Abstract
Insertion of membrane proteins into the lipid bilayer is a crucial step during their biosynthesis. Eukaryotic cells face many challenges in directing these proteins to their predestined target membrane. The hydrophobic signal peptide or transmembrane domain (TMD) of the nascent protein must be shielded from the aqueous cytosol and its target membrane identified followed by transport and insertion. Components that evolved to deal with each of these challenging steps range from chaperones to receptors, insertases, and sophisticated translocation complexes. One prominent translocation pathway for most proteins is the signal recognition particle (SRP)-dependent pathway which mediates co-translational translocation of proteins across or into the endoplasmic reticulum (ER) membrane. This textbook example of protein insertion is stretched to its limits when faced with secretory or membrane proteins that lack an amino-terminal signal sequence or TMD. Particularly, a large group of so-called tail-anchored (TA) proteins that harbor a single carboxy-terminal TMD require an alternative, post-translational insertion route into the ER membrane. In this review, we summarize the current research in TA protein insertion with a special focus on plants, address challenges, and highlight future research avenues.
Update on different pathways and candidates that facilitate membrane insertion of tail-anchored proteins in eukaryotes with a special emphasis on recent insights in plant research.
Diversity of membrane proteins
Roughly, one-third of the average eukaryotic proteome comprises integral membrane proteins (IMPs) that act, for example, as channels, transporters, or receptors (Hegde and Keenan, 2011). IMPs are found in all organellar membranes within the cell. They reside in the lipid bilayers of the endomembrane system (endoplasmic reticulum [ER], Golgi, trans Golgi network [TGN], multivesicular body [MVB], vacuole, peroxisomes, and plasma membrane [PM]) as well as in the membranes of the semiautonomous cell organelles of chloroplast and mitochondria. To maintain membrane integrity and cellular function, correct targeting and insertion of newly synthesized IMPs have to be guaranteed. For this purpose, dedicated signal sequences and insertion pathways have evolved.
ADVANCES
Shared features of all IMPs are strongly hydrophobic transmembrane domains (TMDs); yet, these vary in their sequence, number, and final topology, and thereby define different types of membrane proteins (Guna and Hegde, 2018). However, all IMPs face three fundamental challenges in their biogenesis:
Research in the last decade revealed several different targeting routes for TA protein transport and translocation into the ER and organellar membranes of eukaryotes.
The GET pathway described for TA protein insertion in yeast and mammals is partially conserved in Arabidopsis, where loss of function leads to defects in root hair growth.
Absence of the coreceptor for TA protein docking and insertion at the ER membrane in the context of the GET pathway in plants phenocopies other get lines.
Sequence information confirms conservation of alternative yeast pathways in plants, while functional data currently remain elusive.
TA protein import into the ER membrane was mainly studied in yeast and mammalian cell culture, but plants have proven to be ideal models to gain a deeper understanding of these pathways in an organismal context and to study their functional impact on multicellular systems.
the nascent protein including its nonpolar TMD(s) must navigate through the aqueous cytosolic environment before reaching the membrane. As exposure of the lipophilic TMDs within the cytosol would lead to premature aggregation, chaperoning proteins are needed which recognize and shield the TMDs until their insertion into the hydrophobic bilayer;
IMPs with varying numbers of TMDs and either luminally or cytosolically facing peptide stretches require membrane-bound receptors that aid in the insertion process and guarantee correct orientation within the membrane;
finally, targeting sequences (e.g. retention motifs) within the protein need to be recognized to facilitate delivery to the corresponding target membrane (ER and secretory pathway versus organellar membranes; Pedrazzini et al., 1996).
Of signal recognition and translocons
To cope with the challenges mentioned above, various strategies evolved in eukaryotes were described by scientists in the past decades. Günter Blobel was awarded the 1999 Nobel Prize in Physiology or Medicine “For the discovery that proteins have intrinsic signals that govern their transport and localization in the cell” (Celebrating 20 years of cell biology, 2019). Together with David Sabatini, Blobel had postulated the “signal hypothesis” some 30 years earlier (Sabatini and Blobel, 1970). Although a hypothesis at first and rejected by many at the time, it turned out to be correct and found its way into the textbooks. The majority of secretory proteins or IMPs utilize this signal recognition particle (SRP)-dependent pathway and enter the ER through the Sec61 translocon which was later discovered and similarly earned its discoverer Randy Schekman a Nobel prize (Novick et al., 1980; Deshaies et al., 1991) shared with James Rothman and Thomas Südhof “for their discoveries of machinery regulating vesicle traffic, a major transport system in our cells” (Wickner, 2013). The pathway is also referred to as “co-translational” as it targets and inserts proteins into the ER during their synthesis (Anderson et al., 1982).
Translocation starts with the extrusion of a nascent polypeptide chain from the ribosome exit channel. SRP recognizes ribosomes with either an N-terminal signal sequence or TMD of a nascent protein (Ogg and Walter, 1995; Shao and Hegde, 2011). Subsequent binding of SRP to the ribosome transiently arrests protein synthesis by blocking further tRNA entry (Lakkaraju et al., 2008; Richter and Coller, 2015). Targeting to the ER membrane of the SRP/ribosome–nascent chain (RNC) complex is induced by the binding to the SRP receptor (SR) in a GTP-dependent manner (Gilmore et al., 1982a, 1982b ). Subsequent conformational changes lead to interaction with the Sec61 translocon, unloading of RNC from SRP to Sec61 and determine the duration of the translational pause. GTP hydrolysis triggers the disassembly of SRP from SR and recycling of the components for additional rounds of protein targeting (Song et al., 2000; Shao and Hegde, 2011).
During co-translational insertion, two mechanisms protect the TMD from the aqueous cytosol:
early targeting of the TMD by SRP and maintenance of this connection until docking at the Sec61 channel to ensure minimal exposure to the cytosol before integration and
translational slowdown that prevents translation of additional, subsequent TMDs into the cytosol (Walter and Blobel, 1981; Pechmann et al., 2014).
Little is known about an Archaeplastida Sec61 translocon, although such a fundamental mechanism is undoubtedly conserved in plants. Three homologs of each, the central pore Sec61α as well as the two subunits Sec61β and Sec61γ, are encoded in the Arabidopsis (Arabidopsis thaliana) genome. While functional data are lacking, physical interaction of AtSec61α1 with AtSec61β1 and AtSec61γ1 was shown by our group (Mehlhorn et al., 2018). In addition, the translocon-associated proteins AtSec62 and Sec63 (AtErdjA and AtErdj2B) are conserved as well (Mitterreiter et al., 2020). Together with the tetratricopeptide repeat protein AtTPR7, both are probably involved in a chaperone-assisted post-translational import of small peptides in Arabidopsis (Schweiger et al., 2012).
Tail-anchored proteins
The SRP/Sec61 co-translational pathway reaches its limits, though, when signal sequences or TMDs are lacking within the N-terminal part of the protein. This is in particular the case for type II-orientated membrane proteins that feature a TMD close to their C-terminal end and are referred to as tail-anchored (TA) proteins (Borgese et al., 2003). To distinguish these from other type II proteins, the C-terminal (after translocation: luminal) stretch should by definition be no longer than approximately 30 amino acids (Borgese et al., 2003). This is roughly the length of a peptide stretch within the ribosomal exit channel (Voss et al., 2006). Proteins with such feature are released from the ribosome when their TMD is disclosed to the cytosolic environment. To prevent aggregation of the hydrophobic TMD within the aqueous cytosol, immediate action of chaperones is required aiding in shuttling and post-translational translocation (Pedrazzini, 2009; Johnson et al., 2013).
TA proteins make up to approximately 3%–5% of all IMPs and can be found in almost all cellular membranes (Abell and Mullen, 2011). In Arabidopsis, around 500 TA proteins were predicted in silico (Kriechbaumer et al., 2009). They play key roles in many vital processes such as vesicle trafficking, apoptosis, translocation of other proteins, ubiquitination, signal transduction, enzymatic reactions, or regulation of transcription (Borgese et al., 2003; Kriechbaumer et al., 2009). Some TA proteins even take part in translocation of other membrane proteins as subunits of translocation machineries such as the Sec61β subunit of the SEC61 translocon, or Translocase of outer membrane 22 (Tom22) and Translocase of chloroplast 33 (Toc33) of the mitochondrial and chloroplast import machineries. Additionally, most of the soluble N-ethylmaleimide-sensitive factor attachment receptors (SNAREs) which facilitate vesicle fusion in eukaryotic cells, are TA proteins (Neveu et al., 2020). Their prominent role in many physiological processes is reflected by the dramatic phenotypes associated with their loss-of-function lines, ranging from conditional sensitivity toward pathogens to embryonic lethality (Lipka et al., 2007).
