Abstract
The epidemiologic relatedness of methicillin-resistant Staphylococcus aureus (MRSA) isolates is currently determined by analysis of chromosomal DNA restriction patterns by pulsed-field gel electrophoresis (PFGE). We have evaluated an alternative typing system (MicroSeq StaphTrack Kit; Perkin-Elmer Biosystems) based on the sequence analysis of the chromosomally encoded polymorphic repeat X region of the S. aureus protein A (spa) gene. A total of 69 clinical MRSA isolates were divided into 18 groups according to the number and nucleotide sequences of the spa repeats. Molecular typing results obtained both by spa sequencing and from the PFGE patterns were concordant except for one group, which contained 20 isolates recovered over a 2-year period from hospitalized patients at the Mayo Clinic. Although the spa typing patterns were indistinguishable for those isolates, PFGE analysis yielded seven related but distinguishable patterns. Further coagulase gene sequence analysis subtyped those 20 strains into four groups which followed distinct temporal and geographic distributions. During a 2-year epidemic period there were up to 7 fragment changes in PFGE patterns among epidemiologically related isolates, suggesting that PFGE may be unsuitable for long-term typing of strains involved in epidemics. Although more limited than PFGE in discriminatory power, spa sequencing analysis could be used as a screening method for typing of MRSA strains because of the shorter turnaround time, ease of use, and the inherent advantages of sequence analysis, storage, and sharing of information.
Since its first identification in the early 1960s (12), methicillin-resistant Staphylococcus aureus (MRSA) has become one of the most significant nosocomial pathogens throughout the world and is capable of causing a wide range of hospital infections (1, 17). It continues to spread through new communities wherever the methods and institutions of modern medical practice are adopted, while it regularly causes epidemics in places where it has been endemic for a decade or more (1, 35). Consequently, analysis of the dissemination of MRSA isolates has been a research focus for decades; more recently, the tools used to discriminate MRSA isolates have themselves disseminated (31, 34).
Analysis of genetic relatedness by molecular typing techniques is important for defining epidemics. Accurate and rapid typing of MRSA is crucial for the control of nosocomial outbreaks, and numerous methods have been described (4, 7, 10, 11, 18, 22, 24–26, 33). Pulsed-field gel electrophoresis (PFGE) has proven to be highly discriminatory for MRSA isolates, and it has been suggested that it is superior to other genotyping techniques (2, 23, 26, 29). However, this method is costly and technically complex, and there is a paucity of agreed-upon criteria for the interpretation of banding patterns (15, 31). Furthermore, interlaboratory standardization of PFGE is still problematic (4, 34) and it is not suited for the screening of large numbers of isolates simultaneously (20, 34). Thus, there is an ongoing need for rapid and definitive genotyping systems, especially ones that can be applied in clinical laboratories.
Protein A is a cell wall constituent of S. aureus. Nucleotide sequence analysis of region X of the staphylococcal protein A (spa) gene has demonstrated various numbers of degenerate 24-bp repeats (9). The repetitive region is highly polymorphic; the number and sequence of individual repeats may differ among strains, providing an alternative molecular method for the typing of MRSA strains (5, 6, 28). An MRSA typing system based on amplification and sequencing of the spa gene has been developed and distributed by Perkin-Elmer Biosystems (Foster City, Calif.). The MicroSeq StaphTrack Kit allows rapid typing of MRSA within 2 working days. The X region of the spa gene is amplified by PCR and is then sequenced by a fluorescent DNA sequencing technology. The relatedness of a group of MRSA strains is determined by variation of both the numbers and the sequences of individual repeats in region X of the spa gene (28). We report here on an evaluation of this system for its ability to type 69 clinical MRSA isolates to determine their relatedness in reference to the currently used PFGE technique. Epidemiologic data were collected for resolution of discrepant results.
(This study was presented in part at the 99th General Meeting of the American Society for Microbiology, Chicago, Ill., 30 May to 3 June 1999.)
MATERIALS AND METHODS
Bacterial strains.
