Abstract
The genome of Mycoplasma hyopneumoniae encodes several immunodominant proteins, including a cytosolic protein (p36), three membranous proteins (p46, p65, and p74), and an adhesin (p97). Cross-reactions with M. flocculare and M. hyorhinis reduce the specificity of conventional serological detection methods. However, certain antigenic determinants of the p36 and p46 proteins have been shown to be specific for M. hyopneumoniae. In the present study, pairs of oligonucleotide primers were designed to permit PCR amplification of entire p36 and p46 genes and of internal fragments of these genes. Specific amplicons could be obtained with as low as 0.5 to 50 pg of extracted chromosomal DNA. No amplification product was obtained when testing p36 and p46 primer pairs with genomic DNA or RNA from other mycoplasma species, bacteria, and viruses commonly associated with respiratory diseases in pigs. By using the single p36-PCR method, a positive reaction was demonstrated in 100% (30 of 30) of lungs from pigs that developed typical lesions associated with an M. hyopneumoniae infection, and no false-positive results were detected when 62 apparently normal lungs were tested. On the other hand, with the single p46-PCR method a sensitivity of 86.6% (26 of 30) and a specificity of 96.7% (60 of 62) were obtained in comparison with the necropsy findings. A mixed infection with M. hyorhinis was diagnosed in 13.3% (4 of 30) of the cases by using species-specific primers for the heterologous p37 gene. The sensitivity of the single p36-PCR method for the detection of M. hyopneumoniae, when tested on tracheobronchial swabs, was 100% (20 positive samples), with a specificity of 93.3% (14 of 15 negative samples), compared to the necropsy findings. Both expected amplicons were obtained with 86.6% (26 of 30) positive lungs when p36 and p46 primers were used simultaneously (multiplex PCR) to further increase the specificity of the PCR assay.
Mycoplasma hyopneumoniae is the primary agent of enzootic pig pneumonia (20, 29), a chronic respiratory disease found at pig farms worldwide that is characterized by high morbidity rates and low mortality rates. It causes considerable economic losses through retarded growth, poor food conversion, and increased susceptibility of pigs to infection by other organisms (25, 34).
The diagnosis of M. hyopneumoniae is usually done by cultivation of the organism or by immunofluorescence tests performed on frozen thin lung sections with polyclonal antibodies (1, 25, 27, 34); A. A. Feenstra, V. Sorensen, N. F. Friis, N. E. Jensen, and V. Bille-Hansen, Proc. 13th Int. Pig Vet. Soc. Congr., 1994). However, due to the fastidious nature of M. hyopneumoniae, its culture and serological identification may take up to 1 month. Moreover, other mycoplasmas, especially M. hyorhinis, easily overgrow and contaminate M. hyopneumoniae cultures (25, 34). Current serological detection methods are further hampered by cross-reactions which have been reported between M. hyopneumoniae, M. hyorhinis, and M. flocculare (2, 16).
With the advances made in molecular biology during the last few years, more is known about M. hyopneumoniae genes. Hence, other methods can be used as diagnostic tools for this organism. One such alternative is the PCR, which has been proven to be a highly specific, sensitive, and rapid technique (6). Several investigators have demonstrated the efficacy of the PCR to detect 16S rRNA, as well as conserved regions of genomic DNA, of M. hyopneumoniae in different clinical specimens (3, 4, 7, 9, 22, 30, 36–38), but none of the previously described methods targeted genes encoding species-specific immunodominant proteins. Studies on the antigenic properties of reference and field isolates of M. hyopneumoniae revealed the presence of several immunodominant proteins, including a cytosolic protein with lactate dehydrogenase activity (p36) (17, 21, 39), three membranous proteins (p46, p65, and p74) (8, 24, 32), and an adhesin (p97) (43). Comparative studies with other mycoplasmas commonly found in pigs demonstrated that the p36 and p46 proteins carry highly conserved species-specific antigenic determinants for M. hyopneumoniae (31, 40).
In the present study, different sets of primers were designed to permit specific amplification of variable-length fragments of both p36 and p46 genes of M. hyopneumoniae. Furthermore, to further increase the specificity for the detection of M. hyopneumoniae, a multiplex PCR approach was designed to simultaneously amplify different portions of the p36 and p46 genes. Both single PCR methods and the multiplex PCR assay were tested on reference mycoplasma strains and clinical specimens. Differential diagnosis with M. hyorhinis infection was achieved by using specific primers of the homologous p37 gene.
MATERIALS AND METHODS
Microorganisms and growth conditions.
The ATCC 25934 strain of M. hyopneumoniae, obtained from the American Type Culture Collection, Rockville, Md., was used as the reference strain in this study. Other mycoplasma strains, walled bacteria, and viruses used in this work are listed in Table 1. The different mycoplasmas and Acholeplasma laidlawii were grown in modified Friis medium (18) containing 20% porcine serum (GIBCO BRL) and fresh yeast extract (GIBCO BRL). The cells were harvested by centrifugation at 12,000 × g for 30 min at 4°C, washed three times, and suspended in sterile 1 × PBS (0.1 M phosphate-buffered saline, pH 7.4). Gram-positive and gram-negative bacteria commonly colonizing the porcine respiratory tract were kindly provided to us by S. Messier, Faculty of Veterinary Medecine, University of Montreal, St-Hyacinthe, Quebec, Canada. The viruses used were the IAF-Klop strain of porcine reproductive and respiratory syndrome virus (PRRSV), the Purdue strain of porcine transmissible gastroenteritis virus (TGEV; ATCC VR-763), the IAF-Q890 strain of porcine encephalomyocarditis virus (EMCV), and the A/Sw/H1N1/Quebec/5393/91 (swQC-91) strain of swine influenza virus. The origins, cultivation, and purification procedures for these reference viruses have been described elsewhere (12, 13, 14, 28).
TABLE 1.
