Abstract
A recombinant rabies virus phosphoprotein fusion product (GST-P) was used to generate a series of monoclonal antibodies (MAbs) with anti-P reactivity. Competitive binding assays classified 27 of these MAbs into four groups (I to IV), and 24 of them were deemed to recognize linear epitopes, as judged by their reaction in immunoblots. The linear epitope recognized in each case was mapped by using two series of N- and C-terminally deleted recombinant phosphoproteins. Assessment of the reactivities of representative MAbs to a variety of lyssavirus isolates by an indirect fluorescent antibody test indicated that group I MAbs, which recognized a highly conserved N-terminal epitope, were broadly cross-reactive with all lyssaviruses assayed, while group III MAbs, which reacted with a site overlapping that of group I MAbs, exhibited variable reactivities and group IV MAbs reacted with most isolates of genotypes 1, 6, and 7 only. In contrast, group II MAbs, which recognized an epitope located within a highly divergent central portion of the protein, were exquisitely strain specific. These anti-P MAbs are potentially useful tools for lyssavirus identification and discrimination.
The negative-sense RNA genome of lyssaviruses, of which rabies virus (RV) is the prototype, is organized into five coding regions, N, P, M, G, and L (24). Viral transcription and replication are catalyzed by the viral polymerase complex, composed of the L and P proteins, by mechanisms typical of the Rhabdoviridae (reviewed in reference 1). Lyssavirus diagnosis targets, by an indirect fluorescent antibody (IFA) test (6), the highly expressed and well-conserved N protein (25). Moreover, large panels of anti-N monoclonal antibodies (MAbs) successfully discriminate between many distinct RV strains which circulate in specific host reservoirs in discrete geographical regions (21). Panels of MAbs directed to the RV surface glycoprotein (G) are also sometimes used for RV strain discrimination (21), and both anti-N and anti-G MAbs are reported to discriminate between the various serotypes comprising the lyssavirus genus (22). However, low and variable G protein expression in infected cells, especially in cell culture, can sometimes confound interpretation of anti-G MAb reactivities. Many genetic analyses of lyssaviruses have targeted the N (2, 14, 23) and G (26) genes, thereby complementing the available antigenic data. N gene analysis distinguished six lyssavirus genotypes corresponding to prior serological distinctions within the genus (2). Thus, serogenotypes 1 to 4 include classical RV, Lagos bat virus (LBV), Mokola virus (MOKV), and Duvenhage virus (DUVV), while two groups of European bat lyssaviruses (EBL-1 and EBL-2) were assigned to genotypes 5 and 6, respectively. Recently discovered Australian bat lyssaviruses (ABLs) have been tentatively assigned to genotype 7 (11). Moreover, the RV lineage was subdivided into several discrete clades corresponding to geographically separated and host-specific viral populations. Despite the sensitivity afforded by genotyping strategies, the application of reverse transcription-PCR (RT-PCR) technology and subsequent sequence characterization of its products (16) requires technical facilities, expertise, and considerable care to avoid spurious results. These restrictions may impose difficult, if not impossible, constraints on laboratories, especially in developing countries where rabies enzootics pose a significant threat to human health.
The relatively nonconserved P protein (25), which is produced by the infected cell in significant quantities, represents a potentially useful alternative antigen for lyssavirus discrimination. Historically, this target was poorly characterized antigenically due to limitations in generating monospecific anti-P antibodies from animals receiving whole virus, and detailed genetic information was available for only a small number of RV strains (5, 18, 19, 27) and for MOKV (2). A recent study on genetic variability of the lyssavirus P locus has indicated the presence of both conserved and highly divergent domains within the P protein (S. A. Nadin-Davis et al., manuscript in preparation). Such regions are potentially interesting targets for the diagnosis and serological typing of lyssaviruses, respectively. In this report the use of several recombinant rabies P proteins to generate and characterize a series of anti-P MAbs with potential typing utility is described.
MATERIALS AND METHODS
Construction of a GST-P expression cassette.
The rabies P gene (Ontario arctic fox strain, Type 1) was amplified from total RNA extracted from infected brain tissue by RT-PCR essentially as described elsewhere (17, 18). To facilitate product subcloning, the positive sense primer (5′-GTCGGAGATCTATATGAGCAAAATCTTT-3′) directed to the start of the P gene open reading frame (ORF) (in boldface) incorporated a 5′-terminal BglII restriction endonuclease site (underlined); the negative-sense primer [5′-GAGG(GA)TTTTTGAGTGTCCTCGTC-3′] was directed close to the start of the neighboring M gene. The purified PCR product was digested with BglII and EcoRI, the latter site being present downstream of the P gene ORF, and cloned into the pGEX-3X vector (Amersham Pharmacia Biotech, Baie d'Urfe, Quebec, Canada) restricted with BamHI and EcoRI. A selected clone (pGEX-RabP) was employed for isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression of a GST-P fusion protein.
Nucleotide sequencing.
The sequences of all clones generated in these studies were confirmed by manual sequencing using plasmid DNA templates prepared with a Wizard plasmid isolation kit (Promega Corp., Madison, Wis.), 32P-labeled primers and the fmol Cycle Sequencing Kit (Promega).
Expression of the GST-P product and generation of murine hybridomas.
