Abstract
Under anaerobic stress, Arabidopsis thaliana induces the expression of a collection of core hypoxia genes that encode proteins for an adaptive response. Among these genes is NIP2;1, which encodes a member of the “Nodulin 26-like Intrinsic Protein” (NIP) subgroup of the aquaporin superfamily of membrane channel proteins. NIP2;1 expression is limited to the “anoxia core” region of the root stele under normal growth conditions, but shows substantial induction (up to 1,000-fold by 2–4 h of hypoxia) by low oxygen stress, and accumulation in all root tissues. During hypoxia, NIP2;1-GFP accumulates predominantly on the plasma membrane by 2 h, is distributed between the plasma and internal membranes during sustained hypoxia, and remains elevated in root tissues through 4 h of reoxygenation recovery. In response to hypoxia challenge, T-DNA insertion mutant nip2;1 plants exhibit elevated lactic acid within root tissues, reduced efflux of lactic acid, and reduced acidification of the external medium compared to wild-type plants. Previous biochemical evidence demonstrates that NIP2;1 has lactic acid channel activity, and our work supports the hypothesis that NIP2;1 prevents lactic acid toxicity by facilitating release of cellular lactic acid from the cytosol to the apoplast, supporting eventual efflux to the rhizosphere. In evidence, nip2;1 plants demonstrate poorer survival during argon-induced hypoxia stress. Expressions of the ethanolic fermentation transcript Alcohol Dehydrogenase1 and the core hypoxia-induced transcript Alanine Aminotransferase1 are elevated in nip2;1, and expression of the Glycolate Oxidase3 transcript is reduced, suggesting NIP2;1 lactic acid efflux regulates other pyruvate and lactate metabolism pathways.
Introduction
As obligate aerobes, land plants require a continuous supply of oxygen to support energy demands. Oxygen deprivation stress from flooding or poor soil aeration depresses cellular respiration, leading to an energy crisis that triggers a variety of genetic, metabolic, and developmental adaptation strategies (Voesenek and Bailey-Serres, 2013, 2015). In Arabidopsis (Arabidopsis thaliana), low oxygen stress triggers the preferential transcription and translation of a collection of “core” hypoxia-induced genes that encode glycolytic/fermentation enzymes, other metabolic proteins, and various signaling proteins and transcription factors involved in the adaptive response (Mustroph et al., 2009, 2010). Among the hypoxia-induced core response transcripts is an aquaporin-like membrane channel protein, Nodulin 26-like intrinsic protein (NIP) 2;1 (Choi and Roberts, 2007).
NIPs are a terrestrial plant-specific subfamily of the aquaporin-superfamily that show structural and functional homology to the Glycine max root nodule-specific protein nodulin 26 (Roberts and Routray, 2017). NIPs possess the canonical aquaporin “hourglass” fold, but have diverged as multifunctional transporters of a wide variety of substrates including glycerol, ammonia, boric acid, silicic acid, and toxic metalloid hydroxides (Roberts and Routray, 2017; Pommerrenig et al., 2020). Based on structural modeling, NIPs can be segregated into three “pore families” that have conserved amino acids within their aromatic-arginine (ar/R) selectivity filters (Roberts and Routray, 2017). NIP2;1, along with NIP1;1, NIP1;2, NIP3;1, NIP4;1, and NIP4;2, belongs to the Arabidopsis NIP I subgroup (Johanson et al., 2001; Roberts and Routray, 2017). NIP I proteins have an ar/R amino acid composition similar to nodulin 26 (Wallace and Roberts, 2004), and show a transport selectivity for water, glycerol, and ammonia as natural substrates (Roberts and Routray, 2017), as well as the ability to be permeated by toxic metalloids (e.g. arsenite and antimonite; Kamiya et al., 2009; Kamiya and Fujiwara, 2009; Xu et al., 2015; Diehn et al., 2019). Biochemical analysis of Arabidopsis NIP2;1 in Xenopus (Xenopus laevis) oocytes showed that it differs from those NIP I transport properties, and instead shows preference for transport of lactic acid (Choi and Roberts, 2007).
Lactic acid is the end product of lactic acid fermentation, one of the fermentative pathways employed by plants to sustain energy production under oxygen limiting conditions and other stress conditions in which respiration is repressed (Drew, 1997; Gibbs and Greenway, 2003). Accumulation of lactic acid increases the acid load in the cytosol, and is among the factors that contribute to the cellular acidification observed during low oxygen stress (Davies et al., 1974; Roberts et al., 1984; Felle, 2005). Hypoxia-induced fermentation is accompanied by the acquired ability of plant roots to efflux lactic acid to the rhizosphere (Xia and Saglio, 1992; Rivoal and Hanson, 1993; Xia and Roberts, 1994; Dolferus et al., 2008). In maize (Zea mays) root tips, this lactic acid/lactate efflux mechanism accompanies reduced susceptibility to low oxygen stress from acclimation, and is proposed to be an adaptive mechanism to reduce cytosolic acidification or other toxic effects of cellular lactic acid/lactate accumulation (Xia and Saglio, 1992; Xia and Roberts, 1994).
The previous finding that NIP2;1 is selectively permeable to lactic acid upon expression in Xenopus oocytes, together with its identification as a core-hypoxia gene product, has led to the hypothesis that it mediates the efflux and/or compartmentation of lactic acid during the Arabidopsis response to low oxygen stress (Choi and Roberts, 2007). However, this has yet to be investigated in planta. The objective of this research was to test this hypothetical function of NIP2;1 during the Arabidopsis hypoxia response. In this study, it is shown that a T-DNA insertional nip2;1 mutant exhibits poor tolerance to low oxygen stress compared to wild-type (WT). Further, it is shown that the efflux of lactic acid from hypoxia-stressed roots to the media is reduced in nip2;1 mutant plants. NIP2;1-GFP in complementation seedlings showed broad expression throughout the root in response to hypoxia, predominant localization to the plasma membrane, and persistence throughout the hypoxia time course and during reoxygenation recovery. Lastly, evidence is presented that the hypoxia-induced levels of transcripts encoding fermentation and lactate/alanine metabolism enzymes are altered in nip2;1 mutant plants compared to WT plants. Overall, the data support NIP2;1 as a lactic acid efflux channel involved in the release of this metabolite from the root to the rhizosphere, and is essential for plant survival during the Arabidopsis hypoxia stress response.
Results
During hypoxia, NIP2;1 expression is induced early and primarily in root tissue
Hypoxia treatment of 2-week-old Arabidopsis seedlings resulted in a rapid increase in NIP2;1 expression in root tissues, with reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis revealing a >1,000-fold increase in NIP2;1 transcript levels within 2 h of the onset of anaerobiosis (Figure 1A). NIP2;1 transcript levels then showed a sharp decline by 12 h but still remained over 100-fold elevated relative to normoxic controls before leveling off at 24 h (Figure 1A). While NIP2;1 is predominantly a root transcript, hypoxia also induced NIP2;1 expression in shoot tissues, but with a lower overall expression (40-fold relative to basal levels) compared to roots (Figure 1A, inset). In addition, the time course of accumulation in shoots was delayed compared to roots and the expression did not peak until 12-h treatment.
Figure 1.
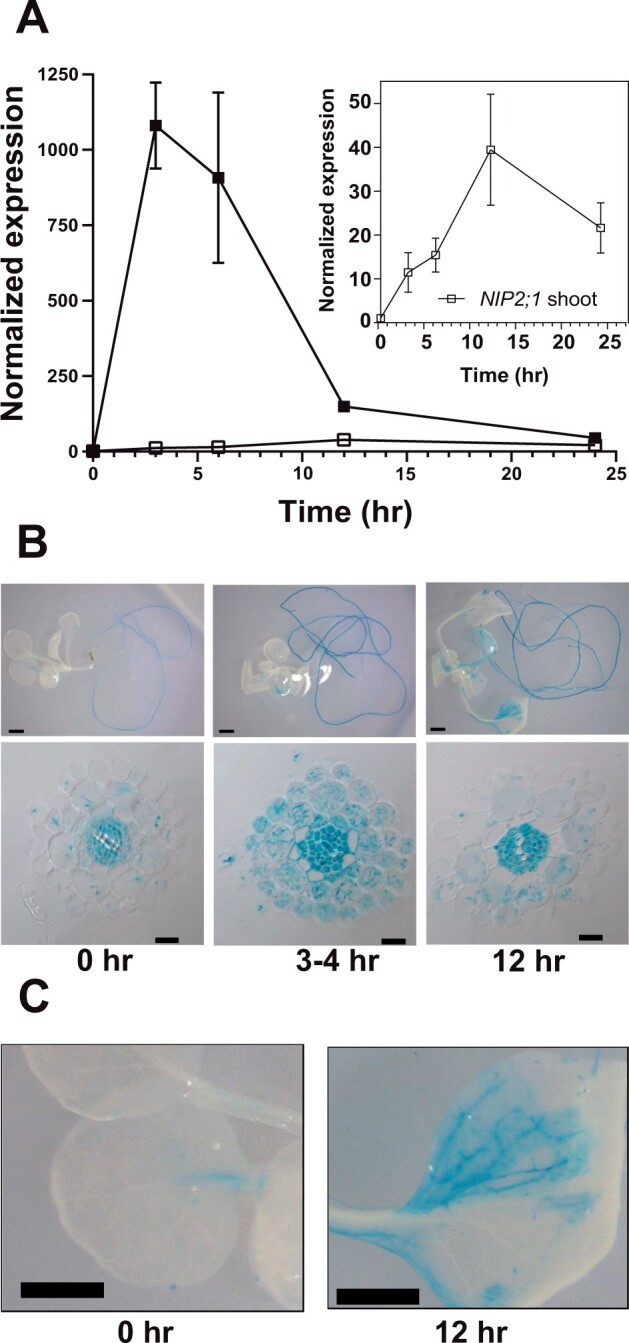
NIP2;1 expression in Arabidopsis seedlings in response to oxygen deficit. A, RT-qPCR analysis of NIP2;1 transcripts in root (filled squares) and shoot (open squares) tissues during a hypoxia time course of 2-week-old Arabidopsis seedlings. The ΔCt value of NIP2;1 obtained from the 0 h shoot sample was used as the calibrator for expression level normalization. The graph in the insert shows a rescaled plot of NIP2;1 expression in shoot tissue. Error bars indicate the standard deviation (sd) of three biological experiments with three technical replicates each. B, GUS staining analysis of 2-week-old NIP2;1pro::GUS Arabidopsis seedlings subjected to the oxygen deprivation conditions as for (A). Top part represents whole seedlings while the bottom part shows root cross-sections. Scale bars are 1.0 mm for the top part and 20 μm in the bottom part. C, Close up of GUS-stained leaves from (B). Scale bar = 1.0 mm.
Similar patterns of NIP2;1 expression were observed with 2-week-old NIP2;1 promoter::GUS fusion plants subjected to the same oxygen deprivation regime (Figure 1B). Analysis of the cellular localization of beta-glucuronidase (GUS) staining under normoxic conditions showed that expression is principally limited to roots with little staining detected in shoot tissues (Figure 1B). Cross-sections of unstressed (normoxic) roots showed that GUS staining was principally observed in the cells of the stele (pericycle, phloem, and procambium), with little or no GUS signal apparent in endodermal, cortical, and epidermal cells (Supplemental Figure S1; Figure 1B). At 4 h after the induction of hypoxia, root tissues showed an increase in the intensity of the GUS staining in the stele, as well as the appearance of the GUS signal in the cortex, epidermis, and root hairs (Figure 1B). Similar to the RT-qPCR result, this staining peaked at 4-h post-hypoxic treatment and then decreased by 12-h post-treatment (Figure 1B), although the expression at these later time points was still much higher than the basal expression in the unstressed roots (Figure 1B). In comparison to roots, increases in GUS expression in shoots were less acute and appeared more slowly, and the expression was mainly restricted to the vascular tissues of leaves (Figure 1C).
