Abstract
We examined 11 naturally occurring isolates of Ehrlichia equi in horses and two human granulocytic ehrlichiosis agent isolates in California for sequence diversity in three genes. Ehrlichia equi isolates were from Sierra (n = 6), Mendocino (n = 3), Sonoma (n = 1), and Marin (n = 1) counties, and human granulocytic ehrlichiosis (HGE) agent isolates were obtained from Humboldt county. PCR with specific primers for 16S rRNA, 444 Ep-ank and groESL heat shock operon genes successfully produced amplicons for all 13 clinical samples. The 444 Ep-ank gene of the HGE agent and E. equi isolates from northern California is different from the eastern U.S. isolates BDS and USG3. The translated amino acid sequence of the groESL heat shock operon gene fragment is identical among E. equi, the HGE agent, and E. phagocytophila, with the exception of the northern Californian equine CASOLJ isolate. Microheterogeneity was observed in the 16S rRNA gene sequences of HGE agent and E. equi isolates from northern California. These results suggest that E. equi and the HGE agent found in California are similar or identical but may differ from the isolates of equine and human origin found in the eastern United States.
Human granulocytic ehrlichiosis (HGE) is a severe or potentially fatal rickettsial infection which is emerging in the upper midwestern and northeastern United States (1, 3, 5). The disease is characterized by fever, severe headaches, and myalgia, as well as occasionally other signs, including respiratory compromise, gastrointestinal disturbance, organ failure, and possibly increased susceptibility to opportunistic infections such as candidiasis (6). There is a greater risk of severe disease in older patients and the case fatality rate has been estimated to be as high as 5% in some populations (7). HGE is caused by an unnamed ehrlichial pathogen which is serologically and morphologically indistinguishable from E. equi, the agent of equine granulocytic ehrlichiosis (EGE) (14).
EGE typically presents as fever, dependent edema, icterus, and ataxia (13). Infections were initially described in horses in northern California but are also found throughout the United States in habitats infested with the tick vectors Ixodes pacificus west of the Rocky Mountains and I. scapularis in the eastern United States (4, 11). The same ticks have been found to be vectors of the HGE agent. Experimental inoculation of horses with the HGE strain produced a clinical syndrome which was indistinguishable from naturally occurring EGE (16).
We and others have hypothesized that the agent of HGE is in fact Ehrlichia equi. This hypothesis was tested in the current study by comparing the sequences of three genes from horse isolates and two recent human cases in northern California (8).
MATERIALS AND METHODS
Field samples.
Table 1 shows the origins of the E. equi isolates obtained from Sierra, Mendocino, Sonoma, and Marin counties and the HGE agent isolates obtained from Humboldt county (8) in California (Fig. 1). Briefly, Lily (CAMELI) was a 12-year-old quarterhorse mare from Mendocino county with icterus, petechial hemorrhages, lethargy, and a rectal temperature at the time of examination of 41.7°C. Lea Jubilee (CASOLJ) was a 10-year-old mare with mild edema bilaterally at the cannon-fetlock region, fever, icterus, and slight petechiae apparent on the vaginal mucosa and sclera. E. equi morulae were observed in a buffy coat smear stained with Giemsa. Clinical hematology indicated thrombocytopenia, anemia, and leukopenia. Blood samples from the remaining horses with clinical signs of EGE were collected during 1996 to 1998 in northern California. The blood samples were tested for E. equi by 16S rRNA nested PCR (6), and the DNA samples were stored until use. Samples from the human patients were obtained 1998 as described previously (8).
TABLE 1.
Clinical cases of horse E. equi and HGE from northern California
| Sample | Name of horseb | Date of sample (mo/day/yr) | County |
|---|---|---|---|
| CASIGR | Garth Robertson | 1/16/96 | Sierra |
| CASIPE | Perle | 1/29/96 | Sierra |
| CASIJA | Jasper | 2/2/96 | Sierra |
| CASICH | Chip Howard | 2/12/96 | Sierra |
| CASIRN | Rind Norton | 4/17/96 | Sierra |
| CASITLa | Topper Leroy | 4/20/96 | Sierra |
| CAMEBSa | Boy Scout | 12/17/96 | Mendocino |
| CAMELI | Lily | 12/20/96 | Mendocino |
| CAMEDO | Doreen | 6/12/97 | Mendocino |
| CASOLJa | Lea Jubilee | 2/28/96 | Sonoma |
| CAMAWIa | Wiliekei | 6/11/98 | Marin |
| CAHU-HGE1a | Human1b | 7/15/98 | Humboldt |
| CAHU-HGE2a | Human2b | 7/28/98 | Humboldt |
Sequenced isolate.
