Abstract
Paracoccidioidomycosis (PCM) is a life-threatening systemic fungal infection acquired after inhalation of Paracoccidioides propagules from the environment. The main agents include members of the P. brasiliensis complex (phylogenetically-defined species S1, PS2, PS3, and PS4) and P. lutzii. DNA-sequencing of protein-coding loci (e.g., GP43, ARF, and TUB1) is the reference method for recognizing Paracoccidioides species due to a lack of robust phenotypic markers. Thus, developing new molecular markers that are informative and cost-effective is key to providing quality information to explore genetic diversity within Paracoccidioides. We report using new amplified fragment length polymorphism (AFLP) markers and mating-type analysis for genotyping Paracoccidioides species. The bioinformatic analysis generated 144 in silico AFLP profiles, highlighting two discriminatory primer pairs combinations (#1 EcoRI-AC/MseI-CT and #2 EcoRI-AT/MseI-CT). The combinations #1 and #2 were used in vitro to genotype 165 Paracoccidioides isolates recovered from across a vast area of South America. Considering the overall scored AFLP markers in vitro (67–87 fragments), the values of polymorphism information content (PIC = 0.3345–0.3456), marker index (MI = 0.0018), effective multiplex ratio (E = 44.6788–60.3818), resolving power (Rp = 22.3152–34.3152), discriminating power (D = 0.5183–0.5553), expected heterozygosity (H = 0.4247–0.4443), and mean heterozygosity (Havp = 0.00002–0.00004), demonstrated the utility of AFLP markers to speciate Paracoccidioides and to dissect both deep and fine-scale genetic structures. Analysis of molecular variance (AMOVA) revealed that the total genetic variance (65-66 %) was due to variability among P. brasiliensis complex and P. lutzii (PhiPT = 0.651–0.658, P < 0.0001), supporting a highly structured population. Heterothallism was the exclusive mating strategy, and the distributions of MAT1-1 or MAT1-2 idiomorphs were not significantly skewed (1:1 ratio) for P. brasiliensis s. str. (χ2 = 1.025; P = 0.3113), P. venezuelensis (χ2 = 0.692; P = 0.4054), and P. lutzii (χ2 = 0.027; P = 0.8694), supporting random mating within each species. In contrast, skewed distributions were found for P. americana (χ2 = 8.909; P = 0.0028) and P. restrepiensis (χ2 = 4.571; P = 0.0325) with a preponderance of MAT1-1. Geographical distributions confirmed that P. americana, P. restrepiensis, and P. lutzii are more widespread than previously thought. P. brasiliensis s. str. is by far the most widely occurring lineage in Latin America countries, occurring in all regions of Brazil. Our new DNA fingerprint assay proved to be rapid, reproducible, and highly discriminatory, to give insights into the taxonomy, ecology, and epidemiology of Paracoccidioides species, guiding disease-control strategies to mitigate PCM.
Key words: AFLP, AMOVA, Endemic mycosis, Genetic diversity, Mating-type, Paracoccidioides, Paracoccidioidomycosis
Introduction
Paracoccidioidomycosis (PCM) is a life-threatening systemic fungal infection first described in Brazil by Lutz and Splendore in 1908 (Lutz 1908). Its description was shortly followed by reported infections throughout South America (Ferguson & Upton 1947, Nino 1950) and, later, Latin America (Gonzalez Ochoa & Esquivel 1950). Following inhalation of soil-born Paracoccidioides propagules, patients may develop primary pulmonary foci, subsequently disseminating to other host organs and systems (Brummer et al. 1993, Hahn et al. 2014) by hematogenic or lymphatic pathway (Restrepo et al. 2008). The classical clinical forms of PCM-disease are divided into two groups (Franco et al. 1987). The first group includes an acute or subacute form ("juvenile"), predominant in children, adolescents, and young adults, and is depicted by tropism of the fungus to the monocyte-phagocyte system. The second and significant group corresponds to the chronic form, found in 80 to 95 % of the total cases, and men between 30 and 50 years of age are the most affected patients (Nery et al. 2021a, 2021b). Brazil accounts for up to 80 % of cases of PCM in Latin America, with an incidence of 7.99 cases per 1 000 hospitalizations based on the overall hospital admissions notified to the Ministry of Health in the year 2011 (Giacomazzi et al. 2016). The incidence and severity of PCM increase with the progression of human immunodeficiency virus (HIV) infection and reduction in CD4 counts. Therefore, the course of the disease in the HIV-infected patient is similar to those observed in the acute presentation of endemic PCM as it tends to be disseminated and more rapidly progressive (Almeida et al. 2017, de Almeida et al. 2018, Macedo et al. 2018).
PCM is caused by the thermodimorphic fungi classified in the order Onygenales, family Ajellomycetaceae and genus Paracoccidioides (Bocca et al. 2013). The etiological agent was first described in 1912 as Zymonema brasiliensis and later named Paracoccidioides brasiliensis as asserted by Floriano P. de Almeida (Almeida 1930). Historically the taxonomy of Paracoccidioides is inconsistent with the description of several names that were found to be invalid. A few examples include Paracoccidioides antarcticus (Gezuele 1989), Paracoccidioides cerebriformis (Moore 1935), and Paracoccidioides tenuis (Moore 1938). All these changes were reduced or described as a synonym of P. brasiliensis (Splendore) de Almeida, aiming to describe species names that reflect a natural classification system and a real need for communication among scientists (Almeida 1930, Del Negro et al. 1993, Garcia et al. 1993).
For decades Paracoccidioides was considered to be a monotypic taxon. Phylogenetic relationships in Paracoccidioides based on the internal transcribed spacer (ITS) region, and partial regions of the mitochondrial genome (COB2, ATP6, COX3, RNS, RNL), or nuclear genome including coding (GP43, PRP8, CHS2, ARF, CDC42, FKS) and non-coding regions (ORN1, 11b12b, 15b16b, AB, KL, MN, R56, TUB, III-IV, and XI-XII) revealed a great deal of diversity among clinical and environmental isolates, supporting the existence of other species beyond P. brasiliensis (Feitosa et al. 2003, Carrero et al. 2008, Salgado-Salazar et al. 2010). Using multilocus sequence analysis (MLSA), two distinct biological species P. brasiliensis sensu lato (containing at least four cryptic phylogenetic species: S1, PS2, PS3, and PS4) and P. lutzii (originally named Pb01-like) were identified (Matute et al. 2006, Teixeira et al. 2009, Theodoro et al. 2012, Turissini et al. 2017). The phylogeographical distribution is diverse in Paracoccidioides, and species 1 (S1) is the predominant agent of human PCM recovered from Argentina, Brazil, Paraguay, Peru, and Venezuela. Phylogenetic species 2 (PS2) was found in Brazil, Venezuela, and Uruguay, while the remaining phylogenetic species PS3 and PS4 appear to be restricted to Colombia and Venezuela, respectively (Theodoro et al. 2012). Paracoccidioides lutzii (formerly Pb01-like) is prevalent in central-west Brazil, mainly in Mato Grosso state, with scattered cases outside this area (Nery et al. 2021b).
Fungal taxonomy has undergone a significant transformation in recent decades as a method for inferring evolutionary relationships and defining species boundaries, especially with the introduction of molecular data in phylogenetic studies (Lücking et al. 2021). Recently, five species were proposed within Paracoccidioides. The P. brasiliensis complex includes the classical agent P. brasiliensis sensu stricto (formerly S1) in addition to the newly described P. americana (formerly PS2); P. restrepiensis (formerly PS3), and P. venezuelensis (formerly PS4) (Turissini et al. 2017, Teixeira et al. 2020). Paracoccidioides lutzii (formerly Pb01-like) is presented as a monophyletic group in phylogenetic analyses (Teixeira et al. 2014c). Nevertheless, there is no consensus among genetic, morphological, and clinical data (Shikanai-Yasuda et al. 2017, de Macedo et al. 2019, Hahn et al. 2019). The morphological markers for the recognition of different Paracoccidioides are scarce due to the overlapping phenotypic features. In this diverging scenario, whole-genome sequencing appears as an essential tool for elucidating relationships and resolving Paracoccidioides taxonomy (Muñoz et al. 2016).
Judging from a clinical perspective, preliminary studies found no significant clinical differences between the disease caused by members of the P. brasiliensis complex (de Macedo et al. 2019) or even between P. brasiliensis s.l. and P. lutzii (Hahn et al. 2019, Pereira et al. 2020). On the one hand, from a molecular epidemiological perspective, we can benefit from recognizing different genotypes of Paracoccidioides (Pinheiro et al. 2020, 2021). Thus, to explore intraspecific variation, it is necessary to develop and apply molecular tools that are highly discriminatory at an affordable price. To meet this need, amplified fragment length polymorphisms (AFLP) can recognize genetic variations between any two Paracoccidioides genomes using a combination of restriction enzyme digestion of DNA, PCR amplification, and separation by capillary electrophoresis (Vos et al. 1995). AFLP has already been used successfully to study genetic variability in fungi, such as Aspergillus fumigatus (Warris et al. 2003), Candida spp. (Borst et al. 2003), Coccidioides species (Duarte-Escalante et al. 2013), Cryptococcus spp. (Hagen et al. 2015), Fonsecaea spp. (Najafzadeh et al. 2011), Histoplasma spp. (Rodrigues et al. 2020a), and Sporothrix spp. (de Carvalho et al. 2020).
We took advantage of the whole-genome sequences now available for Paracoccidioides in GenBank and conducted extensive bioinformatic analyses to screen for markers that were appropriate to address questions about the epidemiology and genetic diversity. These markers were subsequently evaluated in vitro to explore a vast collection of Paracoccidioides samples. Here, we report the AFLP primer combinations as an essential step to characterize specimens, species, and genotypes to complement PCM epidemiology with quality data.