NIP2;1 enhances plant survival under low oxygen conditions
Core hypoxia-response gene loss-of-function mutants generally result in reduced survival or increased sensitivity to low oxygen stress (Ismond et al., 2003; Kursteiner et al., 2003; Licausi et al., 2010; Giuntoli et al., 2014; Sorenson and Bailey-Serres, 2014; Lokdarshi et al., 2016). To investigate whether NIP2;1 is necessary for hypoxia stress survival, a T-DNA insertion mutant (WiscDsLox233237_22K; referred to here as nip2;1) line was studied. The nip2;1 line contains a T-DNA insertion within the promoter region (−30) between a cluster of Anaerobic Response Elements (AREs) and the transcriptional start site (Figure 2A). Consistent with the position of this insertion, hypoxia treatment (4 h) of nip2;1 mutants resulted in poor expression of NIP2;1 compared to WT (Figure 2B). Western blot analysis of WT roots with site-specific anti-NIP2;1 antisera showed the hypoxia-induced appearance of an immunoreactive band that was not detected under the same conditions in nip2;1 mutant roots (Figure 2C). Overall, the data confirm that this T-DNA insertional mutant shows a severe defect in NIP2;1 expression and does not produce a detectable protein product.
Figure 2.
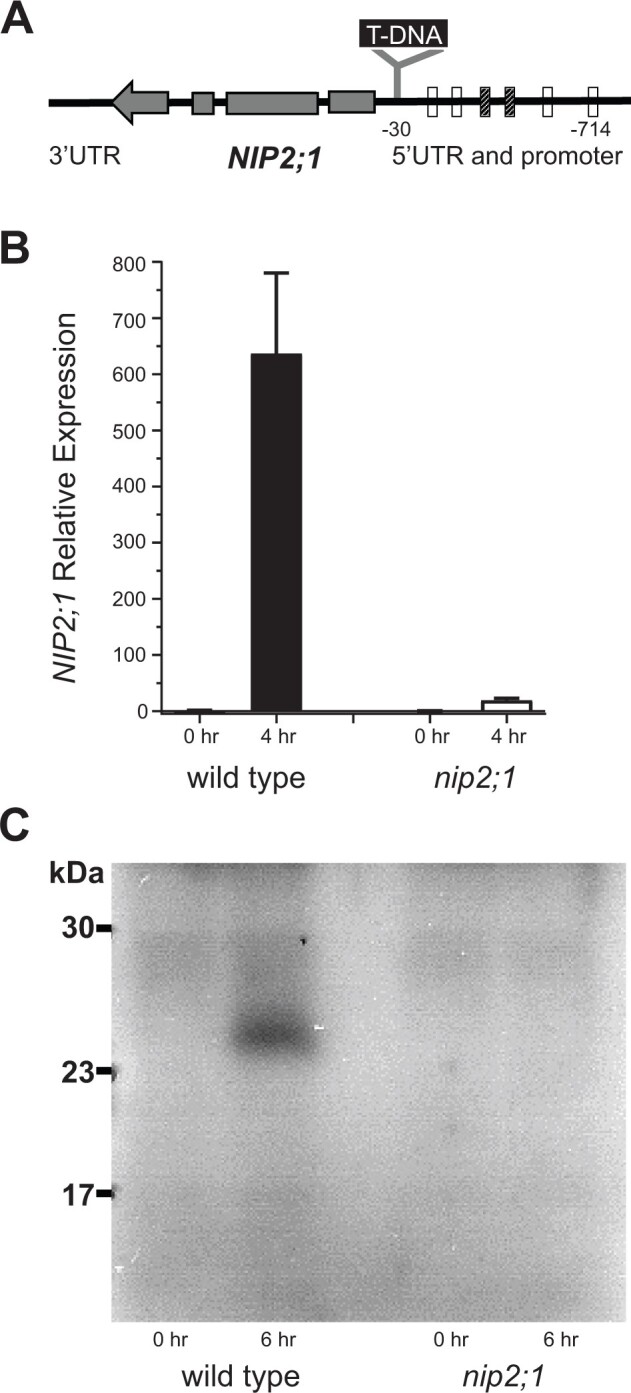
Characterization of nip2;1 T-DNA insertional mutant seedlings. A, Schematic diagram showing the site of T-DNA insertion in the nip2;1 mutant line. The positions of AREs in the promoter region, based on Olive et al. (1990) (cross hatched bar) or Dolferus et al. (1994) (open bars), relative to the site of T-DNA insertion are shown. The gray bars within the NIP2;1 gene indicate the positions of exons with the arrow indicating the direction of transcription. B, RT-qPCR results for NIP2;1 expression in the roots of 7-d-old WT (Col-0) and nip2;1during hypoxic treatment. The ΔCt value of NIP2;1 expression in normoxic WT roots was used as a calibrator for expression normalization. Error bars show the sd of three biological experiments with three technical replicates each. C, Root extracts (10 μg protein/lane) were analyzed by Western blot with site-directed NIP2;1 antibodies. 0 h, normoxic control; 6 h, 6-h hypoxia-treated seedlings.
To determine the effects of low oxygen stress on nip2;1 plants, their growth and survival under normoxic and hypoxic conditions were compared (Figure 3). While WT and nip2;1 seedlings showed little difference in growth under normoxic conditions (Supplemental Figure S2), nip2;1 seedlings showed higher sensitivity to hypoxia treatment (Figure 3). After exposure to argon gas-induced hypoxia, followed by transfer back to normoxic conditions for recovery, nip2;1 seedlings exhibited a higher incidence of chlorosis and seedling death (Figure 3A). Comparison of the overall survival frequency of WT and nip2;1 seedlings showed that the mutant exhibited significantly poorer survival to hypoxic stress (Figure 3C).
Figure 3.
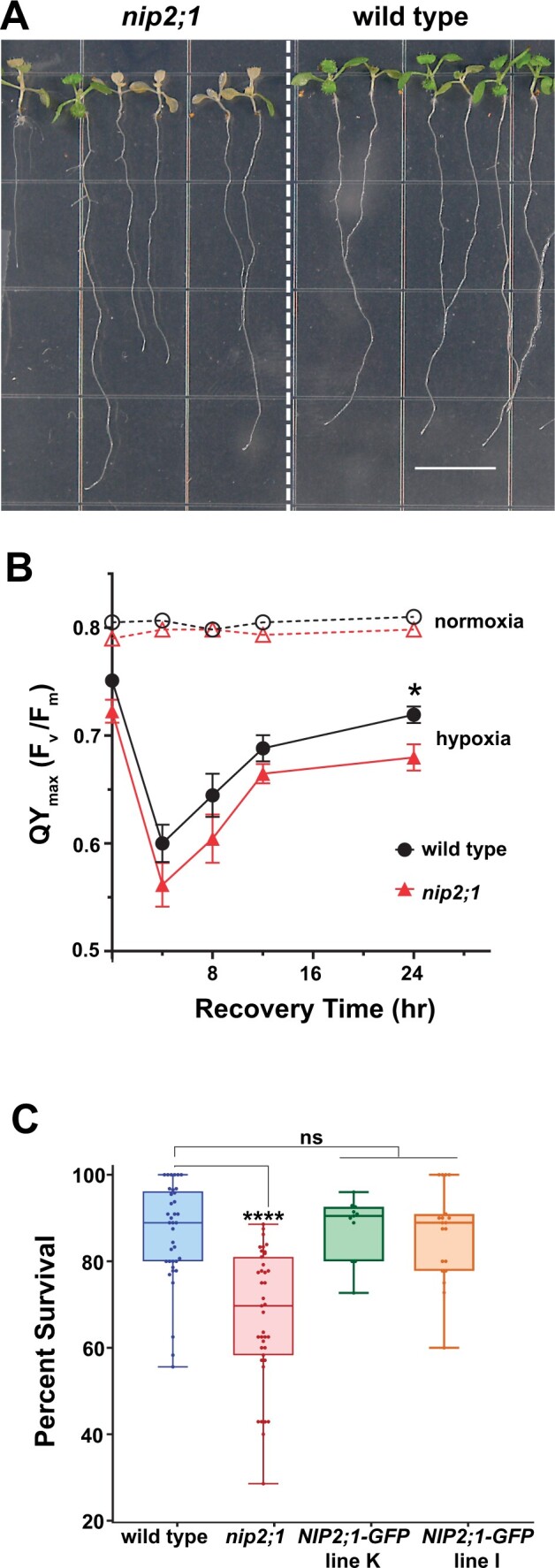
Effects of oxygen deprivation on the survival of nip2;1 T-DNA insertional mutant seedlings. A, Seven-day-old, vertically grown seedlings of WT (Col-0) and nip2;1 were subjected to a 8 h of argon gas treatment and were allowed to recover under normal oxygen conditions for 72 h prior to assessing survival. Scale bar = 1 cm. B, PS II maximal quantum yield [QYmax (Fv/Fm)] was calculated from chlorophyll fluorescence analysis of 7-d-old WT (Col-0, black circles) and nip2;1 mutant (red triangles) seedlings treated with 8 h argon (hypoxia, solid lines) or air control (normoxia, dashed lines). Error bars represent the standard error of mean (sem) of five biological experiments. *P < 0.05 based on Welch’s unpaired t test. C, Box and whiskers plot showing the survival of 7-d-old WT, nip2;1, and two NIP2;1:GFP complementation seedling lines after 8 h argon treatment, represented as percentage seedling survival as described by (Licausi et al., 2010). Each data point represents one biological replicate with the median value indicated as a solid line within in each box. Statistical significance was assessed by multiple comparisons by one-way ANOVA analysis. (****P < 0.0001 for nip2;1 compared to WT and both complementation lines; ns, not significant).
The sensitivity of the nip2;1 mutant to hypoxia was further assessed by measuring the chlorophyll fluorescence properties and calculating the maximum potential quantum efficiency (QYmax or Fv/Fm) of photosystem (PS) II of nip2;1 and WT seedlings under normal and low oxygen stress conditions. Chlorophyll fluorescence measurement is a common technique used to assess the photosynthetic efficiency of PS II, which is an index of the susceptibility of plants to different environmental stressors (Murchie and Lawson, 2013). Under normoxic conditions, WT and nip2;1 seedlings were not significantly different, with Fv/Fm values that fell within the optimum range (0.78–0.8; Murchie and Lawson, 2013). This suggests that the lack of NIP2;1 expression in the mutant line does not exhibit any detectable adverse effect on this parameter under standard growth conditions. However, upon hypoxia treatment, both WT and nip2;1 seedlings showed a steep reduction in photosynthetic efficiency, with the Fv/Fm ratio declining to ˂0.6 within 4 h of the recovery period (Figure 3B). At all time points, the Fv/Fm ratio is lower for nip2;1 than for WT. At later time points, the quantum efficiency of PS II starts to recover for both WT and nip2;1 seedlings (Figure 3B), but the nip2;1 Fv/Fm ratio remains significantly lower than WT after 24 h of recovery.
To confirm that the increased sensitivity of nip2;1 plants to hypoxia is the result of the loss of NIP2;1 gene, two complementation lines containing a NIP2;1pro::NIP2;1-GFP transgene (NIP2;1-GFP plants) in the nip2;1 background were analyzed (Figure 3C). Both complementation lines showed enhanced tolerance to hypoxia challenge compared to nip2;1 seedlings, and were not statistically different from WT seedlings with respect to survival frequency (Figure 3C). Taken together, the nip2;1 phenotype data indicate that NIP2;1 is a hypoxia core response protein that participates in the hypoxia adaptation response, and that reduction in expression of NIP2;1 lowers the ability of Arabidopsis to survive this stress. One NIP2;1-GFP complementation line (line K) was chosen for further study.
