Abstract
Stomata allow CO2 uptake by leaves for photosynthetic assimilation at the cost of water vapor loss to the atmosphere. The opening and closing of stomata in response to fluctuations in light intensity regulate CO2 and water fluxes and are essential for maintaining water-use efficiency (WUE). However, a little is known about the genetic basis for natural variation in stomatal movement, especially in C4 crops. This is partly because the stomatal response to a change in light intensity is difficult to measure at the scale required for association studies. Here, we used high-throughput thermal imaging to bypass the phenotyping bottleneck and assess 10 traits describing stomatal conductance (gs) before, during and after a stepwise decrease in light intensity for a diversity panel of 659 sorghum (Sorghum bicolor) accessions. Results from thermal imaging significantly correlated with photosynthetic gas exchange measurements. gs traits varied substantially across the population and were moderately heritable (h2 up to 0.72). An integrated genome-wide and transcriptome-wide association study identified candidate genes putatively driving variation in stomatal conductance traits. Of the 239 unique candidate genes identified with the greatest confidence, 77 were putative orthologs of Arabidopsis (Arabidopsis thaliana) genes related to functions implicated in WUE, including stomatal opening/closing (24 genes), stomatal/epidermal cell development (35 genes), leaf/vasculature development (12 genes), or chlorophyll metabolism/photosynthesis (8 genes). These findings demonstrate an approach to finding genotype-to-phenotype relationships for a challenging trait as well as candidate genes for further investigation of the genetic basis of WUE in a model C4 grass for bioenergy, food, and forage production.
Dynamic stomatal responses to a decrease in light, measured in 659 Sorghum bicolor accessions, revealed substantial, heritable trait variation, associated with genes involved in water-use efficiency.
Introduction
Water availability is a major limiting factor to agriculture worldwide (Boyer, 1982) and is predicted to become even more limiting due to rising demand for water resulting from increasing atmospheric vapor pressure deficit (VPD) with climate change (Lobell et al., 2008; WWAP, 2015; FAO et al., 2018). Greater crop water use associated with greater above-ground biomass has also been implicated in past and future increases in crop yield (Sinclair et al., 1984; Ray et al., 2013; Lobell et al., 2014; Ort and Long, 2014; Koester et al., 2016; DeLucia et al., 2019). Therefore, unless water-use efficiency (WUE) is enhanced, agricultural systems in the near future will be increasingly threatened by drought, and unsustainable practices such as over-irrigation may result (DeLucia et al., 2019; Leakey et al., 2019).
WUE is a key to terrestrial plant growth due to the inherent tradeoff between net photosynthetic carbon assimilation (A) and water loss through transpiration (Wong et al., 1979). Most leaf water and CO2 fluxes pass through stomatal pores (Kerstiens, 1996; Hetherington and Woodward, 2003). Stomatal aperture is dynamically regulated by the movement of stomatal guard cells (Assmann and Jegla, 2016), which respond to extrinsic and intrinsic signals to optimize CO2 uptake relative to water vapor loss (Medlyn et al., 2011). The synchrony of stomatal opening and closing with fluctuations in photosynthetic CO2 assimilation varies within and among species, with significant consequences for intrinsic WUE (iWUE, the ratio of A to stomatal conductance to water vapor [gs]) (Lawson and Blatt, 2014; Kaiser et al., 2015). If conditions are favorable for A, but gs is low, A will be limited by CO2 supply. Conversely, if A is low, but gs is high, unnecessary transpiration will occur with no corresponding benefit to A, substantially decreasing iWUE. This is important in fluctuating light environments such as field crop canopies, where the movement of clouds and leaves cause frequent and abrupt changes in photosynthetic photon flux density (PPFD) at the leaf level (Pearcy, 1990; Zhu et al., 2004; Way and Pearcy, 2012; Wang et al., 2020).
Generally, gs responds an order of magnitude more slowly than A to a decline in PPFD, declining to a new steady state over the course of several minutes (Hetherington and Woodward, 2003; McAusland et al., 2016). This results in a de-synchronization of A and gs and loss of iWUE. Ensuring that A and gs are synchronized in their response to fluctuating light, by accelerating stomatal movement, could yield substantial benefits to iWUE of 20%–30% (Lawson and Blatt, 2014). Proof-of-concept from transgenic manipulation of the stomatal light-sensing mechanism has led to accelerated stomatal movement and improved iWUE in fluctuating light in Arabidopsis (Arabidopsis thaliana) and Nicotiana benthamiana (Papanatsiou et al., 2019). Still, accelerating stomatal movement by breeding or biotechnology for improved iWUE remains an open challenge in cereal crops (Faralli et al., 2019).
The genetic basis for natural variation in traits describing stomatal opening/closing is poorly understood, in part because such traits are difficult to phenotype at the scale required for association studies. The response of gs to fluctuating light is lengthy, with some species needing >30 min to transition from steady state at one light intensity to another (Lawson and Blatt, 2014; McAusland et al., 2016; Deans et al., 2019). The standard measurement of gs using gas exchange chambers or porometers requires a single instrument for each leaf, so is costly in terms of personnel and equipment. Using thermal imaging to track changes in leaf temperature resulting from stomatal movement, with the leaf warming up as stomata close due to a proportionate reduction of cooling by transpiration, provides a high-throughput phenotyping method in which numerous leaves can be measured simultaneously (Jones et al., 2002; Guilioni et al., 2008; Vialet-Chabrand and Lawson, 2019). Thermal imaging has been used to monitor stomatal closure (Jones et al., 2002) and stress response (Grant et al., 2007) in a grapevine field, to identify Arabidopsis mutants with altered gs (Merlot et al., 2002), and coupled with chlorophyll fluorescence measurements to screen Arabidopsis plants for iWUE (McAusland et al., 2013). In a broad validation experiment, thermal measurements predicted gs for a range of species exposed to different experimental treatments (Grant et al., 2006). Finally, thermal imaging was used to perform high-throughput phenotyping of the response of hundreds of ecotypes of Arabidopsis to changing light and [CO2] (Takahashi et al., 2015). However, the ability of thermal imaging to produce trait estimates with sufficiently high heritability to support the quantitative genetic investigation of genotype-to-phenotype relationships for stomatal opening/closing remains unclear.
Genome-wide association studies (GWAS) are widely used to identify the genetic basis for natural variation in agronomic, developmental, physiological, and biochemical traits in plant species, including sorghum (Sorghum bicolor) (Casa et al., 2008; Morris et al., 2013; Burks et al., 2015; Ortiz et al., 2017). The benefits and limitations of the approach have been reviewed in many contexts (Liu and Yan, 2019; Tam et al., 2019; Zhou and Huang, 2019). GWAS of stomatal movement can be challenging because biophysical tradeoffs can limit the extent of natural variation in the trait, for instance, because the speed of stomatal movement is constrained by the dimensions and patterning of stomata, which in turn are linked to leaf development and photosynthetic capacity. Measurements may also be slow, sensitive to environmental conditions, and lack accuracy, precision, or both. Together, these factors can reduce the variance that can be ascribed to genotype, i.e. reduce heritability, and constrain the size of the mapping population that can be studied, resulting in low statistical power. Recently, combining GWAS with a transcriptome-wide association study (TWAS), which identifies significant associations between trait variation and RNA transcript abundance across all expressed genes in tissue, has been shown to improve the identification of genes underlying trait variation (Kremling et al., 2019).
This study aimed to demonstrate how phenotyping by thermal imaging could be used in conjunction with integrated GWAS/TWAS to quantify natural diversity in stomatal/opening closing and identify genotype-to-phenotype relationships in a model C4 crop. S. bicolor ((L.) Moench) is a model species used to study photosynthesis, abiotic/biotic stress, and canopy architecture, as well as applied investigations in the context of food, fuel, and forage production (Paterson et al., 2009; Morris et al., 2013). It has particular importance in semi-arid conditions due to its high productivity and drought tolerance (Regassa and Wortmann, 2014; Hadebe et al., 2017). Sorghum is an especially interesting model in which to study stomatal movement because it has fast stomatal responses to decreasing light relative to other species (McAusland et al., 2016; Pignon et al., 2021). High-throughput thermal imaging was used to measure 10 traits describing the response of gs to a decrease in PPFD in over 2,000 plants of 659 accessions in a sorghum association mapping population. Results were validated against photosynthetic gas exchange measurements. Phenotypic trait correlations were used to identify general patterns in stomatal behavior. GWAS and TWAS were performed to identify phenotype to genotype associations, along with an ensemble approach combining GWAS and TWAS results using the Fisher’s combined test (FCT) (Kremling et al., 2019), followed by a gene ontology (GO) enrichment analysis. The resulting list of 239 candidate genes identified with the greatest confidence was enriched in putative orthologs of genes implicated in stomatal and photosynthetic traits in Arabidopsis and maize.
Results
Leaf temperature is affected by gs because leaves with increased gs will undergo greater evaporative cooling and have a lower temperature as a result. However, leaf temperature may also be affected by environmental factors including air temperature, airflow, incident light, and leaf angle. To minimize these confounding effects, thermal imaging was performed in a temperature-controlled chamber, and leaf angle was standardized across all plants. Wet and dry reference materials were used to account for environmental effects on leaf temperature. Temperatures of leaves and reference materials were used to estimate gs from thermal imaging, i.e. gsthermal.
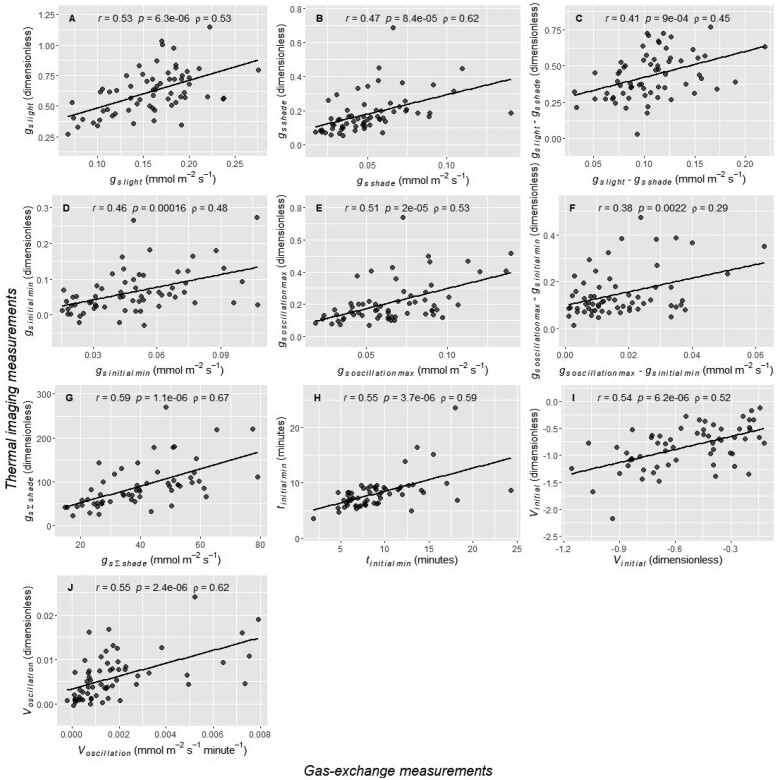
The response of gsthermal to a reduction in PPFD from 750 to 75 µmol m−2 s−1 was measured in 659 sorghum accessions (Table 1 and Figure 1). On a subset of 64 plants, gs estimates were also obtained from gas exchange measurements to validate gsthermal as a proxy for gs. Both gsthermal and gs predicted a similar pattern of stomatal closure upon a decrease in PPFD, sometimes followed by re-opening at low PPFD (Figure 2). All traits derived from gsthermal were significantly and positively correlated with their equivalents from gs (P < 0.005, Pearson’s r ranging from 0.38 to 0.59, Spearman’s rank-order ρ ranging from 0.29 to 0.66, Figure 3).