A total of 69 MRSA strains were included in the study. Sixty-three were isolated from patients seen at the Mayo Clinic, and six were isolated from horses and veterinarians at Oregon State University. Each isolate was from an individual subject, and no repeat isolates were included. All isolates were coagulase positive, as determined by a rapid slide agglutination procedure (Murex Diagnostics, Inc., Norcross, Ga.). Methicillin resistance was determined by the presence of a zone of ≤10 mm around a 1-μg oxacillin disk (BBL, Becton Dickinson Microbiology System, Sparks, Md.) after 24 h of incubation at 35°C on a Mueller-Hinton agar plate (21).
Determination of epidemiologic relatedness.
The Mayo Clinic Infection Control Unit staff performed active surveillance and data collection for MRSA-colonized or -infected patients during this study period. A tracking log was used to identify epidemiologic relatedness between patients in order to select isolates for testing and thus document clusters. The key data used to determine relatedness were patient care unit or room, primary service, surgical procedure dates and the associated operating room, and other invasive procedures. When clusters were identified, control measures were then implemented for the patient population and the units or departments involved.
Genomic DNA analysis by PFGE.
The genomic DNA was extracted from logarithmic-phase S. aureus cultures grown in brain heart infusion broth (BBL) as described previously (19, 36). The extracted DNA was prepared in low-melting-point agarose plugs and was digested with the SmaI enzyme (New England Biolabs, Beverly, Mass.), as described previously (23). The DNA size standards used were a bacteriophage lambda ladder consisting of concatemers starting at 48.5 kbp and increasing to approximately 1,000 kbp (Bio-Rad Laboratories, Hercules, Calif.). Electrophoresis was performed with a Bio-Rad CHEF DR II system (Bio-Rad). Run conditions were 200 V with switching from 10 to 50 s for 15 h at 14°C (3). The gels were stained with ethidium bromide, rinsed, and photographed under UV light. Each strain was classified as indistinguishable, closely related, possible related, or different if the number(s) of fragment differences compared with the number of fragments for reference strain was zero, two to three, four to six, or seven or more, respectively, according to the criteria recommended previously (32).
Coagulase (coa) gene sequencing.
Extraction of bacterial genomic DNA from MRSA cultures was performed as described previously (30). A primer set (Coa1509F [5′-TGC TGG TAC AGG TAT CCG TGA AT-3′] and Coag2143R [5′-AGA AGC ACA TAG AAT GCA TGA-3′]) was designed to encompass the entire 3′ tandem repeat region of the coa gene (13) (GenBank accession no. X16457). PCR amplification was accomplished by adding 50 μl of genomic DNA extract to 50 μl of 2× master mixture buffer (0.5 μM each primer, 2.5 U of Ampli Taq Gold DNA polymerase, 2.0 mM MgCl2, 350 μM total deoxynucleoside triphosphates, 25 mM KCl). Thermal cycling parameters included an initial 10 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 60°C, and 45 s at 72°C; and a final extension at 72°C for 10 min. The PCR products (≥635 bp, on the basis of the number of repeats) were purified and sequenced as described previously (30). The primers used for sequencing were the same as those used for PCR. The coa sequence sample files were assembled, and the final consensus sequences were analyzed with MicroSeq Analysis Software (30).
spa gene sequencing.
Sequence determination and analysis of the spa gene were performed with the MicroSeq StaphTrack Kit provided by Perkin-Elmer Biosystems. The MicroSeq StaphTrack Kit consists of a PCR module and a sequencing module which contain all reagents necessary for PCR amplification and sequencing of the polymorphic X region of the spa gene (28).
Genomic DNA was isolated from bacterial cultures by using the PrepMan extraction reagent from Perkin-Elmer Biosystems by the method provided in the kit for the extraction of DNA from gram-positive bacterial cells. PCR amplification of the spa gene was performed by adding 25 μl of diluted genomic DNA to 25 μl of the PCR master mixture. The samples were placed in a GeneAmp PCR System 9600 (Perkin-Elmer Biosystems). Thermal cycling parameters included an initial 10 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 60°C, and 45 s at 72°C; and a final extension at 72°C for 10 min.