Anchoring in the ER membrane
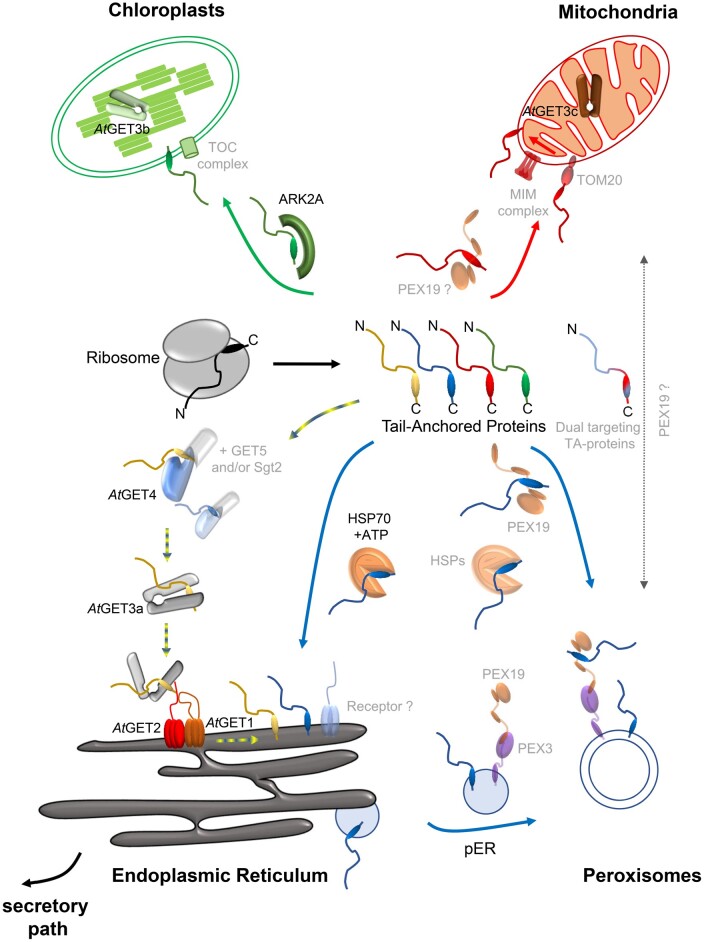
The seemingly textbook example for post-translational membrane insertion of TA proteins into the ER is the Guided Entry of Tail-anchored (GET) proteins pathway (Figure 1) which was initially identified in mammals and yeast (Stefanovic and Hegde, 2007; Schuldiner et al., 2008).
Figure 1.
Graphical summary depicting translocation pathways of TA-proteins in plants. Detailed description of the pathways can be found in the text. ER-destined TA proteins (yellow), peroxisomal TA proteins (blue), mitochondrial TA proteins (red), chloroplastidic TA proteins (green), and dual-targeted TA proteins (mitochondria/peroxisomes, dashed arrow, red-blue TA-protein). Opaqueness generally refers to proposed mechanisms/proteins/complexes which may be involved in TA protein translocation in plants but still require experimental validation.
In yeast, nascent TA proteins are recognized immediately after emergence from the ribosomal exit tunnel through a tripartite pretargeting complex consisting of small glutamine-rich tetratricopeptide cochaperone 2 (Sgt2), Get4, and Get5 (Chang et al., 2010; Wang et al., 2010). A functional mammalian homolog of Get4/5 is the B-cell lymphoma 2 (BCL2)-associated athanogene cochaperone 6 (BAG6) complex comprising BAG6, TMD recognition complex 35 (TRC35) and ubiquitin-like domain (UBL)-containing protein 4A (UBL4A), which works in cooperation with small glutamine-rich tetratricopeptide repeat-containing protein α, the mammalian Sgt2 ortholog (Mariappan et al., 2010; Johnson et al., 2013). While Sgt2 alone is ineffective in binding TA proteins, Get4/5 assists this process by bridging and facilitating TA protein transfer from Sgt2 to the cytosolic ATPase Get3 (in mammals TRC40 or Asna1; Suloway et al., 2009; Simpson et al., 2010; Chartron et al., 2011; Gristick et al., 2014). BAG6 triages nascent TA proteins in either an insertion competent fraction or destined for proteasomal degradation (Leznicki and High, 2012; Shao et al., 2017). Recent work has now demonstrated that polyubiquitinated TA proteins can circumvent recognition by BAG6 and still be inserted via the (mammalian) TRC40 pathway and subsequent deubiquitination (Culver and Mariappan, 2021).
Key component of the pathway is the dimeric ATPase Get3. Its subunit interaction is stabilized by a Zn2+ ion coordinated by a CxxC motif (Mateja et al., 2009; Simpson et al., 2010). Get3 consists of a nucleotide-binding pocket and a TA protein-binding domain and undergoes conformational changes dependent on its nucleotide-binding state (Wereszczynski and McCammon, 2012). In a nucleotide-free state, Get3 is in an open conformation while binding of ATP leads to a closed dimer, thereby creating a hydrophobic groove that binds and shields the TMD of TA proteins (Mateja et al., 2009; Wereszczynski and McCammon, 2012; Mateja et al., 2015). It was demonstrated that, unlike SRP, Get3 does not associate with ribosomes (Stefanovic and Hegde, 2007). Get3 shuttles the client protein to the ER membrane receptors consisting of a heteromeric complex of Get1 (WRB in mammals; Vilardi et al., 2011; McDowell et al., 2020) and Get2 (CAML in mammals; Yamamoto and Sakisaka, 2012; Vilardi et al., 2014). The long cytosolic N-terminal domain of Get2 mediates the tethering of the Get3–TA protein complex (Mariappan et al., 2011; Wang et al., 2011). Interaction of Get2 only takes place with a nucleotide-bound Get3 which is also compatible with TA protein binding (Denic et al., 2013). Hydrolysis of ATP opens the Get3 dimer. This conformational change disrupts the hydrophobic groove releasing the bound TA protein and providing it for insertion by the Get1–Get2 insertase (Wang et al., 2014; Zalisko et al., 2017). Intriguingly, Get1 and Get2 compete for Get3 binding via overlapping binding sites (Stefer et al., 2011; Denic et al., 2013), although the interaction between Get3 and a coiled-coil domain of Get1 occurs only with an open, nucleotide-free Get3 (Mariappan et al., 2011). Rebinding of ATP returns Get3 into a closed conformation, thereby weakening the Get3–Get1 interaction which leads to dissociation of Get3 from the membrane and recycling for another round of TA protein loading via the pre-targeting complex Sgt2/Get4/Get5 (Stefer et al., 2011; Suloway et al., 2012).
It is noteworthy that TA protein recognition from the ribosome to the membrane is assisted by heat-shock proteins (Rabu et al., 2008; Craig, 2018). Recently, the involvement of J-domain proteins involved in the TA protein handover from Hsp70 to Sgt2 in yeast has been demonstrated (Cho and Shan, 2018; Cho et al., 2021).
It GET’s complicated in plants
In Arabidopsis, a high degree of conservation was presumed from an in silico search of GET components (Abell and Mullen, 2011; Duncan et al., 2013). Four years later, the existence and function of a plant GET pathway were demonstrated by two groups, independently (Srivastava et al., 2017; Xing et al., 2017) although some of its components still remain elusive. While a functional Get4 ortholog (At5g63220) was identified in plants, its partner proteins within a putative pre-targeting complex could not be determined as too many potential candidates exist. Based on sequence similarities, there are multiple putative Sgt2 and Get5 orthologs in Arabidopsis, the latter features a ubiquitin-like domain which is present in a wide range of proteins (Paul et al., 2013; Srivastava et al., 2017; Xing et al., 2017). BAG6, the protein that bridges the interaction of Ubl4A to TRC35 within a mammalian pre-targeting complex, is lacking in yeast (Leznicki et al., 2013). Interestingly, a putative BAG6 ortholog (Table 1) exists in Arabidopsis and is involved in triggering autophagy in response to pathogen attack (Li et al., 2016); however, at present, an involvement in a plant GET pathway remains elusive.
Table 1.
