Abstract
We have developed a quantitative real-time PCR (TaqMan) assay aimed at measuring the cellular human herpesvirus 8 (HHV-8) DNA load in various clinical samples. Standard curves were obtained by serial dilutions of a control plasmid containing both HHV-8 (ORF73 gene) and the cellular target (human albumin gene). The assay appeared to be very sensitive (100% detection rate for at least 10 copies per well) and specific and was easily reproducible (less than 3% intra-assay variability, 5% interassay variability). This method allowed us to quantify precisely the average HHV-8 copy number per cell in various persistently HHV-8-infected cell lines (BBG-1 cells, n = 200; BC-1 cells, n = 59; BCBL-1 cells, n = 70). A retrospective study was also conducted to assess the HHV-8 DNA load in 12 human immunodeficiency virus-infected patients with either Kaposi's sarcoma (KS; seven patients monitored over a 3-month period) or multicentric Castleman's disease (MCD; five patients). The HHV-8 DNA load ranged from 0 to 9,171 copies/106 cells in low-risk KS patients (T0, I0, S0 according to the classification of the AIDS Clinical Trials group). We also measured the viral loads in MCD patients either during symptomatic periods or during remission. The results are in agreement with previously published data, with high viral loads correlating with clinical symptoms (1.3 × 106 copies/106 cells) and low viral loads correlating with asymptomatic periods (less than 5,000 copies/106 cells).
Human herpesvirus 8 (HHV-8), also known as Kaposi's sarcoma (KS)-associated herpesvirus, is a new member of the subfamily Gammaherpesvirinae that has been associated with all forms of KS (8, 9, 12), multicentric Castleman's disease (MCD) (22), and body cavity-based lymphomas (6). HHV-8 DNA sequences have been detected by PCR in KS lesions, peripheral blood mononuclear cells (PBMCs), and different types of samples (e.g., saliva, semen, and prostatic tissue). The use of semiquantitative techniques has led to the suggestion that the viral load in PBMCs and KS plaques is correlated with the spread and gravity of these lesions (13). It has also been shown that HHV-8 DNA becomes undetectable after resolution of KS lesions in patients infected with human immunodeficiency virus (HIV) and treatment with highly active antiretroviral therapy (5, 14). Immune reconstitution seems to be the main mechanism by which this therapy leads to an improvement in patients with KS. Similarly, the exacerbation of symptoms in HIV-infected patients with MCD is associated with an increase of the viral load in PBMCs (10). So far, only a few data that can be used to assess the relationship between the viral DNA load in PBMCs and the presence of KS or MCD are available. In addition, the studies from which those data were obtained were conducted by semiquantitative and/or competitive PCR techniques, which are known to be time-consuming, require post-PCR handling, and are not suitable for large-scale investigations.
Reproducible, sensitive, and specific quantitative techniques are needed to assess various hypotheses regarding the HHV-8 DNA load and its correlation with different clinical conditions. We have therefore developed a highly sensitive and specific real-time PCR assay for the quantification of the HHV-8 genomes in PBMCs. This approach was based on the recently described TaqMan technology (11). The HHV-8 PCR assay was sensitive enough to detect an average of one copy of a synthetic target diluted in water or in human DNA and did not cross-amplify other human herpesviruses, HIV type 1 (HIV-1), or human T-cell leukemia virus type 1 (HTLV-1). Intra- and interassay variabilities have been evaluated. Finally, this paper presents preliminary results concerning the quantitative measurement of HHV-8 in PBMCs from seven patients with KS and from five patients with MCD.
MATERIALS AND METHODS
Patients and samples.
Seven HIV-infected patients with CD4 cell counts of ≥200 × 106/liter and low-risk KS (T0 I0 S0 according to the classification of the AIDS Clinical Trials Group) who had not previously been treated with systemic anti-KS agents and who had at least four measurable lesions were prospectively included in this study. The clinical features of these patients have been described elsewhere (21). The HHV-8 load was determined at days (D) D0, D15, D30, and D90 after the beginning of treatment with all-trans retinoic acid. As controls, seven HIV-seronegative blood donors were evaluated for HHV-8 loads. Another subgroup of five HIV-infected patients with symptomatic MCD was included. One of the patients in that group was evaluated during the acute clinical phase of the disease and during the remission period.