The PCR products were purified from excess PCR primers and nucleotides prior to sequencing with Microcon-100 microconcentrator columns (Millipore Corp., Bedford, Mass.). Forward and reverse cycle sequencing reaction mixtures were prepared by adding 3 μl of the PCR product to 13 μl of each sequencing master mixture and 4 μl of water. Cycle sequencing reactions were run on a GeneAmp PCR System 9600 (Perkin-Elmer Biosystems) by using the recommended thermal profile. The sequencing reaction mixtures were purified from excess dye-labeled terminators with Centri-Sep spin columns according to the manufacturer's instructions. Samples were electrophoresed on an ABI Prism 377 automated fluorescent sequencer. The sequence data were analyzed by using the MicroSeq analysis software package to determine the numbers and the sequences of the 24-bp repeats in the X region of the spa gene.
RESULTS
Sequence analysis of the X region of the spa gene resulted in 18 distinct groups. The number of 24-bp repeats within the gene varied between 3 and 11, corresponding to PCR products ranging in size from 226 to 418 bp (Table 1). According to convention, degenerate repeats are assigned alphabetical letters for distinct motifs. Eighteen such motifs were identified in this study; a new sequence (5′-GAA GAC AAC AAA AAG CCT GGC AAA-3′) was designated motif W, raising the total number of described motifs to 26 (5).
TABLE 1.
spa characteristics of 69 MRSA strains included in the study
| spa group | No. of isolates | No. of 24-bp repeats | Amplicon size (bp) | spa patterna | Length of outbreak (wk) | PFGE result | Epidemiologic relatedness |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 9 | 370 | Z B C H R H C C H | NAb | Dissimilarc | Unrelatedc |
| 2 | 1 | 9 | 370 | A A W H E H H J K | NA | Dissimilar | Unrelated |
| 3 | 6 | 9 | 370 | A C W H E I E J K | 1 | Indistinguishable | Related |
| 4 | 2 | 9 | 370 | M F M D D H E M E | 7 | Indistinguishable | Related |
| 5 | 2 | 8 | 346 | Z K H R H P M E | 3 | Indistinguishable | Related |
| 6 | 2 | 8 | 346 | M F M D H E M E | 1 | Indistinguishable | Related |
| 7 | 1 | 10 | 394 | A A C B H E I E J K | NA | Dissimilar | Unrelated |
| 8 | 5 | 10 | 394 | A A B W H E I E J K | 61 | Indistinguishable | Related |
| 9 | 20 | 10 | 394 | E O H E H R H C H M | 107 | See Fig. 1 | See Fig. 1 |
| 10 | 1 | 11 | 418 | A A C T W H E I E J K | NA | Dissimilar | Unrelated |
| 11 | 1 | 11 | 418 | E O H E H R H C H H M | NA | Dissimilar | Unrelated |
| 12 | 15 | 11 | 418 | B C M F M F K H I I I | 82 | Indistinguishable | Related |
| 13 | 1 | 3 | 226 | C H M | NA | Dissimilar | Unrelated |
| 14 | 1 | 5 | 274 | D M C O E | NA | Dissimilar | Unrelated |
| 15 | 7 | 6 | 298 | E O H C H M | 60 | Indistinguishable | Related |
| 16 | 1 | 7 | 322 | E H G C H M | NA | Dissimilar | Unrelated |
| 17 | 1 | 7 | 322 | E G H C H M M | NA | Dissimilar | Unrelated |
| 18 | 1 | 7 | 322 | B C D F K H I | NA | Dissimilar | Unrelated |
For alphanumeric spa patterns, see reference 5. A newly assigned code, W, which was identified in the present study, represents DNA sequence 5′-GAA GAC AAC AAA AAG CCT GGC AAA-3′.
NA, not applicable.
Compared to those in other groups.
Typing results for the spa sequences were compared to the results obtained by the currently used PFGE typing method. Complete concordance, as well as epidemiologic relatedness, was found between the two methods for all except one group (designated group 9), which contained 20 isolates recovered from an unusual hospital effort at surveillance of MRSA isolates conducted over a 2-year period (Table 1). Although the majority of the latter isolates had identical gel patterns, PFGE analysis yielded seven related but distinguishable patterns, with up to seven fragment changes observed (Fig. 1).