Isolates CAHU-HGE1 and -HGE2 were obtained from humans.
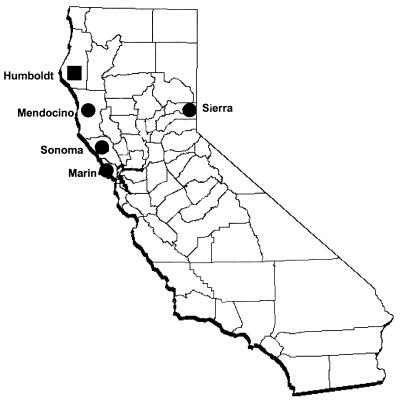
FIG. 1.
Map of California showing the counties where equine and human isolates were collected for PCR to detect E. equi (●) and HGE agent (■).
DNA preparation.
Genomic DNA was extracted from the buffy coat ACD-anticoagulated whole blood of horses as previously described (4). Briefly, the buffy coat was removed after centrifugation and kept at −20°C overnight. The erythrocytes in the buffy coat were lysed by adding 6 ml of 0.2% NaCl for 5 min and then adding 6 ml of 1.2% NaCl. The cell mixture was washed with phosphate-buffered saline, and the final pellet was diluted in 50 to 200 μl of lysis buffer (10 mM Tris-HCl, pH 8.3; 0.45% NP-40, 0.45% Tween 20, 100 μg of proteinase K per ml) and incubated in a 56°C water bath for 3 h. Finally, the DNA samples were incubated at 97°C for 15 min for inactivation of proteinase K and denaturation. The isolation of DNA from EDTA-anticoagulated blood from human cases was as previously described by Foley et al. (8). Briefly, DNAs were extracted from 100 μl of whole blood by using lysis buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA; and 1% [wt/vol] sodium dodecyl sulfate) at 37°C for 1 h. The lysate was incubated at 37°C for 30 min with RNase A (10 μg/ml) and then incubated overnight at room temperature with proteinase K (50 μg/ml). Genomic DNAs were purified with use of Phase Lock Gel I Light (5 Prime → 3 Prime, Boulder, Colo.). The positive control for PCR was BDS strain HGE agent (2); the negative control DNA was obtained from an E. equi-free horse.
PCR amplification.
The nested-PCR amplification for 16S rRNA gene fragment was done as previously described (4) (Table 2). Outer primers were used EE1 and EE2 for the first-round product (1,433 bp) and inner primers were used EE3 and EE4 for second-round product (928 bp). Primers EE5F and EE6R were designed to obtain an additional 517-bp fragment at the 3′ end of the 16S rRNA gene (Table 2). The forward primer LA6 and reverse primer LA1 were used to amplify the 444 Ep-ank gene fragment in the ankyrin repeat region (Table 2) (P. Caturegli, K. Asanovich, J. Walls, J. Bakken, J. Madigan, and J. Dumler, submitted for publication). Cycling conditions were denaturation for 4 min at 94°C, followed by 94°C for 30 s, 62°C for 30 s, and 72°C for 30 s. The annealing temperature was stepped down four times by 2°C every two cycles. The final annealing temperature used was 54°C for 28 cycles, followed by a final extension for 5 min at 72°C.
TABLE 2.
