Abstract
Plant plasma membrane H+-ATPases and Ca2+-ATPases maintain low cytoplasmic concentrations of H+ and Ca2+, respectively, and are essential for plant growth and development. These low concentrations allow plasma membrane H+-ATPases to function as electrogenic voltage stats, and Ca2+-ATPases as “off” mechanisms in Ca2+-based signal transduction. Although these pumps are autoregulated by cytoplasmic concentrations of H+ and Ca2+, respectively, they are also subject to exquisite regulation in response to biotic and abiotic events in the environment. A common paradigm for both types of pumps is the presence of terminal regulatory (R) domains that function as autoinhibitors that can be neutralized by multiple means, including phosphorylation. A picture is emerging in which some of the phosphosites in these R domains appear to be highly, nearly constantly phosphorylated, whereas others seem to be subject to dynamic phosphorylation. Thus, some sites might function as major switches, whereas others might simply reduce activity. Here, we provide an overview of the relevant transport systems and discuss recent advances that address their relation to external stimuli and physiological adaptations.
The regulation of plasma membrane H+-ATPases and autoinhibited Ca2+-ATPases exhibits a complex and dynamic network of posttranslational regulation. The regulation of plasma membrane H+-ATPases and autoinhibited Ca2+-ATPases exhibits a complex and dynamic network of posttranslational regulation.
P-type ATPases are found in all domains of life and constitute a large superfamily of membrane-bound pumps that share a common machinery, including a reaction cycle that involves catalytic phosphorylation of an Asp, resulting in a phosphorylated intermediate (reviewed in Palmgren and Nissen, 2011; (hence the name P-type; Box 1). The catalytic phosphoryl-aspartate intermediate is not to be confused with regulatory phosphorylation, which occurs on Ser, Thr, and Tyr residues. Five major families of P-type ATPases have been characterized (P1–5), each of which is divided into a number of subfamilies (named with letters). Plasma membrane H+-ATPases are classified as P3A ATPases, whereas Ca2+ pumps constitute P2A and P2B ATPases. In plants, these pumps are best characterized in the model plant Arabidopsis thaliana (Arabidopsis).
Box 1.
Enzymology of P-type ATPases.
P-type ATPases (reviewed in Palmgren and Nissen, 2011) alternate between two extreme conformations during their catalytic cycle: a high-affinity (with respect to ATP and the ion to be exported) Enzyme1 (E1) state, and a low-affinity Enzyme2 (E2) state. Many P-type ATPases are autoinhibited by built-in molecular constraints, namely their C- and N-terminal (for plasma membrane H+-ATPases; Palmgren et al., 1999) or N-terminal (for P2B Ca2+-ATPases; Malmström et al., 1997) regulatory (R) domains of approximately 100 amino acid residues, which act as brakes by stabilizing the pumps in a low-affinity conformation (Palmgren and Nissen, 2011), most likely E2. Neutralizing the R domain results in a shift in conformational equilibrium towards a high-affinity state, likely E1. In this way, the R domains of plasma membrane H+-ATPases and Ca2+-ATPases allow posttranslational modification events to control the turnover numbers of these pumps. A structure of a plasma membrane H+-ATPase (from the distantly related yeast S. cerevisiae) in its autoinhibited state has been solved (Heit et al., 2021). Its R domain is situated adjacent to the P domain, which would suggest that the R domain functions to restrict the conformational flexibility of the pump. Normally, the hydrolysis of ATP and transport are tightly coupled in P-type ATPases. Therefore, P-type ATPases hydrolyze bound ATP as soon as their ligand-binding site(s) in the membrane region are occupied, but not before. Thus, increasing the ligand affinity of an ATPase simultaneously increases its turnover number, provided that the concentration of ATP is not limiting, which is rarely the case in cells. A specific feature of plasma membrane H+-ATPases is that in the autoinhibited state, ATP hydrolysis is only loosely coupled to H+ pumping, whereas pump activation results in tight coupling, with one H+ pumped per ATP split (Pedersen et al., 2018).
In response to internal and/or external cues, plasma membrane H+-ATPase and Ca2+-ATPase activities are controlled by intracellular concentrations of H+ and Ca2+, respectively, via interacting proteins, through posttranslational modification by phosphorylation, and by regulated trafficking of the pump to and from the plasma membrane. Their regulation sometimes involves changes in gene expression and turnover, although this is rare, perhaps because both processes are time- and energy-consuming (Haruta et al., 2018).
The Arabidopsis P-type H+-ATPase gene family
Arabidopsis contains 11 P3A plasma membrane H+-ATPases, named autoinhibited H+-ATPase (AHA) 1–11 (Baxter et al., 2003) An exception among AHAs is AHA10, which is not a plasma membrane pump, but has an N-terminal vacuolar targeting signal and is expressed in the vacuolar membrane. All P3A ATPases are equipped with a C-terminal autoinhibitory domain that likely interacts with the rest of the pump protein in order to restrict conformational flexibility (Heit et al., 2021) and keep the pump in a partially uncoupled state (Pedersen et al., 2018). Different AHA isoforms are expressed in different cells at different times during development, but their biochemical properties are remarkably similar to each other (Hoffmann et al., 2020). AHAs are subject to posttranslational regulation by phosphorylation and dephosphorylation (as detailed below), but are also regulated by the pH, membrane potential, K+, and redox status of the cell (Kurkdjian and Guern, 1989; Buch-Pedersen et al., 2006; Reyer et al., 2020; Welle et al., 2021; (Box 2).
Box 2.
Maintenance of cellular and physical equilibrium by AHAs.
The plant cell cytoplasm is strictly maintained at ∼pH 7.5. The basal activity of plasma membrane H+-ATPases is minimal at this pH, but even a slight increase in [H+] causes the pump to increase turnover dramatically, with a maximum activity obtained at ∼pH 6.5. As H+ ions are pumped out, the activity declines again. At pH 7.5 (0.03 µM H+), the E2 conformation predominates, whereas an increase in [H+] shifts the conformational equilibrium towards the E1 conformation, which predominates at ∼pH 6.5 (0.3 µM H+). Thus, any increase in [H+]cyt increases the overall H+ affinity of the pump population, and plasma membrane H+-ATPases operate as biophysical pH stats that maintain cytoplasmic pH at a constant level (Kurkdjian and Guern, 1989). Plasma membrane H+-ATPases have a low-affinity K+ binding site in their cytosolic P-domains, which saturates at K+ concentrations well above 100 mM. K+ bound to this site is not transported, suggesting that the site has a regulatory role. In the plasma membrane H+-ATPase AHA2, K+ binding at this position induces dephosphorylation of the E1P state before H+ pumping has occurred, implying that high K+ levels uncouple the pump (Buch-Pedersen et al., 2006). Uncoupling of plasma membrane H+-ATPases by treatment with high concentrations of K+ might serve to depolarize the membrane, preventing further cation accumulation. Plasma membrane H+-ATPases are electrogenic: since H+ pumping is not followed by a negative charge and is not countered by the opposite transport of a positive charge, it establishes a membrane potential. This membrane energization underlies the secondary active transport of solutes into and out of the cell. Optogenetics analysis revealed that plasma membrane H+-ATPases re-establish membrane potential following membrane depolarization (Reyer et al., 2020). Pump activity increases with plasma membrane depolarization and vice versa. Thus, plasma membrane H+-ATPases function as both pH and voltage stats. The mechanism by which plasma membrane H+-ATPases sense voltage is unclear. P-type H+ and Ca2+ pumps both contain a conserved cysteine residue two residues upstream of the essential aspartate residue that is phosphorylated during the catalytic cycle. Conservative substitutions of this residue in the plasma membrane H+-ATPase AHA2 (Cys327) do not alter the catalytic properties of the pump, but increase its sensitivity to inhibition by reactive oxygen species (ROS) and Cu2+, which also generates ROS (Welle et al., 2021). Thus, Cys327 appears to function as a built-in antioxidant in plasma membrane H+-ATPases and perhaps also in other P-type ATPases.
Regulation of AHAs by phosphorylation
Plasma membrane H+-ATPase activity is regulated by phosphorylation at multiple sites (Table 1, Figures 1 and 2). Twelve in vivo phosphorylation sites have been identified in AHA2 alone (Table 1), many of which are conserved among AHAs (including Thr881, Ser904, Thr924, Ser931, Tyr936, and Thr947; all AHA2 nomenclature; (Falhof et al., 2016). More phosphorylation sites may be present, as the identity of the phosphopeptides recovered to date depends strongly on the method used for their isolation (Rudashevskaya et al., 2012). Strikingly, even in the absence of sequence homology between AHA2 phosphomotifs and yeast proteins, many of the sites (including the penultimate residue) are also phosphorylated when AHA2 is heterologously expressed in the yeast Saccharomyces cerevisiae (Rudashevskaya et al., 2012). This observation can be interpreted as evidence that the phosphosites are promiscuous, i.e. that each site is recognized rather nonspecifically by several protein kinases. The detection of phosphorylation in vivo in the absence of specific signals suggests that protein phosphatases play a prominent role in regulating the phosphorylation status of each residue.
Table 1.