Microorganisms used for testing the specificity of M. hyopneumoniae primers
| Mycoplasma sp., bacterium, or virus | Strain | PCR amplificationa |
|---|---|---|
| Mycoplasma hyopneumoniae | ATCC 25934 | + |
| Mycoplasma hyopneumoniae | ATCC 25095 | + |
| Mycoplasma hyosynoviae | Field | − |
| Mycoplasma flocculare | ATCC 27399 | − |
| Mycoplasma arginini | ATCC 23838 | − |
| Mycoplasma hyorhinis | ATCC 17981 | − |
| Acholeplasma laidlawii | ATCC 23206 | − |
| Escherichia coli | Field | − |
| Haemophilus parasuis | Field | − |
| Streptococcus suis | Field | − |
| Actinobacillus pleuropneumoniae serotypes 1 and 5 | Field | − |
| Bordetella bronchiseptica | Field | W |
| Pasteurella multocida | Field | W |
| PRRSV | IAF-KLOP | − |
| TGEV | Purdue | − |
| EMCV | IAF-Q890 | − |
| Influenza H1N1 | A/SW/Qc91 | − |
+, Amplification; −, no amplification (using the five sets of primers directed against either the p36 or p46 genes; (see Materials and Methods). W, weak reaction eliminated at a higher annealing temperature (55°C).
DNA extraction.
Genomic DNA from the mycoplasmas was obtained after an incubation of 1 h at 37°C with 50 μg of final RNase (Boehringer Mannheim, Laval, Quebec, Canada) per ml and 10% sodium dodecyl sulfate, followed by another incubation of 1 h at 37°C in the presence of 50 μg of proteinase K (Boehringer Mannheim) per ml and 0.5 M EDTA (Sigma-Aldrich Canada, Oakville, Ontario, Canada). The extraction solutions were prepared in water. DNA purification was completed by a phenol-chloroform extraction and precipitation with 100% ethanol according to the method of Sambrook et al. (35).
The bacterial DNAs were obtained by both the method described above and by disrupting colonies in 50 μl 1 × sterile PBS and boiling the mixture for 10 min (crude DNA preparations). Following an incubation period of 10 min at −20°C, the samples were centrifuged at 12,000 × g for 5 min.
The viruses used in this work were already purified and concentrated by isopycnic ultracentrifugation on sucrose or cesium chloride density gradients (12, 13, 14, 28). Genomic DNA or RNA was extracted by using the Trizol kit according to the manufacturer's instructions (GIBCO BRL). Briefly, nucleic acids were extracted from 10- to 100-μl aliquots of the purified viral stocks by using guanidinium isothiocyanate-acid phenol reagent (10), precipitated with isopropanol, and suspended in 20 μl of diethyl pyrocarbonate-treated water containing 2 μl of RNase inhibitor (RNA Guard; Pharmacia LKB, Baie d'Urfé, Pointe-Claire, Quebec, Canada).
Clinical specimens.
Lungs and bronchoalveolar and nasal swabs of 12- to 20-week-old pigs suffering from respiratory problems accompanied by progressive weight losses were obtained from farrow-to-finish operations in Southern Quebec and from Animal Pathology Laboratories of the Ministry of Agriculture in Quebec, Quebec, Canada. Tracheobronchial swabs were taken from 20-week-old pigs at slaughter houses and kindly provided to us by André Broes, Centre pour le Développement du Porc du Québec, Quebec, Canada.
Genomic DNA from clinical samples was extracted using the TriPure isolation reagent according to the manufacturer's instructions (Boehringer Mannheim). Briefly, 0.1 g of fresh or frozen lung specimens was homogenized and incubated in guanidinium isothiocyanate-phenol solution at room temperature for 5 min. Chloroform (Fisher Scientific) was added to separate the mixture into three distinct phases: the lower RNA phase, the organic interphase, and the upper aqueous phase containing the DNA. The upper phase was removed, and the DNA was precipitated with 100% ethanol. After three washes with 0.1 M sodium citrate in 10% ethanol, the DNA pellet was suspended in a 600-μl volume of 8 mM NaOH (Fisher Scientific), and the pH of the sample was adjusted to 8.4 by using 0.1 M HEPES (GIBCO BRL) in order to optimize the DNA for the PCR amplification.
Primers.
The oligonucleotide primers were selected from previously published sequences for the specific immunodominant genes of M. hyopneumoniae ATCC 25934, and the p37 gene of M. hyorhinis ATCC 17981. The GenBank accession numbers were X67286 (21) and D16682 (19) for the p36 and p46 genes of M. hyopneumoniae, respectively, and X14140 (15) for the p37 gene of M. hyorhinis. The sequence analysis for the primer selection was performed by using the McVector 3.5 (International Biothechnologies) and Gene Works 2.2 (IntelliGenetics, Inc., Mountain View, Calif.) programs. The oligonucleotide primers were synthesized in an automated Gene Assembler DNA synthesizer (Pharmacia LKB). The primer designations, nucleotide sequences, and sizes of the amplified products are depicted in Table 2.
TABLE 2.
Oligonucleotide primers used for the single and multiplex PCR assays
| Primer | Sensea | Sequence | Product size (bp) |
|---|---|---|---|
| INT36F | + | 5′-CC GATTAGTGTCTCCCGTTATG-3′ | 300 |
| IR(1) p36 | − | 5′-AGATGAATCACCATGTTCACCCAT-3′ | |
| INT36F | + | 5′-CCGATTAGTGTCTCCCGTTATG-3′ | 600 |
| IR(2) p36 | − | 5′-GGGCCGATGAAACCTATTAAAATAGCT-3′ | |
| FSp36 | + | 5′-GGGCCGATGAAACCTATTAAAATAGCT-3′ | 948 |
| RSp36 | − | 5′-GCCGCGAAATTAAATATTTTTAATTGCATCCTG-3′ | |
| P46IntFS | + | 5′-GCAGCAGGTTGTGGACAGAC-3′ | 580 |
| P46IntRS | − | 5′-CAGCATTTTCGCCTTCAGGAG-3′ | |
| FISp46 | + | 5′-GGGCCGATGAAAAAAATGCTTAGAAAA-3′ | 1,260 |
| RISp46 | − | 5′-GGGCCGTTAGGCATCAGGATTATCAACATT-3′ | |
| Sp37 | + | 5′-GTAGTCAAGCAAGAGGATGT-3′ | 346 |
| Asp37 | − | 5′-GCTGGAGTTATTATACCAGGA-3′ |
+, Sense; −, antisense.