Blood taken from patients was treated with EDTA, and PBMCs and plasma were separated with Ficoll-Paque. Cells were washed twice with phosphate-buffered saline, pelleted, and frozen at −80°C until extraction (106 cells per vial). For PCR assays, DNA was purified by conventional phenol-chloroform extraction as described previously (1), with slight modifications. Briefly, 106 cells were resuspended in 200 μl of lysis buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 25 mM EDTA, 0.5% sodium dodecyl sulfate, 0.1 mg of proteinase K per ml), and the mixture was incubated at 56°C for 18 h, followed by a 10-min incubation at 94°C. The cell lysate was submitted to two consecutive extractions with phenol-chloroform-isoamyl alcohol, and the DNA was precipitated by centrifugation (15,000 × g; 10 min) in the presence of sodium acetate (0.3 M) and ethanol (70%). It was then briefly washed in the presence of 70% ethanol, dried, resuspended in 200 μl of distilled water, and stored at −80°C.
Cell lines.
BBG-1 is a malignant cell line that has been established from the PBMCs of an AIDS patient with a cutaneous lymphoma associated with Epstein-Barr virus (EBV) and HHV-8 (16). BC-1 and BCBL-1 have been described previously (7, 19). Cell cultures were propagated at 37°C in the presence of 5% CO2 in RPMI 1640 medium supplemented with 10% fetal calf serum and antibiotics.
Plasmids.
A 140-bp DNA fragment derived from the human albumin gene was generated from human DNA by conventional PCR with phosphorylated primers 5′-AAACTCATGGGAGCTGCTGGT-3′ and 5′-GCTGTCATCTCTTGTGGGCTG-3′. The PCR product was cloned into pcDNA3.1/HisC (Invitrogen), leading to pcDNA3.1/HisC-alb. A 675-bp PCR product encompassing nucleotides +2827 to the stop codon of the ORF73 gene was generated with primers 5′-TTGCACGGATCCTCATCCGAG-3′ and 5′-GAGAGGTGAAGCTTTTATGTC-3′ from BBG-1 DNA. This product was digested with BamHI and HindIII and inserted into pcDNA3.1/HisC-alb. The resulting plasmid, pcDNA3.1/HisC-alb-HHV8, was used as a standard to quantify the HHV-8 genome and albumin gene copy number. It was purified with the Qiagen plasmid Maxi kit (Qiagen) and was sequenced by the dideoxynucleotide chain termination method according to the manufacturer's recommendations (ABI Prism dRhodamine terminator cycle sequencing ready mix; Applied Biosystems). The DNA concentration was assessed by spectrophotometry at 260 nm and was determined as an average of three measurements.
Sample preparation and real-time quantitative PCR.
The PCR primers and experimental procedure used to quantify the human albumin gene copy number have been described elsewhere (3).
The PCR primers used for HHV-8 quantification were selected from the ORF73 gene. The upstream and downstream primer sequences were 5′-CCGAGGACGAAATGGAAGTG-3′ and 5′-GGTGATGTTCTGAGTACATAGCGG-3′, respectively. A fluorogenic probe located between the PCR primers was synthesized by GENSET [5′-(6FAM) ACAAATTGCCAGTAGCCCACCAGGAGA (TAMRA)-3′, where 6FAM is 6-carboxy-fluorescein and TAMRA is 6-carboxy-tetramethyl-rhodamine]. The amplification was performed in a 50-μl reaction mixture with a PCR core reagent (Perkin Elmer, Foster City, Calif.). The reaction mixture contained 10 μl of DNA solution, 5 μl of 10× TaqMan buffer, 5 μl of a deoxynucleoside triphosphate solution (2 mM each dATP, dCTP, and dGTP and 4 mM dUTP), 0.5 μl of each primer (20 μM), 0.5 μl of probe (10 μM), 0.5 U of Amp Erase uracil N-glycosylase (UNG), and 0.25 μl of Taq Gold.
