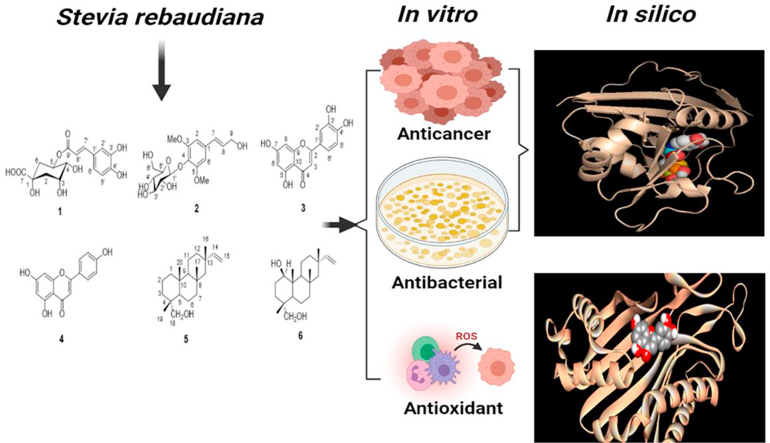
Abstract
The current study was designed to isolate and characterize some bioactive secondary metabolites by using repeated chromatographic and spectroscopic techniques, targeting their anticancer, antimicrobial, and antioxidant properties through in vitro and in silico approaches. Six compounds were isolated and analyzed by thin layer chromatographic technique and the compounds were identified as 5-O-caffeoyl quinic acid (1), syringin (2), luteolin (3), apigenin (4), jhanol (5), and jhanidiol (6) based on spectroscopic methods. The cytotoxic effect of each compound was dose-dependent, and compound 1 showed a higher anti-proliferative effect (IC50 = 181.3 μg/ml) than other compounds (compound 2, 4, 5, and 6). Besides, compound 1 showed the most promising antibacterial activity with a zone of inhibition ranges from 12–15 mm against different strains compared to ciprofloxacin (14–22 mm). In contrast, compound 3 exerted the highest scavenging property against DPPH free radical. Finally, the in vitro bioactivities were also supported by molecular docking studies. The computational study demonstrated that the isolated compounds exerted stronger affinity compared to the standard drugs towards the binding sites of dihydrofolate reductase (DHFR), glutathione reductase, and urase oxidase.
Keywords: Stevia rebaudiana, Phenolic constituents, Cytotoxicity, Antimicrobial, Antioxidant, Molecular docking
Graphical abstract
Stevia rebaudiana; Phenolic constituents; Cytotoxicity; Antimicrobial; Antioxidant; Molecular docking.
1. Introduction
Since thousands of years ago, human civilization has been using various plants as therapeutic weapons to treat numerous diseases based on safe consumption. Nevertheless, screening of different medicinal plants and their application in the treatment of multiple illnesses increases with scientific development [1].
Stevia (Stevia rebaudiana Bertoni), a perennial shrub from the Asteraceae family, is native to South America. However, the plant is cultivated worldwide, including in Bangladesh [2]. This plant has drawn economic and scientific interest due to its natural, non-toxic, and nonmutagenic sweetener properties. Due to the presence of several steviol glycosides in its leaf, its sweetening potency is 200–300 times more than that of sucrose, worth of use as a sugar substitute in the food and drug industry [2, 3]. The leaves possess three main sweetening constituents namely stevioside, rebaudioside A and C; those have ent-kaurene diterpene steviol skeleton common in their structures [4, 5, 6]. Besides, some other glycosides such as staviolbioside, isostaviol, labdane-type diterpenes sterebins E, H, N and I were also found in this plant [7, 8]. In addition to glycosides, several phenolic compounds, the key components for antioxidant property, were also reported [9, 10]. The polyphenol compounds isolated from the plant leaves are chlorogenic acids, esters molecules of polyphenols, including hydroxycinnamic acids with quinic acid, which have excellent free radical scavenging properties, and other numerous health benefits [11]. Apart from its industrial applications, stevia has been found to be effective in the management of diabetes, obesity, cancer, hypertension, dental carries and infectious diseases [12, 13, 14]. Significant effect in alleviating kidney and liver damage was reported earlier along with increments of insulin level in Streptozotocin-induced diabetic rats [15]. A comprehensive review on the plant has been reported mentioning its role in blood pressure regulation, inflammatory bowel disease, renal function, glucoregulation, tumor and obesity management [16]. Although several bioactivities of S. rebaudiana extracts and some of the glycoside constituents have been reported, studies on antimicrobial and anticancer potentials are very limited. Only very few reports are found addressing antimicrobial and anticancer potentials of the crude extracts [17, 18, 19]. In this regard, we have also recently reported antimicrobial and antioxidant properties of different extracts of Stevia rebaudiana [20].
To the best of our knowledge, none of the above study has isolated the bioactive components that may exert these actions. Herein, we report the isolation and characterization of some bioactive secondary metabolites by using repeated chromatographic and spectroscopic techniques, respectively targeting their antimicrobial, anticancer, and antioxidant properties. Docking studies of the isolates were also performed with the active site of dihydrofolate reductase (DHFR), glutathione reductase, and urase oxidase to support the in vitro bioactivities.
2. Method and materials
2.1. Materials and instruments
HELA cell, DME medium, and 10% fetal bovine serum were purchased from authorized suppliers and methotrexate, ciprofloxacin, fluconazole, and ascorbic acid were obtained from Eskyf Pharmaceutical Limited, Bangladesh. Biological biosafety cabinet (NU-400E, Nuaire, USA), CO2 incubator (Nuaire, USA), trinocular microscope with camera (Olympus, Japan), and hemocytometer (Nexcelom, USA) were used to execute the cytotoxicity test. A very safety environment was maintained to handle all instruments and chemicals. Fresh leave of Stevia rebaudiana were collected from BRAC cultivation center, Dhaka, Bangladesh in March 2018. The exsiccated plant samples were identified by Principal Scientific Officer, Bangladesh National Herbarium, Mirpur, Dhaka where a voucher specimen has been deposited for future reference (accession number: DACB-38588).
The 1H and 13C NMR spectra were measured on a Burker AMX-400 operating at 400 MHz and 100 MHz for 1H and 13C, respectively. Chemical shifts are expressed in δ (ppm) scale using tetramethylsilane as internal standard and coupling constants (J) are expressed in hertz (Hz). Analytical thin layer chromatography (TLC) was carried out on pre-coated (TLC Silica gel 60 F254- MerkKGa) plates. TLC spots were screened under UV light (254 nm) and spraying with 10% H2SO4 and anisaldehyde reagents followed by heating. Column chromatography was carried out with powdered silica gel (Kieselgel 60, 230–400 mesh, Merck KGaA, Dermstadt, Germany) and primarily packed in a glass column with a dimension of 5 × 60 cm. Preparative TLC plates were prepared on a glass (20 × 20 cm) surface coated with slurry of silica gel.
2.2. Isolation of compounds
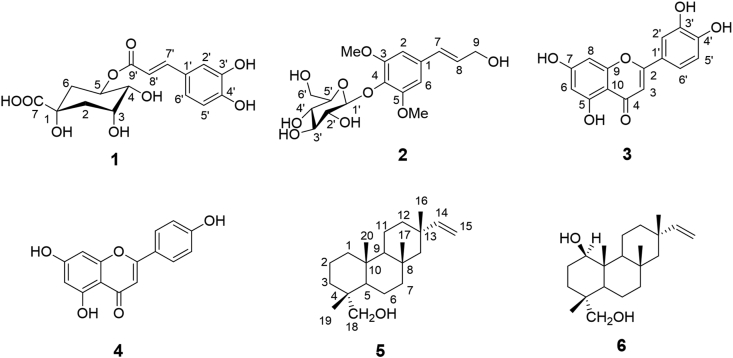
The dried and powdered fresh leaves (1 kg) were extracted with 70% methanol with occasional shaking and stirring. The whole mixer was then filtered through clean, white cotton followed by Whatman filter paper no. 1. The filtrate was concentrated with a rotary evaporator. An aliquot (100 g) of concentrated methanol extracts were fractionated by n-hexane, dichlormethane and ethyl acetate and the resultant fractions were evaporated to dryness to yield n-hexane Stevia rebaudiana (HSR), dichloromethane Stevia rebaudiana (DSR) and ethyl acetate Stevia rebaudiana (EtASR) soluble materials having 8%, 10% and 15% (w/w) yield, respectively. The dried residues were then stored in a refrigerator until further investigation. Based on the preliminary antimicrobial screening [20], EtASR was chosen for further isolation and purification. The EtASR fraction (3 g) was dissolved in methanol and mixed with small amount of silica gel (mesh size 60–120, 3 g) to make a dry sample for convenient application in column chromatography (CC). The dry sample was then loaded on a clean and dry glass column (3 × 40 cm) packed with silica gel (60–120 mesh size) and eluted with a mixture of solvents ethyl acetate: acetic acid: water (8:1:1) with increasing polarity and 10 ml elute was collected and monitored by thin layer chromatography (TLC). After TLC pooling, a total 15 fractions were obtained. Fractions 5–10 were mixed together and repeated preparative Thin Layer Chromatography (PTLC) was carried out to yield 5-O-caffeoyl quinic acid (1) 16 mg, syringin (2) 15 mg, luteolin (3) 8 mg and apigenin (4) 14 mg. Again, the n-hexane fraction (HSR) was subjected to silica column chromatography using the solvent system of n-hexane and ethyl acetate at a ratio 9:1 for elution. The test tubes number 46–53 gave same spot (5) and 60–72 numbers have identical spot (6) on TLC plate which were identified as jhanol (5) 30 mg and jhanidiol (6) 25 mg, respectively.