Escherichia coli cells (TG1 strain) transformed with pGEX-RabP were used for GST-P fusion product expression as described elsewhere (GST gene fusion manual, Pharmacia Biotech, Baie d'Urfe, Quebec, Canada) except that IPTG induction was performed at room temperature in the absence of glucose. For large-scale production, cells of a 100-ml culture were harvested, resuspended in 15 ml of ice-cold 1× phosphate-buffered saline (PBS) containing 67 μg of phenylmethylsulfonyl fluoride per ml and lysed using a French press at 1,500 lb/in2. Particulate material was removed by low-speed centrifugation, (200 × g) for 15 min, and GST-P was affinity purified by using glutathione-Sepharose 4B resin in a batch procedure (GST gene fusion manual [Pharmacia Biotech]). The final eluate was concentrated and exchanged into PBS (pH 7.2) using a Centricon 10 unit (Amicon, Beverly, Mass.), and the protein concentration was estimated by obtaining optical density readings at 280 nm; the yield was generally 5 mg/liter of bacterial culture.
BALB/c mice were initially immunized with crude extracts of E. coli cells, in which GST-P fusion protein had been expressed and mixed 1:1 with TiterMax (Vaxcel, Inc., Norcross, Ga.); the animals were subsequently boosted with purified GST-P fusion protein prior to spleen collection for hybridoma production.
Screening of hybridomas for anti-P reactivity.
Hybridoma supernatants were screened by two methods: (i) an IFA test performed according to the method of Lafon (15) and employing RV-infected (arctic fox strain) BHK21 C13 cells and (ii) an indirect enzyme-linked immunosorbent assay (ELISA), utilizing affinity-purified glutathione S-transferase (GST) or GST-P fusion protein as antigen. Only those MAbs found to be positive by IFA and reacting specifically with the GST-P protein, but not GST, by ELISA were selected for further characterization.
Generation of a series of C-terminally truncated His6-tagged phosphoprotein fusions.
A positive-sense primer (5′-GGTACCCGGGATGAGCAAAATCTTTGTCAATC-3′) paired with a pGEX-directed negative-sense primer were used in a PCR on pGEX-RabP to generate a new P construct incorporating a SmaI site (underlined) immediately upstream of the initiating codon. This product was digested with the restriction endonucleases SmaI and EcoRI, blunt ended, and inserted into vector pQE-32 (Qiagen, Inc., Mississauga, Ontario, Canada) restricted at the SmaI site. A plasmid (pQE-RabP) incorporating the P insert in correct orientation encoded an IPTG-inducible His6-tagged phosphoprotein. 3′-terminal deletions of the P gene were generated in pQE-RabP by using the Henikoff protocol (13), and selected clones were employed for fusion product expression. Depending upon the alignment of the reading frame and the stop codon contributed by the vector downstream of the multiple cloning site, these encoded proteins contained a small number (≤6) of additional C-terminal residues. One additional construct was generated by PCR in which the positive-sense primer was paired with a custom negative-sense primer, corresponding to the complementary sequence of codons for amino acids 134 to 140 and incorporating a downstream translational stop signal and a HindIII restriction site.
Generation of a series of N-terminally deleted His6-tagged phosphoproteins.
N-terminally truncated P protein constructs were generated by PCR using a negative-sense primer flanking the pQE-32 multiple cloning site and eight different primers, each corresponding to 20 nucleotides of positive-sense internal P gene sequence and bearing an upstream SacI site. This series of 3′ coterminal P gene fragments was subcloned into the pQE-30 vector at SacI and PstI sites. All selected clones encoded a truncated P protein bearing a 16-amino-acid N-terminal addition within which the His6 tag was contained.
Expression of His6-tagged P proteins.
All pQE-30/32 recombinant vectors were transformed into the E. coli M15, and individual clones were picked for growth and overnight induction by IPTG. Cell extracts were recovered in a denaturing buffer (8 M urea, 0.1 M sodium phosphate, 0.01 M Tris-HCl, pH 8.0), and His6-tagged protein was purified using Ni-nitriloacetic acid resin (QIAexpressionist manual, Qiagen, Inc., Chatsworth, Calif.).
Western blotting.
Immunoblots were performed by standard procedures (12). Protein preparations were subjected to electrophoresis through sodium dodecyl sulfate (SDS)–15% polyacrylamide gels and transferred to nitrocellulose by semidry transfer. Blots were processed for colorimetric detection of the primary MAb by using a goat anti-mouse horseradish peroxidase (HRP) conjugate (Jackson Immunoresearch) with diaminobenzidine and H2O2.
Indirect ELISA screening of hybridoma supernatants.
Ninety-six-well plates (Nunc 269260; Canadian Life Technologies, Inc.) were coated with 0.25 μg of purified recombinant protein per well in bicarbonate coating buffer overnight at 4°C. Plates were washed five times with PBS containing 0.05% Tween 20 (PBST) using a Bio-Tek Automated Plate Washer Model EL403 (Mandel Scientific, Guelph, Ontario, Canada), blocked with 2% skim milk in PBST for 1 h at 37°C, and then rewashed. Hybridoma supernatants were applied and incubated at 37°C for 1.5 h. After rewashing the plates with PBST, HRP-conjugated goat anti-mouse immunoglobulin G (IgG; Jackson Immunoresearch) was applied as secondary antibody, and this was detected by using 1 mM peroxide and 4 mM chromogen [2′,2-azinobis (3-ethylbenzthiazol-6-sulfonic acid) [ABTS]) in citrate buffer (pH 4.0). After a 10-min incubation, the absorbance was read at 414 nm by using a Titertek Multiscan MCC MKII plate reader (Labsystem, Helsinki, Finland).
Classification of MAbs by competitive ELISA.
