Rice resistance proteins share a conserved defense signal transduction pathway to confer resistance to rice blast disease.
Abstract
Resistance (R) proteins are important components of plant innate immunity. Most known R proteins are nucleotide-binding site leucine-rich repeat (NLR) proteins. Although a number of signaling components downstream of NLRs have been identified, we lack a general understanding of the signaling pathways. Here, we used the interaction between rice (Oryza sativa) and Magnaporthe oryzae to study signaling of rice NLRs in response to blast infection. We found that in blast resistance mediated by the NLR PIRICULARIA ORYZAE RESISTANCE IN DIGU 3 (PID3), the guanine nucleotide exchange factor OsSPK1 works downstream of PID3. OsSPK1 activates the small GTPase OsRac1, which in turn transduces the signal to the transcription factor RAC IMMUNITY1 (RAI1). Further investigation revealed that the three signaling components also play important roles in disease resistance mediated by the distantly related NLR protein Pi9, suggesting that the OsSPK1–OsRac1–RAI1 signaling pathway could be conserved across rice NLR-induced blast resistance. In addition, we observed changes in RAI1 levels during blast infection, which led to identification of OsRPT2a, a subunit of the 19S regulatory particle of the 26S proteasome. OsRPT2a seemed to be responsible for RAI1 turnover in a 26S proteasome-dependent manner. Collectively, our results suggest a defense signaling route that might be common to NLR proteins in response to blast infection.
Introduction
In addition to physical and chemical barriers, plants rely on their innate immune system to defend them from insects and microbial pathogens. The plant innate immune system can be divided into two layers, depending on the immune receptors (Jones and Dangl, 2006; Dodds and Rathjen, 2010; Wang et al., 2020). One layer is basal defense, defined by pattern recognition receptors (PRRs) that perceive molecules conserved across pathogen species (also termed pathogen-associated molecular patterns). PRRs are located at the cell surface, sense signals in the apoplast, and induce defense to slow pathogens’ intracellular spread. The other immunity layer is defined by Resistance (R) genes. R proteins are intracellular receptors, recognizing strain-specific effectors (also termed avirulence proteins, Avr) to trigger rapid and enhanced immune responses, including induction of defense-related genes, a burst of reactive oxygen species (ROS), a sustained increase in cytosolic Ca2+, and even local programmed cell death known as the hypersensitive response. Defenses induced by R genes generally result in strong resistance to invaders; hence, R genes are important genetic resources for crop breeding (Dangl et al., 2013).
In plants, nucleotide-binding site (NBS) leucine-rich repeat (NLR) proteins are encoded by large gene families. For example, about 160 NLR genes are annotated in Arabidopsis (Arabidopsis thaliana), >500 genes in rice, and over 2,000 genes in bread wheat (Triticum aestivum; Gu et al., 2015; van Wersch and Li, 2019). The NLR gene family includes the majority of R genes known in plants, indicating their close relationship to plant immunity. NLR proteins have a central NBS, varying numbers of C-terminal leucine-rich repeats (LRRs), and a variable N-terminal region (Maekawa et al., 2011). Based on their N-terminal regions, NLRs can be roughly categorized as Toll/interleukin 1 receptor (TIR) or coiled-coil (CC) NLRs. TIR-NLRs exist exclusively in dicots and CC-NLRs are distributed widely in vascular plants (McHale et al., 2006).
Activation or inactivation of NLR proteins, in the presence or absence of the cognate effectors, involves complex molecular and intramolecular interactions (Qi and Innes, 2013). The NBS domain serves as a molecular switch that determines the “on” or “off” state of a given NLR. In the “off” state, the NBS is preferentially bound to adenosine diphosphate (ADP), and intramolecular interactions with the LRR domain help to stabilize the “closed” structure. In the “on” state, changes in the intramolecular interactions lead to the release of ADP from the NBS domain and facilitate the binding of adenosine triphosphate (ATP), presenting an “open” structure. The N-terminal regions, especially the CC domains, are usually associated with defense signaling and interact with defense signaling components (Shen et al., 2007; Zhu et al., 2010; Bernoux et al., 2011; Inoue et al., 2013; Wang et al., 2015, 2018).
Although a good number of components vital to NLR functions have been identified since the first NLR gene was cloned, the downstream signaling pathways are still obscure (Qi and Innes, 2013; Monteiro and Nishimura, 2018; Sun et al., 2020). This problem is particularly pronounced in rice. For this important cereal crop, more than 23 major NLR genes have been cloned (Liu et al., 2014), in efforts to genetically manage rice blast, a serious fungal disease caused by Magnaporthe oryzae (Dean et al., 2005, 2012). Almost all of these NLR genes encode CC-NLRs, and many occur in gene clusters (http://www.ricedata.cn/gene/gene_pi.htm). However, work in the past decades has discovered only a few immediate downstream signaling components, and our understanding of the signaling processes remains limited. We speculate that NLRs may share a limited set of downstream signaling components and reexamination of known signaling components might yield insight into the signaling events induced by rice NLRs.
To date, notable signaling components involved in rice NLR-mediated blast resistance include PigmR-INTERACTING AND BLAST RESISTANCE PROTEIN 1 (PIBP1), which is an RNA recognition motif-containing transcription factor required for the disease resistance of NLR genes such as PigmR and Piz-t from the Pi-z cluster (Deng et al., 2017; Zhai et al., 2019); OsWRKY45, a WRKY regulator required for the disease resistance of Pb1, an atypical CC-NLR conferring panicle blast resistance (Hayashi et al., 2010; Inoue et al., 2013); OsRac1, a Rac/Rop GTPase required for Pit and Pi-a functions (Kawasaki et al., 1999; Chen et al., 2010; Kawano et al., 2010); and OsSPK1, which shows guanine nucleotide exchange factor (GEF) activity toward OsRac1 and participates in Pit- and Pi-a-induced blast resistance (Wang et al., 2018). Our earlier work also underscored the importance of OsRac1 in PIRICULARIA ORYZAE RESISTANCE IN DIGU 3 (PID3)-mediated blast resistance (Zhou et al., 2019). In the same study, we identified RAC IMMUNITY1 (RAI1), a key bHLH-type transcription factor working downstream of OsRac1 (Zhou et al., 2019). RAI1 was initially found to be activated by OsRac1 in the PRR OsCERK1-induced basal defense (Kim et al., 2012), showing its key roles in rice innate immunity. These discoveries hint that OsRac1, acting as a molecular switch, might be a conserved regulator of NLR immunity; thus, research that builds on OsRac1 is warranted.
In this study, we reexamined roles of the known OsRac1-related signaling components. Our results suggest that a signaling pathway composed of OsSPK1, OsRac1, and RAI1 is shared by two NLRs that are distantly related. We also characterized OsRPT2a, a subunit of the 19S regulatory particle required for RAI1 turnover. Our work reveals some commonalities in the signaling events after NLR activation.
Results
OsSPK1 contributes to PID3-mediated blast resistance
We previously described a signaling pathway for PID3-mediated blast resistance, in which OsRac1 acts downstream of PID3 and relays defense signals to RAI1 (Zhou et al., 2019). To learn whether other components are involved in this signaling pathway, we examined OsSPK1, which activates OsRac1 (Wang et al., 2018). We generated OsSPK1 knockdown plants using RNA interference (RNAi) in the background of a PID3 transgenic line (PID3-TL), designated OsSPK1-kd/PID3-TL. Evaluation of OsSPK1 transcripts with reverse transcription-quantitative PCR (RT-qPCR) showed that the target gene was suppressed in several T1 generation lines (Figure 1A). We then used a M. oryzae isolate, RB-22, to perform blast resistance tests. Though the Avr gene to PID3 (AvrPID3 as a tentative name) is unknown yet, PID3-TL plants show complete resistance to RB-22 while cultivar TP309, the acceptor material of PID3-TL, is susceptible with this fungus, indicating there could be a AvrPID3 gene in RB-22. The test results revealed that RB-22 was incompatible with the PID3-TL plants (resistant) while was compatible with three OsSPK1-kd/PID3-TL lines (#7, #8, #14, susceptible), which all showed reduced OsSPK1 transcript levels (Figure 1, A and B). The results suggest that OsSPK1 is important to PID3-mediated disease resistance. We also examined the expression of defense-related genes in the loss-of-function OsSPK1-kd/PID3-TL lines. Of them, three important genes controlled by RAI1, including OsLOX3, OsPR5, and OsWRKY45 (Zhou et al., 2019), displayed lower transcript levels in all the examined lines than in PID3-TL plants (Figure 1C), indicating that OsSPK1 has an effect on immune responses.
Figure 1.