Table 1.
Descriptive statistics of stomatal conductance traits across 659 accessions of sorghum
| Trait | Description | Mean | SD | Min | Max | h2 |
|---|---|---|---|---|---|---|
| gs light | Steady-state gsthermal at high PPFD | 0.58 | 0.08 | 0.31 | 0.89 | 0.31 |
| gs shade | Steady-state gsthermal at low PPFD | 0.17 | 0.07 | 0.05 | 0.49 | 0.70 |
| (gs light − gsshade) | Amplitude of overall decline in gsthermal from high to low PPFD | 0.41 | 0.08 | 0.11 | 0.66 | 0.31 |
| gs initial min | Minimum gsthermal reached after the PPFD decrease | 0.05 | 0.04 | −0.03 | 0.24 | 0.71 |
| gs oscillation max | Maximum gsthermal reached as stomata re-opened at low PPFD | 0.20 | 0.07 | 0.08 | 0.53 | 0.67 |
| (gsoscillation max − gsinitial min) | Amplitude of oscillation of gsthermal at low PPFD | 0.15 | 0.05 | 0.05 | 0.43 | 0.49 |
| t initial min | Time for gsthermal to reach 110% of gsinitial min | 5.2 | 1.6 | 1.8 | 13.4 | 0.68 |
| gs Σ shade | Area beneath the gsthermal curve following the PPFD decrease | 84 | 32 | 28 | 231 | 0.72 |
| V initial | Exponential rate of decline in gsthermal after the PPFD decrease | −0.89 | 0.27 | −2.23 | −0.08 | 0.67 |
| V oscillation | Linear rate of increase in gsthermal as stomata re-opened at low PPFD | 0.0052 | 0.0028 | 0.0006 | 0.022 | 0.24 |
h2 is genomic heritability. tinitial min is in minutes, all other traits are dimensionless. Graphical description of these traits is given in Figure 1.
Figure 1.
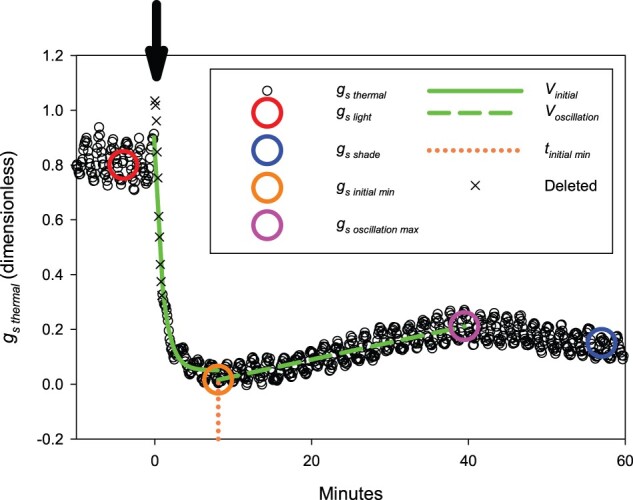
Schematic of gsthermal analysis method, where gsthermal is a proxy for stomatal conductance to water vapor (gs) that is derived from thermal imaging. Each response was measured on a single leaf of sorghum, exposed to PPFD = 750 µmol m−2 s−1 for 40 min. At t = 0, indicated by an arrow, PPFD was reduced by 90%. Black circles show gsthermal. Crosses show measurements from t = 0 to t = 0.9 min which were removed because they consistently showed an anomalous spike. gslight, the steady-state high-PPFD value of gsthermal, was the mean gsthermal from t =-5 to 0 min (red circle). gsshade, the steady-state low-PPFD value of gsthermal, was the mean gsthermal from t = 52 to 60 min (blue circle). gs Σshade was the area under the curve from t = 0 to 60 min. gsinitial min was the minimum of gsthermal reached immediately after the decrease in PPFD (orange circle). gsoscillation max was the maximum of gsthermal reached during stomatal re-opening at low PPFD (pink circle). The time at which gsthermal reached 110% of gsinitial min was recorded as t initial min (dotted orange line). Vinitial, the initial rate of decline in gsthermal after the decrease in PPFD, was the exponential rate of decline of gsthermalt =-0.1 min to t =t initial min (solid green line). Voscillation was the linear rate of increase in gsthermal during stomatal reopening at low PPFD (dashed green line).
Figure 2.
Representative responses of gsthermal and gs to a 90% drop in PPFD for two accessions: PI455257 and PI525728. gsthermal was measured from thermal imaging as a proxy for gs. Each gsthermal response curve was measured on a single leaf, acclimated to PPFD = 750 µmol m−2 s−1 for 40 min, then PPFD was reduced by 90% for 60 min at t = 0, indicated by an arrow. On a subset of plants, including the two shown here, the protocol was immediately repeated to measure gs using the gas exchange.
Figure 3.
Correlation scatterplots between stomatal conductance traits derived from gsthermal measurements and their counterparts derived from gs measurements. A, gslight, B, gsshade, C, gslight − gsshade, D, gsinitial min, E, gsoscillation max, F, gsoscillation max − gsinitial min, G, gs Σshade, H, t initial min, I, Vinitial, J, Voscillation. Data are for a subset of plants that were first imaged using a thermal camera to obtain gsthermal, then immediately measured with gas exchange to obtain gs. Pearson’s r and the associated P-value, along with Spearman’s rank-order ρ, are also given.
Genetic variation, heritability, and trait correlations
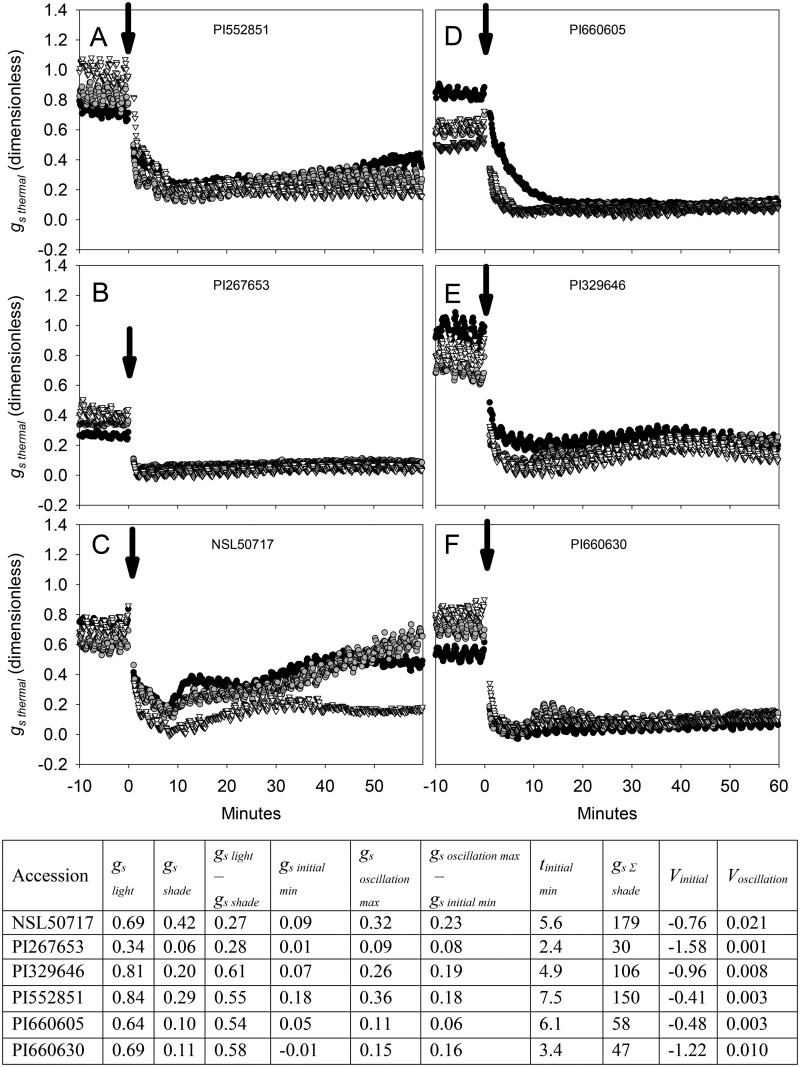
Variation among accessions was 3-fold, 10-fold, 6-fold, and 8-fold, respectively, for steady-state gsthermal at high PPFD (gslight), steady-state gsthermal at low PPFD (gsshade), the amplitude of decline in gsthermal from high to low PPFD (gslight − gsshade), and integrated gsthermal throughout the low-PPFD period (gs Σshade; Table 1). For example, gslight in accession PI552851 was more than double that of accession PI267653 (Figure 4, A and B). After the decrease in PPFD, gsthermal declined at an exponential rate (Vinitial), then reached a minimum (gsinitial min), and the time to reach this minimum was recorded (tinitial min). Variation among accessions was 28-fold, 9-fold, and 7-fold, respectively, for Vinitial, gsinitial min, and tinitial min, respectively (Table 1). For example, the decline of gsthermal was faster in PI267653, which displayed a strongly negative Vinitial and low tinitial min relative to slower accessions such as PI552851 (Figure 4, A and B). Stomatal re-opening often occurred after the initial decline causing a dampened oscillation in gsthermal during adjustment to low PPFD. The variation among accessions was 37-fold, 7-fold, and 9-fold, respectively, for the linear increase in gsthermal (Voscillation), gsthermal at peak stomatal re-opening (gsoscillation max), and the amplitude of oscillation (gsoscillation max − gsinitial min). For example, the oscillation of gsthermal was more pronounced in NSL50717, which displayed high Voscillation and high oscillation amplitude (gsoscillation max − gsinitial min), relative to PI660605, which showed no stomatal re-opening (Figure 4, C and D). Oscillation of gsthermal occurred on different timescales, e.g. more protracted in PI329646 than PI660630 (Figure 4, E and F). Variation in these traits among genotypes was reproducible across replicates (Figure 4, A–F).
Figure 4.
Representative responses of gsthermal to a 90% decrease in PPFD in six accessions, selected to highlight the diverse stomatal responses to decreasing PPFD measured in the sorghum diversity panel. A, PI552851, B, PI267653, C, NSL50717, D, PI660605, E, PI329646, F, PI660630. Different symbols and shades of gray show distinct replicate plants for each accession. Each gsthermal response curve was measured on a single leaf, acclimated to PPFD = 750 µmol m−2 s−1 for 40 min, then PPFD was reduced by 90% for 60 min at t = 0, indicated by an arrow. Values in the table are the mean (n = 3–4 plants) of stomatal conductance traits for each accession. tinitial min is in minutes, all other traits are dimensionless.
Genomic heritability (h2) was highest (0.67–0.72) in traits describing gsthermal at low PPFD, including gsinitial min, gsoscillation max, gsshade, and gs Σshade (Table 1). Traits describing the speed of change in gsthermal after a decrease in PPFD (Vinitial, tinitial min) also had moderately high h2 (0.67–0.68). Traits describing the oscillation in gsthermal during stomatal re-opening at low PPFD (Voscillation, gsoscillation max − gsinitial min) had low to intermediate h2 (0.24–0.49). h2 was low (0.31) in traits describing gsthermal at high PPFD and the amplitude of overall decline in gsthermal from high to low PPFD: gslight and gslight − gsshade, respectively.