Single PCR assays.
The PCR was performed on mycoplasmas, walled bacteria, and viruses by using 2.5 μl of genomic DNA or RNA. The amplification was performed in a 100-μl reaction mixture containing each deoxynucleoside triphosphate at a concentration of 0.2 mM, plus 50 pmol of each forward and reverse primer, 20 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2 (GIBCO BRL), and 1.0 U of Taq DNA polymerase (GIBCO BRL). The tubes were subjected to 35 amplification cycles of denaturation at 94°C for 60 s, primer annealing at 50°C for 60 s, and elongation at 72°C for 90 s. There was a final extension step at 72°C for 10 min. The PCR amplifications were performed in a DNA Engine Thermocycler (MJ Research model PTC-100, with hot bonnet). Aliquots of 10 μl of the amplified products were analyzed by electrophoresis on 1% agarose gels (Boehringer Mannheim) in TAE buffer (0.04 M Tris-acetate [pH 8.5], 0.002 M EDTA) in the presence of ethidium bromide at 100 V for 1 h and then viewed under UV illumination.
PCR on clinical specimens.
The reaction mixtures and conditions were performed as described above with 5 μl of DNA. Products were electrophoresed on agarose gels and viewed under UV illumination.
Multiplex PCR assay.
The PCR conditions were the same as for the single p36- and p46-PCR amplification methods. The mixtures required 5 μl of DNA, 50 pmol of each p46 primer, and 85 pmol of the p36 primers. Products were electrophoresed and viewed under UV illumination.
(This research was taken in part from a dissertation to be submitted by J. Caron to the INRS-Institut Armand-Frappier, Université du Québec, in partial fulfillment of the requirements for the M.Sc. degree.)
RESULTS
Electrophoretic profiles of PCR amplicons obtained for genes encoding the p36 and p46 immunodominant proteins of M. hyopneumoniae and the specificity of the PCR assays.
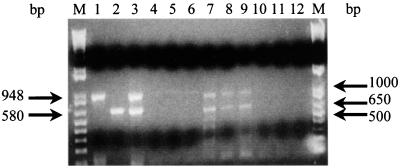
Based on the reported sequences of the immunodominant p36 (948-bp) and p46 (1,260-bp) genes of M. hyopneumoniae (ATCC 25934), primers were designed to permit amplification of the entire open reading frames, as well as internal fragments, of the respective genes. The primer pairs INT36F-IR(1) p36, INT36F-IR(2) p36, and FSp36-RSp36 yielded DNA fragments of 300, 600, and 948 bp of the p36 gene, respectively (Fig. 1A). The primer pairs P46IntFS-P46IntRS and FISp46-RISp46 were used to amplify fragments of 580 and 1,260 bp of the p46 gene (Fig. 1B). All of these primers were also tested for their capacity to amplify DNA fragments from the p36 and p46 genes from two reference strains of M. hyopneumoniae: ATCC 25934 and ATCC 25095. Expected PCR amplification products from the p36 and p46 genes were obtained with both strains. No reactivity was obtained when the PCR was performed with the DNA of other mycoplasma and acholeplasma species that infect pigs. Similarly, no amplified product was observed from genomic DNA of viruses and crude or purified genomic DNA preparations of walled bacteria known to colonize the porcine respiratory tract, except for weak reactions with Bordetella bronchiseptica and Pasteurella multocida (Table 1). The weak signals observed with these two gram-negative bacteria were eliminated by increasing the annealing temperature to 55°C.
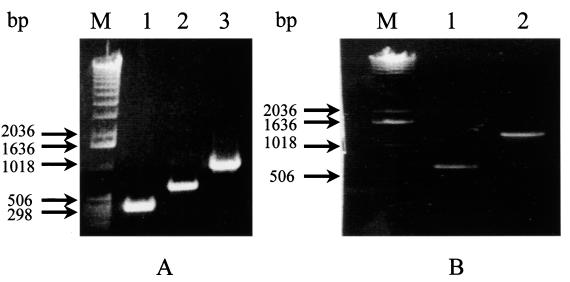
FIG. 1.
Electrophoretic profiles of DNA fragments amplified from the p36 and p46 genes. (A) Different amplified fragments of the p36 gene. Lane 1, 300 bp; lane 2,600 bp; lane 3, 948 bp. (B) Different amplified fragments of the p46 gene. Lane 1, 580 bp; lane 2, 1,260 bp; lane M, 1-kb DNA ladder.
Sensitivity of the single PCR assays.
To assess to sensitivity of the single p36- and single p46-PCR assays, serial dilutions of a preparation of extracted genomic DNA (initial concentration, 200 ng/μl) from M. hyopneumoniae (ATCC 25934) were made and tested for enzymatic amplification with selected p36 and p46 primer pairs. A total of 50 pg of DNA was detected when the entire open reading frame (948 bp) of the p36 gene was amplified (Fig. 2), whereas 0.5 pg of the p36 DNA was detected when the 300-bp fragment was amplified. A lower sensitivity was observed for the single p46-PCR since expected DNA amplicons (580 and 1,260 bp) could not be obtained with dilutions corresponding to less than 0.5 ng of genomic DNA (Fig. 3). Based on the fact that the genome of M. hyopneumoniae is approximately 1,140 kbp, which is equivalent to 1.2 fg of DNA (9), our single p36-PCR detected 4.17 × 102 to 4.17 × 104 organisms for the primers amplifying the 300- and 948-bp fragments, respectively, whereas the single p46-PCR could detect 4.17 × 105 organisms.