Material and Methods
Fungal strains and DNA extraction
This study used one hundred sixty-five clinical and environmental strains of Paracoccidioides spp., recovered from Argentina, Brazil, Colombia, Guadeloupe Island, Peru, Uruguay, and Venezuela (Supplementary Table S1). These isolates are deposited in the Laboratory of Emerging Fungal Pathogens culture collection at the Federal University of São Paulo (UNIFESP), São Paulo, Brazil. Yeast cells were grown on Fava-Netto agar at 37 °C and co-cultured every seven days (Fava-Netto 1961, Fava-Netto et al. 1969). DNA extraction was performed from a 14-d-old yeast culture using the FastDNA kit (MP Biomedicals, Solon, OH, USA) as previously described (Rodrigues et al. 2014). The genomic DNA concentration and purity (A260/A280 nm > 1.8) were analysed by spectrophotometry (NanoDrop 2000; Thermo Fisher Scientific, Waltham, MA, USA), and samples were stored at -20 °C.
Identifying Paracoccidioides by TUB1-RFLP
The PCR-restriction fragment length polymorphism (RFLP) of the α-tubulin gene (TUB1-RFLP) was performed using the protocol previously described (Roberto et al. 2016). Briefly, the α-tubulin gene was amplified from genomic DNA with the primers α-TubF and α-TubR (Table 1) (Kasuga et al. 2002). The PCR was incubated in a Mastercycler (Eppendorf, Hamburg, Germany) with an initial denaturation step of 5 min at 95 °C, followed by 35 cycles of 1 min at 94 °C, 45 s at 48 °C, 1 min at 68 °C, and a final extension of 10 min at 68 °C. For RFLP analysis, 3 μL of the TUB1-PCR product were digested with 2 μl 10× fast digest buffer, 1 μl BclI endonuclease (10 U/μl; Thermo Fisher Scientific), 1 μl MspI endonuclease (10 U/μl; Thermo Fisher Scientific) and ultrapure water to a final volume of 20 μl. Tubes were incubated at 37 °C for 2 h, and the double-digested products were analysed by electrophoresis at 100 V on 2.5 % (w/v) agarose gels for 120 min in the presence of GelRed (Biotium, Fremont, CA, USA). The 50-bp DNA Step Ladder (Promega, Madison, WI, USA) was used as a size marker. The fragments were visualized using the L-Pix Touch imaging system under UV illumination (Roberto et al. 2016).
Table 1.
Primers used in this study for generic amplification, sequencing, and genotyping.
| Locus/Region | Primer | Primer sequence 5′ to 3′ | Tm (°C) | Sense | Amplicon (bp) | Reference |
|---|---|---|---|---|---|---|
| TUB1 | α-TubF | CTGGGAGGTATGATAACACTGC | 48 °C | Forward | 263 | Kasuga et al. 2002 |
| α-TubR | CGTCGGGCTATTCAGATTTAAG | 48 °C | Reverse | Kasuga et al. 2002 | ||
| ITS | ITS1 | TCCGTAGGTGAACCTTGCGG | 52 °C | Forward | 620 | White et al. 1990 |
| ITS4 | TCCTCCGCTTATTGATATGC | 52 °C | Reverse | White et al. 1990 | ||
| MAT1-1 | GMAT1-1 F | GCAATTGTCTATTTCCATCAGT | 56 °C | Forward | 1 455 | Torres et al. 2010 |
| GMAT1-1 R | CTAGATGTCAAGGTACTCGGTA | 56 °C | Reverse | Torres et al. 2010 | ||
| MAT1-1 | MAT1-1 EST2-1 F | GGCATTTAACAAATCTTTACG | 52 °C | Forward | 400 | Torres et al. 2010 |
| MAT1-1 EST2-1 R | CCCAGTTTGTAGCAATGAGT | 52 °C | Reverse | Torres et al. 2010 | ||
| MAT1-2 | GMAT1-2 F | TTCGACCGTCCACGCCTATCTC | 56 °C | Forward | 1 208 | Torres et al. 2010 |
| GMAT1-2 R | TCATTGCGAAAAGGTGTCAAG | 56 °C | Reverse | Torres et al. 2010 | ||
| MAT1-2 | MAT1-2 EST G-F | CATGTCTCTGTCATTGTTCCA | 52 °C | Forward | 1 000 | Torres et al. 2010 |
| MAT1-2 EST G-R | GGAACAAGGAGGTTGAAGTT | 52 °C | Reverse | Torres et al. 2010 |
In silico AFLP analyses
Whole-genome sequence of nine Paracoccidioides isolates covering all the phylogenetic species described so far (Table 2) were in silico analysed to predict AFLP markers in the range of 50–500 bp. In silico AFLP analysis was performed using the software AFLPinSilico v. 2 (Rombauts et al. 2003) (available at http://bioinformatics.psb.ugent.be/webtools/aflpinsilico/). Briefly, Paracoccidioides genomes were retrieved from GenBank and in silico digested with EcoRI (a six-base cutter) and MseI (a four-base cutter) restriction enzymes. Afterward, a total of 16 combinations containing two selective bases (EcoRI+2 and MseI+2) were used to mine a subset of fragments. Combinations were selected based on the AFLP Microbial Fingerprinting kit (Applied Biosystems, Foster City, CA, USA). Finally, to accurately simulate the AFLP technique, we determined the length of all fingerprints with the addition of the adaptor and primer lengths. The diversity of the fragments was used to create a matrix of amplicons, and data were visualized using the software Heatmapper (Babicki et al. 2016). Hierarchical cluster analysis based on average linkage and Euclidean distance was applied to each row cluster.
Table 2.
Genomes of Paracoccidioides species retrieved from NCBI Genome database (https://www.ncbi.nlm.nih.gov/genome) for in silico analysis.
| Strain | Species | Source | Origin | INSDC1 (WGS) | Total length (Mb) | BioProjects | Reference |
|---|---|---|---|---|---|---|---|
| T16B1 | P. brasiliensis | Dasypus novemcinctus | Brazil | SRR4024730 | 29.1 | PRJNA322632 | Muñoz et al. 2016 |
| Pb18 | Chronic PCM | Brazil | ABKI02 | 29.5 | PRJNA28733 | Desjardins et al. 2011 | |
| Pb03 | P. americana | Chronic PCM | Brazil | ABHV02 | 28.8 | PRJNA27779 | Desjardins et al. 2011 |
| Pb262 | Dog food | Brazil | SRR4024732 | 28.9 | PRJNA322632 | Muñoz et al. 2016 | |
| Pb339 | P. restrepiensis | PCM | Brazil | SRR4024750 | 28.7 | PRJNA322632 | Muñoz et al. 2016 |
| CNH | Chronic PCM | Colombia | LYUC01 | 29.4 | PRJNA288047 | Muñoz et al. 2016 | |
| Pb300 | P. venezuelensis | Soil | Venezuela | LZYO01 | 29.4 | PRJNA287815 | Muñoz et al. 2016 |
| Pb01 | P. lutzii | PCM | Brazil | ABKH02 | 32.6 | PRJNA28731 | Desjardins et al. 2011 |
| PlEE | PCM | Brazil | SRR4024735 | 32.3 | PRJNA322632 | Muñoz et al. 2016 |
International Nucleotide Sequence Database Collaboration (INSDC; http://www.insdc.org/)
AFLP fingerprinting
The AFLP fingerprinting analysis was conducted in duplicate with the AFLP Microbial Fingerprinting kit (Applied Biosystems). Digestion of Paracoccidioides DNA, adapter ligation, non-selective and selective amplification was performed following the manufacturer's recommendations, with minor modifications (Najafzadeh et al. 2011). Briefly, Paracoccidioides genomic DNA (200 ng) was digested in vitro using EcoRI (GˆAATTC) and MseI (TˆTAA) restriction enzymes (New England Biolabs, Ipswich, MA) and ligated to EcoRI and MseI adapters simultaneously. A preselective PCR was performed with EcoRI+0 and MseI+0 primers (Vos et al. 1995). Fluorescent AFLP was performed with 6-carboxyfluorescein (FAM) or NED fluorescent dye-labelled EcoRI primer with two bases selection (5′-GAC TGC GTA CCA ATT CNN-3′) and unlabelled MseI primer with two bases selection (5′-GAT GAG TCC TGA GTA ACT -3′). Two different combinations were chosen to evaluate the potential for genetic characterization of Paracoccidioides isolates (combination #1 FAM-EcoRI-AC/MseI-CT or #2 NED-EcoRI-AT/MseI-CT). All oligonucleotides were provided by Applied Biosystems in the AFLP Microbial Fingerprinting kit. AFLP fragments were determined by capillary electrophoresis with an ABI3730 Genetic Analyzer alongside a GeneScan 500 ROX internal size standard (35–500 bp; Applied Biosystems) at the Human Genome and Stem Cell Research Centre Core Facility (University of São Paulo, São Paulo, Brazil) under previously described conditions (de Carvalho et al. 2020). Electropherograms are representative of two independent experiments.
The selection of amplicons was automated, and only robust and high-quality amplicons were considered. Each electropherogram was carefully inspected to exclude doubtful peaks, setting the minimum threshold at 100 relative fluorescence units (RFU) and considering only peaks with sizes in the range of 50 and 500 base pairs. The size and diversity of the AFLP fragments were determined with BioNumerics v. 7.6 software (Applied Maths, Sint-Martens-Latem, Belgium). AFLP fragments were converted to the dominant presence (1) or absence (0) at probable fragment positions.
Pairwise genetic distances were calculated using the band-based Dice similarity coefficient (Dice 1945) combined with a "Fuzzy logic" option. Dendrograms were built using the unweighted pair group mean arithmetic method (UPGMA). To assess the consistency of a given cluster, we calculated the cophenetic correlation coefficient and its standard deviation, which determines the linear correlation coefficient between the cophenetic distances obtained from the tree and the dendrogram-derived similarities. Therefore, it is a measure of how accurately the AFLP-dendrogram represents the similarities among observations.