2.3. MTT cell viability assay
The cell proliferation studies of compounds 1, 2, 4, 5 and 6 were examined in Centre for Advanced Research in Sciences (CARS), Dhaka, Bangladesh against HeLa cell line where Methotrexate was used as a positive control with a series of concentration (62.5, 125 and 250 μL/mL), DMEN (Dulbeco's modified eagles' medium) containing 1% penicillin-streptomycin (1:1), 0.2% gentamycin, and 10% fetal bovine serum (FBS) were maintained for HeLa cell, a human cervical carcinoma cell line. HeLa cells were implanted onto a 96-well plate with 2 × 104 cells per well (100 μL) and incubated at 37 °C under a humidified atmosphere of 5% of CO2 for 24 h. Therefore, the five components (1, 2, 4, 5 and 6) and positive control (methotrexate) were dissolved in 2.5% DMSO where DMSO was used as negative control. Components with a series of concentration (62.5, 125, 250 and 500 μL/mL) and positive control at the concentrations (25, 50 and 100 μL/mL) were added and incubated for 48 h. After incubation using cell counting Kit-8 (CCK-8), a non-radioactive colorimetric Cell Proliferation and Cytotoxicity assay kit (Sigma-Aldrich, USA) was used for measuring cell viability. Duplicate wells were used for each sample [21].
2.4. Antimicrobial assay
The initial screening of antimicrobial activity and minimum inhibitory concentration (MIC) determination for the tested compounds were performed by using disc diffusion technique with different strains [22]. Thirteen bacterial standard strains and three fungal strains among which five Gram-positive and eight Gram-negative strains were obtained from the Biomedical research laboratory, University of Dhaka, Bangladesh. The gram (+)ve bacteria were Bacillus cereus, Bacillus megaterium, Bacillus subtilis, Staphylococcus aureus, Sarcina lutea and the Gram (-)ve were Escherichia coli, Pseudomonas aeruginosa, Salmonella paratyphi, Salmonella typhi, Shigella boydii, Shigella dysenteriae, Vibrio mimicus, Vibrio parahemolyticus, and three unicellular fungi were Candida albicans, Aspergillus niger, Sacharomycescerevacae used. Test samples were prepared using sterile blank discs (6 mm) which were loaded with the test samples dissolved in methanol at concentration of 250 μg/disc by a micropipette. Ciprofloxacin (5 μg/disc) and fluconazole (5 μg/disc) were used as positive controls for antibacterial and antifungal activities, respectively. The experiment was carried out in thrice and the mean diameter was taken.
2.5. DPPH free radical scavenging assay
The antioxidant activity of the isolated compounds was assessed by utilizing the 2,2-diphenyl-1-picrylhydrazyl (DPPH) free radical scavenging assay method explained by Brand-Williams et al. [23]. However, for the quantitative assay described by Sultana et al. [20], the diluted stock solutions were spotted on a TLC plate, which was then run in the proper solvent system for the appearance of the compounds at distinct positions on the plate. Then the plate was sprayed with 0.02% (w/v) DPPH in ethanol at room temperature. An apparent color change was observed as DPPH was cleared out by the effect of these compounds, detecting the potential antioxidant property of these compounds.
The spectroscopic method was further applied for quantitative measurement of antioxidant property of these compounds by utilizing DPPH free radical scavenging assay [20]. Initially, 2 ml of each sample (and ascorbic acid as standard) concentrations ranged from 500 μg/mL to 1 μg/mL by serial dilution (500, 100, 50, 10, 5, 1 μg/mL) were mixed with 2 ml freshly prepared DPPH solution (0.004% w/v). The resulting solutions were mixed in the test tubes carefully. After 30 min incubation in a dark place at room temperature, the UV absorbance of these mixes was recorded precisely against the blank solution (ethanol) at 517 nm wavelength. The percentage of DPPH scavenging or % inhibition was measured by using the following equation:
| (1) |
Where Ablank = absorbance of the DPPH solution only prepared by methanol as solvent and Asample = Absorbance of the plant extract (test sample) and ascorbic acid (standard) after reaction with DPPH solution. The measurement was done in triplicate for accuracy. The % scavenging of free radical was measured by utilizing equation number 1. A graph of inhibition percentages (I%) against sample concentrations was plotted. From the graph, 50% inhibitory concentration (IC50 value) was estimated for each sample.
2.6. Molecular docking study
The widely used software packages (PyRx, PyMoL 2.3, Discovery Studio 4.5, and Swiss PDB viewer) were utilized for computational docking of the six isolated compounds.
2.6.1. Target protein selection
Docking analysis of the isolated compounds were accomplished to understand the antibacterial, anticancer and antioxidant activities. For studies of antibacterial and anticancer activities, dihydrofolate reductase (DHFR) has been chosen as the target protein based on the biochemical pathways and previous evidence [24, 25]. The three-dimensional (3D) structure of the dihydrofolate reductase (DHFR) enzyme was retrieved from the protein data bank (https://www.rcsb.org) with PDB ID 4M6J. Besides, two other proteins with PDB ID 3GRS (Glutathione reductase) and PDB ID 1R4U (Urase oxidase) were downloaded from the same database (https://www.rcsb.org) for computational docking based on previous evidence to evaluate the potential antioxidant property [26]. All the proteins/receptors were saved in PDB format. The collected proteins were made water and any unwanted ligand/residue free by running in PyMoL 2.3. Then all the biomolecules were arranged by adding non-polar hydrogen atoms and kept to the lowest energy state by applying for an energy minimization program by Swiss PDB viewer for further analysis.
2.6.2. Ligand preparation
All the stated compounds (1 to 6) in Figure 1, namely, 5-O-caffeoyl quinic acid, syringin, luteolin, apigenin, jhanol, and jhanidiol, respectively, were searched and found in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Then the ligands with their corresponding standard drugs, including ciprofloxacin, methotrexate, and ascorbic acid, for studying the comparative binding affinity to the target macromolecules, were downloaded and saved in 3D SDF format. These ligands were loaded in the discovery studio 4.5 serially, including their PubChem CID. It is noted that all the phytochemicals were optimized via Pm6 semi-empirical method for enhancing the docking accuracy [27, 28].
Figure 1.
Isolated constituents from Stevia rebaudiana (1–6).
2.6.3. Ligand-protein interaction
The current computer-aided ligand-protein interaction has been sketched for predicting the possible binding profiling of the isolated compounds with their binding affinities to the target molecules [28]. A highly advanced PyRxAutodock Vina was utilized for this molecular drug-protein linking process, where semiflexible modeling was applied for the molecular docking. Firstly, the protein has been loaded and formatted to target macromolecule, and the literature-based amino acids with their ID were selected for ascertaining target-specific binding of the ligands. For 4M6J target, Ala9, Ile16, Leu93, Ser92, Arg91, Arg77, Glu78, Ser76, Leu75, Lys54, Val120, Ser119, Lys55, Thr56, Ser 118, Gly117, and for 1R4U, Arg 176, Val 227, Gln 228, Asn254, His 256 amino acid were picked for site targeted docking [26, 29]. Besides, Val 102, Lys 127, Asn 129, Lys 143, Gly 148, Ser 145, Ser 147, Asp 183, Val 130, Gln 131, and Thr 185 were selected for site-specific docking of these isolated natural compounds with 3GRS protein for predicting antioxidant effects [26]. Secondly, all the PDB files of the ligands were imported and subsequently minimized into pdbqt format with Open Bable tool in the PyRxAutoDock Vina software to fit the best optimal hit during the docking against these chosen macromolecules. Thirdly, the grid box was generated by keeping the active binding sites of the protein within the center of the box where the grid mapping was set as center: X = 7.4850, Y = 7.3500, Z = −18.6398, and dimension: X = 43.8582, Y = 51.2793, Z = 48.8257 during docking with DHFR protein. Besides, the grid mapping was fixed center: X = 27.4790, Y = 50.2748, Z = 40.1613, and dimension: X = 31.7158, Y = 37.2601, and Z = 42.9295 for 1R4U protein while the grid map was kept for 3GRS center: X = 74.0693, Y = 51.4437, Z = 25.0217, and dimension: X = 24.7046, Y = 25.0, Z = 29.9240. The rest of the parameters were set to be default settings during the docking process. Then, the computer-aided molecular docking of the ligands was performed under maintaining all necessary conditions by utilizing AutoDock Vina (version 1.1.2) [29]. Finally, all the docking analysis for extrapolating the best-fitted figures with 2D and 3D configurations was conceived by using BIOVIA Discovery Studio version 4.5.
2.7. Statistical analysis
The in-silico experiments were done triplicate, and the obtained results in the study were presented and explained as mean ± standard error of the mean. The standard errors for all the analyses were below 1%. Besides, the data found from the in vitro antibacterial test were represented as mean ± standard deviation (SD). All the statistical analyses were performed in Microsoft Excel 2019 version.
