One sentence summary: Editing of brassinosteroid signaling family genes is a feasible approach to diversify plant agronomic traits for screening of desired plants to meet various requirements.
Abstract
Brassinosteroids (BRs) regulate various agronomic traits such as plant height, leaf angle, and grain size in rice (Oryza sativa L.); thus, BR signaling components are promising targets for molecular rational design. However, genetic materials for BR-signaling genes or family members remain limited in rice. Here, by genome editing using clustered regularly interspaced short palindromic repeats (CRSPR)/Cas9 tools, we generated a panel of single, double, triple, or quadruple mutants within three BR signaling gene families, including GSK3/SHAGGY-LIKE KINASE1 (GSK1)–GSK4, BRASSINAZOLE-RESISTANT1 (OsBZR1)–OsBZR4, and protein phosphatases with kelch-like (PPKL)1–PPKL3, under the same background (Zhonghua11, japonica). The high-order mutants were produced by either simultaneously targeting multiple sites on different genes of one family (GSKs and PPKLs) or targeting the overlapping sequences of family members (OsBZRs). The mutants exhibited a diversity of plant height, leaf angle, and grain morphology. Comparison analysis of the phenotypes together with BR sensitivity tests suggested the existence of functional redundancy, differentiation, or dominancy among the members within each family. In addition, we generated a set of transgenic plants overexpressing GSK2, OsBZR1/2, and PPKL2, respectively, in wild-type or activated forms with fusion of different tags, and also verified the protein response to BR application. Collectively, these plants greatly enriched the diversity of important agronomic traits in rice. We propose that editing of BR-related family genes could be a feasible approach for screening of desired plants to meet different requirements. Release of these materials as well as the related information also provides valuable resources for further BR research and utilization.
Introduction
Brassinosteroids (BRs) play diverse roles in various aspects of plant growth and development. Rapid progress has been made on understanding BR synthesis and signaling pathways in recent years, particularly in Arabidopsis (Arabidopsis thaliana). BRs are perceived by BR-INSENSITIVE1 (BRI1) receptor kinase and the co-receptor BRI1-ASSOCIATED RECEPTOR KINASE1 on plasma membrane, and the signal is transduced to the central transcriptional factors BRASSINAZOLE-RESISTANT1 (BZR1) and bri1-EMS-SUPPRESSOR1 (BES1)/BZR2 through a series of phosphorylating or dephosphorylating events involving BR-SIGNALING KINASE1 (BSK1), BRI1-SUPPRESSOR1 (BSU1), and BRASSINOSTEORID INSENSITIVE2 (BIN2). Among them, BIN2, belonging to GSK3/SHAGGY-like kinases, plays central negative roles in controlling BR signaling. Increased BRs induce BIN2 degradation to de-repress BZR1 and BES1, which activate BR response by targeting and regulating downstream BR-responsive genes (Nolan et al., 2020). Most counterparts of these BR signaling components in rice (Oryza sativa) have been identified, suggesting that BR signaling pathway is conserved in different species (Tong and Chu, 2018). Each component actually represents a family of genes that are thought to have redundant functions. However, due to lack of related mutants, direct evidence supporting this speculation is still absent in rice. In addition, within each family, whether the members have differential or dominant functions remain largely unclear.
GSK3-like kinase family contains 10 members in Arabidopsis and 9 in rice, which are classified into 4 clades (Yoo et al., 2006). Clade II, containing three members in Arabidopsis (BIN2 and BIN2-LIKE proteins, BIL1 and BIL2) and four in rice (GSK1–GSK4, GSKs), plays a major role in regulating BR signaling. Genetic screenings have identified a number of gain-of-function alleles of bin2 (bin2-D; Li et al., 2001; Choe et al., 2002; Li and Nam, 2002; Perez-Perez et al., 2002). Further investigations have demonstrated that BIN2 and BILs function redundantly in regulating BR signaling, but BIN2 plays a dominant role among them (Yan et al., 2009). In rice, plants overexpressing the activated forms of GSK2 (mimic of bin2-D mutation) show typical BR-defective phenotypes (Tong et al., 2012). However, overexpression of wild-type GSK2 fails to produce obvious phenotype. Simultaneously knockdown of GSK1–GSK4 by RNA interference (RNAi) enhances BR signaling, suggesting that the four members play a redundant role in rice (Tong et al., 2012).
BZR1-like protein family contains at least six members in Arabidopsis and four in rice (OsBZR1–OsBZR4, OsBZRs; Bai et al., 2007). The importance of these members in mediating BR responses in Arabidopsis has been demonstrated recently by construction of high-order knockout mutants (Chen et al., 2019). In rice, knockdown of OsBZR1 by RNAi leads to erect leaves (Bai et al., 2007). Similar to its orthologs in Arabidopsis at molecular level (He et al., 2002), OsBZR1 can be phosphorylated by GSK2 (Tong et al., 2012), and shuttles between cytoplasm and nucleus in response to BRs (Bai et al., 2007).
BSU1-like genes encode a family of protein phosphatases with kelck-like repeat domains (PPKLs). PPKL family contains four members in Arabidopsis (BSU1 and BSU1-LIKE proteins, BSL1–BSL3) and three in rice (PPKL1–PPKL3; Maselli et al., 2014). In Arabidopsis, the family members can interact with each other, forming oligomers to function (Kim et al., 2016). However, it has also been suggested that BSU1 and BSL1 play a dominant role in BR signaling, whereas BSL2 and BSL3 could have BR-independent roles (Maselli et al., 2014). It should be noted that BSU1 is a diverged member only found in the Brassicaceae. Given the lack of BSU1 counterpart in rice, it is highly intriguing to know how PPKLs regulate BR signaling in rice. Surprisingly, a recent study suggests that PPKL1 negatively regulates BR signaling (Gao et al., 2019).
In rice, a typical BR-deficient mutant morphologically shows reduced plant height, decreased leaf angle, and small grain size. The regulation on these important agronomic traits suggests that BRs have great potentials in agriculture improvement, as has been demonstrated by a number of studies (Sakamoto et al., 2006; Wu et al., 2008; Zhang et al., 2013). However, it should be noted that different traits could be negatively correlated each other, which makes BR application not very easy. For example, BRs enhance grain yield but enlarge leaf angles, conferring loose plant architectures that are usually not preferable to breeders and farmers. To deal with this problem, we have proposed that utilization of genes with functional specificities for molecular design could avoid those negative effects (Tong and Chu, 2018). In support of this idea, GRAIN LENGTH2/GRAIN SIZE2, a downstream gene of BR response specifically enhances grain size but does not affect plant architecture, can significantly improve rice yield (Che et al., 2015). Recently, we have showed that knockout of SERK2 encoding a somatic embryogenesis receptor kinase, but not its homologous gene OsBAK1, results in compact plant architecture, without negative effect on grain size (Dong et al., 2020), suggesting that BR signaling gene families could be valuable resources for screening specific members for crop improvement.
Protein or gene family usually contains members having both functional redundancy and differentiation. This feature enables the production of mutants having various phenotypes with different combinations, which could be used for screening desired traits to meet different requirements. Given the significant roles of BRs in regulating many important agronomic traits, the BR signaling gene families could be suitable candidates for implementing this strategy. CRISPR/Cas9 genome editing technology provides a feasible way for rapidly producing knockout mutants. Importantly, most available CRISPR/Cas9 tools allow one vector simultaneously targeting multiple sites. By this approach, here, we produced a number of GSK-, BZR-, and PPKL-related mutants with different combinations. We also generated a set of transgenic plants overexpressing several representative members. We described the details of the phenotypes, genotypes, and molecular features about these plants. The materials and information presented here will certainly provide valuable resources for both BR study and utilization.