To estimate the existence of topological congruence between AFLP dendrograms and their associated confidence level, we determined the congruence index (Icong) (de Vienne et al. 2007), based on maximum agreement subtrees (MAST). The correlation between experiments was calculated using the Pearson product-moment correlation coefficient (Pearson correlation) (Schober et al. 2018). A scatter plot was used to plot each pair of similarity values as one dot in a similarity plot between two experiment types. Especially for extensive data sets resulting in dense scatter plots, we used a histogram displaying a multi-colour scale ranging from white over blue, green, yellow, orange, and red to black.
Minimum spanning trees (MSTs) were calculated to explore the evolutionary relationships among all the observed genotypes of Paracoccidioides. MSTs characterize a set of edges (connections) that connect nodes (isolates) so that the summed distance of all branches is the shortest possible (Vauterin & Vauterin 2006). All figures were exported and treated using Corel Draw X8 (Corel, Ottawa, Canada).
Genetic diversity analysis
To calculate the potential of the two selective primer combinations evaluated here, the following polymorphism indices for dominant markers were calculated: polymorphic information content (PIC) (Botstein et al. 1980), expected heterozygosity (H) (Liu 1998), effective multiplex ratio (E) (Powell et al. 1996), arithmetic mean heterozygosity (Havp) (Powell et al. 1996), marker index (MI) (Powell et al. 1996, Varshney et al. 2007), discriminating power (D) (Tessier et al. 1999), and resolving power (Rp) (Prevost & Wilkinson 1999).
Dimensioning analysis
Alternative grouping approaches such as principal component analysis (PCA) and multidimensional scaling (MDS) were employed to create three-dimensional plots according to their similarity. The optimization and position tolerances for choosing fragments were set to 0.10 %, and automated fragment matching was performed with a minimum profiling of 5 %. Default settings were applied for PCA and MDS, subtracting the average for characters. In addition, the Self-Organizing Map (SOM), a robust artificial neural network algorithm in the unsupervised learning category, was employed to classify AFLP entries in a two-dimensional space (map) according to their likeliness (Kohonen 2001). The Kohonen map size was set to 100 (i.e., the number of neural network nodes in each direction). All figures were exported and treated using Corel Draw X8.
Structure analysis
Analysis of AFLP data in STRUCTURE (v. 2.3.4) (Pritchard et al. 2000) was performed using the admixture model, allowing alpha to be inferred and assuming correlated allele frequencies, using a burn-in period of 10 000 Markov chain Monte Carlo (MCMC) replications followed by 10 000 sampling replications, with 20 independent runs performed for K values one to twenty. All data were analysed using the method of Evanno and colleagues as implemented in StructureHARVESTER (v. 0.6.94) (Evanno et al. 2005, Earl & vonHoldt 2012) to determine the optimal number of clusters (K). Consensus population distributions were obtained with CLUMPP (v. 1.1.2) (Jakobsson & Rosenberg 2007), using the full search for the AFLP data. Final plots were generated using ggplot2 (Wickham 2016) in R (The R Core Team 2014).
Recombination analysis
To explore the relationships among Paracoccidioides species, a split network (Neighbor-Net) was constructed using the software SplitsTree v. 5.0.0 alpha (Huson & Bryant 2006) on AFLP profiles. For the construction of networks, we used the Hamming distances method (Hamming 1950) with the Neighbor-Net algorithm (Bryant & Moulton 2004) adapted for binary sequences (Huson & Kloepper 2005).
Analysis of molecular variance (AMOVA)
The AFLP data was transformed into a binary matrix of the presence/absence of each allele for each individual (Peakall & Smouse 2006, 2012). The genetic differentiation among populations was determined using PhiPT (ΦPT, an analogue of FST). This measure allows intra-individual variation to be suppressed and is therefore ideal for comparing binary data with 9 999 permutations (Teixeira et al. 2014a). Analysis of molecular variance among and within populations was performed using GenAlex v. 6.5 (Excoffier et al. 1992).
Statistical analysis
We calculated Cohen's kappa coefficient (κ) and its 95 % confidence interval (CI) to determine the degree of concordance between AFLP typing and TUB1-RFLP (Roberto et al. 2016). Kappa values were read as follows: 0.00–0.20, poor agreement; 0.21–0.40, fair agreement; 0.41–0.60, moderate agreement; 0.61–0.80, good agreement; 0.81–1.00, very good agreement (Altman 1991). A P-value ≤ 0.05 was considered significant. All statistical calculations were performed with MedCalc Statistical Software v. 20.013 (MedCalc Software, Ostend, Belgium; http://www.medcalc.org; 2021). We calculated Simpson’s diversity (Simpson 1949) and Shannon’s diversity (Shannon 1948) for each organism/genetic group with the relative abundances estimated with frequency data.
Characterization of the mating-type idiomorphs
PCR primers targeting the MAT1-1 or the MAT1-2 region were used to determine the mating-types idiomorphs, as described before (Torres et al. 2010). Approximately 50 ng of genomic DNA was used for PCR with two sets of oligonucleotide primers: GMAT1-1 F and GMAT1-1 R, which amplify a 1 455 bp fragment from the α box region of the MAT1-1 idiomorph, and GMAT1-2 F and GMAT1-2 R, which amplify a 1 208 bp fragment from the HMG domain gene, present in the MAT1-2 idiomorph (Torres et al. 2010) (Table 1). PCRs were performed with PCR Master Mix buffer (Promega) as described above under the following conditions: 4 min at 95 °C; followed by 35 cycles of 1 min at 94 °C, 1 min at 56 °C, and 1 min at 72 °C; and a final step of 10 min at 72 °C. Samples were visualized on 1.2 % agarose gels as described above.
Results
TUB1-RFLP
We conducted a retrospective molecular epidemiological study using the largest collection of Paracoccidioides strains from Latin America (n = 165), preserved in our institution for more than 50 years (1970–2021). The TUB1 gene was amplified followed by double digestion using the BclI and MspI endonucleases, which produced four different electrophoretic patterns corresponding to 92 P. brasiliensis s. str. (S1; fragments of 155 bp and 108 bp), 22 P. americana (PS2; fragments of 62 bp, 93 bp, and 108 bp), 14 P. restrepiensis (PS3; amplicon remained intact with 263 bp) and 37 P. lutzii (Pb01-like; fragments of 62 bp and 204 bp). TUB1-RFLP did not allowed the recognition of P. venezuelensis (PS4) (Supplementary Table S1).
Development of AFLP markers for Paracoccidioides
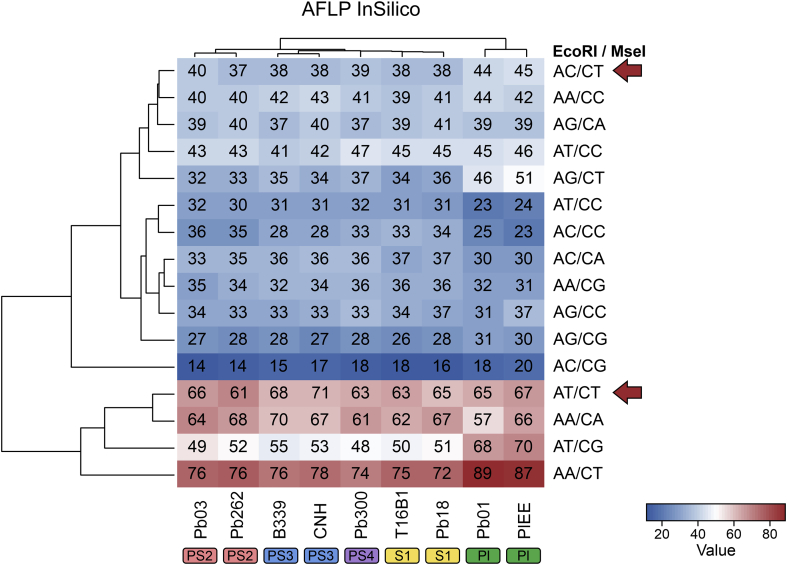
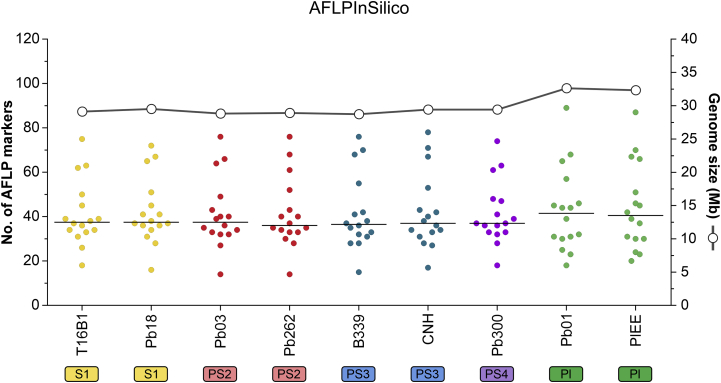
The first step in our approach involved the in silico characterization of nine Paracoccidioides genomes retrieved from NCBI, comprising all medically relevant members described so far. AFLPinSilico was used to inspect restriction spots for EcoRI (GˆAATTC) and MseI (TˆTAA). Subsequently, a group of modified genomic fragments was generated by adding adaptor sequences, and an enriched group of modified genetic fragments was chosen based on two selective bases for EcoRI+2 and MseI+2 primers. Thus, we produced a matrix of 144 in silico AFLP profiles, which are shown as a heatmap in Fig. 1. A significant diversity of fragments was generated, ranging from 14–87 in 16 combinations evaluated in AFLPinSilico (Fig. 2). Paracoccidioides lutzii presented the largest genome core (∼32.6 Mb), and we observed the most significant number of AFLP markers in all combinations, supported by a strong positive correlation (Pearson correlation = 0.926, r2 = 0.9623, P = 0.000337) (Fig. 2).