3. Results
3.1. Isolation and characterization of compounds
Dried powder leaves of Stevia rebaudiana was extracted with 70% methanol to isolate secondary metabolites having potential antimicrobial property. Significant antimicrobial activity was observed in the crude extract which led to the further purification through partitioning into different solvents (hexane, dichloromethane and ethylacetate) according to their polarities. Among the fractions, EtASR fraction showed prominent antibacterial activity in preliminary investigation which was subsequently purified. Repeated chromatographic techniques yielded total six compounds (Figure 1). The 1D and 2D spectral data of all the isolates were compared with established values and were identified as 5-O-caffeoyl quinic acid (1), syringin (2), luteolin (3), apigenin (4), jhanol (5) and jhanidiol (6) [30, 31, 32, 33]. The spectra obtained from NMR analysis of the compounds and the single spot purity check-in TLC plate with retardation factor (Rf) values were added in the supplementary file (Figure S1 and Table S1, respectively).
3.2. Spectral data
5-O-Caffeoyl quinic acid (1): White amorphous powder, 1H NMR (400 MHz, CD3OD): δ 7.58 (1H, d, J = 15.9 Hz, H-7′), 7.04 (1H, d, J = 2.0 Hz, H-2′), 6.92 (1H, dd, J = 8.0, 2.0 Hz, H-6′), 6.77 (1H, d, J = 8.0 Hz, H-5′), 6.24 (1H, d, J = 15.9 Hz, H-8′), 5.34 (1H, brs, OH), 3.74 (1H, ddd, J = 7.7, 2.9, 2.9 Hz, H-3), 3.66 (1H, dd, J = 6.9, 2.9 Hz, H-4), 2.05 (1H, m, H-2ax), 2.01 (1H, m, H-6ax), 1.96 (1H, m, H-6eq), 1.90 (1H, m, H-2eq). 13C NMR (100 MHz, CD3OD): δ 173.8 (C-7), 167.0 (C-9′), 148.2 (C-4′), 145.7 (C-3′), 145.4 (C-7′), 126.2 (C-1′), 121.6 (C- 6′), 115.1 (C-5′), 114.3 (C-2′), 113.8 (C-8′), 71.2 (C-1), 70.7 (C-5), 69.3 (C-4), 67.7 (C-3), 36.4 (C-2), 34.7 (C-6).
Syringin (2): White crystalline solid, 1H NMR (400 MHz, CD3OD): δ 6.65 (2H, s, H-2 & 6), 6.42 (1H, d, J = 16.0 Hz, H-7), 6.23 (1H, dt, J = 16 and 6.0 Hz, H-8), 4.78 (1H, d, J = 8.0 Hz, H-1′), 4.11 (1 H, dd, J = 6.0 and 2.0 Hz, H-9), 3.75 (6H, s, OCH3-3 &-5), 3.65 (1H, dd, J = 12 and 6.0 Hz, H-6′), 3.5 (1H, dd, J = 12 and 2.0 Hz, H-6′), 3.48 (1H, m, H-4′), 3.41 (1H, m, H-3′), 3.32 (1H, m, H-5′) and 3.1 (1H, m, H-2′).
Luteolin (3): Pale yellow needle like crystal, 1H NMR (400 MHz, CD3OD): δ 7.30 (1H, m, overlapped, H-6′), 7.30 (1H, m, overlapped, H-2′), 6.94 (1H, d, J = 7.5 Hz, H-5′), 6.68 (1H, s, H-3), 6.44 (1H, s, H-8), 6.11 (1H, s, H-6).
Apigenin (4): Pale yellow needle like crystal, 1H NMR (400 MHz, CD3OD): δ 7.30 (1H, m, overlapped, H-6′), 7.30 (1H, m, overlapped, H-2′), 6.94 (1H, d, J = 7.5 Hz, H-5′), 6.68 (1H, s, H-3), 6.44 (1H, s, H-8), 6.11 (1H, s, H-6).
Jhanol (5): Needle like solid, 1H NMR (400 MHz, CDCl3): δ 5.91 (1H, dd, J = 17 and 12 Hz, H-14), 5.17 (1H, dd, J = 18 and 2 Hz, H-15), 4.93 (1H, q, J = 12 and 2 Hz, H-15), 3.43 (1H, d, J = 11, H-19), 3.13 (1H, d, J = 11, H-19), 2.34 (2H, m, H-12), 0.75 (3H, s, H-20), 0.85 (3H, s, H-18), 1.31 (3H, s, H-16), 1.35 (3H, s, H-17). 13C NMR (400 MHz, CDCl3): δ 37.6(C-1), 17.2(C-2), 36.9(C-3), 35.8(C-4), 49.9(C-5), 19.7(C-6), 42.4(C-7), 73.3(C-8), 55.6(C-9), 35.2(C-10), 17.8(C-11), 35.9(C-12), 72.0(C-13), 147.5(C-14), 110.4(C-15), 27.5(C-16), 25.5(C-17), 74.9(C-18), 15.8(C-19), 15.2(C-20).
Jhanidiol (6): Needle like solid, 1H NMR (400 MHz, CDCl3): δ 5.95 (1H, dd, J = 17 and 12 Hz, H-14), 5.15 (1H, dd, J = 17 and 2 Hz, H-15), 4.95 (1H, q, J = 12 and 2 Hz, H-15), 4.2 (1H, t, J = 4.8, H-1), 3.45 (1H, d, J = 11, H-19), 3.14 (1H, d, J = 11, H-19), 1.35 (3H, s, H-16), 1.31 (3H, s, H-17), 0.88 (3H, s, H-20), 0.80 (3H, s, H-18). 13C NMR (400 MHz, CDCl3): δ 38.5(C-1), 17.2(C-2), 36.1(C-3), 36.8(C-4), 50.2(C-5), 20.03(C-6), 42.66(C-7), 75.8(C-8), 56.1(C-9), 36.9(C-10), 19.1(C-11), 35.9(C-12), 73.3(C-13), 148.7(C-14), 111.3(C-15), 77.8(C-16), 26.1(C-17), 66.9(C-18), 18.2(C-19), 15.7(C-20).
3.3. Evaluation of anticancer property
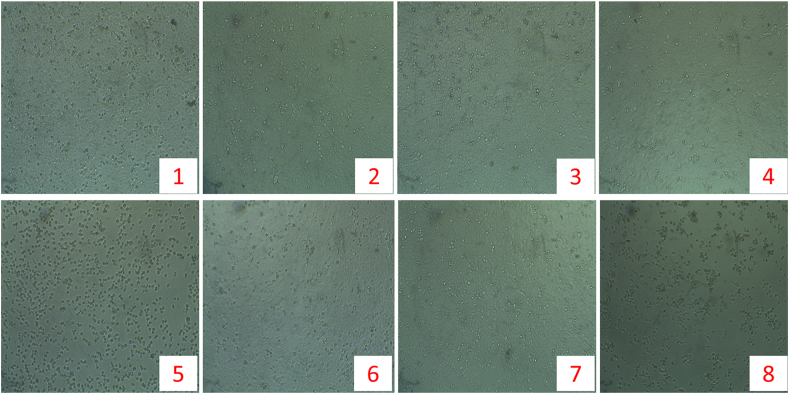
MTT assay was utilized to assess the anti-proliferative effect of the five compounds on HeLa cells. The serially diluted concentrations of the samples and the corresponding percentage of inhibitions against in HeLa cell line were tabulated in Table 1. As representative examples, the cell mortality at 500 mg/mL of each sample has been shown in Figure 2. The IC50 values of the compounds and standard drug (methotrexate) enumerated by using the regression line equation (log of concentration vs. percentage of cell mortality) were also enlisted in Table 1. Among the bioactive samples (1, 2, 4, 5, and 6), compound 1 exhibited the highest in vitro cytotoxic effect (IC50 = 181.3 μg/mL) against the cervical cancer cell line followed by compound 2 (IC50 = 194.4 μg/mL). The IC50 values of compounds 4, 5, 6, and methotrexate were found 252.14, 298.86, 265.07, and 36.50 μg/mL, respectively.
Table 1.
In-vitro anti-proliferative effects with IC50 values of the isolated compounds (1, 2, 4, 5, and 6) on HeLa cell line.
| Compound No. | Concentration (μg/mL) | % of inhibition against in HeLa cell line | IC50 value (μg/mL) |
|---|---|---|---|
| Methotrexate | 62.5 | 30.3 | 36.5 |
| 125 | 53.4 | ||
| 250 | 94.8 | ||
| 1 | 62.5 | 21.45 | 181.3 |
| 125 | 26.68 | ||
| 250 | 70.14 | ||
| 500 | 72.86 | ||
| 2 | 62.5 | 5.28 | 194.4 |
| 125 | 28.53 | ||
| 250 | 67.84 | ||
| 500 | 83.3 | ||
| 4 | 62.5 | 3.38 | 252.14 |
| 125 | 4.17 | ||
| 250 | 63.63 | ||
| 500 | 73.47 | ||
| 5 | 62.5 | 1.88 | 298.86 |
| 125 | 4.87 | ||
| 250 | 34.55 | ||
| 500 | 74.49 | ||
| 6 | 62.5 | 1.58 | 265.07 |
| 125 | 9.15 | ||
| 250 | 60.54 | ||
| 500 | 68.75 | ||
| Control (DMSO) | 2.5 | – | – |
Note: DMSO = Dimethyl sulfoxide.