Phosphosites identified in Arabidopsis AHAs
| Phosphorylation sites detected in AHA isoforms | Cytosolic domain | Motifa | Effect of Asp substitution | Notes |
|---|---|---|---|---|
| Ser2-AHA1 | N terminus |
|
||
| Ser3-AHA2 | N terminus | MSpSLEDIKNETVDLEK (1) | No detectable effect (1) | |
| Ser3-AHA3 | N terminus |
|
||
| Thr35-AHA1 | N terminus | EGLTpTQEGEDR (3) | ||
| Thr305-AHA1/AHA2 and Ser328-AHA1/AHA2; Thr316-AHA3 and Ser329-AHA3; Thr323-AHA4 and Ser336-AHA4; Thr319-AHA11 and Ser332-AHA11 (peptide also conserved in AHA7/AHA8/AHA9) | P domain | MpTAIEEMAGMDVLCpSDK (4) | ||
| Ser535-AHA2 | P domain | LGMGTNMYPSpSALLGTHK (2) | ||
| Ser544-AHA1 | P domain |
|
Oligogalacturonides induce phosphorylation (6). | |
| Ser548-AHA1 | P domain | DSNIApSIPVEELIEK (2) | ||
| Ser548-AHA2 | P domain | DANLApSIPVEELIEK (2,5) | ||
| Ser549-AHA11 | P domain | DEpSIGALPIDDLIEK (2) | ||
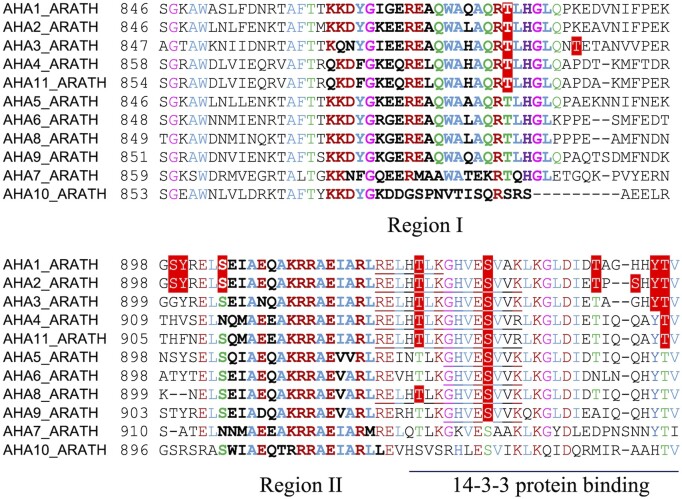
| Thr881-AHA1/AHA2 | C terminus, R-I | pTLHGLQPK (2,7,8,9,10,11,12,,13) | Decreases in response to flg22 (7); decreases in response to the addition of and K+ to nitrogen-starved plants (but not to the addition of )(13). | |
| Thr881-AHA1 | C terminus, R-I |
|
||
| Thr881-AHA2 | C terminus, R-I |
|
Activates (1); overrides regulation via Thr947 (9) | Phosphorylated in ammonium-adapted (but not nitrate-adapted) plants (16). |
| Thr882-AHA3 | C terminus, R-I |
|
Slight decrease in response to flg22 (7). | |
| Thr889-AHA11 | C terminus, R-I |
|
||
| Thr889-AHA3 | C terminus | TLHGLQNpTETANVVPER (2) | ||
| Ser899-AHA1/AHA2 | C terminus | GpSYRELSEIAEQAK (2,4,5,6,20) | ||
| Ser899-AHA1 | C terminus |
|
Occurs in response to flg22 (7) | |
| Ser899-AHA2 | C terminus |
|
Reduces activity (1) | Oligogalacturonides induce phosphorylation (6); increases in response to flg22 (7). |
| Tyr900-AHA1/AHA2 | C terminus | GSpYRELSEIAEQ (20) | ||
| Ser904-AHA1/AHA2 | C terminus, R-II |
|
Phosphorylated by SnRK2.4 in an in vitro kinase reaction of mannitol-treated Arabidopsis (2). | |
| Ser904-AHA2 | C terminus, R-II | EDVNIFPEKGSYRELpSEIAEQAK (2) | Activates (1) | |
| Thr924-AHA1/AHA2; Thr925-AHA3/AHA4; Thr923-AHA8; Thr931-AHA11 | C terminus | LRELHpTLK (2) | Inhibits (1); prevents binding of 14-3-3 protein (22) | |
| Ser931-AHA2; Ser932-AHA3; Ser931-AHA5; Ser931-AHA6; Ser930-AHA8; Ser936-AHA9 | C terminus | GHVEpSVVK (1,2,23) | Inhibits (1); prevents binding of 14-3-3 protein (22) | |
| Ser942-AHA4; Ser938-AHA11 | C terminus | GHVEpSVVR (2) | ||
| Thr942-AHA1 | C terminus | GLDIDpTAGHHYTV (2,11,20) | ||
| Thr942-AHA2 | C terminus | GLDIEpTPSHYTV (2,11) | ||
| Ser944-AHA2 | C terminus |
|
Inhibits (1) | Decreases in response to flg22 (23). |
| Tyr947-AHA1 | C terminus |
|
Decreases during Fe deficiency (10). | |
| Tyr946-AHA2 | C terminus |
|
Inhibitsb (1) | |
| Tyr947-AHA3 | C terminus | GLDIETAGHpYTV (2,12) | ||
| Thr948-AHA1 | C terminus |
|
Increases in response to sucrose (9); phosphorylated in ammonium-adapted (but not nitrate-adapted) plants (16); decreases in response to flg22 (7,23); slightly deregulated in response to auxin (11); upregulated in response to ethylene (25); MPK3 indirectly regulates phosphorylation (6); increased phosphorylation in the BSK8S213D& mutant background (26); upregulated under high C/low N-nutrient conditions (27). | |
| Thr947-AHA2 | C terminus |
|
Phosphorylation required for binding of 14-3-3 protein; protein cannot be activated in its absence (1,28,29) |
|
| Thr948-AHA3 | C terminus |
|
||
| Thr948-AHA6 | C terminus | GLDIDNLNQHYpTV (30) | ||
| Thr959/955-AHA4/11 | C terminus | GLDIETIQQAYpTV (1,17,24) LKGLDIETIQQAYpTV (7,14) | Upregulated in response to ABA or dehydration stress (24). |
References: (1) Rudashevskaya et al. (2012); (2) Wang et al. (2020); (3) Wu et al. (2013); (4) Lin et al. (2015); (5) Mattei et al. (2016); (6) Rayapuram et al. (2018); (7) Nühse et al. (2007); (8) Reiland et al. (2009); (9) Niittylä et al. (2007); (10) Lan et al. (2012); (11) Zhang et al. (2013); (12) Bhaskara et al. (2017); (13) Engelsberger and Schulze (2012); (14) Hoehenwarter et al. (2013); (15) Van Leene et al. (2019); (16) Menz et al. (2016); (17) Wang et al. (2013a); (18) Jones et al. (2009); (19) Whiteman et al. (2008); (20) Sugiyama et al. (2008); (21) Nühse et al. (2003); (22) Fuglsang et al. (2003); (23) Benschop et al. (2007); (24) Umezawa et al. (2013); (25) Yang et al. (2013); (26) Wu et al. (2014); (27) Li et al. (2020); (28) Svennelid et al. (1999); (29) Fuglsang et al. (1999); (30) Mayank et al. (2012); (31) Chen et al. (2010); (32) Xue et al. (2013).
Glu substitution.
Bold letters indicate phosphorylated amino acid residues, “p” denotes phosphorylation.
Figure 1.
Phosphosites in the C-terminal regions of AHAs. In vivo phosphosites are highlighted in red boxes with white text. Underlined sequences represent phosphopeptides that cannot be associated with a unique isoform (see Table 1). Residues in the autoinhibitory regions (Regions I and II) are marked in bold. Conserved amino acid residues are colored according to side chain properties (brown, charged; green, polar; blue, hydrophobic; pink, no side chain).
Figure 2.
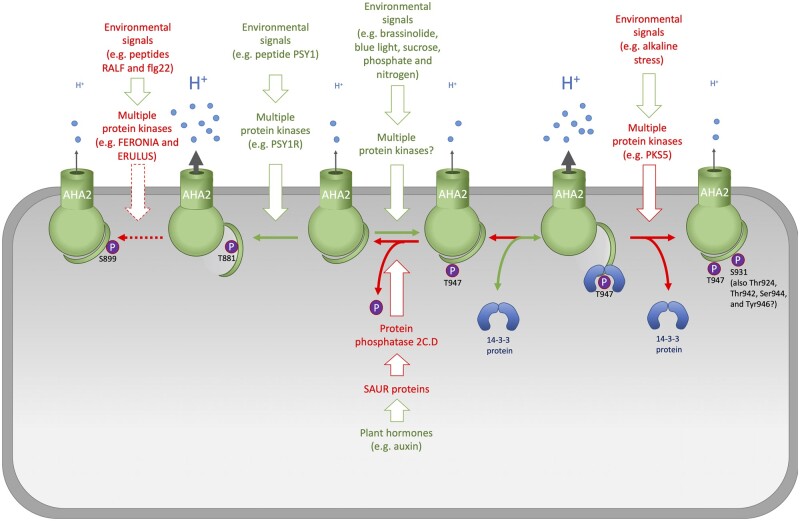
Model for the regulation of AHAs. Arabidopsis AHA2 is used as an example. In response to environmental and developmental signals, multiple protein kinases phosphorylate AHA2 at multiple residues, most of which are located in the C-terminal autoinhibitory R domain. Phosphorylation of Thr947, the penultimate residue, generates a binding site for activating 14-3-3 protein. Phosphorylation at other sites can lead to both activation and de-activation of AHA2 activity, e.g. by preventing binding of 14-3-3 protein. Protein kinase-mediated phosphorylation is countered by protein phosphatases, and the combined action of protein kinases and phosphatases dictates the activation status of AHA2. Colors of fonts and arrows: red, negative regulation; green, positive regulation. Solid arrows, known candidates; Dashed arrows, suggested candidates. PSY1R, PSY1 receptor; PKS5, sos2-like protein kinase 5.
Regulation by 14-3-3 protein. More than 20 years ago, an essential regulatory mechanism for the activation of the pumps was identified, namely, the binding of the regulatory 14-3-3 proteins to the residues at the very end of the C terminus of the R domain (Jahn et al., 1997). 14-3-3 proteins are a large group of soluble proteins involved in regulating the activities of many proteins (Bridges and Moorhead, 2005). The first phosphosite identified in a plasma membrane H+-ATPase (and since then the most commonly observed) is the C-terminal penultimate residue (a Thr; Thr947 in AHA2; Olsson et al., 1998). Subsequently, the phosphorylation of Thr947 was shown to generate the binding site for 14-3-3 proteins (Fuglsang et al., 1999; Svennelid et al., 1999).
The phosphorylation event that creates the 14-3-3 protein binding site occurs spontaneously in highly purified plasma membranes when ATP and Mg2+ are added (Svennelid et al., 1999; Fuglsang et al., 2006), suggesting that the residue corresponding to Thr947 in AHA2 is phosphorylated by a plasma membrane-bound protein kinase that is constitutively active. If this is true, the phosphorylation of Thr947 may in turn be opposed by constitutively active protein phosphatase(s). In this manner, disturbance of the equilibrium between phosphorylation and dephosphorylation, e.g. by the specific inhibition of a protein phosphatase, would provide a mechanism for the rapid regulation of plasma membrane H+-ATPase activity (Falhof et al., 2016). More than one protein kinase may be involved in phosphorylating Thr947. In a mutant of Brassinosteroid Signaling Kinase 8 (BSK8) that mimics the overexpression of BSK8, the penultimate residues of both AHA1 and AHA2 have reduced phosphorylation (Wu et al., 2014). As BSK8 likely activates the Kelch-protein phosphatase Bri1 Suppressor Protein1 (BSU1)-Like2 (BSL2) (Wu et al., 2014), BSL2 represents a candidate negative regulator of AHA1 and AHA2. Mitogen-activated Protein Kinase3 (MPK3) appears to be a regulator of phosphorylation of the C-terminal Thr in AHA1, as deletion of MPK3 reduces its phosphorylation status, even though the Thr947 phosphosite does not match the canonical MPK3 signature (Rayapuram et al., 2018). Plasma membrane-bound protein kinase transmembrane kinases are emerging as important signal transduction components that interact with both abscisic acid (ABA; Li et al., 2021) and auxin (Huang et al., 2019) signaling pathways and should also be considered as having Thr947 as one of their (direct or indirect) targets.
The 14-3-3 protein binds to plasma membrane H+-ATPase as a dimer; the contact region involves the 28 C-terminal residues of the pump (Fuglsang et al., 2003; Ottmann et al., 2007). In this region, phosphorylation of residues other than Thr947 can be predicted to abolish 14-3-3 protein binding even when Thr947 is phosphorylated, which would allow for crosstalk between different signaling pathways. Four conserved residues (Thr924, Ser931, Thr941, and Tyr946) among the dominant AHAs (AHA1, AHA2, AHA3, AHA4, and AHA11) are in vivo phosphorylated in at least one of these AHAs (Figure 1). Mutating Thr924 or Ser931 (Fuglsang et al., 2003, Fuglsang et al., 2007; Rudashevskaya et al., 2012) to Asp (a phosphomimic) abolished the interaction with 14-3-3 protein, even when Thr947 was phosphorylated. The same may be true for Thr942 and Tyr946.
The phosphorylation of residues close to or within the autoinhibitory regions (Regions I and II) activates the pump. These residues include Thr881 in Region I (Niittylä et al., 2007; Rudashevskaya et al., 2012) and Ser904 in Region II (Rudashevskaya et al., 2012). The phosphorylation of Thr881 (in AHA1 or AHA2) is reduced under conditions that result in plasma membrane depolarization (Nühse et al., 2007; Engelsberger and Schulze, 2012). Ser904 in AHA1 or AHA2 functions as a substrate of Sucrose Non-Fermenting 1-related protein kinase2.4 (SnRK2.4) in response to osmotic stress caused by mannitol treatment (Wang et al., 2020).
AHAs control stomatal movements in response to blue light, hormones, and pathogens
Swelling of stomatal guard cells results in opening of the stomatal pore and is a process energized by the plasma membrane H+-ATPase. As opening of stomatal pores allows the plant to take up CO2 and at the same time increases loss of water from the leaf interior, the process has to be tightly regulated. Bacterial pathogens gain entry into leaves through wounds or natural openings such as stomata. Thus, an additional function of guard cells is as active immune signaling cells that rapidly close upon sensing microbe-associated molecular patterns (MAMPs), such as bacterial flagellin, effectively blocking the entry of pathogens into the leaf interior (Melotto et al., 2006). The proton ATPase translocation control 1 (PATROL1) protein regulates secretion of AHA1 to the plasma membrane of stomatal guard cells (Hashimoto-Sugimoto et al., 2013).