NIP2;1 is expressed on the plasma membrane, as well as on internal membranes, during hypoxia and reoxygenation recovery
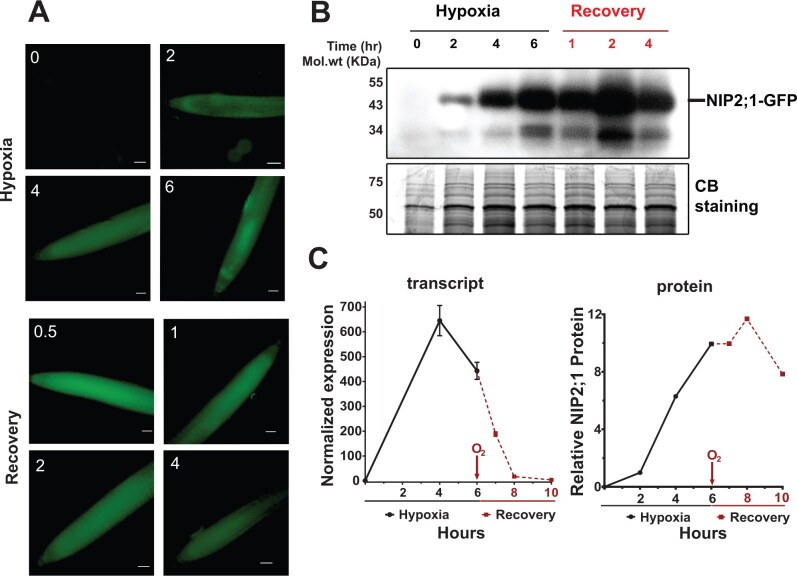
To investigate the dynamics of NIP2;1 expression and its subcellular localization, NIP2;1-GFP expression in the complementation line was analyzed. Similar to WT, RT-qPCR analysis shows that the NIP2;1-GFP transgene transcripts are acutely induced by hypoxia, with a peak at 4 h followed by a decline (Figure 4C). As has been documented with other core hypoxia transcripts (Branco-Price et al., 2008), reoxygenation resulted in a rapid decline of the transcript to basal levels. Analysis of NIP2;1-GFP protein accumulation during hypoxia and re-oxygenation was done by using epifluorescence microscopy and Western blot analysis at different time points of hypoxia stress and reoxygenation recovery (Figure 4, A and B). Microscopy revealed that the GFP signal first appeared within 2 h of the onset of hypoxia (Figure 4A), and increased as hypoxia proceeded. Return to normal oxygen conditions resulted in a substantial increase in the fluorescence intensity at 30 min that remained throughout the recovery period (Figure 4A).
Figure 4.
NIP2;1-GFP expression in the roots of NIP2;1:GFP seedlings during hypoxia and reoxygenation. Ten-day-old NIP2;1:GFP seedlings were subjected to an argon-induced hypoxia time course, with oxygen resupplied at Hour 6. A, Representative epifluorescence images of the primary root of NIP2;1:GFP seedlings at the indicated times (h) of hypoxia treatment or reoxygenation recovery. Scale bars = 50 μm. B, Anti-GFP Western blot showing NIP2;1-GFP protein accumulation (upper). The expected position of NIP2;1-GFP is indicated. Bottom, Coomassie blue stained loading control gel. C, Comparison of root NIP2;1-GFP transcript levels (normalized to 0 h) by RT-qPCR (left, n = 8 biological experiments, error bars show sem) and NIP2;1-GFP protein (right) based on densitometry of the Western blot in (B).
Western blot analysis showed a similar pattern for protein accumulation as that observed with fluorescence microscopy, with the protein levels increasing during hypoxia, and remaining elevated during reoxygenation (Figure 4, B and C). Two bands were observed in Western blot analyses, a major band migrating as expected for the NIP2;1-GFP fusion, and a second minor band with a mobility similar to free GFP, likely representing a degradation product. While transcript levels declined rapidly to non-detectable levels (Figure 4C), the fluorescence intensity and Western blot analyses indicate the persistence of the NIP2;1 protein for hours after return to normal oxygen conditions (Figure 4, A and B).
Different NIP proteins show varied subcellular localization, ranging from polarized plasma membrane localization for the root boric acid channel NIP5;1 (Wang et al., 2017) to specific localization on subcellular organelles, such as the soybean symbiosome membrane protein, nodulin 26 (Weaver et al., 1991). The determination of NIP2;1 localization under native conditions in response to low oxygen stress is important to understand the potential path of lactic acid transport. To investigate its subcellular localization under native conditions during hypoxia and reoxygenation, the roots of NIP2;1-GFP seedlings were examined more closely by using the confocal microscopy (Figure 5;Supplemental Figures S3 and S4).
Figure 5.
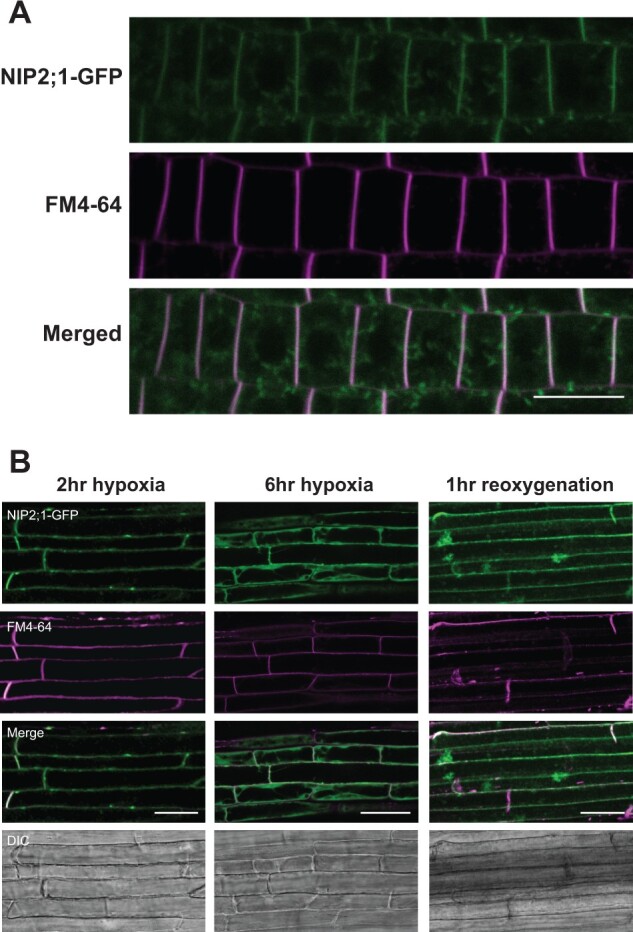
Subcellular localization of NIP2;1-GFP in the roots of hypoxia challenged NIP2;1:GFP complementation seedlings. A, Seven-day-old vertically grown NIP2;1:GFP seedlings were subjected to anaerobic stress by root submergence under argon gas treatment for 2 h, and the appearance of NIP2;1-GFP was monitored by confocal fluorescence microscopy. NIP2;1-GFP (top), FM4-64 (middle) and merged (bottom). Scale bar = 20 μm. B, NIP2;1:GFP seedlings were subjected to argon gas treatment at the times indicated followed by return to normoxic conditions at 6 h. Bars = 50 μm. DIC, differential interference contrast images.
Analysis of NIP2;1-GFP at 2 h after the onset of hypoxia revealed accumulation of GFP fluorescence throughout the root tip and the elongation and maturation zones (Supplemental Figure S3A). Closer examination and co-localization analyses with FM4-64 shows that NIP2;1-GFP is predominantly localized to the plasma membrane, although a lower level of signal within internal structures is also apparent (Figure 5A;Supplemental Figure S3, B and C). Simple examination of the confocal micrograph images suggested that the intensity of the NIP2;1-GFP signal differs between the apical and basal ends of the cells (Figure 5A). Some NIPs, such as the boric acid channel NIP5;1, have been documented to have polarized localization to the plasma membrane, which aids in the directional flow of substrate across the root (Wang et al., 2017). To determine if a similar situation is apparent for NIP2;1, the relative distribution of NIP2;1-GFP fluorescence signal across the radial and longitudinal axes of multiple root cells was quantified and standardized to the FM4-64 plasma membrane signal by the method of (Wakuta et al., 2015; Wang et al., 2017). The analysis indicated that NIP2;1 is evenly distributed across the plasma membrane, and that the ratio of NIP2;1/FM4-64 staining did not differ significantly from unity (polarity indices of 1.17 for radial distribution and 0.97 for longitudinal distribution; Supplemental Figure S3D), suggesting that NIP2;1 does not show polarized expression on selected surfaces of the plasma membrane.
As hypoxia proceeds, and more protein accumulates, there is a stronger accumulation of NIP2;1-GFP signal internally, although plasma membrane localization is also still apparent (Figure 5B, 6-h time point; Supplemental Figure S4). Following 1 h of reoxygenation, the predominant localization of NIP2;1 on the plasma membrane appears again (Supplemental Figure S4). Overall, while the data show some changes in the degree of surface vs. internal localization of NIP2;1 during the hypoxia/recovery response, the protein shows strong localization to the plasma membrane at all phases of hypoxia and early recovery, where it presumably mediates the transport of substrate from the cell into the apoplastic space.
NIP2;1 participates in lactic acid efflux and media acidification during hypoxia
Based on the specificity of NIP2;1 as a lactic acid channel from biochemical analyses (Choi and Roberts, 2007), its localization in part to the surface of root cells, and the observation that hypoxia triggers release of lactic acid/lactate from roots into the external media (Xia and Saglio, 1992; Dolferus et al., 2008; Engqvist et al., 2015), it is hypothesized that NIP2;1 may participate in the excretion of lactic acid during the low oxygen stress response in planta. In support of this, in comparison to WT seedlings, nip2;1 seedlings accumulated significantly higher levels of lactic acid/lactate level within root tissues during a hypoxia time course (Figure 6B), consistent with the inability to efflux this fermentation end product.
Figure 6.