Primer sequences for PCR and sequencing of 16S rRNA, 444 Ep-ank and groESL heat shock operon genes of the E. phagocytophila genogroup
| Gene and primer name | Functions | Nucleotide sequences of primers (5′–3′) | Source or reference |
|---|---|---|---|
| 16S rRNA | |||
| EE1 | PCR, outer primer set | TCCTGGCTCAGAACGAACGCTGGCGGC | 4 |
| EE2 | AGTCACTGACCCAACCTTAAATGGCTG | ||
| EE3 | PCR, inner primer set | GTCGAACGGATTATTCTTTATAGCTTGC | |
| EE4 | CCCTTCCGTTAAGAAGGATCTAATCTCC | ||
| EE5F | PCR, sequencing, end part of 16S rRNA primer set (517 bp) | ACCTTACCACTCCTTGACATGG | 8) |
| EE6R | CACCCTAGTCACTGACCCAA | ||
| EE7F | Sequencing, internal primers | AATTATTGGGCGTAAAGGGCA | |
| EE8R | TTAAGCCCTGGCATTTCACC | ||
| 444 Ep-ank | |||
| LA6 | PCR, sequencing | GAGAGATGCTTATGGTAAGAC | Caturegli et al., submitted |
| LA1 | CGTTCAGCCATCATTGTGAC | ||
| groESL heat shock operon | |||
| EEgro1F | PCR | GAGTTCGACGGTAAGAAGTTCA | Newly designed primers |
| EEgro2R | CAGCGTCGTTCTTACTAGGAAC | ||
| EEgro3F | Sequencing, internal primers | GCGAATGGAGACAAGAACATA | |
| EEgro4R | AGTTGCGTCTTTTGTGATTCG | ||
| EEgro5F | AGCGAAGTTGAGGTGAAGGA | ||
| Eegro6R | CGCTTCCTTAGCCTTGAGAA |
The PCR primers EEgro1F and EEgro2R (Table 2) for amplification of the groESL heat shock operon of the Ehrlichia species were designed from conserved regions on the basis of a multiple sequence alignment of groESL heat shock operon sequences from GenBank database (accession numbers U96727, U72628, U96728, AF033101, U96735, U96729, and U96730). The PCR conditions were as described for the Ep-ank gene, with a final annealing temperature of 55°C for 28 cycles. All PCRs were performed in a Thermal Cycler 480 (Perkin-Elmer Cetus, Norwalk, Conn.), and PCR products were visualized after 1.5% agarose gel electrophoresis with fluorescence of ethidium bromide under a UV transilluminator.
Cloning and sequencing.
The 16S rRNA or groESL gene amplicons were ligated into the plasmid vector pCR 2.1-TOPO and TOP10 One Shot competent cells transformed according to the manufacturer's recommendations (TOPO Cloning Kit; InVitrogen, San Diego, Calif.). The recombinant clones were verified by colony PCR amplification. Two clones of each isolate were arbitrarily chosen for sequencing the forward and reverse strands. Plasmid DNA for sequencing was prepared by using the Quantum Plasmid Miniprep Kit (Bio-Rad, Hercules, Calif.) according to manufacturer's instructions. Amplicons of 444 Ep-ank and the 3′ end of 16S rRNA gene fragments were prepared for direct sequencing by using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany).
A primer complementary to the T7 promoter region of the plasmid vector and a series of internal primers EE7F and EE8R for the 16S rRNA gene (Table 2), EEgro3F, EEgro4R, EEgro5F, and Eegro6R for the groESL heat shock operon (Table 2), were used for sequencing the complete forward and reverse strands according to the protocol with the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA polymerase FS (Perkin-Elmer/ABI, Foster City, Calif.) and a PE Biosystems Prism 377 DNA Sequencer.
Nucleotide and translated amino acid sequence analyses.
The sequence data were collected using ABI Prism Data Collection software (version 2.1), and the sequence data were analyzed using ABI Prism Sequence analysis software (version 2.1.1) and Chromas software (version 1.51) (Technelysium Pty., Ltd., Mt. Gravatt Plaza, Queensland, Australia).
Sequence homology searches were made using the National Center for Biotechnology Information (National Institute of Health) BLAST network service. Nucleotide sequences were translated to amino acid sequence by the ExPASy translation tool of the Swiss Institute of Bioinformatics (http://expasy.hcuge.ch /tools/dna.html). The nucleotide and translated amino acid sequences were aligned and compared by using Multalin (version 5.3.3 [14]) (http://www.toulouse.inra.fr /lgc/multalin/multalin.html) and edited by using the GeneDoc Multiple Sequence Alignment Editor and Shading Utility (version 2.3.000 [32]). Calculations of sequence identities were performed by using ALINE (GenStream, Montpellier France; http://www2.igh.cnrs.fr/home.html).