Figure 2.
Anticancer potentialities of the isolated compounds (1 to 6) and the standard drug methotrexate under inverted light microscope at the dose of 500 (μg/ml).
3.4. Evaluation of antimicrobial activity
All the compounds exhibited mild to moderate activity against both gram positive and gram-negative bacteria as compared with standard ciprofloxacin. Among the isolates, compound 1 displayed most promising result with a range of zone of inhibition 12–15 mm. Compound 2 also showed comparable antibacterial activity and other compound also exhibited mild to moderate antibacterial activity against some strains. However, no compound was effective against the fungus strains in comparison with standard fluconazole (18–28 mm) (Table 2).
Table 2.
Antimicrobial activity of the compounds (1–6) against gram positive and gram negative bacteria.
| Pathogens | Zone of Inhibition (mean ± SD; mm) |
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | Ciprofloxacin | |
|
Gram positive | |||||||
| Bacillus cereus | 14.7 ± 0.11 | 12.7 ± 0.3 | – | 10.2 ± 0.11 | – | 10.3 ± 0.18 | 17.2 ± 0.23 |
| Bacillus megaterium | 15.3 ± 0.48 | 12.4 ± 0.23 | 10.0 ± 0.66 | 11.1 ± 0.32 | – | 10.8 ± 0.21 | 16.8 ± 0.14 |
| Bacillus subtilis | 12.7 ± 0.52 | – | – | – | – | – | 15.8 ± 0.44 |
| Bacillus aureus | 14.1 ± 0.16 | 13.5 ± 0.19 | – | – | – | – | 18.1 ± 0.14 |
| Sarcina lutea |
13.2 ± 0.12 |
12.5 ± 0.29 |
11.5 ± 0.44 |
– |
– |
– |
20.0 ± 0.18 |
|
Gram negative | |||||||
| Escherichia coli | 15.8 ± 0.33 | 12.6 ± 0.65 | – | 12.0 ± 0.52 | – | – | 21.2 ± 0.23 |
| Pseudomonas aeruginosa | – | – | 10.7 ± 0.32 | – | 10.5 ± 0.26 | – | 14.4 ± 0.14 |
| Salmonella paratyphi | 14.2 ± 0.22 | 11.8 ± 0.55 | 8.2 ± 0.23 | 10.2 ± 0.44 | 10.8 ± 0.19 | 10.8 ± 0.52 | 18.5 ± 0.44 |
| Salmonella typhi | 12.1 ± 0.33 | 12.7 ± 0.66 | 9.3 ± 0.32 | 10.8 ± 0.44 | – | – | 17.7 ± 0.14 |
| Shigella boydii | 15.3 ± 0.11 | – | – | – | – | 11.2 ± 0.67 | 18 ± 0.18 |
| Shigella dysenteriae | 14.4 ± 0.26 | 11.5 ± 0.45 | – | – | – | – | 19.5 ± 0.33 |
| Vibrio mimicus | 13.3 ± 0.77 | – | 10 ± 0.74 | 11.5 ± 0.53 | – | – | 18.3 ± 0.42 |
| Vibrio parahemolyticus | – | 10.1 ± 0.45 | – | 12.1 ± 0.23 | – | – | 20.0 ± 0.41 |
3.5. Evaluation of antioxidant property
All the phytochemicals except compound 2 showed significant antioxidant properties. Compounds 3, 6, and 4 exhibited the most promising antioxidant activities against DPPH free radical with IC50 values 8.93, 17.63, and 21.54 μg/mL, respectively, compared to the standard drug ascorbic acid with IC50 = 6.05 μg/mL (Table 3).
Table 3.
Antioxidant properties of the isolated compounds (1 to 6) and the standard drug ascorbic acid with their IC50 (μg/mL) values.
| Compound Code | R2 | IC50 (μg/mL) |
|---|---|---|
| 1 | 0.9905 | 23.33 |
| 2 | 0.9859 | 174.16 |
| 3 | 0.9171 | 8.93 |
| 4 | 0.9258 | 21.54 |
| 5 | 0.9905 | 23.33 |
| 6 | 0.9791 | 17.63 |
| Ascorbic acid | 0.9142 | 6.05 |
3.6. Molecular docking studies
3.6.1. Dihydrofolate reductase (DHFR) inhibition
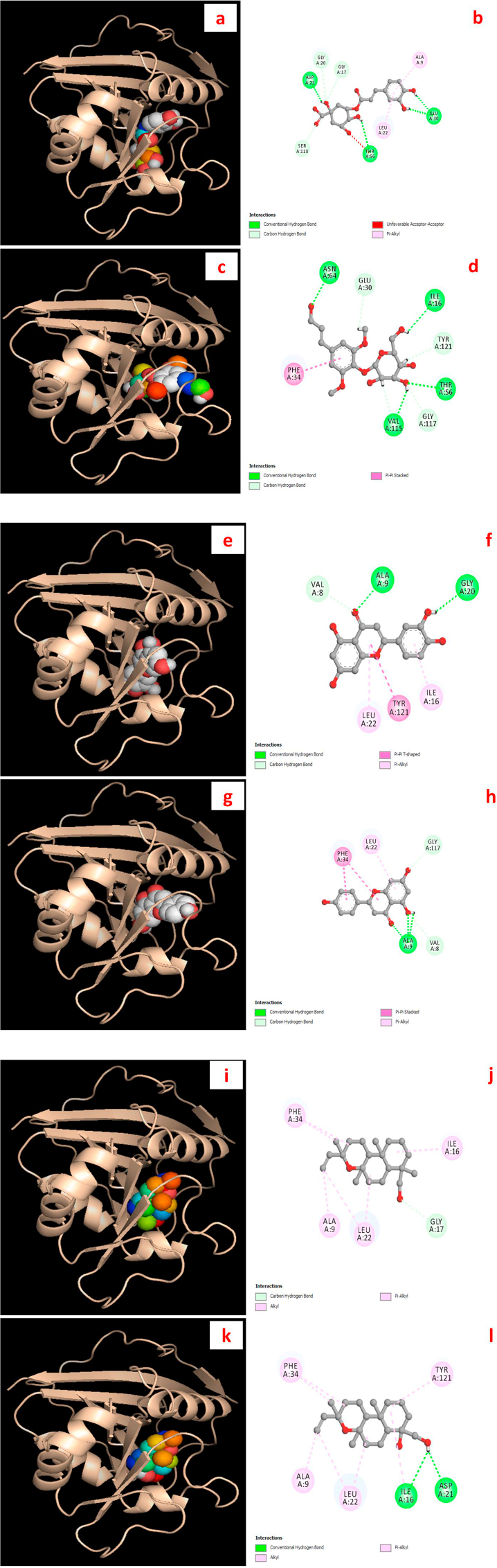
For a clear and better understanding of the molecular effects of these isolated natural products on the above-mentioned biological target, the computer-designed molecular modeling study was performed by utilizing suitable tools. The DHFR protein has a significant role in DNA synthesis in the bacterial or human cell development process. To comprehend antibacterial and anticancer potentialities, we have docked these promising drug candidates/ligands with DHFR enzyme and found surprising docking profiles with high binding affinity compared to their respective standard molecules (ciprofloxacin and methotrexate). Apparently, the lower the binding energy, the higher the binding strength of the ligands has been postulated. Besides, the extrapolated interaction profile having a null RMSD (root mean square deviation) value represents the best docking prediction [29].