All traits were correlated (r from −0.29 to 0.94) with one another (P < 0.05; Figure 5; Supplemental Figure S1). In particular, accessions with high gsthermal at high PPFD also had greater overall gsthermal at low PPFD (positive correlation of gslight and gs Σshade, P < 0.0001, r = 0.58, Figure 6A), took longer to adjust to the decrease in PPFD (positive correlation of gslight and tinitial min, P < 0.0001, r = 0.3; positive correlation of gslight and Vinitial, P < 0.0001, r = 0.34, Figure 6, B and C), and had more pronounced stomatal re-opening at low PPFD (positive correlation of gslight and Voscillation, P < 0.0001, r = 0.21, Figure 6D).
Figure 5.
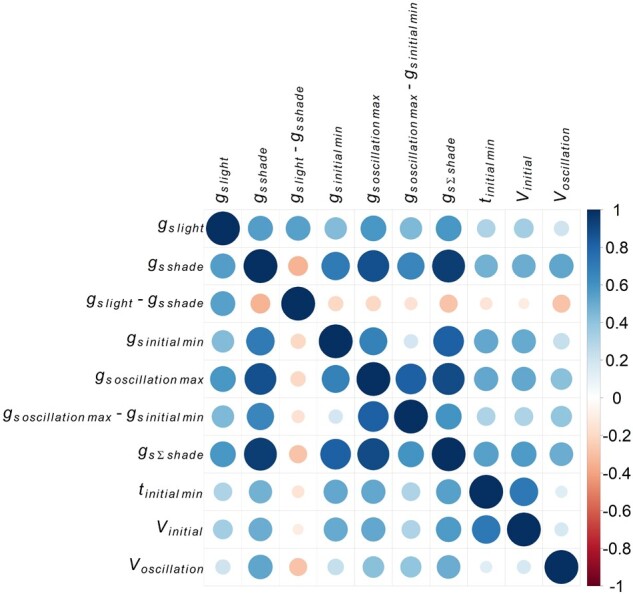
Pearson’s correlation coefficients (r) between BLUPs of stomatal conductance traits. The size of circles indicates the strength of correlation, while color indicates whether the pairwise relationship was negative (r < 0, red) or positive (r > 0, blue). All corresponding scatterplots are given in Supplemental Figure S1.
Figure 6.
Correlation scatterplots between BLUPs for stomatal conductance traits. A, gs Σshade versus gslight, (B) t initial min vs. gslight, (C) Vinitial vs. gslight, (D) Voscillation vs. gslight. Pearson’s r and the associated P-value are also given.
GWAS, TWAS, FCT, and GO enrichment analysis
For each gsthermal trait, genes were initially identified as of potential interest if they were in linkage disequilibrium (LD) (Supplemental Table S1) with the top 0.1% strongest associated SNPs from GWAS (approximately 600 genes per trait; Supplemental Tables S2 and S9); among the top 1% strongest associated genes from TWAS for leaf or shoot tissues (169 and 199 genes per trait, respectively; Supplemental Table S3); or among the top 1% strongest associated genes from FCT for leaf or shoot tissues (150 genes per trait; Supplemental Table S4). In addition to individual traits, multi-trait associations were performed with two trait groups: G1) traits describing the speed of change in gsthermal after a decrease in PPFD (Vinitial, tinitial min), and G2) traits describing overall values of gsthermal (gslight, gsinitial min, gsoscillation max, gsshade, gs Σshade). The compilation of these results (Supplemental Table S5) indicated that there was a substantial overlap in genes identified for different gsthermal traits, with 37% of genes being identified for two or more traits (Table 2).
Table 2.
Number of genes overlapping the top results of GWAS, TWAS, and/or FCT in multiple traits
| No. of overlapping traits | No. of genes |
|---|---|
| 1 | 4,575 |
| 2 | 1,463 |
| 3 | 689 |
| 4 | 281 |
| 5 | 150 |
| 6 | 97 |
| 7 | 38 |
| 8 | 11 |
| 9 | 6 |
| 10 | 1 |
Follow-up analyses were also performed to identify a subset of “higher confidence” genes, i.e. genes for which there was evidence for an association of trait variation with DNA sequence variation (GWAS) and RNA transcript abundance (TWAS) or genes identified in tests from both of the two independent approaches to sampling developing leaf tissue, i.e. from the third leaf or shoot section containing the growing point. Therefore, genes overlapping the top hits for multiple analyses/tissues, i.e. GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot (no. of overlaps ≥2 in Supplemental Table S5), were further investigated (Supplemental Figures S2–S12). Across all traits, 1,548 candidate genes were identified consistently across two or more of the individual tests. Taking Vinitial as a representative trait, of the 1,007 top hits, 180 were identified from at least two analyses/tissues (Figure 7).
Figure 7.
GWAS, TWAS, and FCT results for gslight. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B, Manhattan plots of GWAS, TWAS and FCT results. Top hits are highlighted if they correspond to putative orthologs of known Arabidopsis stomatal genes (FMA, POLAR, and EPF2, Table 3). Top hits are also highlighted if they are among the highest confidence genes identified by GWAS, TWAS, FCT, and subsequent GO enrichment analysis (Supplemental Table S7). Equivalent figures for all other traits are in Supplemental Figures S2–S12.
GO enrichment analysis on Arabidopsis putative orthologs of the “higher confidence” genes identified 153 significantly enriched biological processes (FDR-adjusted P < 0.05), nested within 34 broad categories (Supplemental Table S6). Among these, 22 categories of biological processes (e.g. regulation of histone H3-K27 methylation, hydrogen peroxide metabolic process, cell cycle DNA replication, plant epidermis morphogenesis, lipid catabolic process, and plant-type cell wall biogenesis) were enriched by >2.5-fold (Figure 8). A total of 239 unique genes contained within these GO categories were considered the strongest candidates to underlie variation in stomatal conductance traits studied here. A survey of the literature on their putative orthologs in Arabidopsis, maize, and rice revealed a large proportion of genes (32%) had functions related to stomatal opening/closing (24 genes), stomatal/epidermal cell development (35 genes), leaf/vasculature development (12 genes), or chlorophyll metabolism/photosynthesis (8 genes) (Supplemental Table S7). The positive or negative relationship between transcript abundance and trait variation was indicated by the TWAS (Supplemental Table S3).
Figure 8.
Results of GO enrichment analysis of higher confidence genes. Bars gives the number of genes included in each of the categories of GO biological processes that were significantly and >2.5-fold enriched. Fold-enrichment is shown in the label beneath each bar. Full GO enrichment analysis results are in Supplemental Table S6.
Discussion
Stomatal responses to changes in PPFD influence WUE of plants in fluctuating light environments (Lawson and Blatt, 2014). This study met the objectives of: (1) demonstrating how high-throughput thermal imaging can be used to rapidly phenotype variation in stomatal closure responses to PPFD across a diverse population of C4 plants and (2) using that data to perform an integrated GWAS/TWAS that identified a compelling set of candidate genes for further investigation of stomatal opening and closing in C4 species. Thermal measurements were validated against classical gas exchange, and phenotypic correlations revealed relationships between steady state (e.g. gslight) and dynamic (e.g. Vinitial) stomatal conductance traits. Results showed substantial, heritable variation in dynamic responses of stomata to a reduction in PPFD. This study presents important information on sorghum, a model system with rapid stomatal movement compared with other species (McAusland et al., 2016), and addresses a major knowledge gap that exists for C4 species, despite their agricultural and ecological importance (Edwards et al., 2010; Leakey et al., 2019).
Validation of gsthermal as a high-throughput proxy for gs
Although dynamic gs responses have increasingly studied in the past few years (McAusland et al., 2016; Deans et al., 2019; Acevedo-Siaca et al., 2020; De Souza et al., 2020; Pignon et al., 2021), measurements have not yet been deployed at a scale amenable to association mapping (e.g. GWAS). Here, gsthermal was a useful high-throughput proxy for gs, enabling simultaneous measurement of 18 plants in a single imaging frame. Previous studies have reported a near-perfect correlation between gsthermal and gs when gs varied by more than an order of magnitude as a result of combining data from multiple PPFDs, species, genotypes and mutants (Spearman’s rank-order ρ = 0.96) (McAusland et al., 2013). In contrast, the present study focused on testing the correlations between estimates of individual traits measured by photosynthetic gas exchange versus thermal imaging when considering only natural genetic variation within a single species. For example, thermal imaging was highly significant (P < 0.001) in capturing variation in gs measured by the gas exchange at a single PPFD (ρ ranging from 0.29 to 0.66, Figure 3) and the thermal estimate of gsshade had a heritability of 0.70, making it suitable for association mapping. It is also notable that correlations between the two measurement approaches would have been weakened because gsthermal and gs were not measured simultaneously in the manner achieved by McAusland et al. (2013). For example, consistency between the two approaches to gs measurements was likely reduced by differences in measurement conditions and the immediate history of environmental conditions experienced by leaves as they moved from thermal measurements to the gas exchange chamber. Boundary layer conductance was likely lower while measuring gsthermal than gs due to the air mixing fan used in gas exchange equipment (Grant et al., 2006). The light quality was equal parts red/blue/green for gsthermal versus 90% red 10% blue for gs. This likely affected stomatal opening, which is induced by blue, and to a lesser extent, red light (Assmann and Shimazaki, 1999; Shimazaki et al., 2007; Lawson et al., 2011; Assmann and Jegla, 2016). For these reasons, the significant correlations between all traits estimated from gsthermal and their counterparts estimated from gas exchange measurements were considered a validation of the methods used.
There is potential for further advancing the rapid phenotyping of photosynthesis and water relations under dynamic environments. In a precisely controlled environment, measurements of thermal transients could easily be scaled beyond the 18 plants measured simultaneously here, either by using multiple cameras or by using higher-resolution cameras placed higher above the plants. There is significant value in simultaneously imaging chlorophyll fluorescence and leaf temperature (McAusland et al. 2013). Although great care and possibly technical advances will be needed to insure homogeneous illumination across all samples when scaling up either imaging modality. Transient thermal measurements may also complement other imaging tools used to assess plant water status in controlled environments, such as analysis of plant shape and color using RGB imaging (Fahlgren et al., 2015). In general, effective screening of stomatal dynamics is most practical in controlled environment settings due to greater control of environmental conditions and also how plants are pre-conditioned before data collection. But, thermal cameras mounted on aircraft, gantries, and cable-systems, or used from boom lifts, have enabled reliable field-scale measurements of plant temperature (Deery et al., 2016, 2019; Sagan et al., 2019), including for identification of QTL that overlaps with stomatal density QTL (Prakash et al., 2021), but in these settings, the changes in leaf temperature caused by dynamic stomatal changes coincide with the transient effects of wind, leaf shading, and leaf angle. So, evaluating dynamic stomatal responses to varying PPFD in the field remains a daunting prospect.
Sorghum shows varied, heritable stomatal responses to a decrease in PPFD
Within-species diversity in stomatal light responses has been documented for C3 species including Arabidopsis (Takahashi et al., 2015), poplar (Durand et al., 2019), rice (Acevedo-Siaca et al., 2020), cassava (De Souza et al., 2020), and soybean (Soleh et al., 2016). Expanding the scale of investigation to 659 accessions of sorghum revealed substantial variation within the C4 model species, despite the fact that C4 species generally have lower gs than C3 species (Taylor et al., 2010). The range of tinitial min shown here (1.8–13.4 min, Table 1) overlapped with similar measurements in sorghum (∼2–8 min) (McAusland et al., 2016; Pignon et al., 2021), the closely related C4 grasses miscanthus and maize (∼8 min), the C3 grass rice (∼10 min) (McAusland et al., 2016), and the semi-aquatic rhizomatous fern Marsilea drummondii A. Braun (∼9 min) (Deans et al., 2019). Some C3 dicots such as Arabidopsis and sunflower (∼18 min) were roughly comparable with the slowest accession shown here (tinitial min =13.4 min), while many other species appeared considerably slower (∼30 min) (McAusland et al., 2016; Deans et al., 2019). The faster stomata of sorghum might be related to the unique structure of graminaceous stomata, composed of dumbbell-shaped guard cells flanked by subsidiary cells, which have been linked to rapid movement relative to other forms (Franks and Farquhar, 2007; Lawson et al., 2011; Serna, 2011; McAusland et al., 2016; Lawson and Vialet-Chabrand, 2019). Stomata of C4 plants tend to be smaller and more sensitive to environmental change than their C3 counterparts (Lawson et al., 2011; McAusland et al., 2016).