RESULTS
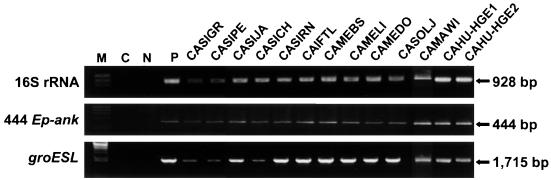
The 16S rRNA (two fragments of 928 and 517 bp), 444 Ep-ank (444 bp), and groESL (1,715 bp) genes were amplified from 13 GE isolates; 11 E. equi isolates from Sierra, Mendocino, Sonoma, and Marin counties; and two HGE agent isolates from Humboldt county in California (Fig. 1 and 2). Amplicons of 6 of the 13 isolates (1 isolate from each county) were selected from E. equi and compared with the two HGE agent isolates. The two HGE agent isolates had identical 16S rRNA gene sequences (1,395 bp) and were identical to the equine isolates from Mendocino and Marin counties. Two other equine isolates, CASITL and CASOLJ, have a C and G in nucleotide positions 34 and 180, respectively (Table 3). In comparison to the U.S. isolate sequences, which have an A at position 40, European isolates from horse, dog, cat, and human sources have a G at the same position. The overall sequence homology in the 16S rRNA gene among E. equi, the HGE agent, and E. phagocytophila is 99.9%.
FIG. 2.
PCR amplicons of 16S rRNA (928 bp), 444 Ep-ank (444 bp), and groESL heat shock operon (1,715 bp) genes from naturally infected with granulocytic ehrlichiae in horses from Sierra (CASIGR, CASIPE, CASIJA, CASICH, CASIRN, and CASITL), Mendocino (CAMEBS, CAMELI, and CAMEDO), Sonoma (CASOLJ), and Marin (CAMAWI) counties, California, and in a human from Humboldt county, California (CAHU-HGE1 and CAHU-HGE2). M, Molecular marker; C, double-distilled water; P, positive HGE agent control.
TABLE 3.
Comparison of nucleotide sequence difference positions of the 16S rRNA gene from the E. phagocytophila genogroup
| Ehrlichia spp. | Host, isolate or strain | Origin | Nucleotide difference positionsd
|
GenBank accession no. or reference | |||||
|---|---|---|---|---|---|---|---|---|---|
| 30 | 34 | 40 | 180 | 345 | 843 | ||||
| E. equi | CAMEBS | Calif. | A | A | A | T | G | G | AF172165 |
| CAMAWI | Calif. | • | • | • | • | • | • | AF172164 | |
| CASITL | Calif. | • | G | • | • | • | • | AF172166 | |
| CASOLJ | Calif. | • | • | • | C | • | • | AF172167 | |
| Anderson | Calif. | • | • | • | • | • | − | M73223 | |
| Alice | Calif. | • | • | • | • | • | • | AF036645 | |
| Merettricious | Calif. | G | • | • | • | • | • | AF036647 | |
| Atempo | Calif. | • | G | • | • | C | • | AF036646 | |
| HGE agent | CAHU-HGE1 | Calif. | • | • | • | • | • | • | AF093788 |
| CAHU-HGE2 | Calif. | • | • | • | • | • | • | AF093789 | |
| USG3 | R.I. | • | • | G | • | • | • | U020521 | |
| NCH-1a | Nantucket Island | / | / | / | • | • | • | U23038 | |
| E. phagocytophila | Ovine, OS strain | Scotland | • | • | • | • | • | − | M73220 |
| Goat, FG strain | Scotland | • | • | • | • | • | − | M73224 | |
| Closely related E. phagocytophila genogroup | Horse | Switzerland | • | • | G | • | • | • | AF057707 |
| Horse | Switzerland | • | • | G | • | • | • | U77389 | |
| Dog, Rosa isolate | Sweden | • | • | G | • | • | • | U10873 | |
| Cat, | Sweden | • | • | G | • | • | • | 8 | |
| Mouseb, PL1559 | Minn. | • | • | G | • | • | • | U72878 | |
| Mouseb, PL505 | Minn. | • | • | G | • | • | • | U72879 | |
| Tick (Ixodes ricinus) | Switzerland | • | • | G | • | • | • | AF084907 | |
| Tick, NCH-1/530-N8c | Nantucket Island | / | / | / | • | • | • | U23039 | |
Not identified in nucleotide position numbers 143, 144, 149, 168, 173, 192, and 195 based on CAHU-HGE1 sequence position compared with CAHU-HGE1.