Following activation of the UNG (2 min, 50°C) and activation of the AmpliTaq Gold for 10 min at 95°C, 45 cycles (15 s at 95°C and 1 min at 65°C) were performed with an ABI 7700 sequence detector system (Perkin-Elmer). The principle of the real-time PCR has been described elsewhere (11). Briefly, fluorescence measurements were taken every 7 s, and a threshold cycle (CT) value for each sample was calculated by determining the points at which the fluorescence exceeded a threshold limit (10 times the standard deviation of the baseline). The positive control consisted of a plasmid, pcDNA3.1/HisC-alb-HHV8, that contained the targeted sequences. A standard graph of the CT values obtained from serial dilutions (10 to 106 copies) of the plasmid was constructed for both HHV-8 and the human albumin gene. The CT values from unknown samples were plotted on the standard curves, and the ratio of the number of HHV-8 genomes per cell was calculated. For each sample, undiluted and diluted (1:10) DNA extracts were analyzed in duplicate. As a control for cross-contamination, a sample consisting of distilled water was also subjected to the DNA extraction procedure and the resulting extract was amplified in duplicate. Samples were considered negative if the CT values exceeded 45 cycles. In addition, several conditions had to be fulfilled for experimental validation. First, the amplification yield, as deduced from the slope of the standard curve, was expected to be equal or superior to 85%. Second, the absence of inhibitors was assessed by verifying that the copy number calculated for the pure DNA extract decreased linearly when the extract was diluted 1:10. Third, only data from experiments in which the yield of the DNA extraction was ≥20% (as deduced from the initial number of cells and from the final genome copy number measured by albumin PCR) were analyzed.
RESULTS
Design of a real-time PCR assay for quantification of HHV-8 DNA.
Our goal was to design a quantitative PCR-based assay suitable for quantification of the HHV-8 load in human cells. Two independent, quantitative PCR methods were used, and these methods were performed with two separate aliquots of the same DNA extract. The first PCR technique has already been described and has proved to be efficient for quantification of the human albumin gene copy number (3). The second technique was developed to measure the HHV-8 genome copy number. It is based on the amplification of a 143-bp region located within the ORF73 gene, a gene that is unique in the viral genome and that is essential for maintenance of the virus in latently infected cells (2). The sequences of the two primers as well as that of the probe were chosen from a region of ORF73 that is not variable, on the basis of previously published sequences (17, 20) and partial sequencing of ORF73 in the BBG-1 cell line (data not shown). In addition, an extensive search of several databases, including the EMBL and GenBank databases, indicated that neither the primers nor the probe shared significant homology with other known nucleotide sequences.
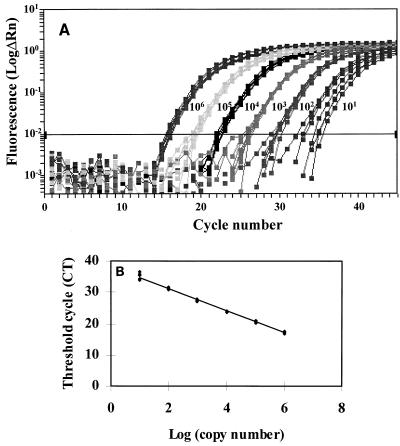
Standard curves were established with a control plasmid, pcDNA3.1/HisC-alb-HHV8, that contained both the target human albumin gene and the HHV-8 sequences. The control plasmid was diluted in water from 106 to 1 copies per sample. Each sample was submitted to the HHV-8 real-time PCR, and amplifications were repeated eight times for each dilution. A standard curve of the CT values plotted against the logarithm of the copy number was constructed. As shown in Fig. 1, HHV-8 quantification proved to be linear over a wide range (from 10 to 106 copies per well). The detection rate was 100% when the copy number was ≥10 copies per well and was 75% for 1 copy per well, which is an agreement with the values that can be estimated from the Poisson probabilities (Table 1). The amplification yield and detection rates were comparable when plasmid dilutions were submitted to the albumin gene PCR (data not shown).
FIG. 1.