Arabidopsis orthologs of yeast and mammalian TA protein insertion pathways
| GET pathway | ||||
|---|---|---|---|---|
| Yeast/mammalian | AGI Code | Gene namea | Protein localization | References |
| GET1/WRB | At4g16444 | AtGET1 | ER membraneb | Srivastava et al., 2017; Xing et al., 2017 |
| GET2/CAML | At4g32680 | AtGET2 | ER membraneb | Asseck et al., 2021 |
| GET3/TRC40 | At1g01910 | AtGET3a | Cytosolb | Srivastava et al., 2017; Xing et al., 2017 |
| At3g10350 | AtGET3b | Chloroplast stromab | ||
| At5g60730 | AtGET3c | Mitochondria matrixb | ||
| GET4/TRC35 | At5g63220 | AtGET4 | Cytosolb | Srivastava et al., 2017; Xing et al., 2017 |
| GET5/UBL4A | At1g55060 | UBQ12 | Cytosolc | Srivastava et al., 2017 |
| SGT2/SGTA | At4g08320 | TPR8 | Nucleusc | Srivastava et al., 2017 |
| BAG6 | At2g46240 | BAG6 | Nucleusc | PSI-BLAST, TAIR |
|
| ||||
| SND pathway | ||||
|
| ||||
| SND1 | Not found | – | – | PSI-BLAST |
| SND2 | At4g30500At2g23940 | AtSND2a | Plasma membranec | PSI-BLAST |
| AtSND2b | ER membranec | |||
| SND3 | Not found | – | – | PSI-BLAST |
|
| ||||
| ER Membrane Complex (EMC) | ||||
|
| ||||
| EMC1 | At5g11560 | PNET5 | ER membranec | PSI-BLAST, TAIR |
| EMC2 | At3g04830 | AtEMC2a | ER membranec | PSI-BLAST, TAIR |
| At5g28220 | AtEMC2b | Cytosolc | ||
| EMC3 | At4g12590 | AtEMC3 | ER membranec | PSI-BLAST, TAIR |
| EMC4 | At5g10780 | AtEMC4 | ER membranec | PSI-BLAST, TAIR |
| EMC5 | At5g03345 | PRCE2 | ER membranec | PSI-BLAST, TAIR |
| EMC6 | At5g49540 | AtEMC6 | Plasma membranec | PSI-BLAST, TAIR |
| EMC7 | At2g25310 | AtEMC7a | ER membranec | PSI-BLAST, TAIR |
| At4g32130 | AtEMC7b | ER membranec | ||
| EMC8/9 | At5g55940 | EMB2731 | ER membranec | PSI-BLAST, TAIR |
| EMC10 | Not found | – | – | PSI-BLAST |
Found annotated at TAIR (Arabidopsis.org) or—if designated as unknown protein—our suggestion for future use.
Experimentally validated, see referenced publication for details.
Predicted using SUBA (suba.live).
PSI, Position-specific iterative
Other than Get1/WRB, Get2/CAML has no sequence ortholog in plants. However, only recently, a functional Get2/CAML homolog has been identified in Arabidopsis using affinity purification-mass spectrometry (Asseck et al., 2021). Despite low sequence similarity, the overall structure comprising three TMDs and a cytosolic N-terminal stretch of basic amino acid residues seem to be evolutionarily conserved to maintain a common function (Asseck et al., 2021). Position-specific iterative- basic local alignment search tool (BLAST) analysis of the human CAML sequence revealed co-selection of the two functional domains, allowing the identification of orthologous genes also in distant phyla (Borgese, 2020; Asseck et al., 2021). In mammals, the two subunits of the GET receptor complex have been shown to depend on each other for expression and are degraded in the absence of the binding partner (Carvalho et al., 2019; Inglis et al., 2020). Similarly, Get1 deficiency in yeast leads to a reduced protein level of Get2 and vice versa, demonstrating reciprocal regulation of these two proteins (Schuldiner et al., 2008; Stefer et al., 2011). In Arabidopsis, however, the relationship between the receptor components seems to be distinct from that in Opisthokonts. In the absence of its co-receptor, AtGET2 is still expressed but no longer interacts with the targeting factor AtGET3a (Asseck et al., 2021).
There are additional, intriguing differences among Archaeplastida GET components such as three different GET3 proteins that were identified in Arabidopsis (namely AtGET3a, AtGET3b, and AtGET3c). In silico comparison of these three paralogs revealed two distinct clades (GET3a and GET3bc) present in the Archaeplastida and SAR supergroup but not in Opisthokonts and Amoebozoa, indicating a duplication event in the evolution of eukaryotes (Xing et al., 2017; Farkas et al., 2019). However, orthologs of AtGET3c seem to be Brassicaceae-specific, whereas several copies of AtGET3b orthologs can exist in other plant species (Bodensohn et al., 2019). Obvious differences between Archaeplastida GET3 proteins and Opisthokont Get3 include:
the conserved CxxC motif necessary for the coordination of a zinc ion and dimer formation (see above) is lacking in GET3a but not in the GET3bc clade despite AtGET3a retaining the ability to form dimers (Xing et al., 2017). Instead, in GET3a an ExxE motif and additional acidic residues adjacent to the site that usually bears the CxxC motif in other species’ sequences may take over metal ion coordination and dimer stabilization (Farkas et al., 2019);
an approximately 30 amino acid long, strongly charged extension was only found in the GET3a clade and suggested to be involved in dimerization (Farkas et al., 2019);
AtGET3a is targeted to the cytosol and probably recruited to the ER membrane as it can be found in microsomal fractions (Srivastava et al., 2017; Bodensohn et al., 2019), which might represent the receptor-bound state. AtGET3b, however, is located within the stroma of chloroplasts and AtGET3c in the matrix of mitochondria (Xing et al., 2017). Their organellar function is currently not understood (Zhuang et al., 2017; Bodensohn et al., 2019); and
while all three orthologs possess the ATPase motif, GET1 and GET4 binding residues are only conserved in AtGET3a. Consistent with this finding, only AtGET3a interacts with AtGET4 and AtGET1 but neither AtGET3b nor AtGET3c (even in truncated, cytosolic forms; Xing et al., 2017). This suggests that only the cytosolic AtGET3a plays a role in a canonical ER GET pathway in plants.
GETting knocked out—phenotypic consequences
But there remain more mysteries. So far only two TA proteins have been identified that show reduced membrane insertion in Atget mutants, the pollen-specific SNARE protein SYP72 (Srivastava et al., 2017), and the root-hair-specific SNARE protein SYP123 (Xing et al., 2017). The GET pathway is considered as the main route for post-translational TA protein insertion into the ER. Contrary to such an implied vital role, yeast loss-of-function strains are viable under normal growth conditions (Schuldiner et al., 2008) and the lethality under oxidative stress likely relates to the function of ScGet3 as a chaperone for unfolding soluble proteins (Voth et al., 2014). Later analysis of yeast TA proteins revealed that only 2 out of 46 potential client proteins show dependency on the presence of an intact GET pathway. Nonetheless, knockout of the mammalian ortholog TRC40 leads to embryo lethality in mice (Mukhopadhyay et al., 2006) and severe organ defects in induced get mutants (Lin et al., 2016; NorLin et al., 2016; Vogl et al., 2016). One could conclude from this that among multicellular Opisthokonts, an intact GET pathway became indispensable for survival.
The data in other multicellular organisms such as plants, however, rule out such a general conclusion. In Arabidopsis, loss of GET pathway function clearly causes effects such as increased ER-stress levels (Srivastava et al., 2017) and reduced root hair length (Xing et al., 2017), yet no pleiotropic phenotypes, let alone seedling or embryo lethality, was observed. Such strong phenotypes, however, should be expected considering that certain vital TA proteins such as the cytokinesis-specific SNARE KNOLLE (Lauber et al., 1997) do not reach their target membrane.
With the implication that the GET pathway is the major hub for TA protein insertion in the ER, the question is justified whether this can hold true with respect to such mild phenotypes and whether or not backup systems have evolved. An alternative explanation would be that a plant GET pathway evolved additional/alternative function(s) instead/apart from TA protein insertion. The latter suggestion is supported by an immunoprecipitation mass spectrometry (IP-MS) analysis where only 23 TA proteins interacted with AtGET3a-GFP (Xing et al., 2017) which is <5% of all predicted TA proteins in Arabidopsis (Kriechbaumer et al., 2009). Thus, it seems that in plants, the GET pathway might not play a—not to mention the—major role in TA protein insertion; or at least that plants have evolved alternative mechanisms to secure TA protein insertion in case one route breaks down.
GET alternatives
The dispensability of Arabidopsis GET components for general plant growth and survival with merely an effect on root hair growth (Xing et al., 2017; Asseck et al., 2021) allows speculation regarding the existence of a yet-undiscovered alternative insertion pathways in plants that might redundantly substitute TA protein insertion into the ER membrane.