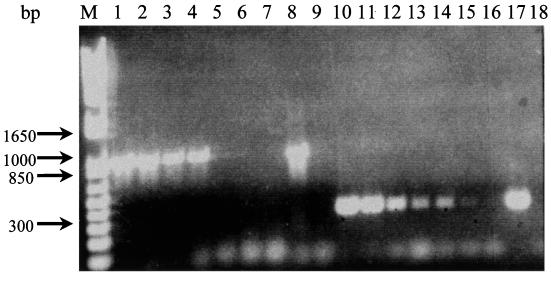
FIG. 2.
Sensitivity of the single p36-PCR procedure in detecting 10-fold dilutions of extracted genomic M. hyopneumoniae ATCC 25934 DNA at an initial concentration of 200 ng/μl by using two sets of primer pairs. Lanes 1 to 9, primer pair FSp36-RSp36; lanes 10 to 18, primer pair INT36F-IR(1) p36. Concentrations: lanes 1 and 10, 10−1; lanes 2 and 11, 10−2; lanes 3 and 12, 10−3; lanes 4 and 13, 10−4; lanes 5 and 14, 10−5; lanes 6 and 15, 10−6; lanes 7 and 16, 10−7; lanes 8 and 17, nondiluted DNA; lanes 9 and 18, negative control. Lane M, 1-kb plus DNA ladder.
FIG. 3.
Sensitivity of the single PCR procedure in detecting 10-fold dilutions of extracted genomic DNA at an initial concentration of 200 ng/μl by using two different primer pairs for the p46 gene. Lanes 1 to 9, primer pair P46IntFS-P46IntRS; lanes 10 to 18, primer pair FISp46-RISp36. Concentrations: lanes 1 and 10, 10−1; lanes 2 and 11, 10−2; lanes 3 and 12, 10−3; lanes 4 and 13, 10−4; lanes 5 and 14, 10−5; lanes 6 and 15, 10−6; lanes 7 and 16, 10−7; lanes 8 and 17, nondiluted DNA; lanes 9 and 18, negative control. Lane M, 1-kb plus DNA ladder.
Detection of M. hyopneumoniae in tracheobronchiolar swabs of pigs with clinical signs of chronic pneumonia.
The efficacy of the single p36-PCR method was first evaluated on 20 tracheobronchiolar swabs collected from pigs at the slaughterhouse with (15) or without (5) lung lesions compatible with a diagnosis of enzootic pneumonia. The DNA of these specimens was extracted with the TriPure isolation reagent. When the PCR was performed with the primer pair INT36F-IR(1) p36 that amplifies the 300-bp fragment of the p36 gene, a positive reaction was obtained with only 33.3% (6 of 15) of the cases with lung lesions. However, when the primer pair FSp36-RSp36 was used, 14 of 15 (93.3%) specimens with typical gross lesions yielded the expected 948-bp amplicon. No positive reaction was obtained with both primer pairs with DNA extracted from the tracheobronchiolar swabs (0 of 5) corresponding to the clinically normal lungs.
Detection of M. hyopneumoniae from lung homogenates.
To study the efficacy of the PCR method, a total of 92 lung specimens, obtained from different pig farms in Southern Quebec, were processed and tested. The data obtained showed that within 2 days, all of the 30 specimens showing typical lesions of enzootic pneumonia were confirmed as positive for an M. hyopneumoniae infection by PCR when using the FSp36-RSp36 primers (yielding a 948-bp fragment). A total of 66.6 and 86.6% of these cases, respectively, were also confirmed as positive for this agent when using primer pairs that permit enzymatic amplification of the 300-bp fragment of the p36 (20 of 30) and the 580-bp fragment of the p46 (26 of 30) genes. An absence of reactivity was noticed when the M. hyopneumoniae FSp36-RSp36 primers were used with DNA extracted from apparently healthy lungs (0 of 62); therefore, a lack of false-positive results was demonstrated. However, a weakly positive reaction (i.e., the presence of amplified bands with an approximate size of 580 bp) was obtained with 2 of the 62 healthy lungs (specificity of 96.7%), when using the p46 specific primer pairs, and in only one case (1 of 62) was a positive signal obtained with the p37 primer pair specific for M. hyorhinis.
Differentiation of M. hyopneumoniae and M. hyorhinis by PCR.
To differentiate lesions caused by M. hyopneumoniae and M. hyorhinis and to detect a mixed infection by the two agents, a primer set was designed and synthesized to amplify a 346-bp fragment of the p37 gene of M. hyorhinis. The Sp37-Asp37 pair was shown to be specific to M. hyorhinis, since no amplicon was obtained from M. hyopneumoniae genomic DNA (Fig. 4). When they were evaluated on clinical specimens, only 4 of 30 (13.3%) lungs with lesions of enzootic pneumonia were found to be positive for M. hyorhinis by PCR. On the other hand, only 1 positive case (1.6%) of M. hyorhinis was detected from the 62 apparently normal lungs tested.
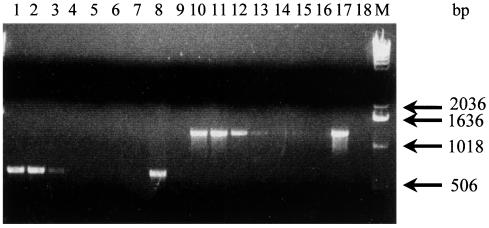
FIG. 4.
Differentiation between M. hyopneumoniae and M. hyorhinis after PCR amplification of extracted genomic DNA with specific p36 and p37 primer pairs, respectively. Lanes 1 and 2, M. hyopneumoniae primers amplifying M. hyopneumoniae and M. hyorhinis DNA, respectively; lanes 3 and 4, M. hyorhinis primers amplifying M. hyorhinis and M. hyopneumoniae, respectively. Lanes M, 1-kb plus DNA ladder.
Detection of M. hyopneumoniae by multiplex PCR.