Amplification plots (A) and standard curve (B) obtained with the control plasmid. Serial 10-fold dilutions with 106 to 10 copies per reaction well were made in water. Amplification was repeated eight times for each dilution. The normalized reporter signal (Rn) is calculated by dividing the amount of fluorescence emitted by the reporter by the amount of fluorescence emitted by a passive reporter. ΔRn is the amount of the normalized reporter signal minus the amount of the reporter signal before PCR.
TABLE 1.
Sensitivity and intraexperimental variability of HHV-8 DNA quantification
| Copy no. | Control plasmid diluted in watera
|
Control plasmid diluted in human DNAb
|
||||||
|---|---|---|---|---|---|---|---|---|
| Detection rate (%) | Mean CT | SD of CT | CVc (%) | Detection rate | Mean CT | SD of CT | CV (%) | |
| 106 | 100 | 17.16 | 0.22 | 1.3 | ||||
| 105 | 100 | 20.66 | 0.30 | 1.5 | ||||
| 104 | 100 | 23.88 | 0.26 | 1.1 | ||||
| 103 | 100 | 27.48 | 0.21 | 0.7 | ||||
| 102 | 100 | 31.03 | 0.22 | 0.7 | 100 | 31.91 | 0.21 | 0.6 |
| 101 | 100 | 35.16 | 1.02 | 2.9 | 100 | 35.76 | 1.19 | 3.3 |
| 100 | 75 | 36.40d | 0.83d | 2.3 | 75 | 37.76d | 1.03d | 2.7 |
Measurements were repeated eight times in the same experiment.
Measurements were repeated four times in the same experiment.
CV, coefficient of variation.
Calculated for samples with CT values of <45.
A quantification method based on serial dilutions of a standard plasmid in water might not reflect the complex environment of DNA extracted from PBMCs, thus leading to an overestimation of the sensitivity of the assay. To address this question, we performed a real-time PCR with the control plasmid diluted in human DNA extracted from PBMCs of a healthy volunteer. Preliminary experiments indicated that no HHV-8 sequences could be detected in this DNA either by real-time PCR assay or by conventional PCR assays (data not shown). The control plasmid was diluted from 100 to 1 copies per well, and each dilution was submitted to four independent PCRs. As shown in Table 1, the sensitivity and performance of the amplification were only moderately affected by the presence of 2 μg of purified human DNA. In addition, the HHV-8 real-time PCR appeared to be highly reproducible, since the coefficient of variation ranged from 0.6 to 3.3% in intra-assay variability measurements (Table 1).
Specificity and interassay variability.
Various HHV-8-infected and noninfected cells were subjected to real-time PCR. PCR was performed with HHV-8-negative DNA extracted from healthy donor PBMCs (7 patients) and from DG75 (uninfected), B95.8 (EBV), Raji (EBV), 8E5 (HIV-1), and MT2 (HTLV-1) cell lines. DNA extracted from Vero cells cocultured with clinical isolates of HSV-1 or HSV-2 and MRC5 cells infected with clinical isolates of human cytomegalovirus or varicella-zoster virus was also submitted to HHV-8 amplification. In all instances, the PCR reproducibly scored negative for HHV-8 detection. In contrast, when various cell lines that were infected with HHV-8 only (BCBL-1) or coinfected with EBV (BBG-1, BC-1) were assayed in triplicate, the results indicated that the average viral load in these cells was 200 (BBG-1), 59 (BC-1), and 70 (BCBL-1) copies per cell, which is in agreement with previous works (7, 18).
To estimate the interexperimental variability of the viral load measurements in biological samples, PBMCs from a patient with MCD were collected during the acute clinical phase, aliquoted, and stored at −80°C. Each aliquot was submitted to DNA extraction on a different day, and each DNA extract was subjected twice to real-time PCR for HHV-8 and human albumin gene quantification. This analysis was conducted twice per experiment with five independent aliquots. In each experiment measurements were performed both with a pure DNA extract and with a 1:10 dilution. This experiment allowed the overall variability of the viral load measurement to be estimated by real-time PCR. Notably, this includes variations associated with the extraction of DNA, preparation of the PCR mixture, and reaction and analysis procedures. As indicated in Table 2, the coefficient of variation was only 5% when PCR was performed with an undiluted DNA sample and 10% when the extract was diluted.