Fig. 1.
High-resolution maps based on 16 AFLP fingerprints simulated using nine up-to-date sequenced Paracoccidioides genomes available at GenBank. The numbers inside the squares represent the expected numbers of amplicons after in silico digestion with EcoRI and MseI endonucleases following ligation of adapters and selective amplification using selective primer EcoRI (5′-GAC TGC GTA CCA ATT CNN-3′) and MseI (5′-GAT GAG TCC TGA GTA ANN-3′), as indicated.
Fig. 2.
A total of 16 combinations of selective EcoRI+2 and MseI+2 primer pairs were employed to generate 144 virtual AFLP profiles AFLPinSilico. The dots located on the left X-axis represented the number of fragments generated for each combination and were colour-coded according to their genetic groups. The bold bar represents the average of AFLP markers obtained for all combinations. The white dots located on the right X-axis represent the genome size, estimated by whole-genome sequencing.
We highlighted two combinations (#1 EcoRI-AC/MseI-CT or #2 EcoRI-AT/MseI-CT) to be evaluated in vitro, which revealed the highest number of polymorphic markers (i.e., number and size) with the potential to speciate Paracoccidioides (Supplementary Table S2). A total of 154 polymorphic fragments were amplified in vitro using the selective primers EcoRI+2 and MseI+2, among them 67 and 87 loci, for combinations #1 and #2, respectively.
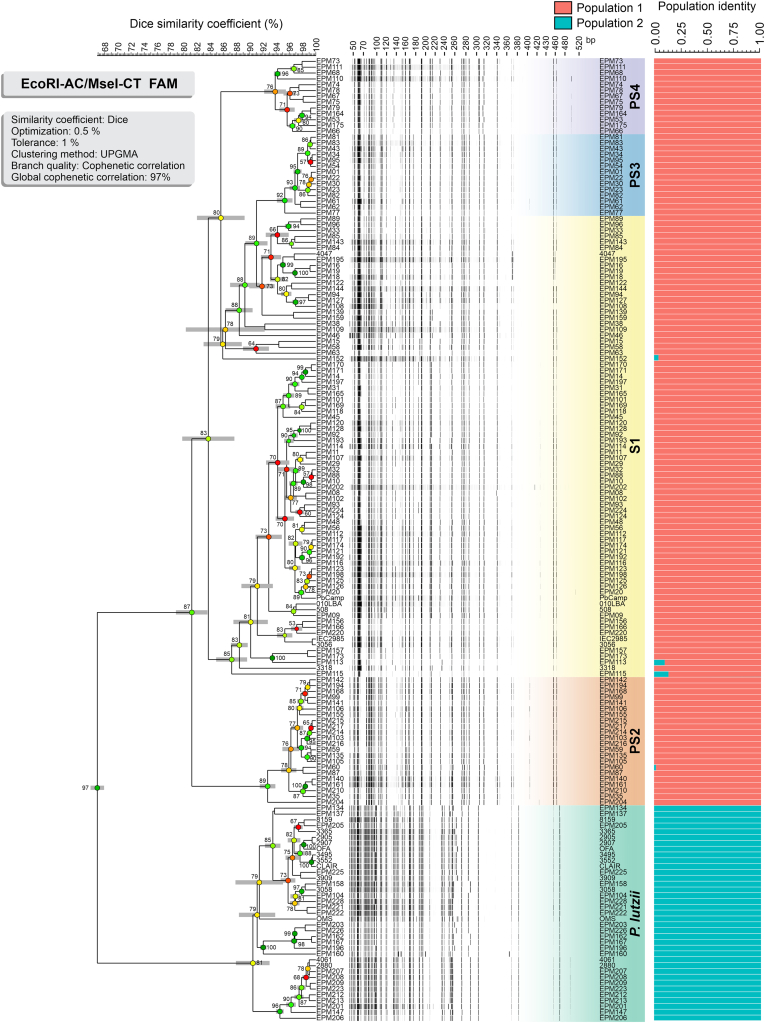
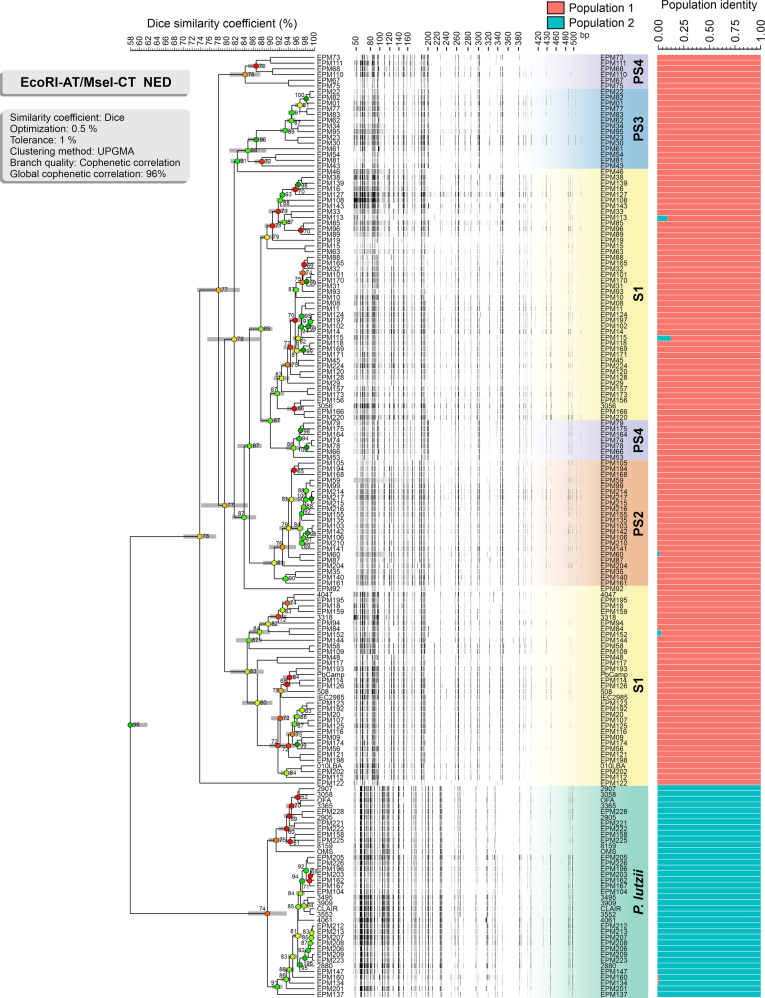
The dendrograms generated based on Dice’s similarity coefficient are depicted in Fig. 3, Fig. 4. Clustering analysis shows five well-supported clades with a global similarity level ranging between 55.84 % ± 3.53 % and 66.86 % ± 1.49 %. The global cophenetic correlation coefficient between the dendrogram and the original similarity matrix was significant (96–97 %) for both markers supporting a reasonable degree of confidence in the association obtained for 165 isolates of Paracoccidioides (Supplementary Table S3). This AFLP clustering profile agrees with the broadly applied GP43-based classification (Morais et al. 2000).
Fig. 3.
The UPGMA dendrogram, based on AFLP fingerprint, generated with a total of four selective bases (FAM-EcoRI-AC/MseI-CT) for 165 Paracoccidioides spp. originated from Latin America. The dendrogram shows cophenetic correlation values (circles, which are represented by colour ranges between green-yellow-orange-red according to decreasing cophenetic correlation) for a given clade and its standard deviation (grey bar). For pairwise genetic distances calculation, the Dice coefficient was used. The cophenetic correlation of the dendrogram is 97 %. Bayesian cluster analyses with STRUCTURE (k = 2) of 165 Paracoccidioides spp. based on AFLP. Each vertical bar represents one individual and its probabilities of being assigned to clusters. Further information about isolate sources can be found in Supplementary Table S1.
Fig. 4.
The UPGMA dendrogram, based on AFLP fingerprint, generated with a total of four selective bases (NED-EcoRI-AT/MseI-CT) for 165 Paracoccidioides spp. originated from Latin America. The dendrogram shows cophenetic correlation values (circles, which are represented by colour ranges between green-yellow-orange-red according to decreasing cophenetic correlation) for a given clade and its standard deviation (grey bar). For pairwise genetic distances calculation, the Dice coefficient was used. The cophenetic correlation of the dendrogram is 96 %. Bayesian cluster analyses with STRUCTURE (k = 2) of 165 Paracoccidioides spp. based on AFLP. Each vertical bar represents one individual and its probabilities of being assigned to clusters. Further information about isolate sources can be found in Supplementary Table S1.
The AFLP fingerprints revealed that 128 out of 165 isolates were embedded within the P. brasiliensis complex (cophenetic correlation values: #1 87 % and #2 78 %), with 79 isolates (48 %) classified as P. brasiliensis s. str. (AFLP S1), 22 isolates (13 %) as P. americana (AFLP PS2), 14 isolates (9 %) as P. restrepiensis (AFLP PS3), and 13 isolates (8 %) as P. venezuelensis (AFLP PS4). The second significant genetic cluster refers to 37 out of 165 isolates (22 %) which were classified as P. lutzii (cophenetic correlation values: #1 81 %, and #2 74 %) (Fig. 3, Fig. 4). The AFLP clusters classification was confirmed by TUB1-RFLP genotyping. To determine the level of concordance of the results of the TUB1-RFLP and any AFLP assay, we calculated the kappa statistic and its 95 % confidence interval (CI). A very good agreement was observed for P. brasiliensis (κ = 0.843 ± 0.041, 95 % CI 0.762–0.923), P. americana (κ = 1.0, 95 % CI 1.000–1.000), P. restrepiensis (κ = 1.0, 95 % CI 1.000–1.000) and P. lutzii (κ = 1.0, 95 % CI 1.000–1.000), but poor agreement for P. venezuelensis (κ = 0.0, 95 % CI -2.8859 × 10-8–2.8859 × 10-8). Although TUB1-RFLP could not distinguish P. venezuelensis (PS4), the AFLP fingerprinting could cluster the isolates into this group, considered closely related to P. brasiliensis s. str. and P. americana under both markers (#1 and #2). In this case, the AFLP PS4 group was identified based on the reference strains EPM67 (Pb300/V1) and EPM73 that were characterized as P. venezuelensis in previous studies (Salgado-Salazar et al. 2010, Muñoz et al. 2014, Turissini et al. 2017, Pinheiro et al. 2021).