Blue light controls the opening of stomata by activating plasma membrane H+-ATPases via the phosphorylation and binding of 14-3-3 proteins (Kinoshita and Shimazaki, 1999). Several protein kinases involved in the blue light response have been identified, but a protein kinase that directly phosphorylates Thr947 in the proton pump has not been identified. The initial receptors of blue light are the protein kinases PHOTOTROPIN 1 (PHOT1) and PHOT2 (Kinoshita et al., 2001). Following activation by blue light, phototropins phosphorylate another protein kinase, BLUE LIGHT SIGNALING 1 (BLUS1; Takemiya et al., 2013). PHOT1 phosphorylates BLUS1 more efficiently than PHOT2 in stomatal guard cells, but the target of BLUS1 remains uncertain; nonetheless, the end result of blue light-dependent activation of phototropins is phosphorylation and activation of the downstream target, the plasma membrane H+-ATPase (Takemiya and Shimazaki, 2016). In a recent study, Inoue et al. (2020) searched specifically for protein kinases that interact with phototropins and identified a calcineurin B-like protein (CBL)-interacting protein kinase 23 (CIPK23) that interacts with both PHOT1 and PHOT2; however, CIPK23 was not phosphorylated by PHOT1 in vitro. Mutants lacking CIPK23 exhibited impaired stomatal opening in response to blue light. Although plasma membrane H+-ATPase activity was not affected in cipk23 mutants, blue light-induced activation of inward-rectifying K+ channels was impaired in mutant guard cells, as was observed in the blus1 mutant (Takemiya et al., 2013). These findings suggest that CIPK23 promotes stomatal opening by activating K+ channels, most likely in concert with BLUS1, via a mechanism different from that of plasma membrane H+-ATPase activation. Two other kinases, Raf-like kinases Convergence of Blue Light and CO2 1 and 2 (CBC1 and CBC2), were originally identified as positive regulators of both blue light- and low CO2-induced stomatal opening (Hiyama et al., 2017). However, a recent study showed that the phosphorylation level of the C-terminal Thr of plasma membrane H+-ATPase was higher in cbc1 cbc2 guard cells than in those of the wild-type and that stomata in the cbc1 cbc2 epidermis opened partially in the dark and opened further in response to light (Hayashi et al., 2020). Thus, CBC1 and CBC2 play redundant roles in negatively regulating stomatal opening in both the dark and blue light.
A recent addition to our understanding of the regulation of the plasma membrane H+-ATPase in guard cells comes from the finding that an auxin regulatory module consisting of the regulatory protein small auxin-up RNA (SAUR) and its target protein phosphatase 2C.D (PP2C.D) modulate stomatal opening and the phosphorylation status of Thr947 in AHA2/1. Specific PP2C.Ds (2, 5, and 6) act redundantly to inhibit stomatal opening, and SAUR56 and SAUR60 play partially redundant roles in opening stomata in response to light (Wong et al., 2021). These findings suggest that not only auxin, but also other stimuli use this regulatory module.
Roles of AHAs in regulating cell elongation and root growth
Roots (especially root hairs) facilitate nutrient uptake by the plant. Root hair growth is regulated by the plant hormone auxin and depends on the localized biosynthesis, secretion, and modification of the root hair tip cell wall (Lee and Cho, 2013).
Root hair tip growth
Since AHA7 is highly expressed in both pollen and root hairs, its role in tip growth was recently investigated (Hoffmann et al., 2019). aha7 does not exhibit a visible phenotype in any of these cell types, which suggests a redundant function with other pumps. aha2 and aha2 aha7 single and double mutants have longer root hairs than the wild-type but cannot be distinguished from each other. AHA7 contains an unusual domain that causes its activity to be inhibited at low apoplastic pH (Hoffmann et al., 2019), but the exact physiological role of this feature is unknown.
Another root hair phenotype related to these pumps is observed in the erulus (eru) mutant, which has very short root hairs (Schoenaers et al., 2018). ERULUS (a CrRLK1L kinase), which is localized to the apical root hair plasma membrane, regulates cell wall composition in root hairs and modulates pectin dynamics by negatively regulating pectin methylesterase activity. Phosphopeptides from FERONIA and AHA1/2 are present at significantly lower levels in eru plants than in the wild-type (FERONIA causes apoplastic alkalinization upon the binding of Rapid Alkalization Factor [RALF] peptides; see below). Among the phosphopeptides was an AHA1 or AHA2 derived peptide in which Ser904 was phosphorylated, which is a post-translational modification previously suggested to activate the pump (Rudashevskaya et al., 2012). This would indicate that ERU activates the pump and causes acidification of the apoplast. Auxin supplementation caused a decrease in the abundance of two other phosphopeptides, an AHA1/2-derived peptide phosphorylated at Ser899 and one from AHA2 phosphorylated at Thr947 (Schoenaers et al., 2018). As phosphorylation at these two sites activates the pump, it is challenging to interpret these data; perhaps other auxin-stimulated processes interfere with this activation.
Regulation by growth-promoting hormones
According to the acid growth theory (Rayle and Cleland, 1970; Hager et al., 1971; Rayle, 1973), auxin triggers the activation of plasma membrane H+-ATPases, and the resulting acidification of the apoplast softens the rigid cell wall and allows for cell expansion. This theory has been debated, especially since there are inconsistencies in the way in which leaves and roots respond to auxin, e.g. external application of auxin to roots leads to alkalinization of the apoplast (Monshausen et al., 2011; Gjetting et al., 2012). Using a combination of the external probe 8‐hydroxypyrene‐1,3,6‐trisulfonic acid and two auxin response reporter lines, Barbez et al. (2017) demonstrated that endogenous auxin in roots triggers cell wall acidification that leads to cellular expansion. Reductions in auxin levels, perception, or signaling abolish both extracellular acidification and cellular expansion (Barbez et al., 2017).
Auxin enhances the phosphorylation level of Thr947 in Arabidopsis hypocotyls within 10 min after addition, without altering the amount of the plasma membrane H+-ATPase, and concomitant with activation of hypocotyl elongation (Takahashi et al., 2012). This effect need not occur via stimulation of a protein kinase. Among many other effects, auxin induces the expression of SAUR genes (McClure and Guilfoyle, 1987; Hagen and Guilfoyl, 2002). The identification of SAUR proteins as inhibitors of PP2C.D1, a type 2C protein phosphatase, and the observation that PP2C.D1 dephosphorylates Thr947 in plasma membrane H+-ATPase (Spartz et al., 2014) have taken our understanding of the roles of auxin in regulating plasma membrane H+-ATPase activity to unprecedented heights and led to a wave of studies in the field of auxin-regulated growth.
The phytohormone brassinolide also regulates the activity of plasma membrane H+-ATPases. In 2011, a model based on studies of the interaction between BRASSINOSTEROID INSENSITIVE 1 (BRI1) and AHA1 was proposed describing a rapid brassinolide L-regulated signal response pathway containing BRI1 and a plasma membrane H+-ATPase that regulates cell wall expansion (Caesar et al., 2011). BL-induced cell elongation and phosphorylation of plasma membrane H+-ATPases were subsequently shown to exhibit the same dependency on brassinolide concentration (Minami et al., 2019). This phosphorylation is dependent on BRI1, which is regulated by SAUR proteins and leads to the phosphorylation of Thr947, the penultimate residue in plasma membrane H+-ATPase.
The response of AHAs to nutrient supply
As plasma membrane H+-ATPases drive several nutrient transport processes, a number of laboratories have systematically tested the phospho-proteome in response to changes in nutrient availability. An early study showed that the addition of sucrose upon starvation results in increased phosphorylation of the Thr947 residue of plasma membrane H+-ATPase (Niittylä et al., 2007). Similarly, high C/low N nutrient stress increases the phosphorylation levels of Thr947 and 14-3-3 binding in AHA1/2 (Li et al., 2020). Also, photosynthetic activity influences the plasma membrane H+-ATPase activity via phosphorylation of Thr947 (Okumura et al., 2016). This is dependent on the activity of SUCROSE PROTON SYMPORTER2 (SUC2) and could be part of the same response pathway observed upon addition of sucrose (Niittylä et al., 2007). The overexpression of the plasma membrane H+-ATPase traffic regulator PATROL1 leads to faster stomatal opening responses and elevated photosynthetic rates (Kimura et al., 2020). Finally, the overexpression of the rice (Oryza sativa) plasma membrane H+-ATPase 1 (OSA1) results in increased contents of N, P, and K compared with wild-type plants (Zhang et al., 2021), which is caused by a combination of increased photosynthesis rate and stronger root-mediated acidification of the growth medium.
Another example of the role of plasma membrane H+-ATPases in the nutrient response is related to root branching (Meier et al., 2020). Auxin-mediated root branching was found to be determined by the form of available nitrogen. The authors concluded that apoplastic acidification in response to ammonium uptake is mediated by plasma membrane H+-ATPases, including AHA2, and increases auxin diffusion and lateral root branching.
Another aspect of plasma membrane H+-ATPase activity in nutrient transport is the influence of individual isoforms: Do they all respond similarly to a stimulus if present in the same tissue? In rice (O. sativa), silencing of OsA2 affected the ability of the plant to adapt to low concentrations, even when other isoforms were expressed in the same tissue (Loss Sperandio et al., 2020). This finding is in contrast to the results of Haruta et al. (2010), who reported that the effects of single mutations in genes encoding different isoforms are mostly masked by overlapping patterns of expression and redundant functions, as well as by compensation at the posttranslational level.
Regulation of AHAs by signaling peptides
A topic that has recently drawn much interest is the roles of signaling peptides in regulating plant growth. Plant peptide containing sulfated tyrosine 1 (PSY1; Amano et al., 2007) and phytosulfokine (PSK; Matsubayashi and Sakagami, 1996) function as hormones in cell elongation activity in the elongation/differentiation zone of the root (Kutschmar et al., 2009; Matsubayashi, 2011) and hypocotyl (Stührwohldt et al., 2011; Fuglsang et al., 2014). The receptors for PSY1 and PSK (PSY1R and PSKR1/PSKR2) are leucine-rich repeat receptor kinases. PSY1R phosphorylates the C-terminal domain of AHA2 at Thr881, thus activating this plasma membrane H+-ATPase. Proton pumping is observed upon the addition of PSY1 peptide and is dependent on the presence of PSY1R, but occurs independently of 14-3-3 binding (Fuglsang et al., 2014). In the receptor mutant psy1r, Fuglsang et al. (2014) observed a decreased cell length in etiolated seedlings, whereas Amano et al. (2007) reported a more pronounced effect on root length.
Interestingly, the growth-promoting effects of PSK do not require extracellular acidification by plasma membrane H+-ATPases (Stührwohldt et al., 2011). PSKR1 failed to interact with AHA2 in a bimolecular fluorescence complementation assay (Fuglsang et al., 2014), but the two proteins do co-localize in the plasma membrane (Ladwig et al., 2015) suggesting that they are part of the same plasma membrane domain.
RALF peptides constitute another family of signaling peptides that inhibit plant growth and downregulate plasma membrane H+-ATPase activity. The receptor kinase FERONIA binds to RALF1 peptides, triggering a signal transduction cascade that ends with the phosphorylation of Ser899 on the C-terminal domain of AHA2. This phosphorylation is considered to inhibit the pump, thus reducing cellular expansion (Haruta et al., 2014). Intriguingly, this is in contrast to the results obtained when the aha2 Ser899Asp mutant protein is expressed in yeast; here, the Ser to Asp mutation (phospho-mimicking) results in higher plasma membrane H+-ATPase activity (Rudashevskaya et al., 2012). FERONIA is not the only receptor of RALFs in the root, as other receptor kinase(s) perceive RALF peptides and initiate the Ca2+ signature required to reduce plasma membrane H+-ATPase activity (Gjetting et al., 2020).