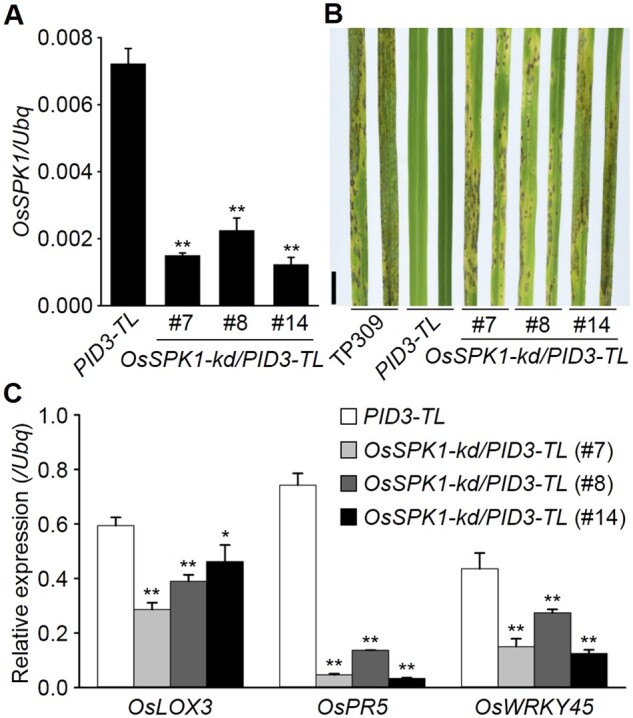
OsSPK1 contributes to PID3-mediated blast resistance. A, Determination of OsSPK1 transcripts in rice plants. B, Blast resistance tests. The T1 generation of the OsSPK1-kd/PID3-TL lines, growing in greenhouses, was used for the assay with a spraying inoculation method. TP309 was used as a susceptibility control. Pictures were taken 5 d after the inoculation. Scale bar, 1 cm. C, Induction of defense-related genes in rice plants. In A and C, Reverse transcription-quantitative PCR (RT-qPCR) data are shown as means ± sd (n = 3), and the significance of differences was analyzed based on comparison to PID3-TL. *P < 0.05; **P < 0.01 (Student’s t test).
OsSPK1 works downstream of PID3 and affects OsRac1 activity
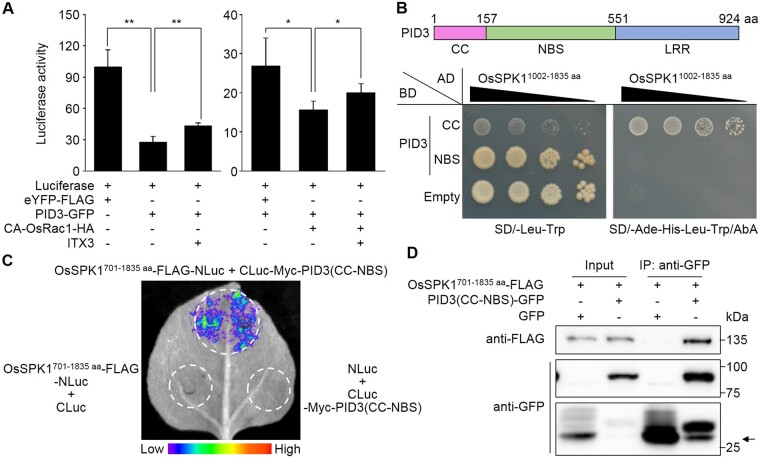
OsSPK1 might be attracted by the NLR Pit to activate plasma membrane-anchored OsRac1 (Wang et al., 2018). To test this, we used a luciferase reporter system (Cesari et al., 2014) to study the relationships between OsSPK1, OsRac1, and PID3. Immune responses usually cause cell death, leading to a reduced luciferase activity which can be measured by luciferase reporter system. Compared with enhanced YELLOW FLUORESCENT PROTEIN (eYFP), expressing PID3 in rice protoplasts led to a reduction in luciferase activity (Figure 2A, left; Supplemental Figure S1), indicating PID3-induced immune responses.
Figure 2.
OsSPK1 mediates OsRac1 activation by PID3. A, Cell death induction in rice protoplasts as detected with a luciferase reporter system. Means ± sd of relative luciferase activity values is shown (n ≥ 4). *P < 0.05; **P < 0.01 (Student’s t test). ITX3, inhibitors of GEF activity. Dimethyl sulphoxide was used as a negative control for ITX3. B, Yeast two-hybrid (Y2H) assay. OsSPK1 was fused to the carboxyl end of the GAL4 AD and PID3 was fused to the carboxyl end of the GAL4 DNA BD. Full-length PID3 and its LRR domain could not be sufficiently detected by Western blotting analysis, so the both were absent from this assay. C, Split-luciferase complementation (split-LUC) assays. Nicotiana benthamiana leaves were co-infiltrated with Agrobacterium tumefaciens-containing construct combinations as indicated. The luminescence images were captured using a CCD imaging apparatus. D, Co-immunoprecipitation (co-IP) assay. The OsSPK1701–1,835 aa and PID3(CC-NBS) proteins were transiently co-expressed in N. benthamiana leaves and total proteins were extracted for analysis. PID3(CC-NBS) was immunoprecipitated by anti-GFP antibody, followed by immunoblot analysis with anti-FLAG antibody to monitor the presence of OsSPK1701–1,835 aa in the precipitate. Arrow indicates the expected position of GFP blots. kDa, kilo-Dalton. The experiments were repeated 3 times with similar results.
ITX3 is a nontoxic inhibitor of GEF activity, which is used to selectively block Rac1 activation (Bouquier et al., 2009). ITX3 treatment could partly suppress the immune responses induced by PID3 in comparison with treatment with the solvent dimethyl sulphoxide (Figure 2A, left; Supplemental Figure S1). The results, together with those of the blast resistance tests (Figure 1), suggest that OsSPK1 works downstream of PID3. Expressing a constitutively active OsRac1 (CA-OsRac1) also caused immune responses, which exaggerated the immune responses induced by PID3, but additional ITX3 treatment also suppressed the immune responses (Figure 2A, right; Supplemental Figure S1). These data suggest that OsRac1 is involved in PID3-induced immune responses in an OsSPK1 (GEF)-dependent manner. Therefore, our results suggest that OsSPK1 works between PID3 and OsRac1 and may be recruited by PID3 to activate OsRac1.
Our preliminary evidence suggested that the CC domain of PID3, rather than the NBS, shows a binding affinity for OsSPK11,002–1,835aa in yeast two-hybrid (Y2H) assays (Figure 2B). In a split-luciferase complementation (split-LUC) assay performed in Nicotiana benthamiana, we also detected an interaction between PID3(CC-NBS) and OsSPK1701–1,835 aa (Figure 2C; Supplemental Figure S2). We conducted co-immunoprecipitation (co-IP) assays by transiently expressing PID3 (CC-NBS) and OsSPK1701–1,835 aa in N. benthamiana leaves. When GREEN FLUORESCENT PROTEIN (GFP)-tagged PID3(CC-NBS) was precipitated by anti-GFP antibody, OsSPK1701–1,835 aa-FLAG was co-precipitated as detected by anti-FLAG (Figure 2D). These findings indicate an intimate association between PID3 and OsSPK1. In addition, we observed that the T0 generation of OsSPK1-kd/PID3-TL plants was poor in seed setting compared to PID3-TL plants, in spite of a comparable number of grains per plant (Supplemental Figure S3), which resembles the phenotype when OsRac1 was suppressed in PID3-TL (Zhou et al., 2019), implying that OsSPK1 and OsRac1 may work in the same signaling pathway.
These results in combination with our earlier study suggest that OsSPK1, OsRac1, and RAI1 constitute a signaling pathway for PID3.
Both OsSPK1 and OsRac1 are important for blast resistance induced by Pi9
Next, we asked whether the OsSPK1–OsRac1–RAI1 signaling pathway discovered for PID3 is shared by other NLR proteins. We chose to check Pi9, for this NLR gene is well known for its broad-spectrum blast resistance (Qu et al., 2006). Pi9 and PID3 are distantly related (Supplemental Figure S4A) and share low identity (∼27%) in amino acid sequence (Supplemental Figure S4B).
We generated OsSPK1 knockdown plants in the background of a Pi9 transgenic line (Pi9-TL), named OsSPK1-kd/Pi9-TL, and confirmed the suppression of OsSPK1 in the T2 generation by RT-qPCR (Figure 3A). We used the isolate RB-22 to perform blast resistance tests, as this isolate harbors the Avr gene AvrPi9 (Supplemental Figure S5) and hence is theoretically avirulent to Pi9-TL plants (Wu et al., 2015). The inoculation results showed that Pi9-TL plants were completely incompatible with RB-22 as expected, while the OsSPK1-kd/Pi9-TL plants developed typical blast symptoms of susceptibility (Figure 3B). These observations suggest that OsSPK1 is required for Pi9-induced disease resistance. Similarly, we found that those RAI1-regulated defense-related genes showed lower transcript levels in the susceptible OsSPK1-kd/Pi9-TL plants than in Pi9-TL plants (Figure 3C). Besides, OsSPK1701–1,835 aa associated with Pi9(CC-NBS) in N. benthamiana as detected by split-LUC assays (Supplemental Figure S6).