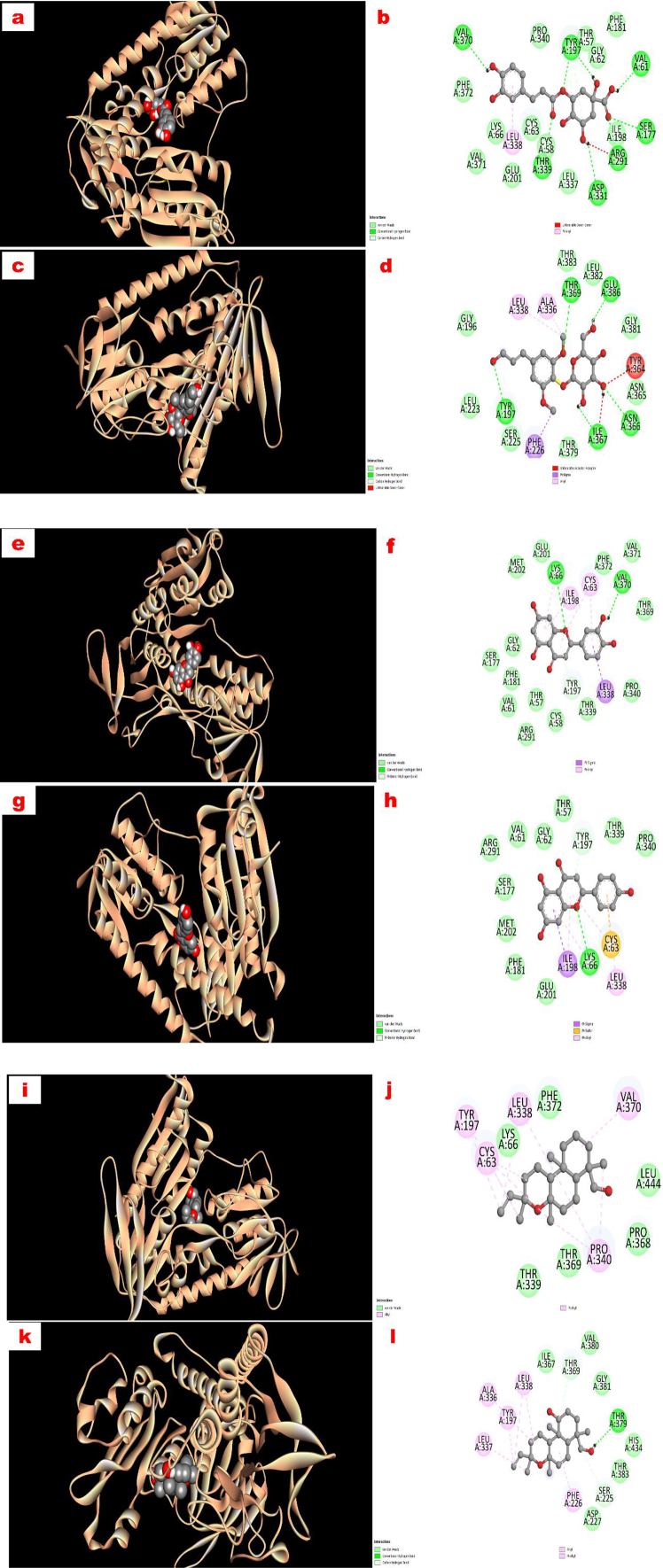
All the drug candidates displayed higher binding affinity to the DHFR protein with docking score −10.7 to −15.3 kcal/mol in comparison with ciprofloxacin (−8.4 kcal/mol) and methotrexate (−9.0 kcal/mol). Notably, 5-O-caffeoyl quinic acid exhibited the highest DHFR enzyme affinity (−15.3 kcal/mol) among all the ligands involved in the study. The order of the binding affinity might be noted here as like 5-O-caffeoyl quinic acid > syringin > apigenin > luteolin > jhanidiol > jhanol > methotrexate > ciprofloxacin (Table 4). The active binding sites of compound 1 (5-O-caffeoyl quinic acid) were ASP 21, GLY 20, GLY 17, ALA 9, GLU 30, LEU 22, THR 56, and SER 118, where compound 2 (syringin) showed its affinity towards GLU 30, ASN 64, ILE 16, TYR 16, THR 56, GLY 117, VAL 115, PHE 34 in chain A. Similarly, all the binding sites of the DHFR protein where these ligands interacted were shown in Figure 3 (a-l).
Table 4.
Binding affinity or in silico docking scores (kcal/mol) of the isolated compounds (1 to 6) from Stevia rebaudiana (Bert.) and the corresponding standard drugs during molecular interaction with dihydrofolate reductase (PDB ID 4M6J), glutathione reductase (PDB: 3GRS), and urase oxidase (1R4U) enzymes for determining antibacterial, anticancer, and antioxidant potentialities.
| Compound No. | Compound Name | PubChem CID | Binding affinity (kcal/mol) |
||
|---|---|---|---|---|---|
| 4M6J | 3GRS | 1R4U | |||
| 1 | 5-O-caffeoyl quinic acid | 5280633 | −15.3 | −7.8 | −7.7 |
| 2 | Syringin | 5316860 | −13.6 | −6.4 | −7.3 |
| 3 | Luteolin | 5280445 | −12.6 | −7.8 | −8.3 |
| 4 | Apigenin | 5280443 | −12.7 | −7.7 | −8.1 |
| 5 | Jhanol | 13996014 | −10.7 | −7.2 | −7.7 |
| 6 | Jhanidiol | 15226315 | −11.2 | −7.4 | −7.3 |
| Standard | Ciprofloxacin | 2764 | −8.4 | ||
| Methotrexate | 126941 | −9.0 | |||
| Ascorbic acid | 54670067 | −5.7 | −5.3 | ||
Figure 3.
Graphical representation of the molecular interactions of the isolated compounds with dihydrofolate reductase (DHFR; PDB ID: 4M6J) enzyme with 3D and 2D visualization (compound 1 = a and b, compound 2 = c and d, compound 3 = e and f, compound 4 = g and h, compound 5 = i and j, and compound 6 = k and l). It is noted here that the docking visualization for the standard drugs were not displayed in the figure.
3.6.2. Glutathione reductase and urase oxidase inhibition
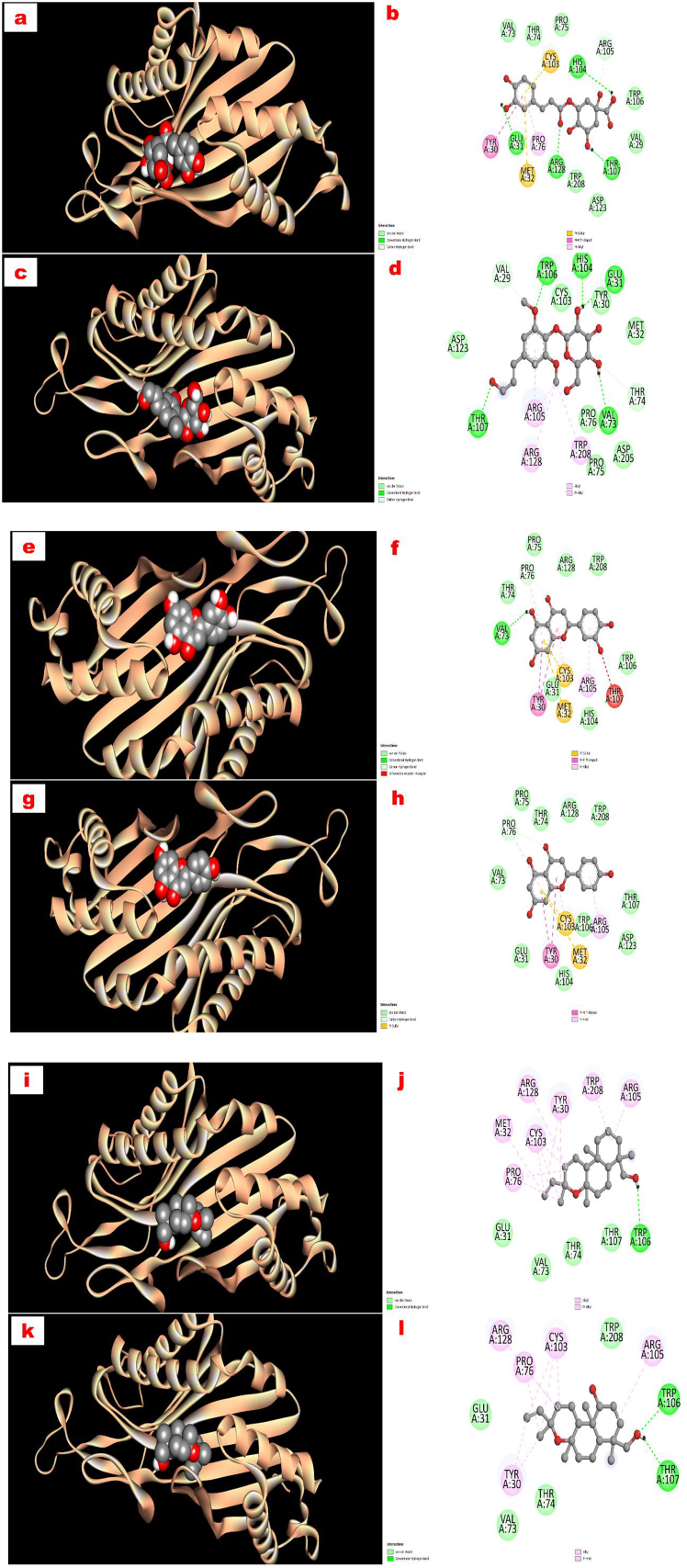
All the compounds exhibited much better binding affinity (−6.4 to −7.8 kcal/mol towards glutathione reductase and −7.3 to −8.3 kcal/mol towards urase oxidase enzyme) than the standard drug ascorbic acid (−5.7 and −5.3 kcal/mol towards glutathione reductase and urase oxidase enzyme, respectively). Compounds 1 (−7.8 kcal/mol) and 3 (−7.8 kcal/mol) showed the most binding affinity towards the glutathione reductase enzyme, while compounds 3 (−8.3 kcal/mol) and 4 (−8.1 kcal/mol) exhibited the highest docking score during interacting with urase oxidase enzyme. The active binding sites of the glutathione reductase for compound 1 were VAL 370, PRO 340, TYR 197, THR 57, GLY 62, PHE 181, VAL 61, SER 177, ILE 198, ARG 291, ARG 291, ASP 331, LEU 337, CYS 58, THR 339, GLU 201, CYS 63, LEU 338, LYS 66, VAL 371, PHE 372 and for compound 3 were MET 202, GLU 201, LYS 66, ILE 198, CYS 63, PHE 372, VAL 371, THR 369, PRO 340, LEU 338, THR 339, TYR 197, CYS 58, THR 57, ARG 291, VAL 61, PHE 181, SER 177, GLY 62 (Figure 4). Similarly, the active binding sites of the glutathione reductase enzyme for all the compounds were shown in Figure 4(a-l). Moreover, the binding sites of the urase oxidase enzyme for the compound 3 were VAL 73, THR 74, PRO 76, PRO 75, ARG 128, TRP 208, TRP 106, THR 107, ARG 105, HIS 104, CYS 103, MET 32, GLU 31, TYR 30 (Figure 5). Likewise, the active binding sites of the urase oxidase enzyme for all the compounds were shown in Figure 5(a-l).