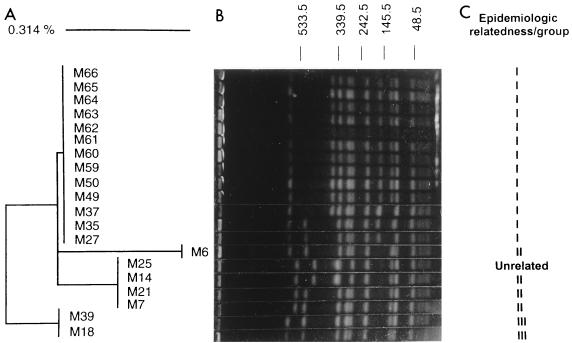
FIG. 1.
Variations in coagulase sequences and PFGE patterns in a 20-strain MRSA group with identical spa sequence characteristics. (A) Neighbor-joining analysis of the coagulase gene sequences. The scale reflects relative phylogenetic distance. (B) PFGE pattern. Molecular masses (in kilobases) are displayed at the top. (C) Three epidemiologically related subgroups determined by epidemiologic data. M25 was identified as a community-acquired MRSA isolate on the basis of its temporal and geographic distributions.
Although only 6 nucleotide sequence differences were found in the coa gene (<1%), 20 MRSA isolates from group 9 could be further divided into four groups (Fig. 1). The validities of the typing results from the PFGE, spa, and coa patterns were also compared with the epidemiologic data. Both M35 and M61 were recovered from two patients residing in the same long-term-care facility. Although both spa and coa sequence patterns were identical, PFGE analysis yielded two band changes (Fig. 1). In addition, M27, M35, and M49 were isolated from three patients treated from the same nursing station within a 2-week period; however, the PFGE patterns of M27 and M35 were different from that of M49 by three significant fragment changes (Fig. 1).
In an investigation of the group 9 isolates, there was no evidence of epidemiologic relatedness between isolates M7 and M59 or between isolates M18 and M50. However, the epidemiologic unrelatedness of these strains was confirmed by coa sequence analysis (Fig. 1). For the isolates other than the group 9 isolates spa sequence analysis placed epidemiologically distinct strains into the different groups in complete concordance with the groups into which they were placed by PFGE. Taken together, these data suggest that the combined sequence analysis of spa and coa genes provided greater concordance with epidemiologic information than PFGE.
DISCUSSION
Rapid and accurate differentiation of bacterial strains associated with nosocomial infections is central to epidemiologic surveillance and hospital infection control. We have shown that detection of genetic polymorphisms in the X region of the spa gene can be used as a typing method to determine the epidemiologic relatedness of MRSA isolates. Protein A is a component of the S. aureus cell wall and is covalently bound to the peptidoglycan. The spa gene is approximately 2,150 bp and contains three distinct regions: the Fc portion, the X region, and the C terminus. The polymorphic X region contains various numbers of 24-bp repeats with various sequences. The number of repeats varies between 3 and 15, and 26 different DNA sequences of repeats have been described (5). Previous studies have demonstrated that the X region of the spa gene is stable enough both in vitro and in vivo to discriminate between different clones (5). Our data indicate that the organization of the spa gene is stable among epidemiologically related MRSA strains recovered over a period of 2 years. The variability and stability of this gene indicate that sequence analysis of the spa gene could be used as an alternative system for the molecular typing of MRSA isolates.
PFGE has been used as a typing method in many clinical laboratories, including that at the Vanderbilt University Medical Center and the Mayo Clinic (2, 3, 23, 26, 29). PFGE was developed for the resolution of large (50- to 700-kb) fragments of DNA obtained by digesting genomic DNA with restriction enzymes that cleave DNA infrequently (27). Because it randomly samples the entire genome, genomic macrorestriction analysis by PFGE possesses a relatively high discriminatory power. However, PFGE is a complicated and time-consuming procedure. It involves growth of the organisms, usually overnight in broth, imbeddment of the washed cells in agarose, lysing of the cells in situ, digestion of the DNA with restriction enzyme in situ, loading of the agar block into gel, and then separation of the fragments on the gel by pulsed-field electrophoresis. Excluding culture, the whole procedure requires 2 to 5 days and approximately 6 to 8 h of technologist time. In addition, although the gel images can be scanned into computer systems and several commercial systems are available to facilitate comparison of isolates tested at different times or in different laboratories, interlaboratory standardization is still problematic, and problems of interpretation still exist.