Figure 3.
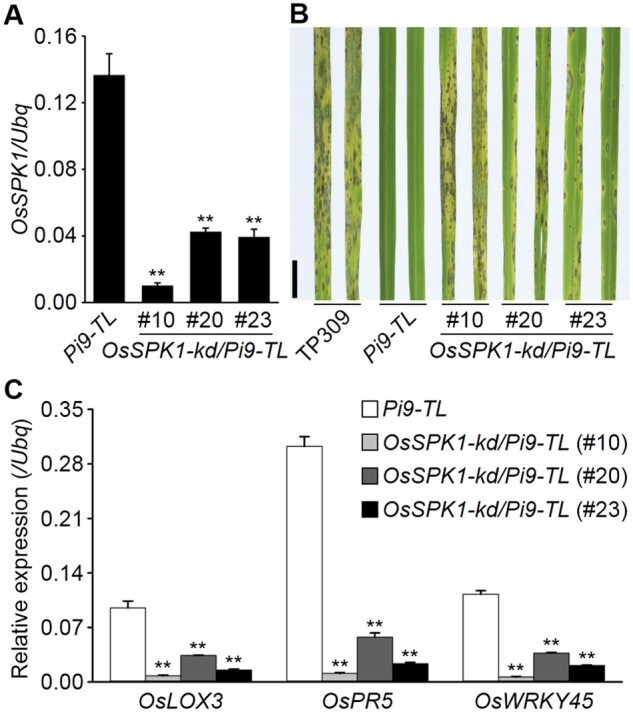
OsSPK1 contributes to Pi9-induced blast resistance. A, Determination of OsSPK1 transcripts in rice plants. B, Blast resistance tests. The T2 generation of the OsSPK1-kd/Pi9-TL lines, growing in greenhouses, was used for the assay with a spraying method. TP309 was used as a susceptibility control. Pictures were taken 5 d after the inoculation. Scale bar, 1 cm. C, Induction of defense-related genes in rice plants. In (A) and (C), RT-qPCR data are shown as means ± sd (n = 3), and the significance of differences was analyzed based on comparison to Pi9-TL. **P < 0.01 (Student’s t test).
In susceptible OsSPK1-kd/Pi9-TL plants, OsRac1 transcripts were also decreased (Supplemental Figure S7), implying OsRac1 might be involved in Pi9-induced immunity. To test, we generated OsRac1 knockdown lines in the background of Pi9-TL and confirmed the suppression of OsRac1 in the T1 generation of the obtained plants, named OsRac1-kd/Pi9-TL (Figure 4A). Blast inoculation tests with RB-22 showed OsRac1-kd/Pi9-TL plants were susceptible to the fungal isolate, unlike Pi9-TL plants (Figure 4B). Again, those genes regulated by RAI1 showed lower transcript levels in all the OsRac1-kd/Pi9-TL plants than in Pi9-TL plants (Figure 4C). In addition, a strong in planta interaction was observed between OsRac1 and Pi9 by split-LUC assays (Supplemental Figure S8). The results suggest that OsRac1 is also required for Pi9-induced blast resistance. Interestingly, suppression of OsRac1 in the background of Pi9-TL also led to poor fertility, reminiscent of the case in PID3-TL (Zhou et al., 2019).
Figure 4.
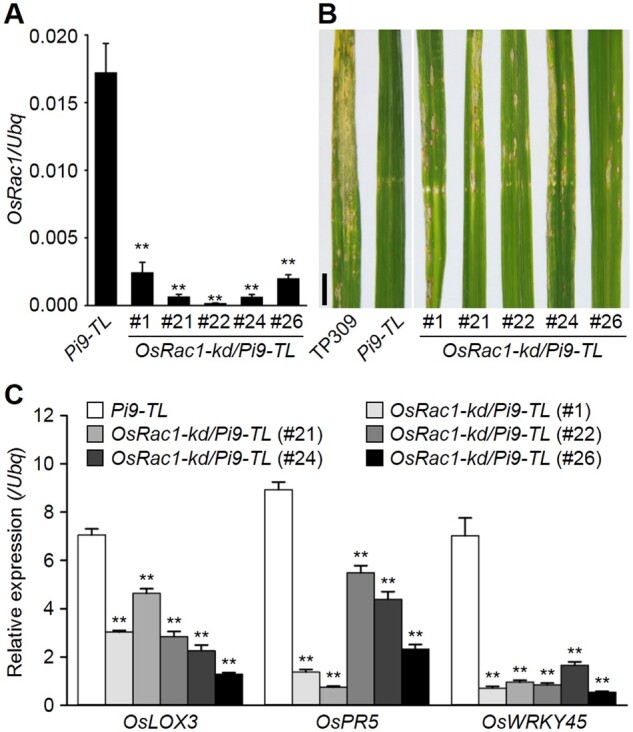
OsRac1 knockdown compromises Pi9-induced blast resistance. A, Determination of OsRac1 transcripts in rice plants. B, Blast resistance tests. T1 generation of the OsRac1-kd/Pi9-TL lines, at a tillering stage and growing in paddy fields, was used for the tests. TP309 was used as a susceptibility control. Pictures were taken 6 d after inoculation. Scale bar, 1 cm. C, Induction of defense-related genes in rice plants. In (A) and (C), the RT-qPCR data are shown as means ± sd (n = 3), and the significance of differences was analyzed based on comparison to Pi9-TL. **P < 0.01 (Student’s t test).
RAI1 acts as a positive regulator of Pi9-induced disease resistance
Then we examined the roles of RAI1 in Pi9-induced disease resistance. RAI1 knockdown lines in the Pi9-TL background were generated and named RAI1-kd/Pi9-TL. As the basal level of RAI1 transcripts was low, we used a semi-quantitative RT-PCR method to estimate RAI1 transcript levels and confirmed the suppression of RAI1 in at least two RAI1-kd/Pi9-TL homozygous lines (Figure 5A). Blast inoculation tests with RB-22 revealed that the two RAI1-kd/Pi9-TL lines were susceptible to the fungus, unlike Pi9-TL plants (Figure 5B). RT-qPCR results showed that the defense-related genes mentioned above were transcriptionally lower in the knockdown lines than in Pi9-TL (Figure 5C). Therefore, these results suggested that RAI1 is required for Pi9-induced blast resistance.
Figure 5.
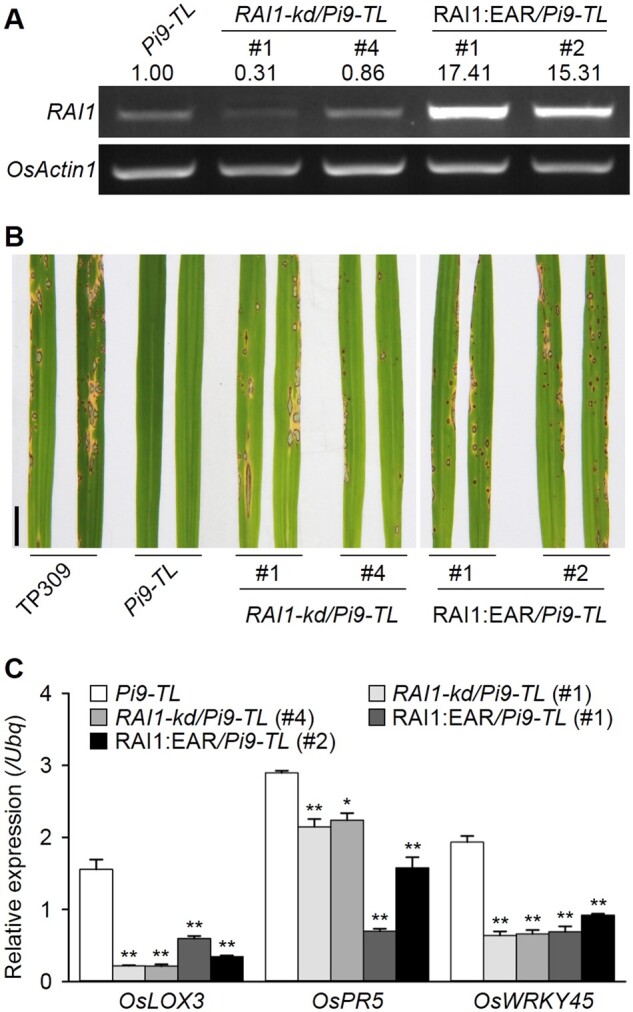
RAI1 acts as a positive regulator of Pi9-induced blast resistance. A, Determination of RAI1 transcripts in rice plants by semi-quantitative RT-PCR. Numbers indicate RAI1 transcripts normalized to OsActin1 using ImageJ software. B, Blast resistance tests. Four-week-old homogeneous seedlings growing in greenhouses were subject to the tests, with a spraying inoculation method. Pictures were taken 5 d after the inoculation. Scale bar, 1 cm. C, Induction of defense-related genes in rice plants, measured by RT-qPCR. Data are shown as means ± sd (n = 3), and the significance of differences was analyzed based on comparison to Pi9-TL. *P < 0.05; **P < 0.01 (Student’s t test).