Topographical epitope mapping of the RV P protein was performed by competitive binding assays (7, 8) which employed both unlabeled MAbs (as cell culture supernatant) and biotin-labeled (10) versions of MAbs purified from mouse ascitic fluids. Each biotinylated MAb was titrated against purified GST-P protein to determine the optimal volume required. For competitive ELISAs (cELISAs), equal volumes of unlabeled MAb and appropriately diluted biotinylated MAb were allowed to compete for antigen binding for 1 h at room temperature. After five washes with PBST, streptavidin-HRP conjugate (Kirkegaard and Perry Laboratories Gaithersburg, Md.), diluted 1:1,500 in PBST, was applied and incubated for 1 h at room temperature. After another wash with PBST, ABTS solution was added, and absorbances at 414 nm were read after 10 min. The results were expressed as the mean percent inhibition observed for the unlabeled MAb compared to control wells containing diluent in the place of hybridoma supernatant.
Epitope mapping by cELISA with synthetic peptides.
Synthetic peptides were obtained from Southwest Scientific Resources (Alberquerque, N.Mex.). The sequences were as follows: PVEP1, N′-CSTQTTGRELKKETTS-C′; PVEP2, N′-CGRELKKETTSISSQRD-C′; and PVEP3, N′-CKKETTSISSQRDSQSSKA-C′; the N-terminal cys residue had been included to allow conjugation to a carrier protein if required. These peptides, which correspond to residues 145 to 159 (PVEP1), 150 to 165 (PVEP2), and 154 to 171 (PVEP3), respectively, of the RV (arctic fox strain) P protein were employed in a cELISA in which various amounts of peptides (1 ng to 20 μg) were used in place of labeled MAbs. Primary antibody binding was visualized by using HRP-conjugated goat anti-mouse IgG and ABTS chromogen as described for indirect ELISA.
RESULTS
Generation of a series of antiphosphoprotein MAbs.
IPTG induction of E. coli cells transformed with vector pGEX-RabP (see Materials and Methods) resulted in high-level expression of a 55-kDa protein; this size was close to the expected value for the GST-P fusion product of 59.7 kDa. Furthermore, this 55-kDa product, but not the 27-kDa GST protein produced in cells transformed with the pGEX-3X plasmid, was reactive against an anti-P MAb (16AD8) generated previously from the inoculation of mice with whole-virus preparations of the HEP RV laboratory strain (data not shown). These findings confirmed the authenticity of the fusion product. Mice were inoculated with preparations of the GST-P protein and subsequently used for hybridoma generation as described above. Hybridoma screening yielded several independent IgG class anti-P MAbs, of which 27 have been extensively characterized here.
Anti-P MAb classification by a competitive binding assay.
cELISAs were performed with all 27 anti-P MAbs and with biotin-labeled versions of 7 of them. The MAbs were grouped according to the magnitude of their reciprocal inhibition, with values of >80% indicating that the two antibodies are either directed against the same epitope or that the epitopes are spatially close to each other within the same antigenic site. The percent inhibition results shown in Table 1 clearly divide the epitopes represented by the 27 MAbs into four antigenic sites. Some overlap is observed between sites I and III, but sites II and IV appear to be discrete and nonoverlapping. MAbs were distributed fairly evenly among these four groups (eight in I, six in II, eight in III, and five in IV). Complete inhibition of M960 by group II MAbs was not observed. M960 could not be efficiently labeled with biotin; therefore, complete saturation of the antigenic sites with biotinylated M960 was never possible.
TABLE 1.
Classification of anti-P MAbs by cELISAa
| Unlabeled MAb | % Inhibition of biotin-labeled MAbs
|
Site | ||||||
|---|---|---|---|---|---|---|---|---|
| M953 | M957 | M974 | M960 | M962 | M965 | M971 | ||
| M953 | 99 | 91 | 98 | 29 | 70 | 63 | 7 | I |
| M954 | 94 | 75 | 83 | 22 | 47 | 38 | 9 | |
| M955 | 59 | 50 | 30 | 13 | 26 | 25 | 0 | |
| M956 | 99 | 91 | 99 | 7 | 49 | 48 | 0 | |
| M957 | 99 | 93 | 99 | 10 | 47 | 44 | 0 | |
| M973 | 99 | 96 | 99 | 29 | 66 | 58 | 12 | |
| M974 | 97 | 84 | 94 | 29 | 54 | 50 | 16 | |
| M975 | 99 | 92 | 99 | 2 | 42 | 31 | 0 | |
| M958 | 20 | 11 | 11 | 70 | 17 | 23 | 4 | II |
| M960 | 20 | 21 | 7 | 70 | 16 | 17 | 0 | |
| M961 | 17 | 14 | 10 | 67 | 16 | 24 | 2 | |
| M966 | 20 | 2 | 7 | 55 | 19 | 21 | 0 | |
| M976 | 25 | 6 | 11 | 64 | 19 | 24 | 13 | |
| M977 | 20 | 13 | 14 | 67 | 20 | 13 | 13 | |
| M951 | 24 | 17 | 10 | 0 | 54 | 59 | 0 | III |
| M959 | 45 | 35 | 12 | 0 | 99 | 99 | 0 | |
| M962 | 41 | 27 | 22 | 4 | 99 | 99 | 0 | |
| M964 | 52 | 27 | 24 | 6 | 98 | 99 | 0 | |
| M965 | 43 | 27 | 18 | 0 | 98 | 99 | 0 | |
| M967 | 45 | 28 | 17 | 7 | 99 | 99 | 0 | |
| M970 | 42 | 20 | 11 | 0 | 98 | 99 | 0 | |
| M972 | 54 | 28 | 21 | 22 | 99 | 99 | 7 | |
| M963 | 16 | 0 | 9 | 24 | 21 | 21 | 98 | IV |
| M968 | 27 | 11 | 5 | 15 | 21 | 26 | 97 | |
| M969 | 5 | 1 | 0 | 12 | 25 | 22 | 98 | |
| M971 | 24 | 0 | 12 | 15 | 21 | 15 | 96 | |
| M978 | 11 | 0 | 7 | 0 | 16 | 24 | 88 | |
Each value represents the percent inhibition of the biotin-labeled MAb by the unlabeled MAb. Boxed areas enclose those MAbs recognizing a single antigenic site.