In a pioneering effort, an SRP-independent targeting (SND) pathway consisting of three genetically linked proteins localizing to the cytosol (Snd1) or ER membrane (Snd2 and Snd3) was identified in yeast (Aviram et al., 2016). Here, cytosolic Snd1 is predicted to interact with ribosomes, co-translationally capturing nascent proteins, whereas Snd2 and Snd3 associate with the Sec61 translocon acting as putative receptors. The SND pathway was initially described as a pathway for IMPs harboring an internal TMD, and its loss leads to mislocalisation of these proteins. It was shown that all three Snd proteins act in the same pathway and it additionally serves as a safeguard for both SRP-dependent insertion and the GET pathway. As for the get knockouts, SND deletion did not affect the viability of Saccharomyces under normal growth conditions. Interestingly, double knockouts between SND and GET are nonviable, suggesting a compensatory role of TA protein delivery to the ER (Aviram et al., 2016).
In mammals, homologs of Snd1 and Snd3 have not been found, yet an Snd2 homolog (TMEM208 or hSND2) was identified and its localization to the ER confirmed (Hassdenteufel et al., 2017). In two independent studies, the function of hSND2 in TA protein biogenesis shown as deletion leads to decreased TA protein insertion (Casson et al., 2017; Hassdenteufel et al., 2017). Interestingly, loss of hSND2 is compensated by upregulation of the SRP receptor SRα, which was shown to aid in an SRP-dependent post-translational insertion of some client TA proteins (Casson et al., 2017; Hassdenteufel et al., 2017). In Arabidopsis, two sequence paralogs for Snd2 can be identified via BLAST search, but no obvious orthologs for Snd1 or Snd3 (Table 1). It remains to be seen whether an SND pathway is functionally conserved and which proteins pair up with SND2 in plants to facilitate such function.
Another recently discovered post-translational insertase for ER-destined TA proteins with TMDs of moderate-to-low hydrophobicity is the ER membrane complex (EMC). In semi-permeabilized cells silenced for EMC components, integration of the mammalian ER-resident enzyme squalene synthase and four other TA proteins with similar hydrophobic TMD characteristics failed. Calmodulin seems to play a role as a chaperone in this pathway (Guna et al., 2018; Volkmar et al., 2019).
Putative orthologs for all 9–10 components of the mammalian EMC can be found in plants through sequence homology (Table 1). Whether a similar function is associated with these proteins in Arabidopsis or which other proteins are involved within a putative plant EMC complex is currently unresolved. It is noteworthy, that EMC3 as well as Get1 are ER-resident homologs of the Oxa1/Alb3/YidC family of insertases that facilitate co- and post-translational insertion of transmembrane proteins into the inner mitochondrial membrane (Oxa1), the thylakoid membrane (Alb3), and the inner membrane of bacteria (YidC), respectively (Anghel et al., 2017; Samuelson et al., 2000).
The SEC61 translocon and its auxiliary proteins SEC62/SEC63 make use of heat-shock proteins to provide an additional post-translation pathway (Abell et al., 2007; Wu et al., 2019). In Arabidopsis, AtTPR7 together with the translocon-associated proteins AtSec62 and AtErdj2 (AtSEC63) seems to facilitate heat-shock protein-mediated delivery of proteins for post-translational translocation (Schweiger et al., 2012; Schweiger and Schwenkert, 2013). Loss of AtSec62 impairs plant growth and reduces male fertility (Mitterreiter et al., 2020), yet it remains to be dissected whether the causative effect of this phenotype is an impairment in translocation or an interference in ER-phagy (FumagalLi et al., 2016; Hu et al., 2020).
Insertion of TA proteins in other organelles
Translocation to the ER may be the major route for most TA proteins, yet post-translational insertion requires recognition of the target membrane ahead of distribution. This is even more challenging for plants with one additional endomembrane compared with other eukaryotic cells. To distinguish between different destination membranes, targeting information is required within the TA protein.
More than two decades of research in TA proteins has unveiled properties and motives that seem important for endomembrane distinction; however, many candidates still seem to be exempt from rules (Borgese et al., 2001, 2019). These rules comprise targeting signals encoded in the hydrophobicity of the TMD as well as charge and length of the adjacent C-terminal element (CTE; Beilharz et al., 2003; Borgese et al., 2007; Abell and Mullen, 2011; Marty et al., 2014; Rao et al., 2016; Costello et al., 2017).
For ER targeting, the consensus motif seems to be a long and hydrophobic TMD followed by nonpolar, negative, or no residues in the CTE (Rao et al., 2016).
It is currently proposed that TA proteins of the outer mitochondrial membrane (OMM) show less hydrophobic and shorter TMDs with reduced helical content compared to TA proteins destined to the ER or secretory pathway (Kriechbaumer et al., 2009; Lee et al., 2014; Rao et al., 2016; Chio et al., 2017).
Targeting of some mitochondrial TA proteins to the OMM is also conducted by a moderately positively charged CTE (Marty et al., 2014; Rao et al., 2016). For Fis1, it could be demonstrated that a minimum of four basic residues are needed for mitochondrial localization while mutation of the basic residues in the CTE of some OMM TA proteins changes their destination (Rao et al., 2016). For example, mammalian ER-localized cytb5 with a negatively charged CTE localizes to the OMM when artificially reverted to a positive net charge. This same construct expressed in plant cells, however, is directed to the chloroplast highlighting the challenges associated with the discrimination of multiple destination membranes (Maggio et al., 2007). It was also demonstrated that two cytochrome b5 (cytb5) isoforms—both with positive net charges in their CTE, but a number of putative phosphorylation sites—localize to either the ER or the chloroplast outer envelope (Maggio et al., 2007), which leads to the speculation of phosphorylation as a cue to aid in discriminating target membranes through reversion of a positive net charge. Mitochondrial targeting is also dependent on the distance between TMD and CTE (Marty et al., 2014). Another potent indicator of plant OMM TA proteins is found in the dibasic motif adjacent to the C-terminal part of the TMD (Marty et al., 2014).
In mammals and yeast, no unambiguous amino acid motif for TA protein targeting had been found so far. A recent study in Arabidopsis, however, showed that some plastid outer envelope membrane (OEM) TA proteins harbor a CTE with an RK/ST sequence motif. OEP7.2, which localizes to the OEM, was used for swapping experiments with CTEs of other TA proteins with and without this motif. Only CTE with RK/ST motifs was functionally interchangeable. Thus, they concluded that for a subset of OEM TA proteins, there is a conserved element for plastid targeting (Moog, 2019; Teresinski et al., 2019).
Overall, it seems that the length and hydrophobicity of the TMD with a combination of charge dictates the localization of TA proteins within the cell, while plant OEM TA proteins with a specific motif might be more of an exception.
However, dually targeted TA proteins such as AtPMD1 to mitochondria and peroxisomes (Aung and Hu, 2011), AtPAP2 to chloroplast and mitochondria (Sun et al., 2012), or proteins which display multiple targeting [chloroplast, mitochondria, and peroxisomes] as AtFIS1A (Ruberti et al., 2014), highlight that topogenic information (alone) cannot suffice to discern targeting routes. Nonetheless, the specificity of targeting motifs is interlinked with the binding properties of different chaperones that shepherd their substrate to their destination membrane.
Potentially as a consequence of ambiguous signals, mistargeting occurs against which fail-safe mechanisms evolved: in yeast, the AAA-ATPase Msp1 (Okreglak and Walter, 2014; Wang et al., 2020) recognizes TA proteins wrongly delivered to the OMM and either hands them over for proteasome-mediated degradation or extracts them for correct rerouting (thoroughly reviewed in Wang and Walter, 2020). While such dislocase function also exists in animals (ATAD1, Chen et al., 2014a) a similar function has not been found in plants where a large number of AAA-ATPases exist (Ogura and Wilkinson, 2001).
Insertion into chloroplasts
The translocation mechanism of TA proteins into the OEM of chloroplasts is currently not well understood. Unassisted insertion dependent on the lipid composition of the membrane and the TA protein CTE has been observed (Qbadou et al., 2003; Pedrazzini, 2009; Dhanoa et al., 2010). Additionally, a cytosolic OEM chaperone, ankyrin repeat-containing protein (AKR2a) was found to play a role for the targeting of some TA proteins to chloroplasts and the delivery of dual-targeted ascorbate peroxidase (APX3) to peroxisomes (Bae et al., 2008; Shen et al., 2010). This observation would argue against its role as a specific chloroplast TA protein insertion factor indicating AKR2a as a rather unspecific chaperone.