The roles of AHAs in abiotic stress responses
ABA plays an important role in plant drought responses. ABA inhibits plasma membrane H+-ATPase activity, thus promoting stomatal closure under drought stress, and reducing water loss. The underlying mechanism, described only recently, includes internalization of the pump and soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins (Xia et al., 2019). SNARE proteins play an essential role in vesicle trafficking in eukaryotes by facilitating the fusion of vesicles with their target membranes. Thus, the Qa-SNARE protein Syntaxin of Plants 132 (SYP132), the expression of which is tightly regulated by auxin, reduces the amount of plasma membrane H+-ATPase protein at the plasma membrane. ABA induces the SNARE protein Vesicle-associated Membrane Protein 711 (VAMP711) to interact with AHA1 and AHA2 (Xue et al., 2018). The inhibitory interaction of VAMP711 with plasma membrane H+-ATPases occurs at the R domain (see Box 1). Deletion of VAMP711 in Arabidopsis results in higher H+-ATPase activity and slower stomatal closure in response to ABA or drought. In addition, overexpressing VAMP711 partially rescues the drought-sensitive phenotype of open stomata 2 (ost2; Xue et al., 2018), which harbors a mutation in AHA1 that results in the constitutive activation of this plasma membrane H+-ATPase (Merlot et al., 2007). An internalization mechanism also underlies auxin-mediated regulation of plasma membrane H+-ATPase activity (Xia et al., 2019). The physiological consequences of SYP132 overexpression include reduced apoplast acidification and suppressed vegetative growth (Xia et al., 2019). The same phenomenon is observed in response to dim/blue light (Haruta et al., 2018). In this study, intracellular accumulation of AHA2 occurs specifically in the transition zone and is dependent on a functional FERONIA-receptor. It suggests that developmental factors influence root growth via re-localization of the plasma membrane H+-ATPase, potentially in response to receptor kinase-mediated signaling. Thus, a common scheme is emerging: In addition to posttranslational modifications, plasma membrane H+-ATPase activity is regulated by processes including internalization and recycling of these proteins.
Key regulators of ABA signaling are the Protein Phosphatase 2Cs (PP2Cs). PP2Cs suppress the kinase activity of SnRK2s through dephosphorylation of the kinase activation loop and thereby repress ABA signaling. Abscisic acid insensitive 1 (ABI1; a PP2C) directly interacts with the R domain of AHA2 and dephosphorylates Thr947, which decreases H+ extrusion and negatively regulates primary root growth (Miao et al., 2021). Thereby both auxin and ABA responses are regulated by members of the protein phosphatase 2C family; in each case the PP2Cs are from different clades but common among them is that they target the same residue, Thr947.
The sos2-like protein kinase 5 (pks5/cipk11) mutant was originally isolated based on its tolerance to high pH (Fuglsang et al., 2007). PKS5 phosphorylates Ser931 in AHA2 and is regulated by the calcium-binding protein SCaBP3/CBL7 and the chaperone J3 (Yang et al., 2010). A recent study revealed that PKS5 kinase is involved in the mechanistic response to saline–alkaline soil (Yang et al., 2019). A model was proposed in which SCaBP3/CBL7 interacts with the C terminus of AHA2 under nonstressed conditions, allowing for its PKS5-mediated phosphorylation. Under saline–alkaline stress conditions, Ca2+ influx into the cytoplasm is triggered, and the binding of Ca2+ to ScaBP3 releases its inhibitory interaction with the C terminus of AHA2. When the ScaBP3–PKS5 complex is less stable, Ser931 is no longer phosphorylated, allowing for 14-3-3 protein binding and the activation of AHA2.
The roles of AHAs in biotic stress responses
Plant pathogens modulate the apoplastic pH to facilitate infection. Thus, the addition of elicitors leads to the rapid alkalinization of the growth medium, most likely by inhibiting plasma membrane H+-ATPase activity (Saijo et al., 2018).
The fungus Fusarium oxysporum induces the rapid acidification of the apoplast, which modulates cell wall structure and root growth by reducing cellulose synthesis. Infection resulted in the phosphorylation of Thr947 and, notably, treatment with elicitors from F. oxysporum had the same effect (Kesten et al., 2019). This is in contrast to the effect of RALF-like peptides secreted by F. oxysporum. These F-RALFs trigger alkalization of the apoplast due to inhibition of plasma membrane H+-ATPase activity via a FERONIA-dependent pathway (Masachis et al., 2016).
Bacterial pathogens sometimes mimic plant regulatory peptides. For example, the biotrophic pathogen Xanthomonas oryzae pv. oryzae (Xoo) produces a sulfated peptide named RaxX, which shares similarity with peptides in the PSY family (Pruitt et al., 2017). RaxX serves as a molecular mimic of PSY peptides to facilitate Xoo infection and is recognized by the receptor kinase-like protein Xanthomonas resistance 21 (XA21), which has evolved the ability to recognize and respond specifically to the microbial form of the peptide and not the plant version. This was further demonstrated by binding studies showing that RaxX peptides bind to the ectodomain of XA21, in contrast to PSY1 (Luu et al., 2019).
The peptide flg22, a MAMP derived from bacterial flagellin, leads to alkalinization of the apoplast and changes the phosphosites on AHA1/AHA2. In response to flg22, Thr881 and Thr947 phosphorylation decreases, whereas Ser899 phosphorylation increases (Nühse et al., 2007). De-phosphorylation of Thr881 and Thr947, as well as phosphorylation of Ser899, all reduce plasma membrane H+-ATPase activity (Nühse et al., 2007; Table 1).
Another fungus that specifically targets plant plasma membrane H+-ATPases is Stemphylium loti. This fungus secretes tenuazonic acid, which inhibits plasma membrane H+-ATPase activity via a mechanism involving its C-terminal domain, resulting in necrotic cells (Bjørk et al., 2020).
Biotic interactions can benefit plants and plasma membrane H+-ATPases also function in this interaction. Trichoderma species have long been known to have beneficial effects on plant growth (Yedidia et al., 2001; Contreras-Cornejo et al., 2014). Although the underlying mechanism is unknown, plasma membrane H+-ATPase activity is modulated by Trichoderma ssp. (López-Coria et al., 2016; Guo et al., 2020), which suggests that activation of the plasma membrane H+-ATPase is part of the mechanism underlying the beneficial effects.
The autoinhibited P-type Ca2+-ATPase gene family in Arabidopsis
Arabidopsis P-type Ca2+-ATPases belong to two subfamilies: P2A ATPases (four endomembrane Ca2+-ATPases named ER-type Ca2+-ATPases [ECA]1, 2, 3, and 4) and P2B ATPases (10 so-called autoinhibited Ca2+-ATPases [ACA]1, 2, 4, 7, 8, 9, 10, 11, 12, and 13) (Box 3; Baxter et al., 2003; Palmgren et al., 2020). For a recent review on plant Ca2+-ATPases, the reader is referred to García Bossi et al. (2020).
Box 3.
The two Ca2+-ATPase families in plants.
Two families of P-type Ca2+-ATPases exist in plants, P2A and P2B, both of which have an ancient origin and exist in all life forms. Whereas P2B ATPases are regulated by the binding of Ca2+/calmodulin to an autoinhibitory terminal domain in the pump, the simpler P2A ATPases do not have such a domain. P2A ATPases are divided into two groups, P2A-I and P2A-II, which separated from each other in an early eukaryotic cell before the emergence of plants, animals, fungi, stramenopiles, alveolata, and rhizaria (Palmgren et al., 2020). In Arabidopsis, ECA3 is a P2A-I ATPase (like animal sarco-endoplasmic reticulum Ca2+-ATPases; SERCA pumps), whereas ECA1, ECA2, and ECA4 are P2A-II ATPases. As each group has an ancient origin, one would expect P2A-I and P2A-II ATPases to have properties distinct from each other. Strikingly, however, functional differences between members of each group have not been identified so far. Both the Arabidopsis eca1 and eca3 mutants exhibit phenotypes related to Ca2+ and Mn2+ sensitivity, suggesting that both pumps play physiological roles related to supplying intracellular compartments with Ca2+ and Mn2+. Thus far, there is little evidence that the activities of P2A ATPases are regulated by external cues (reviewed in García Bossi et al., 2020). In animal cells, P2B ATPases are exclusively expressed in the plasma membrane, whereas in plant cells, each P2B ATPase isoform is expressed in either the plasma membrane or intracellular membranes. In Arabidopsis, ACA8 and ACA10 are the predominant plasma membrane Ca2+ pumps, whereas ACA4 and ACA11 are the major Ca2+ pumps in the vacuolar membrane. ACA1, ACA2, and ACA7 are expressed in the ER (Ishka et al., 2021). Among ACAs, only ACA12 (and likely also ACA13) is constitutively active and is not activated by calmodulin, even though it is able to interact with calmodulin (Limonta et al., 2014). ACAs are tightly regulated in response to external cues, as discussed in detail in the main text.
P2A ATPases: ECAs
P2A ATPases supply organelles with Ca2+, but there is little evidence that they regulate cytoplasmic Ca2+ concentrations. P2A ATPases transport two Ca2+ or Mn2+ ions per ATP split and have short N- and C-terminal sequences.
P2B ATPases: ACAs–
P2B ATPases maintain cytoplasmic Ca2+ at very low (sub-micromolar) concentrations and, in plant cells, load internal stores with Ca2+. P2B ATPases transport a single Ca2+ per ATP split and are equipped with an N-terminal (in plants) or C-terminal (in animals) autoinhibitory domain that binds calmodulin. When activated, P2B ATPases have a higher affinity for Ca2+ than P2A ATPases and are therefore more efficient in removing Ca2+ from the cytoplasm.
Why are there different isoforms of ACAs?
The presence of 10 ACA isoforms suggests that their gene products have different properties. However, there is little evidence for this and, despite structural differences between ACAs they appear to be functionally alike. Arabidopsis aca10 mutants have a marked compact inflorescence (cif) phenotype that is not observed in aca8 or aca9 mutants. Nonetheless, ACA8 can complement the cif phenotype when overexpressed in the aca10 background (George et al., 2008), suggesting that ACA8 might have the same biochemical function as ACA10. By equipping the plasma membrane-localized ACA8 with the vacuolar targeting signal of ACA4, ACA8 was successfully relocalized to the tonoplast (Hilleary et al., 2020) and, at this location, rescued the phenotype of the aca4 aca11 mutant, which has an elevated Ca2+ response to flg22. Thus, specific biochemical or regulatory properties of ACA4 and ACA11 do not determine their ability to modulate the Ca2+ signal induced by flg22, but instead, the vacuolar location of any ACA appears to be sufficient for this response. In this case it may be reasonable to assume that isoform diversity is merely a means for controlling the tissue-specific distribution of ACAs within the cellular space and over time.
Regulation of ACAs by Ca2+/calmodulin
The Ca2+ affinity of plant P2B ATPases is regulated by their N-terminal regulatory domains. In response to an increase in [Ca2+]cyt, Ca2+ forms a complex with calmodulin (encoded by seven genes in Arabidopsis), which can then bind to the N-terminal domains of P2B ATPases. Binding neutralizes the inhibitory effect of the domain on the pump, which results in an active pump state with high affinity for Ca2+. Thus, Ca2+ not only acts as a transported ligand of ACAs, but also increases the affinity of ACAs for Ca2+. The N-terminal domain of ACA8 also interacts with Ca2+-CML36, a calmodulin-like protein (CML), and other ACAs also likely interact with Ca2+-CMLs (encoded by 50 genes in Arabidopsis; Astegno et al., 2017).