Results
Editing of GSK1–GSK4 by simultaneously targeting multiple genomic sites
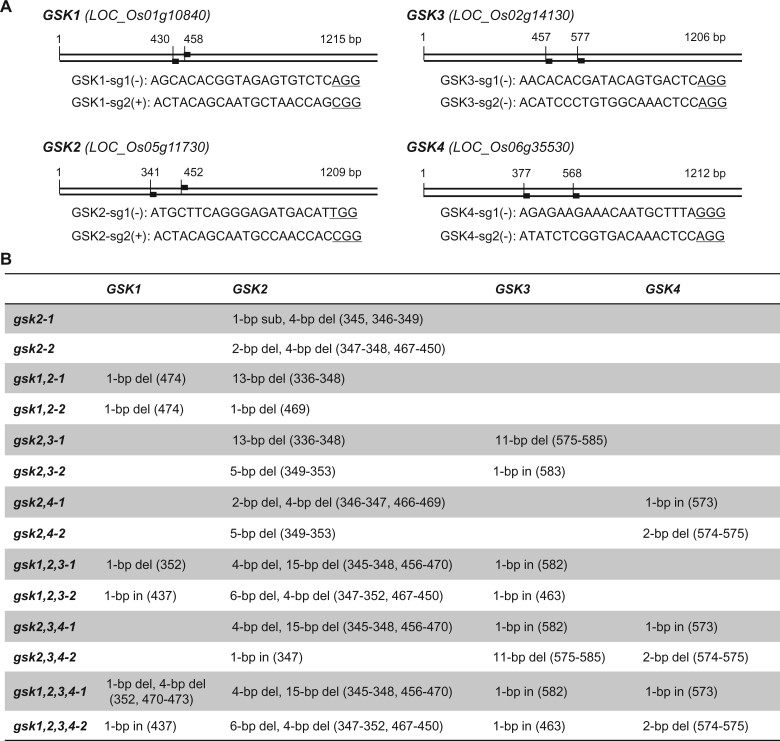
To efficiently generate mutants of GSK1–GSK4, we simultaneously edited the four genes by utilizing CRISPR/Cas9 system. In order to enhance the editing efficiency, we designed two independent targeting sites for each gene (Figure 1A). All the sites were distributed in the front region of the genes corresponding to the center of protein kinase domain. In the end, one vector carrying eight guiding ribonucleic acids (RNAs) (sgRNAs), with two for each gene (sg1 and sg2), was introduced into Zhonghua11 (ZH11, a japonica cultivar used as wild-type) for the editing. As expected, this strategy enabled us to rapidly generate high-order mutants. Actually, we have obtained a quadruple mutant line (gsk1,2,3,4) at T0 generation, although it eventually died after transferring to the field. Besides, a number of additional mutants with various combinations of different mutations on different genes were also obtained. Our intense polymerase chain reaction (PCR) sequencing revealed that mutations have occurred at seven of eight targeting sites, except GSK4-sg1 where no editing events have ever been detected.
Figure 1.
Targeting sites and mutant information of GSK-related mutants. A, Targeting sites on the coding sequence of each gene with the full length and the starting sites were indicated. The guiding RNA sequences were listed. The protospacer-adjacent motifs (PAMs) were underlined. B, The mutation information of a set of gsk2-centered mutants. in, insertion; del, deletion; sub, substitution.
We previously have demonstrated the roles of GSK3-like kinases in BR signaling by using GSK2 as the representative member (Tong et al., 2012). With two more generations of selfing and segregation (T3), we successfully generated a set of gsk2-centered pyramiding mutant lines, including gsk2, gsk1,2, gsk2,3, gsk2,4, gsk1,2,3, gsk2,3,4, and gsk1,2,3,4 (Figure 1B, with two alleles shown for each). All these lines contained homozygous mutations, which can be stably inherited after clearance of T-DNA insertions. All the mutations caused frame-shift transcription around the targeting sites that could disrupt the kinase activity thus should be knockout alleles. Since the phenotypes among different alleles were highly consistent each other according to our observation, we selected one for each of the mutants at T5 generations for the following analysis (the first one listed in Figure 1B).
GSK-related mutants showed diverse plant height, leaf angle, and grain size
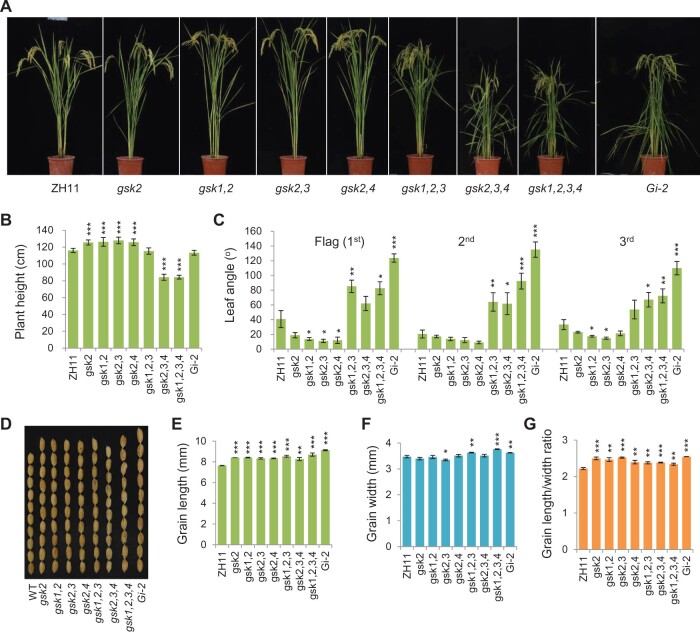
BRs predominantly regulate plant height, leaf angle, and grain size in rice. We thus evaluated these three agronomic traits, respectively, in the mutants. Compared to ZH11, all the single and double mutants (gsk2, gsk1,2, gsk2,3, gsk2,4) exhibited slightly increased plant height, to a basically similar extent (∼5%), suggesting that BR signaling has been enhanced in these plants (Figure 2, A and B). In contrast, both gsk2,3,4 and gsk1,2,3,4 showed greatly decreased plant height, also to a very similar extent (∼30%), whereas gsk1,2,3 had no change in this aspect (Figure 2, A and B). The alterations of plant height in these pyramiding mutants are consistent with our previous reports that sufficiently high BR levels would turn to inhibit plant growth (Tong et al., 2014). While most of the mutants exhibited little change of tiller number, the gsk1,2,3,4 showed slightly increased tillers (Supplemental Figure S1A), consistent with a recent report that BRs promote tillering (Fang et al., 2020). In term of leaf angle, we surprisingly found that all the single and double mutants had slightly decreased lamina inclinations on average according to statistical analyses (Figure 2C). Nevertheless, the triple and quadruple mutants had greatly enlarged leaf angles (Figure 2C), demonstrating the negative roles of GSK-family members in BR responses.
Figure 2.
Phenotypic analysis of GSK-related mutants. A, Gross morphology of the mutants, with a GSK2-RNAi line (Gi-2) shown for comparison. B, Statistical data of plant height of the mutants and Gi-2 at the harvesting stage. Bars indicate standard deviation (sd, n = 10). ***P < 0.001 generated by t test. C, Statistical data of leaf angle of the plants at the harvesting stage. Bars indicate standard error of the mean values (sem, n = 8). *P < 0.05, **P < 0.01, and ***P < 0.001 generated by t test. D, Grain morphology of the plants. E−G, Statistical data of the grain length (E), width (F), and length/width ratios (G). Bars indicate sd of three measurements. About 50 − 100 seeds were used for each measurement. *P < 0.05, **P < 0.01, and ***P < 0.001 generated by t test.