Recently, another putative chaperone was detected in the green algae Chlamydomonas reinhardtii. Here, an arsenite transporter (CrArsA1) binds Toc34 and delivers it to chloroplasts (Maestre-Reyna et al., 2017). Intriguingly, two ArsA paralogous genes can be found in the C. reinhardtii genome, CrArsA1 and CrArsA2. Both are homologs of the cytosolic targeting factors, TRC40 and Get3 (Formighieri et al., 2013). CrArsA1 and CrArsA2 have a discrete ligand preference, with CrArsA1 supposedly carrying TA proteins to the OEM and CrArsA2 to the ER (Maestre-Reyna et al., 2017). The subcellular localization of ArsA1 homologs in chlorophytes is a matter of debate. While Formighieri et al. (2013) propose CrArsA1 to be cytoplasmic, its protein sequence clearly features an organellar transit peptide at the N-terminus (Xing et al., 2017; Farkas et al., 2019). Its sequence also suggests a high similarity to other GET3bc clade homologs of Archaeplastida, which are also organellar localized (Xing et al., 2017; Lin et al., 2019). In addition, a recent affinity purification mass spectrometry of the chloroplastic ribosome interactome of Chlamydomonas reinhardtii revealed CrArsA1 lending further support to its stroma rather than cytosolic localization (Westrich et al., 2021).
The Arabidopsis homolog of CrArsA1 is AtGET3b, which also features an N-terminal transit peptide and localizes to the stroma of chloroplasts (Xing et al., 2017). However, localizing within the stroma precludes a possible involvement in TA protein insertion at the OEM. One could speculate that AtGET3b is involved in TA protein targeting the inner envelope membrane or thylakoids (Anderson et al., 2019; Bodensohn et al., 2019). While AtGET3b does not bind to ER-resident AtGET1 (Xing et al., 2017), interaction assays should first elucidate whether AtGET3b could potentially bind to the Get1 ortholog Alb3 (At2g28800) or Alb4 (At1g24490), which facilitates membrane protein biogenesis in endosymbiontic organelles (Anghel et al., 2017; McDowell et al., 2021).
Insertion into mitochondria
Mitochondria have a small semi-autonomous genome, although most of the mitochondrial proteins are encoded by the nuclear genome, synthesized by cytosolic ribosomes, and transported post-translationally into the mitochondria (Neupert, 1997; Pfanner and Geissler, 2001). There are many mitochondrial TA proteins, yet the pathway(s) responsible for their insertion are not clear. It had been reported that insertion of mitochondrial TA proteins depended on the unique lipid composition of the OMM, especially the ergosterol levels (Setoguchi et al., 2006; Kemper et al., 2008; Krumpe et al., 2012) and with the help of peroxisome import factor Pex19 (Cichocki et al., 2018). Moreover, translocation of TA proteins was moderately affected with hampered mitochondrial import complex (MIM) or Tom20 receptors (Thornton et al., 2010; Doan et al., 2020). It is conceivable that Tom20 acts as a receptor, while the MIM complex mediates insertion (Drwesh and Rapaport, 2020). Also, N-terminally GFP-labeled OMM protein Mcp3 mislocalizes to the ER in wild-type yeast but not in get knockout strains (Vitali et al., 2018). Apparently, when the mitochondrial import is compromised, TA proteins intended for the OMM are mistargeted to the ER membrane by the GET pathway. This implies that in yeast insertion pathways may compete for client delivery.
AtGet3c, a homolog of Get3 is found in the mitochondrial matrix of Arabidopsis. Whether or not it is involved in TA protein insertion into the inner membrane of mitochondria is currently unknown. However, its loss-of-function line seems to show no obvious growth or cellular defects (Xing et al., 2017). It was speculated that the GET3c variants are Brassicaceae-specific, while some GET3b homologs (that localize to chloroplasts in Arabidopsis) were mitochondria localized in the Fabidae (Bodensohn et al., 2019). Similar to chloroplasts, a Get1 ortholog is present in the mitochondrial inner membrane (Oxa1). As discussed above, the GET3bc clade lacks the GET1 binding motif (Anghel et al., 2017; Farkas et al., 2019) and has not been demonstrated to interact with or depend on Oxa1 so far.
Insertion into peroxisomes
Peroxisomes are single membrane, multifunctional organelles with essential roles in development such as scavenging of reactive oxygen species or peroxides, photorespiration, glycolate cycle, and fatty acid β-oxidation (Aung and Hu, 2011; Kao et al., 2018). In contrast to chloroplasts and mitochondria, they neither contain DNA nor possess protein-synthesizing machinery. Peroxisomes are discussed to be ER-derived and early acting peroxin (PEX) proteins such as PEX3, PEX16, and PEX19 help in the peroxisomal genesis but also a division by fission is possible (Kao et al., 2018). Therefore, the acquisition of protein delivery machineries is of great importance for peroxisomal identity.
In mammals and yeast, it was shown that peroxisomal-targeted TA proteins can take two distinct routes, (1) directly from the cytosol or (2) via the ER (Borgese et al., 2019). Both ways depend on the peroxisomal import proteins Pex19 and Pex3. Cytosolic Pex19 binds nascent peroxisomal TA proteins within a hydrophobic groove, thereby stabilizing them. Recognition occurs via the TMD and basic CTE of the TA proteins (Halbach et al., 2006; Yagita et al., 2013; Chen et al., 2014b). The binding of its membrane receptor Pex3 leads to direct insertion into the membrane (Cichocki et al., 2018).
ER-dependent insertion is partially carried out by the GET machinery. For instance, yeast Pex15 is ER-inserted via the GET pathway (Schuldiner et al., 2008; van der Zand et al., 2010). Here, a specialized subdomain within the ER is formed, the so-called peroxisomal ER (pER). Localized budding of peroxisomal vesicles carrying TA proteins and subsequent fusion to existing peroxisomes requires Pex3, Pex19, ATP, and additional yet unidentified cytosolic factors (van der Zand et al., 2010; Lam et al., 2011). Studies on these events proposed a dual functionality of Pex3. Its luminal sequence harbors a sorting signal for delivering Pex3 to the pER, whereas the TMD of Pex3 is important for later directing of the vesicles to peroxisomes (Tam et al., 2005; Fakieh et al., 2013; Chio et al., 2017).
In plants, the peroxisomal-targeted TA protein APX was shown to insert post-translationally dependent on ATP, Hsp70, and an additional, unknown receptor via pER (Mullen and Trelease, 2000). Unassisted insertion can also be observed for some peroxisomal TA proteins as MDAR4 (Lisenbee et al., 2005; Abell and Mullen, 2011). A conserved mechanism for translocation of plant TA proteins as seen in Opisthokonts is conceivable; however, exact information is lacking (Cross et al., 2016).
Future perspectives
The most puzzling discovery in TA protein insertion in plants is certainly with a rather mild phenotype associated with GET loss-of-function lines (see Advances). How can this be reconciled with the notion that the GET pathway is universally conserved and acts as the textbook pathway for TA protein insertion into the ER? A nonlethal phenotype of a plant that lacks a general membrane insertion pathway of an important subclass of membrane proteins would surely lead to more pleiotropic growth defects. Failure to insert TA proteins—among them the trafficking facilitating SNARE proteins which are required for polar growth and cytokinesis—should lead to embryo lethality “at best”, or developmental arrests in earlier stages such as compromised pollen tube growth. Their absence suggests one or more backup system(s) in place. Existence, identity, and conservation of such systems (e.g. SND, EMC, Table 1) are a major avenue for future research as well as the identification of further GET pathway substrates which may also aid in understanding additional function(s) of a plant GET pathway.
Another obvious question is the precise targeting and distinction of TA proteins to their various destination membranes. A complex combination of physicochemical properties or as in the case of some plant OEM TA proteins, a specific motif (Teresinski et al., 2019) might be the answer. Yet, how exactly dual-targeted TA proteins are sorted is still not clear and a simple solution is unlikely.
A puzzling observation is the additional GET3 paralogs in Archaeplastida (Xing et al., 2017; Farkas et al., 2019). While clade a GET3 appears to be functionally related to yeast Get3 and mammalian TRC40, the roles of clade bc GET3 remain elusive. All plants likely possess at least one copy of a chloroplast GET3b which might be involved in TA protein targeting to the inner envelope or thylakoids. However, the mitochondrial GET3c seems absent in most plant species, which begs questions about its functional role and evolution (Bodensohn et al., 2019).
These are just some points that require addressing in future research and there is a lot to learn in terms of TA protein insertion in plants (see Outstanding Questions). Other fundamental homeostatic pathways such as cytokinesis (Jurgens, 2005) have significantly diverged among Opisthokonts and Archaeplastida—an evolutionary divide of more than 1.5 billion years—and validated the importance of research into different model species. Nonetheless, evidence for functional conservation of important fundamental processes such as membrane protein insertion remains limited in plants. The vast amount of data gained from research in single-celled models such as bacteria, yeast, and cell culture should be used to inform hypothesis-driven research in plants. Especially, the model plant Arabidopsis and the palette of modern genomic tools established therein will allow a more organismal-focused, phenotypic analyses of these pathways in the context of a multicellular organism.
OUTSTANDING QUESTIONS
Which additional pathways for TA protein insertion exist in plants?