Mapping of linear epitopes recognized by anti-P MAbs.
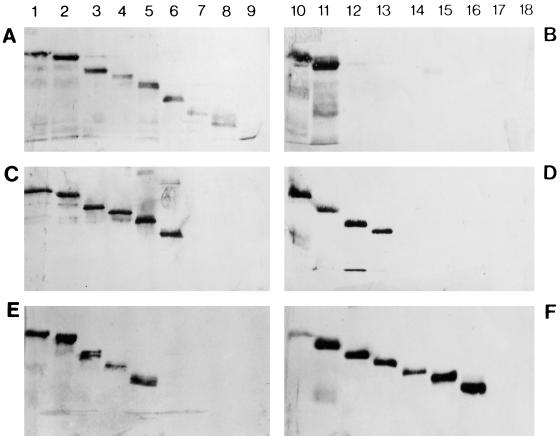
Two series of nested P gene deletions were generated and employed to express a total of 16 His6-tagged P products, of which 8 were C-terminally truncated and 8 were N-terminally truncated. Proteins smaller than the 11.2-kDa product, P1-66, did not accumulate to any measurable level and could not, therefore, be included in the study. All truncated proteins migrated through SDS-polyacrylamide gels with observed sizes close to, but slightly larger than, those predicted; this was possibly due to the positive charge provided by the His6 tag.
Preliminary studies demonstrated that three of the anti-P MAbs (M951, M955, and M978) did not react to the full-length P protein or to several of the truncated products on Western analysis; it is likely that these MAbs recognize conformational epitopes. The remaining 24 MAbs were assessed for their reactivity against all truncated P proteins. Table 2 presents the reactivity patterns observed for representatives of all MAb groups, and representative blots are shown in Fig. 1. Table 3 summarizes the location of each epitope site for all MAbs that could be analyzed in this manner. While the epitopes recognized by MAbs of groups I and III were all located close to amino acids 37 to 66 and could not thus be distinguished by this method, in other respects these mapping data closely matched the classification scheme presented in Table 1. The epitope site for group II MAbs appeared to be localized within residues 145 to 165 (but see below), while group IV MAbs bound to a region encompassing amino acids 82 to 140, with one MAb of this group (M969) mapped more precisely (residues 82 to 113).
TABLE 2.
Reactivity patterns of representative anti-P MAbs against C and N terminally truncated rabies P proteinsa
| MAb | Entire P, residues 1–297 | C terminally truncated P proteins, residues:
|
N terminally truncated P proteins, residues:
|
Group | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–258 | 1–215 | 1–193 | 1–165 | 1–140 | 1–113 | 1–101 | 1–66 | 20–297 | 37–297 | 53–297 | 82–297 | 105–297 | 130–297 | 145–297 | 170–297 | |||
| M957 | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | I |
| M974 | + | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | I |
| M960 | + | + | + | + | + | − | − | − | − | + | + | + | + | + | + | + | − | II |
| M977 | + | + | + | + | + | − | − | − | − | + | + | + | + | + | + | + | − | II |
| M964 | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | III |
| M968 | + | + | + | + | + | + | − | − | − | + | + | + | + | − | − | − | − | IV |
| M969 | + | + | + | + | + | + | + | − | − | + | + | + | + | − | − | − | − | IV |
Residue numbers refer to the residues of the native P protein that are retained in each recombinant P protein.
FIG. 1.
Western blots illustrating the distinctive reactions of anti-P MAbs with truncated P proteins. (A, C, and E) Blots prepared with C-terminally truncated proteins. (B, D, and F) Blots prepared with N-terminally truncated proteins. Lanes were loaded as follows: 1, P1-297; 2, P1-258; 3, P1-215; 4, P1-193; 5, P1-165; 6, P1-140; 7, P1-113; 8, P1-101; 9, P1-66; 10, P20-297; 11, P37-297; 12, P53-297; 13, P82-297; 14, P105-297; 15, P130-297; 16, P145-297; 17, P170-297; 18, none (negative control comprising cell extracts transformed with the plasmid pQE-32). The sizes of the products were confirmed by marker proteins coelectrophoresed on each gel (data not shown). The MAbs tested were M973 (group I) (A and B), M969 (group IV) (C and D), and M958 (group II) (E and F).
TABLE 3.
Summary of antigenic sites mapped
| Group | MAbs in group | Antigenic site (aa)a | Total no. of MAbs |
|---|---|---|---|
| I and III | M953, M954, M956, M957, M959, M962, M964, M965, M967, M970, M972, M973, M974, M975 | 37–66 | 14 |
| II | M958, M960, M961, M966, M976, M977 | 145–165 | 6 |
| IV | M969 | 82–113 | 1 |
| IV | M963, M968, M971 | 82–140 | 3 |
| Unclassified | M951, M955, M978 | 3 | |
| Total | 27 |
Numbers refer to the positions of amino acids (aa) in the native P protein.