Regarding the grain size, all the mutants from single to quadruple had enhanced grain length (Figure 2, D and E). It was noted that most of the mutants had a comparably increased grain length except the quadruple mutant, which had further enlarged grains. This observation is quite different to the cases in terms of plant height and leaf angles. Apparently, different tissues had distinctive BR sensitivities. However, the grain width could either be increased or decreased compared to ZH11 (Figure 2F), implying a complicated regulation of BRs on grain width. As results, the grain length/width ratios were generally increased (Figure 2G). Taken together, these analyses demonstrated the existence of functional redundancy among GSK members, and also suggested the importance of BR balance for plant growth and development. Notably, the beneficial traits shown in the single or double mutants, including the enhanced grain size accompanied with decreased leaf angles, suggested the potential of this strategy for crop improvement.
Comparison of the quadruple mutant and the knockdown plant
Gi-2, a GSK2-RNAi line yet with GSK1-GSK4 simultaneously repressed, showed typical BR-enhanced phenotypes (Tong et al., 2012), thus was included for the comparison. Intriguingly, even the quadruple mutant gsk1,2,3,4 had far less increased leaf angle as well as grain size than Gi-2 (Figure 2, C−E). One possibility is that the RNAi has nonspecifically repressed additional GSK members that could be involved in regulating these traits, such as GSK5, which has been suggested to regulate grain size (Hu et al., 2018; Xia et al., 2018; Ying et al., 2018). On the other hand, however, gsk1,2,3,4 had much more decreased plant height than Gi-2 (Figure 2, A and B). Actually, compared to Gi-2, gsk1,2,3,4 exhibited poor growth vigor accompanied with many more additional growth defections. In opposite to the elongated dropping leaves in Gi-2, gsk1,2,3,4 had shortened leaf elongation (Figure 2A). In addition, both Gi-2 and gsk1,2,3,4 had delayed leaf senescence and seed maturation but the symptoms were much more obvious in gsk1,2,3,4. This result suggested that knockout of GSK1–GSK4 had a more severe effect on plant growth and development than knockdown of the genes.
BR sensitivity of GSK-related mutants
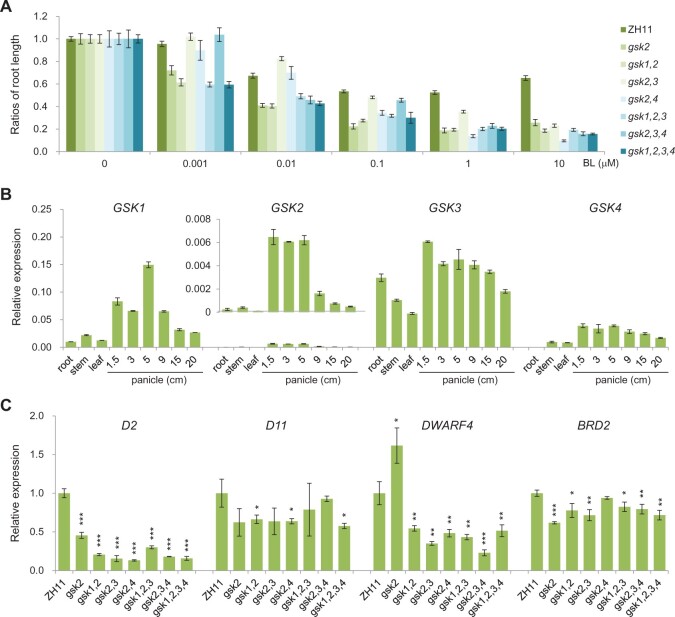
We further evaluated the BR sensitivities of all the mutants in the root, as this tissue is most feasible for quantification analysis. Without BR treatment, most of the mutants have increased root length on average compared to ZH11 (Supplemental Figure S1B). Application of brassinolide (BL), one of the active BR forms, can greatly suppress the root elongation in both ZH11 and the mutants (Figure 3A). However, at high BL concentrations (such as 1 µM and 10 µM), the inhibitory extents were much more substantial in the mutants than in ZH11 (Figure 3A). Clearly, all the mutants had enhanced BR sensitivities. This result is also supported by coleoptile elongation analysis, which showed that the mutants more or less had increased BR sensitivities and the high-order mutants tended to be more sensitive in term of the elongation ratios (Supplemental Figure S2).
Figure 3.
Evaluation of BR sensitivities in GSK-related mutants. A, BR sensitivity test in root. For each genotype, the ratios of root lengths with BL on those without BL were shown. Bars indicate sem (n = 8–30). B, Expression of GSKs in different tissues. Root, stem, and leaf were sampled at 60 d after germination. Bars indicate sd (n = 3). C, Expression of BR-synthetic genes in the shoots of 1-week-old seedlings of different plants. Bars indicate sd (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001 generated by t test.
We further performed leaf bending assay to assess the mutants’ response to BRs. 28-homobrassinolide (HBL), another active form of BRs (Takatsuto et al., 1983), was directly supplied in culture medium for the analysis. Compared to that in ZH11, HBL (1 µM)-induced leaf bending were significantly enhanced in the triple and quadruple mutants but not obvious in others when evaluating the angles of third leaves (Supplemental Figure S3A). Intriguingly, we failed to observe this difference when evaluating the response in the second leaves (Supplemental Figure S3B), indicating a rather more complicated role of the GSKs in different leaves. Combined with the mutant phenotypes, these analyses altogether demonstrated the pivotal but redundant roles of GSK1–GSK4 in negatively regulating BR signaling in rice.
Expression of GSK1–GSK4 and BR synthetic genes
Reverse transcription quantitative PCR (RT-qPCR) revealed that all the four genes were highly expressed in young panicles, relatively less in elder panicles, further less in other tissues such as leaf, root, and culm (Figure 3B). This expression pattern suggested a substantial role of the genes in determining grain size, somewhat consistent with the above phenotype analysis as grain size was more markedly affected even in single mutant. It was noted that GSK3 has relatively minor tissue specificity compared to others. In addition, GSK3 has the highest expression abundance among the four genes with an order GSK3 > GSK1 > GSK4 > GSK2 (Figure 3B). These results implied a basal role of GSK3 in controlling BR signaling among the family members.
It is well-known that enhanced BR signaling could inhibit BR synthesis in a feedback manner. We detected the expression levels of four BR synthetic genes, DWARF2 (D2), DWARF11 (D11), DWARF4, and BR-DEFICIENT DWARF2 (BRD2). Indeed, although to a quite different extent, all the genes, in most cases, had decreased expression in the mutants (Figure 3C).