Given that RAI1 may act as a transcription activator, we additionally created a chimeric RAI1 repressor by fusing an ERF-ASSOCIATED AMPHIPHILIC REPRESSION (EAR) motif to the carboxyl end of RAI1. We introduced this plasmid construct, driven by a constitutive promoter, into Pi9-TL and generated chimeric RAI1 repressor plants, named RAI1:EAR/Pi9-TL. Semi-quantitative PCR showed a sharp increase of RAI1 transcripts in two of the homozygous transgenic lines (Figure 5A), indicating constitutive expression of the chimeric gene. Blast inoculation tests showed that RAI1:EAR/Pi9-TL plants were susceptible to the isolate RB-22, resembling those RAI1-kd/Pi9-TL plants (Figure 5B). Consistent with the loss of disease resistance, the expression of those RAI1-regulated defense-related genes was also downregulated in the RAI1:EAR/Pi9-TL lines (Figure 5C). These results together with the above suggest that RAI1 could be a positive regulator for Pi9-induced blast resistance.
Functioning Pi9 affects RAI1 accumulation pattern
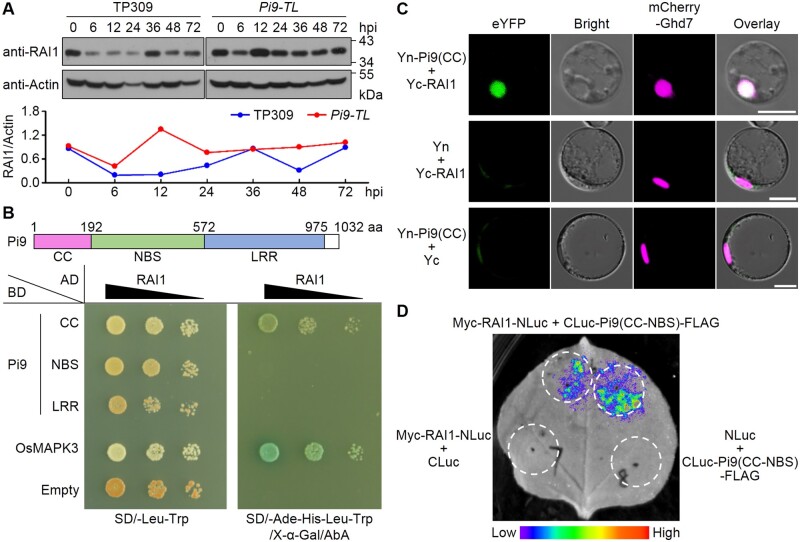
RAI1 showed transactivation activity in yeast cells and in a dual-luciferase reporter system (Supplemental Figures S9 and S10), implying that it might be recruited to regulate the expression of defense-related genes. To test how RAI1 varies upon blast infection, we examined RAI1 levels in blast-infected rice plants in a time course with developed antiserum of RAI1 (anti-RAI1) (Supplemental Figure S11). This revealed that in RB-22-infected Pi9-TL plants, RAI1 level was low at 6 h post-inoculation (hpi), then rapidly increased (12 hpi) and remained high until 24 hpi (Figure 6A). In contrast, in infected TP309 plants (the background cultivar of Pi9-TL), RAI1 levels remained low after the inoculation, especially between 6 hpi and 24 hpi (Figure 6A), an interval during which early molecular interactions between blast fungi and rice hosts are thought to occur (Li et al., 2006; Vergne et al., 2007). From 36 hpi on, RAI1 levels in TP309 plants were comparable to those in Pi9-TL plants (except at 48 hpi). The results suggest that RAI1 could be stabilized upon Pi9 activation in early M. oryzae–rice interactions.
Figure 6.
Pi9 affects RAI1 accumulation pattern during blast infections. A, Determination of RAI1 levels in blast fungus-infected rice plants by immunoblotting. RAI1 levels are normalized to Actin using ImageJ software and shown in a line chart (TP3090 hpi = 1.0). B, Y2H assay. RAI1 was fused to the carboxyl end of the GAL4 AD. Functional domains of Pi9 were each fused to the GAL4 DNA BD as indicated. Interaction between OsMAPK3 and RAI1 was used as a positive control in the assay. C, Bimolecular fluorescence complementation (BiFC) assays in rice protoplasts. The CC domain of Pi9 was fused to the carboxyl end of the N-terminal half of eYFP (Yn), and RAI1 was fused to the carboxyl end of the C-terminal half of YFP (Yc). The mCherry-Ghd7 fusion was used to indicate the location of nuclei. Fluorescent signals were captured with a confocal fluorescence microscope. Bars, 10 μm. D, Split-LUC assays. N. benthamiana leaves were co-infiltrated with A. tumefaciens-carrying constructs as indicated. The luminescence images were captured using a CCD imaging apparatus. The experiments were repeated 3 times with similar results.
We also observed that the CC domain of Pi9 possessed a binding affinity for RAI1 in Y2H and bimolecular fluorescence complementation (BiFC) assays (Figure 6, B and C). Split-LUC assays further confirmed the association of Pi9(CC-NBS) with RAI1 in N. benthamiana (Figure 6D; Supplemental Figure S12).
OsRPT2a mediates RAI1 degradation in a 26S proteasome-dependent manner
To see how RAI1 is regulated, we identified RAI1 interactors by Y2H screening from a rice complementary DNA library and this led to identification of OsRPT2a. OsRPT2a was initially recognized as an ATPase subunit of the 19S regulatory particle of the 26S proteasome complex (Shibahara et al., 2002), but its biological function remains unknown. We confirmed that the carboxyl end of OsRPT2a (364–448 aa) determined its affinity with RAI1 in yeast (Supplemental Figure S13A), and split-LUC assays in N. benthamiana proved this association (Supplemental Figure S13, B and C).
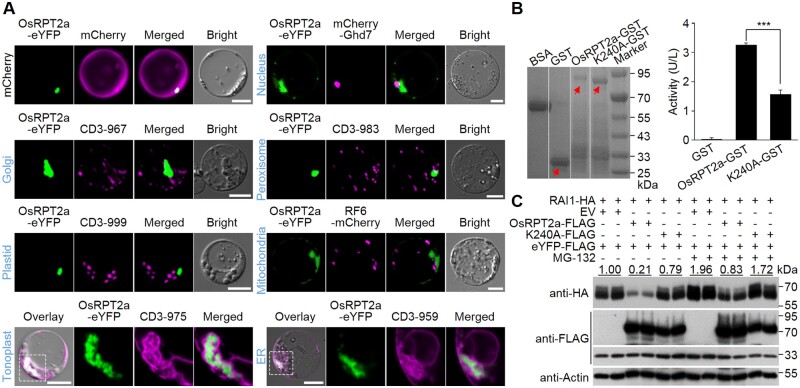
In rice protoplasts, the fluorescent signals for OsRPT2a-eYFP fusion predominantly overlapped with those for the tonoplast or endoplasmic reticulum markers but were absent from the endomembrane or nucleus (Figure 7A). OsRPT2a has ATP hydrolysis activity, related to a key residue (Lys240) in the Walker A motif, as a Lys240-to-Ala (K240A) substitution impaired the enzymatic activity (Figure 7B). We observed that the K240A substitution seemed not to affect OsRPT2a’s subcellular localization (Supplemental Figure S14A).
Figure 7.