Assessment of the reactivity of all anti-P MAbs against a variety of lyssaviruses.
MAbs representative of all four groups were assessed by IFA for their reactivities to a variety of RV strains and to several lyssaviruses of other genotypes. Table 4 summarizes a representative selection of these data. Clearly, the group I MAbs are broadly cross-reactive with all lyssavirus P proteins, including those of the most divergent lyssaviruses LBV and MOKV, while the group IV MAbs were reactive with most isolates except for MOKV, LBV, DUV, and EBL-1. Group III MAbs reacted against most street (wild-type) RVs but exhibited greater variability in their reaction with laboratory strains of RV and other lyssavirus serotypes. The most striking pattern was displayed by group II MAbs, which, as expected, generally reacted strongly with the arctic fox RV strain used for their generation but not to any other lyssavirus isolate. Notably, none of these MAbs reacted substantially to the two nonlyssavirus rhabdoviruses (Adelaide and Manitoba viruses) included in the study.
TABLE 4.
Reactivity of anti-P MAbs against several lyssaviruses
| Viral strain | Reactivitya of MAb (group and no.):
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I
|
II
|
III
|
IV
|
||||||||
| M953 | M974 | M960 | M977 | M951 | M962 | M965 | M972 | M963 | M968 | M971 | |
| RVs (terrestrial) | |||||||||||
| FX:CANARCTIC | +++ | +++ | +++ | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| FX:ECANADA | +++ | +++ | +++ | var | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| SK:WCANADA | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| SK:WCANADA-V | +++ | +++ | − | − | − | − | − | − | +++ | +++ | +++ |
| RAC:US-MIDATLA | +++ | +++ | − | − | + | +++ | +++ | +++ | +++ | +++ | +++ |
| MG:PUERTORICO | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| DG:PERU | +++ | +++ | − | − | +++ | +++ | +++ | +++ | |||
| NPPOLEN | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| PIEMONTE | +++ | +++ | − | − | − | +++ | +++ | +++ | +++ | +++ | +++ |
| DG:SRILANKA | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| MG:AFRICA | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| CAN:AFRICA | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| DG:NIGERIA | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| American bats | |||||||||||
| BB1:CANADA | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| MY:CANADA | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| LB:CANADA | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | + | +++ |
| SH:CANADA | +++ | +++ | − | − | +++ | +++ | +++ | +++ | var | +++ | +++ |
| LA:CANADA | +++ | +++ | − | − | var | +++ | +++ | +++ | var | var | +++ |
| VAMP:ARGENTINA | +++ | +++ | − | − | +++ | +++ | +++ | +++ | var | + | +++ |
| VAMP:MEXICO | +++ | +++ | − | − | +++ | +++ | +++ | +++ | + | +++ | +++ |
| Laboratory rabies strains | |||||||||||
| SAG | +++ | +++ | − | − | − | + | + | + | +++ | +++ | +++ |
| ERA | +++ | +++ | − | − | − | var | var | + | +++ | +++ | +++ |
| LEP | +++ | +++ | − | − | − | +++ | +++ | +++ | +++ | +++ | +++ |
| CVS | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Rabies-related viruses | |||||||||||
| LBV | +++ | +++ | − | − | − | +++ | +++ | +++ | − | − | − |
| MOKV | +++ | +++ | − | − | − | − | − | − | − | − | − |
| DUVV | +++ | +++ | − | − | + | + | +++ | +++ | − | − | − |
| EBL-1 | +++ | +++ | − | − | var | − | var | + | − | − | − |
| EBL-2 | +++ | +++ | − | − | − | +++ | +++ | +++ | +++ | +++ | +++ |
| ABL | +++ | +++ | − | − | var | var | + | + | +++ | +++ | +++ |
| ABL-V | +++ | +++ | − | − | − | − | − | + | +++ | +++ | +++ |
| Nonlyssavirus rhabdoviruses | |||||||||||
| ADELAIDE | − | − | − | − | − | − | − | + | − | − | − |
| MANITOBA | − | − | − | − | − | − | − | − | − | − | − |
Reactivities were scored from +++ (strong) through + (weak) to − (negative). var, variable results between isolates of the same strain. Street RVs are described by using the following host species codes: BB, big brown bat; CAN, canid; DG, dog; FX, fox; LA, Lasiurus bat species; LB, little brown bat; MG, mongoose; MY, Myotis bat species; RAC, raccoon; SH, silver-haired bat; SK, skunk; VAMP, vampire bat; the country or continent of origin is also indicated. Variants are denoted numerically or by “-V.”
Use of synthetic peptides to further delineate the epitope site of group II MAbs.