Figure 4.
Graphical representation of the molecular interactions of the isolated compounds with glutathione reductase (PDB ID: 3GRS) enzyme with 3D and 2D visualization (compound 1 = a and b, compound 2 = c and d, compound 3 = e and f, compound 4 = g and h, compound 5 = i and j, and compound 6 = k and l). It is noted here that the docking visualization for the standard ascorbic acid was not displayed in the figure.
Figure 5.
Graphical representation of the molecular interactions of the isolated compounds with urase oxidase (PDB ID: 1R4U) enzyme with 3D and 2D visualization (compound 1 = a and b, compound 2 = c and d, compound 3 = e and f, compound 4 = g and h, compound 5 = i and j, and compound 6 = k and l). It is noted here that the docking visualization for the standard ascorbic acid was not displayed in the figure.
4. Discussion
A large number of potential efforts have been conducted to identify the novel scaffolds from the plants that may help alleviate the global threat of infectious disease. Plant contains a variety of secondary metabolites among which phenolic compounds are considered of great interests for their wide applications. Phenolics such as coumaric acid, caffeic acid, ferulic acid, sinapic acid, tannins, flavonoids, glycosides etc. are synthesized from the phenylpropanoid pathway in plants and demonstrated antimicrobial properties [34, 35, 36]. Stevia rebaudiana contains a wide variety of diterpene glycosides among which stevioside (5–13%), rebaudioside A (2–4%), and rebaudioside C (1–2%) are the main constituents imparting sweetening properties [37]. Apart from these, minor constituents such as flavonoids, sterebins, chlorogenic acids, triterpenes, volatile oils etc. have also been reported [38]. Present study also added some valuable number of secondary metabolites in those lists. To the best of our knowledge, this is the first time reporting of a phenylpropanoid glycoside syringin (2) and jhanidiol (6) bearing diterpene skeleton from Stevia leaves. In terms of total phenolic constituents, the study finding is similar to a previous study where 80% ethanolic crude extracts of S. rebaudiana leaves contains total phenolic concentration with a range of 25–65 mg/g gallic acid equivalent [39].
In the cytotoxicity study (MTT cell viability assay), it has been indicated that the compounds 1, 2, 4, 5, and 6 depicted growth inhibition against HeLa cells in a dose dependent manner. Among the isolated compounds, the compound 1 exhibited most profound effect on the HeLa cell viability indicating its anticancer activity. Although the effect is much lower compared to the standard drug Methotrexate, structural modifications might lead to development of more potent analogues. In some reported previous studies, aqueous and methanolic extracts of Stevia rebaudiana and the diterpene glycosides along with their synthetic congeners and their aglycone part steviol, isosteviol showed prominent antimicrobial and antitubercular properties [40, 41, 42]. In our study, all the isolates (1–6) displayed a significant zone of inhibition against a variety of pathogens, which postulates the findings observed in the previous studies. The present results further stipulate that the activity observed by the extracts in the previous studies might be due to the presence of the isolated constituents (1–6) from Stevia. To validate the resulting pharmacological effect and to discern the exact mechanistic pathway, molecular docking was also performed for the isolated compounds. Interestingly, all the isolates showed significant binding affinity towards the folate synthesizing enzyme DHFR.
More than 24 different chlorogenic acid derivatives have been isolated from this plant [43]; among them 5-O-caffeoylquinic acid displayed significant anti-proliferative and antimicrobial activities. The in vitro study revealed that compound 1 showed the highest growth inhibition against the HeLa cell line (IC50 value = 181.3 μg/mL). Besides, compound 1 also exhibited maximum binding affinity in the molecular modeling study (binding energy = −15.3 kcal/mol) against DHFR, indicating potential anti-proliferative and antibacterial activity. Likewise, Sultana et al. [20] found promising antibacterial and antioxidant activities of this similar plant leaf's n-hexane and ethyl acetate extracts. The effects of the isolated compounds against bacteria, cancer cell lines and free radicals were consistent to the effects of the leaf extracts. This might be assumed and predicted by due to dipole-dipole and H-bond interaction with amino acids of targeted protein DHFR. It has been further confirmed by the ribbon model of protein where compound 1 bound deeply inside the pocket of DHFR, which depicted its anticancer and antimicrobial activities through the anti-metabolite pathway. A large number of non-covalent H-bond and Pi-sigma interactions (Pi-alkyl) which largely involves charge transfer help in intercalating the drug in the binding site. These findings suggest high structural stability and may lead to high inhibitory activity of compound 1 [44].
Syringin (2), a common phenylpropanoid glycoside was isolated earlier from different plants [45, 46] has been isolated from this plant for the first time. This compound exhibited remarkable in vitro anti-cancer effect (IC50 = 194.40 μg/mL, Table 1) and antimicrobial activity against a variety of pathogens which has been further evinced by in silico study (binding affinity −13.6 kcal/mol). In a previous report, it was found that syringin isolated from Foeniculum vulgare showed significant in vitro anti-proliferative effect against HeLa cell line [47]. As depicted in Figure 2, compound 2 interacted strongly with its binding sites through three conventional H-bonds and two pi-alkyl interactions.
Luteolin (3) and apigenin (4), have already exhibited antibacterial potentiality [48]. The luteolin (3) involved two non-covalent H bonds and two Pi bonds, while apigenin (4) has three H-bonds, two Pi-alkyl bonds and one T shaped Pi-Pi stacking interaction (Figure 2). This result indicates that apigenin (4) has more binding affinity towards DHFR than luteolin (3) though the docking score in case of both compounds were almost same (−12.6 kcal/mol and −12.7 kcal/mol for compound 3 and 4, respectively). The higher inhibitory effect of compound 4 compared to compound 3 might be attributed due to the high binding affinity and deep binding pocket [49].
Jhanol (5), a labdane-type hydroxyditerpene was reported earlier from the flower part of Stevia plants [41] possessed mild cytotoxicity and antibacterial activity in in vitro studies. The binding energy towards DHFR was −10.7 kal/mol which could be due to non-covalent interaction like Pi-Pi interaction. Meanwhile, the binding affinity of compound 6 was satisfactory with an estimated free binding energy of −11.2 kal/mol (Figure 3) which might be responsible for its anticancer and antibacterial activity. Interestingly, labdane type diterpenes 5 and 6 bound to the same pocket of DHFR (Figure 3) and showed similar binding energy because of their structural similarity.
It has been shown that apigenin (3) gave most promising antioxidant effect (IC50 = 8.93) than other compounds. This might be due to the presence of multiple hydroxyl group as reported earlier in a previous study [50]. Some sort of antioxidant activity was reported by 5-O-caffeol quinic acid, luteolin and syringin (compound 1, 2, and 4) in some previous studies [51, 52, 53]. Our results also re-establish the previous results; however, we found less potent antioxidant action of syringin by DPPH scavenging assay method compared to the reported data by Shen and co-authors [52]. Likewise, we found potent antioxidant effect against DPPH free radical in case of 5-O-caffeol-quinic acid compared to the previous result reported by Kim et al. (IC50 = 23.33 μg/mL vs. 37 μM) [53]. Jhanol and jhanidiol also showed potent antioxidant activity. This is the first DPPH scavenging report of these compounds. The capability to scavenge DPPH could be accredited to the content of polyphenols, flavonoids as well as alkaloids as accentuated in several reports [54, 55]. Redox properties are mainly responsible for this activity which could be due to adsorbing and neutralizing free radicals, quenching singlet and triplet oxygen or decomposing peroxides [56]. The in vitro antioxidant activity was further supported by the docking study towards glutathione reductase and urase oxidase enzyme as stated in Table 4.
5. Conclusion
Among the isolated molecules, compound 1 showed significant anti-proliferative activity against HeLa cell and antibacterial activities against various gram positive and gram-negative microorganisms. Other isolated compounds also showed moderate to mild cytotoxicity and antibacterial effects. On the other hand, compound 4 (apigenin) exerted strong antioxidant effect compared to ascorbic acid. Compound 1 (5-O- caffeol quinic acid), 3 (luteolin) and 6 (janidiol) showed moderate free radical scavenging activity. This research work also demonstrated that the secondary metabolites isolated from this plant have different degrees of affinity towards binding sites of target protein, DHFR, glutathione reductase and urase oxidase responsible for anticancer and antimicrobial actions and antioxidant effects, respectively. The results of in-vitro cytotoxicity and antibacterial activity were clearly justified by computational studies predicting plausible binding modes of the six compounds in the active site of DHFR. Therefore, this study has established the plant species as the potential source of anticancer, antibacterial and free radical scavenging agents for the first time. Based on this study, we may assume that the isolated compounds might be the lead compounds for development of chemotherapy and antibacterial therapeutic agents though the exact mechanism is not underlying yet and, the research should be carried for improvement of semi-synthetic derivatives or better drugs to combat cancer as well as various microbial infection.
