strain was classified into the group of ribodeme 1 (4), HE87-1 strain was also classified into the ribodeme 1 group in this study. However, the RFLP pattern of strain HJ96A-29 digested with HinfI did not fit any previous RFLP patterns of ribodemes 1 to 7 (4). Therefore, HJ96A-29 was classified into a new group, i.e., ribodeme 8 (Table 4). According to the banding patterns, strains HJ96B-1, HJ96B-4, HJ96B-6, and HV93-13 were coincident with ribodeme 2. In contrast, the HJ96AS-1 and B strains showed quite different riboprinting patterns compared to ribodemes 1 to 7. Therefore, the HJ96AS-1 and B strains were classified into the ribodeme 9 and ribodeme 10 groups, respectively (Table 4).
FIG. 4.
Restriction enzyme profiles of SSU rDNA digested with HinfI (A) and RsaI (B). The HinfI restriction enzyme produced five patterns, while the RsaI restriction product showed four kinds of patterns. Strains HJ96A-26, HJ96B-1, HJ96B-4, HJ96B-6, and HV93-13 showed the same RFLP pattern with both HinfI and RsaI enzymes. HJ96A-29 strain showed the same RFLP pattern with Nand II and HE87-1 strains with RsaI enzyme, while with HinfI enzyme the HJ96A-29 strain showed two bands of 180 and 100 bp instead of a band of 280 bp as with the Nand II and HE87-1 strains. Strains HJ96AS-1 and B also showed distinct banding patterns. Lanes: MM, molecular marker of a 100-bp ladder; 1, HJ96A-26; 2, HJ96A-29; 3, HJ96AS-1; 4, HJ96B-1; 5, HJ96B-4; 6, HJ96B-6; 7, HV93-13; 8, Nand II; 9, HE87-1; 10, B strain.
TABLE 4.
Correlation of genetic diversity between subtypes and ribodeme classification with special references of the base-pair size of the RFLP pattern of SSU rDNA restricted by HinfI and RsaI enzymes
| Subtype | Ribodeme type | Strains tested | Banding pattern of RFLP of SSU rDNA (bp)
|
|
|---|---|---|---|---|
| HinfI | RsaI | |||
| 1 | 1 | Nand II, HE87-1 | 900, 530, 280, 70 | 550, 380, 170, 130, 60 |
| 1 variant | 8 | HJ96A-29 | 900, 530, 180, 100, 70 | 550, 380, 170, 130, 60 |
| 2 | 10 | B | 1,200, 600, 290, 250, 70 | 360, 320, 230, 200, 170, 150, 60 |
| 3 | 2 | HV93-13, HJ96A-26, HJ96B-1, HJ96B-4, HJ96B-6 | 1,500, 1,200, 270, 100, 70 | 1,300, 1,150, 510, 170, 60 |
| 4 | 9 | HJ96AS-1 | 900, 530, 400, 270, 250, 70 | 550, 510, 150, 60 |
The approximate band sizes are listed in Table 4 with the subtype reference and ribodeme classifications. It is evident that the subtype classification correlated with the ribodeme classification based on the difference of the RFLP profiles of SSU rDNA.
DISCUSSION
Identification of B. hominis in humans has been traditionally accomplished by analysis of fecal samples or cultured organisms. However, it was evident that a great deal of morphological variation occurred in the organisms (1, 10). Recently, the genetic diversity among B. hominis strains has been ascertained by using different techniques (2, 4, 6, 12). Although at present the name Blastocystis hominis is used for strains isolated from humans, many B. hominis-like organisms were isolated from a wide variety of animals (1, 14, 15). Since these organisms were indistinguishable from B. hominis by morphological criteria, it is difficult to judge whether B. hominis-like organisms isolated from various animals are identical to or different from B. hominis. Although the transmission route of B. hominis has not been conclusively determined, the spread of infection between family members and institutionalized patients and in communities without adequate sanitary facilities has been reported (5, 7, 8). Therefore, it has been assumed that B. hominis is transmitted by the fecal-oral route in the same manner as common gastrointestinal protozoa (10). However, systematic studies of the possibility of human-to-human transmission of B. hominis infection have not yet been carried out. In addition, the presence of zoonotic strains isolated from a chicken and a guinea pig shows a human-to-animal transmission route (4, 12, 13). Recently, we developed PCR-based methodology to classify or identify B. hominis intraspecific variations by PCR primers (12, 13). This technique successfully subcategorized three subtypes among nine B. hominis strains isolated from different geographical areas by using the diagnostic primers. In addition, this method revealed the effectiveness of identifying zoonotic strains of nonhuman origin. These studies reveal that B. hominis populations are highly genotypically polymorphic.