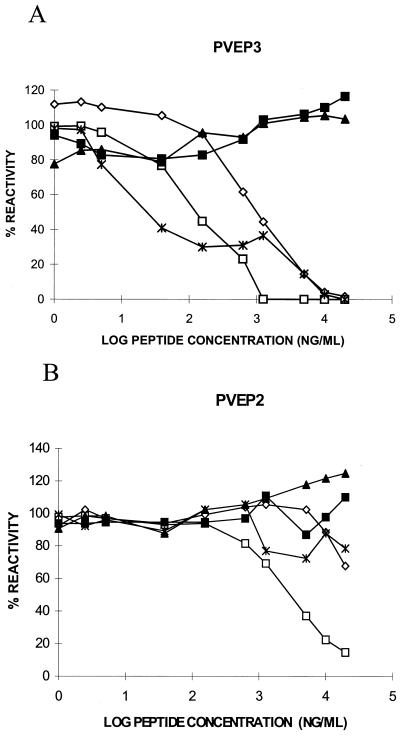
Three synthetic peptides denoted PVEP (P variable epitope) bearing overlapping sequences corresponding to P protein residues 145 to 159 (PVEP1), 150 to 165 (PVEP2), and 154 to 171 (PVEP3) were evaluated for their ability to inhibit MAb binding to the complete P antigen in a cELISA. Five MAbs were selected for testing in this assay: M972, a member of group III, and M963, a representative of group IV, were used as negative controls, while M958, M960, and M977 were representative of group II. The only peptide which interfered with binding of all group II MAbs but not to M972 and M963 was PVEP3 (Fig. 2A). Although the efficiency of this inhibitory effect apparently differed slightly between the group II MAbs, complete inhibition was achieved in each case at a peptide concentration of 10 μg/ml. In contrast, over the peptide concentration range tested, PVEP2 was able to inhibit M960 only (Fig. 2B) and with far less efficiency than PVEP3, indicating that PVEP2 most likely lacked a complete epitope site and hence competed poorly with the antigen for MAb binding. PVEP1 did not inhibit antigen binding by any of the five MAbs tested (data not shown). The observation that the binding of MAbs M972 and M963 to P antigen was unaffected by the presence of any of the three peptides was consistent with previous data mapping the positions of their epitopes elsewhere in the protein. These analyses suggest that residues 154 to 171 contain the epitope site for group II MAbs and that, interestingly, this site is not completely retained within residues 150 to 165.
FIG. 2.
Inhibition of the binding of anti-P MAbs to P antigen by peptides PVEP3 (A) and PVEP2 (B) in a cELISA. Five MAbs were used at the following optimal dilutions: M963, 1:50; M972, 1:5,000; M960, 1:100; M958, 1:100; and M977, 1:4. All absorbance readings were corrected for background levels obtained in the absence of antigen and normalized to a percent reactivity value, with 100% representing the binding of antigen to MAb in the absence of peptide. Symbols: ◊, M958; □, M960; ▴, M963; ■, M972; ✻, M977.
DISCUSSION
The RV P protein, which is functionally analogous to its vesicular stomatitis virus P counterpart, is an RNA polymerase cofactor which binds to both the L and N viral proteins. The domains responsible for these interactions have been examined; N binding requires two distinct N- and C-terminal P protein domains (3, 9), while interaction with the L protein was mapped more precisely to the first 19 amino acids of P (4). This functional requirement of the P N terminus may place considerable constraints on variation in this region of the protein and, indeed, the first 50 amino acids are the most conserved of the entire P protein (S. A. Nadin-Davis, manuscript in preparation). The physically close antigenic sites I and III identified by cELISA, which were mapped to amino acids 37 to 66, overlap this conserved region. While linear epitope mapping could not distinguish these two sites, the differential reactivities of group III MAbs to certain lyssaviruses suggest their target more precisely. For example, of the five ABL isolates examined, group III MAbs reacted weakly or variably with four of them, whereas most were unreactive to the human isolate (ABL-V), which has been noted as segregating distinctly by phylogenetic analysis (Nadin-Davis et al., in preparation). Comparison of the first 66 amino acids of the five ABL P proteins indicated identity over the first 49 residues but differences at several positions between residues 50 and 66. It is therefore likely that group III MAbs recognize a site which includes the less-conserved amino acids downstream of residue 50. Further support for this conclusion was provided by a variant isolate of the Western Canada skunk strain (SK:WCANADA-V [see Table 4]) which, in contrast to other isolates of this strain (SK:WCANADA), was unreactive with all group III MAbs. Sequence comparison of the normal and variant isolates of this strain over the first 66 codons revealed a single coding change; the conserved Met53 was replaced by Thr in the variant form (data not shown). This observation indicates the important contribution of residue 53 to the binding of group III MAbs. In contrast, since group I MAbs bind to a strictly conserved lyssavirus sequence, this site is most likely contained within residues 37 to 50.
Group II MAbs, in contrast, exhibited a very high degree of strain specificity, which can be explained by their binding to a site contained within a highly variable central region (Nadin-Davis et al., in preparation). While the nested protein deletions had suggested this site was contained within residues 145 to 165, cELISA using synthetic peptides located the site at residues 154 to 171. A possible explanation for this discrepancy between the two methods may be considered. The native P protein encodes a serine at position 168; a serine residue, encoded by vector cloning junction sequence, is retained in this position within the truncated P1-165 product. We speculate that Ser168 is an important component of antigenic site II and that the fortuitous retention of this residue in P1-165 facilitates MAb binding. From a combination of all of the data, it can be inferred that site II is probably contained within residues 154 to 168. However, the differential reactivity of peptide PVEP2 with M960, M958, and M977 suggests that the residues involved differ slightly for individual MAbs. Indeed, M960 is also distinguishable from the M958 and M977 MAbs by its ability to bind, in Western analysis, the P protein of the ONT T2 RV variant (unpublished data), which, within the variable epitope, differs from the ONT T1 variant protein used throughout these studies by a single substitution, i.e., Ile160 (T1) versus Val160 (T2). Since M958 and M977 are unreactive to the T2 variant protein as seen by Western analysis (data not shown), Ile160 is clearly implicated as an essential component of their epitopes. These MAbs may therefore facilitate antigenic discrimination of these two Ontario virus variants, which, to date, have been distinguished only by genetic methods (17). Indeed, the failure of M977 to recognize some isolates of the arctic fox strain by IFA (see Table 4) might be an indication of this discriminatory capability, although the variant nature of the viral isolates employed in these studies was not examined.