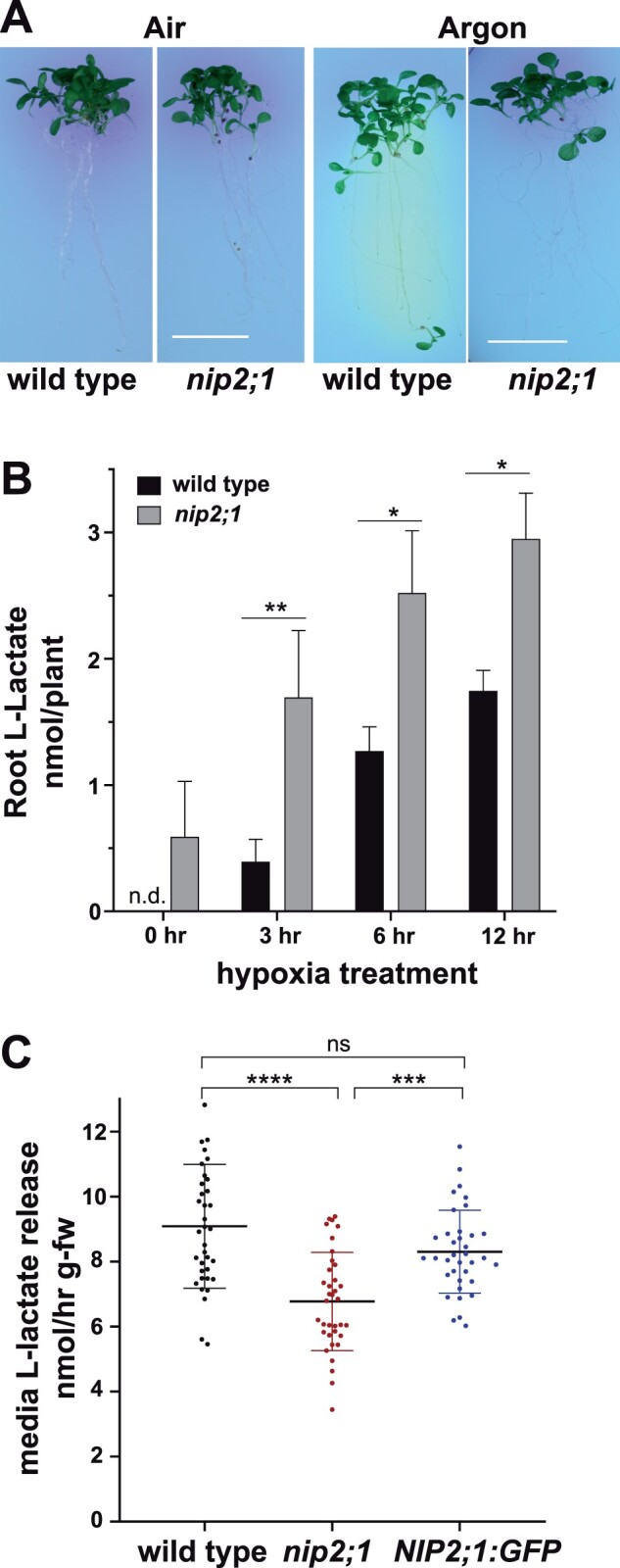
Media acidification and lactic acid efflux in hypoxia-challenged nip2;1. A, Ten-day-old nip2;1 and WT (Col-0) seedlings were transferred to pH indicator plates containing bromocresol purple, and were subjected to 8 h treatment of hypoxia induced by argon gas (Argon). Air indicates a normoxic control. A color change from purple to yellow indicates a decrease in the pH of the environment. Scale bars = 1 cm. B, Quantitation of lactic acid/lactate in seedling roots based on enzymatic analysis during hypoxia. Values are the average from three biological experiments at each time point, with the error bars showing the sd. C, The rate of media lactic acid/lactate release from the seedlings of indicated lines during a hypoxia time course. Each value represents an individual determination from samples taken over an 8-h hypoxia time course, as described in the “Materials and methods,” with the error bars showing the sd. Asterisks in (B) and (C) indicate statistically significant differences based on one-way ANOVA analysis with multiple comparisons (B) or a paired Student’s t test analysis (C). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To test this hypothesis further, the media pH and rates of lactic acid/lactate efflux from the roots of WT and nip2;1 seedlings challenged with hypoxia were examined. To compare the hypoxia-induced acidification of the external medium, 10-d-old WT and nip2;1 seedlings were subjected to argon gas treatment on media containing the pH-sensitive dye, bromocresol purple (Figure 6A). Upon transfer from normoxic to hypoxic conditions, WT seedlings showed substantial yellowing of the media surrounding the roots, suggesting a decrease in the pH and acidification of the media, while nip2;1 plants showed no difference in bromocresol purple staining between normoxic and hypoxic conditions (Figure 6A).
Further comparison of hypoxia-challenged WT and nip2;1 seedlings reveal that WT roots show a significantly higher rate of lactic acid/lactate release into the medium compared with the roots of nip2;1 plants (Figure 6C). Assay of a NIP2;1-GFP complementation line shows that its rate of lactic acid/lactate efflux is restored to WT levels (Figure 6C), verifying that the reduction in lactic acid efflux in nip2;1 plants is due to the loss of NIP2;1 protein. These results, combined with previous functional studies (Choi and Roberts, 2007), suggest that NIP2;1 participates in lactic acid transport, homeostasis, and efflux from roots during low oxygen stress.
The loss of NIP2;1 function affects the expression of pyruvate and lactate metabolic enzymes
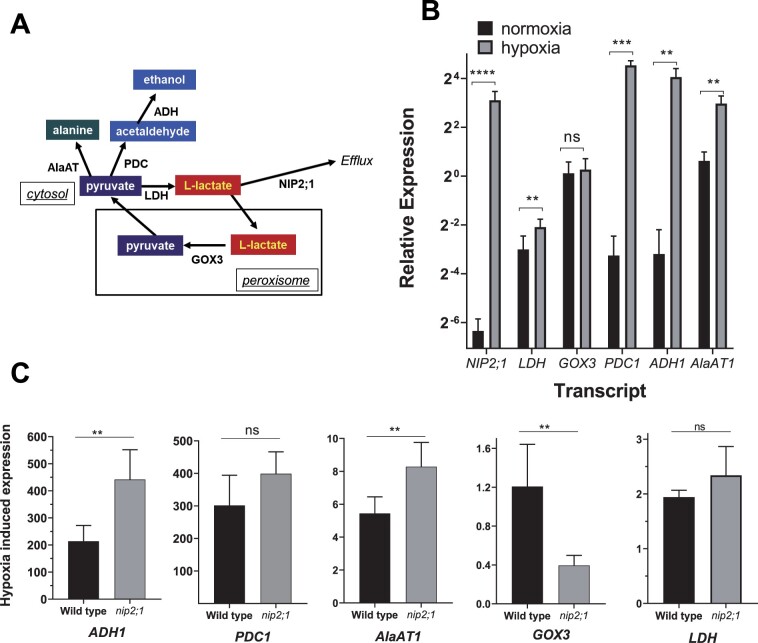
Anaerobic metabolism of pyruvate during oxygen limitation in plants occurs through three conserved pathways: lactic acid fermentation, ethanolic fermentation, and alanine synthesis (Figure 7). While all three pathways use pyruvate as a substrate, lactic acid and ethanolic fermentation regenerate NAD+, whereas alanine synthesis serves as a mechanism to store nitrogen and carbon for reoxygenation (Sato et al., 2002; Ricoult et al., 2005). Genes that encode enzymes in these pathways (such as ALCOHOL DEHYDROGENASE1 [ADH1], PYRUVATE DEHYDROGENASE1 [PDC1], LACTATE DEHYDROGENASE [LDH], and ALANINE AMINOTRANSFERASE1 [AlaAT1]) are among the “core hypoxia response” genes that are induced in Arabidopsis roots during hypoxia (Mustroph et al., 2009; Lee et al., 2011; Mustroph et al., 2014). Conversely, L-lactate produced via LDH is proposed to be converted back to pyruvate in peroxisomes (Figure 7A) by the root-specific glycolate oxidase 3 (GOX3) enzyme (Engqvist et al., 2015). Unlike other members of this enzyme family that participate in the metabolism of glycolate, GOX3 is specific for L-lactate and utilizes oxygen as an electron acceptor to oxidize lactate, producing hydrogen peroxide and pyruvate as end products (Engqvist et al., 2015). GOX3 is not a hypoxia-induced transcript and is, rather, proposed to regulate the concentration of lactate in a coordinated fashion with LDH under aerobic conditions (Engqvist et al., 2015; Maurino and Engqvist, 2015).
Figure 7.
Comparison of transcripts of lactate and pyruvate metabolic enzymes in hypoxia treated nip2;1 and WT roots. A, Schematic showing the principal pathways of pyruvate and lactate metabolism during fermentation. B, RT-qPCR analysis of the indicated transcripts in the roots of 7-d-old WT seedlings grown under normoxic conditions (black bars) or in response to 4 h of argon-induced hypoxia (gray bars). The data are normalized to the transcript levels of GOX3 under normoxic conditions. C, Comparison of the hypoxia-induced changes of selected transcripts from the roots of WT and nip2;1 seedlings. The data are normalized to the expression levels of the indicated transcript under normoxic conditions. Error bars in (B) and (C) show the sd (three biological experiments each with three technical replicates), and asterisks indicate statistically significant differences based on an unpaired Student’s t test analysis. NS indicates not significant; *P > 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
RT-qPCR analysis shows that hypoxia response transcripts (ADH1, PDC1, LDH, and AlaAT1) show induction in both WT and nip2;1 mutant root tissues during 4 h of argon gas (Figure 7, B and C). However, closer analysis of WT and nip2;1 roots reveals different levels of selective transcripts. While LDH and PDC1 transcript levels show no statistical differences between WT and nip2;1 roots, ADH1, and AlaAT1 are significantly elevated in nip2;1 compared to WT roots (Figure 7C). Conversely, GOX3, which is expressed at the same level under normal and hypoxic conditions in WT roots, is substantially reduced in hypoxic nip2;1 roots. Overall, the data suggest that the alterations in lactic acid homeostasis within the nip2;1 mutant affect the expression of enzymes in pyruvate and lactate metabolic pathways, with transcripts encoding fermentation enzymes in ethanol (ADH1) and alanine (AlaAT1) producing pathways being enhanced, whereas the transcript that encodes the lactic acid metabolizing enzyme GOX3 is suppressed.
Discussion
NIP2;1-mediated lactic acid efflux promotes Arabidopsis survival during low oxygen stress
In response to low oxygen conditions resulting from flooding or submergence stress, plants switch to anaerobic fermentation pathways to maintain glycolytic flux and energy production. In Arabidopsis, enzymes of ethanolic (ADH and PDC) and lactic acid (LDH) fermentation pathways are necessary for optimal survival under low oxygen stress (Ellis et al., 1999; Ismond et al., 2003; Kursteiner et al., 2003; Dolferus et al., 2008). The levels of lactic acid/lactate increase 14-fold within the root during the first 2 h of hypoxia challenge (Mustroph et al., 2014), suggesting that lactic acid fermentation is induced during the initial stages of hypoxia. As lactic acid/lactate accumulates, hypoxia-stressed Arabidopsis plants (Dolferus et al., 2008), similar to other plant lineages (Xia and Saglio, 1992; Rivoal and Hanson, 1993; Xia and Roberts, 1994), efflux this fermentation end product from the roots to the external media/rhizosphere. Lactic acid efflux mechanisms in plant roots may assist in mitigating cellular acidification or other toxic effects of lactic acid accumulation during anaerobic stress (Xia and Roberts, 1994; Greenway and Gibbs, 2003).
To alleviate potential cellular acidification from lactic acid accumulation, the pathways for the efflux of lactic acid must transport either the protonated form (lactic acid) or co-transport lactate with a proton (reviewed in Greenway and Gibbs, 2003). In the case of animal cells, lactate is effluxed or taken up by members of the SLC16 subgroup of the major facilitator superfamily known as Monocarboxylate Transporters (MCTs; Counillon et al., 2016; Sun et al., 2017). These proteins are symporters that co-transport lactate with H+ in a bidirectional fashion. They participate in the efflux of excess lactic acid during anaerobic fermentation, and also serve as a reuptake mechanism of extracellular lactate for further metabolism (Sun et al., 2017). Land plants lack members of the SLC16/MCT transporter family, and the molecular identity of the transporters or channels that mediate the efflux of lactic acid/lactate produced during anaerobic fermentation has remained unclear. In this study, cellular, genetic, and physiological evidence is provided that indicates, together with previous protein functional analyses (Choi and Roberts, 2007), that the aquaporin-like NIP2;1 assists in lactic acid efflux from Arabidopsis roots.