In the present study, 32 strains isolated from the patients and/or staff members of two long-term health care facilities were tested with three diagnostic PCR primer sets that successfully classified the strains into two groups of subtypes (13). However, only HJ96A-29 strain was amplified with the SB83 primer set, while it was negative with the SB82 primer set. Since both primer sets were developed from the specific sequence of the Nand II strain, the HJ96A-29 strain was designated as a variant of subtype 1 (Table 2). Therefore, additional diagnostic PCR primers were developed to classify more genotypes among the newly isolated strains. Interestingly, two new diagnostic primer sets (SB227 and SB228) developed from the HV93-13 strain, amplified 30 of 32 strains isolated from the patients and a staff member of two health care facilities. Strain HV93-13 was isolated from a patient living in Tokyo, Japan, in 1993 and was confirmed a variant subtype of subtype 1 and subtype 2 (13). An additional PCR primer set (SB229) was constructed from strain HJ96A-26 for the confirmation of genomic homology between HV93-13 strain and the 30 strains that could be amplified with SB227 and SB228. As expected, the primer set SB229 amplified all 30 strains and the HV93-13 strain. Therefore, a total of 30 strains isolated in facilities A and B amplified by three diagnostic primer sets, SB227, SB228, and SB229, were classified as subtype 3 among the B. hominis population with the HV93-13 strain (Table 2). Since strain HJ96AS-1 was not amplified by these three primer sets, an additional diagnostic PCR primer set (SB332) was developed from the HJ96AS-1 strain. This diagnostic primer set only amplified the primer source strain and did not amplify all of the other strains, including the Nand II, HE87-1, B, and HV93-13 strains. Therefore, strain HJ96AS-1 was classified as subtype 4 among the B. hominis population. Based on the present results and our previous report, the studied strains of B. hominis were classified into four subtypes based on the amplification with the diagnostic PCR primers, as shown in Table 2.
In this study, it is evident that 24 strains isolated from facility A and all 6 strains isolated from facility B were identical at the genomic level to strain HV93-13, which had been isolated from a Vietnamese patient in Tokyo, Japan (Tables 2 and 3). Although, it is difficult to ascertain whether the origin of the HV93-13 strain is Vietnamese or Japanese, it is of interest that the results show that diagnostic primer sets SB227, SB228, and SB229 only amplified the strains isolated from Japanese people (H. Yoshikawa, unpublished data). All six strains isolated from the patients of facility B were identical to the B. hominis subtype 3 (Tables 2 and 3), which constituted the majority of strains of facility A (24 of 26 [92.3%]). Therefore, B. hominis infection at facility B may have been transmitted by two patients who had previously resided in facility A. On the other hand, one patient and a staff member of facility A might have been infected by different routes, since HJ96A-29 and HJ96AS-1, isolated from a patient and a staff member, respectively, had genotypes different from the other 30 strains (Tables 2 and 3). In contrast, another staff member of facility A that might have transmitted the organism had become infected by patients in facility A, because HJ96AS-2 strain shared the same genotype (subtype 3) as a majority of the strains isolated from the patients of facility A (Tables 2 and 3).
Analysis of RFLP profiles of SSU rDNA, termed riboprinting (4), was performed for the evaluation of the subtype and the route of transmission between two health care facilities using diagnostic PCR primers. Applying this technique, seven ribodeme groups had been classified by combining the two restriction enzymes HinfI and RsaI among 30 strains of B. hominis (4). In this study, therefore, these two restriction enzymes were tested against 10 strains selected from different subtypes to evaluate the classification of subtypes and the estimation of transmission routes of B. hominis between the two health care facilities A and B. The four subtypes, including a subtype-variant classified from the results of PCR primers, completely corresponded to the different riboprinting patterns (Table 4). When two strains of subtype 1 were examined using riboprinting, strain HE87-1 showed the same banding pattern with the Nand II strain, which had been designated ribodeme 1 (4). When five strains of subtype 3 were examined, all five strains, including strains HV93-13, HJ96A-26, HJ96B-1, HJ96B-4, and HJ96B-6, showed the same banding pattern which corresponded to ribodeme 2. However, the riboprinting pattern of the HJ96A-29, HJ96AS-1, and B strains were quite different from any of the seven ribodemes reported (Table 4). Therefore, these three strains were designated as new ribodeme groups, i.e., ribodemes 8 to 10.
In light of the genetic similarities and differences observed among human and nonhuman Blastocystis isolates, it is reasonable to speculate that the transmission of B. hominis organisms occurs either by human-to-human or animal-to-human routes. The latter transmission route was demonstrated by the presence of zoonotic strains identified from a chicken and a guinea pig (4, 12, 13). In the present study, the majority of B. hominis infections among two health care facilities were shown to be subtype 3 (30 of 32 [94%]), thus confirming that these cases could be attributed to human-to-human transmission (Table 3). However, the other two cases (2 of 32 [6%]) had different subtypes. In our laboratory, although a total of 47 isolates of human origin and 21 isolates of animal origin were examined with the different diagnostic PCR primer sets, SB332 successfully amplified strain QQ93-3 isolated from a Japanese quail (Yoshikawa, unpublished data). In addition, SB155 developed from the B strain isolated from a patient in Singapore (13) also amplified other strains of Japanese quail origin (QQ98-4 and QQ98-7) (Yoshikawa, unpublished data). Therefore, it is reasonable to speculate that some B. hominis strains originate in birds. The present study is the first report of a systematic comparison between various methods of subclassification in B. hominis populations and of the molecular epidemiology of B. hominis infection in human-to-human transmission.