What alternative functions have evolved for the GET pathway components in Arabidopsis or more generally in plants?
Why did Archaeplastida evolve organellar variants of the GET3 ATPase and what is (are) their function(s)?
Is a post-translational pretargeting complex conserved in archaeplastida?
Acknowledgments
We thank Sonja Mehlhorn for detailed comments on this review.
Funding
This work was supported by a PhD grant from the Carl-Zeiss Stiftung (to D.G.M) and grants from the Deutsche Forschungsgemeinschaft (DFG GR4251/2-1 and SFB1101-A06; to C.G.).
Conflict of interest statement. None declared.
D.G.M. and C.G. wrote the manuscript with input from L.Y.A.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Christopher Grefen (christopher.grefen@rub.de).
References
- Abell BM, Mullen RT (2011) Tail-anchored membrane proteins: exploring the complex diversity of tail-anchored-protein targeting in plant cells. Plant Cell Rep 30:137–151 [DOI] [PubMed] [Google Scholar]
- Abell BM, Rabu C, Leznicki P, Young JC, High S (2007) Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J Cell Sci 120:1743–1751 [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Walter P, Blobel G (1982) Signal recognition protein is required for the integration of acetylcholine receptor delta subunit, a transmembrane glycoprotein, into the endoplasmic reticulum membrane. J Cell Biol 93:501–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Singhal R, Fernandez DE (2019) Membrane-specific targeting of tail-anchored proteins SECE1 and SECE2 within chloroplasts. Front Plant Sci 10:1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anghel SA, McGilvray PT, Hegde RS, Keenan RJ (2017) Identification of Oxa1 homologs operating in the eukaryotic endoplasmic reticulum. Cell Rep 21:3708–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asseck LY, Mehlhorn DG, Monroy JR, Ricardi MM, Breuninger H, Wallmeroth N, Berendzen KW, Nowrousian M, Xing S, Schwappach B, et al. (2021) Endoplasmic reticulum membrane receptors of the GET pathway are conserved throughout eukaryotes. Proc Natl Acad Sci U S A 118: e2017636118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K, Hu J (2011) The Arabidopsis tail-anchored protein PEROXISOMAL AND MITOCHONDRIAL DIVISION FACTOR1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell 23:4446–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviram N, Ast T, Costa EA, Arakel EC, Chuartzman SG, Jan CH, Hassdenteufel S, Dudek J, Jung M, Schorr S, et al. (2016) The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 540:134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W, Lee YJ, Kim DH, Lee J, Kim S, Sohn EJ, Hwang I (2008) AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat Cell Biol 10:220–227 [DOI] [PubMed] [Google Scholar]
- Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T (2003) Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem 278:8219–8223 [DOI] [PubMed] [Google Scholar]
- Bodensohn US, Simm S, Fischer K, Jaschke M, Gross LE, Kramer K, Ehmann C, Rensing SA, Ladig R, Schleiff E (2019) The intracellular distribution of the components of the GET system in vascular plants. Biochim Biophys Acta Mol Cell Res 1866:1650–1662 [DOI] [PubMed] [Google Scholar]
- Borgese N (2020) Searching for remote homologs of Caml among eukaryotes. Traffic 21:647–658 [DOI] [PubMed] [Google Scholar]
- Borgese N, Brambillasca S, Colombo S (2007) How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol 19:368–375 [DOI] [PubMed] [Google Scholar]
- Borgese N, Colombo S, Pedrazzini E (2003) The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J Cell Biol 161:1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Coy-Vergara J, Colombo SF, Schwappach B (2019) The ways of tails: the GET Pathway and more. Protein J 38:289–305 [DOI] [PubMed] [Google Scholar]
- Borgese N, Gazzoni I, Barberi M, Colombo S, Pedrazzini E (2001) Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol Biol Cell 12:2482–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho HJF, Del Bondio A, Maltecca F, Colombo SF, Borgese N (2019) The WRB subunit of the Get3 receptor is required for the correct integration of its partner CAML into the ER. Sci Rep 9:11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson J, McKenna M, Hassdenteufel S, Aviram N, Zimmerman R, High S (2017) Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J Cell Sci 130:3851–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celebrating 20 years of cell biology. (2019) Nat Cell Biol 21:1. [DOI] [PubMed] [Google Scholar]
- Chang YW, Chuang YC, Ho YC, Cheng MY, Sun YJ, Hsiao CD, Wang C (2010) Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem 285:9962–9970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Gonzalez GM, Clemons WM Jr. (2011) A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. J Biol Chem 286:34325–34334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pieuchot L, Loh RA, Yang J, Kari TM, Wong JY, Jedd G (2014b) Hydrophobic handoff for direct delivery of peroxisome tail-anchored proteins. Nat Commun 5:5790. [DOI] [PubMed] [Google Scholar]
- Chen YC, Umanah GK, Dephoure N, Andrabi SA, Gygi SP, Dawson TM, Dawson VL, Rutter J (2014a) Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J 33:1548–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio US, Cho H, Shan SO (2017) Mechanisms of tail-anchored membrane protein targeting and insertion. Annu Rev Cell Dev Biol 33:417–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Shan SO (2018) Substrate relay in an Hsp70-cochaperone cascade safeguards tail-anchored membrane protein targeting. EMBO J 37: e99264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Shim WJ, Liu Y, Shan SO (2021) J-domain proteins promote client relay from Hsp70 during tail-anchored membrane protein targeting. J Biol Chem 296:100546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki BA, Krumpe K, Vitali DG, Rapaport D (2018) Pex19 is involved in importing dually targeted tail-anchored proteins to both mitochondria and peroxisomes. Traffic 19:770–785 [DOI] [PubMed] [Google Scholar]
- Costello JL, Castro IG, Camoes F, Schrader TA, McNeall D, Yang J, Giannopoulou EA, Gomes S, Pogenberg V, Bonekamp NA, et al. (2017) Predicting the targeting of tail-anchored proteins to subcellular compartments in mammalian cells. J Cell Sci 130:1675–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA (2018) Hsp70 at the membrane: driving protein translocation. BMC Biol 16:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross LL, Ebeed HT, Baker A (2016) Peroxisome biogenesis, protein targeting mechanisms and PEX gene functions in plants. Biochim Biophys Acta 1863:850–862 [DOI] [PubMed] [Google Scholar]
- Culver JA, Mariappan M (2021) Deubiquitinases USP20/33 promote the biogenesis of tail-anchored membrane proteins. J Cell Biol 220: e202004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Dotsch V, Sinning I (2013) Endoplasmic reticulum targeting and insertion of tail-anchored membrane proteins by the GET pathway. Cold Spring Harb Perspect Biol 5:a013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R (1991) Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349:806–808 [DOI] [PubMed] [Google Scholar]
- Dhanoa PK, Richardson LG, Smith MD, Gidda SK, Henderson MP, Andrews DW, Mullen RT (2010) Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS One 5:e10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan KN, Grevel A, Martensson CU, Ellenrieder L, Thornton N, Wenz LS, Opalinski L, Guiard B, Pfanner N, Becker T (2020) The mitochondrial import complex MIM functions as main translocase for alpha-helical outer membrane proteins. Cell Rep 31:107567. [DOI] [PubMed] [Google Scholar]
- Drwesh L, Rapaport D (2020) Biogenesis pathways of alpha-helical mitochondrial outer membrane proteins. Biol Chem 401:677–686 [DOI] [PubMed] [Google Scholar]
- Duncan O, van der Merwe MJ, Daley DO, Whelan J (2013) The outer mitochondrial membrane in higher plants. Trends Plant Sci 18:207–217 [DOI] [PubMed] [Google Scholar]
- Fakieh MH, Drake PJ, Lacey J, Munck JM, Motley AM, Hettema EH (2013) Intra-ER sorting of the peroxisomal membrane protein Pex3 relies on its luminal domain. Biol Open 2:829–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas A, De Laurentiis EI, Schwappach B (2019) The natural history of Get3-like chaperones. Traffic 20:311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formighieri C, Cazzaniga S, Kuras R, Bassi R (2013) Biogenesis of photosynthetic complexes in the chloroplast of Chlamydomonas reinhardtii requires ARSA1, a homolog of prokaryotic arsenite transporter and eukaryotic TRC40 for guided entry of tail-anchored proteins. Plant J 73:850–861 [DOI] [PubMed] [Google Scholar]
- Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, Fregno I, Galli C, Loi M, Solda T, et al. (2016) Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol 18:1173–1184 [DOI] [PubMed] [Google Scholar]
- Gilmore R, Blobel G, Walter P (1982a) Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol 95:463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R, Walter P, Blobel G (1982b) Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol 95:470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristick HB, Rao M, Chartron JW, Rome ME, Shan SO, Clemons WM Jr. (2014) Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4. Nat Struct Mol Biol 21:437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guna A, Hegde RS (2018) Transmembrane domain recognition during membrane protein biogenesis and quality control. Curr Biol 28:R498–R511 [DOI] [PubMed] [Google Scholar]