Peromyscus leucopus.
Not identified in nucleotide position numbers 97, 234, 235, 261, 410, 458, 503, 517, 621, 681, 687, and 692 based on CAHU-HGE1 sequence position.
/, Not identified; −, gap by Multalin sequence alignment program; •, conserved position.
The sequences of 444 Ep-ank from all four E. equi isolates and both HGE isolates were 100% identical (Table 4). The 444 Ep-ank nucleotide sequences from California isolates differed from the BDS strain (GenBank accession number AF047897) from the eastern United States at positions 326 and 398, leading to predicted amino acid changes at positions 109 and 133 (Table 4). The California isolates differed from the USG3 strain (GenBank accession number AF020521) (from Rhode Island) in 16 nucleotide positions, leading to 9 predicted amino acid differences (Table 4).
TABLE 4.
Comparison of nucleotide and translated amino acid sequence difference positions of 444 Ep-ank from the E. phagocytophila genogroup
| Ehrlichia spp. | Host and isolate or strain (origin) | GenBank accession no. | Nucleotide difference positionsa
|
Translated amino acid difference positionsa
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 13 | 18 | 66 | 68 | 70 | 101 | 107 | 108 | 113 | 157 | 223 | 224 | 225 | 326 | 398 | 3 | 23 | 34 | 36 | 38 | 52 | 75 | 109 | 133 | |||
| E. equi | CASITL | AF172156 | T | A | T | A | A | T | A | G | C | T | C | G | − | − | − | G | G | Y | L | A | L | H | K | − | A | A |
| CAMEBS | AF172155 | • | • | • | • | • | • | • | • | • | • | • | • | − | − | − | • | • | • | • | • | • | • | • | − | • | • | |
| CASOLJ | AF172157 | • | • | • | • | • | • | • | • | • | • | • | • | − | − | − | • | • | • | • | • | • | • | • | − | • | • | |
| CAMAWI | AF172154 | • | • | • | • | • | • | • | • | • | • | • | • | − | − | − | • | • | • | • | • | • | • | • | − | • | • | |
| HGE agent | CAHU-HGE1 | AF172152 | • | • | • | • | • | • | • | • | • | • | • | • | − | − | − | • | • | • | • | • | • | • | • | − | • | • |
| CAHU-HGE2 | AF172153 | • | • | • | • | • | • | • | • | • | • | • | • | − | − | − | • | • | • | • | • | • | • | • | − | • | • | |
| BDS strain (Wis.) | AF047897 | • | • | • | • | • | • | • | • | • | • | • | • | − | − | − | T | A | • | • | • | • | • | • | − | S | T | |
| USG3 strain (R.I.) | AF020521 | A | G | G | G | G | A | G | C | T | G | G | T | T | T | T | • | A | R | M | P | C | D | N | F | A | T | |
−, Gap by Multalin sequence alignment program; •, conserved positions.
The groESL heat shock operon gene sequences of CASITL, CAMEBS, CAMAWI, and CAHU-HGE1 isolates were identical, but the sequences from CASOLJ and CAHU-HGE2 isolates had four and two nucleotide differences, respectively (Table 5). This nucleotide sequence difference was not predicted to result in amino acid differences between CAHU-HGE1 and CAHU-HGE2 but would result in expected amino acid differences at three positions when compared with CASOLJ. Similarly, up to 12 nucleotide differences were detected in comparing these northern California isolates with other HGE and E. phagocytophila isolates, but none except CASOLJ would be predicted to alter amino acid translation (Table 5).
TABLE 5.