NIPs are a plant-specific subgroup of the aquaporin superfamily that have diversified structurally and functionally during land plant evolution, and which have acquired solute transport activities beyond canonical aquaporin water transport (Roberts and Routray, 2017). NIP transport functions range from glycerol to ammonia to metalloids such as boric and silicic acid. However, biophysical and biochemical analyses of Arabidopsis NIP2;1 in Xenopus oocytes (Choi and Roberts, 2007) indicate that it is an outlier from other classical NIP proteins and is impermeable to water and all traditional NIP solute substrates, and instead shows specific bidirectional permeability to lactic acid. Several observations in the present work provide strong support that lactic acid transport and efflux are the biological functions of NIP2;1. First, the expression of NIP2;1 in response to hypoxia coincides with the appearance of lactic acid/lactate in the external medium; second, genetic mutation of the NIP2;1 via T-DNA insertion results in the reduction of lactic acid efflux from hypoxic roots into the external medium and a concomitant increase in the accumulation of lactic acid/lactate within root tissue; and third, nip2;1 mutants show reduction in the acidification of the media surrounding hypoxic roots.
nip2;1 mutant seedlings show poorer survival to argon-induced low oxygen stress compared to WT, presumably because of the over accumulation of toxic levels of lactic acid due to a reduced ability to efflux this end product from roots. As noted above, cytosolic lactic acid generation would increase the acid load of the cytosol that could contribute to acidosis (Davies et al., 1974; Roberts et al., 1984; Felle, 2005). Additionally, the accumulation of lactic acid/lactate could also contribute to reduced glycolytic flux by affecting NAD+ regeneration by altering the equilibrium of the LDH reaction, or potentially through product feedback inhibition mechanisms. For example, recent studies in yeast and mammals show that over accumulation of lactate leads to the production of the toxic side product 2-phospholactate catalyzed by pyruvate kinase. This side product of lactate blocks the production of fructose-2-6 bisphosphate, leading to the inhibition of the key glycolytic enzyme phosphofructokinase-1 (Collard et al., 2016). The production of similar toxic lactate metabolites side products could conceivably occur in plant tissues as well.
NIP2;1 expression during normoxia, hypoxia stress, and recovery
NIP2;1 expression is predominately limited to root tissues with a precise pattern of transcript and protein expression during normoxic, hypoxic, and reoxygenation conditions. Under normoxic conditions, NIP2;1 promoter activity is restricted to cells within the stele of the mature root. The cells of the stele are hypoxic even under well-aerated growth conditions due to the low rate of lateral oxygen diffusion across the mature differentiated root (Armstrong et al., 1994; Gibbs and Greenway, 2003). “Anoxic cores” in the root stele are proposed to aid in hypoxia sensing and acclimation, potentially by the communication of low oxygen or energy signals (ethylene, metabolites, low pH, and Ca2+) between hypoxic and well-aerated cells (Armstrong et al., 2019). The roots of nip2;1 mutants show increased accumulation of lactic acid/lactate under normoxic conditions. This suggests that LDH is active in anoxic core tissues, even under aerobic conditions, and that NIP2;1 basal expression is necessary for maintaining low lactic acid/lactate accumulation.
NIP2;1 RT-qPCR and GUS data show the characteristics of a core hypoxia response transcript, with acute induction of NIP2;1 expression in roots within 1 h of the initiation of hypoxia stress, followed by a peak and eventual decline to a reduced but elevated steady-state level that is sustained during hypoxia. Interestingly, examination of the cell-specific translatome atlas based on the work of (Mustroph et al., 2009) shows that the expression of NIP2;1 during hypoxia parallels the expression of the two lactate metabolizing enzyme transcripts, LDH and GOX3 (Supplemental Figure S5). All three transcripts are predominantly, if not exclusively, expressed in root tissue (Dolferus et al., 2008; Mustroph et al., 2014; Engqvist et al., 2015), and accumulate to the highest levels in the root cortex, as well as to high levels within the epidermal and vascular tissues, but are absent or poorly expressed in leaf tissues. The root-predominant expression pattern of NIP2;1, LDH1, and GOX3 is consistent with the distinct lactate metabolic properties and responses of roots and shoots to low oxygen stress (Ellis et al., 1999; Mustroph et al., 2014). Indeed, lactic acid fermentation and accumulation is predominantly restricted to root tissues during hypoxia in Arabidopsis (Mustroph et al., 2014). Based on the model of (Engqvist et al., 2015), these three gene products are proposed to coordinate root lactic acid/lactate homeostasis through its production (LDH), its recovery back to pyruvate (GOX3), and the excretion of lactic acid from the cell when it is overproduced during low oxygen stress (NIP2;1).
Similar to other hypoxia-induced genes (Branco-Price et al., 2008), reoxygenation results in suppression of NIP2;1 mRNA expression and a return to low basal levels within 2 h of recovery. In contrast, NIP2;1 protein levels increase during early reoxygenation and remain elevated for several hours post-recovery, suggesting that the activity of the protein is also required during recovery. In addition to excretion of lactic acid to the media, Arabidopsis roots take up L-lactate from the media and metabolize it (Dolferus et al., 2008; Engqvist et al., 2015). Since NIP2;1 mediates the bidirectional flux of lactic acid (Choi and Roberts, 2007), it could assist in the recovery of excreted lactic acid to trigger its metabolism to pyruvate and entry into thecitric acid cycle as part of the replenishment of cycle intermediates that takes place during post-anoxic recovery (Branco-Price et al., 2008; Tsai et al., 2014; Yeung et al., 2019).
NIP2;1 localization during hypoxia stress and recovery
Previous work with NIP2;1 expression in heterologous systems with non-native promoters under unstressed conditions showed conflicting results, with localization to the plasma membrane (Choi and Roberts, 2007), internal membranes (Mizutani et al., 2006), or a mixture of both locations (Wang et al., 2017) observed in different experiments. By using the complementation lines with a NIP2;1-GFP transgene under the native promoter, we were able to establish more clearly the localization of the protein during hypoxia stress and recovery. This work shows substantial, if not exclusive, localization of NIP2;1-GFP to the plasma membrane, both during hypoxia as well as during the first hours of reoxygenation recovery. This observation suggests that the protein is involved in the efflux of lactic acid from the site of production (cytosol) to the apoplastic space. Additional details on the subsequent pathway of lactic acid movement, including the participation of other transporters that ultimately lead to the release of toxic fermentation product from the root to the media, remain undetermined. By analogy to boric acid homeostasis, which involves the collaboration of NIP channels and BOR transporters (Takano et al., 2008), NIP2;1 may be part of a larger transport network that coordinates lactic acid efflux and release.
The processes that govern spatio-temporal localization of NIP2;1 to internal compartments versus the plasma membrane remains an open question. The trafficking of plant aquaporins to various target membranes through endocytic and redistribution pathways is regulated based on metabolic need and stress physiology (reviewed by Chevalier and Chaumont (2015) and Takano et al. (2017)). For example, PIP2;1 aquaporins are dynamically cycled between the internal membranes and the plasma membrane (Li et al., 2011), with regulation via phosphorylation (Boursiac et al., 2008; Prak et al., 2008) or other factors (Santoni, 2017; Takano et al., 2017) leading to preferential surface expression or internalization, which regulates the hydraulic conductivity of the cell. The dual localization of NIP2;1 could reflect a similar dynamic distribution and trafficking between internal membranes and the cell surface to regulate lactic acid efflux. In the case of some plant (Prak et al., 2008), as well as mammalian (Noda and Sasaki, 2006) aquaporins, preferential trafficking to the plasma membrane is controlled by the phosphorylation of serine within the cytosolic carboxyl terminal domain. Proteins of the NIP I subgroup are phosphorylated on a homologous serine within the carboxyl terminal domain (Wallace et al., 2006; Santoni, 2017), which is catalyzed by CDPK/CPK kinases (Weaver et al., 1991). This phosphorylation motif is conserved in NIP2;1 (Ser 278). In addition, phosphoproteomic analysis reveals that NIP2;1 is also phosphorylated in the N-terminal domain at Ser 5 by an unidentified protein kinase (Vialaret et al., 2014). Whether phosphorylation, or other regulatory factors, control trafficking or distribution of NIP2;1 in response to hypoxia or recovery signals to regulate lactic efflux or uptake merits further investigation.
Lactic acid and ethanolic fermentation pathways
Ethanolic fermentation through the PDC-catalyzed decarboxylation of pyruvate, followed by subsequent production of ethanol from acetaldehyde via ADH, is proposed to be the major anaerobic catabolism pathway (Gibbs and Greenway, 2003). However, lactic acid fermentation is also carried out in most plant species, and in many cases may precede, and regulate, the transition to ethanolic fermentation (Gibbs and Greenway, 2003). The reason for initial reliance on lactic acid fermentation during hypoxia prior to a shift to ethanolic fermentation is not clear (Gibbs and Greenway, 2003). However, this pathway, unlike ethanolic fermentation, could allow recovery of the fermentation end product. Additionally, lactate production via LDH occurs under aerobic conditions in response to other abiotic and biotic stresses that could affect energy metabolism (Dolferus et al., 2008; Maurino and Engqvist, 2015). In animal systems, the physiological role of lactic acid/lactate transcends serving as an end product for anaerobic glucose metabolism, and its larger role as a metabolic regulator has emerged, including G-protein signaling as well as transcriptional regulation through histone modification (Latham et al., 2012; Sun et al., 2017; Zhang et al., 2019).
The possibility of lactic acid/lactate as a signal in plant systems remains largely unexplored. Nevertheless, there is evidence that the balance between the two fermentation pathways is regulated. For example, in classical studies of maize root tips, plants subjected to hypoxia stress initially engage in lactic acid fermentation, followed by a switch to primarily ethanolic fermentation that is proposed to be driven by cellular acidification (by lactic acid accumulation or other means) that results in subsequent pH-dependent activated ethanolic fermentation (the “pH stat” model [Davies et al., 1974; Roberts et al., 1984]). In the case of Arabidopsis, overexpression of LDH results in an increase in the activities of some ethanol fermentation enzymes (Dolferus et al., 2008), suggesting that increased lactic acid fermentation induces this alternative fermentation pathway. Conversely, adh1 null plants induce higher levels of lactic acid production to apparently compensate for reduced flux through the ethanolic fermentation pathway (Ismond et al., 2003). In nip2;1 mutants, the accumulation of higher levels of tissue lactic acid/lactate may produce an effect similar to LDH1 overexpression. Higher transcript levels encoding enzymes within alternative pathways of the metabolism of pyruvate (e.g. ADH1 and AlaAT1) may be an adaptive response to the accumulation of lactic acid in nip2;1 roots.
The reason for the selective reduction of GOX3 inhibition in hypoxic nip2;1 roots is less clear. As pointed out by Engqvist et al. (2015), the proposed role of this enzyme is to convert lactate back to pyruvate within the peroxisome, which would serve to reduce lactic acid levels within the cell. Notably, however, this conversion occurs with the production of a reactive oxygen end product (hydrogen peroxide). ROS production is a major contributor to reoxygenation stress and is associated with poor tolerance to hypoxia and recovery (Yeung et al., 2019). If cytosolic lactic acid/lactate levels are elevated in nip2;1 mutants, GOX3 (which uses oxygen as a co-substrate) could trigger greater ROS production upon reoxygenation.
Aquaporins as lactic acid channels in other plant and microbial systems
In addition to Arabidopsis NIP2;1, select aquaporins with lactic acid permeability and efflux function have been described in other systems. For example, the Lactobacillales, which produce large quantities of lactic acid through fermentation, possess isoforms of the glycerol facilitator encoded by GlpF1 and GlpF4 that facilitate lactic acid efflux (Bienert et al., 2013). The human trematode pathogen, Schistosoma mansoni, which performs lactic acid fermentation during the pathogenic part of its life cycle, possesses a lactic acid permeable plasma membrane aquaporin SmAQP that is proposed to release this end product (Faghiri et al., 2010). More pertinent to this study, recent work (Mateluna et al., 2018) has identified other NIP I proteins, PruavNIP1;1 and PrucxmNIP1;1, that are induced during low oxygen stress in hypoxia-tolerant Prunus root stocks, and which are proposed to be lactic acid permeable proteins based on yeast lactate auxotroph assays. These observations suggest that a subset of the NIP I subfamily may have a biological function in lactic acid efflux.