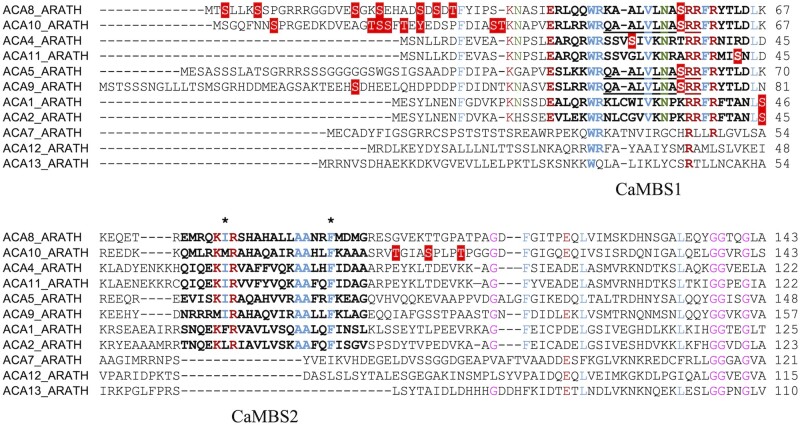
The structure of the N-terminal domain of AtACA8 in complex with calmodulin appears as a long alpha-helix with two calmodulin molecules bound to it (Tidow et al., 2012). The two sites, named calmodulin binding sites 1 and 2 (CaMBS1 and CaMBS2), bind Ca2+-calmodulin with different, but physiologically relevant, affinities and provide the pump with a high- and a low-affinity sensor for Ca2+, respectively. CaMBS1 overlaps with an autoinhibitory sequence, which is neutralized by the binding of Ca2+-calmodulin (Baekgaard et al., 2006). CaMBS2 likewise overlaps with a less-defined autoinhibitory stretch of residues (Tidow et al., 2012). Mathematical network modeling demonstrated that such a system with two binding sites is ready for steep activation above a basal Ca2+ concentration, meaning that the pump is inactive below a certain level and rapidly activates as soon as Ca2+ concentrations exceed this level (Tidow et al., 2012).
Among ACAs, ACA7, ACA12, and ACA13 have degraded calmodulin-binding sites (Figure 3), which explains why ACA12 is constitutively activated but still able to bind Ca2+/calmodulin (Limonta et al., 2014).
Figure 3.
Phosphosites in the N-terminal regions of ACAs. Red boxes with white text represent phosphosites observed in vivo, except for the ACA2 phosphosite, which was phosphorylated in vitro. Underlined sequences represent phosphopeptides that cannot be associated with a unique isoform (see Table 2). Hydrophobic anchor points involved in calmodulin binding are marked with asterisks. Residues in CaMBS1 and CaMBS2 are marked in bold. Conserved amino acid residues are colored according to side chain properties (brown, charged; green, polar; blue, hydrophobic; pink, no side chain).
Regulation of ACAs by phosphorylation
The N-terminal autoinhibitory domain of ACAs can undergo posttranslational modification by phosphorylation, which can weaken the binding capacity of calmodulin or CML proteins (resulting in a decreased Ca2+ affinity) or relieve autoinhibition in the absence of these proteins (resulting in an increased Ca2+ affinity).
The N-terminal residues upstream of CaMBS1 in the closely related plasma membrane Ca2+-ATPases ACA8 and ACA10 are hotspots of phosphorylation, and several can be phosphorylated simultaneously, but phosphosites are not conserved between the two pumps (Figure 3). Mutant ACA8 pumps in which such upstream residues have been substituted with the phosphomimic Asp appear to be partially activated (Giacometti et al., 2012). Ser29-ACA8 and Ser22-ACA10 are targets of Oxidative Signal-Inducible 1 (OXI1) in response to H2O2 (Wang et al., 2020).
Further downstream, in CamBS1, Ser57 in the sequence motif NASRRF has also been found to be phosphorylated, but less often. Whether this phosphorylation event promotes or decreases pump activity is unknown, but it is predicted to interfere with the binding of Ca2+-calmodulin. In ACA2, an Asp substitution of Ser45, which is not homologous to ACA8 Ser57 but is likewise situated in CamBS1, reduces calmodulin-induced stimulation of the recombinant ACA2 produced in yeast. This observation suggests that mutations in this region interfere with calmodulin stimulation.
The CDPK isoform CPK1, a Ca2+-dependent protein kinase, phosphorylates Ser45 of ACA2. Phosphorylation inhibits the binding of calmodulin and does not occur when calmodulin has already bound to ACA2 (Hwang et al., 2000).
The calcium-dependent protein kinase CPK16 phosphorylates the N terminus of ACA8 at Ser19 and Ser22 (Giacometti et al., 2012). ACA8 is phosphorylated at multiple sites within its N terminus by CBL and CIPK complexes (Costa et al., 2017). The phosphorylation state of specific residues is enhanced by in vivo treatment with ABA and gibberellins (Ser27 and Ser29; Chen et al., 2010), or with the elicitor flagellin (Ser27 and Ser99 situated just downstream of CamBS2; Nühse et al., 2003, 2004, 2007; Benschop et al., 2007), and is downregulated in response to sucrose in cultured cells (Ser22; Niittylä et al., 2007).
ACAs and Ca2+ signaling
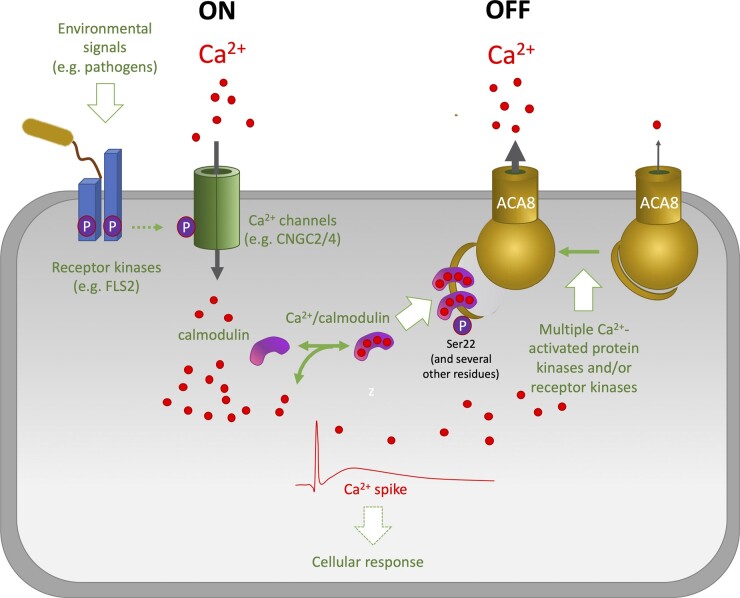
Ca2+-ATPases maintain cytoplasmic [Ca2+] at sub-micromolar levels even though external [Ca2+] is typically in the millimolar range. When cytoplasmic Ca2+ concentrations increase in response to external cues, ACAs work together with H+/Ca2+-antiporters to lower Ca2+ concentrations again, thereby attenuating the signal (Figure 4). The magnitude of the Ca2+ spike can provide important information about the nature of the signal that elicited a particular response and provide a language for how to respond to the signal. A recent study provided an excellent example of this concept (Suda et al., 2020). Touching a sensory hair cell on the surface of a Venus flytrap (Dionaea) leaf elicits a Ca2+ spike without a detectable physiological response, while touching the same hair again a few seconds later elicits a new spike that augments the previous one. If the second stimulus causes [Ca2+]cyt to exceed a certain threshold level, movement of the leaf is triggered, trapping the prey. The elevated [Ca2+]cyt decreases two-phase exponentially after the first stimulus (Suda et al., 2020), demonstrating that the process is the result of the sum of fast and slow exponential decay. This prompts the question: How do ACAs contribute to shaping such Ca2+ spikes?
Figure 4.
Model for the regulation of ACAs. Arabidopsis ACA8 is used as an example. In Ca2+ signaling, an “on” period, which is induced by external signals that provoke a rapid influx and surge in [Ca2+]cyt, is always followed by an “off” period, when Ca2+ is removed from the cytoplasm again (Berridge et al., 2000, 2003). This results in a Ca2+ spike, which can be repeated rhythmically to produce Ca2+ oscillations or can proceed throughout the plant as a Ca2+ wave. Both the influx and efflux of Ca2+ involve multiple transport proteins whose combined activity determines the amplitude and frequency of the Ca2+ signal (Kudla et al., 2018; Tian et al., 2020). Furthermore, it has been proposed (Hwang et al., 2000; Sze et al., 2000) that the opposite actions of calmodulin and phosphorylation allow for crosstalk between different Ca2+ signaling pathways that modulate the initial calmodulin activation kinetics to shape a specific Ca2+ signature. The brown structure interacting with the receptor kinase to the left depicts a bacterium with its flagellum. Colors of fonts and arrows: red, negative regulation; green, positive regulation. Solid arrows, known candidates; Dashed arrows, suggested candidates.CNGC2/4, Cyclic Nucleotide Gated Channel 2 and 4.
Even though aca4 aca11 plants have an elevated Ca2+ signal in response to flg22, Ca2+ concentrations in both wild-type and aca4 aca11 plants return to basal levels concurrently. This observation suggests that, in response to flg22, ACA4 and ACA11 are important for regulating the maximal magnitude of the Ca2+ spike, while the duration of the spike is controlled by efflux systems other than ACA4 and ACA11 (Hilleary et al., 2020). In the absence of ACAs, cytoplasmic Ca2+ levels increase, which leads to severe stress symptoms, as seen in aca4 aca11 double and aca1 aca2 aca7 triple mutants, whose leaves show lesions resulting from the induction of programmed cell death (Hilleary et al., 2020; Ishka et al., 2021).
ACAs and plant defense
MAMPs such as flg22 function as elicitors that, following binding to cell surface receptors, trigger changes in Ca2+ levels inside the plant cell. Ca2+-based signaling can then induce pattern-triggered immunity (PTI), which can lead to systemic acquired resistance (Ranf et al., 2011). The aca4 aca11 double mutant has a high frequency of hypersensitive response-like lesions, which result from the activation of a salicylic acid-dependent programmed cell death pathway (Boursiac et al., 2010), and aca4 aca11 plants exhibit a further increase in Ca2+ signal in response to flg22 (Hilleary et al., 2020), suggesting that ACA4 and ACA11 contribute to the “off” mechanism in response to MAMPs. Loss of another Arabidopsis Ca2+ efflux system, the antiporter H+/Ca2+ exchange 1 (CAX1), which is not related to Ca2+-ATPases, also results in the activation of plant defense and accelerated senescence (Zhang et al., 2020).
Mutant aca8 aca10 plants have an increase in basal [Ca2+]cyt, but in response to flg22 show a less prominent Ca2+ signal and a relatively minor burst of reactive oxygen species (ROS) that is not followed by spontaneous lesion formation (Frei dit Frey et al., 2012). This finding argues against a direct role of these ACAs in PTI. Analysis of the cif growth phenotype of aca10 suggests that ACA10 is a negative regulator of disease resistance and that the growth retardation observed in the mutant results from a constitutive immune response (Yang et al., 2017). In accordance with this notion, the aca10 single mutant, as well as aca8 aca10 and aca10 aca13, shows increased resistance to the virulent pathogen Pseudomonas syringe pv. tomato (Pst) DC3000 compared to the wild-type (Yu et al., 2018).
Sensing of pathogen-derived elicitors and effectors leads to posttranslational modification of ACAs. AvrRpt2 is a pathogen effector protein from Pseudomonas syringae pv. tomato (Pto). When AvrRpt2 expression was induced in Arabidopsis, phosphorylation of AtACA8 at Ser-22 was observed (Kadota et al., 2019), an event predicted to lead to a partially activated pump (Giacometti et al., 2012). Treatment with flg22 or fungal xylanase leads to phosphorylation of AtACA10 at the N-terminal residue Thr-20 (Benschop et al., 2007). Likewise, treating plants with flg22 leads to phosphorylation of AtACA8 at the N-terminal residue Ser-27 (Nühse et al., 2007) and is also predicted to lead to a partially activated pump (Giacometti et al., 2012). AtACA8 interacts directly with the flg22 receptor FLAGELLIN SENSING2 (FLS2) in planta (Frei dit Frey et al., 2012). Whether FLS2, which has an intracellular protein kinase domain, phosphorylates AtACA8 directly is unknown, but an intriguing possibility is that FLS2-mediated sensing of a bacterial MAMP directly primes the “off” mechanism for export of Ca2+. In such a scenario, sensing of a specific “on” signal would be directly linked to a specific “off” mechanism and together could generate a signal-specific Ca2+ signature.