Editing of OsBZR1–OsBZR4 by targeting two overlapping sequences
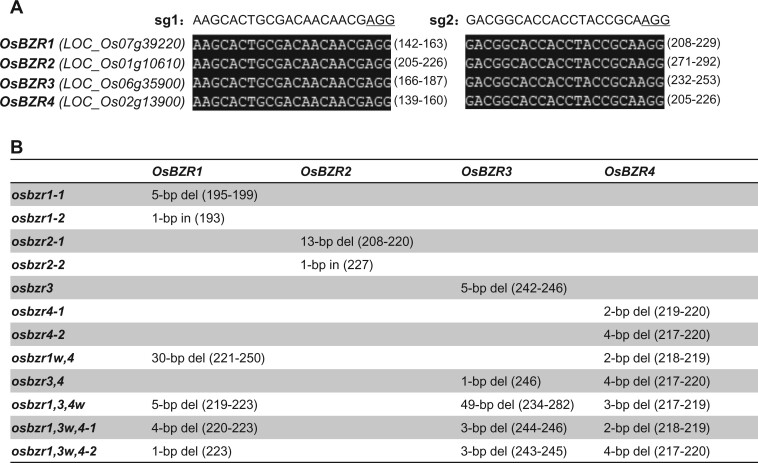
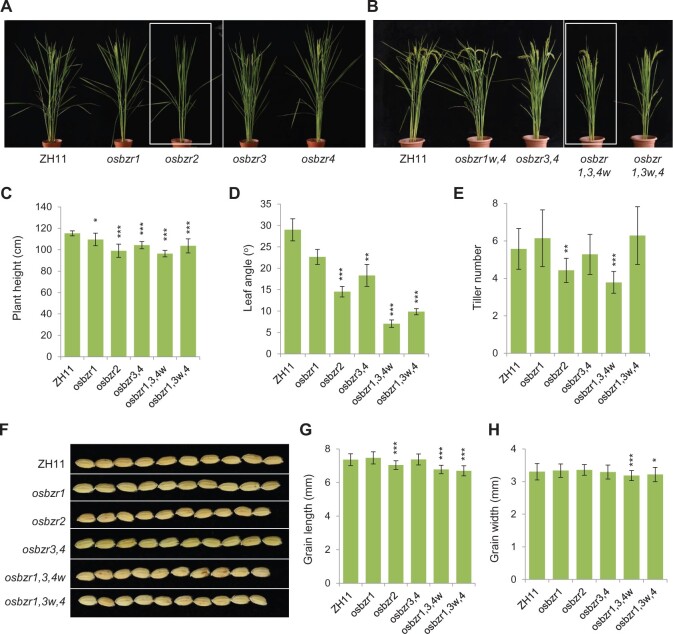
Sequence alignment of OsBZR1–OsBZR4 identified two overlapping segments that could be used for guiding the simultaneous editing of the four genes (Figure 4A). We thus constructed two editing vectors targeting the two sites, respectively, but mixed them for transgenes in order to enhance the probability of the occurrence of high-order mutants. Since the two targeting sites are close, the editing events at the two sites could be detected by single PCR sequencing. Besides, editing of only OsBZR1 has been carried out earlier by targeting a unique site (188–210 of coding sequence). As results, we obtained mutants containing various combinations of mutations (Figure 4B). Intriguingly, our efforts failed to identify any homozygous quadruple mutant. In addition, the homozygous triple knockout mutants with all genes containing frame-shift mutations have been identified but are mostly sterile, and we eventually failed to maintain the lines. However, a few triple mutants with one member containing nonframe-shift mutation have been obtained (labeled with w indicating weak allele; Figure 4B). Despite some complicated sequencing results that required further analysis, we obtained a set of stably inherited mutants with ascertained mutations, including osbzr1, osbzr2, osbzr3, osbzr4, osbzr1w,4, osbzr3,4, osbzr1,3,4w, and osbzr1,3w,4. For those having multiple alleles, the phenotypes among different alleles were consistent with each other according to our observation. Therefore, we selected one allele for each at T5 generations for the following analysis (the first one listed in Figure 4B).
Figure 4.
Targeting sites and mutant information of OsBZR-related mutants. A, Two overlapping targeting sequences on the four genes, with the positions on the coding sequences of each gene were respectively indicated. PAMs were underlined. B, The information of the mutations in different mutants.
Marked phenotype of osbzr2 single mutant and triple mutants
Among the four single mutants, osbzr2 showed distinctive phenotypes, whereas osbzr1, osbzr3, and osbzr4 had relatively small morphological change (Figure 5A). This result suggested a dominant role of OsBZR2 in the family, and also provided a possible explanation for our failure to obtain a high-order mutant containing osbzr2. Nevertheless, both osbzr1,3,4w and osbzr1,3w,4, the two triple mutants, also showed marked phenotypes, whereas both osbzr1w,4 and osbzr3,4 did not (Figure 5B). Statistical data revealed that most of the mutants had decreased plant height and leaf angle, but to a higher extent in the three mutants with more marked phenotypes (Figure 5, C and D). In addition, the tiller number of both osbzr2 and osbzr1,3,4w were also reduced, but not in osbzr1,3w,4 (Figure 5E). Interestingly, the grain length was decreased in all the three mutants, but the grain width was only decreased in the two triple mutants but not in osbzr2 (Figure 5, F−H). Taking all the traits for consideration, osbzr1,3,4w clearly showed the most severe, but still mild, phenotype among all the mutants, representing a typical BR-signaling-deficient mutant that could be suitable for BR-related studies.
Figure 5.
Phenotypic analysis of OsBZR-related mutants. A and B, Gross morphology of the mutants. Two genotypes with evident phenotype were boxed. C–E, Statistical data of the plant height, tiller number, and the flag leaf angles of the plants at the harvesting stage. Bars indicate sd for C and D, sem for E (n = 10, 14, 15, respectively). *P < 0.05, **P < 0.01, and ***P < 0.001 generated by t test. F, Grain morphology of the plants. G and H, Statistical data of the grain length (G) and grain width (H). Bars indicate sd (n = 90). *P < 0.05 and ***P < 0.001 generated by t test.
BR sensitivity of OsBZR-related mutants
To testify whether the phenotypes were related to BR signaling, we assayed the BR sensitivities of the mutants. In coleoptile elongation analysis, while no difference was observed between the single and double mutants and ZH11, osbzr1,3,4w showed obviously decreased BR sensitivity (Supplemental Figure S4A), consistent with the most obvious phenotype of the triple mutant. In root inhibition analysis, surprisingly, all the mutants appeared to have increased BR sensitivities with osbzr1,3,4w having most significant changes (Supplemental Figure S4B). This intriguing result suggested the complicated roles of OsBZRs in mediating BR regulation on root development. In lamina bending assay, the triple mutant showed most obviously decreased BR sensitivity (Supplemental Figure S4C), consistent with the coleoptile elongation test. Interestingly, osbzr1 also exhibited lower BR sensitivity than ZH11, whereas osbzr2 did not. Taken together, these analyses supported the canonical roles of OsBZRs in mediating BR signaling. On the other hand, the results implied the differential roles of the family members in BR signaling or other unknown processes.
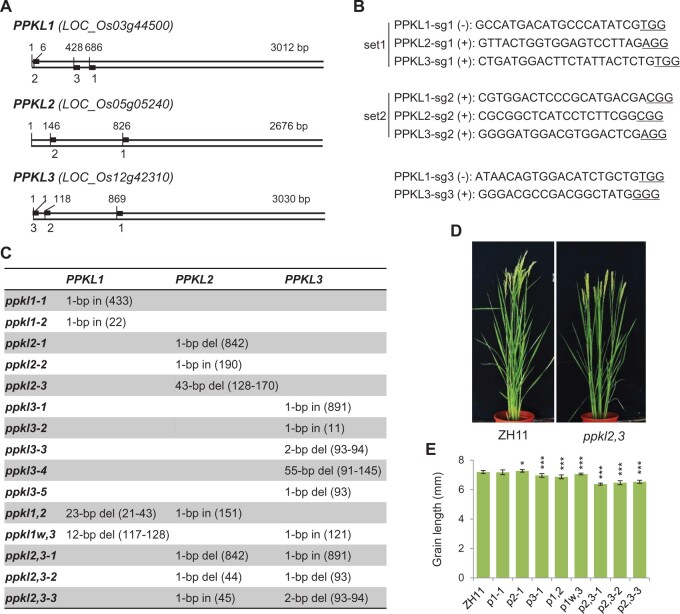
Editing of PPKL1–PPKL3 and the obtained mutants
For simultaneously editing of PPKL1–PPKL3, we constructed two editing vectors expressing two sets of guiding RNAs, with each set contains three guiding RNAs targeting the three genes, respectively (set1 and set2; Figure 6, A and B). The two vectors were then, respectively, introduced into ZH11 for the editing. We have also performed the single editing of PPKL1 and PPKL3, respectively, and the obtained mutants were also included for description (sg3; Figure 6, A and B). As results, we obtained a number of single and double mutants (Figure 6C). However, our efforts failed to identify a triple mutant. In addition, we failed to identify a double-mutant of ppkl1 and ppkl3 with both genes containing frame-shift mutations but can only obtain a weak allele ppkl1w,3 (Figure 6C). These results suggested that simultaneously knockout of PPKL1 and PPKL3 or all the three genes could somehow be lethal to rice. Unless else indicated, we selected one allele for each of the mutants at T7 generations for the following analysis (the first one listed in Figure 6C).