Comparison of the hydrophilicity profiles of the central variable region of lyssavirus phosphoproteins shows that despite the divergent nature of its primary sequence this domain is highly hydrophilic and has a high probability for exposure on the protein surface, a finding consistent with its highly immunogenic properties. Again by analogy to the VSV P product (see reference 1), it is hypothesized that this variable central region may act as a molecular hinge by which the N and L binding domains on opposite ends of the molecule may interact. This linker function may explain the relative lack of sequence constraints apparently operating on this domain.
Three MAbs generated in these studies—M951, M955, and M978—were deemed to bind conformational epitopes based on their nonreactivity with P protein by immunoblotting. These same three MAbs gave reduced levels of inhibition in competitive binding assays but were nevertheless placed into groups III, IV, and I, respectively. These conformational epitopes may physically overlap the linear epitopes represented by the other MAbs and hence explain their lack of ability to fully inhibit binding of the labeled MAbs. The nature of these conformational epitopes could be more fully understood by carrying out two-way competitive binding assays.
There has been one other report describing the generation of a panel of anti-P MAbs using recombinant P protein generated from the PV laboratory RV strain (20). The sites bound by these MAbs were also examined, and the results can be summarized as follows: of 36 MAbs examined in detail, 3 mapped to the first 19 N-terminal residues, 7 bound to a region defined by residues 20 to 82, 21 bound to an antigenic site located in amino acids 83 to 172, and 5 reacted with residues 177 to 297 at the C terminus. In many respects these results are in accord with ours except that Raux et al. (20) did obtain MAbs directed to the phosphoprotein C terminus; our panel lacked MAbs targeting this region (residues 170 to 297). It appears that, at least in the GST-P fusion product, this area was poorly antigenic. Since Raux et al. employed a His6-tagged protein for MAb generation, it is possible that the different affinity tags employed influenced the presentation of the C-terminal portion of the protein to the mouse immune system. Apart from documenting differential reactivities of certain of these MAbs with the PV and CVS rabies strains, the discriminatory potential of the MAb panel of Raux et al. was not explored (20).
The MAb panel described here has excellent potential utility for lyssavirus strain identification and classification. The broadly cross-reactive MAbs (group I) apparently identify all members of this genus, group IV MAbs recognize the phylogenetically most closely related lyssaviruses (genotypes 1, 6, and 7), while several group III MAbs were reactive to isolates of genotypes 2 and 4, poorly reactive to viruses of genotypes 5 and 7, and unreactive to genotype 3 isolates. Additional anti-P MAbs which recognize genotype 3 isolates have recently been generated (unpublished data), and these will be highly complementary to this MAb panel. The availability of all of these MAbs, especially to laboratories examining Old World isolates, would allow their further evaluation with additional field isolates and may eventually facilitate epidemiological investigations into the prevalence of nonrabies lyssaviruses. The highly strain-specific group II MAbs described here are of clear value in detailed epidemiological studies of rabies in areas in which the arctic fox strain cocirculates with strains in other host reservoirs, e.g., insectivorous bats and raccoons. Moreover, the generation of additional strain-specific MAbs may be entertained through the immunological presentation of this variable antigenic site using either recombinant P protein fragments or synthetic peptides. If successful, this would allow relatively rapid production of additional highly specific tools for the surveillance of areas invaded by multiple, spatially overlapping rabies enzootics. These highly specific reagents could be more advantageous than the large anti-N MAbs now in current use. Finally, the delineation of the binding sites of these MAbs makes them valuable tools for future structure-function analyses of the P protein.
ACKNOWLEDGMENTS
This work was supported in part by a contract from the Ontario Ministry of Natural Resources.
We thank W. Huang for excellent technical support and C. Elmgren for expert assistance in hybridoma production and generation of the anti-P MAbs.