Concluding remarks
Both plasma membrane H+-ATPases and Ca2+-ATPases are targets for multiple protein kinases responding to cellular development and environmental cues (Tables 1 and 2). This points to a very sensitive apparatus that allows the plant to react differentially to multiple environmental signal received simultaneously. Thus, dependent on the number and localization of residues being phosphorylated, and their stoichiometric relationships, responses with respect to H+ and Ca2+ dynamics are being moderated in a dynamic and continuous fashion. Interacting regulatory proteins serve to fine-tune these responses. As more molecular players are identified, the signaling network is becoming increasingly complex (Figures 2 and 4). When considering cause and effects, it is important to keep in mind that even when a phosphorylation event is detected in vivo, the site in question need not be phosphorylated in the whole pump population. Detecting the different combinations of sites phosphorylated in response to abiotic and biotic stimuli, in addition to mapping the spatiotemporal separation of these sites in the plant material studied, will be required in order to better understand the dynamics caused by multiple phosphorylation events. The current challenge is to couple the ends of this network; which stimulus leads to specific phosphorylation or dephosphorylation events and who are the players on the way? We propose that the regulation is based on more or less constitutively active protein kinases that, in concert with more specific protein phosphatases, cause a dynamic phosphorylation equilibrium that dictates the immediate activation status in response to multiple environmental cues. Testing this hypothesis together with answering other essential questions are important future challenges (see Outstanding questions).
Table 2.
Phosphosites identified in Arabidopsis P2B ACAs. For references see Table 1
| Phosphorylation sites detected in ACA isoforms | Cytosolic domain | Motif | Effect of Asp substitution | Notes |
|---|---|---|---|---|
| Ser3-ACA8 | N terminus | TpSLLKpSSPGR (3) | ||
| Ser7-ACA8 | N terminus | (ac)TSLLKpSSPGR (3) | Reduced phosphorylation level in sirk1 mutant (3) | |
| Ser8-ACA10 | N terminus |
|
||
| Thr20-ACA10 | N terminus | GEDKDVEAGpTSSFTEYEDSPFDIASTK (23) | Increases after xylanase treatment (23); Increases after flg22 treatment (23) | |
| Ser21-ACA10 | N terminus | DVEAGTpSSFTEYEDSPFDIASTK (2) | ||
| Ser22-ACA10 | N terminus | DVEAGTSpSFTEYEDSPFDIASTK (2,31) | Increases after ABA (31), IAA (31); and JA treatment (31); Phosphorylated by OXI1 in an in vitro kinase reaction in H2O2-treated Arabidopsis (2) | |
| Thr24-ACA10 | N terminus | DVEAGTSSFpTEYEDSPFDIASTK (2) | ||
| Tyr26-ACA10 | N terminus | DVEAGTSSFTEpYEDSPFDIASTK (2) | ||
| Ser35-ACA10 | N terminus | DVEAGTSSFTEYEDSPFDIApSTK (2) | ||
| Thr36-ACA10 | N terminus | DVEAGTSSFTEYEDSPFDIASpTK (2) | ||
| ACA10 hotspot | DVEAGTSpS(0.511)FpT(0.434)EYEDSP FDIASTK (5) | |||
| Ser28-ACA4 | N terminus | SSVpSIVK (2) | ||
| Ser23-ACA8 | N terminus |
|
||
| Ser26-ACA8 | N terminus |
|
Decreases in response to sucrose (9) | |
| Ser27-ACA8 | N terminus |
|
Transiently phosphorylated in nitrate-adapted (but not ammonium-adapted) plants (16); Slight decrease in response to auxin (11); Decreases in response to mannitol (32); Increases in response to ABA (31) and GA (31) | |
| Ser29-ACA8 | N terminus |
|
Decreases in response to mannitol (32); increases in response to ABA (31) and GA (31); Phosphorylated by OXI1 in an in vitro kinase reaction in H2O2-treated Arabidopsis (2) | |
| Thr31-ACA8 | N terminus |
|
||
| ACA8 hotspot | N terminus |
|
||
| Ser32-ACA9 | N terminus | TEEHpSDHEELQHDPDDPFDIDNTK (5,17) | ||
| Ser41-ACA11 | N terminus | MIpSNLDK (2) | ||
| Ser46-ACA1 | N terminus | IVKNPKRRFRFTANLpSKRSEAEAIRRSNQEK (15) | ||
| Ser57-ACA8/ACA10; Ser60-ACA5; Ser71-ACA9 | N terminus; CamBS1 |
|
||
| Ser57-ACA8 | N terminus; CamBS1 | KAALVLNApSR (17) | ||
| Ser57-ACA10; Ser60-ACA5; Ser71-ACA9 | N terminus; CamBS1 |
|
||
| Thr100-ACA10 | N terminus; CamBS2 | VpTGIASPLPTPGGGDFGIGQEQIVSISR (2) | ||
| Ser104-ACA10 | N terminus; CamBS2 | VTGIApSPLPTPGGGDFGIGQEQIVSISR (2) | ||
| Thr108-ACA10 | N terminus; CamBS2 | VTGIASPLPpTPGGGDFGIGQEQIVSISR (2) | ||
| Thr104-ACA8 | N terminus; CamBS2 | pTTGPATPAGDFGITPEQLVIMSK (2) | ||
| Ser586-ACA13 | N domain | ILAMCSTFCDGpSGVVREMK (2) | ||
| Ser1073-ACA8 | C terminus | NSSGEGpSL (17) |
Bold letters indicate phosphorylated amino acid residues, “p” denotes phosphorylation.
Advances box
AHAs are regulated at multiple, largely conserved phosphosites, some leading to the activation of these pumps and others to their downregulation.
Inhibition of phosphatase 2C.D by SAUR regulatory proteins is an important mechanism for the activation of AHAs.
Activation of AHAs by auxin and other phytohormones occurs via SAUR proteins.
ACAs are regulated by the phosphorylation of residues in the vicinity of or within calmodulin-binding regions, but phosphosites are not conserved between isoforms.
Differential phosphorylation of ACAs likely modulates the kinetics of pump activation in response to external stimuli, which in turn provides a mechanism for generating stimulus-specific Ca2+ response signatures.
Outstanding questions box
Are many, if not all, phosphosites in plant plasma membrane H+-ATPases and Ca2+-ATPases constitutively phosphorylated by protein kinases and dephosphorylated by protein phosphatases?
Does the equilibrium between phosphate addition and removal dictate protein activity?
In H+ signaling, do “on” components (including receptor kinases) and “off” components (like protein phosphatases) that regulate plasma membrane H+-ATPase activity form functional complexes?
In Ca2+ signaling, do “on” components (such as signal receptors and Ca2+ channels) and “off” components (such as Ca2+-ATPases) form functional complexes that together dictate the Ca2+ signature?
Is internalization of plasma membrane H+-ATPase protein a common regulatory mechanism?
What are the roles of the less-studied plasma membrane H+-ATPases and Ca2+-ATPases?
Acknowledgments
We acknowledge the members of our groups for their contributions and critical discussions of the hypotheses put forward in this review.
Funding
We thank the Danish National Research Foundation (PumpKin; A.T.F. and M.P.), the Innovation Fund Denmark (LESSISMORE; M.P.), Danish research council (COMBAT; project number DFF–4184-00548; ATF), the Carlsberg Foundation (RaisingQuinoa; project number CF18-1113; M.P.), and the Novo Nordisk Foundation projects (NovoCrops; project number 2019OC53580; M.P.) and (PlantsGoImmune; project number NNF19OC0056457; A.T.F) for financial support.
Conflict of interest statement. None declared.
A.T.F. and M.P. performed the literature search and wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Michael Palmgren (palmgren@plen.ku.dk).
References
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y (2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA 104: 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astegno A, Bonza MC, Vallone R, La Verde V, D'Onofrio M, Luoni L, Molesini B, Dominici P (2017) Arabidopsis calmodulin-like protein CML36 is a calcium (Ca2+) sensor that interacts with the plasma membrane Ca2+-ATPase isoform ACA8 and stimulates its activity. J Biol Chem 292: 15049–15061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekgaard L, Luoni L, De Michelis MI, Palmgren MG (2006) The plant plasma membrane Ca2+ pump ACA8 contains overlapping as well as physically separated autoinhibitory and calmodulin-binding domains. J Biol Chem 281: 1058–1065 [DOI] [PubMed] [Google Scholar]
- Barbez E, Dünser K, Gaidora A, Lendl T, Busch W (2017) Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc Natl Acad Sci USA 114: E4884–E4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132: 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O’Flaherty M, Heck AJ, Slijper M, Menke FL (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Roderick HL (2003) Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol 4: 517–529 [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD (2000) The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol 1: 11–21 [DOI] [PubMed] [Google Scholar]
- Bhaskara GB, Wen TN, Nguyen TT, Verslues PE (2017) Protein phosphatase 2Cs and Microtubule-Associated Stress Protein 1 control microtubule stability, plant growth, and drought response. Plant Cell 29: 169–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørk PK, Rasmussen SA, Gjetting SK, Havshøi NW, Petersen TI, Ipsen JØ, Larsen TO, Fuglsang AT (2020) Tenuazonic acid from Stemphylium loti inhibits the plant plasma membrane H+-ATPase by a mechanism involving the C-terminal regulatory domain. New Phytol 226: 770–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac Y, Lee SM, Romanowsky S, Blank R, Sladek C, Chung WS, Harper JF (2010) Disruption of the vacuolar calcium-ATPases in Arabidopsis results in the activation of a salicylic acid-dependent programmed cell death pathway. Plant Physiol 154: 1158–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D, Moorhead GB (2005) 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE 2005: re10. [DOI] [PubMed] [Google Scholar]
- Buch-Pedersen MJ, Rudashevskaya EL, Berner TS, Venema K, Palmgren MG (2006) Potassium as an intrinsic uncoupler of the plasma membrane H+-ATPase. J Biol Chem 281: 38285–38292 [DOI] [PubMed] [Google Scholar]
- Caesar K, Elgass K, Chen Z, Huppenberger P, Witthöft J, Schleifenbaum F, Blatt MR, Oecking C, Harter K (2011) A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. Plant J 66: 528–540 [DOI] [PubMed] [Google Scholar]
- Chen Y, Hoehenwarter W, Weckwerth W (2010) Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J 63: 1–17 [DOI] [PubMed] [Google Scholar]
- Contreras-Cornejo HA, Macías-Rodríguez L, Alfaro-Cuevas R, López-Bucio J (2014) Trichoderma spp. Improve growth of Arabidopsis seedlings under salt stress through enhanced root development, osmolite production, and Na+ elimination through root exudates. Mol Plant Microbe Interact 27: 503–514 [DOI] [PubMed] [Google Scholar]
- Costa A, Luoni L, Marrano CA, Hashimoto K, Köster P, Giacometti S, De Michelis MI, Kudla J, Bonza MC (2017) Ca2+-dependent phosphoregulation of the plasma membrane Ca2+-ATPase ACA8 modulates stimulus-induced calcium signatures. J Exp Bot 68: 3215–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsberger WR, Schulze WX (2012) Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J 69: 978–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falhof J, Pedersen JT, Fuglsang AT, Palmgren M (2016) Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol Plant 9: 323–337 [DOI] [PubMed] [Google Scholar]
- Frei dit Frey N, Mbengue M, Kwaaitaal M, Nitsch L, Altenbach D, Häweker H, Lozano-Duran R, Njo MF, Beeckman T, Huettel B, et al. (2012) Plasma membrane calcium ATPases are important components of receptor-mediated signaling in plant immune responses and development. Plant Physiol 159: 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Borch J, Bych K, Jahn TP, Roepstorff P, Palmgren MG (2003) The binding site for regulatory 14-3-3 protein in plant plasma membrane H+-ATPase: involvement of a region promoting phosphorylation-independent interaction in addition to the phosphorylation-dependent C-terminal end. J Biol Chem 278: 42266–42272 [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ -ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Kristensen A, Cuin TA, Schulze WX, Persson J, Thuesen KH, Ytting CK, Oehlenschlæger CB, Mahmood K, Sondergaard TE, et al. (2014) Receptor kinase-mediated control of primary active proton pumping at the plasma membrane. Plant J 80: 951–964. [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Tulinius G, Cui N, Palmgren MG (2006) Protein phosphatase 2A scaffolding subunit A interacts with plasma membrane H+‐ATPase C‐terminus in the same region as 14‐3‐3 protein. Physiol Plant 128: 334–340 [Google Scholar]
- Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG (1999) Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J Biol Chem 274: 36774–36780 [DOI] [PubMed] [Google Scholar]
- García Bossi J, Kumar K, Barberini ML, Domínguez GD, Rondón Guerrero YDC, Marino-Buslje C, Obertello M, Muschietti JP, Estevez JM (2020) The role of P-type IIA and P-type IIB Ca2+-ATPases in plant development and growth. J Exp Bot 71: 1239–1248 [DOI] [PubMed] [Google Scholar]
- George L, Romanowsky SM, Harper JF, Sharrock RA (2008) The ACA10 Ca2+-ATPase regulates adult vegetative development and inflorescence architecture in Arabidopsis. Plant Physiol 146: 716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti S, Marrano CA, Bonza MC, Luoni L, Limonta M, De Michelis MI (2012) Phosphorylation of serine residues in the N-terminus modulates the activity of ACA8, a plasma membrane Ca2+-ATPase of Arabidopsis thaliana. J Exp Bot 63: 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjetting KS, Ytting CK, Schulz A, Fuglsang AT (2012) Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J Exp Bot 63: 3207–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjetting SK, Mahmood K, Shabala L, Kristensen A, Shabala S, Palmgren M, Fuglsang AT (2020) Evidence for multiple receptors mediating RALF-triggered Ca2+ signaling and proton pump inhibition. Plant J 104: 433–446 [DOI] [PubMed] [Google Scholar]
- Guo K, Sui Y, Li Z, Huang Y, Zhang H (2020) Trichoderma viride Tv-1511 colonizes Arabidopsis leaves and promotes Arabidopsis growth by modulating the MAP Kinase 6-mediated activation of plasma membrane H+-ATPase. J Plant Growth Regul 39: 1261–1276 [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Hager A, Menzel H, Krauss A (1971) Versuche und Hypothese zur Primärwirkung des Auxins beim Streckungswachstum. Planta 100: 47–75. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Sugimoto H, Takahashi H, Seki M, Shinozaki K, Sawasaki T, Kinoshita T, Inoue SI (2020) Raf-like kinases CBC1 and CBC2 negatively regulate stomatal opening by negatively regulating plasma membrane H+-ATPase phosphorylation in Arabidopsis. Photochem Photobiol Sci 19: 88–98 [DOI] [PubMed] [Google Scholar]
- Haruta M, Burch HL, Nelson RB, Barrett-Wilt G, Kline KG, Mohsin SB, Young JC, Otegui MS, Sussman MR (2010) Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J Biol Chem 285: 17918–17929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Tan LX, Bushey DB, Swanson SJ, Sussman MR (2018) Environmental and genetic factors regulating localization of the plant plasma membrane H+-ATPase. Plant Physiol 176: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Sugimoto M, Higaki T, Yaeno T, Nagami A, Irie M, Fujimi M, Miyamoto M, Akita K, Negi J, Shirasu K, et al. (2013) A Munc13-like protein in Arabidopsis mediates H+-ATPase translocation that is essential for stomatal responses. Nat Commun 4: 2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heit S, Geurts MGM, Murphy BJ, Corey RA, Mills DJ, Kühlbrandt W, Bublitz M (2021) Structure of the hexameric fungal plasma membrane proton pump in its auto-inhibited state. bioRxiv 442159. doi: 10.1101/2021.04.30.442159 (April 30, 2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama A, Takemiya A, Munemasa S, Okuma E, Sugiyama N, Tada Y, Murata Y, Shimazaki KI (2017) Blue light and CO2 signals converge to regulate light-induced stomatal opening. Nat Commun 8: 1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleary R, Paez-Valencia J, Vens C, Toyota M, Palmgren M, Gilroy S (2020) Tonoplast-localized Ca2+ pumps regulate Ca2+ signals during pattern-triggered immunity in Arabidopsis thaliana. Proc Natl Acad Sci USA 117: 18849–18857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehenwarter W, Thomas M, Nukarinen E, Egelhofer V, Röhrig H, Weckwerth W, Conrath U, Beckers GJ (2013) Identification of novel in vivo MAP kinase substrates in Arabidopsis thaliana through use of tandem metal oxide affinity chromatography. Mol Cell Proteomics 12: 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann RD, Olsen LI, Ezike CV, Pedersen JT, Manstretta R, López-Marqués RL, Palmgren M (2019) Roles of plasma membrane proton ATPases AHA2 and AHA7 in normal growth of roots and root hairs in Arabidopsis thaliana. Physiol Plant 166: 848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann RD, Portes MT, Olsen LI, Damineli DSC, Hayashi M, Nunes CO, Pedersen JT, Lima PT, Campos C, Feijó JA, et al. (2020) Plasma membrane H+-ATPases sustain pollen tube growth and fertilization. Nat Commun 11: 2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Zheng R, He J, Zhou Z, Wang J, Xiong Y, Xu T (2019) Noncanonical auxin signaling regulates cell division pattern during lateral root development. Proc Natl Acad Sci USA 116: 21285–21290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sze H, Harper JF (2000) A calcium-dependent protein kinase can inhibit a calmodulin-stimulated Ca2+ pump (ACA2) located in the endoplasmic reticulum of Arabidopsis. Proc Natl Acad Sci USA 97: 6224–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue SI, Kaiserli E, Zhao X, Waksman T, Takemiya A, Okumura M, Takahashi H, Seki M, Shinozaki K, Endo Y, et al. (2020) CIPK23 regulates blue light-dependent stomatal opening in Arabidopsis thaliana. Plant J 104: 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishka MR, Brown E, Rosenberg A, Romanowsky S, Davis JA, Choi W-G, Harper JF (2021) Arabidopsis Ca2+-ATPases 1, 2, and 7 in the endoplasmic reticulum contribute to growth and pollen fitness. Plant Phys 185: 1966–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T, Fuglsang AT, Olsson A, Brüntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C (1997). The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 9: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, MacLean D, Studholme DJ, Serna-Sanz A, Andreasson E, Rathjen JP, Peck SC (2009) Phosphoproteomic analysis of nuclei-enriched fractions from Arabidopsis thaliana. J Proteomics 72: 439–451 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Liebrand TWH, Goto Y, Sklenar J, Derbyshire P, Menke FLH, Torres MA, Molina A, Zipfel C, Coaker G, et al. (2019) Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol 221: 2160–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesten C, Gámez-Arjona FM, Menna A, Scholl S, Dora S, Huerta AI, Huang HY, Tintor N, Kinoshita T, Rep M, et al. (2019) Pathogen-induced pH changes regulate the growth-defense balance in plants. EMBO J 38: e101822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Hashimoto-Sugimoto M, Iba K, Terashima I, Yamori W (2020) Improved stomatal opening enhances photosynthetic rate and biomass production in fluctuating light. J Exp Bot 71: 2339–2350 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki Ki (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18: 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K (2001) Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660 [DOI] [PubMed] [Google Scholar]
- Kudla J, Becker D, Grill E, Hedrich R, Hippler M, Kummer U, Parniske M, Romeis T, Schumacher K (2018) Advances and current challenges in calcium signaling. New Phytol 218: 414–431 [DOI] [PubMed] [Google Scholar]
- Kurkdjian A, Guern J (1989) Intracellular pH: measurement and importance in cell activity. Annu Rev Plant Physiol Plant Mol Biol 40: 271–303 [Google Scholar]
- Kutschmar A, Rzewuski G, Stührwohldt N, Beemster GT, Inzé D, Sauter M (2009) PSK-α promotes root growth in Arabidopsis. New Phytol 18: 820–831 [DOI] [PubMed] [Google Scholar]
- Ladwig F, Dahlke RI, Stührwohldt N, Hartmann J, Harter K, Sauter M (2015) Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell 27: 1718–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Schmidt W (2012) Quantitative phosphoproteome profiling of iron-deficient Arabidopsis roots. Plant Physiol 159: 403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RD, Cho HT (2013) Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front Plant Sci 4: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li B, Zhu S, Wang L, Song L, Chen J, Ming Z, Liu X, Li X, Yu F (2021). TMK4 receptor kinase negatively modulates ABA signaling by phosphorylating ABI2 and enhancing its activity. J Integr Plant Biol. doi: 10.1111/jipb.13096. [DOI] [PubMed] [Google Scholar]
- Li X, Sanagi M, Lu Y, Nomura Y, Stolze SC, Yasuda S, Saijo Y, Schulze WX, Feil R, Stitt M, et al. (2020) Protein phosphorylation dynamics under carbon/nitrogen-nutrient stress and identification of a cell death-related receptor-like kinase in Arabidopsis. Front Plant Sci 11: 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limonta M, Romanowsky S, Olivari C, Bonza MC, Luoni L, Rosenberg A, Harper JF, De Michelis MI (2014) ACA12 is a deregulated isoform of plasma membrane Ca2+-ATPase of Arabidopsis thaliana. Plant Mol Biol 84: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LL, Hsu CL, Hu CW, Ko SY, Hsieh HL, Huang HC, Juan HF (2015) Integrating phosphoproteomics and bioinformatics to study brassinosteroid-regulated phosphorylation dynamics in Arabidopsis. BMC Genomics 16: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Coria M, Hernández-Mendoza J L, Sánchez-Nieto S (2016) Trichoderma asperellum Induces maize seedling growth by activating the plasma membrane H+-ATPase. Mol Plant Microbe Interact 29: 797–806 [DOI] [PubMed] [Google Scholar]
- Loss Sperandio MV, Santos LA, Huertas Tavares OC, Fernandes MS, de Freitas Lima M, de Souza SR (2020) Silencing the Oryza sativa plasma membrane H+-ATPase isoform OsA2 affects grain yield and shoot growth and decreases nitrogen concentration. J Plant Physiol 251: 153220. [DOI] [PubMed] [Google Scholar]
- Luu DD, Joe A, Chen Y, Parys K, Bahar O, Pruitt R, Chan LJG, Petzold CJ, Long K, Adamchak C, et al. (2019) Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc Natl Acad Sci USA 116: 8525–8534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmström S, Askerlund P, Palmgren MG (1997) A calmodulin-stimulated Ca2+-ATPase from plant vacuolar membranes with a putative regulatory domain at its N-terminus. FEBS Lett 400: 324–328 [DOI] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D, Leon-Ruiz M, Fürst U, El Ghalid M, Leonard G, López-Berges MS, Richards TA, Felix G, et al. (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat Microbiol 1: 16043. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y (1996) Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA 93: 7623–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y (2011) Posttranslational modifications in secreted peptide hormones in plants. Plant Cell Physiol 52: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei B, Spinelli F, Pontiggia D, De Lorenzo G (2016) Comprehensive analysis of the membrane phosphoproteome regulated by oligogalacturonides in Arabidopsis thaliana. Front Plant Sci 7: 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayank P, Grossman J, Wuest S, Boisson-Dernier A, Roschitzki B, Nanni P, Nühse T, Grossniklaus U (2012) Characterization of the phosphoproteome of mature Arabidopsis pollen. Plant J 72: 89–101 [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T (1987) Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol 9: 611–623 [DOI] [PubMed] [Google Scholar]