Figure 6.
Targeting sites, mutant information, and phenotype analysis of PPKL-related mutants. A and B, Targeting sites on the coding sequence of each gene with the full length and the starting sites were indicated. The guiding RNA sequences were listed with PAM underlined. Two sets of sgRNAs (set1 and set2) were used for simultaneously editing of the three genes. C, The mutation information. D, Gross morphology of ppkl2,3 double-mutant. E, Statistical data of grain length of selected plants. Bars indicate sd (n = 20). *P < 0.05 and ***P < 0.001 generated by t test.
Phenotype and BR sensitivity of PPKL-related mutants
In field conditions, most of the mutants showed little morphological changes, except ppkl2,3 which showed a marked phenotype with decreased plant height, leaf angle, and grain size, resembling a mild BR-deficient plant (Figure 6, D and E). Statistical data revealed that the grain size, plant height, tiller number of ppkl2,3 were all reduced, much more obvious than others which exhibited little or only slight decreases (Figure 6E;Supplemental Figure S5, A and B). Surprisingly, in agar medium, we found that the root length of the ppkl2 as well as ppkl1,2 was significantly elongated compared to others, whereas those of ppkl2,3 were not changed (Supplemental Figure S5C). However, coleoptile elongation test failed to detect altered BR sensitivities in most mutants including ppkl2,3 (Supplemental Figure S6A). Even more intriguingly, except ppkl1, all the other mutants showed greatly enhanced BR sensitivities in root, and the inhibitory effects were much more obvious in the double mutants (Supplemental Figure S6B). This result is somewhat reminiscent of those OsBZRs-related mutants, implying the complicated roles of BRs or these BR signaling components (PPKLs and OsBZRs) in root development.
We further conducted leaf bending assay to test the mutants’ responses to BR, and found that most of them, including ppkl2,3, had relatively decreased BR sensitivities compared to ZH11 (Supplemental Figure S7). However, ppkl2,3 did not exhibit a more severe change as expected. We also analyzed the expression of BR synthetic genes in the mutants, and found all the four genes tests (D2, D11, DWARF4, and BRD2) had more or less increased expression in most of the mutants (Supplemental Figure S8). Therefore, PPKLs should be indeed involved in BR response, but the detailed roles required further investigation.
Overexpression of GSK2, OsBZR1/2, and PPKL2 in various forms
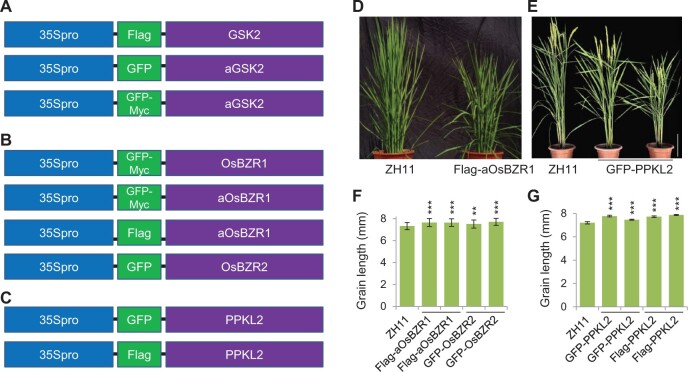
We previously have reported the overexpression plants of GSK2 in both its wild-type form fused with GFP tag and its activated form (called aGSK2 here) without tag fusion (Tong et al., 2012). To facilitate the molecular research, we further produced three additional transgenic plants overexpressing Flag-GSK2, GFP-aGSK2, and GFP-Myc-aGSK2 respectively (Figure 7A). For OsBZR1, we generated GFP-Myc-OsBZR1, GFP-Myc-aOsBZR1, and Flag-aOsBZR1 overexpression plants. Since OsBZR2 was suggested to be the dominant member in the family, we also produced a GFP-OsBZR2 overexpression line (Figure 7B). PPKL2 is most closely related to BSL1, thus could most possibly be the family member dominantly regulating BR signaling in rice. We therefore generated both GFP-PPKL2 and Flag-PPKL2 overexpression plants (Figure 7C). CaMV 35S promoter (35Spro) was adopted for all these expressions. For each construct, we usually produced up to 10 independent lines and only those showing consistent phenotypes were described below.
Figure 7.
Information of the overexpression plants. A–C, Schematic show of the expression cassette of different genes in different forms, with CAMV 35S promoter (35Spro), tag fusion (GFP, Flag, or GFP-Myc), and desired overexpressing genes indicated. D, A representative line overexpressing Flag-aOsBZR1, showing typical BR-enhanced phenotypes. E, Representative lines overexpressing GFP-PPKL2. The images were digitally extracted for comparison. F and G, Statistical data of the grain length of the indicated plants. Bars indicate sd(n = 90 in F, = 20 in G). **P < 0.01 and ***P < 0.001 generated by t test.
Phenotype and BR sensitivity of the overexpression plants
Severely dwarfism, small grains, and twisted dark-green leaves were frequently observed in the strong lines overexpressing either GFP-aGSK2 or GFP-Myc-aGSK2, similar as those reported without tag fusion (Tong et al., 2012). In addition, overexpression of Flag-aOsBZR1 led to a typical BR-enhanced phenotype, featured with enlarged leaf angles but reduced plant height (Figure 7D). Overexpression of GFP-OsBZR2 produced a weak BR-enhanced phenotype, and overexpression of GFP-PPKL2 produced a mild BR-enhanced phenotype, with reduced plant height but increased leaf angle (Figure 7E). Grain length was accordingly increased in these plants (Figure 7, F and G). Statistical analysis detected slight alteration of tiller number in GFP-PPKL2 and Flag-aOsBZR1 but not in others (Supplemental Figure S9A). It should be mentioned that a certain protein form, when fused with different tags, could lead to different results. For example, when overexpressing GFP-Myc-aOsBZR1 and Flag-PPKL2, we failed to observe such a clear phenotype as those overexpressing Flag-aOsBZR1 and GFP-PPKL2. Similar phenomena have also been reported previously (Hurst et al., 2018; Yin et al., 2020), reminding the utilization of suitable materials for research.
We further analyzed the BR sensitivities of several representative transgenic plants. In coleoptile elongation test, GFP-aGSK2 showed basically no response to BL treatment (Supplemental Figure S9B). In contrast, Flag-aOsBZR1 exhibited enhanced BR sensitivity compared to ZH11, whereas GFP-PPKL2 showed normal response (Supplemental Figure S9B). In root inhibition assay, however, both GFP-aGSK2 and GFP-PPKL2 showed normal responses, whereas Flag-aOsBZR1 was highly sensitive to BL (Supplemental Figure S9C). In lamina bending assay, all the four plants tested, including Flag-aOsBZR1, GFP-OsBZR2, GFP-PPKL2, and Flag-PPKL2, clearly showed enhanced BR sensitivities, with Flag-aOsBZR1 and GFP-OsBZR2 much more obvious than others (Supplemental Figure S10). Taken together, these analyses supported the negative roles of GSK2 but the positive roles of OsBZR1/2 as well as PPKL2 in BR signaling.