OsRPT2a promotes RAI1 degradation. A, Subcellular distribution of OsRPT2a. OsRPT2a was fused to the amino end of eYFP and expressed in rice protoplasts. mCherry-Ghd7 was used as a nuclear marker, CD3-967 as a golgi marker, CD3-983 as a peroxisome marker, CD3-999 as a plastid marker, RF6-mCherry as a mitochondria marker, CD3-975 as a tonoplast marker, and CD3-959 as an endoplasmic reticulum (ER) marker. Dashed boxes were magnified for closer views using a 63× oil objective. Pictures were taken using a confocal fluorescence microscope. Scale bars, 10 μm. B, ATPase activity assay. Purified protein samples were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and visualized by CBB staining (left, red arrows). ATPase activities are shown on the right. Data are shown as means ± sd (n = 5). ***P < 0.001 (Student’s t test). C, In vivo degradation of RAI1 by OsRPT2a. Rice protoplasts were co-transfected with plasmid combinations as indicated and total proteins were extracted for immunoblotting analysis. eYFP-FLAG levels were used to estimate the transfection efficiency of each treatment. Numbers indicate RAI1-HA levels normalized to eYFP-FLAG, means of two replicates. Protein levels were measured by ImageJ software. Actin levels are shown as equal loading controls. EV, empty vector. The experiments were repeated 3 times with similar results.
We next explored the significance of the RAI1-OsRPT2a association. When RAI1 was co-expressed with OsRPT2a in rice protoplasts, its accumulation was reduced in comparison with that when it was co-expressed with a control plasmid (empty), suggesting that OsRPT2a may promote RAI1 degradation (Figure 7C). Further, the ATP hydrolysis activity of OsRPT2a seemed to be required for the degradation, because expression of the K240A variant led to an increased RAI1 level (Figure 7C). Treatment with MG-132, an inhibitor of the proteolytic activity of the 26S proteasome, greatly improved RAI1 accumulation levels for the RAI1 and OsRPT2a combination (lane 9/10 versus lane 3/4, approximately four-fold), in contrast to the combination of RAI1 and an empty control (lane 7/8 versus lane 1/2, approximately two-fold; Figure 7C). These results suggest that RAI1 degradation by OsRPT2a is 26S proteasome-dependent. As supportive evidence, we observed that the eYFP fusions of OsRPT2a or the K240A variant had a striking overlap with Myc-RAI1-mCherry in respect to their fluorescent signals, when both were co-expressed in rice protoplasts (Supplemental Figure S14, B and C), indicating subcellular localization of OsRPT2a changes, supposed to be affected by the nucleus-localized RAI1.
Discussion
In this study, we unraveled a downstream signaling pathway consisting of a GEF (OsSPK1), a GTPase (OsRac1), and a transcription factor (RAI1) for the NLR protein PID3. Genetic functions of the three signaling components in blast resistance were validated for another, distantly related NLR protein. Although more evidence, especially biochemical, is necessary to confirm that the OsSPK1–OsRac1–RAI1 cascade is a common signaling route, this pathway seems reasonable for various rice CC-NLRs.
Pivotal roles of the OsSPK1–OsRac1 cascade in defense signaling
In the proposed working model (Figure 8), OsRac1 occupies a central position. OsRac1 functions in rice immunity have long been known, including those for PRR- and NLR-induced immunity (Kawano and Shimamoto, 2013; Kawano et al., 2014). The importance of this GTPase is determined by its role as a molecular switch, cycling between a GDP-bound “off” and a GTP-bound “on” state. Moreover, OsRac1 enhances ROS production by activating NADPH oxidase activity and suppressing a ROS scavenger (Wong et al., 2004, 2007). To date, OsRac1 has been reported to participate in blast resistance induced by various NLRs such as Pit (Kawano et al., 2010), Pi-a (Chen et al., 2010), PID3 (Zhou et al., 2019), and Pi9 (this study). Additionally, OsRac1 shows binding affinities for Pib, as well as Xa1, a rice bacterial blight-resistant NLR protein (Kawano et al., 2010). These NLRs belong to different gene clusters (Supplemental Figure S4). Given the functional similarities for clustered genes (i.e. genes from the Pi-z locus [Pi9, Piz-t, PigmR] have a resistance spectrum broader than other NLRs; Qu et al., 2006; Zhou et al., 2006; Deng et al., 2017), it is supposed that OsRac1 might be recruited by other NLRs to transduce defense signals. Thus, OsRac1 most likely serves as a common signaling component for rice NLRs. In this sense, it is conceivable that pathogens would target OsRac1 during their coevolution with rice hosts. Blast races harboring the effector AvrPiz-t provide such an example. In the absence of Piz-t recognition, AvrPiz-t acts as a virulence factor by interacting with OsRac1 to interfere with OsRac1-mediated ROS production, thereby allowing intracellular growth of fungi (Park et al., 2012; Bai et al., 2019).
Figure 8.
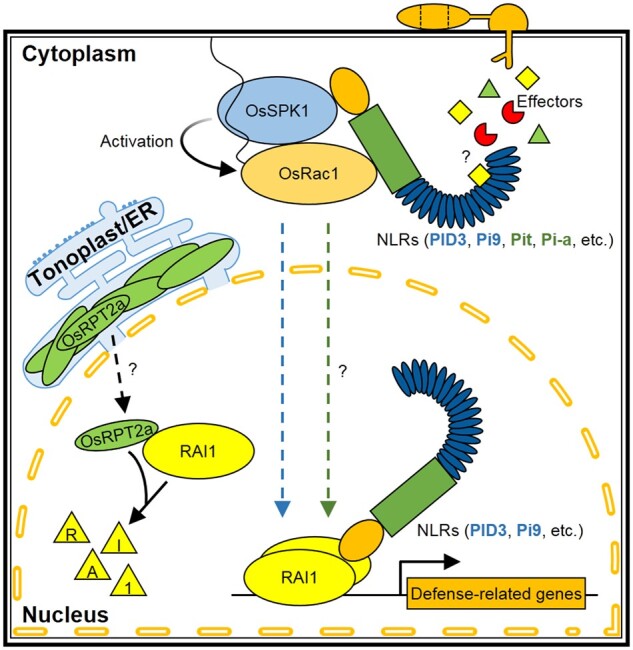
A working model for rice NLR protein-induced defense signaling pathway. After perceiving cognate effectors from blast fungi, rice NLR proteins, including PID3, Pi9, Pit, and Pi-a, transduce their signals to the downstream component OsSPK1, a GEF, by directly binding with it. OsSPK1 facilitates the conversion of the GTPase OsRac1 at the plasma membrane from an inactive GDP-bound state to an active GTP-bound one. Activated OsRac1 leads to the activation of the transcription factor RAI1 in the nucleus, which in turn drives transcriptional reprogramming of defense-related genes. Whether RAI1 participates in Pit- or Pi-a-induced blast resistance remains to be answered (marked with a question mark). Both PID3 and Pi9 show affinity with RAI1 in the nucleus, presumed to protect RAI1 from the degradation by 26S proteasome. Tonoplast/ER-localized component OsRPT2a provides a solution to fine-tuning RAI1 accumulation in vivo. By a yet unknown way, OsRPT2a translocates into the nucleus, where it associates with RAI1, leading to reduced RAI1 accumulation in a proteasome-dependent manner.
Because GEFs facilitate the release of GDP from GTPases and promote the binding of GTP, they are viewed as activators of GTPases. In studying the mechanisms of Pit-induced immunity, three types of GEFs were examined and only OsSPK1, of the DOCK family of GEFs, showed a physical affinity to the CC domain of Pit (Wang et al., 2018). Importantly, OsSPK1 shows GEF activity toward OsRac1 and is required for disease resistance induced by Pit and Pi-a as well (Wang et al., 2018). OsRacGEF1, a PRONE-type GEF, contributes to PRR OsCERK1-induced basal defense (Akamatsu et al., 2013). Although OsRacGEF1 also shows GEF activity toward OsRac1, it does not participate in Pit-induced immunity (Wang et al., 2018). These observations lead to the hypothesis that GEFs have diverged between PRR- and NLR-induced signaling pathways.
We added evidence here that OsSPK1 is required for blast resistance induced by PID3 and Pi9, besides Pit and Pi-a mentioned above. The results imply that OsSPK1, as a member of the OsSPK1–OsRac1 cascade, also serves as a common signaling component for rice NLRs. OsSPK1 localizes mainly in the endomembranes but not in the plasma membrane (Wang et al., 2018). And, OsRac1 is anchored in the plasma membrane (Kawano et al., 2010). Therefore, NLR proteins potentially serve as a platform for combining OsSPK1 with OsRac1. In the case of Pit, the mutated CC domain impairs its binding affinity for OsSPK1, which in turn reduces the GEF activity of OsSPK1 toward OsRac1 on the plasma membrane (Wang et al., 2018). We previously observed a stronger binding affinity of PID3 for OsRac1 via the NBS domain (Zhou et al., 2019) and binding of the CC domain of PID3 to OsSPK1 in yeast and in planta (Figure 2, B–D, this study). These observations suggest that PID3, resembling Pit, might form a multicomponent complex by which OsSPK1 is recruited to activate OsRac1, resulting in induction of disease resistance. The present bottleneck in purifying sufficient PID3 or Pi9 is a hindrance to further biochemical experiments, however.