In order to increase the specificity of our PCR assay, a multiplex PCR approach was developed; it combines the simultaneous amplification of selected p36 and p46 fragments with primer pairs FSp36-RSp36 and P46IntFS-P46IntRS. From the extracted genomic DNA of the ATCC 25934 strain of M. hyopneumoniae, the multiplex PCR amplified two fragments of 948 and 580 bp from the p36 and p46 genes, respectively (Fig. 5, lanes 1 to 3). The multiplex PCR assay also clearly amplified these two genomic fragments from all lung homogenates (26 of 26) from sick pigs previously found to be positive for the p46 580-bp product in the single p46 PCR (Fig. 5, lanes 7 to 9). The multiplex PCR didn't amplify DNA from 20 normal lungs tested (Fig. 5, lanes 10 to 12) or M. hyorhinis genomic DNA (Fig. 5, lanes 4 to 6).
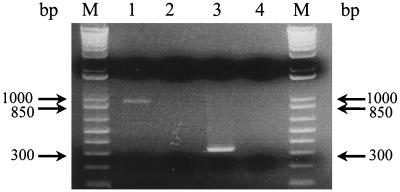
FIG. 5.
Electrophoretic pattern of p36 and p46 amplicons obtained by the multiplex PCR assay. Two fragments of 948 and 580 bp were amplified for the p36 and p46 genes of M. hyopneumoniae ATCC 25934, respectively (lanes 1 and 2). Simultaneous amplification of the p36 and p46 gene fragments from M. hyopneumoniae ATCC 25934 genomic DNA is also shown (lane 3). Both p36 and p46 primer pairs were used to amplify M. hyorhinis genomic DNA by single PCR (lanes 4 and 5) and by multiplex PCR (lane 6). Simultaneous amplification of the p36 and p46 fragments from M. hyopneumoniae field isolates is shown in lanes 7 to 9. Lack of amplification when using both primer pairs in the multiplex PCR assay on healthy lung homogenates is shown in lanes 10 to 12. Lanes M, 1-kb plus DNA ladder.
Cultivation of M. hyopneumoniae from lung homogenates.
To compare the sensitivity of the single and multiplex PCR assays with cultivation, 15 lungs found to be positive for M. hyopneumoniae by the single p36-PCR were processed for cultivation in modified Friis medium. Subpassages were done every 3 to 4 days. After four to five consecutive passages, 13 of the 15 cultures showed evidence of positive growth, as suggested by changes of turbidity and acidification of the medium. Mycoplasma cells were pelleted, and genomic DNA was extracted and processed for confirmation by the single PCR methods for the presence of M. hyopneumoniae and/or M. hyorhinis. Ten cultures (66.6%) were found to contain M. hyopneumoniae only, and in two cultures (13.3%) a mixed infection with M. hyopneumoniae and M. hyorhinis was diagnosed. The remaining culture (6.6%) was contaminated by M. hyorhinis only; no M. hyopneumoniae genomic DNA was amplified by PCR.
DISCUSSION
With the recent advances made in molecular biology, new techniques are becoming readily available for the detection of fastidious microorganisms, such as M. hyopneumoniae. In this research we used the PCR to detect M. hyopneumoniae in lung homogenates and in tracheobronchiolar lavages of clinically infected pigs.
Efforts to control the infection caused by M. hyopneumoniae have been hampered due to the inefficacy of the vaccines currently on the market and to the lack of adequate treatment. Moreover, the major source of infection among pig herds is the healthy carriers. The costs to have separate rooms for pigs of various ages, to have farms sterilized, or to have the piglets by caesarian section are considerable (25, 34). Furthermore, the usual detection method of cultivating and identifying the mycoplasma is extremely time-consuming, taking up to 6 weeks in some cases. The other serological methods are not always appropriate due to the cross-reactions that occur between M. hyopneumoniae, M. hyorhinis, and M. flocculare (2, 16); Feenstra et al., Proc. 13th Int. Pig Vet. Soc. Congr.). Hence, PCR has become a very interesting potential tool for the diagnosis of M. hyopneumoniae.
In this study, the p36 and p46 genes of M. hyopneumoniae were targeted as the PCR templates. From what is reported in the literature, the p36 protein is immunodominant and highly conserved among different strains of M. hyopneumoniae; it has also been reported to carry antigenic determinants that are species specific (17, 21, 39, 40). The p46 protein also elicits an early immune response in swine and is specific to M. hyopneumoniae (31, 32). When the PCR was tested against the various mycoplasmas, walled bacteria, and viruses found in the porcine respiratory tract, expected amplified products could be obtained only with DNA extracted from the M. hyopneumoniae strains tested; thus, the FSp36-RSp36 and P46IntFS-P46IntRS primer pairs for the p36 and p46 genes, respectively, did not allow detection of genomic relationships or cross-reactions between the various mycoplasma species tested. The single p36-PCR method was also found to be very sensitive, detecting as little as 0.5 to 50 pg of mycoplasma genomic DNA. The p46 primer pairs were slightly less sensitive, with no amplification reaction detected with <0.5 ng of genomic DNA. The discrepancy between the sensitivity of the p36 primers when amplifying extracted genomic DNA versus DNA from clinical specimens was probably due to the high GC content at the 5′ end of the antisense RSp36 primer (948 bp) as opposed to the lower GC content of antisense primer IR(1) p36 (300 bp). The RSp36 primer has a higher melting temperature and adheres more strongly to the template DNA; hence, it is more sensitive when the number of organisms present in the samples is lower, as in lung homogenates and tracheobronchial swabs. The p46 primer pairs were less sensitive, detecting 0.5 ng of genomic DNA, a threshold level which is comparable to that previously described by others (3, 5, 7, 22, 30).