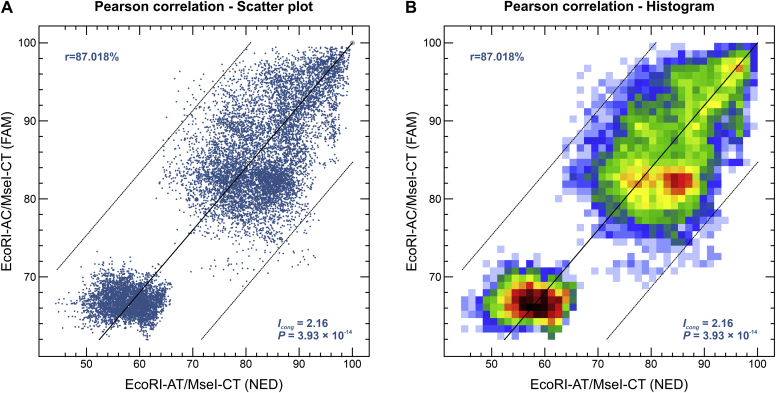
To assess the existence of topological correspondence between the two AFLP dendrograms, we used the congruence index (Icong) (de Vienne et al. 2007), and the Pearson product-moment correlation coefficient (Pearson correlation). A comparable and constant clustering signature was noted in pairwise comparisons, as demonstrated by the great congruence index value and their significant associated P-value (Icong = 2.16; P = 3.93 × 10-14), as well as a strong positive correlation for the Pearson product-moment correlation coefficient (r = 87.018 %, P < 0.00001) (Fig. 5). Thus, our AFLP dendrograms are more congruent than expected by chance, supporting the use of new AFLP markers to speciate Paracoccidioides and to explore both deep and fine-scale genetic structures.
Fig. 5.
The correlation between AFLP experiments evaluated for 165 Paracoccidioides isolates. A similarity plot for two experiments EcoRI-AC/MseI-CT and EcoRI-AT/MseI-CT was assessed using (A) the Pearson correlation coefficient (scatter plot) to plot each pair of similarity values as one dot, and (B) the Pearson correlation coefficient (histogram) representing the average the number of dots in each area. A multi-colour scale ranges continuously from white over blue, green, yellow, orange, and red to black.
The averages of fragments for combination #1 (EcoRI-AC / MseI-CT) varied per species between 37–49 for P. brasiliensis s. str. (Median = 44; CV = 5.79 %); 44–47 for P. americana (Median = 46; CV = 1.90 %); 37–41 for P. restrepiensis (Median = 40; CV = 2.38 %); 41–46 for P. venezuelensis (Median = 43; CV = 3.35 %); and 44–50 for P. lutzii (Median = 49; CV = 4.18 %). A greater number of fragments was observed for combination #2 (EcoRI-AT / MseI-CT) varying between 43–69 for P. brasiliensis s. str. (Median = 58; CV = 10.61 %); 62–68 for P. americana (Median = 66; CV = 2.79 %); 61–67 for P. restrepiensis (Median = 63.5; CV = 2.41 %); 63–69 for P. venezuelensis (Median = 67; CV = 2.79 %); and 41–66 for P. lutzii (Median = 58; CV = 11.06 %) (Supplementary Table S3). Table 3 shows the features of marker attributes for AFLP primer combinations #1 and #2.
Table 3.
Summary of polymorphism statistics calculated for two different pairs of selective primers (EcoRI+2 and MseI+2) for Paracoccidioides species.
|
#1 EcoRI-AC/MseI-CT | ||||||||
|
Species |
Scored bands |
H |
PIC |
E |
Havp |
MI |
D |
Rp |
| S1 (n = 79) | 53 | 0.2854 | 0.2447 | 43.8608 | 0.0001 | 0.0030 | 0.3152 | 10.6076 |
| PS2 (n = 22) | 50 | 0.1472 | 0.1364 | 46.0000 | 0.0001 | 0.0062 | 0.1537 | 2.3636 |
| PS3 (n = 14) | 41 | 0.0705 | 0.0680 | 39.5000 | 0.0001 | 0.0049 | 0.0719 | 1.5714 |
| PS4 (n = 13) | 48 | 0.1841 | 0.1671 | 43.0769 | 0.0003 | 0.0127 | 0.1948 | 4.1538 |
| P. lutzii (n = 37) | 56 | 0.2366 | 0.2086 | 48.3243 | 0.0001 | 0.0055 | 0.2554 | 7.1892 |
| Overall (n = 165) |
67 |
0.4443 |
0.3456 |
44.6788 |
0.00004 |
0.0018 |
0.5553 |
22.3152 |
|
#2 EcoRI-AT/MseI-CT | ||||||||
|
Species |
Scored bands |
H |
PIC |
E |
Havp |
MI |
D |
Rp |
| S1 (n = 79) | 77 | 0.3558 | 0.2925 | 59.1772 | 0.0001 | 0.0035 | 0.4094 | 22.0253 |
| PS2 (n = 22) | 71 | 0.1462 | 0.1355 | 65.3636 | 0.0001 | 0.0061 | 0.1525 | 6.7273 |
| PS3 (n = 14) | 63 | 0.2098 | 0.1878 | 55.5000 | 0.0002 | 0.0132 | 0.2240 | 11.8571 |
| PS4 (n = 13) | 65 | 0.2265 | 0.2008 | 56.5385 | 0.0003 | 0.0152 | 0.2435 | 15.8462 |
| P. lutzii (n = 37) | 74 | 0.2300 | 0.2036 | 64.1892 | 0.0001 | 0.0054 | 0.2476 | 12.3784 |
| Overall (n = 165) | 87 | 0.4247 | 0.3345 | 60.3818 | 0.00002 | 0.0018 | 0.5183 | 34.3152 |
D = discriminating power; E = effective multiplex ratio; H = expected heterozygosity; Havp = mean heterozygosity; MI = marker index; PIC = polymorphism information content; Rp = resolving power.
The PIC established for each primer pair was dissimilar among species but comparable between markers. In general, PIC values varied from low polymorphism in P. americana (PIC = 0.1355–0.1364), P. restrepiensis (PIC = 0.0680–0.1878), and P. venezuelensis (PIC = 0.1671–0.2008) to average polymorphism in P. brasiliensis s. str. (PIC = 0.2447–0.2925), and P. lutzii (PIC = 0.2086–0.2036), and both markers presented high discriminating power (D = 0.5183–0.5553). The highest overall PIC value was observed for primer combination #1 (PIC = 0.3456), and the lowest was noted for primer combination #2 (PIC = 0.3345), supporting good diversity among the studied Paracoccidioides. Overall, P. brasiliensis s. str. and P. lutzii showed slightly higher PIC values than the remaining phylogenetic species (Table 3).
The global usefulness of each marker system was estimated using the marker index (MI), which was obtained as a product of polymorphic information content and effective multiplex ratio. Equal overall MI values (MI = 0.0018) were obtained for both combinations. A moderate positive correlation was observed between MI and PIC values (combination #1 Pearson correlation = 0.6797, r2 = 0.462, P = 0.206831; combination #2 Pearson correlation = 0.6896, r2 = 0.4755, P = 0.197644). The resolving power (Rp), which is the ability of each primer combination to detect level of variation among individuals was found to be higher in primer combination #2 (Rp = 34.3152) and lower for primer combination #1 (Rp = 22.3152) (Table 3). The Rp values were not correlated with MI for combination #1 (Pearson correlation = 0.4111, r2 = 0.169, P = 0.491713), but for combination #2 (Pearson correlation = 0.9336, r2 = 0.8716, P = 0.020334).
We assessed the expected heterozygosity (H), which is the probability that an individual in the population is heterozygous for the locus. The expected heterozygosity relates to Nei’s unbiased gene diversity (HS), as adapted for dominant markers under the assumptions of Hardy-Weinberg equilibrium and the Lynch-Milligan model (Lynch & Milligan 1994). The overall average expected heterozygosity for Paracoccidioides species ranged between 0.4247–0.4443 (Table 3). The high values for expected heterozygosity among P. brasiliensis (H = 0.2854–0.3558) and P. lutzii isolates (H = 0.2300–0.2366) supports high genetic diversity in these groups (Muñoz et al. 2016, Teixeira et al. 2020). The remaining species, such as P. americana (H = 0.1462–0.1472), P. restrepiensis (H = 0.0705–0.2098), and P. venezuelensis (H = 0.1841–0.2265), showed discrete variation, which is in accordance with a prevalently clonal population (Table 3). Moreover, the application of a concordant genotyping method to Paracoccidioides species, for which the relationship between the number of AFLP markers used and the estimated genetic diversity converged to the expected variation, further confirms that the approach described here allows assessment of the accuracy of inferences on the genetic diversity of prevalently clonal organisms derived using combinations #1 (EcoRI-AC/MseI-CT) and #2 (EcoRI-AT/MseI-CT) (Arnaud-Haond et al. 2005).