Molecular characterization of GSK2, OsBZR1, and PPKL2 in response to BR
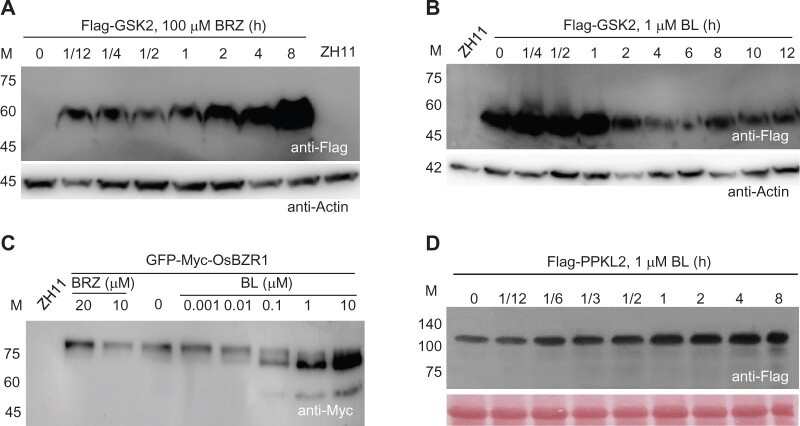
Consistent with our previous report (Tong et al., 2012), overexpression of Flag-GSK2 did not result in a clear phenotype. This could be due the instability of the wild-type GSK2 in plant. Indeed, we can hardly detect the protein in most lines using the tag antibody. However, when the plants were treated using BRZ (100 µM), an inhibitor of BR synthesis, the fusion protein was rapidly accumulated in minutes and reached to a very high level in 8 h (Figure 8A). In addition, when the BRZ pre-treated plants were further treated with 1 µM BL (in presence of BRZ), the Flag-GSK2 protein level was gradually decreased (Figure 8B). These analyses demonstrated that the fusion protein is highly sensitive to BR alteration. Similarly, overexpression of GFP-Myc-OsBZR1 also led to little phenotype, although the fusion protein can be evidently detected in the transgenic plants using anti-Myc antibody. However, the detected protein size (>75 kDa) was much larger than the calculated formula weight (∼60 kDa), indicating that the fusion protein has been inactivated by phosphorylating modification in plants (Figure 8C). We thus treated the plants with BL in a gradient of concentrations, and found that high BL obviously induced the fast-shift of the proteins, corresponding to the dephosphorylating forms of OsBZR1 (Figure 8C). For Flag-PPKL2, BL can obviously induce the protein accumulation in a time-dependent manner (Figure 8D). Thus, Flag-GSK2 and GFP-Myc-OsBZR1 as well as Flag-PPKL2 could be used as sensitive BR markers for studying upstream signaling events.
Figure 8.
Responses of the fusion proteins to BL and/or BRZ treatments. One-week-old seedlings were treated with different concentrations of BL and/or BRZ for different times as indicated. In (B), the plants were pre-treated with BRZ (20 µM) for 18 h, and then BL was added for the treatment in presence of BRZ. M, maker size (kDa). Blotting against Actin was shown as references in (A) and (B). Ponceau S staining was shown in (D) to indicate equal loading.
Discussions
BR signaling pathway could be one of the best characterized biological pathways in plants. However, it should be noted that several critical BR signaling components, including BIN2, BZR1/BES1, and BSU1, have been early identified from gain-of-function mutants in Arabidopsis. The redundant functions of BIN2- and BZR1-family genes in BR signaling were further demonstrated by construction and analysis of high-order mutants (Yan et al., 2009; Chen et al., 2019). Similar attempt has been carried out regarding BSU1-family genes, and an unexpected role of two members (BSL2/3) other than BR signaling has been implicated (Maselli et al., 2014). In rice, except OsBRI1, which was cloned from a loss-of-function mutant by forward genetics (Yamamuro et al., 2000), most other primary BR signaling genes (OsBAK1, OsBSK1, GSK2, and OsBZR1) were firstly identified by homology-based cloning followed with transgenic validation (Bai et al., 2007; Li et al., 2009; Tong et al., 2012; Zhang et al., 2015). Mutant evidence showing typical BR signaling-related characterizations remains absent regarding GSK-family and OsBZR-family genes. Although PPKL1 was first identified as the large grain-associated locus (qGL3 or GL3.1) by forward genetics (Hu et al., 2012; Qi et al., 2012; Zhang et al., 2012), its role in BR signaling was confirmed in a following study, which surprisingly found that PPKL1 is a negative regulator of BR signaling (Gao et al., 2019). Whether other members positively or negatively regulate BR signaling remains unclear. Given most of the single mutants are phenotypic silent due to functional redundancy, construction and analysis of high-order mutants are crucial for addressing these concerns.
In addition to the materials described above, our group also generated a number of single mutants of other BR signaling components, including OsSERK-family genes (Dong et al., 2020) and OsBSK-family genes. OsBRI1 has also been included for analysis but the knockout mutant cannot be maintained due to the sterility and severe phenotype as reported (Nakamura et al., 2006). This could also be the case for OsBZRs and PPKLs as we failed to obtain the highest-order mutants of both the family genes. In Arabidopsis, it has been shown that BZRs have a BR signaling-independent regulatory role in anther development (Chen et al., 2019). While whether OsBZRs are also involved in this process remain to be examined, this could be the cause for the failure of getting strong osbzr1,3,4 alleles or the quadruple mutants. Nevertheless, the evident BR-defective phenotype of the triple mutant osbzr1,3,4w demonstrated the positive roles of the family genes, at least some members, in BR signaling. In contrast, the gsk1,2,3,4 quadruple mutant showed a typical BR-enhanced phenotype, demonstrating the negative roles of the family genes. However, our comparison analysis of gsk1,2,3,4 and GSKs-knockdown plants suggested the roles of additional GSK members other than Clade II in regulating grain size and leaf angle. For example, GSK5 has been shown to regulate grain size via modulating auxin signaling (Hu et al., 2018; Xia et al., 2018; Ying et al., 2018).
The extensive BR sensitivity tests of the mutants as well as the overexpression plants revealed that one certain genotype could confer plants different or even opposite BR responses in different tissues (coleoptile, root, lamina joint) or at different development stages. Particularly in root, it was unexpected that both osbzr1,3,4w and ppkl2,3 showed greatly enhanced BR sensitivities, in contrast to the cases in coleoptile or lamina joint where the plants showed decreased or unaltered BR sensitivities. Although the detailed reason remains unclear, the complicated roles of BRs in root as well as in other tissues or processes, which appear conflicting each other, have been reported and discussed in a number of literatures (Singh and Savaldi-Goldstein, 2015; Tong and Chu, 2018). Regarding the roles of PPKLs in BR signaling, although our analyses provide some clues, whether PPKLs are involved in other biological process, as has been implied in Arabidopsis (Maselli et al., 2014), remained open to be investigated. The genetic materials as well as some intriguing observations presented in this study could be a starting point for further functional dissection of BRs and the related signaling components.
BRs regulate many important agronomic traits thus have great potential in crop improvement. However, BRs have high activity and the hormone functions in regulating different traits are complicated. High levels of BRs tend to cause negative effects on plant, and different traits could be negatively associated each other (Tong and Chu, 2018). At molecular level, overexpression or knockout of a certain BR signaling component could simultaneously confer beneficial effect on one trait but adverse effect on another trait. These features make BR utilization for molecular design not very easy. Our study exemplified a strategy for BR utilization by editing BR-related gene family. Gene family usually contains genes with functional differentiation, dominancy, and redundancy. Simultaneously editing thus could produce different combinations of various mutants. Theoretically, a family of four mutant genes could constitute four single mutants, six double mutants, four triple mutants, and one quadruple mutant. Given each gene could have multiple alleles and even more if edited by targeting different sites, the genotype numbers could be largely increased. By this approach, the agronomic traits could be largely diversified, producing a population containing various combinations of different traits, which enables to screening out the desired mutants to meet different requirements.