Comparison of nucleotide and translated amino acid sequence difference positions of groESL heat shock operon from the E. phagocytophila genogroup
| Ehrlichia spp. | Host, isolate or strain (origin) | GenBank accession no. | Nucleotide difference positionsa
|
Translated amino acid difference positionsa | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 166 | 347 | 400 | 424 | 496 | 565 | 775 | 847 | 889 | 955 | 964 | 1048 | 1084 | 1087 | 1111 | 1117 | 1228 | 1243 | 1303 | 1315 | 1448 | 1474 | 1483 | 1496 | 1546 | 110 | 477 | 493 | |||
| E. equi | CASITL | AF172162 | G | T | A | A | T | G | C | A | G | C | A | C | G | A | T | T | G | C | A | A | C | G | C | T | G | S | R | S |
| CAMEBS | AF172161 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | |
| CAMAWI | AF172160 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | |
| CASOLJ | AF172163 | • | C | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | G | • | T | • | • | C | • | P | C | P | |
| California horse | U96727 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | |
| HGE agent | CAHU-HGE1 | AF172158 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| CAHU-HGE2 | AF172159 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | A | • | A | • | • | • | |
| WI-1 strain (Wis.) | U72628 | • | • | G | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | T | • | A | • | • | • | • | • | • | |
| Patient (N.Y.) | U96728 | • | • | G | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | / | / | / | / | / | • | / | / | |
| Slovenia | AF033101 | A | • | • | • | • | • | T | • | • | • | G | • | A | C | C | • | A | T | • | G | / | / | / | / | / | • | / | / | |
| E. phagocytophila | Horse (Switzerland) | U96735 | A | • | • | C | • | • | • | • | • | • | G | • | A | C | C | • | A | T | • | G | / | / | / | / | / | • | / | / |
| Goat, FG strain (Scotland) | U96729 | • | • | • | • | C | A | • | • | • | T | G | T | A | C | C | • | • | • | • | G | / | / | / | / | / | • | / | / | |
| Ovine, OS strain (Scotland) | U96730 | • | • | • | • | • | A | • | G | T | T | G | • | A | C | C | C | A | T | • | G | / | / | / | / | / | • | / | / | |
/, Not identified; •, conserved positions.
DISCUSSION
We and others have suggested that the E. phagocytophila genogroup is a tightly related group of strains of ehrlichiae that share tick vectors, cross host species, and should be differentiated primarily on the basis of geographical and not host species origin. There are numerous lines of evidence suggesting that the HGE agent is a strain within this group.
The HGE agent is antigenically indistinguishable from E. equi by immunofluorescence assay and on Western blots (2, 7). The HGE agent and E. equi are geographically and seasonally coincident and are transmitted by the same tick. They are indistinguishable ultrastructurally, cause indistinguishable clinical conditions in animal models (9, 12, 16), and are cross-protective (19). Munderloh et al. (17) reported that, based on PCR and immunocytology analyses, the EGE agent isolated in tick cell culture was closely related or identical to the HGE agent.
Evidence from genetic analyses of E. equi and HGE agent strains supports the theory that the ehrlichiae are conspecific. Our group previously showed that there was variability in the DNA sequence of the 16S rRNA gene among tick-exposed horses in California (18). Also, equine isolates in New England have 16S rRNA gene sequences which are identical to the HGE agent (14). Our present results indicate that the 16S rRNA gene sequences from Mendocino (CAMEBS) and Marin (CAMAWI) county horse isolates were identical to two HGE isolates. However, other isolates had one nucleotide difference, and a previously isolated EGE from a horse (Alice) in California (18) was identical to CAMEBS, CAMAWI, and the two human isolates from California in the homologous fragment sequences available for analysis. There was a suggestion that geographical origin could account for some of the observed variability, in that horse and human isolates from California and E. phagocytophila (ovine and caprine) from Scotland had an A in position 40 (Table 3), but isolates of human, mice, and tick origin from the eastern and middle United States had a G in that position. However, DNA of horse, dog, cat, and tick origin from Europe also contained a G at position 40. The microheterogeneity consisting of one or two unique nucleotide changes among E. equi strains, HGE agent, and E. phagocytophila strains suggests that it is not possible to classify isolates solely on the basis of their 16S rRNA gene nucleotide sequences.
Comparison of DNA sequences of the groESL operon in the E. phagocytophila genogroup also indicated that variability was associated with geographic origin. In a previous study which also examined groESL variability, found slightly more variability was found in the 16S rRNA gene and European origin isolates tended to differ from isolates from the United States (21). DNA sequencing of the 444 Ep-ank gene also revealed that the E. equi and HGE agent isolates from northern California were identical but that California isolates differed significantly from the eastern U.S. BDS and NCH-1 strains. Moreover, there was predicted protein diversity in this gene product between western and eastern U.S. isolates. Another reasonable target for genetic analysis would be the 44-kDa major outer membrane protein of the HGE agent and E. equi (22).