From a phylogenetic perspective, the NIP2;1 subfamily is mostly restricted to plant lineages of the Brassicaceae family (Supplemental Figure S6). Since Arabidopsis NIP2;1 retains the ar/R pore constriction properties of other lactic acid-impermeable NIP aquaglyceroporins (e.g. nodulin 26; Choi and Roberts, 2007), the substrate preference for lactic acid over other substrates likely includes pore structural features besides the canonical selectivity filter. Additionally, some closely related NIP2;1 orthologs in other Brassicaceae lineages, such as Brassica napus, readily transport metalloids, such as hydroxides of arsenite and boron, and these transport functions cannot be excluded for other NIP2;1 subfamily members (Diehn et al., 2019). Additional analyses are required to determine whether NIP2;1 orthologs in Brassicaceae are core hypoxia-response genes and share the lactic acid permeability properties of the Arabidopsis NIP2;1.
Materials and methods
Plant growth conditions and stress treatments
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used in all experiments. Seeds were sterilized and stratified at 4°C for 2 d, and were germinated as in (Choi and Roberts, 2007). Seedlings were grown vertically on in Murashige–Skoog (MS) media supplemented with 1% (w/v) sucrose and 0.8% (w/v) Phyto-agar (plantMedia) with a long-day (LD) cycle of 16 h of light (100 μmol m−2 s−1) and 8 h of dark (LD conditions). Hypoxia treatment was administered at the end of the light cycle by the argon-treatment protocol described by (Lokdarshi et al., 2016). For normoxic controls, seedlings were treated simultaneously under identical conditions except in the presence of air instead of argon gas. For reoxygenation, the seedlings were returned to normoxic LD conditions at the end of the hypoxia time course. Phenotype analysis for hypoxia survival was conducted by established stress/recovery protocols for Arabidopsis (Licausi et al., 2010; Sorenson and Bailey-Serres, 2014; Lokdarshi et al., 2016) with modifications. Seven-day-old seedlings grown vertically as described above were administered 8 h of argon gas-induced hypoxia (treatment) or air (normoxia control) in darkness, and were returned to normoxic LD growth conditions. The survival frequency (the absence of chlorosis and shoot meristem death as described by Sorenson and Bailey-Serres, 2014) was scored after 3 d of recovery from stress treatment.
Photosynthetic efficiency measurement
The maximum quantum yield of PS II [QYmax = Fv/Fm] was measured with a FluorCam 800MF instrument (Photon Systems Instruments) by the general method of (Murchie and Lawson, 2013). Seven-day-old seedlings were administered 8 h hypoxia, and QYmax was measured at different recovery time points upon return to LD conditions. For the first time point (time = 0), seedlings were removed from hypoxia and were subjected to a saturating pulse of 1800 µEin m−2s−1 for 0.8 s (Fm). Variable fluorescence (Fv) was calculated as the difference between Fo and Fm to calculate the maximum quantum yield [Fv/Fm]. For subsequent measurements, seedlings were dark adapted for 2 min (F0) prior to application of the saturating pulse and conducting measurements.
T-DNA insertion mutant nip2;1 and complementation lines
A sequence-tagged T-DNA insertion line within the NIP2;1 gene (WiscDsLox233237_22k) from the WiscDs-Lox T-DNA collection (Woody et al., 2007) was obtained from Arabidopsis Biological Resource Center at the Ohio State University. The mutant, herein named nip2;1, was selected on MS media supplemented with 15 μg/mL Basta. The genotype of mutant plants was assessed by a PCR-based genotyping protocol as described at (http://signal.salk.edu/tdnaprimers.2.html). For this purpose, genomic DNA was extracted from 2-week-old seedlings using the Wizard Genomic DNA purification kit by the manufacturer’s instructions (Promega, Madison, WI, USA) and was subjected to PCR analysis using two AtNIP2;1 gene-specific primers and the left border T-DNA primer (Supplemental Table S1). The precise site of T-DNA insertion was verified by a cloning of the PCR product into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA, USA) followed by automated DNA sequencing. All sequencing was performed at the University of Tennessee Molecular Biology Resource Facility. T4 homozygous mutant seedlings were used for all phenotyping and other analyses in this study.
For complementation of nip2;1, as well as to localize NIP2;1 protein by fluorescence microscopy, transgenic lines containing a construct consisting of a NIP2;1-GFP translational fusion under the control of the NIP2;1 promoter were generated in the nip2;1 background. The promoter region of the NIP2;1 gene (from the translational start site to a site 2,000-bp upstream) was amplified by PCR from Arabidopsis genomic DNA with gene-specific primers with added KpnI and AatII restriction sites (Supplemental Table S1) to facilitate its insertion into the binary vector pKGW_RedRoot_OCSA, replacing the ubiquitin promoter and dsRed CDS (Niyikiza et al., 2020). The modified destination vector was named pKGW_OCSA_NIP2;1Pro. The NIP2;1 coding sequence was amplified with gene-specific primers with XbaI and EcoRI sites (Supplemental Table S1) using a template of cDNA prepared from total RNA from 4-h hypoxic Arabidopsis seedlings. The resulting product was cloned into the Gateway entry vector CD3-1822 (Wang et al., 2013) to generate a construct encoding NIP2;1 as an in-frame carboxyl-terminal fusion with GFP separated by a 3X Gly linker. The NIP2;1-GFP construct was then recombined into the pKGW_OCSA_NIP2;1Pro vector by using a gateway LR reaction (Invitrogen) to generate the final binary vector with NIP2;1pro::NIP2;1-GFP. The constructs were sequenced and verified by using Snap Gene 4.2.11 software. Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986) was transformed with the final construct by electroporation with a Bio-Rad Gene Pulser Xcell Electroporation system. Colonies carrying the correct construct were verified by PCR, and were used to transform Arabidopsis nip2;1 plants by using floral dip method (Clough and Bent, 1998). Transgenic lines were selected on MS media supplemented with 25 μg/mL kanamycin, and were confirmed by PCR-based genotyping with transgene-specific primers (Supplemental Table S1). Twelve transgenic lines were generated and the two with the highest NIP2;1 transcript abundance with expression levels similar to WT under hypoxia stress (lines K and I) were selected for further analysis. T2 generation homozygous complementation lines were used for further studies.
RNA purification and qPCR
Total RNA was isolated from plant tissues by grinding on liquid nitrogen followed by extraction with the PureLink Plant RNA Reagent (Invitrogen). RNA isolation and DNase treatment were carried out with a Direct-zol RNA MiniPrep Plus Kit by using the manufacturer’s protocol (Zymo Research, Irvine, CA, USA). Each RNA isolate for all RT-qPCR experiments was generated from biological material from 50 seedlings. RT-qPCR was performed on a Bio-Rad IQ5 real-time PCR detection system by using iTaq Universal SYBR Green One-Step RT-qPCR kit (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions with the following program: 50°C for 10 min, 95°C for 1 min, and 40 cycles of 95°C for 10 s and 60°C for 30 s. This amplification protocol was followed by an additional thermal denaturation cycle (65–95°C with 0.5°C increments), which was performed to generate melting curves to validate the amplification specificity. All primer sets produced a single amplification product with the expected Tm. All gene-specific primers used in this study are listed in Supplementary Table S1. PCR amplification cycle efficiencies for all primer pairs were greater than 95%.
Quantitative expression analysis was calculated by the comparative threshold cycle (Ct) method (Pfaffl, 2001) modified to take into account two separate reference genes as described in Hellemans et al. (2007). In this study, two transcripts that are commonly used as references for the hypoxia-induced gene expression, UBQ10 (Choi and Roberts, 2007; Giuntoli et al., 2014; Schmidt et al., 2018) and ACTIN2 (Loreti et al., 2020) were used. The stability of both reference genes was assessed using GeNorm software; both showing the M value (0.354) lower than the threshold of 1.5, suggesting their stable expression and suitability for the tested samples (Vandesompele et al., 2002). The relative expression of genes was calculated by normalizing the data to the geometric mean of relative quantity of the reference genes, as described here.
where ΔCt = Ctcalibrator – Ctsample; GOI is the gene of interest; and Geomean refers to the geometric mean of 2ΔCt of UBQ10 and ACTIN2. The specific calibrators are described in the figure legends.
Histochemical and microscopy techniques
GUS staining and clearing of A. thaliana lines with the NIP2;1pro:GUS reporter transgene was done by the protocol of (Choi and Roberts 2007), and stained tissues were imaged with a LEICA MZ16FA microscope (Leica Microsystems, Wetzlar, Germany). For analysis of NIP2;1 promoter activity in root cross-sections, GUS-stained roots were dehydrated in ethanol and embedded in Technovit 7100 resin by the manufacturer’s (Kulzer GmbH, Hanau, Germany) protocol. Cross-sections (2.5-µm thickness) were generated from the mature differentiated region of the primary root with a Reichert OMV3 microtome equipped with a glass knife, and were mounted in 50% (w/v) glycerol. Cross-sections were imaged with a Nikon ECLIPSE E600 microscope equipped with MicroPublisher version 3.3 and QCapture version 2.60 software (QImaging Corporation, Surrey, Canada).
Epifluorescence imaging of NIP2;1-GFP seedlings was captured with an Axiovert 200M microscope (Zeiss, Oberkochen, Germany) equipped with filters for GFP fluorescence (Zeiss; filter set 38 HE) and a digital camera (Hamamatsu Orca-ER; Hamamatsu Corp., Hamamatsu, Japan) controlled by the Openlab software (Improvision, Coventry, UK). Subcellular localization analysis under hypoxia and reoxygenation was done with a Leica SP8 white laser confocal microscope system at the Advanced Microscopy and Imaging Center at The University of Tennessee, Knoxville. To stain the plasma membrane, hypoxia-treated seedlings were removed from the plate and were incubated in 4 µM FM4-64 (Invitrogen) under hypoxic conditions in darkness for 10 min. For reoxygenation, seedlings were returned to aerobic conditions under light for 1 h before staining and visualization. The 488-nm excitation filter was used, and the emission filter for detection was set to 495–550 for GFP and 580–650 nm for FM4-64. Confocal micrographs were captured with the Leica LASX software and uncompressed images were exported and analyzed in ImageJ version 1.53a (Schneider et al., 2012), to adjust the brightness and contrast of images, and to generate merged images.
Fluorescence intensities were calculated and co-localization analysis of FM4-64 and NIP2;1-GFP was performed by analysis of confocal images by using the ImageJ software (NIH, http://imagej.nih.gov/ij/). The determination of the polarity index of NIP2;1 expression was generated from the FM4-64 normalized image data from 10 cells from two NIP2;1-GFP complementation lines by using the approach described by Wang et al. (2017) for Arabidopsis NIP5;1. The polarity index is defined as the ratio of NIP2;1-GFP to FM4-64 on the stele vs. epidermal side of the cell in the traverse direction, and the root tip vs. the base sides of the cell in the longitudinal direction (Wakuta et al., 2015; Wang et al., 2017). A polarity index of 1 indicates non-polar localization.