Implications of natural diversity in stomatal responses to decreasing PPFD
Variation within and among accessions in gsthermal could be observed at different stages of the light to dark transition (Figure 4), which was captured effectively by the steady state and dynamic traits extracted from thermal transients (Figure 1) resulting in generally high h2 (Table 1). Compared with steady-state gs under high PPFD, traits describing the dynamic change in gsthermal after a decrease in PPFD had greater variability and h2, making them more tractable targets for association studies (Table 1). Additionally, variation in traits describing the speed of change in gsthermal after a decrease in PPFD (Vinitial, tinitial min) might be leveraged to accelerate stomatal responses even in species with “fast” stomata such as sorghum, potentially improving coordination of gs with photosynthetic carbon assimilation (A) and resulting in improved iWUE (Lawson and Blatt, 2014). Other traits (Voscillation, gsoscillation max − gsinitial min) described stomatal oscillations similar to those observed in other species, especially when blue light is applied (Ballard et al., 2019) and when stomatal apertures are low (Kaiser and Kappen, 2001). However, the stomatal oscillations observed in our study occurred on a lengthy timescale (up to 1 h), and are unlikely to substantially impact iWUE of field crop stands, where most light fluctuations last on the order of seconds (Kaiser et al., 2018).
The relationships among traits observed across the genetic variation surveyed here are consistent with biophysical tradeoffs driven by structure–functional relationships, as well as selection for trait combinations that favor carbon gain versus water savings to differing degrees in different environments. The finding that accessions with greater gslight took longer to adjust to decreasing PPFD (Figure 6, B and C) is supported by similar patterns across diverse species (McAusland et al., 2016), and is consistent with greater gslight being associated with greater stomatal apertures requiring more time to close (Lawson and Blatt, 2014). However, it is worth noting that variation in stomatal opening/closing is also associated with guard cell physiology, including ion transport processes (Lawson and Blatt 2014). Adaptation to different environments may also contribute to the observed trait correlations, with high gslight and slow stomatal closure working in concert to favor A and rapid growth in environments where water is not limiting. In contrast, low gslight and rapid stomatal closure combine to prioritize WUE and conservative but sustained growth in water-limited environments (Vico et al., 2011). Since more rapid stomatal closure after a decrease in PPFD would increase iWUE, there is significant interest in identifying more genes underpinning structural and functional components of stomatal movements, as well as their interactions with steady-state gas exchange and leaf development and physiology more broadly (Lawson and Blatt 2014).
The number of stomata per unit leaf area, dimensions of stomata, extent of opening, and arrangement of stomata on the leaf surface all influence gs (Dow et al., 2014; Faralli et al., 2019). Three of these four factors are anatomical features that are fixed during leaf development (Serna, 2011; Nunes et al., 2020). In addition, guard cell movement allows variation of the stomatal aperture in response to environmental signals including light intensity and spectral composition (Assmann and Jegla, 2016), [CO2] (Assmann and Jegla, 2016) and leaf-to-air vapor pressure deficit (Ball et al., 1987; McAdam and Brodribb, 2015). Therefore, many biological processes, pathways, and genes can influence gs. This study advanced understanding of these factors through a GWAS/TWAS approach used to identify the most influential genes underlying phenotypic variation in gsthermal traits.
GWAS/TWAS identifies genes enriched in stomatal, leaf developmental, and photosynthetic functions
Gene candidates putatively associated with genetic variation in stomatal closure in sorghum were identified using GWAS and TWAS integrated with FCT, followed by GO enrichment analysis. This approach has identified known causal variants more efficiently than GWAS and TWAS alone (Kremling et al., 2019), while also increasing the consistency in results observed when testing was repeated across different conditions (Ferguson et al., 2021). The present study reinforced these prior reports, with an order of magnitude more genes being consistently identified by FCT versus TWAS across the two independent tissue sampling strategies used (Supplemental Table S5). GO enrichment analysis of the Arabidopsis putative orthologs of these genes revealed 22 GO biological processes that were significantly and >2.5-fold enriched (Supplemental Table S6; Figure 8). The 239 genes belonging to these 22 categories were selected as the greatest confidence candidate genes (Figure 8; Supplemental Table S7). A large proportion (32%) of these genes has putative orthologs in Arabidopsis, maize or rice that is already implicated in regulating traits related to stomata or WUE. While it is unlikely that such enrichment would occur by random chance, the function of the genes identified here will require confirmation by follow-up reverse genetic studies of transgenic or mutant plants. Examples of genes discovered through the GWAS/TWAS are described below in three categories corresponding to genes involved in (1) guard cell signaling, metabolism or transport; (2) stomatal or epidermal patterning; and (3) overall leaf development, vasculature, and photosynthesis.
Twenty-three putative orthologs of genes implicated in signaling, metabolism, or transporters in guard cells belong to enriched GO terms including hydrogen peroxide metabolic process, response to disaccharide, response to heat, and lipid catabolic process (Supplemental Table S7). For example, loss of ASCORBATE PEROXIDASE 1 (APX1) and the RESPIRATORY BURST OXIDASE HOMOLOG PROTEIN F (RBOHF) influence redox oxygen species (ROS) to alter stomatal responses to light, [CO2] or abscisic acid (ABA; Pnueli et al., 2003; Chater et al., 2015; Sierla et al., 2016). The TWAS revealed that lower APX1 transcript abundance in developing leaf tissues in a sorghum accession was associated with slower stomatal closure (APX1 putative ortholog SOBIC.001G410200 in traits Vinitial min and tinitial min, Supplemental Table S3), which is a more subtle version of the severe impairment of stomatal movements observed in knockout-APX1 plants of Arabidopsis (Pnueli et al., 2003). Meanwhile, the MYB60 transcription factor is required for light-induced opening of stomata in Arabidopsis. It is expressed exclusively in guard cells, with expression increasing and decreasing in accordance with conditions that promote stomatal opening and closing, respectively (Cominelli et al., 2005). This is consistent with the observation that sorghum accessions with greater MYB60 transcript abundance in the shoot growing point closed their stomata more rapidly after a decrease in PPFD (MYB60 putative ortholog SOBIC.005G155900 in trait tinitial min, Supplemental Table S3). Other genes identified that have putative orthologs in Arabidopsis that are involved in guard cell metabolism or signaling included PHOSPHOLIPASE Dα1, which along with its lipid product phosphatidic acid, impacts ABA induction of ROS production and stomatal closure (Zhang et al., 2009). Mutants of ABC TRANSPORTER G FAMILY MEMBER 40 (ABCG40) shut more slowly in response to ABA (Kang et al., 2010), while its sorghum putative ortholog was variously associated with five different gs thermal traits in GWAS, TWAS, and FCT tests.
Recently, lipid metabolism of guard cells has discovered to be important as an energy source for light-induced stomatal opening (McLachlan et al., 2016). Five sorghum genes associated with variation in gsthermal traits were putative orthologs of genes involved in triacylglyceride mobilization and expressed in guard cells of Arabidopsis (enoyl-coa delta isomerase 1 and 3, ECI1 and ECI3; peroxisomal fatty acid beta-oxidation multifunctional protein; acyl-coenzyme A oxidases 2 and 4, ACX2 and ACX4; McLachlan et al., 2016). Notably, wrinkled1, a transcription factor that regulates metabolic genes in a manner that promotes carbon allocation to fatty acid synthesis, was also identified (Cernac and Benning, 2004). Overall, a substantial proportion of the highest confidence candidate genes identified by the integrated GWAS/TWAS is plausibly involved in signaling, metabolism, and transport functions in guard cells. Further study will be needed to determine if the candidate genes identified here play specific roles in stomatal closure, or if the associations observed in this study are partly a product of the strong correlations between rates of stomatal opening and closing (Lawson and Blatt, 2014). Current understanding of lipid metabolism guard cells would seem to suggest the latter option is more likely, but this area of study is still relatively nascent.
The functions of putative orthologs of the highest confidence sorghum candidate genes are also consistent with the importance of stomatal size, density, and distribution to the speed of stomatal opening and closing. Thirty-five putative orthologs of genes implicated in stomatal or epidermal cell patterning belong to enriched GO terms including plant epidermis morphogenesis, cell cycle DNA replication, plant-type cell wall biogenesis, and plastid organization (Supplemental Table S7). Arabidopsis genes known to impact stomatal development or patterning which had sorghum putative orthologs found in the highest confidence candidates for gsthermal traits included:Mitogen-activated protein (MAP) kinase kinase kinase 4 (YODA, Bergmann et al., 2004); the FAMA basic helix-loop-helix (bHLH)-type transcription factor (Ohashi-Ito and Bergmann, 2006), cyclin-dependent kinase B1;1 (Boudolf et al., 2004); phytochrome interacting factor 1 (Klermund et al., 2016), somatic embryogenic receptor kinasE 1 (Meng et al., 2015), DNA-directed RNA polymerase II subunit 2 (NRPB2; Chen et al., 2016), chromatin regulator enhanced downy mildew 2 (EDM2, Wang et al., 2016), protein phosphatase 2A (Bian et al., 2020), GATA, nitrate-inducible, carbon metabolism-involved (GNC, Klermund et al., 2016), extra-large GTP-binding protein (XLG3, Chakravorty et al., 2015), and ADP-ribosylation factor (ARF) guanine-nucleotide exchange factor (Le et al., 2014). Sorghum accessions with lower NRPB2 transcript abundance in developing leaf tissue had greater gs at high light (NRPB2 putative ortholog SOBIC.001G155000 in trait gslight; Supplemental Table S3), which is consistent with a positive correlation between stomatal density and gs (Pignon et al., 2021) and NRPB2 loss-of-function mutants having greater stomatal density (Chen et al., 2016). Meanwhile, stomata closed more rapidly in accessions with a lower abundance of EDM2 transcripts in developing leaf tissue (EDM2 putative ortholog SOBIC.002G154000 in trait tinitial min, Supplemental Table S3). This is consistent with bigger stomata having slower movements, occurring when stomatal density is low (Pignon et al., 2021) and EDM2 loss-of-function mutants having greater stomatal density (Wang et al., 2016).
Focusing on putative orthologs in other grasses, SOBIC.010G277300 shares 96% predicted protein sequence similarity with GRMZM2G057000 in maize. The nana plant2 (na2) mutant of this gene displays alterations in brassinosteroid synthesis and the morphology of stomatal complexes (Best et al., 2016). Similarly, SOBIC.004G116400 shares 74% predicted protein sequence similarity with Os02g15950 (ERECT PANICLE 3, EP3) in rice (Oryza sativa). Loss-of-function mutants of EP3 have smaller stomata, which appeared to drive reductions in gs and A (Yu et al., 2015). Given the substantial evidence for links between gas exchange, stomatal complex size, and stomatal density (Lawson and Blatt 2014; Xie et al., 2021), there is potential for variations in the sequence and expression of these genes to drive variation in the gsthermal traits measured in this study. A number of other genes identified by the integrated GWAS/TWAS have been implicated in the development of epidermal cells in general (Supplemental Table S7). Cross talk between development pathways for stomata and other types of epidermal cells (Kim and Dolan, 2011; Raissig et al., 2016) creates the opportunity for such genes to influence gs and its response to PPFD.