These results for 16S rRNA and for 444 Ep-ank and groESL heat shock operon gene sequences suggest that the CAHU-HGE1 isolate and E. equi strains from Mendocino and Marin counties are invariant and that at least some HGE agent strains are conspecific with E. equi. However, the species designation should also incorporate data regarding infectivity or pathogenicity, and such studies are incomplete. Experimental manipulation of the equine model supports the hypothesis that HGE isolates do not differ in pathogenicity from strains of E. equi (16).
Our results indicate that the HGE agent and E. equi are conspecific, which is a very important finding because the risk of HGE to humans cannot be adequately evaluated without considering infections in domestic animals. HGE is an emerging infection in people, partly because of increased scrutiny, and yet E. equi is well established, suggesting that the banks of tick vectors in areas where horse, dog, and hoofstock disease is endemic are contaminated and pose risks to people in the area. In a seroepidemiological survey in 1985 and 1986 for E. equi in northern California, 10.4% of 335 horses were found to be positive for antibodies and 38 horses were diagnosed as having EGE based on clinical signs and the observation of inclusion bodies in neutrophils in blood smears (15). Antibodies to E. equi have been detected in dogs from Oklahoma (20), the midwestern United States (10), and California (J. Foley, unpublished data). Thus, surveillance for human disease will need to incorporate dynamics of infections in domestic animals as well.
ACKNOWLEDGMENTS
We thank Elfriede DeRock, University of California, Davis, for technical assistance throughout this study.
This work was supported in part by grant A14213 from the National Institutes of Health.
REFERENCES
- 1.Aguero-Rosenfeld M, Horowitz H, Wormser G, McKenna D, Nowakowski J, Munoz J, Dumler J. Human granulocytic ehrlichiosis: a case series from a medical center in New York State. Ann Intern Med. 1996;125:904–908. doi: 10.7326/0003-4819-125-11-199612010-00006. [DOI] [PubMed] [Google Scholar]
- 2.Asanovich K, Bakken J, Madigan J, Aguero-Rosenfeld M, Wormser G, Dumler J. Antigenic diversity of granulocytic Ehrlichia isolates from humans in Wisconsin and New York and a horse in California. J Infect Dis. 1997;176:1029–1034. doi: 10.1086/516529. [DOI] [PubMed] [Google Scholar]
- 3.Bakken J S, Krueth J, Wilson-Nordskog C, Tilden R L, Asanovich K, Dumler J S. Clinical and laboratory characteristics of human granulocytic ehrlichiosis. J Am Med Assoc. 1996;275:199–205. [PubMed] [Google Scholar]
- 4.Barlough J, Madigan J, DeRock E, Bigornia L. Nested polymerase chain reaction for detection of Ehrlichia equi genomic DNA in horses and ticks (Ixodes pacificus) Vet Parasitol. 1996;63:319–329. doi: 10.1016/0304-4017(95)00904-3. [DOI] [PubMed] [Google Scholar]
- 5.Dumler J, Bakken J. Human granulocytic ehrlichiosis in Wisconsin and Minnesota: a frequent infection with the potential for persistence. J Infect Dis. 1996;173:1027–1030. doi: 10.1093/infdis/173.4.1027. [DOI] [PubMed] [Google Scholar]
- 6.Dumler J S, Bakken J S. Human ehrlichioses: newly recognized infections transmitted by ticks. Ann Rev Med. 1998;49:201–213. doi: 10.1146/annurev.med.49.1.201. [DOI] [PubMed] [Google Scholar]
- 7.Dumler S, Asanovich K, Bakken J, Richter P, Kimsey R, Madigan J. Serologic cross-reactions among Ehrlichia equi, Ehrlichia phagocytophila, and human granulocytic ehrlichia. J Clin Microbiol. 1995;33:1098–1103. doi: 10.1128/jcm.33.5.1098-1103.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foley J, Crawford-Miksza L, Dumler J, Glaser C, Chae J-S, Yeh E, Schnurr D, Madigan J. Two cases of human granulocytic ehrlichiosis in northern California. Clin Infect Dis. 1999;29:388–392. doi: 10.1086/520220. [DOI] [PubMed] [Google Scholar]