When used with clinical specimens, the extraction method yielded a large amount of pure DNA in less than a day; thus, along with the PCR reaction, the results could be obtained within a period of only 2 days. The data obtained by PCR, from the lung homogenates and tracheobronchial swabs, agreed with serological tests performed in parallel (data not shown). With lung homogenates, positive results always correlated with the presence of pneumonic lesions affecting mainly the apical and cardiac lobes; the positive slaughtered pigs originated from farms where several pigs manifested clinical signs of typical M. hyopneumoniae infection (25, 34). As well, 15 of the 30 cases found to be M. hyopneumoniae positive by PCR were tested in culture. As expected, the cultivation process took almost 2 months for some of the field isolates but, in general, a 3-week period was required before confirmation of the isolation in culture was obtained. Only 8 of 12 positive cultures grew well, and the presence of M. hyopneumoniae was either confirmed by PCR or serologically by Western blotting with polyclonal rabbit hyperimmune serum (data not shown). Two of these eight cultures were contaminated by M. hyorhinis, and one culture was found to contain only M. hyorhinis. None of the three contaminated samples was positive for M. hyorhinis by PCR when the original lung samples were tested. M. hyorhinis is less fastidious than M. hyopneumoniae and can be easily cultivated in vitro; consequently, the single p37-PCR method for M. hyorhinis was apparently less sensitive than the cultivation method, which appeared not to be true for M. hyopneumoniae.
In the literature, the simultaneous amplification of mycoplasmal (11, 42), bacterial (26), and viral (33) genes using multiplex PCR assays has been described. These multiplex PCR assays have been used to detect microorganisms in clinical specimens and to increase the specificity of the tests, either by eliminating cross-reactions between strains of the same pathogen or by differentiating between different species. In the present study, a multiplex PCR approach was designed to permit the simultaneous amplification of desired fragments of the p36 and p46 genes of M. hyopneumoniae. Of the 86.6% of the cases which originally tested positive for M. hyopneumoniae by the single p46-PCR, 100% were found to be positive for both genes by the multiplex PCR assay, often with more intense signals. No false-positive results were ever obtained. This assay, albeit less sensitive than the single p36-PCR method, will be very useful for testing clinical specimens that are highly contaminated with other mycoplasmas, especially with M. hyorhinis. The multiplex PCR method should eliminate risks of obtaining false-positive amplified reactions due to cross-reactions with other mycoplasma or bacterial species; since both the p36 and p46 genes have been shown to be highly specific to M. hyopneumoniae, the simultaneous amplification of both expected amplicons will undoubtedly confirm a specific result for this microorganism.
PCR has been used since the early 1990s to detect M. hyopneumoniae (22), but the assay wasn't evaluated under field conditions. Verdin et al. (41) designed a nested-PCR approach that detected M. hyopneumoniae in tracheobronchiolar lavages; this method was found to be more sensitive than serological and immunofluorescent assays during the early stages of infection (42%). The sensitivity of the PCR in that study in fattening animals was comparable to the results obtained in the present study (93.9 to 94.6%). Baumeister et al. (5) developed a PCR detecting M. hyopneumoniae in bronchoalveolar lavage fluids. From pigs with pneumonic lung lesions, they obtained only 30 and 55% positive results with the DNA extracted from bronchial washings and bronchoalveolar lavage fluids, respectively. The single p36-PCR assay described here appears to be more sensitive, since it permitted the detection of 93.3% of the positive cases with tracheobronchial swabs and 100% of the positive lung specimens. On the other hand, the single p46 and multiplex p36-p46 PCR approaches yielded an 86.6% sensitivity.
The detection by PCR of M. hyopneumoniae from live pigs would provide a useful tool for identifying healthy carriers and preventing the spread of the disease. Two studies have reported the detection by PCR of this microorganism from nasal swabs (9, 30). In both cases, primers were selected so as to permit the amplification of species-specific 16S rRNA gene fragments. Mattson et al. (30) used only a limited number of pigs in their experiment and detected the microorganism in only one of five pigs 3 weeks postinfection. The second study (9) demonstrated the use of a nested-PCR assay to detect the presence of M. hyopneumoniae in the nasal cavity. A detection rate of 61% was obtained, but no time period postinfection was mentioned as to when the animals were shedding the mycoplasma in their nasal cavity. In a preliminary study to evaluate the efficacy of the single p36-PCR method to detect the agent in live pigs, approximately 100 nasal swabs were received from various pig farms in Quebec, Canada, either from healthy or infected herds, and all the samples were determined to be negative by PCR (data not shown). From an experimental infection, the presence of M. hyopneumoniae could be detected in nasal swabs at 2 weeks postinfection only (data not shown). These results suggest that M. hyopneumoniae is present in small quantities in the nasal cavity and only for a limited time. More research is needed to confirm these findings. Since the technique was found to be very sensitive for detecting the p36 gene in tracheobronchial swabs taken at slaughterhouses, it should be possible to use such specimens for determining infections in live animals.
In conclusion, both the single and the multiplex PCR assays described here were found to be highly effective tools for detecting the presence of M. hyopneumoniae in different clinical specimens, including samples from live animals (tracheobronchial swabs). With these techniques, the infection can be detected at an early stage, and suitable measures can be taken to limit the damage done to pig farms.
ACKNOWLEDGMENTS
We thank Louise Wilson and Hélène Drolet for technical assistance. We also thank pathologists from the Ministère de l'Agriculture, des Pêcheries, et de l'Alimentation du Quebec for their assistance in obtaining clinical samples from sick pigs. The collaboration of Serge Messier, Department of Microbiology and Pathology, Faculty of Veterinary Medecine, St-Hyacinthe, Quebec, Canada, in providing the different bacterial species was greatly appreciated. We also thank André Broes, Centre de Développement du Porc du Québec, Inc., for providing tracheobronchiolar swabs from slaughterhouse pigs.
This research was partly funded by the Conseil de Recherches en Pêche et Agro-Alimentaire du Québec (grant 4600); Bio Vet Research, Inc., St-Hyacinthe, Quebec, Canada, and the Fédération des Producteurs de Porcs du Québec.