Overexpression analysis is widely used to study the function of genes, particularly those with functional redundancy. In addition, overexpression provides feasibility to study protein features at molecular level in plants, particularly those with poor abundance in wild-type plants. For GSK2 and OsBZR1, we generated their overexpression plant by fusing the proteins, either in wild-type forms or in stabilized forms, with either GFP or GFP-Myc or Flag tags. These tags are widely adopted for protein analysis and isolation in molecular biology research, providing great conveniences for various analyses of a protein regulation by using different techniques developed based on the tag antibodies. In addition, the GFP tag also enables the observation of protein in living cells. In Arabidopsis, the phosphorylation status of BZR1 and BES1 is usually used to indicate the output strength of upstream BR signaling. We also confirmed that both Flag-GSK2 and GFP-Myc-OsBZR1 are hypersensitive to BR alteration. Thus, the two proteins might be used as an effective reporter of BR levels or BR signaling.
Although our physiological analysis revealed the involvement of these family members in BR signaling, the solid biochemical evidence remains absent. Since the effective native antibodies against OsBZR1 are currently unavailable in our lab, we will introduce the above-mentioned fusion construct into the mutants to evaluate whether GSKs and PPKLs indeed confer impact on OsBZR1 phosphorylation status or protein stability, which is under our investigation. Considering GSK2, OsBZR1/2, and PPKL2 are representative members of their respective family, the generation of their overexpression plants in various forms will benefit the further investigation of the protein properties. Together with a large number of mutants created in this study, the release of these materials as well as the related information will greatly facilitate BR application and research in crop plants.
Materials and methods
Plant growth conditions and evaluation of agronomic traits
Rice (O. sativa) ZH11, a japonica cultivar, was used as wild-type for all the editing. Plants were grown in experimental stations located either in Beijing (40°06′N) or in Hainan province (18°30′N) under regular field conditions. Agronomic traits including plant height, tiller number, and leaf angle of osbzrs and gsks were measured at mature stage in Beijing, and of ppkls in Hainan. Grain size was measured after harvesting using a SC-G system (WSeen). For seedling analysis, plants were grown in a growth chamber (GXZ-800D, Ningbo, China) with settings: 14-h/10-h light/dark cycle, 30°C/28°C day/night temperature cycle, light intensity 600 µmol m−2 s−1. Half-strength Murashige and Skoog (1/2 MS) basal salt mixture was supplied as nutrients.
Genome editing and mutant identification
Editing of PPKLs using set1 sgRNAs as well as those editing of any single gene was performed in a company (Biogle, Changzhou, China). The CRISPR/Cas9 system as well as the detailed method has been described previously (Lu et al., 2017). See the previous reports for the system and methods used for the editing of PPKLs (set2), GSKs, and OsBZRs, respectively (Wang et al., 2015; Ma and Liu, 2016). The binary vectors were introduced into callus by Agrobacterium-mediated method and then regenerated into transgenic plants. Mutations were detected by directly sequencing with the primers designed around the targeting sites (Supplemental Table S1), and the sequencing results were analyzed using a web tool named DSDecodeM (http://skl.scau.edu.cn/). The clearance of T-DNA insertion was confirmed by detection of the selection marker genes by PCR using primers listed in Supplemental Table S1.
Construction of the overexpression vectors
A set of empty binary vectors, namely pEZR(K)-LC (carrying 35S:GFP expression cassette), 1300-35S-Flag, and pEZR(K)-LC-Myc were used for construction of the overexpression vectors. The coding sequences of the genes were amplified and then introduced into the final vectors by in-fusion technology. See Supplemental Table S2 for the primers and restriction enzyme sites used for the sequence insertion. The activated form of GSK2 has been reported previously (Tong et al., 2012). The activated form of OsBZR1 carries a point mutation (P249>L) referring to the Arabidopsis bzr1-D or bes1-D (Wang et al., 2002; Yin et al., 2002). The introduction of the point mutation was generated following a method described in a previous report (Heckman and Pease, 2007). See Supplemental Table S2 for the primers used for the mutation introduction. All the final vectors were sequenced to ensure the fidelity.
Gene expression analysis
For expression analysis of GSKs, root, stem, and leaf tissues were sampled from plants at 60 d after germination in field, and different lengths of panicle were subsequently harvested. One-week-old seedling shoots were used for analysis of BR synthetic genes. The total RNAs were extracted using TRIzol (Invitrogen). Then, 2 µg RNAs were reversely transcribed to cDNA with a reverse transcription kit (Toyobo). RT-qPCR was performed on a LightCycler 96 system (Roche) using SYBR Green PCR mix (Roche) according to product instructions. Ubiquitin was used as reference. Primers were listed in Supplemental Table S3.
Chemical treatment and protein expression analysis
For evaluating the protein response to BRs, different concentrations of BL (Wako) and/or BRZ (TCI) were supplied in the growth medium (1/2 MS) of one-week-old rice seedlings and the shoots were sampled at different time points for further analysis. The samples were ground into powder in liquid nitrogen and then sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer was added according to the weight (3 µL mg−1). The samples were boiled for 10 min and centrifuged, and the supernatants were resolved by SDS–PAGE. Commercial anti-Flag (1:2,000, Sigma) and anti-Myc monoclonal antibody (1:2,000, Abmart) was used to detect Flag-fused protein and GFP-Myc-fused protein, respectively. Immunoblotting was performed using a Trans-Blot Turbo Transfer System (Bio-Rad) according to the manufacturer’s instructions. Blotting against Actin using anti-Actin monoclonal antibody (1:5,000, Abmart) was performed as references.
BR sensitivity test
De-husked seeds were sterilized using 1.5% (w/v) NaClO solution for half an hour, then sowed on solid agar containing 1/2 MS with different concentrations of BL (0, 0.001, 0.01, 0.1, 1, 10 µM). After grown for 10 d under conditions as described above, the root lengths were measured. See a previous report for more detailed procedures (Tong and Chu, 2017). About 15–30 seeds were evaluated for each line and those effective values with normal growth were used for statistical analysis. For leaf bending assay, germinated seeds were cultivated in water with 1/2 MS supplied as nutrients. Since assays of those many different plants require relatively large amount of the hormone, the affordable HBL (28-HBL, Shanghai Yuanye Bio-Technology) was used as an alternative of BL for the analysis. HBL was added to the culture medium after 3-d growth when the second leaves were emerging, and the angles of the second or the third leaves were measured following additional 4-d growth.
Accession numbers
Sequence information of the genes mentioned in this study could be found in the rice genome annotation database (http://rice.plantbiology.msu.edu/) under the accession numbers shown in Figure 1A (GSKs), Figure 4A (OsBZRs), and Figure 6A (PPKLs).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Tiller number and root length of GSK-related mutants.
Supplemental Figure S2. BR sensitivity of GSK-related mutants evaluated by coleoptile elongation test.
Supplemental Figure S3. BR sensitivity of GSK-related mutants evaluated by lamina bending tests of the second and third leaves.
Supplemental Figure S4. BR sensitivity tests of OsBZR-related mutants.
Supplemental Figure S5. Plant height, tiller number, and root length of PPKL-related mutants.
Supplemental Figure S6. BR sensitivity of PPKL-related mutants evaluated by coleoptile elongation test and root inhibition test.
Supplemental Figure S7. BR sensitivity of PPKL-related mutants evaluated by lamina bending test.
Supplemental Figure S8. Expression of BR-synthetic genes in the shoots of 1-week-old seedlings of different plants.
Supplemental Figure S9. Tiller number and BR sensitivities of the representative overexpression plants.
Supplemental Figure S10. BR sensitivity of the overexpression plants evaluated by lamina bending test.
Supplemental Table S1. Primer sequences for mutation identification.
Supplemental Table S2. Primer sequences for vector constructions.