Declarations
Author contribution statement
Most. Chand Sultana Khatun: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Md. Abdul Muhit: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Md. Jamal Hossain: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Muhammad Abdullah Al-Mansur: Performed the experiments; Contributed reagents, materials, analysis tools or data.
S. M. Abdur Rahman: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to thank the Division of Bioorganic and Medicinal Chemistry, Osaka University, Japan for assisting with some NMR analysis of our samples. Besides, MCSK is grateful to Ministry of Science and Technology for providing a fellowship to conduct this research.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Zengin G., Mahomoodally M.F., Sinan K.I., Sadeer N., Maggi F., Caprioli G., Angeloni S., et al. Evaluation of chemical constituents and biological properties of two endemic Verbascum species. Process Biochem. 2021;108:110–120. [Google Scholar]
- 2.Ahmad K., Khan I., Blundell R., Azzopardi J., Mahomoodally M.F. Stevia rebaudiana Bertoni. an updated review of its health benefits, industrial applications and safety. Trends Food Sci. Technol. 2019;100:177–189. [Google Scholar]
- 3.Ghaheri M., Adibrad E., Safavi S.M., Kahrizi D., Soroush A., Muhammadi S., Ghorbani T., Sabzevari A., Ansarypour Z., Rahmanian E. Effects of life cycle and leaves location on gene expression and glycoside biosynthesis pathway in Stevia rebaudiana Bertoni. Cell. Mol. Biol. (Noisy-le-Grand, France) 2018;64(2):17–22. doi: 10.14715/cmb/2018.64.2.4. [DOI] [PubMed] [Google Scholar]
- 4.Lemus-Mondaca, Vega-Gálvez A., Zura-Bravo L., Kong A.H. Stevia rebaudiana Bertoni, source of a high-potency natural sweetener: a comprehensive review on the biochemical, nutritional and functional aspects. Food Chem. 2012;132:1121–1132. doi: 10.1016/j.foodchem.2011.11.140. [DOI] [PubMed] [Google Scholar]
- 5.Esmaeili F., Ghaheri M., Kahrizi D., Mansouri M., Safavi S.M., Ghorbani T., Muhammadi S., Rahmanian E., Vaziri S. Effects of various glutamine concentrations on gene expression and steviol glycosides accumulation in Stevia rebaudiana Bertoni. Cell. Mol. Biol. (Noisy-le-Grand, France) 2018;64(2):1–5. doi: 10.14715/cmb/2018.64.2.1. [DOI] [PubMed] [Google Scholar]
- 6.Kahrizi D., Ghaheri M., Yari Z., Yari K., Bahraminejad S. Investigation of different concentrations of MS media effects on gene expression and steviol glycosides accumulation in Stevia rebaudiana Bertoni. Cell. Mol. Biol. (Noisy-le-Grand, France) 2018;64(2):23–27. doi: 10.14715/cmb/2018.64.2.11. [DOI] [PubMed] [Google Scholar]
- 7.Segura-Campos M., Barbosa- Martin E., Matus-Basto A., Cabrera-Amaro D., Murguia-Olmedo M., Moguel-Ordonez Y., Betacur-Ancona D. Comparison of chemical and functional properties of Stevia rebaudiana (Bertoni) varieties cultivated in Mexican southeast. Am. J. Plant Sci. Vol. 2014;5(3):286–293. [Google Scholar]
- 8.Azzam C.R., Al-Taweel S.K., Abdel-Aziz R.M., Rabea K.M., Abou-Sreea A., Rady M.M., Ali E.F. Salinity effects on gene expression, morphological, and physio-biochemical responses of Stevia rebaudiana Bertoni in vitro. Plants (Basel, Switzerland) 2021;10(4):820. doi: 10.3390/plants10040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim I.S., Yang M., Lee O.H., Kang S.N. The antioxidant activity and the bioactive compound content of Stevia rebaudiana water extracts. LWT - Food Sci. Technol. (Lebensmittel-Wissenschaft -Technol.) 2011;44(5):1328–1332. 2011. [Google Scholar]
- 10.Kahrizi D., Ghari S.M., Ghaheri M., Fallah F., Ghorbani T., Beheshti Ale Agha A., Kazemi E., Ansarypour Z. Effect of KH2PO4 on gene expression, morphological and biochemical characteristics of stevia rebaudiana Bertoni under in vitro conditions. Cell. Mol. Biol. (Noisy-le-Grand, France) 2017;63(7):107–111. doi: 10.14715/cmb/2017.63.7.18. [DOI] [PubMed] [Google Scholar]
- 11.Myint K.Z., Wu K., Xia Y., et al. Polyphenols from Stevia rebaudiana (Bertoni) leaves and their functional properties. J. Food Sci. 2020;85(2):240–248. doi: 10.1111/1750-3841.15017. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Ruiz J.C., Moguel-Ordoñez Y.B., Segura-Campos M.R. Biological activity of Stevia rebaudianaBertoni and their relationship to health. Crit. Rev. Food Sci. Nutr. 2017;57(12):2680–2690. doi: 10.1080/10408398.2015.1072083. [DOI] [PubMed] [Google Scholar]
- 13.Ghaheri M., Miraghaee S., Babaei A., Mohammadi B., Kahrizi D., Saivosh Haghighi Z.M., Bahrami G. Effect of Stevia rebaudiana Bertoni extract on sexual dysfunction in Streptozotocin-induced diabetic male rats. Cell. Mol. Biol. (Noisy-le-Grand, France) 2018;64(2):6–10. doi: 10.14715/cmb/2018.64.2.2. [DOI] [PubMed] [Google Scholar]
- 14.Hashempoor S., Ghaheri M., Kahrizi D., Kazemi N., Muhammadi S., Safavi S.M., Ghorbani T., Rahmanian E., Heshmatpanaah M. Effects of different concentrations of mannitol on gene expression in Stevia rebaudiana Bertoni. Cell. Mol. Biol. (Noisy-le-Grand, France) 2018;64(2):28–31. doi: 10.14715/cmb/2018.64.2.6. [DOI] [PubMed] [Google Scholar]
- 15.Shivanna N., Naika M., Khanum F., Kaul V.K. Antioxidant, anti-diabetic and renal protective properties of Stevia rebaudiana. J. Diabet. Complicat. 2013;27(2):103–113. doi: 10.1016/j.jdiacomp.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Gupta E, Purwar S, Sundaram S, Rai GK. Nutritional and therapeutic values of Stevia rebaudiana: a review. J. Med. Plants Res. 7(46); 3343–3353.