REFERENCES
- 1.Amanfu W, Weng C N, Ross R F, Barnes H J. Diagnosis of mycoplasmal pneumonia of swine: sequential study by direct immunofluorescence. Am J Vet Res. 1984;45:1349–1352. [PubMed] [Google Scholar]
- 2.Armstrong C H, Freeman M J, Sands-Freeman L. Cross-reactions between Mycoplasma hyopneumoniae and Mycoplasma flocculare: practical implications for the serodiagnosis of mycoplasmal pneumonia of swine. Isr J Med Sci. 1987;23:654–656. [PubMed] [Google Scholar]
- 3.Artiushin S, Stipkovits L, Minion F C. Development of polymerase chain reaction primers to detect Mycoplasma hyopneumoniae. Mol Cell Probes. 1993;7:381–385. doi: 10.1006/mcpr.1993.1056. [DOI] [PubMed] [Google Scholar]
- 4.Artiushin S, Minion F C. Arbitrarily primed PCR analysis of Mycoplasma hyopneumoniae field isolates demonstrates genetic heterogeneity. Int J Syst Bacteriol. 1996;46:324–328. doi: 10.1099/00207713-46-1-324. [DOI] [PubMed] [Google Scholar]
- 5.Baumeister A K, Runge M, Gunter M, Feenstra A A, Delbeck F, Kirchhoff H. Detection of Mycoplasma hyopneumoniae in bronchoalveolar lavage fluids of pigs by PCR. J Clin Microbiol. 1998;36:1984–1988. doi: 10.1128/jcm.36.7.1984-1988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bej A K, Mahbubani M H, Atlas R M. Amplification of nucleic acids by polymerase chain reaction (PCR) and other methods and their application. Crit Rev Biochem Mol Biol. 1991;26:301–334. doi: 10.3109/10409239109114071. [DOI] [PubMed] [Google Scholar]
- 7.Blanchard B, Kobisch M, Bové J M, Saillard C. Polymerase chain reaction for Mycoplasma hyopneumoniae detection in tracheobronchiolar washings from pigs. Mol Cell Probes. 1996;10:15–22. doi: 10.1006/mcpr.1996.0003. [DOI] [PubMed] [Google Scholar]
- 8.Brooks E, Faulds D. The Mycoplasma hyopneumoniae 74.5-kD antigen elicits neutralizing antibodies and shares sequence similarity with heat-shock proteins. Vaccines. 1989;89:265–269. [Google Scholar]
- 9.Calsamiglia M, Pijoan C, Trigo A. Application of a nested polymerase chain reaction assay to detect Mycoplasma hyopneumoniae from nasal swabs. J Vet Diagn Investig. 1999;11:246–251. doi: 10.1177/104063879901100307. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Choppa P C, Vojdani A, Tagle C, Andrin R, Magtoto L. Multiplex PCR for the detection of Mycoplasma fermentans, M. hominis and M. penetrans in cell cultures and blood samples of patients with chronic fatigue syndrome. Mol Cell Probes. 1998;12:301–308. doi: 10.1006/mcpr.1998.0186. [DOI] [PubMed] [Google Scholar]
- 12.Dea S, Bilodeau R, Sauvageau R A, Martineau G P. Outbreaks of respiratory and reproductive problems associated with encephalomyocarditis virus in Quebec pig farms. J Vet Diagn Investig. 1991;3:275–282. doi: 10.1177/104063879100300401. [DOI] [PubMed] [Google Scholar]
- 13.Dea S, Garzon S. Identification of coronaviruses by the use of indirect protein A immunogold electron microscopy. J Vet Diagn Investig. 1991;3:297–305. doi: 10.1177/104063879100300405. [DOI] [PubMed] [Google Scholar]
- 14.Dea S, Bilodeau R, Monpetit C, Sauvageau R A, Martineau G P. Antigenic variant of swine influenza virus causing proliferative and necrotizing pneumonia in pigs. J Vet Diagn Investig. 1992;4:380–392. doi: 10.1177/104063879200400403. [DOI] [PubMed] [Google Scholar]
- 15.Dudler R, Schmidhauser C, Parish R W, Wettenhall R E, Schmidt T. A mycoplasma high-affinity transport system and the in vitro invasiveness of mouse sarcoma cell. EMBO J. 1988;7:3963–3970. doi: 10.1002/j.1460-2075.1988.tb03283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman M J, Armstrong C H, Freeman-Sands L L, Lopez-Osuna M. Serological cross-reactivity of porcine reference antisera to Mycoplasma hyopneumoniae, M. flocculare, M. hyorhinis and M. hyosynoviae indicated by the enzyme-linked immunosorbent assay, complement fixation and indirect hemagglutination tests. Can J Comp Med. 1984;48:202–207. [PMC free article] [PubMed] [Google Scholar]
- 17.Frey J, Haldimann A, Kobisch M, Nicolet J. Immune response against the l-lactate dehydrogenase of Mycoplasma hyopneumoniae in enzootic pneumonia of swine. Microb Pathog. 1994;17:313–322. doi: 10.1006/mpat.1994.1077. [DOI] [PubMed] [Google Scholar]
- 18.Friis N F. The pathogenicity of Mycoplasma flocculare. Acta Vet Scand. 1973;14:344–346. doi: 10.1186/BF03547455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Futo S, Seto Y, Mitsuse S, Mori Y, Suzuki T, Kawai K. Molecular cloning of a 46-kilodalton surface antigen (p46) gene from Mycoplasma hyopneumoniae: direct evidence of CGG codon usage for arginine. J Bacteriol. 1995;177:1915–1917. doi: 10.1128/jb.177.7.1915-1917.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodwin R F W, Pomeroy A P, Whittlestone P. Production of enzootic pneumonia in pigs with mycoplasma. Vet Rec. 1965;77:1247–1249. [Google Scholar]
- 21.Haldimann A, Nicolet J, Frey J. DNA sequence determination and biochemical analysis of the immunogenic protein p36, the lactate dehydrogenase (LDH) of Mycoplasma hyopneumoniae. J Gen Microbiol. 1993;139:317–323. doi: 10.1099/00221287-139-2-317. [DOI] [PubMed] [Google Scholar]