Immunochemical techniques
Anti-NIP2;1 antisera were produced against a synthetic peptide (GenScript, Piscataway, NJ, USA) corresponding to the C-terminal sequence of NIP2;1 (CHKMLPSIQNAEPEFSKTGSSHKRV), following the immunization protocol of (Guenther et al., 2003), with the exception that Titermax-Gold was substituted for Freund’s adjuvants. Antibodies were affinity purified on peptide resins as described in (Guenther et al., 2003).
For analysis of NIP2;1 protein in WT and nip2;1 mutant seedlings after hypoxia treatment, Arabidopsis root tissues from seedlings treated with 6 h hypoxia or normoxic controls were extracted, and a membrane microsomal fraction was prepared as described by (Ishikawa et al., 2005). Protein concentrations were determined by using the BCA assay (Pierce Biochemical, Waltham, MA, USA). Thesodium dodecyl sulfate–poly acrylamide gel electrophoresis (PAGE) and Western blot analyses were performed using 10 μg protein from membrane microsomal fractions as previously described (Guenther et al., 2003). For the analysis of NIP2;1-GFP expression in complementation lines, hypoxia, and reoxygenation treatments were conducted as described above, and samples were directly extracted into SDS–PAGE sample buffer (Laemmli, 1970) for Western blot analysis. Rabbit anti-GFP polyclonal antibodies (Abcam, Cambridge, UK) were used for the detection of NIP2;1-GFP.
Media acidification and L-lactate measurements
Media acidification assays with the pH-sensitive indicator bromocresol purple were done by the method of (Silva et al., 2018). Seven-day-old WT and nip2;1 seedlings (20 seedlings per treatment) were transferred from MS media to 1.5% (w/v) agarose plates containing 60 mg L−1 bromocresol purple (Acros Organics, Fair Lawn, NJ, USA) and 1 mM CaSO4 in sterile distilled water, with the pH adjusted to 5.7 using KOH. Seedlings were subjected to argon-induced hypoxia or air (normoxic controls) as described above, and the change in pH indicator color was assessed throughout hypoxia treatment. Images were captured with a DSLR camera (Canon Rebel XS; Canon, Tokyo, Japan).
To determine L-lactic acid/lactate concentration within Arabidopsis roots, vertically grown seedlings (63–72 seedlings per treatment replicate) were submerged in MS media and hypoxia treatment was carried out over a 12 h time course by flushing continuously with nitrogen gas. At each time point, samples were removed from treatment, roots were dissected, and were snap frozen in liquid nitrogen. The frozen tissue was ground in a mortar with a pestle and was extracted in two volumes of 1N perchloric acid, and was neutralized with potassium carbonate on ice prior to assay by the LDH method of (Bergmeyer and Bernt, 1974).Reduced nicotinamide adenine dinucleotide production was assayed by the change in absorbance at 340 nm, with background corrected from duplicate samples treated identically except for no added LDH enzyme.
To quantify the rate of L-lactic acid/lactate efflux from roots to the external media, a modified assay with increased sensitivity was used that utilizes bacterial lactate oxidase (Cell Biolabs, San Diego, CA, USA). Seven-day-old seedlings (60–75 per treatment replicate) were weighed and transferred to each well of a six-well plate with roots submerged in 2 mL of MS liquid media. Argon gas hypoxia was administered over an 8-h time course, the lactate content of media aliquots was assayed hourly, and the rate of root lactate release was standardized to seedling fresh weight.
Computational methods
All statistical analyses were performed in the GraphPad Prism version 8.0.1 with the built-in analysis software. Specific statistical methods are noted in each figure legend. Box and whisker plots were generated by using the “Min to Max show all points” option in Prism with each plot containing a bar indicating the median value and the box limits showing the first and third quartiles. Phylogenetic analyses of NIP2;1 and the identification of orthologs and paralogs from 40 different plant lineages of the Embryophyta was performed by using the PhyloGenes version 3.0 resource available at http://www.phylogenes.org/ (Zhang et al., 2020).
Gene accession information
Accession numbers of the genes used in this study are AT2g34390 (NODULIN26-LIKE INTRINSIC PROTEIN 2;1), AT4g33070 (PYRUVATE DECARBOXYLASE 1), AT4g18360 (GOX3), AT1g17290 (ALANINE AMINOTRANSFERASE 1), AT1g77120 (ALCOHOL DEHYDROGENASE 11), AT3g18780 (ACTIN 2), AT4g05320 (UBIQUITIN 10), and AT4g17260 (LDH1).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cellular localization of AtNIP2;1 promoter activity in the roots under normoxic growth conditions.
Supplemental Figure S2. Complementation of nip2;1 T-DNA mutant with a NIP2;1pro::NIP2;1-GFP construct.
Supplemental Figure S3. Subcellular localization of NIP2;1-GFP in the roots of hypoxia challenged 7-d-old NIP2;1-GFP complementation lines.
Supplemental Figure S4. Comparison of surface and internal localization of NIP2;1-GFP during hypoxia and recovery.
Supplemental Figure S5. Comparison of hypoxia-induced expression of NIP2;1 and lactate metabolizing enzyme transcripts LDH and GOX3.
Supplemental Figure S6. Phylogenetic analysis of Arabidopsis NIP2;1.
Supplemental Table S1. Oligonucleotide primers.
Supplementary Material
Acknowledgments
We appreciate the assistance and valuable contributions of the following undergraduate students of the Biochemistry & Cellular and Molecular Biology Department at the University of Tennessee, Knoxville: Samantha McIntire for helping in phenotyping experiment, and Shivam Ishanpara for helping us in growing and genotyping transgenic plants and preparing media for the experiments; and especially Clayton Nunn who participated in initial localization experiments. We are thankful to Drs Tessa Burch-Smith, Andreas Nebenfuehr, and John Dunlap for assistance and advice on microscopic analyses. We also thank Amanda J.V. Roberts for technical assistance with figures.
Contributor Information
Zachary G Beamer, Department of Biochemistry and Cellular, and Molecular Biology, the University of Tennessee, Knoxville, Tennessee 37996, USA.
Pratyush Routray, Department of Biochemistry and Cellular, and Molecular Biology, the University of Tennessee, Knoxville, Tennessee 37996, USA.
Won-Gyu Choi, Department of Biochemistry and Molecular Biology, The University of Nevada, Reno, Nevada 89557, USA.
Margaret K Spangler, Department of Biochemistry and Cellular, and Molecular Biology, the University of Tennessee, Knoxville, Tennessee 37996, USA.
Ansul Lokdarshi, Department of Biochemistry and Cellular, and Molecular Biology, the University of Tennessee, Knoxville, Tennessee 37996, USA.
Daniel M Roberts, Department of Biochemistry and Cellular, and Molecular Biology, the University of Tennessee, Knoxville, Tennessee 37996, USA.
Optimal response to low oxygen stress requires the Nodulin 26-like Intrinsic Protein 2;1, which releases lactic acid/lactate from roots during hypoxia stress.
Z.B. organized and executed the majority of the experiments, and together with P.R. wrote the manuscript; P.R., co-corresponding senior author who directed the design and supervision of the experiments, generated the complementation lines, conducted experiments on phenotype, and was involved in analyzing the data and organizing the manuscript; W.G.C. initiated the early work, generated and characterized the promoter-GUS lines, and performed early analyses of the nip2;1 T-DNA line; M.S., an undergraduate research student who assisted in phenotype survival analyses; A.L. assisted in localization experiments as well as the QYmax measurements; D.M.R., supervisor of the project, obtained funding for the work, and was involved in the design of experiments, analysis, and interpretation of the data, and co-edited the manuscript with P.R. D.M.R. agrees to serve as the contact, communication, and source for materials emerging from this project.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Daniel M. Roberts (drobert2@utk.edu).
Funding
This work was supported by National Science Foundation Grant MCB-1121465 and support from the Charles Postelle Distinguished Professorship fund to D.M.R.
Conflict of interest statement. We declare no conflicts of interest.
References
- Armstrong W, Beckett PM, Colmer TD, Setter TL, Greenway H (2019) Tolerance of roots to low oxygen: ‘Anoxic’ cores, the phytoglobin-nitric oxide cycle, and energy or oxygen sensing. J Plant Physiol 239:92–108 [DOI] [PubMed] [Google Scholar]
- Armstrong W, Strange ME, Cringle S, Beckett PM (1994) Microelectrode and modeling study of oxygen distribution in roots. Ann Bot 74:287–299 [Google Scholar]
- Bergmeyer HU, Bernt E (1974) Colorimetric assay with l-lactate, NAD phenazine methosulphate and INT. InBergmeyer HU, ed, Methods of Enzymatic Analysis, Ed 2nd. Academic Press, Cambridge, MA, pp 579–582 [Google Scholar]
- Bienert GP, Desguin B, Chaumont F, Hols P (2013) Channel-mediated lactic acid transport: a novel function for aquaglyceroporins in bacteria. Biochem J 454:559–570 [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Prak S, Boudet J, Postaire O, Luu DT, Tournaire-Roux C, Santoni V, Maurel C (2008) The response of Arabidopsis root water transport to a challenging environment implicates reactive oxygen species- and phosphorylation-dependent internalization of aquaporins. Plant Signal Behav 3:1096–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56:743–755 [DOI] [PubMed] [Google Scholar]
- Chevalier AS, Chaumont F (2015) Trafficking of plant plasma membrane aquaporins: multiple regulation levels and complex sorting signals. Plant Cell Physiol 56:819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WG, Roberts DM (2007) Arabidopsis NIP2;1, a major intrinsic protein transporter of lactic acid induced by anoxic stress. J Biol Chem 282:24209–24218 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 [DOI] [PubMed] [Google Scholar]
- Collard F, Baldin F, Gerin I, Bolsee J, Noel G, Graff J, Veiga-da-Cunha M, Stroobant V, Vertommen D, Houddane A, et al. (2016) A conserved phosphatase destroys toxic glycolytic side products in mammals and yeast. Nat Chem Biol 12:601–607 [DOI] [PubMed] [Google Scholar]
- Counillon L, Bouret Y, Marchiq I, Pouyssegur J (2016) Na+/H+ antiporter (NHE1) and lactate/H+ symporters (MCTs) in pH homeostasis and cancer metabolism. Biochim Biophys Acta Mol Cell Res 1863:2465–2480 [DOI] [PubMed] [Google Scholar]
- Davies DD, Grego S, Kenworthy P (1974) The control of the production of lactate and ethanol by higher plants. Planta 118:297–310 [DOI] [PubMed] [Google Scholar]
- Diehn TA, Bienert MD, Pommerrenig B, Liu Z, Spitzer C, Bernhardt N, Fuge J, Bieber A, Richet N, Chaumont F, et al. (2019) Boron demanding tissues of Brassica napus express specific sets of functional Nodulin26-like Intrinsic Proteins and BOR1 transporters. Plant J 100: 68–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Dolferus R, Jacobs M, Peacock WJ, Dennis ES (1994) Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol 105: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R, Wolansky M, Carroll R, Miyashita Y, Ismond K, Good A (2008) Functional analysis of lactate dehydrogenase during hypoxic stress in Arabidopsis. Funct Plant Biol 35:131–140 [DOI] [PubMed] [Google Scholar]
- Drew MC (1997) OXYGEN DEFICIENCY AND ROOT METABOLISM: injury and acclimation under hypoxia and anoxia. Annu Rev Plant Physiol Plant Mol Biol 48:223–250 [DOI] [PubMed] [Google Scholar]