ACKNOWLEDGMENTS
This study was supported by grant-in-aid for scientific research (C) from the Ministry of Education, Sciences, Sports, and Culture of Japan.
We thank Michelle-Becker Hapak for critical reading of the manuscript.
REFERENCES
- 1.Boreham P F L, Stenzel D J. Blastocystis in humans and animals: morphology, biology, and epizootiology. Adv Parasitol. 1993;32:1–70. doi: 10.1016/s0065-308x(08)60206-7. [DOI] [PubMed] [Google Scholar]
- 2.Boreham P F L, Upcroft J A, Dunn L A. Protein and DNA evidence for two demes of Blastocystis hominis from humans. Int J Parasitol. 1992;22:49–53. doi: 10.1016/0020-7519(92)90079-z. [DOI] [PubMed] [Google Scholar]
- 3.Brumpt E. Blastocystis hominis n.sp. et formes voisines. Bull Soc Pathol Exot. 1912;5:725–730. [Google Scholar]
- 4.Clark C G. Extensive genetic diversity in Blastocystis hominis. Mol Biochem Parasitol. 1997;87:79–83. doi: 10.1016/s0166-6851(97)00046-7. [DOI] [PubMed] [Google Scholar]
- 5.Kain K C, Noble M A, Freeman H J, Barteluk R L. Epidemiology and clinical features associated with Blastocystis hominis infection. Diagn Microbiol Infect Dis. 1987;8:235–244. doi: 10.1016/0732-8893(87)90055-1. [DOI] [PubMed] [Google Scholar]
- 6.Mansour N S, Mikhail E M, El Masry N A, Sabry A G, Mohareb E W. Biochemical characterization of human isolates of Blastocystis hominis. J Med Microbiol. 1995;42:304–307. doi: 10.1099/00222615-42-4-304. [DOI] [PubMed] [Google Scholar]
- 7.Nimri L F. Evidence of an epidemic of Blastocystis hominis infections in preschool children in northern Jordan. J Clin Microbiol. 1993;31:2706–2708. doi: 10.1128/jcm.31.10.2706-2708.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nimri L, Batchoun R. Intestinal colonization of symptomatic and asymptomatic schoolchildren with Blastocystis hominis. J Clin Microbiol. 1994;32:2865–2866. doi: 10.1128/jcm.32.11.2865-2866.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silberman J D, Sogin M L, Leipe D D. Human parasite finds taxonomic home. Nature. 1996;380:398. doi: 10.1038/380398a0. [DOI] [PubMed] [Google Scholar]
- 10.Stenzel D J, Boreham P F L. Blastocystis hominis revisited. Clin Microbiol Rev. 1996;9:563–584. doi: 10.1128/cmr.9.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshikawa H, Kuwayama N, Enose Y. Histochemical detection of carbohydrates of Blastocystis hominis. J Eukaryot Microbiol. 1995;42:70–74. doi: 10.1111/j.1550-7408.1995.tb01542.x. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa H, Nagano I, Yap E H, Singh M, Takahashi Y. DNA polymorphism revealed by arbitrary primers polymerase chain reaction among Blastocystis strains isolated from humans, a chicken, and a reptile. J Eukaryot Microbiol. 1996;43:127–130. doi: 10.1111/j.1550-7408.1996.tb04492.x. [DOI] [PubMed] [Google Scholar]
- 13.Yoshikawa H, Nagano I, Wu Z, Yap E H, Singh M, Takahashi Y. Genomic polymorphism among Blastocystis hominis strains and development of subtype-specific diagnostic primers. Mol Cell Probes. 1998;12:153–159. doi: 10.1006/mcpr.1998.0161. [DOI] [PubMed] [Google Scholar]
- 14.Yoshikawa H, Morimoto K, Nagashima M. Isolation and characterization of Blastocystis spp. isolated from anuran amphibians. In: Tada I, Kojima S, Tsuji M, editors. 9th International Congress of Parasitology—1988. Bologna, Italy: International Proceedings Division, International Congress of Parasitology; 1998. pp. 1075–1079. [Google Scholar]
- 15.Zaman V, Ng G C, Suresh K, Yap E H, Singh M. Isolation of Blastocystis from the cockroach (Dictyoptera:Blattidae) Parasitol Res. 1993;79:73–74. [Google Scholar]
- 16.Zierdt C H. Blastocystis hominis—past and future. Clin Microbiol Rev. 1991;4:61–79. doi: 10.1128/cmr.4.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]