TABLE 2.
Interexperimental variability: viral load in five separate aliquots of PBMCs from a patient with MCD
| DNA extract | No. of HHV-8 DNA copies per 106 cells in aliquot:
|
CVa (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | Mean | SD | ||
| Undilutedb | 1,209,724 | 1,291,996 | 1,368,835 | 1,243,433 | 1,343,713 | 1,291,540 | 66,530 | 5.2 |
| 1:10 dilutionb | 1,445,277 | 1,421,853 | 1,463,155 | 1,276,061 | 1,119,156 | 1,345,100 | 146,356 | 10.9 |
CV, coefficient of variation.
Each measurement was carried out in duplicate.
Cellular HHV-8 loads in patients with KS and MCD.
The usefulness of the HHV-8 real-time PCR was evaluated in a preliminary study intended to estimate the viral load in PBMCs of patients with various HHV-8-associated diseases. The viral loads of seven patients with KS were therefore measured over a 3-month period, and the results are summarized in Table 3. In addition, the average viral loads of five HIV-infected patients with MCD were also measured, either during the acute clinical phase or during remission. One patient was investigated both during and after an acute phase of the disease. As indicated in Table 4, the viral loads in these patients were as high as 1.24 × 106 copies/106 cells during the acute clinical phase and much lower (less than 5,000 copies/106 cells) during the remission period.
TABLE 3.
HHV-8 load in seven patients with KS
| Day | No. of HHV-8 DNA copies per 106 cells in PBMCs from patient:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| D0 | 174 | 685 | 524 | 407 | 38 | 9,171 | 83 |
| D15 | 80 | 227 | NDa | 0 | 0 | ND | ND |
| D30 | 56 | 108 | 3,760 | 0 | 0 | 5,030 | 59 |
| D90 | 63 | 2,810 | 2,200 | 0 | 0 | ND | 0 |
ND, not determined.
TABLE 4.
HHV-8 load in 5 patients with MCD
| Clinical symptoms | No. of HHV-8 DNA copies per 106 cells in PBMCs from patient:
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| No | 932 | 0 | 4,800 | 1,138 | NDa |
| Yes | 1.24 × 106 | ND | ND | ND | 6.14 × 105 |
ND, not determined.
DISCUSSION
Our results indicate that a real-time PCR assay that combines quantification of HHV-8 and the albumin gene is a sensitive and specific tool for measuring the HHV-8 load in PBMCs from patients with KS or MCD. Repeated measures performed with serial dilutions of a control plasmid or with DNA extracted from PBMCs of HHV-8-infected patients have shown that the procedure described here is also highly reproducible. Given that this technique allows HHV-8 DNA to be quantified without the time-consuming steps of standard PCR and also limits the contamination associated with post-PCR handling, it could be of use in large-scale clinical investigations. The assay allowed us to quantify precisely the HHV-8 copy number in persistently infected cell lines, and the results were consistent with previously published data (7, 18).