REFERENCES
- 1.Banerjee A K, Barik S. Gene expression of vesicular stomatitis virus genome RNA. Virology. 1992;188:417–428. doi: 10.1016/0042-6822(92)90495-b. [DOI] [PubMed] [Google Scholar]
- 2.Bourhy H, Bachir K, Tordo N. Molecular diversity of the Lyssavirus genus. Virology. 1993;194:70–81. doi: 10.1006/viro.1993.1236. [DOI] [PubMed] [Google Scholar]
- 3.Chenik M, Chebli K, Gaudin Y, Blondel D. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J Gen Virol. 1994;75:2889–2896. doi: 10.1099/0022-1317-75-11-2889. [DOI] [PubMed] [Google Scholar]
- 4.Chenik M, Schnell M, Conzelmann K K, Blondel D. Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J Virol. 1998;72:1925–1930. doi: 10.1128/jvi.72.3.1925-1930.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conzelmann K, Cox J H, Schneider L G, Thiel H. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. [DOI] [PubMed] [Google Scholar]
- 6.Dean D J, Ableseth M K, Atanasiu P. The fluorescent antibody test. In: Meslin F X, Kaplan M M, Koprowski H, editors. Laboratory techniques in rabies. 4th ed. Geneva, Switzerland: World Health Organization; 1996. p. 88-95. [Google Scholar]
- 7.Elmgren L D, Wandeler A I. Competitive ELISA for the detection of rabies virus neutralizing antibodies. In: Meslin F-X, Kaplan M M, Koprowski H, editors. Laboratory techniques in rabies. 4th ed. Geneva, Switzerland: World Health Organization; 1996. p. 200-208. [Google Scholar]
- 8.Elmgren L D. Epitope mapping of lyssavirus structural proteins. Ph.D. thesis. Ottawa, Ontario, Canada: School of Graduate Studies and Research, University of Ottawa; 1999. [Google Scholar]
- 9.Fu Z F, Zheng Y, Wunner W H, Koprowski H, Dietzschold B. Both the N- and the C-terminal domains of the nominal phosphoprotein of rabies virus are involved in binding to the nucleoprotein. Virology. 1994;200:590–597. doi: 10.1006/viro.1994.1222. [DOI] [PubMed] [Google Scholar]
- 10.Goding J W. Monoclonal antibodies: principles and practice. 2nd ed. London, England: Academic Press, Inc.; 1986. [Google Scholar]
- 11.Gould A R, Hyatt A D, Lunt R, Kattenbelt J A, Hengstberger S, Blacksell S D. Characterisation of a novel lyssavirus isolated from Pteropid bats in Australia. Virus Res. 1998;54:165–187. doi: 10.1016/s0168-1702(98)00025-2. [DOI] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. Immunoblotting; p. 471-510. [Google Scholar]
- 13.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 14.Kissi B, Tordo N, Bourhy H. Genetic polymorphism in the rabies virus nucleoprotein gene. Virology. 1995;209:526–537. doi: 10.1006/viro.1995.1285. [DOI] [PubMed] [Google Scholar]
- 15.Lafon M. Techniques for the production, screening and characterisation of monoclonal antibodies. In: Meslin F X, Kaplan M M, Koprowski H, editors. Laboratory techniques in rabies. 4th ed. Geneva, Switzerland: World Health Organization; 1996. pp. 133–144. [Google Scholar]
- 16.Nadin-Davis S A. Polymerase chain reaction protocols for rabies virus discrimination. J Virol Methods. 1998;75:1–8. doi: 10.1016/s0166-0934(98)00106-2. [DOI] [PubMed] [Google Scholar]
- 17.Nadin-Davis S A, Casey G A, Wandeler A. Identification of regional variants of the rabies virus within the Canadian province of Ontario. J Gen Virol. 1993;74:829–837. doi: 10.1099/0022-1317-74-5-829. [DOI] [PubMed] [Google Scholar]
- 18.Nadin-Davis S A, Huang W, Wandeler A I. Polymorphism of rabies viruses within the phosphoprotein and matrix protein genes. Arch Virol. 1997;142:1–14. doi: 10.1007/s007050050133. [DOI] [PubMed] [Google Scholar]
- 19.Poch O, Tordo N, Keith G. Sequence of the 3386 3′ nucleotides of the genome of the AVO1 strain rabies virus: structural similarities in the protein regions involved in transcription. Biochimie. 1988;70:1019–1029. doi: 10.1016/0300-9084(88)90265-9. [DOI] [PubMed] [Google Scholar]
- 20.Raux H, Iseni F, Lafay F, Blondel D. Mapping of monoclonal antibody epitopes of the rabies virus P protein. J Gen Virol. 1997;78:119–124. doi: 10.1099/0022-1317-78-1-119. [DOI] [PubMed] [Google Scholar]
- 21.Rupprecht C E, Dietzschold B, Wunner W H, Koprowski H. Antigenic relationships of lyssaviruses. In: Baer G M, editor. The natural history of rabies. 2nd ed. Boca Raton, Fla: CRC Press; 1991. p. 69-100. [Google Scholar]
- 22.Smith J S, King A A. Monoclonal antibodies for the identification of rabies and non-rabies lyssaviruses. In: Meslin F X, Kaplan M M, Koprowski H, editors. Laboratory techniques in rabies. 4th ed. Geneva, Switzerland: World Health Organization; 1996. pp. 145–156. [Google Scholar]
- 23.Smith J S, Orciari L A, Yager P A, Seidel H D, Warner C K. Epidemiologic and historical relationships among 87 rabies virus isolates as determined by limited sequence analysis. J Infect Dis. 1992;166:296–307. doi: 10.1093/infdis/166.2.296. [DOI] [PubMed] [Google Scholar]
- 24.Tordo N. Characteristics and molecular biology of the rabies virus. In: Meslin F X, Kaplan M M, Koprowski H, editors. Laboratory techniques in rabies. 4th ed. Geneva, Switzerland: World Health Organization; 1996. pp. 28–51. [Google Scholar]
- 25.Tordo N, Kouknetzoff A. The rabies virus genome: an overview. Onderstepoort J Vet Res. 1993;60:263–269. [PubMed] [Google Scholar]
- 26.Tordo N, Badrane H, Bourhy H, Sacramento D. Molecular epidemiology of Lyssaviruses: focus on the glycoprotein and pseudogenes. Onderstepoort J Vet Res. 1993;60:315–323. [PubMed] [Google Scholar]
- 27.Tordo N, Poch O, Ermine A, Keith G, Rougeon F. Walking along the rabies genome: is the large G-L intergenic region a remnant gene? Proc Natl Acad Sci USA. 1986;83:3914–3918. doi: 10.1073/pnas.83.11.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]