Finally, a smaller number of sorghum genes identified in this study have putative orthologs known to influence overall leaf development/vasculature (nine genes) or chlorophyll/photosynthesis (seven genes) (Supplemental Table S7). Leaf vasculature determines the hydraulic capacity of the leaf to deliver water that eventually diffuses out of the leaf as vapor. Consequently, strong traits associations between leaf hydraulics and stomata have been described in a range of contexts (Sack et al., 2003; Bartlett et al., 2016). In that vein, it is plausible that genes known to alter vascular development via effects on polyamine metabolism (5′-methylthioadenosine nucleosidase; Waduwara-Jayabahu et al., 2012), glucuronoxylan synthesis (beta-1,4-xylosyltransferase, IRX9, Pena et al., 2007), and transcriptional regulation (defectively organized tributaries 5, DOT5, Petricka et al., 2008) might be associated with variation in gs thermal traits. Stomata closed more slowly in accessions with a lower abundance of DOT5 transcripts at the shoot growing point (DOT5 putative ortholog SOBIC.002G164700 in tinitial min, Supplemental Table S3). This is consistent with DOT5 mutants having lower vein density (Wang et al., 2016), which is typically associated with lower stomatal density (Brodribb and Jordan 2011). This is also consistent with low-density stomata tending to be larger with slower movements (Pignon et al., 2021).
Similarly, the identification of genes involved in photosynthesis may reflect the tight linkage between A and gs, which is observed in many plant species and is particularly strong in sorghum (Leakey et al., 2019). QTL for traits related to A and gs often overlap, including in sorghum (Ortiz et al., 2017). When compared with other species, sorghum shows exceptional coordination between gs and A following decreases in PPFD, driven by rapid responses in gs (McAusland et al., 2016). Among diverse sorghum accessions, there is significant covariation between the responses of A and gs following decreases in PPFD, i.e. accessions with more rapid declines in A also have more rapid declines in gs, and vice-versa (Pignon, 2017). Most notable was the identification of the sorghum rubisco activase (RCA) by GWAS, TWAS, and FCT across eight different gsthermal traits (Supplemental Table S7). RCA encodes an enzyme that plays a key role in activating Rubisco to perform the key step in photosynthetic CO2 assimilation (Portis 2003), and which is known to limit the rate of photosynthetic induction after an increase in PPFD (Pearcy 1990). One notable component of the results was significantly lower gs under high light in sorghum accessions with greater RCA transcript abundance in developing leaf tissues (RCA putative ortholog SOBIC.005G231500 in gslight, Supplemental Table S3). This would be consistent with a general syndrome of high water use efficiency combining low steady-state gs and fast stomatal movements (Lawson and Blatt 2014; Leakey et al., 2019). Along with a number of the results described above, these findings open the possibility that phenotyping only stomatal closure may have facilitated the identification of associations between genotype or gene expression and both stomatal opening and closing, as a result of the two aspects of stomatal movement being so tightly linked. If functional validation of candidate genes supports that notion, then considerable time can be saved when collecting phenotypic data.
Conclusion
This study demonstrates how high-throughput phenotyping by thermal imaging can be used to assess genetic variation in stomatal closure after a decrease in PPFD at a scale suitable for association mapping. Integrated GWAS/TWAS and FCT were then applied to identify a set of candidate genes, which were enriched in putative orthologs of Arabidopsis, maize and rice genes involved in stomatal opening/closing, epidermal patterning, leaf development and photosynthesis. This is an important proof of concept for methods to break the phenotyping bottleneck for a trait that is important to plant productivity and sustainability but has until now been intractable as a target for study by quantitative genetics. The method described here could also be applied to other species from a variety of plant functional types or grown in different environments to assess G × E of stomatal traits. In addition, the study provides knowledge of trait variation and underlying candidate genes in an important C4 model crop, which is notable for the speed of its stomatal opening/closing. This lays the foundation for future studies to establish gene function and potentially improve crop performance.
Materials and methods
Physiology measurements
Plant material and growing conditions
A random subset of 659 accessions was selected from the biomass sorghum (S. bicolor) diversity panel at the University of Illinois at Urbana-Champaign, as previously described (Valluru et al., 2019). Accession names are provided in Supplemental Table S8. Plants were grown from seed in flats (3×6 sets of 281 mL inserts) containing a peat/bark/perlite-based growing medium (Metro-Mix 900; Sun Gro Horticulture, Agawam, MA, USA) and supplemented with 1 mL slow release 13-13-13 fertilizer (Osmocote Classic, Everris NA, Inc., Dublin, OH, USA). One accession, PI147837, was included in each flat to identify spatial and temporal variation in measurements. Three seeds/inserts were planted and then thinned to 1 seedling/insert. Flats were watered regularly to field capacity and grown in a greenhouse maintained at 27°C day/25°C night, with supplemental lighting to ensure the minimum light intensity of 90 W m−2 during a 13-h d. n = 3–4 plants were assessed per accession.
Experimental conditions and leaf temperature measurement
Once the fourth leaf had fully expanded, as evidenced by ligule emergence, plants were transferred to a growth cabinet overnight (Model PCG20, Conviron, Winnipeg, MB R3H 0R9, Canada). Cabinets were maintained at 14-h/10-h day/night cycle under 1200 μmol photons m−2 s−1PPFD, 30°C daytime/25°C nighttime temperature, and 75% RH. On the day of measurement, the fourth leaf of each plant was laid flat across a frame to standardize leaf angle and incident light interception (Supplemental Figure S13). This presented a 4-cm length of the mid-leaf for measurement. Leaves were not detached from plants. Dry and wet reference materials were prepared as in (McAusland et al., 2013) to correct for the effects of net isothermal radiation and VPD, respectively (Guilioni et al., 2008). A thin coating of petroleum jelly was applied over 1 cm of abaxial and adaxial sides of leaves, providing a dry reference unique to each leaf. Two sections of filter paper, moistened by a water reservoir, were used as a wet reference.
Flats of 18 plants were transferred to a second growth cabinet with conditions identical to the first cabinet, except that light was provided by a 20 × 20 cm LED panel providing equal parts blue, red, and green light, with a combined incident photon flux of 750 µmol m−2 s−1 at the leaf level (LED Light Source SL 3500, Photon Systems Instruments, Brno, Czech Republic). An infrared camera (Thermo Gear Model G100, Nippon Avionics CO., Ltd., Tokyo, Japan) was placed 0.5 m above the leaves without obstructing the light source. Incident photon flux was maintained at 750 µmol m−2 s−1 for 40 min, and then reduced by 90% for an additional 60 min. Images were recorded every 6 s, with emissivity = 0.95 (Jones et al., 2002). The cabinet’s PPFD sensor was used to evaluate spatial heterogeneity of incident photon flux, which was contained to ± 6% variation across the measured area.
Image analysis and stomatal conductance estimation
Analysis of thermal images was performed in ImageJ (ImageJ1.51j8, NIH, USA). Sections of leaf and reference materials of each image were hand-selected to derive profiles of temperature versus experimental time. Leaf and reference temperatures were used to calculate gsthermal:
| (1) |
where Tleaf, Tdry, and Twet are temperatures of the leaf, dry and wet references, respectively. gsthermal is theoretically proportional to gs given constant environmental conditions (Jones, 1999; Jones et al., 2002; Grant et al., 2006; Guilioni et al., 2008; McAusland et al., 2013). Air RH and temperature were controlled by the growth cabinet and assumed constant across all leaf and reference surfaces. Since the cabinet was designed to deliver a uniform airflow, and replicate plantings were randomly positioned to avoid systematic spatial variation, boundary layer conductance was also assumed constant. Differences in gsthermal between leaves and overtime were therefore attributed to gs.
Analysis of gsthermal profiles
Several traits were derived from profiles of gsthermal versus experimental time: gslight, gsshade, gs Σshade, gsinitial min, gsoscillation max, tinitial min, Vinitial, and Voscillation. A graphical description of these traits is given in Figure 1. After the decrease in PPFD, gsthermal declined as stomata closed, often followed by an oscillation in gsthermal as stomata re-opened and then closed again. gslight and gsshade were the averages of gsthermal from t =−5 to 0, and from t = 52 to 60 min, respectively. These gave steady-state gsthermal at PPFD of 750 and 75 μmol m−2 s−1, respectively. gs Σshade was the area beneath the curve following the PPFD change, i.e. from t = 0 to 60 min. gsinitial min was the minimum of gsthermal reached immediately after the decrease in PPFD. gsoscillation max was the maximum of gsthermal reached during the stomatal re-opening phase. The time at which gsthermal reached 110% of gsinitial min was recorded as tinitial min. The amplitude of the overall change in gsthermal from high to low PPFD was (gslight − gsshade), and the amplitude of oscillation in gsthermal at low PPFD was (gsoscillation max − gsinitial min).
V initial was derived from non-linear regression (PROC NLIN, SAS v9.4; SAS Institute, Cary, NC, USA) as the exponential rate of decline of gsthermal from t = −0.1 min to t =tinitial min:
| (2) |
here b and a give estimates of gsthermal at t =−0.1 min and t =tinitial min, respectively, and a more negative Vinitial indicates a more rapid decline in gsthermal. Voscillation was derived from linear regression (PROC GLM, SAS v9.4) as the linear slope of gsthermal versus time during stomatal re-opening at low PPFD. A more positive Voscillation indicates a more rapid stomatal re-opening.
Validation of gs thermal with gas exchange measurements
Validation of gsthermal as a proxy for gs was obtained on a subset of 64 plants. After gsthermal measurements were completed, plants were placed back in the first growth cabinet. The leaf section previously used for gsthermal measurements was placed in the cuvette of a portable gas exchange system incorporating infra-red CO2 and water vapor analyzers (LI-COR 6400; LI-COR, Inc., Lincoln, NE USA). Incident PPFD was set to 750 μmol m−2 s−1, [CO2] to 400 ppm, and leaf-to-air water vapor pressure deficit maintained <2 kPa. PPFD was 10% blue and 90% red light provided by integrated red and blue LEDs. The gsthermal measurement protocol was replicated, i.e. initial PPFD was maintained for 40 min, then reduced by 90% for an additional 60 min, with gs logged every 5 s (von Caemmerer and Farquhar, 1981). Pearson’s correlation (r) at P = 0.05 threshold, along with Spearman’s rank-order correlation (ρ) was tested between equivalent stomatal conductance traits derived from gsthermal and gs using R 3.6.1 (R Core Team, 2017).
When comparing gsthermal measurements to this validation data, an anomalous spike in gsthermal, reaching up to twice the steady-state high-PPFD gsthermal, was consistently recorded from t = 0 to 0.9 min (Figure 1). This was likely due to the different radiative properties of the white wet reference and the green leaf and dry reference. Therefore, gsthermal measurements from t = 0 to 0.9 min were discarded.
Model development for best linear unbiased predictors
A linear mixed model was used to account for spatial and temporal variation using the ASReml-R package (Butler et al., 2009). The best linear unbiased predictors (BLUPs) were obtained for all accessions and traits and were used for subsequent analysis (Supplemental Table S8). The most appropriate model for each trait was chosen in two steps. First, fixed effects with a Wald statistics P >0.05 were excluded from the model. Subsequently, the Akaike information criterion was used to select random effects variables and residual variance–covariance structures for each trait. The full model was:
| (3) |
where is the vector of phenotypes, is a vector of fixed effects including the intercept, a blocking effect, and a cubic smoothing splines terms for leaf position and time of measurement, with design matrix . Here the block term refers to three discrete periods throughout the experiment where the LED light had to be repaired and repositioned within the measurement cabinet. The vector f is the vector of random effects of “flat” within a block with and design matrix . Here the term “flat” refers to a sequential flat number to account for temporal variation between measured flats of plants. The vector l is the vector of random effects of the interaction between row leaf position and column leaf position with and design matrix . The vector is the vector of random genotypic effects of accessions with and design matrix and is the vector of residuals with distribution . The matrix represents the Kronecker product of first-order autoregressive processes across the row and column plant positioning within a flat, respectively, and is the spatial residual variance. The matrices , , , and are the identity matrices of the same dimensions as “flat” within the block, genotypic effects, row leaf position, and column leaf position interaction effect, and block, respectively. Outliers were removed following method 2 of (Bernal-Vasquez et al., 2016), and the Box–Cox power transformation was used on traits with non-normal residuals.