- 9.Foley J E, Lerche N W, Dumler J S, Madigan J E. A simian model of human granulocytic ehrlichiosis. Am J Trop Med Hyg. 1999;60:987–993. doi: 10.4269/ajtmh.1999.60.987. [DOI] [PubMed] [Google Scholar]
- 10.Greig B, Asanovich K M, Armstrong P J, Dumler J S. Geographic, clinical, serologic, and molecular evidence of granulocytic ehrlichiosis, a likely zoonotic disease, in Minnesota and Wisconsin dogs. J Clin Microbiol. 1996;34:44–48. doi: 10.1128/jcm.34.1.44-48.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gribble D. Equine ehrlichiosis. JAVMA. 1969;155:462–469. [PubMed] [Google Scholar]
- 12.Lewis G, Huxsoll D, Ristic M, Johnson A. Experimentally induced infection of dogs, cats, and nonhuman primates with Ehrlichia equi, etiologic agent of equine ehrlichiosis. Am J Vet Res. 1975;36:85–88. [PubMed] [Google Scholar]
- 13.Madigan J. Equine ehrlichiosis. Vet Clin N Am Equine Pract. 1993;9:423–428. doi: 10.1016/s0749-0739(17)30408-x. [DOI] [PubMed] [Google Scholar]
- 14.Madigan J, Barlough J, Dumler J, Schankman N, DeRock E. Equine granulocytic ehrlichiosis in Connecticut caused by an agent resembling the human granulocytic ehrlichia. J Clin Microbiol. 1996;34:434–435. doi: 10.1128/jcm.34.2.434-435.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madigan J, Hietala A, Chambers S. Seroepidemiologic survey of antibodies to Ehrlichia equi in horses in northern California. JAVMA. 1990;196:1962–1964. [PubMed] [Google Scholar]
- 16.Madigan J, Richter P, Kimsey R, Barlough J, Bakken J, Dumler J. Transmission and passage in horses of the agent of human granulocytic ehrlichiosis. J Infect Dis. 1995;172:1141–1144. doi: 10.1093/infdis/172.4.1141. [DOI] [PubMed] [Google Scholar]
- 17.Munderloh U, Madigan J, Dumler S. Isolation of the equine granulocytic ehrlichiosis agent, Ehrlichia equi, in tick cell culture. J Clin Microbiol. 1996;34:664–670. doi: 10.1128/jcm.34.3.664-670.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reubel G, Kimsey R, Barlough J, Madigan J. Experimental transmission of Ehrlichia equi to horses through naturally infected ticks (Ixodes pacificus) from northern California. J Clin Microbiol. 1998;36:2131–2134. doi: 10.1128/jcm.36.7.2131-2134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter P, Kimsey R, Madigan J, Barlough J, Dumler J, Brooks D. Ixodes pacificus (Acari: Ixodidae) as a vector of Ehrlichia equi (Rickettsiales: Ehrlichieae) J Med Entomol. 1996;33:1–5. doi: 10.1093/jmedent/33.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Rodgers S J, Morton R J, Baldwin C A. A serological survey of Ehrlichia canis, Ehrlichia equi, Rickettsia rickettsia, and Borrelia burgdorferi in dogs in Oklahoma. J Vet Diagn Investig. 1989;1:154–159. doi: 10.1177/104063878900100212. [DOI] [PubMed] [Google Scholar]
- 21.Sumner J W, Nicholson W L, Massung R F. PCR amplification and comparison of nucleotide sequences from the groESL heat shock operon of Ehrlichia species. J Clin Microbiol. 1997;35:2087–2092. doi: 10.1128/jcm.35.8.2087-2092.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhi N, Ohashi N, Rikihisa Y, Horowitz H W, Wormser G P, Hechemy K. Cloning and expression of the 44-kilodalton major outer membrane protein gene of the human granulocytic ehrlichiosis agent and application of the recombinant protein to serodiagnosis. J Clin Microbiol. 1998;36:1666–1673. doi: 10.1128/jcm.36.6.1666-1673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]