Supplemental Table S3. Primer sequences for RT-qPCR analysis.
Supplementary Material
Acknowledgments
We thank Prof. Yaoguang Liu (South China Agricultural University), Dr Kejian Wang (China National Rice Research Institute), and Dr Yangwen Qian (Biogle Genome Editing Center) for the help in genome editing.
Funding
This work was supported by National Natural Science Foundation of China (Nos. 31871587, 31722037, 91735302), Central Public-interest Scientific Institution Basal Research Fund (Nos. Y2020XK16, S2021ZD01, S2018PY02), and Natural Science Foundation of Anhui Province (No. 1908085MC93, to S.W.).
Conflict of interest statement. None declared.
D.L. performed PPKLs-related analyses and most sensitivity tests. D.L., H.T., W.Y., G.Z., and X.Z. performed overexpression analyses. Z.Y., H.T., and L.L. performed GSKs-related analyses. G.Z., H.T., and W.Y. performed OsBZR-related analyses. M.N., W.M., N.D., J.L., and Y.Y. helped in phenotype analyses. H.T. conceived the study and designed the experiments. H.T., C.C., and S.W. co-supervised the study.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Hongning Tong (tonghongning@caas.cn).
References
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che R, Tong H, Shi B, Liu Y, Fang S, Liu D, Xiao Y, Hu B, Liu L, Wang H, et al. (2015) Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants 2: 15195. [DOI] [PubMed] [Google Scholar]
- Chen LG, Gao ZH, Zhao ZY, Liu XY, Li YP, Zhang YX, Liu XG, Sun Y, Tang WQ (2019) BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol Plant 12: 1408–1415 [DOI] [PubMed] [Google Scholar]
- Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee MO, Yoshida S, Feldmann KA, Tax FE (2002) Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3β-like kinase. Plant Physiol 130: 1506–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N, Yin W, Liu D, Zhang X, Yu Z, Huang W, Liu J, Yang Y, Meng W, Niu M, et al. (2020) Regulation of brassinosteroid signaling and salt resistance by SERK2 and potential utilization for crop improvement in rice. Front Plant Sci 11: 621859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang ZM, Ji YY, Hu J, Guo RK, Sun SY, Wang XL (2020) Strigolactones and brassinosteroids antagonistically regulate the stability of the D53-OsBZR1 complex to determine FC1 expression in rice tillering. Mol Plant 13: 586–597 [DOI] [PubMed] [Google Scholar]
- Gao XY, Zhang JQ, Zhang XJ, Zhou J, Jiang ZS, Huang P, Tang ZB, Bao YM, Cheng JP, Tang HJ, et al. (2019) Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell 31: 1077–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2: 924–932 [DOI] [PubMed] [Google Scholar]
- Hu Z, Lu SJ, Wang MJ, He H, Sun L, Wang H, Liu XH, Jiang L, Sun JL, Xin X, et al. (2018) A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol Plant 11: 736–749 [DOI] [PubMed] [Google Scholar]
- Hu ZJ, He HH, Zhang SY, Sun F, Xin XY, Wang WX, Qian X, Yang JS, Luo XJ (2012) A kelch motif-containing serine/threonine protein phosphatase determines the large grain QTL trait in rice. J Integr Plant Biol 54: 979–990 [DOI] [PubMed] [Google Scholar]
- Hurst CH, Turnbull D, Myles SM, Leslie K, Keinath NF, Hemsley PA (2018) Variable effects of C-terminal fusions on FLS2 function: not all epitope tags are created equal. Plant Physiol 177: 522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E-J, Youn J-H, Park C-H, Kim T-W, Guan S, Xu S, Burlingame AL, Kim Y-P, Kim S-K, Wang Z-Y, et al. (2016) Oligomerization between BSU1 family members potentiates brassinosteroid signaling in Arabidopsis. Mol Plant 9: 178–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K (2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7: 791–806 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JM, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Lu YM, Ye X, Guo RM, Huang J, Wang W, Tang JY, Tan LT, Zhu JK, Chu CC, Qian YW (2017) Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol Plant 10: 1242–1245 [DOI] [PubMed] [Google Scholar]
- Ma X, Liu YG (2016) CRISPR/Cas9-based multiplex genome editing in monocot and dicot plants. Curr Protoc Mol Biol 115: 31.06.01–31.06.21 [DOI] [PubMed] [Google Scholar]
- Maselli GA, Slamovits CH, Bianchi JI, Vilarrasa-Blasi J, Cano-Delgado AI, Mora-Garcia S (2014) Revisiting the evolutionary history and roles of protein phosphatases with kelch-like domains in plants. Plant Physiol 164: 1527–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, et al. (2006) The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol 140: 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan TM, Vukašinović N, Liu D, Russinova E, Yin Y (2020) Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32: 295–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez JM, Ponce MR, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242: 161–173 [DOI] [PubMed] [Google Scholar]
- Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang LW, Gao JP, Lin HX (2012) The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res 22: 1666–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24: 105–109 [DOI] [PubMed] [Google Scholar]
- Singh AP, Savaldi-Goldstein S (2015) Growth control: brassinosteroid activity gets context. J Exp Bot 66: 1123–1132 [DOI] [PubMed] [Google Scholar]
- Takatsuto S, Yazawa N, Ikekawa N, Morishita T, Abe H (1983) Synthesis of (24R)-28-homobrassinolide analogues and structure-activity relationships of brassinosteroids in the rice-lamina inclination test. Phytochemistry 22: 1393–1397 [Google Scholar]
- Tong H, Chu C (2017) Physiological analysis of brassinosteroid responses and sensitivity in rice. Methods Mol Biol 1564: 23–29 [DOI] [PubMed] [Google Scholar]
- Tong H, Chu C (2018) Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci 23: 1016–1028 [DOI] [PubMed] [Google Scholar]
- Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, Jin Y, Qian Q, Chu C (2014) Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26: 4376–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong HN, Liu LC, Jin Y, Du L, Yin YH, Qian Q, Zhu LH, Chu CC (2012) DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24: 2562–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shen L, Fu Y, Yan C, Wang K (2015) A simple CRISPR/Cas9 system for multiplex genome editing in rice. J Genet Genomics 42: 703–706 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He JX, Chen M, Vafeados D, Yang YL, Fujioka S, Yoshida S, Asami T, et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang JL, Wan JM, Zhai HQ, Takatsuto S, et al. (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Zhou H, Liu RJ, Dan WH, Li PB, Wu B, Chen JX, Wang LQ, Gao GJ, Zhang QL, et al. (2018) GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol Plant 11: 754–756 [DOI] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan ZY, Zhao J, Peng P, Chihara RK, Li JM (2009) BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol 150: 710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Xiao Y, Niu M, Meng W, Li L, Zhang X, Liu D, Zhang G, Qian Y, Sun Z, et al. (2020) ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell 32: 2292–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Ying JZ, Ma M, Bai C, Huang XH, Liu JL, Fan YY, Song XJ (2018) TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol Plant 11: 750–753 [DOI] [PubMed] [Google Scholar]
- Yoo MJ, Albert VA, Soltis PS, Soltis DE (2006) Phylogenetic diversification of glycogen synthase kinase 3/SHAGGY-like kinase genes in plants. BMC Plant Biol 6: 14–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang X, Zhao Z, Wang R, Huang X, Zhu Y, Yuan L, Wang Y, Xu X, Burlingame AL, et al. (2015) OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol 170: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Wang JF, Huang J, Lan HX, Wang CL, Yin CF, Wu YY, Tang HJ, Qian Q, Li JY, et al. (2012) Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA 109: 21534–21539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, Xu J, Liao JY, Wang XJ, Qu LH, Chen F, et al. (2013) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 31: 848–852 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.