PFGE analysis works accurately for the typing of isolates recovered from a localized outbreak of disease within a short period of time. However, individual loci or uncharacterized regions of the genome may be highly variable within the bacterial population; consequently, variation that is observed may be due to a variety of genetic mechanisms, the results of which can be misleading for long-term outbreaks (16). In theory, PFGE may be too discriminatory for long-term epidemiologic investigations because it fails to account for genetic changes within a single, globally distributed, clonal lineage of MRSA. In our study, several epidemiologically related MRSA strains, as well as MRSA strains with identical spa and coa sequences, recovered over 2-year period had up to seven fragment changes in their PFGE patterns. These data suggest that during the course of a nosocomial epidemic of MRSA infection strain evolution occurs, and this may confound the use of PFGE for strain typing. Thus, when PFGE analysis is used for bacterial typing, the duration of the outbreak may need to be considered when patterns are interpreted.
The criteria set forth for determination of the suitability of typing schemes include typeability, reproducibility, discriminatory power, ease of use, and ease of interpretation. A parallel evaluation of up to 12 currently available typing systems with 60 staphylococcal isolates demonstrated that no single typing method, including PFGE, clearly prevailed over the others, and ultimately, a combination of two methods may be most efficacious (31). This has been shown for other organisms such as Escherichia coli O157 (8). Theoretically, one method that would be sensitive enough to identify all potential patients or sources may be used for the screening of isolates early in an epidemiologic evaluation, and another method for detailed strain differentiation may be used later. In our experience, typing systems based on spa sequencing can be completed within 2 days and possess a satisfactory discriminatory power. In addition, driven in part by the human and microbial genome projects, sequencing costs will probably continue their rapid trend downward, bringing this technology within reach of many microbiology laboratories. The potential advantages in terms of data analysis, storage, and sharing of information suggest that spa sequencing may soon become an alternative to PFGE for the typing of MRSA isolates.
Although spa sequencing appeared to be a quicker, unambiguous method for the typing of MRSA isolates, additional procedures may be necessary to provide maximum resolution. The discriminatory power of a single polymorphic gene is usually limited. Since the spa sequence is relatively stable both in vitro and in vivo, strains with different spa types are generally not epidemiologically related (5). However, the converse cannot be concluded; i.e., if strains have the same spa type, this does not imply that they are epidemiologically related. In our study, 20 MRSA isolates possessed an identical spa sequence; however, not all of them were epidemiologically related, as demonstrated by PFGE analysis, coa gene sequencing, and the epidemiologic data themselves. A recent study on molecular typing of meningococci indicated that multilocus sequence typing is a potentially viable approach to the identification of clones within populations of pathogenic microorganisms (16). The combination of two chromosomal genetic elements showed enough discriminatory power to provide a rich source of strain-specific genotypic heterogeneity (14). Thus, in our own clinical context, if spa sequencing fails to demonstrate a difference between strains, especially in the context of a long-term nosocomial outbreak, coa sequencing and PFGE could probably be used to resolve epidemiologically significant groups.
ACKNOWLEDGMENTS
We thank Scott Anderson, Margaret Riehman, Eric Troop, John Bartell, Doug Bost, Deborah Dodge, Dan Chapman, Brenda Dylla, Garry Hall, Patty Schams, Sue Dana-Goodon, Kerri Smidt, Sharlene Allen, Nancy Haukom, Barbara Lecy, Barbara Meline, Karen Olson, and Alan Wright for excellent assistance.