The viral loads of the seven KS patients ranged between 0 and 9,171 copies/106 cells before and after all-trans retinoic acid treatment, and for each patient the loads did not change significantly with time (Table 3). This is not surprising as the treatments that sometimes result in HHV-8 load clearance are mainly the antiretroviral combinations. Indeed, the latter treatments can lead to the cure of clinical KS in some HIV-infected individuals, probably by means of immune reconstitution (5, 14). For three patients (patients 4, 5, and 7), the PCR failed to detect HHV-8 DNA after the start of the treatment with all-trans retinoic acid, a situation that has already been described for HIV-infected patients with KS (23). This probably reflects the low levels of HHV-8 DNA in PBMCs from low-risk KS patients rather than a direct effect of the treatment since, in a previous study, none of the patients treated with all-trans retinoic acid experienced a total remission of their KS (21). At the time of sampling, the HIV-infected patients were not receiving highly active antiretroviral therapy combinations; therefore, immune reconstitution is unlikely to have been sufficient to allow HHV-8 clearance. It should be noted that an asymptomatic MCD patient (patient 4, Table 4) also exhibited evolving KS lesions at the time of sampling. Nevertheless, the HHV-8 load in this patient was low (1,138 copies/106 cells), confirming that KS lesions are not necessarily associated with high viral levels in circulating mononuclear cells. In conclusion our results corroborate those of others regarding the HHV-8 loads in PBMCs from HIV-infected patients with KS, who have relatively small numbers of copies compared to the numbers in symptomatic MCD patients. As for now, there is no explanation regarding the difference between the HHV-8 loads in PBMCs from patients with either symptomatic KS or MCD. Some investigators showed by means of semiquantitative analysis that the HHV-8 DNA load is higher (i) in patients with nodular stage KS than in patients with the patch or plaque stage of KS and (ii) in skin biopsy specimens from patients with multicentric and/or visceral KS involvement than in patients with localized KS involvement (15). However, by a quantitative competitive PCR, Boivin and coworkers (4) failed to detect any correlation between HHV-8 DNA load in leukocytes and the tumor burden. Furthermore, the same investigators found no difference between the HHV-8 DNA load in leukocytes and the presence or absence of clinical KS. Altogether these data raise the question of the usefulness of monitoring the HHV-8 DNA load in patients with KS.
In PBMCs from patients with asymptomatic MSD, the viral loads were in the same range as those measured in KS patients (Table 4). As the symptoms develop, there is a very large increase in viral load to 106 copies per 106 cells. In one patient (patient 1), chemotherapy led to a temporary resolution of symptoms and to a very large viral load decrease (from 1.24 × 106 to 932 copies per 106 cells). These results are in agreement with those of Grandadam and collaborators (10), who showed, by means of semiquantitative PCR, that the exacerbation of clinical symptoms in HIV-infected patients with MCD was accompanied by large increases in the HHV-8 DNA loads in their PBMCs. Whether HHV-8 replication is the cause or the consequence of MCD activation still remains to be determined.
MCD can be controlled in many patients with single-dose chemotherapy every 2 weeks, but in most patients the therapy must be continued. Reliable quantitative viral assays have been shown to be useful in monitoring therapy for other viral infections such as those caused by cytomegalovirus or hepatitis B or C virus. In the advent of future standard therapy against HHV-8, it is possible that a simple and reproducible quantitative assay might be useful as a guide to such treatment. Monitoring of the HHV-8 load might be useful for determination of who could discontinue therapy, but further data will be needed to confirm this.
ACKNOWLEDGMENTS
F. Lallemand and N. Désiré have equally contributed to this work. We thank P. Saiag, N. Dupin and V. Calvez for the gift of some biological samples. We also thank V. Fauveau and C. Dutreuil for sequencing and A. Beaumont for carefully reading this manuscript.
This work was supported by a grant from l'Association de Recherche sur le Cancer and Sidaction (8ème appel d'offre).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 2nd ed. New York, N.Y: Greene Publishing Associates, John Wiley & Sons, Inc.; 1992. Preparation of genomic DNA from mammalian tissue; pp. 2.4–2.12. [Google Scholar]