BLUPs were obtained for all accessions and each trait and added to the grand mean for GWAS and TWAS. Genetic and residual variances estimated from the null GWAS model were used to calculate genomic heritability (h2) as the ratio of genetic variance over phenotypic variance (de los Campos et al., 2015). Phenotypic correlations between all traits were tested using Pearson’s correlation (r) at P = 0.05 threshold using cor.mtest() function in package corrplot (Wei and Simko, 2017).
Genomic data collection for GWAS and TWAS
Genotypic data
Genotyping was performed as previously reported (dos Santos et al., 2020). Briefly, DNA from dark-grown etiolated seedling tissue was extracted and placed in 96-well plates following CTAB protocol (Doyle and Doyle, 1987). The genotyping was done using two pairs of restriction enzymes, PstI-HF/HinP1I and PstI-HF/BfaI (New England Biolabs, Ipswich, MA, USA) with the genotyping-by-sequencing (GBS) protocol (Elshire et al., 2011; Morris et al., 2013). Tag alignment was done with Bowtie2 (Langmead and Salzberg, 2012) using the S. bicolor genome v3.1 (www.phytozome.jgi.doe.gov). SNPs were identified using the TASSEL3 GBS pipeline (Glaubitz et al., 2014). Reads that did not perfectly match a barcode and restriction site were discarded. After barcode trimming, all unique 64-bp sequences present >9 times in the dataset and that mapped uniquely to the sorghum genome were selected as "master tags." These were compared with tags in each individual at each genomic address to identify SNPs. SNPs with >95% missing data or minor allele frequency (MAF)<5% were discarded.
A HapMap of 239 whole-genome-resequenced sorghum accessions containing 5.5M biallelic phased SNPs with MAF > 0.01 (Valluru et al., 2019), was used as a reference panel to impute the GBS data. GBS markers were filtered to only consider markers that were also present in the HapMap. Beagle 4.1 (Browning and Browning, 2016) was used under GT mode with Ne set to 150,000, window = 60,000 SNPs, and overlap = 4,000 SNPs. After imputation, markers with AR2 < 0.3 were removed, resulting in 2,457,023 SNPs. LD pruning using PLINK (Chang et al., 2015) eliminated markers in high LD (r2 > 0.9) within 50-kb windows. The final dataset consisted of 422,897 SNPs.
Gene expression data
One hundred and thirteen sorghum accessions were grown for 3′-RNAseq measurement. Environmental conditions were: 12-h/12-h day/night cycle under 500 µmol m−2 s−1PPFD, 25°C daytime/23°C nighttime temperature, and 75% RH. The Shoot and leaf tissues of 2 cm were sampled at third leaf stage. Samples were processed according to (Kremling et al., 2019). Briefly, RNA was extracted using TRIzol (Invitrogen) with Direct-zol columns (Zymo Research), and 3′-RNA-seq libraries were prepared robotically from 500 ng total RNA in 96-well plates on an NXp liquid handler (Beckman Coulter) using QuantSeq FWD kits (Lexogen) according to the manufacturer’s instructions. Libraries were pooled to 96-plex and sequenced with 90-nt single-end reads using Illumina TruSeq primers on an Illumina NextSeq 500 with v2 chemistry at the Cornell University Sequencing facility.
The first 12 bp and Illumina Truseq adapter remnants were removed from each reading using Trimmomatic version 0.32, following kit marker instructions. The splice-aware STAR aligner v.2.4.2a was used to align reads against the sorghum v3.1.1 reference genome annotations, allowing a read to map in at most 10 locations (-outFilterMultimapNmax 10) with at most 4% mismatches (–outFilterMismatchNoverLmax 0.04), while filtering out noncanonical intron motifs (–outFilterIntronMotifs RemoveNoncanonicalUnannotated). Default settings from STAR v.2.4.2a aligner were used to obtain gene-level counts (–quantModel GeneCounts) from the resulting BAM files.
GWAS, TWAS, and FCT
Traits went through an additional normal quantile transformation, then single and multi-trait associations were performed as in (Zhou and Stephens, 2014). Two groups of multi-trait models were considered: (G1) traits describing the speed of change in gsthermal after a decrease in PPFD (Vinitial, tinitial min) and (G2) traits describing overall values of gsthermal (gslight, gsinitial min, gsoscillation max, gsshade, gs Σshade). Both groups of traits went through a step of multivariate outlier removal (Filzmoser et al., 2005) performed before running GWAS and TWAS.
GEMMA (Zhou and Stephens, 2012) was used for single and multivariate GWAS (Zhou and Stephens, 2014). Population structure was accounted for by using principal components (PCs) as fixed effects. Based on the Scree plot, four PCs obtained from PLINK (Chang et al., 2015) using the full SNP dataset (i.e. not LD-pruned) were included in all models. Relatedness was controlled for by a kinship matrix obtained from TASSEL 5 (Bradbury et al., 2007) using the default method (Endelman and Jannink, 2012).
TWAS was tested in developing leaf and shoot growing point tissues with genes expressed in at least half of the tested plants. Analyses were implemented in R 3.3.3 (R Core Team, 2017) with the lm function used for single-trait TWAS and the MANOVA function for multi-trait TWAS. Similar to (Kremling et al., 2019), 29 Peer factors (Stegle et al., 2010) and 5 multidimensional scaling factors were used as covariates. The effect estimate for genes from TWAS was used to determine if there was a positive or negative relationship between transcript abundance and trait variation.
An ensemble approach combining GWAS and TWAS results was performed using the FCT (Kremling et al., 2019). Briefly, to integrate both the results from GWAS and TWAS, each SNP in the top 10% of GWAS analysis was assigned to the nearest gene. The P-values of genes not tested in the TWAS (genes expressed in less than half of tested plants) were set to one. The GWAS and TWAS P-values for each gene were combined using FCT in metap package in R, producing Fisher’s combined P-values.
Candidate gene analysis
Results of GWAS, TWAS, and FCT were used to identify potential candidate genes driving variation in traits. A threshold set at the 0.1% lowest P-values was used to identify candidates for each SNP–trait association, i.e. 423 marker associations per trait. This threshold was chosen to focus the analysis on a minimum number of large-effect variants and to limit the number of false positives. PLINK (Chang et al., 2015) was used to calculate LD blocks with option–blocks and a window of 200 kb and default values for D-prime’s confidence interval (0.7;0.98) (Supplemental Table S1). Genes within these LD blocks were compiled from the Phytozome database for Sorghum bicolor v3.1.1 (Goodstein et al., 2012). Similarly, the top 1% most strongly associated genes from TWAS and FCT were ascertained for each trait and tissue. Genes were selected for further analysis if they were identified by more than one test of phenotype–genotype associations, i.e. they overlapped the top hit for several analyses/tissues (e.g. overlapping top hits for both GWAS and TWAS leaf). The Arabidopsis (A. thaliana) putative orthologs of these genes were collected with INPARANOID (Remm et al., 2001) and used for GO term enrichment analysis in biological function (GO Ontology database DOI: 10.5281/zenodo.4081749 Released 2020-10-09) (Ashburner et al., 2000; Carbon et al., 2019; Mi et al., 2019). PANTHER overrepresentation test was used with Fisher’s test and FDR-adjusted P-values, with significance declared at α < 0.05. GO biological processes significantly and >2.5-fold enriched were further considered, and the genes contained within these GO categories were considered the strongest candidates to underlie variation in stomatal conductance traits.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers for the top candidate genes are in Supplemental Table S5.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Table S1. LD blocks containing the top 0.1% SNPs from GWAS.
Supplemental Table S2. Top 0.1% strongest GWAS results for each trait, including SNP chromosome and position, marker R2 and minor allele effect size.
Supplemental Table S3. TWAS results from each tissue and trait, including chromosome position, R2, and predicted direction of change (+ or −) in phenotype with increased gene expression, for each gene.
Supplemental Table S4. FCT results from each tissue and trait, including the GWAS and TWAS P-values taken from Supplemental Tables S2 and S3 and used to calculate an FCT P-value for each gene.
Supplemental Table S5. Summary of genes appearing in the top results for GWAS, TWAS leaf, TWAS shoot, FCT leaf, and FCT shoot.
Supplemental Table S6. GO enrichment analysis of Arabidopsis putative orthologs of genes overlapping top hits for multiple analyses/tissues.
Supplemental Table S7. Summary of the most promising candidate genes, selected because they belong to a GO biological process category significantly enriched by >2.5-fold among the subset of genes overlapping the top results for GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot.
Supplemental Table S8. BLUPs of all traits for all accessions.
Supplemental Table S9. Full GWAS results for each trait.
Supplemental Figure S1. Pearson’s correlation coefficients (r, top right), pairwise correlation scatterplots (bottom left) and density plots (diagonal) for BLUPs of stomatal traits. Data are the same as in Figure 5.
Supplemental Figure S2. GWAS, TWAS, and FCT results for gslight. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B, Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S3. GWAS, TWAS, and FCT results for gsshade. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B, Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S4. GWAS, TWAS, and FCT results for gsoscillation max. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B, Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S5. GWAS, TWAS, and FCT results for gsinitial min. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B, Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S6. GWAS, TWAS, and FCT results for gs Σshade. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B, Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S7. GWAS, TWAS, and FCT results for gslight − gsshade. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B, Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S8. GWAS, TWAS, and FCT results for tinitial min. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B: Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S9. GWAS, TWAS, and FCT results for Voscillation. A: Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B: Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S10. GWAS, TWAS, and FCT results for gsoscillation max − gsoscillation min. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B, Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S11. GWAS, TWAS, and FCT results for G2. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B, Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S12. GWAS, TWAS, and FCT results for G1. A, Upset plot showing the number of overlapping genes between the top hits in GWAS, TWAS leaf, TWAS shoot, FCT leaf, and/or FCT shoot. B, Manhattan plots of GWAS, TWAS, and FCT results.
Supplemental Figure S13. Photograph (right) and corresponding thermal image (left) of experimental setup.
Supplemental data. Full reference list from literature review of genes in Supplemental Table S7.
Funding
This research was funded by the Advanced Research Projects Agency-Energy (ARPA-E), U.S. Department of Energy (DOE), under Award Number DE-AR0000661, and the Office of Biological and Environmental Research in the DOE Office of Science (DE-SC0018277).
Conflict of interest statement. None declared.
Supplementary Material
The research was conceived by C.P., S.L., E.B., M.G., and A.L. Experiments were performed by C.P., P.B., and R.V. Data was analyzed and interpreted by C.P., S.F., R.V., N.B., R.L., and A.L. C.P. and A.L. wrote the paper with input from all authors. A.L. agrees to serve as the author responsible for contact and ensuring communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Andrew D. B. Leakey (leakey@illinois.edu).