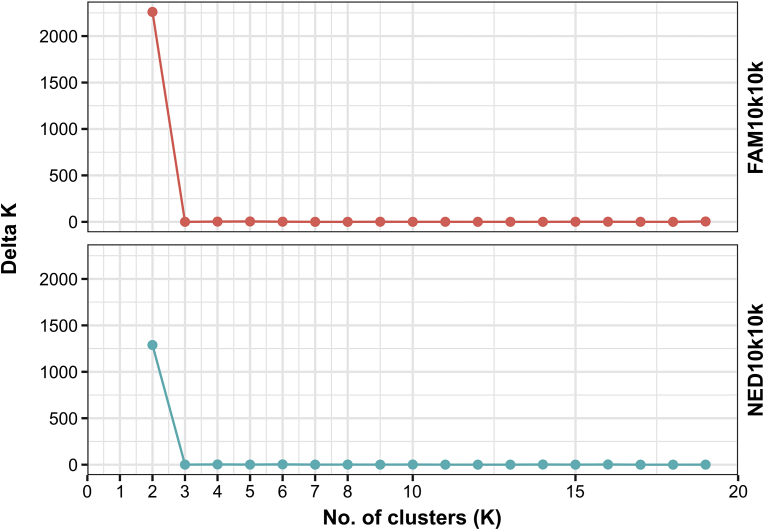
Structure analysis
We found a strong correlation between population structure and Paracoccidioides species. The structure analysis indicated two genetic clusters as the most probable number of genetically distinct populations (Fig. 3, Fig. 4). The Delta K plot (Fig. 6) showed the highest peak at K = 2, supporting the partition into two genetic clusters with no or a very weak signal of admixture (Supplementary Fig. S1). For K = 2, members of the P. brasiliensis complex clustered with the population 1. On the other hand, the second cluster corresponded to the P. lutzii isolates embedded in population 2, originating from endemic areas mainly in mid-west Brazil (Fig. 3, Fig. 4).
Fig. 6.
STRUCTURE Harvester results. The most plausible number of genetic clusters (K) within the complete data set of 165 individuals based on the method depicted by Evanno et al. (2005). Population genetic structure of the estimated ΔK value determined the maximum value at K = 2.
The AFLP profiles were employed to generate pairwise genetic distance matrices based on Dice's similarity coefficient, which were then analysed using PCA. The PCA plots for combinations #1 and #2 are shown in Fig. 7, and the distribution of 165 Paracoccidioides isolates among the three coordinates illustrated a trend similar to cluster analysis. Combination #1 revealed the highest cumulative percentage explained, with 68.2 % of the variation described by the first three components (coordinates X, Y, and Z). PCAs and MDSs analysis indicated considerable intraspecific clustering as well as a large genetic separation between any two taxa (interspecific variation). The structure evidenced by PCA supports the separation of P. brasiliensis complex and P. lutzii, consistent with the higher level of intraspecific variability shown in dendrogram analysis. Importantly, AFLP results agree with those detected using whole-genome sequencing (Muñoz et al. 2016, Teixeira et al. 2020).
Fig. 7.
Principal component analysis (PCA) and Multidimensional scaling (MDS) analysis of the combinations #1 EcoRI-AC/MseI-CT (67 loci) and #2 EcoRI-AT/MseI-CT (87 loci) informative AFLP markers plotted in three-dimensional space coloured according to the genetic groups. (A) PCA, and (B) MDS based on combination #1 EcoRI-AC/MseI-CT (n = 165). (C) PCA, and (D) MDS based on combination #2 EcoRI-AT/MseI-CT (n = 165). PCAs and MDS were created in the software BioNumerics v. 7.6.
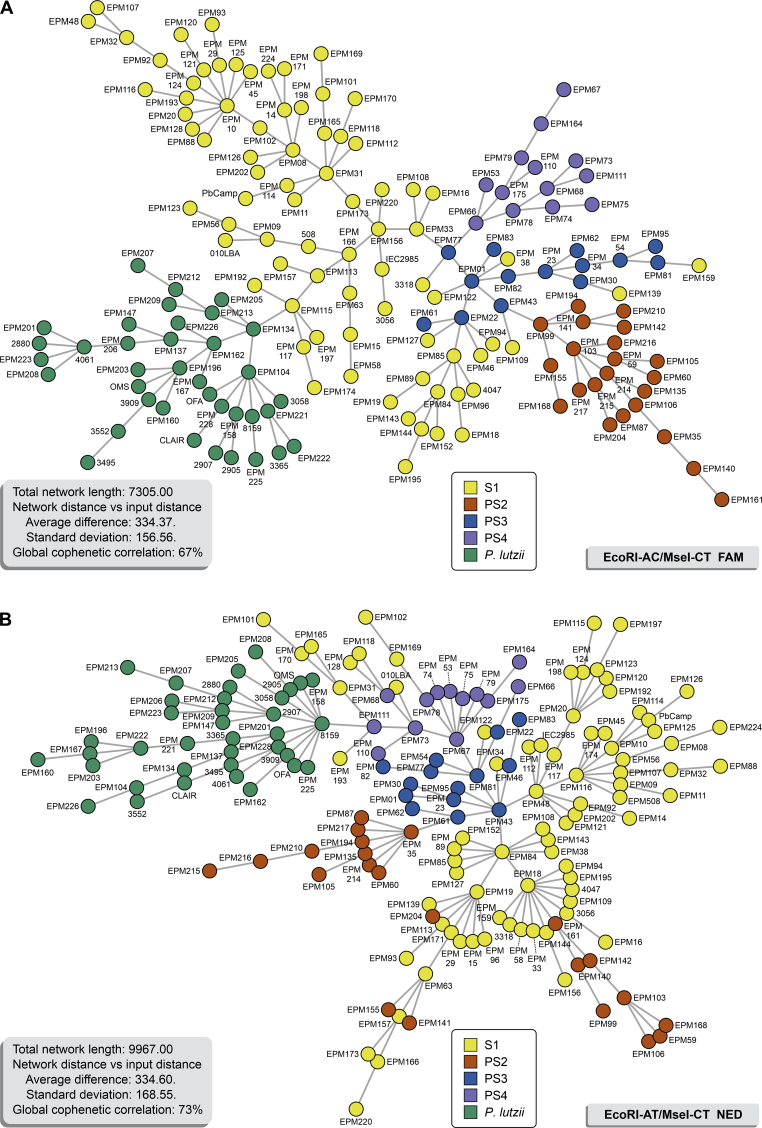
The structure of members of the P. brasiliensis complex and P. lutzii is confirmed by the AFLP-derived MSTs in Fig. 8, with most isolates having a unique genotype. A few isolates in the P. brasiliensis complex were randomly distributed, similar to the dendrogram, particularly for combination #2 (Fig. 8B), suggesting a plausible chain of disease transmission in molecular epidemiology (Salipante & Hall 2011). Many invariable fragments were observed in P. americana, P. restrepiensis, and P. venezuelensis (as evidenced by low PIC values in Table 3), and together with the overall high similarity of > 80 % between the fingerprints, agreed with a monophyletic origin of the isolates.
Fig. 8.
Minimum Spanning Trees (MSTs) showing the genetic relationship between 165 Paracoccidioides genotypes using (A) EcoRI-AC/MseI-CT (Total network length 7 305.00) and (B) EcoRI-AT/MseI-CT (Total network length 9 967.00). Each genotype was considered unique. Isolates were colour-coded according to their genetic groups. The distance between genotypes in the diagram does not reflect any relationship with the genetic distance between genotypes. The annotation of the genetic distance between each edge that connects the nodes is shown in Supplementary Fig. S2 (EcoRI-AC/MseI-CT) and Supplementary Fig. S3 (EcoRI-AT/MseI-CT).
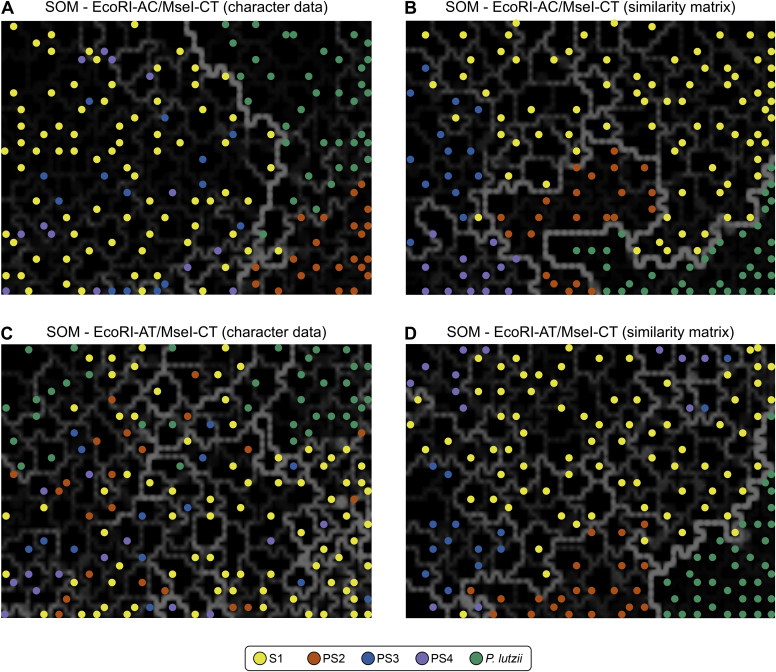
SOMs, an unsupervised artificial neural network, were used to cluster high-dimensional AFLP data by projecting it according to genetic clusters onto a low-dimensional map (Fig. 9). Contrasting to PCA, the distance between entries in the SOMs is not proportional to the taxonomic distance between the entries (Felix et al. 2015). Therefore, in Fig. 9, Paracoccidioides strains with low genetic distance form clusters (typified by black blocks). The relative genetic distance between neighbouring groups (black blocks) is designated by the intensity of white lines separating the clusters, with closely related groups separated by faint dark lines and more distantly related strains separated by increasingly lighter thicker lines. Thus, phylogenetic species displaying slight intraspecific variation, such as P. americana, P. restrepiensis, and P. venezuelensis, tended to remain closer, separated by thinner lines, but bright solid lines were observed separating clusters interspecifically (Fig. 9).
Fig. 9.