RAI1 might be a common signaling component downstream of the OsSPK1–OsRac1 cascade
OsRac1 is involved in ROS accumulation, but it could also enhance the amplitude of immune responses in other ways. In PRR OsCERK1-induced basal defense, activated OsRac1 transduces signals through a mitogen-activated protein kinase cascade to two transcription factors, RAI1 and OsRap2.6, both of which control the expression of defense-related genes (Kim et al., 2012; Wamaitha et al., 2012). Therefore, the discovery of RAI1 as a signaling component downstream of OsRac1 in NLR immunity does not seem unexpected. RAI1 has been suggested to participate in blast resistance induced by PID3 (Zhou et al., 2019) and Pi9 (this study). Interestingly, the CC domains of rice NLRs such as Pi9, PID3, and Pit (Z. Zhou and L. Zhu, unpublished data) all showed binding affinities for RAI1 at nuclei. These observations imply that RAI1 potentially works for various NLRs. However, transcription factors other than RAI1 could also play a major role in rice NLR immunity. Zhai et al. observed that PigmR, Pi9, and Piz-t show binding affinities for PIBP1, an RRM-containing transcription factor, and interestingly, PID3 and Pish are exempted from the binding (Zhai et al., 2019). The WRKY regulator OsWRKY45 also interacts with the CC domain of Pb1 (Inoue et al., 2013). Both PIBP1 and OsWRKY45 are important for disease resistance induced by the NLRs with which they interact (except for Pi9). Further studies are necessary to examine if the OsSPK1–OsRac1–RAI1 cascade is also recruited by NLRs such as PigmR, Piz-t, and Pb1. The answers will help to determine whether the downstream transcription factors are NLR-specific, unique, or multiple-choice.
Regulation of RAI1
Immune responses take energy away from plant growth and thus must be finely governed (Tian et al., 2003; Denance et al., 2013). In this study, we identified OsRPT2a as a RAI1 interactor (Supplemental Figure S13, A–C), which can promote RAI1 degradation in a 26S proteasome-dependent manner (Figure 7C). Although future studies are necessary to clarify how RAI1 is regulated by a subunit of the 19S regulatory particle of the 26S proteasome, the degradation promoted by OsRPT2a seems to be a solution to fine regulation of RAI1-regulated immune responses.
In plants, 26S proteasome-dependent regulation participates in diverse aspects of development and stress responses. OsWRKY45, one of the central regulators of the salicylic acid signaling pathway in rice, is constantly degraded by the 26S proteasome in the absence of pathogen signals (Matsushita et al., 2013). Upon perception of blast fungus signals, the NLR Pb1 protects OsWRKY45 from 26S proteasome degradation, thus allowing OsWRKY45-regulated immune responses (Inoue et al., 2013). In this study, we observed in blast-infected rice leaves that RAI1 displayed a different accumulation pattern in the presence of a functioning Pi9 (Figure 6A), possibly as a result of response to the effector AvrPi9. As OsRPT2a promotes RAI1 turnover, we hypothesize that part of activated PID3/Pi9 might migrate into the nuclei where interactions with RAI1 occur and stabilize RAI1 (Figure 8), especially in the early stages of rice–M. oryzae interactions.
In summary, the present studies show that various rice NLR proteins may follow a conserved defense signaling pathway in response to blast infection. Once activated, the NLR recruits a GEF molecule (OsSPK1) to activate the key GTPase OsRac1, which then transduces the signals to downstream transcription factors such as RAI1 and/or others.
Materials and methods
Experimental materials and blast inoculation tests
The japonica rice (Oryza sativa) variety “TP309” and the transgenic rice line PID3-TL are from Prof. Lihuang Zhu’s lab (Shang et al., 2009). The M. oryzae isolate RB-22 and seeds of Pi9-TL are generous gifts from Prof. Guo-Liang Wang (Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China). Transgenic rice lines mentioned in this study were generated with construct-containing Agrobacterium tumefaciens (strain GV3101) by an Agrobacterium-mediated transformation method (Hiei et al., 1994). All the rice plants were grown in greenhouses or paddy fields in Beijing, China. An injection method was followed to test blast resistance of plants grown in paddy fields (Lv et al., 2013), and a spray method was used for seedlings grown in greenhouses (Zhou et al., 2016).
Plasmid constructs and transformation
We targeted nucleotides 4,691–4,840 of OsSPK1 to design the OsSPK1 RNAi construct. The sense-loop-antisense assembly, flanked with SmaI and SalI restriction sites, was cloned into pCAMBIA2300 (CAMBIA, Canberra, Australia), driven by a constitutive promoter. This construct was transformed into calli derived from PID3-TL or Pi9-TL seeds, leading to the transgenic lines OsSPK1-kd/PID3-TL and OsSPK1-kd/Pi9-TL. RNAi constructs for OsRac1 and RAI1 and the RAI1:EAR construct were constructed by Zhou et al. (2019) and used to generate transgenic rice lines OsRac1-kd/Pi9-TL, RAI1-kd/Pi9-TL, and RAI1:EAR/Pi9-TL.
Transient expression constructs for CA-OsRac1 and RAI1 were described previously (Zhou et al., 2019). Each of them contains a C-terminal hemagglutinin (HA) tag. The coding sequence for PID3 was amplified, with XbaI and BamHI sites appended to both the ends, and then cloned into the modified binary plasmid vector, pZh01-GFP, to establish the PID3-GFP fusion construct. Two adenosine residues at positions 718 and 719 of OsRPT2a were replaced with a guanosine and a cytidine, respectively, using a Gene Site-directed Mutagenesis kit (Biomed Biotech Co., Ltd, Beijing, China), to produce the K240A variant, which, together with those for OsRPT2a and eYFP, was each cloned into the vector pUC19-35S-FLAG-RBS at the SacI and KpnI sites, as described (Li et al., 2005), to generate constructs for FLAG fusions. These constructs, together with those described below, were created based on a homologous DNA recombination method, using the HiFi DNA Assembly Master Mix (New England Biolabs, Beijing, China). Plasmids were produced using HiPure Plasmid Filter Midiprep Kit (Thermo Fisher Scientific Baltics UAB, V.A. Graiciuno, Vilnius, Lithuania), according to the manufacturer’s methods. These plasmids were transiently transformed into rice protoplasts for cell death induction, or in vivo protein degradation assays.
Rice protoplasts were prepared with 7-d-old yellowish seedlings grown in darkness, and unless otherwise indicated, we used seeds of the variety “TP309” for growing these seedlings. We followed previously described procedures (Bart et al., 2006).
BiFC assay and microscopy
For BiFC assays, sequences of interest were amplified, then cloned into the vectors CD3-1102 and CD3-1111 at the EcoRI and BamHI sites, to generate the C-terminal half of YFP (Yc) and the N-terminal half of YFP (Yn) fusion constructs, respectively. For subcellular microscopic examination, the sequences for OsRPT2a and K240A were cloned into the vector CD3-1104, at the EcoRI and BamHI sites, to generate the OsRPT2a-eYFP and K240A-eYFP fusion constructs. The sequence for Myc-RAI1 was cloned from the pGBKT7 recombinant plasmid (see below) and then inserted into a modified CD3-1104, on which eYFP was replaced with mCherry, to generate the Myc-RAI1-mCherry fusion construct. The CD3 series vectors were purchased from the Arabidopsis Biological Resource Center at Ohio State University (Columbus, OH, USA).
After rice protoplasts transformation, live cell imaging was conducted with an AxioImager M2 fluorescence microscope (Carl Zeiss, Germany), with 514 nm of excitation wavelength and 549 nm of emission wavelength for YFP and, 561 nm of excitation wavelength, 637 nm of emission wavelength for mCherry.
RT-qPCR
Methods for total RNA isolation and procedures for RT-qPCR were as described (Zhou et al., 2019).
Co-IP assay and immunoblotting
For co-IP assay, the CC-NBS region of PID3 was amplified and cloned into the modified vector pZh01-GFP at the XbaI and BamHI sites, and the nucleotides 2,101–5,505 of OsSPK1 was cloned into the modified vector pZh01-FLAG also at the XbaI and BamHI sites, to generate fusion constructs. We performed the agroinfiltration procedures according to Zhou et al. (2019), to co-express PID3(CC-NBS)-GFP and OsSPK1701–1,835 aa-FLAG in N. benthamiana leaves. After 3 d, total proteins were extracted with native extraction buffer 1 as described previously (Liu et al., 2010). We used GFP-Trap Agarose (ChromoTek GmbH, Planegg-Martinsried, Germany) to conduct co-IP assay. Details refer to the previous description by Gao et al. (2021).