References
- Acevedo-Siaca LG, Coe R, Wang Y, Kromdijk J, Quick WP, Long SP (2020) Variation in photosynthetic induction between rice accessions and its potential for improving productivity. New Phytol 227:1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM, Jegla T (2016) Guard cell sensory systems: recent insights on stomatal responses to light, abscisic acid, and CO2. Curr Opin Plant Biol 33:157–167 [DOI] [PubMed] [Google Scholar]
- Assmann SM, Shimazaki K (1999) The multisensory guard cell. Stomatal responses to blue light and abscisic acid. Plant Physiol 119:809–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball JT, Woodrow IE, Berry JAB (1987) A model predicting stomatal conductance and its contribution to the control of photosynthesis under different environmental conditions. InBiggins J, ed, Progress in Photosynthesis Research. Springer, Netherlands, pp 221–224 [Google Scholar]
- Ballard T, Peak D, Mott K (2019) Blue and red light effects on stomatal oscillations. Funct Plant Biol 46:146–151 [DOI] [PubMed] [Google Scholar]
- Bartlett MK, Klein T, Jansen S, Choat B, Sack L (2016) The correlations and sequence of plant stomatal, hydraulic, and wilting responses to drought. Proc Natl Acad Sci USA 113:13098–13103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304:1494–1497 [DOI] [PubMed] [Google Scholar]
- Best NB, Hartwig T, Budka J, Fujioka S, Johal G, Schulz B, Dilkes BP (2016) Nana plant2 encodes a maize ortholog of the arabidopsis brassinosteroid biosynthesis gene DWARF1, identifying developmental interactions between brassinosteroids and gibberellins. Plant Physiol 171:2633–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal-Vasquez AM, Utz HF, Piepho HP (2016) Outlier detection methods for generalized lattices: a case study on the transition from ANOVA to REML. Theor Appl Genet 129:787–804 [DOI] [PubMed] [Google Scholar]
- Bian C, Guo XY, Zhang Y, Wang L, Xu TD, DeLong A, Dong J (2020) Protein phosphatase 2A promotes stomatal development by stabilizing SPEECHLESS in Arabidopsis. Proc Natl Acad Sci USA 117:13127–13137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Barroco R, Engler JD, Verkest A, Beeckman T, Naudts M, Inze D, De Veylder L (2004) B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16:945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JS (1982) Plant productivity and environment. Science 218:443–448 [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635 [DOI] [PubMed] [Google Scholar]
- Brodribb TJ, Jordan GJ (2011) Water supply and demand remain balanced during leaf acclimation of Nothofagus cunninghamii trees. New Phytol 192: 437–448 [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR (2016) Genotype imputation with millions of reference samples. Am J Human Genet 98:116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burks PS, Kaiser CM, Hawkins EM, Brown PJ (2015) Genomewide association for sugar yield in sweet sorghum. Crop Sci 55:2138–2148 [Google Scholar]
- Butler DG, Cullis BR, Gilmour AR, Gogel BJ (2009) ASReml-R Reference Manual. The State of Queensland, Department of Primary Industries and Fisheries, Brisbane. [Google Scholar]
- Carbon S, Douglass E, Dunn N, Good B, Harris NL, Lewis SE, Mungall CJ, Basu S, Chisholm RL, Dodson RJ, et al. (2019) The gene ontology resource: 20 years and still going strong. Nucl Acids Res 47:D330–D338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in sorghum. Crop Sci 48:30–40 [Google Scholar]
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40:575–585 [DOI] [PubMed] [Google Scholar]
- Chakravorty D, Gookin TE, Milner MJ, Yu YQ, Assmann SM (2015) Extra-large G proteins expand the repertoire of subunits in Arabidopsis heterotrimeric G protein signaling. Plant Physiol 169:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater C, Peng K, Movahedi M, Dunn JA, Walker HJ, Liang YK, McLachlan DH, Casson S, Isner JC, Wilson I, et al. (2015) Elevated CO2-Induced Responses in Stomata Require ABA and ABA Signaling. Curr Biol 25:2709–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Guan LP, Qian PP, Xu F, Wu ZL, Wu YJ, He K, Gou XP, Li J, Hou SW (2016) NRPB3, the third largest subunit of RNA polymerase II, is essential for stomatal patterning and differentiation in Arabidopsis. Development 143:1600–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier L, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Current Biol 15:1196–1200 [DOI] [PubMed] [Google Scholar]
- Deery DM, Rebetzke GJ, Jimenez-Berni JA, Bovill WD, James RA, Condon AG, Furbank RT, Chapman SC, Fischer RA (2019) Evaluation of the phenotypic repeatability of canopy temperature in wheat using continuous-terrestrial and airborne measurements. Front Plant Sci 10: 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deery DM, Rebetzke GJ, Jimenez-Berni JA, James RA, Condon AG, Bovill WD, Hutchinson P, Scarrow J, Davy R, Furbank RT (2016) Methodology for high-throughput field phenotyping of canopy temperature using airborne thermography. Front Plant Sci 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G, Sorensen D, Gianola D (2015) Genomic heritability: What is it? Plos Genetics 11:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza AP, Wang Y, Orr DJ, Carmo-Silva E, Long SP (2020) Photosynthesis across African cassava germplasm is limited by Rubisco and mesophyll conductance at steady state, but by stomatal conductance in fluctuating light. New Phytologist 225:2498–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans RM, Brodribb TJ, Busch FA, Farquhar GD (2019) Plant water-use strategy mediates stomatal effects on the light induction of photosynthesis. New Phytol 222:382–395 [DOI] [PubMed] [Google Scholar]
- DeLucia EH, Chen S, Guan K, Peng B, Li Y, Gomez‐Casanovas N, Kantola IB, Bernacchi CJ, Huang Y, Long SP, Ort DR (2019) Are we approaching a water ceiling to maize yields in the United States? Ecosphere 10 [Google Scholar]
- dos Santos JPR, Fernandes SB, McCoy S, Lozano R, Brown PJ, Leakey ADB, Buckler ES, Garcia AAF, Gore MA (2020) Novel bayesian networks for genomic prediction of developmental traits in biomass sorghum. G3 Genes Genom Genet 10:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow GJ, Bergmann DC, Berry JA (2014) An integrated model of stomatal development and leaf physiology. New Phytolog 201:1218–1226 [DOI] [PubMed] [Google Scholar]
- Doyle J, Doyle J (1987) A rapid procedure for DNA purification from small quantities of fresh leaf tissue. Phytochem Bull 19:11–15 [Google Scholar]
- Durand M, Brendel O, Bure C, Le Thiec D (2019) Altered stomatal dynamics induced by changes in irradiance and vapour-pressure deficit under drought: impacts on the whole-plant transpiration efficiency of poplar genotypes. New Phytol 222:1789–1802 [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Osborne CP, Stromberg CAE, Smith SA, Bond WJ, Christin PA, Cousins AB, Duvall MR, Fox DL, Freckleton RP, et al. (2010) The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science 328:587–591 [DOI] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLos ONE 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman JB, Jannink JL (2012) Shrinkage estimation of the realized relationship matrix. G3 Genes Genomes Genetics 2:1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Feldman M, Gehan MA, Wilson MS, Shyu C, Bryant DW, Hill ST, McEntee CJ, Warnasooriya SN, Kumar I, et al. (2015) A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in setaria. Mol Plant 8:1520–1535 [DOI] [PubMed] [Google Scholar]
- FAO, IFAD, UNICEF, WFP, and WHO (2018) The State of Food Security and Nutrition in the World 2018. Building climate resilience for food security and nutrition. FAO, Rome, p 202 [Google Scholar]
- Faralli M, Matthews J, Lawson T (2019) Exploiting natural variation and genetic manipulation of stomatal conductance for crop improvement. Curr Opin Plant Biol 49:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson JF, Fernandes SB, Monier B, Miller ND, Allen D, Dmitrieva A, Schmuker P, Lozano R, Valluru R, Buckler ES, et al. (2021) Machine learning enabled phenotyping for GWAS and TWAS of WUE traits in 869 field-grown sorghum accessions. Plant Physiol 10.1093/plphys/kiab346 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filzmoser P, Garrett RG, Reimann C (2005) Multivariate outlier detection in exploration geochemistry. Computers Geosci 31:579–587 [Google Scholar]
- Franks PJ, Farquhar GD (2007) The mechanical diversity of stomata and its significance in gas-exchange control. Plant Physiol 143:78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLos ONE 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucl Acids Res 40:D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant OM, Chaves MM, Jones HG (2006) Optimizing thermal imaging as a technique for detecting stomatal closure induced by drought stress under greenhouse conditions. Physiol Plantarum 127:507–518 [Google Scholar]
- Grant OM, Tronina L, Jones HG, Chaves MM (2007) Exploring thermal imaging variables for the detection of stress responses in grapevine under different irrigation regimes. J Exp Bot 58:815–825 [DOI] [PubMed] [Google Scholar]
- Guilioni L, Jones HG, Leinonen I, Lhomme JP (2008) On the relationships between stomatal resistance and leaf temperatures in thermography. Agricul Forest Meteorol 148:1908–1912 [Google Scholar]
- Hadebe ST, Modi AT, Mabhaudhi T (2017) Drought tolerance and water use of cereal crops: a focus on sorghum as a food security crop in Sub-Saharan Africa. J Agron Crop Sci 203:177–191 [Google Scholar]
- Hetherington AM, Woodward FI (2003) The role of stomata in sensing and driving environmental change. Nature 424:901–908 [DOI] [PubMed] [Google Scholar]
- Jones HG (1999) Use of infrared thermometry for estimation of stomatal conductance as a possible aid to irrigation scheduling. Agricul Forest Meteorol 95:139–149 [Google Scholar]
- Jones HG, Stoll M, Santos T, de Sousa C, Chaves MM, Grant OM (2002) Use of infrared thermography for monitoring stomatal closure in the field: application to grapevine. J Exp Bot 53:2249–2260 [DOI] [PubMed] [Google Scholar]
- Kaiser E, Morales A, Harbinson J (2018) Fluctuating light takes crop photosynthesis on a rollercoaster ride. Plant Physiol 176:977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser E, Morales A, Harbinson J, Kromdijk J, Heuvelink E, Marcelis LFM (2015) Dynamic photosynthesis in different environmental conditions. J Exp Bot 66:2415–2426 [DOI] [PubMed] [Google Scholar]
- Kaiser H, Kappen L (2001) Stomatal oscillations at small apertures: indications for a fundamental insufficiency of stomatal feedback-control inherent in the stomatal turgor mechanism. J Exp Bot 52:1303–1313 [PubMed] [Google Scholar]
- Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E, Lee Y (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107:2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstiens G (1996) Cuticular water permeability and its physiological significance. J Exp Bot 47:1813–1832 [Google Scholar]
- Kim CM, Dolan L (2011) Root hair development involves asymmetric cell division in Brachypodium distachyon and symmetric division in Oryza sativa. New Phytologist 192:601–610 [DOI] [PubMed] [Google Scholar]
- Klermund C, Ranftl QL, Diener J, Bastakis E, Richter R, Schwechheimer C (2016) LLM-domain B-GATA transcription factors promote stomatal development downstream of light signaling pathways in Arabidopsis thaliana hypocotyls. Plant Cell 28:646–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester RP, Nohl BM, Diers BW, Ainsworth EA (2016) Has photosynthetic capacity increased with 80 years of soybean breeding? An examination of historical soybean cultivars. Plant Cell Environ 39:1058–1067 [DOI] [PubMed] [Google Scholar]
- Kremling KAG, Diepenbrock CH, Gore MA, Buckler ES, Bandillo NB (2019) Transcriptome-wide association supplements genome-wide association in zea mays. G3 Genes Genomes Genet 9:3023–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–U354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T, Blatt MR (2014) Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol 164:1556–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T, Vialet-Chabrand S (2019) Speedy stomata, photosynthesis and plant water use efficiency. New Phytol 221:93–98 [DOI] [PubMed] [Google Scholar]