REFERENCES
- 1.Ayliffe G A. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1997;24:S74–S79. doi: 10.1093/clinids/24.supplement_1.s74. [DOI] [PubMed] [Google Scholar]
- 2.Carles-Nurit M J, Christophle B, Broche S, Gouby A, Bouziges N, Ramuz M. DNA polymorphisms in methicillin-susceptible and methicillin-resistant strains of Staphylococcus aureus. J Clin Microbiol. 1992;30:2092–2096. doi: 10.1128/jcm.30.8.2092-2096.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cockerill F R, III, Thompson R L, Musser J M, Schlievert P M, Talbot J, Holley K E, Harmsen W S, Ilstrup D M, Kohner P C, Kim M H, Frankfort B, Manahan J M, Steckelberg J M, Roberson F, Wilson W R. Molecular, serological, and clinical features of 16 consecutive cases of invasive streptococcal disease. Clin Infect Dis. 1998;26:1448–1458. doi: 10.1086/516376. [DOI] [PubMed] [Google Scholar]
- 4.Cookson B D, Aparicio P, Deplano A, Struelens M, Goering R, Marples R. Inter-centre comparison of pulsed-field gel electrophoresis for the typing of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1996;44:179–184. doi: 10.1099/00222615-44-3-179. [DOI] [PubMed] [Google Scholar]
- 5.Frenay H M, Bunschoten A E, Schouls L M, van Leeuwen W J, Vandenbroucke-Grauls C M, Verhoef J, Mooi F R. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur J Clin Microbiol Infect Dis. 1996;15:60–64. doi: 10.1007/BF01586186. [DOI] [PubMed] [Google Scholar]
- 6.Frenay H M, Theelen J P, Schouls L M, Vandenbroucke-Grauls C M, Verhoef J, van Leeuwen W J, Mooi F R. Discrimination of epidemic and nonepidemic methicillin-resistant Staphylococcus aureus strains on the basis of protein A gene polymorphism. J Clin Microbiol. 1994;32:846–847. doi: 10.1128/jcm.32.3.846-847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goh S H, Byrne S K, Zhang J L, Chow A W. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992;30:1642–1645. doi: 10.1128/jcm.30.7.1642-1645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grif K, Karch H, Schneider C, Daschner F D, Beutin L, Cheasty T, Smith H, Rowe B, Dierich M P, Allerberger F. Comparative study of five different techniques for epidemiological typing of Escherichia coli O157. Diagn Microbiol Infect Dis. 1998;32:165–176. doi: 10.1016/s0732-8893(98)00103-5. [DOI] [PubMed] [Google Scholar]
- 9.Guss B, Uhlen M, Nilsson B, Lindberg M, Sjoquist J, Sjodahl J. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur J Biochem. 1984;138:413–420. doi: 10.1111/j.1432-1033.1984.tb07931.x. [DOI] [PubMed] [Google Scholar]
- 10.Hartstein A I, Morthland V H, Eng S, Archer G L, Schoenknecht F D, Rashad A L. Restriction enzyme analysis of plasmid DNA and bacteriophage typing of paired Staphylococcus aureus blood culture isolates. J Clin Microbiol. 1989;27:1874–1879. doi: 10.1128/jcm.27.8.1874-1879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hookey J V, Richardson J F, Cookson B D. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36:1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jevons M P. “Celbenin”-resistant staphylococci. Br Med J. 1961;1:124–125. [Google Scholar]
- 13.Kaida S, Miyata T, Yoshizawa Y, Kawabata S, Morita T, Igarashi H, Iwanaga S. Nucleotide sequence of the staphylocoagulase gene: its unique COOH-terminal 8 tandem repeats. J Biochem. 1987;102:1177–1186. doi: 10.1093/oxfordjournals.jbchem.a122156. [DOI] [PubMed] [Google Scholar]
- 14.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 15.Kumari D N, Keer V, Hawkey P M, Parnell P, Joseph N, Richardson J F, Cookson B. Comparison and application of ribosome spacer DNA amplicon polymorphisms and pulsed-field gel electrophoresis for differentiation of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 1997;35:881–885. doi: 10.1128/jcm.35.4.881-885.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiden M C, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno F, Crisp C, Jorgensen J H, Patterson J E. Methicillin-resistant Staphylococcus aureus as a community organism. Clin Infect Dis. 1995;21:1308–1312. doi: 10.1093/clinids/21.5.1308. [DOI] [PubMed] [Google Scholar]