- Meier M, Liu Y, Lay-Pruitt KS, Takahashi H, von Wirén N (2020) Auxin-mediated root branching is determined by the form of available nitrogen. Nat Plants 6: 1136–1145 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Müller A, et al. (2007) Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J 26: 3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz J, Li Z, Schulze WX, Ludewig U (2016) Early nitrogen-deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and (phospho-) proteome levels between nitrate and ammonium nutrition. Plant J 88: 717–734 [DOI] [PubMed] [Google Scholar]
- Miao R, Yuan W, Wang Y, Garcia-Maquilon I, Dang X, Li Y, Zhang J, Zhu Y, Rodriguez PL, Xu W (2021) Low ABA concentration promotes root growth and hydrotropism through relief of ABA INSENSITIVE 1-mediated inhibition of plasma membrane H+-ATPase 2. Sci Adv 7: eabd4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami A, Takahashi K, Inoue SI, Tada Y, Kinoshita T (2019) Brassinosteroid induces phosphorylation of the plasma membrane H+-ATPase during hypocotyl elongation in Arabidopsis thaliana. Plant Cell Physiol 60: 935–944 [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Miller ND, Murphy AS, Gilroy S (2011) Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J 65: 309–318 [DOI] [PubMed] [Google Scholar]
- Niittylä T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX (2007) Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol Cell Proteomics 6: 1711–1726 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Stensballe A, Jensen ON, Peck SC (2003) Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics 2: 1234–1243 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Bottrill A, Jones AME, Peck SC (2007) Quantitative phosphoproteomics analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J 51: 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura M, Inoue S, Kuwata K, Kinoshita T (2016). Photosynthesis activates plasma membrane H+-ATPase via sugar accumulation. Plant Physiol 171: 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C (1998) A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol 118: 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C, Marco S, Jaspert N, Marcon C, Schauer N, Weyand M, Vandermeeren C, Duby G, Boutry M, Wittinghofer A, et al. (2007) Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol Cell 25: 427–440 [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Nissen P (2011) P-type ATPases. Annu Rev Biophys 40: 243–266 [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C (1991) Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J Biol Chem 266: 20470–20475 [PubMed] [Google Scholar]
- Palmgren M, Sørensen DM, Hallström BM, Säll T, Broberg K (2020) Evolution of P2A and P5A ATPases: ancient gene duplications and the red algal connection to green plants revisited. Physiol Plant 168: 630–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JT, Kanashova T, Dittmar G, Palmgren M (2018) Isolation of native plasma membrane H+-ATPase (Pma1p) in both the active and basal activation states. FEBS Open Bio 8: 774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Joe A, Zhang W, Feng W, Stewart V, Schwessinger B, Dinneny JR, Ronald PC (2017) A microbially derived tyrosine-sulfated peptide mimics a plant peptide hormone. New Phytol 215: 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D (2011) Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J 68: 100–113 [DOI] [PubMed] [Google Scholar]
- Rayapuram N, Bigeard J, Alhoraibi H, Bonhomme L, Hesse AM, Vinh J, Hirt H, Pflieger D (2018) Quantitative phosphoproteomic analysis reveals shared and specific targets of Arabidopsis Mitogen-Activated Protein Kinases (MAPKs) MPK3, MPK4, and MPK6. Mol Cell Proteomics 17: 61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle D, Cleland RE (1970) Enhancement of wall loosening and elongation by acid solutions. Plant Physiol 46: 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL (1973) Auxin-induced hydrogen-ion excretion in Avena coleoptiles and its implications. Planta 114: 63–73 [DOI] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyer A, Häßler M, Scherzer S, Huang S, Pedersen JT, Al-Rascheid KAS, Bamberg E, Palmgren M, Dreyer I, Nagel G, et al. (2020) Channelrhodopsin-mediated optogenetics highlights a central role of depolarization-dependent plant proton pumps. Proc Natl Acad Sci USA 117: 20920–20925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudashevskaya EL, Ye J, Jensen ON, Fuglsang AT, Palmgren MG (2012) Phosphosite mapping of P-type plasma membrane H+-ATPase in homologous and heterologous environments. J Biol Chem 287: 4904–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Loo EP, Yasuda S (2018) Pattern recognition receptors and signaling in plant-microbe interactions. Plant J 93: 592–613 [DOI] [PubMed] [Google Scholar]
- Schoenaers S, Balcerowicz D, Breen G, Hill K, Zdanio M, Mouille G, Holman TJ, Oh J, Wilson MH, Nikonorova N, et al. (2018) The auxin-regulated CrRLK1L kinase ERULUS controls cell wall composition during root hair tip growth. Curr Biol 28: 722–732. [DOI] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stührwohldt N, Dahlke RI, Steffens B, Johnson A, Sauter M (2011) Phytosulfokine-α controls hypocotyl length and cell expansion in Arabidopsis thaliana through phytosulfokine receptor 1. PLoS One 6: e21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda H, Mano H, Toyota M, Fukushima K, Mimura T, Tsutsui I, Hedrich R, Tamada Y, Hasebe M (2020) Calcium dynamics during trap closure visualized in transgenic Venus flytrap. Nat Plants 6: 1219–1224 [DOI] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M (1999) Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11: 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H, Liang F, Hwang I, Curran AC, Harper JF (2000) Diversity and regulation of plant Ca2+ pumps: insights from expression in yeast. Annu Rev Plant Physiol Plant Mol Biol 51: 433–462 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T (2012) Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol 159: 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemiya A, Shimazaki K (2016) Arabidopsis phot1 and phot2 phosphorylate BLUS1 kinase with different efficiencies in stomatal opening. J Plant Res 129: 167–174 [DOI] [PubMed] [Google Scholar]
- Takemiya A, Sugiyama N, Fujimoto H, Tsutsumi T, Yamauchi S, Hiyama A, Tada Y, Christie JM, Shimazaki K (2013) Phosphorylation of BLUS1 kinase by phototropins is a primary step in stomatal opening. Nat Commun 4: 2094. [DOI] [PubMed] [Google Scholar]
- Tian W, Wang C, Gao Q, Li L, Luan S (2020) Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat Plants 6: 750–759 [DOI] [PubMed] [Google Scholar]
- Tidow H, Poulsen LR, Andreeva A, Knudsen M, Hein KL, Wiuf C, Palmgren MG, Nissen P (2012) A bimodular mechanism of calcium control in eukaryotes. Nature 491: 468–472 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6: rs8. [DOI] [PubMed] [Google Scholar]
- Van Leene J, Han C, Gadeyne A, Eeckhout D, Matthijs C, Cannoot B, De Winne N, Persiau G, Van De Slijke E, Van de Cotte B, et al. (2019) Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat Plants 5: 316–327 [DOI] [PubMed] [Google Scholar]
- Wang X, Bian Y, Cheng K, Gu LF, Ye M, Zou H, Sun SS, He JX (2013) A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J Proteomics 78: 486–98 [DOI] [PubMed] [Google Scholar]
- Wang P, Hsu CC, Du Y, Zhu P, Zhao C, Fu X, Zhang C, Paez JS, Macho AP, Tao WA, et al. (2020) Mapping proteome-wide targets of protein kinases in plant stress responses. Proc Natl Acad Sci USA 117: 3270–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welle M, Pedersen JT, Ravnsborg T, Hayashi M, Maaß S, Becher D, Jensen ON, Stöhr C, Palmgren M (2021) A conserved, buried cysteine near the P-site is accessible to cysteine modifications and increases ROS stability in the P-type plasma membrane H+-ATPase. Biochem J 478: 619–632 [DOI] [PubMed] [Google Scholar]
- Whiteman SA, Serazetdinova L, Jones AM, Sanders D, Rathjen J, Peck SC, Maathuis FJ (2008) Identification of novel proteins and phosphorylation sites in a tonoplast enriched membrane fraction of Arabidopsis thaliana. Proteomics 8: 3536–3547 [DOI] [PubMed] [Google Scholar]
- Wong JH, Klejchová M, Snipes SA, Nagpal P, Bak G, Wang B, Dunlap S, Park MY, Kunkel EN, Trinidad B, et al. (2021) SAUR proteins and PP2C.D phosphatases regulate H+-ATPases and K+ channels to control stomatal movements. Plant Physiol 185: 256–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XN, Sanchez Rodriguez C, Pertl-Obermeyer H, Obermeyer G, Schulze WX (2013) Sucrose-induced receptor kinase SIRK1 regulates a plasma membrane aquaporin in Arabidopsis. Mol Cell Proteomics 12: 2856–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Sklodowski K, Encke B, Schulze WX (2014) A kinase-phosphatase signaling module with BSK8 and BSL2 involved in regulation of sucrose-phosphate synthase. J Proteome Res 13: 3397–3409 [DOI] [PubMed] [Google Scholar]
- Xia L, Mar Marquès-Bueno M, Bruce CG, Karnik R (2019) Unusual roles of secretory SNARE SYP132 in plasma membrane H+-ATPase traffic and vegetative plant growth. Plant Physiol 180: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Wang P, Wang L, Renzi E, Radivojac P, Tang H, Arnold R, Zhu JK, Tao WA (2013) Quantitative measurement of phosphoproteome response to osmotic stress in arabidopsis based on Library-Assisted eXtracted Ion Chromatogram (LAXIC). Mol Cell Proteomics 12: 2354–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Yang Y, Yang Z, Wang X, Guo Y (2018) VAMP711 is required for abscisic acid-mediated inhibition of plasma membrane H+-ATPase activity. Plant Physiol 178: 1332–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Guo G, Zhang M, Liu CY, Hu Q, Lam H, Cheng H, Xue Y, Li J, Li N (2013) Stable isotope metabolic labeling-based quantitative phosphoproteomic analysis of Arabidopsis mutants reveals ethylene-regulated time-dependent phosphoproteins and putative substrates of constitutive triple response 1 kinase. Mol Cell Proteomics 12: 3559–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Shi Z, Bao Y, Yan J, Yang Z, Yu H, Li Y, Gou M, Wang S, Zou B, et al. (2017) Calcium pumps and interacting BON1 protein modulate calcium signature, stomatal closure, and plant immunity. Plant Physiol 175: 424–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Qin Y, Xie C, Zhao F, Zhao J, Liu D, Chen S, Fuglsang AT, Palmgren MG, Schumaker KS, et al. (2010) The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22: 1313–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wu Y, Ma L, Yang Z, Dong Q, Li Q, Ni X, Kudla J, Song C, Guo Y (2019) The Ca2+ sensor SCaBP3/CBL7 modulates plasma membrane H+-ATPase activity and promotes alkali tolerance in Arabidopsis. Plant Cell 31: 1367–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia I, Shrivasta AK, Kapulnik Y, Chet I (2001) Effect of Trichoderma harzianum on microelement concentration and increased growth of cucumber plants. Plant Soil 235: 235–242 [Google Scholar]
- Yu H, Yan J, Du X, Hua J (2018) Overlapping and differential roles of plasma membrane calcium ATPases in Arabidopsis growth and environmental responses. J Exp Bot 69: 2693–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhou H, Berke L, Heck AJ, Mohammed S, Scheres B, Menke FL (2013) Quantitative phosphoproteomics after auxin-stimulated lateral root induction identifies an SNX1 protein phosphorylation site required for growth. Mol Cell Proteomics 12: 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang Y, Chen X, Xu F, Ding M, Ye W, Kawai Y, Toda Y, Hayashi Y, Suzuki T, et al. (2021) Plasma membrane H+-ATPase overexpression increases rice yield via simultaneous enhancement of nutrient uptake and photosynthesis. Nat Commun 12: 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Jiang L, Huang J, Ding Y, Liu Z (2020) Loss of proton/calcium exchange 1 results in the activation of plant defense and accelerated senescence in Arabidopsis. Plant Sci 296: 110472. [DOI] [PubMed] [Google Scholar]