- Lawson T, von Caemmerer S, Baroli I (2011) Photosynthesis and stomatal behaviour. InLuttge U, Beyschlag W, Budel B, Francis D, eds, Progress in Botany, Vol 72. Springer-Verlag Berlin, Berlin, pp 265–304 [Google Scholar]
- Le J, Liu XG, Yang KZ, Chen XL, Zou JJ, Wang HZ, Wang M, Vanneste S, Morita M, Tasaka M, et al. (2014) Auxin transport and activity regulate stomatal patterning and development. Nat Commun 5 [DOI] [PubMed] [Google Scholar]
- Leakey ADB, Ferguson JN, Pignon CP, Wu A, Jin Z, Hammer GL, Lobell DB (2019) Water use efficiency as a constraint and target for improving the resilience and productivity of C3 and C4 crops. Annu Rev Plant Biol 70:781–808 [DOI] [PubMed] [Google Scholar]
- Liu HJ, Yan JB (2019) Crop genome-wide association study: a harvest of biological relevance. Plant J 97:8–18 [DOI] [PubMed] [Google Scholar]
- Lobell DB, Burke MB, Tebaldi C, Mastrandrea MD, Falcon WP, Naylor RL (2008) Prioritizing climate change adaptation needs for food security in 2030. Science 319:607–610 [DOI] [PubMed] [Google Scholar]
- Lobell DB, Roberts MJ, Schlenker W, Braun N, Little BB, Rejesus RM, Hammer GL (2014) Greater sensitivity to drought accompanies maize yield increase in the US midwest. Science 344:516–519 [DOI] [PubMed] [Google Scholar]
- McAdam SAM, Brodribb TJ (2015) The evolution of mechanisms driving the stomatal response to vapor pressure deficit. Plant Physiol 167:833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAusland L, Davey PA, Kanwal N, Baker NR, Lawson T (2013) A novel system for spatial and temporal imaging of intrinsic plant water use efficiency. J Exp Bot 64:4993–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAusland L, Vialet-Chabrand S, Davey P, Baker NR, Brendel O, Lawson T (2016) Effects of kinetics of light-induced stomatal responses on photosynthesis and water-use efficiency. New Phytol 211:1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan DH, Lan J, Geilfus CM, Dodd AN, Larson T, Baker A, Horak H, Kollist H, He ZS, Graham I, et al. (2016) The breakdown of stored triacylglycerols is required during light-induced stomatal opening. Curr Biol 26:707–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlyn BE, Duursma RA, Eamus D, Ellsworth DS, Prentice IC, Barton CVM, Crous KY, de Angelis P, Freeman M, Wingate L (2011) Reconciling the optimal and empirical approaches to modelling stomatal conductance. Glob Change Biol 17:2134–2144 [Google Scholar]
- Meng XZ, Chen X, Mang HG, Liu CL, Yu X, Gao XQ, Torii KU, He P, Shan LB (2015) Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol 25:2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Mustilli AC, Genty B, North H, Lefebvre V, Sotta B, Vavasseur A, Giraudat J (2002) Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J 30:601–609 [DOI] [PubMed] [Google Scholar]
- Mi HY, Muruganujan A, Ebert D, Huang XS, Thomas PD (2019) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucl Acids Res 47:D419–D426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, et al. (2013) Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci USA 110:453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes TDG, Zhang D, Raissig MT (2020) Form, development and function of grass stomata. Plant J 101:780–799 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18:2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort DR, Long SP (2014) Limits on Yields in the Corn Belt. Science 344:483–484 [DOI] [PubMed] [Google Scholar]
- Ortiz D, Hu JY, Fernandez MGS (2017) Genetic architecture of photosynthesis in Sorghum bicolor under non-stress and cold stress conditions. J Exp Bot 68:4545–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanatsiou M, Petersen J, Henderson L, Wang Y, Christie JM, Blatt MR (2019) Optogenetic manipulation of stomatal kinetics improves carbon assimilation, water use, and growth. Science 363:1456. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556 [DOI] [PubMed] [Google Scholar]
- Pearcy RW (1990) Sunflecks and photosynthesis in plant canopies. Annu Rev Plant Physiol Plant Mol Biol 41:421–453 [Google Scholar]
- Pena MJ, Zhong RQ, Zhou GK, Richardson EA, O'Neill MA, Darvill AG, York WS, Ye ZH (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19:549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Clay NK, Nelson TM (2008) Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J 56:251–263 [DOI] [PubMed] [Google Scholar]
- Pignon CP (2017) Strategies to improve C4 photosynthesis, water and resource-use efficiency under different atmospheres, temperatures, and light environments. University of Illinois at Urbana-Champaign, Urbana, Illinois [Google Scholar]
- Pignon CP, Leakey ADB, Long SP, Kromdijk J (2021) Drivers of natural variation in water-use efficiency under fluctuating light are promising targets for improvement in sorghum. Front Plant Sci 12:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Liang H, Rozenberg M, Mittler R (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34:187–203 [DOI] [PubMed] [Google Scholar]
- Portis AR (2003) Rubisco activase-Rubisco's catalytic chaperone. Photosynth Res 75:11–27 [DOI] [PubMed] [Google Scholar]
- Prakash PT, Banan D, Paul RE, Feldman MJ, Xie D, Freyfogle L, Baxter I, Leakey ADB (2021) Correlation and co-localization of QTL for stomatal density and canopy temperature under drought stress in Setaria. J Exp Bot 72: 5024–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Australia [Google Scholar]
- Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLos ONE 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Abrash E, Bettadapur A, Vogel JP, Bergmann DC (2016) Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc Natl Acad Sci USA 113:8326–8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regassa TH, Wortmann CS (2014) Sweet sorghum as a bioenergy crop: literature review. Biomass Bioenergy 64:348–355 [Google Scholar]
- Remm M, Storm CEV, Sonnhammer ELL (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314:1041–1052 [DOI] [PubMed] [Google Scholar]
- Sack L, Cowan PD, Jaikumar N, Holbrook NM (2003) The ‘hydrology’ of leaves: co-ordination of structure and function in temperate woody species. Plant Cell Environ 26:1343–1356 [Google Scholar]
- Sagan V, Maimaitijiang M, Sidike P, Eblimit K, Peterson KT, Hartling S, Esposito F, Khanal K, Newcomb M, Pauli D, et al. (2019) UAV-based high resolution thermal imaging for vegetation monitoring, and plant phenotyping using ICI 8640 P, FLIR Vue Pro R 640, and thermoMap cameras. Remote Sens 11 [Google Scholar]
- Serna L (2011) Stomatal development in Arabidopsis and grasses: differences and commonalities. Int J Dev Biol 55:5–10 [DOI] [PubMed] [Google Scholar]
- Shimazaki K, Doi M, Assmann SM, Kinoshita T (2007) Light regulation of stomatal movement. Annu Rev Plant Biol 58:219–247 [DOI] [PubMed] [Google Scholar]
- Sierla M, Waszczak C, Vahisalu T, Kangasjarvi J (2016) Reactive oxygen species in the regulation of stomatal movements. Plant Physiol 171:1569–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair TR, Tanner CB, Bennett JM (1984) Water-use efficiency in crop production. Bioscience 34:36–40 [Google Scholar]
- Soleh MA, Tanaka Y, Nomoto Y, Iwahashi Y, Nakashima K, Fukuda Y, Long SP, Shiraiwa T (2016) Factors underlying genotypic differences in the induction of photosynthesis in soybean Glycine max (L.) Merr. Plant Cell Env 39:685–693 [DOI] [PubMed] [Google Scholar]
- Stegle O, Parts L, Durbin R, Winn J (2010) A bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. Plos Comput Biol 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Monda K, Negi J, Konishi F, Ishikawa S, Hashimoto-Sugimoto M, Goto N, Iba K (2015) Natural variation in stomatal responses to environmental changes among Arabidopsis thaliana ecotypes. Plos ONE 10:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam V, Patel N, Turcotte M, Bosse Y, Pare G, Meyre D (2019) Benefits and limitations of genome-wide association studies. Nat Rev Genet 20:467–484 [DOI] [PubMed] [Google Scholar]
- Taylor SH, Hulme SP, Rees M, Ripley BS, Woodward FI, Osborne CP (2010) Ecophysiological traits in C3 and C4 grasses: a phylogenetically controlled screening experiment. New Phytologist 185:780–791 [DOI] [PubMed] [Google Scholar]
- Valluru R, Gazave EE, Fernandes SB, Ferguson JN, Lozano R, Hirannaiah P, Zuo T, Brown PJ, Leakey ADB, Gore MA, et al. (2019) Deleterious mutation burden and its association with complex traits in sorghum (Sorghum bicolor). Genetics 211:1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialet-Chabrand S, Lawson T (2019) Dynamic leaf energy balance: deriving stomatal conductance from thermal imaging in a dynamic environment. J Exp Bot 70:2839–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vico G, Manzoni S, Palmroth S, Katul G (2011) Effects of stomatal delays on the economics of leaf gas exchange under intermittent light regimes. New Phytol 192:640–652 [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Farquhar GD (1981) Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153:376–387 [DOI] [PubMed] [Google Scholar]
- Waduwara-Jayabahu I, Oppermann Y, Wirtz M, Hull ZT, Schoor S, Plotnikov AN, Hell R, Sauter M, Moffatt BA (2012) Recycling of methylthioadenosine is essential for normal vascular development and reproduction in Arabidopsis. Plant Physiol 158:1728–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Burgess SJ, de Becker EM, Long SHP (2020) Photosynthesis in the fleeting shadows: an overlooked opportunity for increasing crop productivity? Plant J 101:874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CP, Liu S, Dong Y, Zhao Y, Geng AK, Xia XL, Yin WL (2016) PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol J 14:849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Xue XY, Zhu JK, Dong J (2016) Demethylation of ERECTA receptor genes by IBM1 histone demethylase affects stomatal development. Development 143:4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way DA, Pearcy RW (2012) Sunflecks in trees and forests: from photosynthetic physiology to global change biology. Tree Physiol 32:1066–1081 [DOI] [PubMed] [Google Scholar]
- Wei T, Simko V (2017) R package "corrplot": Visualization of a Correlation Matrix. https://cran.r-project.org/web/packages/corrplot/citation.html
- Wong SC, Cowan IR, Farquhar GD (1979) Stomatal conductance correlates with photosynthetic capacity. Nature 282:424–426 [Google Scholar]
- WWAP (2015) The United Nations world water development report 2015: Water for a sustainable world. UNESCO, Paris [Google Scholar]
- Xie J, Fernandes SBMayfield-Jones D, Erice G, Choi M, Lipka AELeakey ADB (2021) Optical topometry and machine learning to rapidly phenotype stomatal patterning traits for maize QTL mapping. Plant Physiol 10.1093/plphys/kiab299 (in press) [DOI] [PMC free article] [PubMed]
- Yu HY, Murchie EH, Gonzalez-Carranza ZH, Pyke KA, Roberts JA (2015) Decreased photosynthesis in the erect panicle 3 (ep3) mutant of rice is associated with reduced stomatal conductance and attenuated guard cell development. J Exp Botany 66:1543–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Zhu HY, Zhang Q, Li MY, Yan M, Wang R, Wang LL, Welti R, Zhang WH, Wang XM (2009) Phospholipase D alpha 1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21:2357–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M (2012) Genome-wide efficient mixed-model analysis for association studies. Nature Genetics 44:821–U136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M (2014) Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat Meth 11:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XY, Huang XH (2019) Genome-wide association studies in rice: how to solve the low power problems? Mol Plant 12:10–12 [DOI] [PubMed] [Google Scholar]
- Zhu XG, Ort DR, Whitmarsh J, Long SP (2004) The slow reversibility of photosystem II thermal energy dissipation on transfer from high to low light may cause large losses in carbon gain by crop canopies: a theoretical analysis. J Exp Bot 55:1167–1175 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.