- 2.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 3.Bieche I, Olivi M, Champeme M H, Vidaud D, Lidereau R, Vidaud M. Novel approach to quantitative polymerase chain reaction using real-time detection: application to the detection of gene amplification in breast cancer. Int J Cancer. 1998;78:661–666. doi: 10.1002/(sici)1097-0215(19981123)78:5<661::aid-ijc22>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 4.Boivin G, Gaudreau A, Toma E, Lalonde R, Routy J P, Murray G, Handfield J, Bergeron M G. Human herpesvirus 8 DNA load in leukocytes of human immunodeficiency virus-infected subjects: correlation with the presence of Kaposi's sarcoma and response to anticytomegalovirus therapy. Antimicrob Agents Chemother. 1999;43:377–380. doi: 10.1128/aac.43.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burdick A E, Carmichael C, Rady P L, Tyring S K, Badiavas E. Resolution of Kaposi's sarcoma associated with undetectable level of human herpesvirus 8 DNA in a patient with AIDS after protease inhibitor therapy. J Am Acad Dermatol. 1997;37:648–649. doi: 10.1016/s0190-9622(97)70188-9. [DOI] [PubMed] [Google Scholar]
- 6.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 8.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 9.Dupin N, Grandadam M, Calvez V, Gorin I, Aubin J T, Havard S, Lamy F, Leibowitch M, Huraux J M, Escande J P, et al. Herpesvirus-like DNA sequences in patients with Mediterranean Kaposi's sarcoma. Lancet. 1995;345:761–762. doi: 10.1016/s0140-6736(95)90642-8. [DOI] [PubMed] [Google Scholar]
- 10.Grandadam M, Dupin N, Calvez V, Gorin I, Blum L, Kernbaum S, Sicard D, Buisson Y, Agut H, Escande J P, Huraux J M. Exacerbations of clinical symptoms in human immunodeficiency virus type 1-infected patients with multicentric Castleman's disease are associated with a high increase in Kaposi's sarcoma herpesvirus DNA load in peripheral blood mononuclear cells. J Infect Dis. 1997;175:1198–1201. doi: 10.1086/593567. [DOI] [PubMed] [Google Scholar]
- 11.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y Q, Li J J, Kaplan M H, Poiesz B, Katabira E, Zhang W C, Feiner D, Friedman-Kien A E. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet. 1995;345:759–761. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- 13.Lebbe C, Blum L, Pellet C, Blanchard G, Verola O, Morel P, Danne O, Calvo F. Clinical and biological impact of antiretroviral therapy with protease inhibitors on HIV-related Kaposi's sarcoma. AIDS. 1998;12:F45–F49. doi: 10.1097/00002030-199807000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Martinelli C, Zazzi M, Ambu S, Bartolozzi D, Corsi P, Leoncini F. Complete regression of AIDS-related Kaposi's sarcoma-associated human herpesvirus-8 during therapy with indinavir. AIDS. 1998;12:1717–1719. [PubMed] [Google Scholar]
- 15.Mendez J C, Procop G W, Espy M J, Paya C V, Smith T F. Detection and semiquantitative analysis of human herpesvirus 8 DNA in specimens from patients with Kaposi's sarcoma. J Clin Microbiol. 1998;36:2220–2222. doi: 10.1128/jcm.36.8.2220-2222.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morand P, Buisson M, Collandre H, Chanzy B, Genoulaz O, Bourgeat M J, Pinel N, Leclercq P, Leroux D, Marechal V, Fritsch L, Ruigrok R, Seigneurin J M. Human herpesvirus 8 and Epstein Barr-virus in a cutaneous B-cell lymphoma and a malignant cell line established from the blood of an AIDS patient. Leukemia Lymphoma. 1999;35(3-4):379–387. doi: 10.3109/10428199909145743. [DOI] [PubMed] [Google Scholar]
- 17.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Neill E, Douglas J L, Chien M L, Garcia J V. Open reading frame 26 of human herpesvirus 8 encodes a tetradecanoyl phorbol acetate- and butyrate-inducible 32-kilodalton protein expressed in a body cavity-based lymphoma cell line. J Virol. 1997;71:4791–4797. doi: 10.1128/jvi.71.6.4791-4797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 20.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saiag P, Pavlovic M, Clerici T, Feauveau V, Nicolas J C, Emile D, Chastang C. Treatment of early AIDS-related Kaposi's sarcoma with oral all-trans-retinoic acid: results of a sequential non-randomized phase II trial. Kaposi's Sarcoma ANRS Study Group, Agence Nationale de Recherches sur le SIDA. AIDS. 1998;12:2169–2176. doi: 10.1097/00002030-199816000-00012. [DOI] [PubMed] [Google Scholar]
- 22.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 23.Viviano E, Vitale F, Ajello F, Perna A M, Villafrate M R, Bonura F, Arico M, Mazzola G, Romano N. Human herpesvirus type 8 DNA sequences in biological samples of HIV-positive and negative individuals in Sicily. AIDS. 1997;11:607–612. doi: 10.1097/00002030-199705000-00008. [DOI] [PubMed] [Google Scholar]