Total proteins from rice leaves or protoplasts were extracted with lysis buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 0.5 mM EDTA, 0.5% [v/v] NP-40, 2×protease inhibitor cocktail). We followed the Western blotting procedures as described previously (Zhou et al., 2019). Commercial antibodies used in this study included anti-β-Actin, anti-HA, anti-FLAG, horseradish peroxidase (HRP)-conjugated goat-anti-mouse immunoglobulin G (IgG), and HRP-conjugated goat-anti-rabbit IgG, all purchased from CoWin Biotech Co., Ltd (Jiangsu, China). Anti-GFP and anti-Myc were purchased from Abmart Shanghai Co., Ltd (Shanghai, China). Anti-RAI1 serum was developed in rabbits with Escherichia coli-expressed RAI1 as the antigen, done by Protein Innovation Co., Ltd (Beijing, China). To visualize the immunoblots, ECL Western Blotting Substrate (Smart-Lifesciences, Jiangsu, China) was used.
Split-LUC assay
Split-LUC assays were performed as described (Zhou et al. 2018). Briefly, pairs of A. tumefaciens (strain GV3101)-containing desired constructs were co-infiltrated into N. benthamiana leaves at 0.1 of OD600 concentration. After 3 d, the leaves were sprayed with 1 mM D-luciferin (Promega Corporation, WI, USA) and then kept in the dark for 6 min. Luminescence images were captured using a cooled CCD imaging system, LB 985 NightSHADE (Berthold Technologies, Bad Wildbad, Germany).
Yeast-related assays
Coding sequences for PID3, Pi9, RAI1, and OsRPT2a and their mutants were each amplified and then inserted into the bait plasmid pGBKT7 at the EcoRI and BamHI sites, to generate GAL4-DNA binding domain (BD)-fused constructs. Full-length sequence of RAI1 and nucleotides 3,004–5,505 of OsSPK1 were each amplified and then inserted into the prey plasmid pGADT7 at the EcoRI and BamHI sites, to generate GAL4 activation domain (AD)-fused constructs. The resulting constructs were used for Y2H or transactivation activity assays. Related yeast strains, plasmids, and procedures were from Clontech Laboratories Inc. (Mountain View, CA, USA).
Dual-luciferase reporter assay
Plasmids used for this assay were kindly provided by Prof. Shou-Yi Chen (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China). The sequence for RAI1 was fused to the 3′-end of the GAL4BD element in the vector pGAL4DBD, as previously described (Wu et al., 2017).
ATPase activity assays
The coding sequences for OsRPT2a and OsRPT2aK240A were each fused to the glutathione S-transferase (GST) tag in the vector pGEX-KG (GenBank accession LT986714.1) at the EcoRI and HindIII sites, to produce the constructs for GST fusions. The constructs were transformed into E. coli strain BL21 competent cells and the target proteins were induced by isopropyl-β-d-thiogalactoside at a concentration of 0.3 mM. The fusion targets were purified through affinity chromatography using Glutathione Sepharose 4B (GE Healthcare, NJ, USA). Details for ATPase activity assays are described in the technical bulletin for MAK113 of Sigma-Aldrich Co. LLC. (St Louis, MO, USA).
Phylogenetic analysis
For phylogenetic analysis, involved sequences were obtained from National Center for Biotechnology Information databases (https://www.ncbi.nlm.nih.gov/genbank/) by accession numbers listed in ACCESSION NUMBERS. ClustalW in MEGA6 software was used to perform the sequence alignment (Gap Opening Penalty, 10; Gap Extension Penalty, 0.2). The phylogenetic tree was generated by MEGA6 using a neighbor-joining method, with 1,000 bootstrap replicates.
Accession numbers
Sequence data for genes mentioned in this study can mostly be found in NCBI databases with the following accession numbers: PID3 (FJ745364), Pi9 (DQ454158), Pb1 (AB570371), Pi1-5/6 (HQ606329), Pi2 (DQ352453), Pi5-1/2 (EU869185/EU869186), Pi7-J-1/2 (KY225903/KY225902), Pi25 (ADO17324.1), Pi35 (FW369328), Pi36 (GU169396), Pi37 (DQ923494), Pi50 (AKS24976.1), Pi63 (BAO79824), Pi-a (AK066950), Pib (AB013449), Pid4 (MG839283), PigmR (KU904633), Pik-1/2 (ADZ48537.1/ADZ48538.1), Pikh (AY914077), Pikm1-TS (AB462324), Pikm2-TS (AB462325), Pikp-1/2 (HM035360), Piks-1 (HQ662329), Pish (AP008207), Pish-J (KY225901), Pit (AB379815), Pi-ta (AF207842), Piz-t (DQ352040), and Xa1 (AB002266); OsRac1 (AK071126), OsRPT2a (P46466), OsSPK1 (XM_015775852), and RAI1 (AK067834); OsLOX3 (AY208920), OsPR5 (X68197), OsWRKY45 (AK103959), OsACTIN1 (Q10DV7), UBIQUITIN (Ubq; P0CH34) and AvrPi9 (KM004023). We checked the reference (Liu et al., 2013) for the sequence of Pi56(t) and the reference (Ma et al., 2015) for that of Pi64. Primers used in this study can be found in Supplemental Table S1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Detection of the expressed proteins in Figure 2A.
Supplemental Figure S2. Detection of the expressed proteins in Figure 2C.
Supplemental Figure S3. Comparison of grain numbers and seed-setting rate between PID3-TL and OsSPK1-kd/PID3-TL plants (T0 generation).
Supplemental Figure S4. Evolutionary relationships of Pi9 with other NLR proteins.
Supplemental Figure S5 Sequence for the AvrPi9 locus identified in the rice blast isolate RB-22.
Supplemental Figure S6. Pi9 interacts with OsSPK1 as detected by LUC assay.
Supplemental Figure S7. OsSPK1 knockdown affects OsRac1 transcripts in rice plants.
Supplemental Figure S8. Pi9 interacts with OsRac1 as detected by LUC assay.
Supplemental Figure S9. Determination of RAI1 transactivation activity in yeast cells.
Supplemental Figure S10. Determination of RAI1 transactivation activity with a dual-luciferase reporter system.
Supplemental Figure S11. Performance of anti-RAI1 in Western blotting analysis.
Supplemental Figure S12. Detection of the expressed proteins in Figure 6D.
Supplemental Figure S13. OsRPT2a associates with RAI1.
Supplemental Figure S14. OsRPT2aK240A has little effect on its subcellular distribution or co-localization with RAI1.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Professor Guo-Liang Wang for providing the above-mentioned experimental materials. We also thank the State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, and Chinese Academy of Sciences for substantial support.
Funding
This work was supported by the Ministry of Agriculture and Rural Affairs of the People’s Republic of China in the name of the Special Program for Transgenic Research (2016ZX08009-003), and the Major Project of New Varieties of Genetically Modified Organism of China (2016ZX08009-001).
Conflict of interest statement. The authors declare no competing financial interests.
L.Z. and Z.Z. designed the study; M.Y. and Z.Z. performed the majority of the experiments with help from D.Yi., X.Li., R.C., and L.L.; D.L., X.Li., S.L., and D.Ya. provided parts of the experimental materials; X.Z. provided management of rice growing in paddy fields; L.Z., Z.Z., D.T., and M.Y. analyzed the data; Z.Z. and M.Y. wrote the manuscript; and D.T. and L.Z. revised it.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Minxiang Yu (mxyu0037@163.com) or Zhuangzhi Zhou (zhouzz@genetics.ac.cn).