- 22.Harasawa R, Koshimizu K, Takeda O, Uemori T, Asada K, Kato I. Detection of Mycoplasma hyopneumoniae by the polymerase chain reaction. Mol Cell Probes. 1991;5:103–109. doi: 10.1016/0890-8508(91)90003-3. [DOI] [PubMed] [Google Scholar]
- 23.Hsu T, Artiushin S, Minion F C. Cloning and functional analysis of the P97 swine cilium adhesin gene of Mycoplasma hyopneumoniae. J Bacteriol. 1997;179:1317–1323. doi: 10.1128/jb.179.4.1317-1323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim M F, Heidari M B, Stull S J, McIntosh M A, Wise K S. Identification and mapping of an immunogenic region of Mycoplasma hyopneumoniae p65 surface lipoprotein expressed in Escherichia coli from a cloned genomic fragment. Infect Immun. 1990;58:2637–2643. doi: 10.1128/iai.58.8.2637-2643.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobisch M, Friis N F. Swine mycoplasmoses. Rev Sci Tech Off Int Epizoot. 1996;15:1569–1606. doi: 10.20506/rst.15.4.983. [DOI] [PubMed] [Google Scholar]
- 26.Lo T M, Ward C K, Inzana T J. Detection and identification of Actinobacillus pleuropneumoniae serotype 5 by multiplex PCR. J Clin Microbiol. 1998;36:1704–1710. doi: 10.1128/jcm.36.6.1704-1710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maes D, Verdonck M, Deluyker H, de Kruif A. Enzootic pneumonia in pigs. Vet Quart. 1996;18:104–109. doi: 10.1080/01652176.1996.9694628. [DOI] [PubMed] [Google Scholar]
- 28.Mardassi H, Athanassious R, Mounir S, Dea S. Porcine reproductive and respiratory syndrome virus: morphological, biological and serological characteristics of Quebec isolates associated to acute and chronic outbreaks of PRSS. Can J Vet Res. 1994;58:55–64. [PMC free article] [PubMed] [Google Scholar]
- 29.Maré C J, Switzer W P. Mycoplasma hyopneumoniae, a causative agent of virus pig pneumonia. Vet Med. 1965;60:841–845. [PubMed] [Google Scholar]
- 30.Mattson J G, Bergstrom K, Wallgren P, Johansson K E. Detection of Mycoplasma hyopneumoniae in nose swabs from pigs by in vitro amplification of the 16S rRNA gene. J Clin Microbiol. 1995;33:893–897. doi: 10.1128/jcm.33.4.893-897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori Y, Hamaoka T, Sato S. Use of monoclonal antibody in an enzyme-linked immunosorbent assay (ELISA) for the detection of antibodies against Mycoplasma hyopneumoniae. Isr J Med Sci. 1987;23:657–662. [PubMed] [Google Scholar]
- 32.Mori Y, Hamaoka T, Sato S, Takeuchi S. Immunoblotting analysis of antibody response in swine experimentally inoculated with Mycoplasma hyopneumoniae. Immunol Immunopathol. 1988;19:239–250. doi: 10.1016/0165-2427(88)90111-0. [DOI] [PubMed] [Google Scholar]
- 33.Ouardani M, Dea S, Wilson L, Jetté R, Montpetit C. Multiplex PCR for detection and typing of porcine circoviruses. J Clin Microbiol. 1999;37:3917–3924. doi: 10.1128/jcm.37.12.3917-3924.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross R F. Mycoplasmal diseases. In: Leman A D, Straw B, Mengeling W, D'Allaire S, Taylor D, editors. Diseases of swine. 7th ed. Ames: Iowa State University Press; 1992. pp. 537–551. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Stärk K D, Nicolet J, Frey J. Detection of Mycoplasma hyopneumoniae by air sampling with a nested PCR assay. Appl Environ Microbiol. 1998;64:543–548. doi: 10.1128/aem.64.2.543-548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stemke G W, Phan R, Young T F, Ross R F. Differentiation of Mycoplasma hyopneumoniae, M. flocculare, and M. hyorhinis on the basis of amplification of a 16S rRNA gene sequence. Am J Vet Res. 1994;55:81–84. [PubMed] [Google Scholar]
- 38.Stemke G W. Gene amplification (PCR) to detect and differentiate mycoplasmas in porcine mycoplasmal pneumonia. Lett Appl Microbiol. 1997;25:327–330. doi: 10.1046/j.1472-765x.1997.00243.x. [DOI] [PubMed] [Google Scholar]
- 39.Stipkovits L, Nicolet J, Haldimann A, Frey J. Use of antibodies against the p36 protein of Mycoplasma hyopneumoniae for the identification of M. hyopneumoniae strains. Mol Cell Probes. 1991;5:451–457. doi: 10.1016/s0890-8508(05)80017-9. [DOI] [PubMed] [Google Scholar]
- 40.Strasser M, Frey J, Bestetti G, Kobisch M, Nicolet J. Cloning and expression of a novel species-specific early immunogenic 36-kilodalton of Mycoplasma hyopneumoniae in Escherichia coli. Infect Immun. 1991;59:1217–1222. doi: 10.1128/iai.59.4.1217-1222.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verdin E, Blanchard B, Kobisch M, Bové J M, Saillard C. Use of nested PCR diagnosis test to detect Mycoplasma hyopneumoniae under field conditions. IOM Lett. 1996;4:101–102. [Google Scholar]
- 42.Wang H, Fadl A A, Khan M I. Multiplex PCR for avian pathogenic mycoplasmas. Mol Cell Probes. 1997;11:211–216. doi: 10.1006/mcpr.1997.0108. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q, Young T, Ross R F. Identification and characterization of a Mycoplasma hyopneumoniae adhesin. Infect Immun. 1995;63:1013–1019. doi: 10.1128/iai.63.3.1013-1019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]