The distribution of the studied AFLP genotypes of 165 Paracoccidioides species originated from Latin America, using self-organizing mapping (SOM). The dimensioning analyses were performed using BioNumerics v. 7.6 to determine the consistency of the differentiation of the populations defined by the cluster analysis. (A) and (B) show the SOM for EcoRI-AC/MseI-CT combination (67 loci) using character data (binary matrix) and similarity matrix, respectively. (C) and (D) show the SOM for EcoRI-AT/MseI-CT combination (87 loci) using character data (binary matrix) and similarity matrix, respectively. The lighter and thicker the line (white, grey) between black blocks, the more distant are those samples contained in the black block from the adjacent black block. Isolates were colour-coded according to their genetic groups.
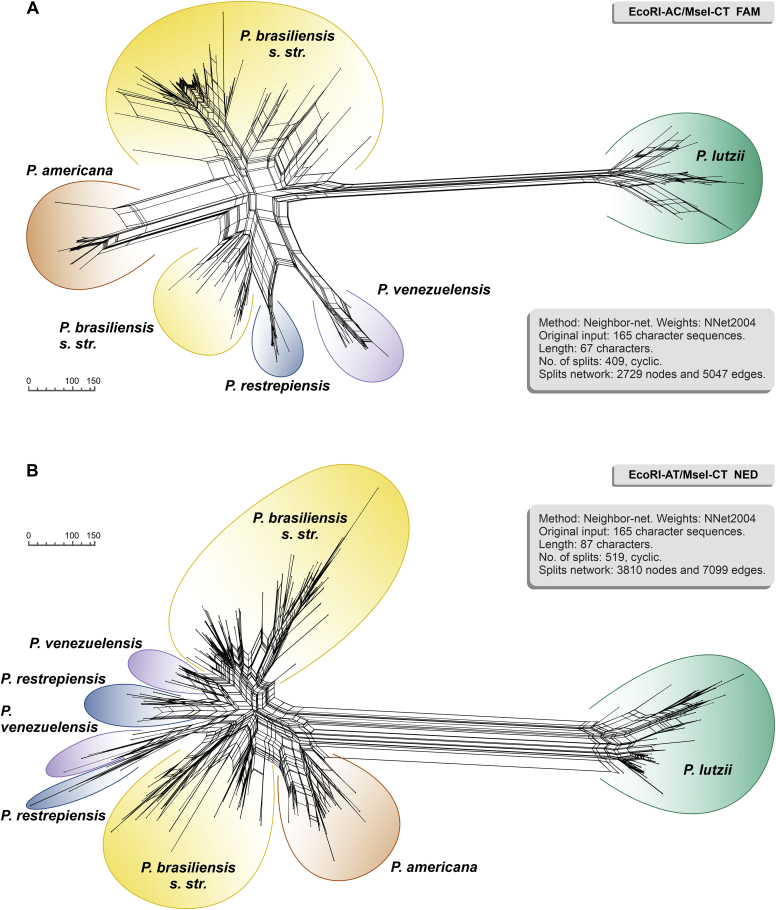
In a phylogenetic network analysis of Paracoccidioides AFLP profiles, we found that members of the P. brasiliensis complex and P. lutzii were the most differentiated from each other in both markers (Fig. 10). Moreover, in the reconstruction of evolutionary history, phylogenetic networks revealed large sets of parallel edges, suggestive of recombination events.
Fig. 10.
Neighbor-Net network showing genetic relationships based on AFLPs among Paracoccidioides species (scale equals genetic distance). (A) EcoRI-AC/MseI-CT and (B) EcoRI-AT/MseI-CT split networks. Analysis was performed using SplitsTree v. 5.0.0_alpha (Huson & Bryant 2006) for binary sequences (Huson & Kloepper 2005), and the original input consisted of 165 standard character sequences.
AMOVA
We used AMOVA to investigate genetic variance among 165 individuals of the two populations in Paracoccidioides (P. brasiliensis complex, n = 128, population 1; P. lutzii, n = 37, population 2). Table 4 shows the AMOVA findings for the population genetic analysis. AMOVAs performed for marker #1 in P. brasiliensis complex and P. lutzii showed that 66 % of the total genetic variance was triggered by variability among populations, whereas 34 % was driven by variability within populations (PhiPT = 0.658, P < 0.0001). A similar trend was observed for marker #2 with 65 % of total variation among populations and 35 % within populations (PhiPT = 0.651, P < 0.0001) (Supplementary Fig. S4). The hierarchical analysis of molecular variance leaves a strong differentiation among the groups, supporting a highly structured population. The results were highly significant (P < 0.0001).
Table 4.
Analysis of molecular variance (AMOVA) shows the partitioning of genetic variation within and between Paracoccidioides species populations.
| Marker | Source of variation | Df | SS | MS | Est. var. | % | P-value |
|---|---|---|---|---|---|---|---|
| #1 EcoRI-AC/MseI-CT | Among Population | 1 | 520.417 | 520.417 | 8.984 | 66 % | 0.0001 |
| Within Population | 163 | 760.516 | 4.666 | 4.666 | 34 % | 0.0001 | |
| #2 EcoRI-AT/MseI-CT | Among Population | 1 | 775.274 | 775.274 | 13.380 | 65 % | 0.0001 |
| Within Population | 163 | 1166.811 | 7.158 | 7.158 | 35 % | 0.0001 |
df = degree of freedom, SS = sum of squares, MS mean squares, Est. var. = estimate of variance, % = percentage of total variation, P-value is based on 9 999 permutations.
Mating-type
A mating type-specific PCR assay was used to amplify the MAT1-1 or the MAT1-2 regions among 165 Paracoccidioides isolates. The MAT1-1 region was detected in 88 isolates, while the MAT1-2 region was detected among 77 isolates (χ2 = 0.733; P = 0.3918). Heterothallism (self-sterility) was the universal mating strategy amongst Paracoccidioides species. The distribution of each sexual idiomorph within molecular species (S1, PS2, PS3, PS4, and P. lutzii) is presented in Table 5. The distributions of MAT1-1 or MAT1-2 idiomorph were not significantly skewed (1:1 ratio) for P. brasiliensis s. str. (χ2 = 1.025; P = 0.3113), P. venezuelensis (χ2 = 0.692; P = 0.4054), and P. lutzii (χ2 = 0.027; P = 0.8694), supporting the presence of random mating within each species. However, a biased distribution was found for P. americana (χ2 = 8.909; P = 0.0028) and P. restrepiensis (χ2 = 4.571; P = 0.0325) with an overwhelming presence of MAT1-1 idiomorphs.
Table 5.
Distribution of mating type alleles determined by PCR with mating-type allele-specific primers in Paracoccidioides isolates.
| Species | No. of isolates | No. of isolates by mating-type |
Chi-square value | P-value | |
|---|---|---|---|---|---|
| MAT 1-1 (%) | MAT 1-2 (%) | ||||
| P. brasiliensis (S1) | 79 | 35 (44.30) | 44 (55.69) | 1.025 | 0.3113 |
| P. americana (PS2) | 22 | 18 (81.81) | 4 (18.18) | 8.909 | 0.0028 |
| P. restrepiensis (PS3) | 14 | 11 (78.57) | 3 (21.42) | 4.571 | 0.0325 |
| P. venezuelensis (PS4) | 13 | 5 (38.46) | 8 (61.53) | 0.692 | 0.4054 |
| P. brasiliensis complex | 128 | 69 (53.90) | 59 (46.09) | 0.781 | 0.3768 |
| P. lutzii | 37 | 19 (51.35) | 18 (48.64) | 0.027 | 0.8694 |
| Overall | 165 | 88 (53.33) | 77 (46.66) | 0.733 | 0.3918 |
P. brasiliensis complex = S1, PS2, PS3, and PS4.
Phylogenetic trends in Paracoccidioides
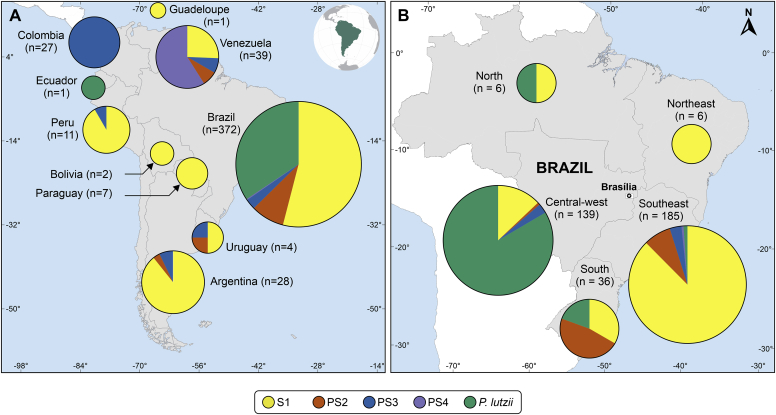
Distribution patterns were explored combining our dataset (n = 165) with data from 333 Paracoccidioides isolates reported in the literature and identified down to species level using molecular methods (e.g., whole-genome sequencing, MLSA, DNA fingerprint) (Matute et al. 2006, Carrero et al. 2008, Teixeira et al. 2009, Salgado-Salazar et al. 2010, Theodoro et al. 2012, Turissini et al. 2017, de Macedo et al. 2019, Hahn et al. 2019, Cocio et al. 2020b, Teixeira et al. 2020, Mattos et al. 2021, Nery et al. 2021b). Phylogenetic trends revealed that the P. brasiliensis complex species, including the four cryptic species, are widely distributed among different countries in Latin America. Most cryptic siblings occur in sympatry, as exemplified by P. brasiliensis s. str. and P. americana, with a clear overlapping distribution. In contrast, P. lutzii is endemic to Brazil (Fig. 11). The source of isolation revealed an overwhelming occurrence of clinical isolates (90.77 %, n = 452 out of 498) followed by animals (e.g., armadillo, dog, and penguin; 7.23 %, n = 36 out of 498), and from the environment (e.g., soil and dog food; 1 %, n = 5 out of 498). The source of isolation was unknown for five strains (1 %). Most molecularly characterized isolates (n = 498) are from Brazil (74.69 %, n = 372), followed by Venezuela (7.83 %, n = 39), Argentina (5.62 %, n = 28) Colombia (5.42 %, n = 27), Peru (2.20 %, n = 11), Paraguay (1.4 %, n = 7), Uruguay (0.8 %, n = 4), Bolivia (0.4 %, n = 2), Ecuador (0.2 %, n = 1) and Guadeloupe Island (0.2 %, n = 1). These data reveal the urgency to increase genetic surveillance in Paracoccidioides-affected areas (Fig. 11A).