References
- Akamatsu A, Wong HL, Fujiwara M, Okuda J, Nishide K, Uno K, Imai K, Umemura K, Kawasaki T, Kawano Y, et al. (2013) An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13: 465–476 [DOI] [PubMed] [Google Scholar]
- Bai P, Park CH, Shirsekar G, Songkumarn P, Bellizzi M, Wang GL (2019) Role of lysine residues of the Magnaporthe oryzae effector AvrPiz-t in effector- and PAMP-triggered immunity. Mol Plant Pathol 20:599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9:200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquier N, Vignal E, Charrasse S, Weill M, Schmidt S, Leonetti JP, Blangy A, Fort P (2009) A cell active chemical GEF inhibitor selectively targets the Trio/RhoG/Rac1 signaling pathway. Chem Biol 16: 657–666 [DOI] [PubMed] [Google Scholar]
- Cesari S, Kanzaki H, Fujiwara T, Bernoux M, Chalvon V, Kawano Y, Shimamoto K, Dodds P, Terauchi R, Kroj T (2014) The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J 33:1941–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Shiotani K, Togashi T, Miki D, Aoyama M, Wong HL, Kawasaki T, Shimamoto K (2010) Analysis of the Rac/Rop small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant Cell Physiol 51:585–595 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341:746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, et al. (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, et al. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986 [DOI] [PubMed] [Google Scholar]
- Denance N, Sanchez-Vallet A, Goffner D, Molina A (2013) Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front Plant Sci 4: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Zhai K, Xie Z, Yang D, Zhu X, Liu J, Wang X, Qin P, Yang Y, Zhang G, et al. (2017) Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355:962–965 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11:539–548 [DOI] [PubMed] [Google Scholar]
- Gao C, Sun P, Wang W, Tang D (2021) Arabidopsis E3 ligase KEG associates with and ubiquitinates MKK4 and MKK5 to regulate plant immunity. J Integr Plant Biol 63: 327–339 [DOI] [PubMed] [Google Scholar]
- Gu L, Si W, Zhao L, Yang S, Zhang X (2015) Dynamic evolution of NBS-LRR genes in bread wheat and its progenitors. Mol Genet Genomics 290: 727–738 [DOI] [PubMed] [Google Scholar]
- Hayashi N, Inoue H, Kato T, Funao T, Shirota M, Shimizu T, Kanamori H, Yamane H, Hayano-Saito Y, Matsumoto T, et al. (2010) Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J 64:498–510 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by agrobacterium and sequence-analysis of the boundaries of the T-DNA. Plant J 6:271–282 [DOI] [PubMed] [Google Scholar]
- Inoue H, Hayashi N, Matsushita A, Xinqiong L, Nakayama A, Sugano S, Jiang CJ, Takatsuji H (2013) Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein–protein interaction. Proc Natl Acad Sci USA 110: 9577–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J, Yao A, Nakashima A, Takahashi H, Yoshida H, Wong H, et al. (2010) Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7:362–375 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kaneko-Kawano T, Shimamoto K (2014) Rho family GTPase-dependent immunity in plants and animals. Front Plant Sci 5:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Shimamoto K (2013) Early signaling network in rice PRR-mediated and R-mediated immunity. Curr Opin Struct Biol 16: 496–504 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K (1999) The small GTP-binding protein Rac is a regulator of cell death in plants. Proc Natl Acad Sci USA 96: 10922–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Oikawa T, Kyozuka J, Wong HL, Umemura K, Kishi-Kaboshi M, Takahashi A, Kawano Y, Kawasaki T, Shimamoto K (2012) The bHLH Rac Immunity1 (RAI1) is activated by OsRac1 via OsMAPK3 and OsMAPK6 in rice immunity. Plant Cell Physiol 53:740–754 [DOI] [PubMed] [Google Scholar]
- Li Q, Chen F, Sun L, Zhang Z, Yang Y, He Z (2006) Expression profiling of rice genes in early defense responses to blast and bacterial blight pathogens using cDNA microarray. Physiol Mol Plant Pathol 68:51–60 [Google Scholar]
- Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, Zhou JM (2005) FLAGellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci USA 102: 12990–12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61:893–903 [DOI] [PubMed] [Google Scholar]
- Liu W, Liu J, Triplett L, Leach JE, Wang GL (2014) Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol 52:1–29 [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu B, Zhu X, Yang J, Bordeos A, Wang G, Leach JE, Leung H (2013) Fine-mapping and molecular marker development for Pi56(t), a NBS-LRR gene conferring broad-spectrum resistance to Magnaporthe oryzae in rice. Theor Appl Genet 126:985–998 [DOI] [PubMed] [Google Scholar]
- Lv Q, Xu X, Shang J, Jiang G, Pang Z, Zhou Z, Wang J, Liu Y, Li T, Li X, et al. (2013) Functional analysis of Pid3-A4, an ortholog of rice blast resistance gene Pid3 revealed by allele mining in common wild rice. Phytopathology 103: 594–599 [DOI] [PubMed] [Google Scholar]
- Ma J, Lei C, Xu X, Hao K, Wang J, Cheng Z, Ma X, Ma J, Zhou K, Zhang X, et al. (2015) Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol Plant Microbe Interact 28:558–568 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12:818–826 [DOI] [PubMed] [Google Scholar]
- Matsushita A, Inoue H, Goto S, Nakayama A, Sugano S, Hayashi N, Takatsuji H (2013) Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J 73:302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro F, Nishimura MT (2018) Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Annu Rev Phytopathol 56:243–267 [DOI] [PubMed] [Google Scholar]
- Park CH, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M, et al. (2012) The Magnaporthe oryzae effector AvrPiz-t targets the ring e3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24:4748–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, Innes RW (2013) Recent advances in plant NLR structure, function, localization, and signaling. Front Immunol 4:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, Wang GL (2006) The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172:1901–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Tao Y, Chen X, Zou Y, Lei C, Wang J, Li X, Zhao X, Zhang M, Lu Z, et al. (2009) Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site–leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 182:1303–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315:1098–1103 [DOI] [PubMed] [Google Scholar]
- Shibahara T, Kawasaki H, Hirano H (2002) Identification of the 19S regulatory particle subunits from the rice 26S proteasome. Eur J Biochem 269:1474–1483 [DOI] [PubMed] [Google Scholar]
- Sun Y, Zhu YX, Balint-Kurti PJ, Wang GF (2020) Fine-tuning immunity: players and regulators for plant NLRs. Trends Plant Sci 25:695–713 [DOI] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423:74–77 [DOI] [PubMed] [Google Scholar]
- van Wersch S, Li X (2019) Stronger when together: clustering of plant NLR disease resistance genes. Trends Plant Sci 24:688–699 [DOI] [PubMed] [Google Scholar]
- Vergne E, Ballini E, Marques S, Mammar BS, Droc G, Gaillard S, Bourot S, DeRose R, Tharreau D, Notteghem JL, et al. (2007) Early and specific gene expression triggered by rice resistance gene Pi33 in response to infection by ACE1 avirulent blast fungus. New Phytol 174:159–171 [DOI] [PubMed] [Google Scholar]
- Wamaitha MJ, Yamamoto R, Wong HL, Kawasaki T, Kawano Y, Shimamoto K (2012) OsRap2.6 transcription factor contributes to rice innate immunity through its interaction with Receptor for Activated Kinase-C 1 (RACK1). Rice (NY) 5: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF, Ji J, Ei-Kasmi F, Dangl JL, Johal G, Balint-Kurti PJ (2015) Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog 11:e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li Y, Ishikawa K, Kosami KI, Uno K, Nagawa S, Tan L, Du J, Shimamoto K, Kawano Y (2018) Resistance protein Pit interacts with the GEF OsSPK1 to activate OsRac1 and trigger rice immunity. Proc Natl Acad Sci USA 115: E11551–E11560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Feng B, Zhou JM, Tang D (2020) Plant immune signaling: advancing on two frontiers. J Integr Plant Biol 62:2–24 [DOI] [PubMed] [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, Kojima C, Yoshioka H, Iba K, Kawasaki T, et al. (2007) Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19: 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HL, Sakamoto T, Kawasaki T, Umemura K, Shimamoto K (2004) Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol 135:1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kou Y, Bao J, Li Y, Tang M, Zhu X, Ponaya A, Xiao G, Li J, Li C, et al. (2015) Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol 206:1463–1475 [DOI] [PubMed] [Google Scholar]
- Wu Q, Liu X, Yin D, Yuan H, Xie Q, Zhao X, Li X, Zhu L, Li S, Li D (2017) Constitutive expression of OsDof4, encoding a C2-C2 zinc finger transcription factor, confesses its distinct flowering effects under long- and short-day photoperiods in rice (Oryza sativa L.). BMC Plant Biol 17:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai K, Deng Y, Liang D, Tang J, Liu J, Yan B, Yin X, Lin H, Chen F, Yang D, et al. (2019) RRM transcription factors interact with NLRs and regulate broad-spectrum blast resistance in rice. Mol Cell 74:996–1009 [DOI] [PubMed] [Google Scholar]
- Zhou B, Qu S, Liu G, Dolan M, Sakai H, Lu G, Bellizzi M, Wang GL (2006) The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant Microbe Interact 19:1216–1228 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Bi G, Zhou JM (2018) Luciferase complementation assay for protein-protein interactions in plants. Curr Protoc Plant Biol 3:42–50 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Pang Z, Li G, Lin C, Wang J, Lv Q, He C, Zhu L (2016) Endoplasmic reticulum membrane-bound MoSec62 is involved in the suppression of rice immunity and is essential for the pathogenicity of Magnaporthe oryzae. Mol Plant Pathol 17:1211–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Pang Z, Zhao S, Zhang L, Lv Q, Yin D, Li D, Liu X, Zhao X, Li X, et al. (2019) Importance of OsRac1 and RAI1 in signalling of nucleotide-binding site leucine-rich repeat protein-mediated resistance to rice blast disease. New Phytol 223:828–838 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y (2010) Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci USA 107: 13960–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.