- 17.Abdel-Fattah S.M., Badr A.N., Seif F.A.H.A., Ali S.M., Hassan R.A. Antifungal and anti-mycotoxigenic impact of eco-friendly extracts of wild stevia. J. Biol. Sci. 2018;18(8):488–499. [Google Scholar]
- 18.Mann T.S., Agnihotri V.K., Kumar D., et al. In vitro cytotoxic activity guided essential oil composition of flowering twigs of Stevia rebaudiana. Nat. Produc. Commun. May 2014 [PubMed] [Google Scholar]
- 19.Voloshina A.D., Sapunova A.S., Kulik N.V., Belenok M.G., Strobykina I.Y., Lyubina A.P., Gumerova S.K., Kataev V.E. Antimicrobial and cytotoxic effects of ammonium derivatives of diterpenoids steviol and isosteviol. Bioorg. Med. Chem. 2021 Feb 15;32:115974. doi: 10.1016/j.bmc.2020.115974. [DOI] [PubMed] [Google Scholar]
- 20.Sultana C., Azad M.A.K., Rahman M.M., Muhit M.A., Rahman S.M.A. Phytochemical and biological investigation of Stevia rebaudiana (Bert.) leaves grown in Bangladesh. Dhaka Univ. J. Pharm. Sci. 2020;19:191–197. [Google Scholar]
- 21.Su Wen-Yi, Pan Rong-Kai, Song Jiang-Li, Li Guo-Bi, Liu Sheng-Gui. Synthesis, crystal structures and cytotoxic activity of two zinc (II) complexes derived from benzimidazole derivatives. Polyhedron. 2019;161:268–275. [Google Scholar]
- 22.Rashid P.T., Ahmed M., Rahaman M.M., Muhit M.A. 14-deoxyandrographolide isolated from Andrographis paniculata (Burm. f) needs growing in Bangladesh and its antimicrobial properties. Dhaka Univ. J. Pharm. Sci. 2018;17(2):265–267. [Google Scholar]
- 23.Brand-Williams W., Cuvelier M.E., Berset C. Use of a free radical method to evaluate antioxidant activity. LWT - Food Sci. Technol. (Lebensmittel-Wissenschaft -Technol.) 1995;28(1):25–30. [Google Scholar]
- 24.Hawser S., Lociuro S., Islam K. Dihydrofolate reductase inhibitors as antibacterial agents. Biochem. Pharmacol. 2006;71(7):941–948. doi: 10.1016/j.bcp.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 25.El-Naggar M., Sallam H.A., Shaban S.S., Abdel-Wahab S.S., E Amr A.E., Azab M.E., Nossier E.S., Al-Omar M.A. Design, synthesis, and molecular docking Study of novel heterocycles incorporating 1,3,4-thiadiazole moiety as potential antimicrobial and anticancer agents. Molecules. 2019;24(6):1066. doi: 10.3390/molecules24061066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alam S., Rashid M.A., Sarker M.M.R., Emon N.U., Arman M., Mohamed I.N., Haque M.R. Antidiarrheal, antimicrobial and antioxidant potentials of methanol extract of Colocasia gigantea Hook. f. leaves: evidenced from in vivo and in vitro studies along with computer-aided approaches. BMC Compl. Med. Ther. 2021;21(1):119. doi: 10.1186/s12906-021-03290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahmud S., Rafi M.O., Paul G.K., et al. Designing a multi-epitope vaccine candidate to combat MERS-CoV by employing an immunoinformatics approach. Sci. Rep. 2021;11(1):15431. doi: 10.1038/s41598-021-92176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bikadi Z., Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J. Cheminf. 2009;11(1):15. doi: 10.1186/1758-2946-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed T., Rahman S.M.A., Asaduzzaman M., Islam A.B.M.M.K., Chowdhury A.K.A. Synthesis, in vitro bioassays, and computational study of heteroaryl nitazoxanide analogs. Pharmacol. Res. Perspect. 2021;9(3):e00800. doi: 10.1002/prp2.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., Jiang H., Shi R. A new acylated querctin glycoside from the leaves of Stevia rebaudiana Bertoni. Nat. Prod. Res. 2009;23(15):1378–1383. doi: 10.1080/14786410802447294. [DOI] [PubMed] [Google Scholar]
- 31.Bajko E., Kalinowska M., Borowski P., Seirgiejezyk L., Lewandowski W. 5-O-Caffeoylquinic acid: a spectroscopic study and biological screening for antimicrobial activity. LWT-Food Sci. Technol. 2016;65:471–479. [Google Scholar]
- 32.Gonzalez A.G., Arteaga J.M., Breton J.L., Fraga B.M. Five new labdane diterpene oxides from Eupatorium Jhani. Phytochemrstry. 1976;4:107–111. [Google Scholar]
- 33.Medimagh S., Hammami S., Faidi K., Hajji N., Abreu P.J.M., Mighri Z. Gallocatechin and Trans syringin from Limoniastrum guyonianum bios growing in Tunisa. J. de la Société Chimique de Tunisie. 2010;12:207–210. [Google Scholar]
- 34.Barber M.S., MeConnel V.S., DeCaux B.S. Antimicrobial intermediates of the general phenylpropanoid and lignin specific pathways. Phytochemistry. 2000;54:53–56. doi: 10.1016/s0031-9422(00)00038-8. [DOI] [PubMed] [Google Scholar]
- 35.Dixon R.A. Natural products and plant disease resistance. Nature. 2001;411:843–847. doi: 10.1038/35081178. [DOI] [PubMed] [Google Scholar]
- 36.Bouarab-Chibani Lynda, Forquet V., Lanteri P., Clement Y., Leonard-Akkari L., Qulahal N., Degraeve P., Bordes C. Antibacterial properties of polyphenols: characterization and QSAR (Quantative structure activity relationship) models. Front. Microbiol. 2019;10:1–23. doi: 10.3389/fmicb.2019.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makapugay H.C., Nanayakkara N.P.D., Kinghorn A.D. Improved high–performance liquid chromatographic separation of the Stevia rebaudiana diterpene glycosides using linear gradient elution. J. Chromatogr. 1984;283:390–395. [Google Scholar]
- 38.Wolwer-Rieck U. The Leaves of Stevia rebaudiana (Bertoni) their constituens and the analysis thereof: a review. J. Agric. Food Chem. 2012;608:86–885. doi: 10.1021/jf2044907. [DOI] [PubMed] [Google Scholar]
- 39.Jahan I.A., Mostafa M., Hossain H., Nimmi I., Sattar A., Alim A., Moeiz S.M.I. Antioxidant activity of Stevia rebaudiana Bert. Leaves from Bangladesh. Bangladesh Pharmaceut. J. 2010;2(13):67–75. [Google Scholar]
- 40.Ullah A., Munir S., Mabkhot Y., Badsha S.L. Bioactivity profile of the diterpene isosteviol and its derivative. Molecules. 2019;24:678. doi: 10.3390/molecules24040678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tadhani M.B., Subhash R. In vitro antimicrobial activity of Stevia rebaudiana Bertoni. Leaves. Trop. J. Pharmaceut. Res. 2006;5(1):557–560. [Google Scholar]
- 42.Tomita T., Sato N., Arai T., Shiraishi H., Sato M., Takeuchi M., Kamio Y. Bactericidal activity of a fermented hot-water extract from Stevia rebaudiana Bertoni towards entero haemorrahagic E. coli 0157: H7 and other Food- Borne pathogenic bacteria. Microbiol. Immunol. 1997;41(12):1005–1009. doi: 10.1111/j.1348-0421.1997.tb01961.x. [DOI] [PubMed] [Google Scholar]
- 43.Karakose H., Jaiswal R., Kuhnert N. Characterization and quantification of hydroxycinnamate derivatives in Stevia rebaudiana leaves by LC-MS. J. Agric. Food Chem. 2011;59:10143–10150. doi: 10.1021/jf202185m. [DOI] [PubMed] [Google Scholar]
- 44.Arthur David Ebuka, Uzairu Adamu. Molecular docking studies on the interaction of NCI anticancer analogues with human Phosphatidylinositol 4, 5-bisphosphate 3-kinase catalytic Subunit. J. King Saud Univ. 2019;2019:1151–1166. 01. 011. [Google Scholar]
- 45.Ono M., Ito Y., Ishikawa T., Kitajima J., Tanaka Y., Niiho Y., Nohara T. Five new monoterpene glycosides and other compounds from Foeniculi fructus (Fruite of Foeniculum vulgare Miller) Chem. Pharm. Bull. 1996;44:337–342. [Google Scholar]
- 46.Caos Thao Quyen, Lee Bo Mi, Jung Yeon Woo, Nguyen Van Thu, Kim Jeong Ah, Min Byung Sun. Cytotoxic activity of compounds from Styrax obassia. Nat. Prod. Commun. 2017;12(2):259–260. [PubMed] [Google Scholar]
- 47.Shak Katrin. Cytotoxicity of diatery flavonoids on different cancer cell types. Phcog. Rev. 2014;8(16):122–126. doi: 10.4103/0973-7847.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adamczak A., Ozarowski M., Karpinski T.M. Antibacterial activity of some flavonoids and organic acids widely distributed in plants. J. Clin. Med. 2020;9:109. doi: 10.3390/jcm9010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukunishi Y., Nakamura H. Prediction of ligand-binding sites of proteins by molecular docking calculation for a random ligand library. Protein Sci. 2011;20(1):95–106. doi: 10.1002/pro.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sánchez-Marzo N., Pérez-Sánchez A., Ruiz-Torres V., Martínez-Tébar A., Castillo J., Herranz-López M., Barrajón-Catalán E. Antioxidant and photoprotective activity of apigenin and its potassium salt derivative in human keratinocytes and absorption in CaCO2 cell monolayers. Int. J. Mol. Sci. 2019;20(9):2148. doi: 10.3390/ijms20092148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashokkumar P., Sudhandiran G. Protective role of luteolin on the status of lipid peroxidation and antioxidant defense against azoxymethane-induced experimental colon carcinogenesis. Biomed. Pharmacother. 2008;62(9):590–597. doi: 10.1016/j.biopha.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 52.Shen Z., Yang C., Zhu P., Tian C., Liang A. Protective effects of syringin against oxidative stress and inflammation in diabetic pregnant rats via TLR4/MyD88/NF-κB signaling pathway. Biomed. Pharmacother. 2020;131:110681. doi: 10.1016/j.biopha.2020.110681. [DOI] [PubMed] [Google Scholar]
- 53.Kim W.R., Kim E.O., Kang K., Oidovsambuu S., Jung S.H., Kim B.S., Nho C.W., Um B.H. Antioxidant activity of phenolics in leaves of three red pepper (Capsicum annuum) cultivars. J. Agric. Food Chem. 2014;62(4):850–859. doi: 10.1021/jf403006c. [DOI] [PubMed] [Google Scholar]
- 54.Madsen H.L., Neilsen B.R., Bertelsen G., Skibsted L.H. Screening of antioxidative activity of spices. A comparison between assays based on ESR spin trapping and electrochemical measurement of oxygen consumption. Food Chem. 1996;57:331–337. [Google Scholar]
- 55.Moller J.K.S., Madsen H.L., Altonen T., Skibsted L.H. Dittany (Origanum dictamnus) as a source of water extractable antioxidants. Food Chem. 1999;64:215–219. [Google Scholar]
- 56.Zheng W., Wang S.Y. Antioxidant activity and phenolic compounds in selected herbs. J. Agric. Food Chem. 2001;49:5165–5170. doi: 10.1021/jf010697n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.