- 18.Mulligan M E, Kwok R Y, Citron D M, John J F, Jr, Smith P B. Immunoblots, antimicrobial resistance, and bacteriophage typing of oxacillin-resistant Staphylococcus aureus. J Clin Microbiol. 1988;26:2395–2401. doi: 10.1128/jcm.26.11.2395-2401.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nada T, Ichiyama S, Osada Y, Ohta M, Shimokata K, Kato N, Nakashima N. Comparison of DNA fingerprinting by PFGE and PCR-RFLP of the coagulase gene to distinguish MRSA isolates. J Hosp Infect. 1996;32:305–317. doi: 10.1016/s0195-6701(96)90041-9. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. 6th ed. 1997. Approved standard M2-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 22.Olmos A, Camarena J J, Nogueira J M, Navarro J C, Risen J, Sanchez R. Application of an optimized and highly discriminatory method based on arbitrarily primed PCR for epidemiologic analysis of methicillin-resistant Staphylococcus aureus nosocomial infections. J Clin Microbiol. 1998;36:1128–1134. doi: 10.1128/jcm.36.4.1128-1134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prevost G, Pottecher B, Dahlet M, Bientz M, Mantz J M, Piemont Y. Pulsed field gel electrophoresis as a new epidemiological tool for monitoring methicillin-resistant Staphylococcus aureus in an intensive care unit. J Hosp Infect. 1991;17:255–269. doi: 10.1016/0195-6701(91)90270-i. [DOI] [PubMed] [Google Scholar]
- 24.Richardson J F, Aparicio P, Marples R R, Cookson B D. Ribotyping of Staphylococcus aureus: an assessment using well-defined strains. Epidemiol Infect. 1994;112:93–101. doi: 10.1017/s0950268800057459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saulnier P, Bourneix C, Prevost G, Andremont A. Random amplified polymorphic DNA assay is less discriminant than pulsed-field gel electrophoresis for typing strains of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:982–985. doi: 10.1128/jcm.31.4.982-985.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlichting C, Branger C, Fournier J M, Witte W, Boutonnier A, Wolz C, Goullet P, Doring G. Typing of Staphylococcus aureus by pulsed-field gel electrophoresis, zymotyping, capsular typing, and phage typing: resolution of clonal relationships. J Clin Microbiol. 1993;31:227–232. doi: 10.1128/jcm.31.2.227-232.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz D C, Cantor C R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 28.Shopsin B, Gomez M, Montgomery S O, Smith D H, Waddington M, Dodge D E, Bost D A, Riehman M, Naidich S, Kreiswirth B N. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Struelens M J, Deplano A, Godard C, Maes N, Serruys E. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J Clin Microbiol. 1992;30:2599–2605. doi: 10.1128/jcm.30.10.2599-2605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Y W, Ellis N M, Hopkins M K, Smith D H, Dodge D E, Persing D H. Comparison of phenotypic and genotypic techniques for identification of unusual pathogenic aerobic gram-negative bacilli. J Clin Microbiol. 1998;46:3674–3679. doi: 10.1128/jcm.36.12.3674-3679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenover F C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hebert G A, Hill B, Hollis R. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Belkum A, Bax R, Peerbooms P, Goessens W H, van Leeuwen N, Quint W G. Comparison of phage typing and DNA fingerprinting by polymerase chain reaction for discrimination of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 1993;31:798–803. doi: 10.1128/jcm.31.4.798-803.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Belkum A, van Leeuwen W, Kaufmann M E, Cookson B, Forey F, Etienne J, Goering R, Tenover F, Steward C, O'Brien F, Grubb W, Tassios P, Legakis N, Morvan A, El Solh N, de Ryck R, Struelens M, Salmenlinna S, Vuopio-Varkila J, Kooistra M, Talens A, Witte W, Verbrugh H. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J Clin Microbiol. 1998;36:1653–1659. doi: 10.1128/jcm.36.6.1653-1659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voss A, Milatovic D, Wallrauch-Schwarz C, Rosdahl V T, Braveny I. Methicillin-resistant Staphylococcus aureus in Europe. Eur J Clin Microbiol Infect Dis. 1994;13:50–55. doi: 10.1007/BF02026127. [DOI] [PubMed] [Google Scholar]
- 36.Wanger A R, Morris S L, Ericsson C, Singh K V, LaRocco M T. Latex agglutination-negative methicillin-resistant Staphylococcus aureus recovered from neonates: epidemiologic features and comparison of typing methods. J Clin Microbiol. 1992;30:2583–2588. doi: 10.1128/jcm.30.10.2583-2588.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]