- Guna A, Volkmar N, Christianson JC, Hegde RS (2018) The ER membrane protein complex is a transmembrane domain insertase. Science 359:470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach A, Landgraf C, Lorenzen S, Rosenkranz K, Volkmer-Engert R, Erdmann R, Rottensteiner H (2006) Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J Cell Sci 119:2508–2517 [DOI] [PubMed] [Google Scholar]
- Hassdenteufel S, Sicking M, Schorr S, Aviram N, Fecher-Trost C, Schuldiner M, Jung M, Zimmermann R, Lang S (2017) hSnd2 protein represents an alternative targeting factor to the endoplasmic reticulum in human cells. FEBS Lett 591:3211–3224 [DOI] [PubMed] [Google Scholar]
- Hegde RS, Keenan RJ (2011) Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol 12:787–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Ye H, Cui Y, Jiang L (2020) AtSec62 is critical for plant development and is involved in ER-phagy in Arabidopsis thaliana. J Integr Plant Biol 62:181–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis AJ, Page KR, Guna A, Voorhees RM (2020) Differential modes of orphan subunit recognition for the WRB/CAML complex. Cell Rep 30: 3691–3698, e3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N, Powis K, High S (2013) Post-translational translocation into the endoplasmic reticulum. Biochim Biophys Acta 1833:2403–2409 [DOI] [PubMed] [Google Scholar]
- Jurgens G (2005) Cytokinesis in higher plants. Annu Rev Plant Biol 56:281–299 [DOI] [PubMed] [Google Scholar]
- Kao YT, Gonzalez KL, Bartel B (2018) Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol 176:162–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper C, Habib SJ, Engl G, Heckmeyer P, Dimmer KS, Rapaport D (2008) Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J Cell Sci 121:1990–1998 [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V, Shaw R, Mukherjee J, Bowsher CG, Harrison AM, Abell BM (2009) Subcellular distribution of tail-anchored proteins in Arabidopsis. Traffic 10:1753–1764 [DOI] [PubMed] [Google Scholar]
- Krumpe K, Frumkin I, Herzig Y, Rimon N, Ozbalci C, Brugger B, Rapaport D, Schuldiner M (2012) Ergosterol content specifies targeting of tail-anchored proteins to mitochondrial outer membranes. Mol Biol Cell 23:3927–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K (2008) SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 133:440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SK, Yoda N, Schekman R (2011) A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci U S A 108:E51–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jurgens G (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol 139:1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim DH, Hwang I (2014) Specific targeting of proteins to outer envelope membranes of endosymbiotic organelles, chloroplasts, and mitochondria. Front Plant Sci 5:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P, High S (2012) SGTA antagonizes BAG6-mediated protein triage. Proc Natl Acad Sci U S A 109:19214–19219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P, Roebuck QP, Wunderley L, Clancy A, Krysztofinska EM, Isaacson RL, Warwicker J, Schwappach B, High S (2013) The association of BAG6 with SGTA and tail-anchored proteins. PLoS One 8:e59590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kabbage M, Liu W, Dickman MB (2016) Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell 28:233–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Vollrath MA, Mangosing S, Shen J, Cardenas E, Corey DP (2016) The zebrafish pinball wizard gene encodes WRB, a tail-anchored-protein receptor essential for inner-ear hair cells and retinal photoreceptors. J Physiol 594:895–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TW, Chen CC, Wu SM, Chang YC, Li YC, Su YW, Hsiao CD, Chang HY (2019) Structural analysis of chloroplast tail-anchored membrane protein recognition by ArsA1. Plant J 99:128–143 [DOI] [PubMed] [Google Scholar]
- Lipka V,, Kwon C,, Panstruga R (2007). SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu Rev Cell Dev Biol 23:147–174 [DOI] [PubMed] [Google Scholar]
- Lisenbee CS, Lingard MJ, Trelease RN (2005) Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J 43:900–914 [DOI] [PubMed] [Google Scholar]
- Maestre-Reyna M, Wu SM, Chang YC, Chen CC, Maestre-Reyna A, Wang AH, Chang HY (2017) In search of tail-anchored protein machinery in plants: reevaluating the role of arsenite transporters. Sci Rep 7:46022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio C, Barbante A, Ferro F, Frigerio L, Pedrazzini E (2007) Intracellular sorting of the tail-anchored protein cytochrome b5 in plants: a comparative study using different isoforms from rabbit and Arabidopsis. J Exp Bot 58:1365–1379 [DOI] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS (2010) A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466:1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M, Mateja A, Dobosz M, Bove E, Hegde RS, Keenan RJ (2011) The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature 477:61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty NJ, Teresinski HJ, Hwang YT, Clendening EA, Gidda SK, Sliwinska E, Zhang D, Miernyk JA, Brito GC, Andrews DW, et al. (2014) New insights into the targeting of a subset of tail-anchored proteins to the outer mitochondrial membrane. Front Plant Sci 5:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A, Paduch M, Chang HY, Szydlowska A, Kossiakoff AA, Hegde RS, Keenan RJ (2015) Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 347:1152–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, Hegde RS, Keenan RJ (2009) The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461:361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell MA, Heimes M, Fiorentino F, Mehmood S, Farkas A, Coy-Vergara J, Wu D, Bolla JR, Schmid V, Heinze R, et al. (2020) Structural basis of tail-anchored membrane protein biogenesis by the GET insertase complex. Mol Cell 80:72–86 e77 [DOI] [PubMed] [Google Scholar]
- McDowell MA, Heimes M, Sinning I (2021) Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily. Nat Struct Mol Biol 28:234–239 [DOI] [PubMed] [Google Scholar]
- Mehlhorn DG, Wallmeroth N, Berendzen KW, Grefen C (2018) 2in1 vectors improve in planta BiFC and FRET analyses. Methods Mol Biol 1691:139–158 [DOI] [PubMed] [Google Scholar]
- Mitterreiter MJ, Bosch FA, Brylok T, Schwenkert S (2020) The ER luminal C-terminus of AtSec62 is critical for male fertility and plant growth in Arabidopsis thaliana. Plant J 101:5–17 [DOI] [PubMed] [Google Scholar]
- Moog D (2019) Higher complexity requires higher accuracy: tail-anchored protein targeting to the outer envelope membrane of plant plastids via a specific C-terminal motif. Plant Cell Physiol 60:489–491 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Ho YS, Swiatek PJ, Rosen BP, Bhattacharjee H (2006) Targeted disruption of the mouse Asna1 gene results in embryonic lethality. FEBS Lett 580:3889–3894 [DOI] [PubMed] [Google Scholar]
- Mullen RT, Trelease RN (2000) The sorting signals for peroxisomal membrane-bound ascorbate peroxidase are within its C-terminal tail. J Biol Chem 275:16337–16344 [DOI] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66:863–917 [DOI] [PubMed] [Google Scholar]
- Neveu E, Khalifeh D, Salamin N, Fasshauer D (2020) Prototypic SNARE proteins are encoded in the genomes of Heimdallarchaeota, potentially bridging the gap between the prokaryotes and eukaryotes. Curr Biol 30:2468–2480 e2465 [DOI] [PubMed] [Google Scholar]
- Norlin S, Parekh VS, Naredi P, Edlund H (2016) Asna1/TRC40 controls beta-cell function and endoplasmic reticulum homeostasis by ensuring retrograde transport. Diabetes 65:110–119 [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R (1980) Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21:205–215 [DOI] [PubMed] [Google Scholar]
- Ogg SC, Walter P (1995) SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell 81:1075–1084 [DOI] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ (2001) AAA+ superfamily ATPases: common structure–diverse function. Genes Cells 6:575–597 [DOI] [PubMed] [Google Scholar]
- Okreglak V, Walter P (2014) The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci U S A 111:8019–8024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P, Simm S, Blaumeiser A, Scharf KD, Fragkostefanakis S, Mirus O, Schleiff E (2013) The protein translocation systems in plants—composition and variability on the example of Solanum lycopersicum. BMC Genomics 14:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Chartron JW, Frydman J (2014) Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol 21:1100–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini E (2009) Tail-anchored proteins in plants. J Plant Biol 52:88–101 [Google Scholar]
- Pedrazzini E, Villa A, Borgese N (1996) A mutant cytochrome b5 with a lengthened membrane anchor escapes from the endoplasmic reticulum and reaches the plasma membrane. Proc Natl Acad Sci U S A 93:4207–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Geissler A (2001) Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol 2:339–349 [DOI] [PubMed] [Google Scholar]
- Qbadou S, Tien R, Soll J, Schleiff E (2003) Membrane insertion of the chloroplast outer envelope protein, Toc34: constrains for insertion and topology. J Cell Sci 116:837–846 [DOI] [PubMed] [Google Scholar]