- Ellis MH, Dennis ES, Peacock WJ (1999) Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol 119:57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist MK, Schmitz J, Gertzmann A, Florian A, Jaspert N, Arif M, Balazadeh S, Mueller-Roeber B, Fernie AR, Maurino VG (2015) GLYCOLATE OXIDASE3, a glycolate oxidase homolog of yeast l-lactate cytochrome c oxidoreductase, supports l-lactate oxidation in roots of Arabidopsis. Plant Physiol 169:1042–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghiri Z, Camargo SM, Huggel K, Forster IC, Ndegwa D, Verrey F, Skelly PJ (2010) The tegument of the human parasitic worm Schistosoma mansoni as an excretory organ: the surface aquaporin SmAQP is a lactate transporter. PLoS One 5:e10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle HH (2005) pH regulation in anoxic plants. Ann Bot 96:519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J, Greenway H (2003) Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism (vol 30, pg 1, 1993). Funct Plant Biol 30: U353–U356 [DOI] [PubMed] [Google Scholar]
- Giuntoli B, Lee SC, Licausi F, Kosmacz M, Oosumi T, van Dongen JT, Bailey-Serres J, Perata P (2014) A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol 12:e1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway H, Gibbs J (2003) Mechanisms of anoxia tolerance in plants. II. Energy requirements for maintenance and energy distribution to essential processes. Funct Plant Biol 30:999–1036 [DOI] [PubMed] [Google Scholar]
- Guenther JF, Chanmanivone N, Galetovic MP, Wallace IS, Cobb JA, Roberts DM (2003) Phosphorylation of soybean nodulin 26 on serine 262 enhances water permeability and is regulated developmentally and by osmotic signals. Plant Cell 15:981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F, Suga S, Uemura T, Sato MH, Maeshima M (2005) Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett 579:5814–5820 [DOI] [PubMed] [Google Scholar]
- Ismond KP, Dolferus R, De Pauw M, Dennis ES, Good AG (2003) Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol 132:1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjovall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol 126:1358–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T, Fujiwara T (2009) Arabidopsis NIP1;1 transports antimonite and determines antimonite sensitivity. Plant Cell Physiol 50:1977–1981 [DOI] [PubMed] [Google Scholar]
- Kamiya T, Tanaka M, Mitani N, Ma JF, Maeshima M, Fujiwara T (2009) NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. J Biol Chem 284:2114–2120 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396 [Google Scholar]
- Kursteiner O, Dupuis I, Kuhlemeier C (2003) The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol 132:968–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ, Hayward L, Langridge-Smith P, Gilbert N, Ramsahoye BH (2012) Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res 40:4794–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LA, Bailey-Serres J (2011) Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 190:457–471 [DOI] [PubMed] [Google Scholar]
- Li X, Wang X, Yang Y, Li R, He Q, Fang X, Luu DT, Maurel C, Lin J (2011) Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23:3780–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62:302–315 [DOI] [PubMed] [Google Scholar]
- Lokdarshi A, Conner WC, McClintock C, Li T, Roberts DM (2016) Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol 170:1046–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Betti F, Ladera-Carmona MJ, Fontana F, Novi G, Valeri MC, Perata P (2020) ARGONAUTE1 and ARGONAUTE4 regulate gene expression and hypoxia tolerance. Plant Physiol 182: 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateluna P, Salvatierra A, Solis S, Nunez G, Pimentel P (2018) Involvement of aquaporin NIP1;1 in the contrasting tolerance response to root hypoxia in Prunus rootstocks. J Plant Physiol 228:19–28 [DOI] [PubMed] [Google Scholar]
- Maurino VG, Engqvist MK (2015) 2-Hydroxy acids in plant metabolism. Arabidopsis Book 13:e0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Watanabe S, Nakagawa T, Maeshima M (2006) Aquaporin NIP2;1 is mainly localized to the ER membrane and shows root-specific accumulation in Arabidopsis thaliana. Plant Cell Physiol 47:1420–1426 [DOI] [PubMed] [Google Scholar]
- Murchie EH, Lawson T (2013) Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications. J Exp Bot 64:3983–3998 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Barding GA Jr., Kaiser KA, Larive CK, Bailey-Serres J (2014) Characterization of distinct root and shoot responses to low-oxygen stress in Arabidopsis with a focus on primary C- and N-metabolism. Plant Cell Environ 37:2366–2380 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang HJ, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J (2010) Cross-Kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152:1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106:18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyikiza D, Piya S, Routray P, Miao L, Kim WS, Burch-Smith T, Gill T, Sams C, Arelli PR, Pantalone V, et al. (2020) Interactions of gene expression, alternative splicing, and DNA methylation in determining nodule identity. Plant J 103: 1744–1766 [DOI] [PubMed] [Google Scholar]
- Noda Y, Sasaki S (2006) Regulation of aquaporin-2 trafficking and its binding protein complex. Biochim Biophys Acta 1758:1117–1125 [DOI] [PubMed] [Google Scholar]
- Olive MR, Walker JC, Singh K, Dennis ES, Peacock WJ (1990) Functional properties of the anaerobic responsive element of the maize Adh1 gene. Plant Mol Biol 15: 593–604 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerrenig B, Diehn TA, Bernhardt N, Bienert MD, Mitani-Ueno N, Fuge J, Bieber A, Spitzer C, Brautigam A, Ma JF, et al. (2020) Functional evolution of nodulin 26-like intrinsic proteins: from bacterial arsenic detoxification to plant nutrient transport. New Phytol 225:1383–1396 [DOI] [PubMed] [Google Scholar]
- Prak S, Hem S, Boudet J, Viennois G, Sommerer N, Rossignol M, Maurel C, Santoni V (2008) Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol Cell Proteomics 7:1019–1030 [DOI] [PubMed] [Google Scholar]
- Ricoult C, Cliquet J-B, Limami AM (2005) Stimulation of alanine amino transferase (AlaAT) gene expression and alanine accumulation in embryo axis of the model legume Medicago truncatula contribute to anoxia stress tolerance. Physiol Plantarum 123:30–39 [Google Scholar]
- Rivoal J, Hanson AD (1993) Evidence for a large and sustained glycolytic flux to lactate in anoxic roots of some members of the halophytic genus limonium. Plant Physiol 101:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DM, Routray P (2017) The nodulin26 intrinsic protein subfamily. InChaumont F, Tyerman SD, eds, Plant Aquaporins From Transport to Signaling. Springer International Publishing, New York, NY, pp 267–296 [Google Scholar]
- Roberts JK, Callis J, Jardetzky O, Walbot V, Freeling M (1984) Cytoplasmic acidosis as a determinant of flooding intolerance in plants. Proc Natl Acad Sci U S A 81:6029–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni V (2017) Plant aquaporin postranslational regulation. InChaumont F, Tyerman SD, eds, Plant Aquaporins From Transport to Signaling. Springer International Publishing, New York, NY, pp 83–105 [Google Scholar]
- Sato T, Harada T, Ishizawa K (2002) Stimulation of glycolysis in anaerobic elongation of pondweed (Potamogeton distinctus) turions. J Exp Bot 53:1847–1856 [DOI] [PubMed] [Google Scholar]
- Schmidt RR, Fulda M, Paul MV, Anders M, Plum F, Weits DA, Kosmacz M, Larson TR, Graham IA, Beemster GTS, et al. (2018) Low-oxygen response is triggered by an ATP-dependent shift in oleoyl-CoA in Arabidopsis. Proc Natl Acad Sci U S A 115: E12101–E12110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AL, Dressano K, Ceciliato PHO, Guerrero-Abad JC, Moura DS (2018) Evaluation of root pH change through gel containing pH-sensitive indicator bromocresol purple. Bio-Protocol 8:e2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson R, Bailey-Serres J (2014) Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc Natl Acad Sci U S A 111:2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SR, Li H, Chen JH, Qian Q (2017) Lactic acid: no longer an inert and end-product of glycolysis. Physiology 32:453–463 [DOI] [PubMed] [Google Scholar]
- Takano J, Miwa K, Fujiwara T (2008) Boron transport mechanisms: collaboration of channels and transporters. Trends Plant Sci 13:451–457 [DOI] [PubMed] [Google Scholar]
- Takano J, Yoshinari A, Luu D-T (2017) Plant aquaporin trafficking. InChaumont F, Tyerman SD, eds, Plant Aquaporins From Transport to Signaling. Springer International Publishing, New York, NY, pp 47–81 [Google Scholar]
- Tsai KJ, Chou SJ, Shih MC (2014) Ethylene plays an essential role in the recovery of Arabidopsis during post-anaerobiosis reoxygenation. Plant Cell Environ 37:2391–2405 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialaret J, Di Pietro M, Hem S, Maurel C, Rossignol M, Santoni V (2014) Phosphorylation dynamics of membrane proteins from Arabidopsis roots submitted to salt stress. Proteomics 14:1058–1070 [DOI] [PubMed] [Google Scholar]
- Voesenek LA, Bailey-Serres J (2013) Flooding tolerance: O2 sensing and survival strategies. Curr Opin Plant Biol 16:647–653 [DOI] [PubMed] [Google Scholar]
- Voesenek LA, Bailey-Serres J (2015) Flood adaptive traits and processes: an overview. New Phytol 206:57–73 [DOI] [PubMed] [Google Scholar]
- Wakuta S, Mineta K, Amano T, Toyoda A, Fujiwara T, Naito S, Takano J (2015) Evolutionary divergence of plant borate exporters and critical amino acid residues for the polar localization and boron-dependent vacuolar sorting of AtBOR1. Plant Cell Physiol 56: 852–862 [DOI] [PubMed] [Google Scholar]
- Wallace IS, Choi WG, Roberts DM (2006) The structure, function and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochim Biophys Acta 1758:1165–1175 [DOI] [PubMed] [Google Scholar]
- Wallace IS, Roberts DM (2004) Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Physiol 135: 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yoshinari A, Shimada T, Hara-Nishimura I, Mitani-Ueno N, Feng Ma J, Naito S, Takano J (2017) Polar localization of the NIP5;1 boric acid channel is maintained by endocytosis and facilitates boron transport in Arabidopsis roots. Plant Cell 29:824–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fan C, Zhang X, Zhu J, Fu YF (2013) BioVector, a flexible system for gene specific-expression in plants. BMC Plant Biol 13:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CD, Crombie B, Stacey G, Roberts DM (1991) Calcium-dependent phosphorylation of symbiosome membrane proteins from nitrogen-fixing soybean nodules: evidence for phosphorylation of nodulin-26. Plant Physiol 95:222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ (2007) The WiscDsLox T-DNA collection: an arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120:157–165 [DOI] [PubMed] [Google Scholar]
- Xia JH, Roberts J (1994) Improved cytoplasmic pH regulation, increased lactate efflux, and reduced cytoplasmic lactate levels are biochemical traits expressed in root tips of whole maize seedlings acclimated to a low-oxygen environment. Plant Physiol 105:651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia JH, Saglio PH (1992) Lactic Acid efflux as a mechanism of hypoxic acclimation of maize root tips to anoxia. Plant Physiol 100:40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Dai W, Yan H, Li S, Shen H, Chen Y, Xu H, Sun Y, He Z, Ma M (2015) Arabidopsis NIP3;1 plays an important role in arsenic uptake and root-to-shoot translocation under arsenite stress conditions. Mol Plant 8:722–733 [DOI] [PubMed] [Google Scholar]
- Yeung E, Bailey-Serres J, Sasidharan R (2019) After the deluge: plant revival post-flooding. Trends Plant Sci 24:443–454 [DOI] [PubMed] [Google Scholar]
- Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al. (2019) Metabolic regulation of gene expression by histone lactylation. Nature 574: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Berardini TZ, Ebert D, Li Q, Mi H, Muruganujan A, Prithvi T, Reiser L, Sawant S, Thomas PD, et al. (2020) PhyloGenes: an online phylogenetics and functional genomics resource for plant gene function inference. Plant Direct 4:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.