- Rabu C, Wipf P, Brodsky JL, High S (2008) A precursor-specific role for Hsp40/Hsc70 during tail-anchored protein integration at the endoplasmic reticulum. J Biol Chem 283:27504–27513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Okreglak V, Chio US, Cho H, Walter P, Shan SO (2016) Multiple selection filters ensure accurate tail-anchored membrane protein targeting. Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Coller J (2015) Pausing on polyribosomes: make way for elongation in translational control. Cell 163:292–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti C, Costa A, Pedrazzini E, Lo Schiavo F, Zottini M (2014) FISSION1A, an Arabidopsis tail-anchored protein, is localized to three subcellular compartments. Mol Plant 7:1393–1396 [DOI] [PubMed] [Google Scholar]
- Sabatini DD, Blobel G (1970) Controlled proteolysis of nascent polypeptides in rat liver cell fractions. II. Location of the polypeptides in rough microsomes. J Cell Biol 45:146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE (2000) YidC mediates membrane protein insertion in bacteria. Nature 406:637–641 [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS (2008) The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134:634–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger R, Muller NC, Schmitt MJ, Soll J, Schwenkert S (2012) AtTPR7 is a chaperone-docking protein of the Sec translocon in Arabidopsis. J Cell Sci 125:5196–5207 [DOI] [PubMed] [Google Scholar]
- Schweiger R, Schwenkert S (2013) AtTPR7 as part of the Arabidopsis Sec post-translocon. Plant Signal Behav 8[AQ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi K, Otera H, Mihara K (2006) Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J 25:5635–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Hegde RS (2011) Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol 27:25–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Rodrigo-Brenni MC, Kivlen MH, Hegde RS (2017) Mechanistic basis for a molecular triage reaction. Science 355:298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G, Kuppu S, Venkataramani S, Wang J, Yan J, Qiu X, Zhang H (2010) Ankyrin repeat-containing protein 2A is an essential molecular chaperone for peroxisomal membrane-bound ascorbate peroxidase 3 in Arabidopsis. Plant Cell 22:811–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson PJ, Schwappach B, Dohlman HG, Isaacson RL (2010) Structures of Get3, Get4, and Get5 provide new models for TA membrane protein targeting. Structure 18:897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Raden D, Mandon EC, Gilmore R (2000) Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell 100:333–343 [DOI] [PubMed] [Google Scholar]
- Srivastava R, Zalisko BE, Keenan RJ, Howell SH (2017) The GET system inserts the tail-anchored protein, SYP72, into endoplasmic reticulum membranes. Plant Physiol 173:1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S, Hegde RS (2007) Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128:1147–1159 [DOI] [PubMed] [Google Scholar]
- Stefer S, Reitz S, Wang F, Wild K, Pang YY, Schwarz D, Bomke J, Hein C, Lohr F, Bernhard F, et al. (2011) Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science 333:758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway CJ, Chartron JW, Zaslaver M, Clemons WM Jr. (2009) Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci U S A 106:14849–14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway CJ, Rome ME, Clemons WM Jr. (2012) Tail-anchor targeting by a Get3 tetramer: the structure of an archaeal homologue. EMBO J 31:707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Carrie C, Law S, Murcha MW, Zhang R, Law YS, Suen PK, Whelan J, Lim BL (2012) AtPAP2 is a tail-anchored protein in the outer membrane of chloroplasts and mitochondria. Plant Signal Behav 7:927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam YY, Fagarasanu A, Fagarasanu M, Rachubinski RA (2005) Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J Biol Chem 280:34933–34939 [DOI] [PubMed] [Google Scholar]
- Teresinski HJ, Gidda SK, Nguyen TND, Howard NJM, Porter BK, Grimberg N, Smith MD, Andrews DW, Dyer JM, Mullen RT (2019) An RK/ST C-terminal motif is required for targeting of OEP7.2 and a subset of other Arabidopsis tail-anchored proteins to the plastid outer envelope membrane. Plant Cell Physiol 60:516–537 [DOI] [PubMed] [Google Scholar]
- Thornton N, Stroud DA, Milenkovic D, Guiard B, Pfanner N, Becker T (2010) Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of alpha-helical outer membrane proteins. J Mol Biol 396:540–549 [DOI] [PubMed] [Google Scholar]
- van der Zand A, Braakman I, Tabak HF (2010) Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol Biol Cell 21:2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardi F, Lorenz H, Dobberstein B (2011) WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J Cell Sci 124:1301–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardi F, Stephan M, Clancy A, Janshoff A, Schwappach B (2014) WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS One 9:e85033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali DG, Sinzel M, Bulthuis EP, Kolb A, Zabel S, Mehlhorn DG, Figueiredo Costa B, Farkas A, Clancy A, Schuldiner M, et al. (2018) The GET pathway can increase the risk of mitochondrial outer membrane proteins to be mistargeted to the ER. J Cell Sci 131: jcs211110 [DOI] [PubMed] [Google Scholar]
- Vogl C, Panou I, Yamanbaeva G, Wichmann C, Mangosing SJ, Vilardi F, Indzhykulian AA, Pangrsic T, Santarelli R, Rodriguez-Ballesteros M, et al. (2016) Tryptophan-rich basic protein (WRB) mediates insertion of the tail-anchored protein otoferlin and is required for hair cell exocytosis and hearing. EMBO J 35:2536–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmar N, Thezenas ML, Louie SM, Juszkiewicz S, Nomura DK, Hegde RS, Kessler BM, Christianson JC (2019) The ER membrane protein complex promotes biogenesis of sterol-related enzymes maintaining cholesterol homeostasis. J Cell Sci 132: jcs223453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Gerstein M, Steitz TA, Moore PB (2006) The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol 360:893–906 [DOI] [PubMed] [Google Scholar]
- Voth W, Schick M, Gates S, Li S, Vilardi F, Gostimskaya I, Southworth DR, Schwappach B, Jakob U (2014) The protein targeting factor Get3 functions as ATP-independent chaperone under oxidative stress conditions. Mol Cell 56:116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G (1981) Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol 91:557–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Brown EC, Mak G, Zhuang J, Denic V (2010) A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell 40:159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Chan C, Weir NR, Denic V (2014) The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature 512:441–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Whynot A, Tung M, Denic V (2011) The mechanism of tail-anchored protein insertion into the ER membrane. Mol Cell 43:738–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Myasnikov A, Pan X, Walter P (2020) Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction. Elife 9: e54031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Walter P (2020) Msp1/ATAD1 in protein quality control and regulation of synaptic activities. Annu Rev Cell Dev Biol 36:141–164 [DOI] [PubMed] [Google Scholar]
- Wereszczynski J, McCammon JA (2012) Nucleotide-dependent mechanism of Get3 as elucidated from free energy calculations. Proc Natl Acad Sci U S A 109:7759–7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrich LD, Gotsmann VL, Herkt C, Ries F, Kazek T, Trosch R, Armbruster L, Muhlenbeck JS, Ramundo S, Nickelsen J, et al. (2021) The versatile interactome of chloroplast ribosomes revealed by affinity purification mass spectrometry. Nucleic Acids Res 49:400–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner WT (2013) Profile of Thomas Sudhof, James Rothman, and Randy Schekman, 2013 Nobel Laureates in Physiology or Medicine. Proc Natl Acad Sci U S A 110:18349–18350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Cabanos C, Rapoport TA (2019) Structure of the post-translational protein translocation machinery of the ER membrane. Nature 566:136–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Mehlhorn DG, Wallmeroth N, Asseck LY, Kar R, Voss A, Denninger P, Schmidt VA, Schwarzlander M, Stierhof YD, et al. (2017) Loss of GET pathway orthologs in Arabidopsis thaliana causes root hair growth defects and affects SNARE abundance. Proc Natl Acad Sci U S A 114:E1544–E1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita Y, Hiromasa T, Fujiki Y (2013) Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J Cell Biol 200:651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Sakisaka T (2012) Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol Cell 48:387–397 [DOI] [PubMed] [Google Scholar]
- Zalisko BE, Chan C, Denic V, Rock RS, Keenan RJ (2017) Tail-anchored protein insertion by a single Get1/2 heterodimer. Cell Rep 20:2287–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Chung KP, Jiang L (2017) Targeting tail-anchored proteins into plant organelles. Proc Natl Acad Sci U S A 114:1762–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]