Fig. 11.
Distribution patterns of 498 Paracoccidioides spp. isolates based on molecular characterization. (A) Distribution patterns observed in South America. (B) Distribution patterns observed in Brazil (n = 372). The sizes of circumferences are roughly proportional to the number of strains included. Codes reported within the pies denote genetic groups. Further information about isolate sources can be found in Supplementary Table S1.
Paracoccidioides brasiliensis s. str. (S1) is predominantly found in southeastern and southern Brazil, Argentina, Peru, Venezuela, Paraguay, Uruguay, Bolivia, and Guadeloupe Island. P. americana has sporadic distribution and is less frequently reported, with human cases described thus far in Brazil, Venezuela, Uruguay, and Argentina. The remaining species, such as P. restrepiensis (PS3) and P. venezuelensis (PS4), are sporadic PCM agents, and cases have been found in Colombia and Venezuela, respectively. Occasional cases related to P. restrepiensis have been found outside Colombia, mainly in Brazil, Argentina, Peru, and Uruguay. Paracoccidioides lutzii, on the other hand, comprises a single species and is primarily distributed in the Midwest and Amazon regions of Brazil. A single P. lutzii strain was reported from Ecuador (Fig. 11A).
In Brazil, an essential difference in the geographical incidence of each phylogenetic species was noted (Fig. 11B). The southeast region corresponds to the majority of PCM agents and presents the highest levels of diversity (Simpson Index = 0.772; Shannon Index = 0.721), with all species being reported. In the central-west region, PCM cases are mainly due to P. lutzii, followed by P. brasiliensis s. str. and P. americana (Simpson Index = 0.576; Shannon Index = 1.124). Paracoccidioides americana was the principal agent in the south region, followed by P. brasiliensis s. str. and P. restrepiensis (Simpson Index = 0.464; Shannon Index = 1.157). The lowest index of diversity was found for the North region (Simpson Index = 0.500; Shannon Index = 0.811), albeit only six isolates were recovered from this region. We did not detect species diversity in Northeast Brazil, with only six isolates characterized as P. brasiliensis s. str. (Fig. 11B).
Discussion
We here present the broadest population genetic study of Paracoccidioides species to date using isolates recovered from across a vast area of South America. Two sets of highly discriminatory AFLP markers were developed and shown to be a promising tool to dissect both deep and fine-scale genetic structures. The typing method proposed here combines robustness, reproducibility, high discriminatory power, and affordability, which is desirable for an important neglected mycosis such as PCM that is usually associated with poverty.
Pathogens with higher genetic diversity, significant effective population size, a mixed reproduction system, and great mutation rates are assumed to possess the highest evolutionary potential (Nath et al. 2013). Therefore, information regarding the current Paracoccidioides population and its evolutionary potential helps make informed disease-control strategies to mitigate PCM. Our AFLP technique demonstrated polymorphisms among closely related Paracoccidioides, which may contribute to resolve local epidemiological patterns as well as broader changes within populations over time and in response to selection pressures imposed by the environment and host resistance (McDonald & Linde 2002).
The availability of complete genome sequences for Paracoccidioides allowed us to predict the DNA fragments that AFLP would generate (Desjardins et al. 2011, Muñoz et al. 2014, 2016, Teixeira et al. 2020). Here, we demonstrated the best combination of restriction enzymes (EcoRI and MseI) by modelling their performance in silico for each species. Our analysis showed that the fragments observed following AFLP with EcoRI-AC / MseI-CT or EcoRI-AT / MseI-CT primers represent the optimal combinations to explore genetic diversity in Paracoccidioides. A similar in silico framework has been successfully applied for medically relevant Sporothrix species (de Carvalho et al. 2020, 2021a), supporting that combining bioinformatics tools and whole-genome sequences can make the AFLP method more predictable instead of, rather, using random combinations of suboptimal endonuclease-combinations (Rombauts et al. 2003, Paris et al. 2010).
Our AFLP dendrograms for Paracoccidioides species complex are compatible with the evolutionary history of the etiological agents of PCM, based on multilocus sequencing of proteins-encoding genes such as ARF, GP43, TUB1, and intein PRP8, or phylogenomic analyses (Morais et al. 2000, Theodoro et al. 2008, 2012, Turissini et al. 2017, Teixeira et al. 2020). Convergence between fingerprints and genomic methods has already been demonstrated for Candida auris using AFLP (Schelenz et al. 2016), short tandem repeat typing (de Groot et al. 2020), and whole-genome sequencing (Lockhart et al. 2017). Phylogenetic studies of Paracoccidioides suggest that P. brasiliensis s. str., P. americana, P. restrepiensis, and P. venezuelensis are closely related taxonomic entities (Muñoz et al. 2016, Turissini et al. 2017), and this clustering profile was recognized in our AFLP dendrograms.
Our dendrograms were more congruent than expected by chance, supported by the Icong value and a positive Pearson correlation, confirming that different markers reveal congruent evolutionary histories. In each case, P. lutzii is basal to members of the P. brasiliensis complex, and our AFLP data indicates that the P. brasiliensis complex members all share a more recent common ancestor with each other than they do with P. lutzii. P. americana, P. restrepiensis, P. venezuelensis and P. brasiliensis s. str. remained as sister species, as previously reported (Theodoro et al. 2012, Muñoz et al. 2016, Turissini et al. 2017, Teixeira et al. 2020).
From the fragment’s profiles observed, P. americana, P. restrepiensis, and P. venezuelensis reveal more invariant fragments than P. brasiliensis s. str., suggesting a more recent differentiation and a monophyletic origin of these lineages. Nearly all P. restrepiensis and P. venezuelensis occur within Colombia and Venezuela, respectively, suggesting that they evolved in these regions. Clusters of strains that presented many invariant fragments were mainly collected at a proximate geographic distance from each other. This finding suggests that vectors of dispersal for Paracoccidioides species are slow, leading to detectable regional diversification. Moreover, it may indicate a founder effect, the species being the most recently emerged taxon in Paracoccidioides, like the patterns found in Fonsecaea species (Najafzadeh et al. 2011). In this scenario, cases reported outside these areas may be regarded as imported cases. Contrasting to the above species, P. brasiliensis s. str. is by far the most diverse taxon in our dataset. Coding and non-coding nuclear markers also support the reciprocal monophyly in members of the P. brasiliensis complex (Turissini et al. 2017). These observations match their close arrangement in the PCAs and MDSs plots. MSTs and Neighbor-Net also capture phylogenetic proximity, an association further supported by our Kohonen maps (SOMs).
Currently, P. lutzii is described as a new biological species (Teixeira et al. 2014c), mainly due to the geographic, antigenic, and genetic differences when compared to the cryptic species of the P. brasiliensis complex (S1, PS2, PS3, and PS4) (Rodrigues et al. 2020b). Studies of evolutionary history suggest that P. lutzii diverged from P. brasiliensis around 22.5 million years ago (Muñoz et al. 2016); however, divergence times between Paracoccidioides species pairs range between 0.03 and 33 million years (Teixeira et al. 2020). This genetic distance between P. lutzii and the four members of the P. brasiliensis complex observed in molecular phylogeny studies was also reflected in our AFLP dendrograms and Neighbor-Net analysis.
The results of our study indicate no or very limited genetic introgression between P. brasiliensis complex and P. lutzii in South America. The assessment of the genetic structure based on two sets of AFLP markers indicate the coexistence of two genetic clusters with no or minimal admixture. This agrees with the results observed by Teixeira et al. which suggests that there is a signature of introgression in only one species pair out of ten possible pairs in Paracoccidioides (Teixeira et al. 2020). This scenario was confirmed using whole-genome sequencing and structure analysis (Muñoz et al. 2016). Further confirmation for this consideration is given by: (1) genetic diversity criterion assessed for the studied Paracoccidioides populations. Although they are not measures of genetic variation, they may indicate a genetic distinctiveness between the P. brasiliensis complex and P. lutzii. Indeed, PCA, MSTs, and SOMs performed for P. brasiliensis complex and P. lutzii combined clearly indicated different genetic clusters; (2) significant genetic differentiation (PhiPT) between P. brasiliensis complex and P. lutzii; and (3) slight genetic differentiation among isolates embedded in the P. brasiliensis complex in Neighbor-Net analysis.
We found high diversity in the P. brasiliensis s. str., suggesting that this lineage has high fitness favouring its dispersion, allowing the survival and adaptation to varied geographic conditions throughout Latin America. Likewise, the epicentre of occurrence for P. lutzii lies in the Mato Grosso state, an area characterized by the biogeographic formations of the Cerrado savannas, Pantanal, and the Amazon rainforest (Simoes et al. 2020). These biomes may have contributed to the geographic isolation and population structure in Paracoccidioides. Currently, with less than 50 % of the native vegetation cover remaining, the deforestation of the Cerrado surpass those in Amazonia (Grecchi et al. 2014, Beuchle et al. 2015, Espírito-Santo et al. 2016), and along with the occupation of the Cerrado lands for mechanized agricultural production may lead to the emergence and expansion of the area of occurrence of P. lutzii. Indeed, Paracoccidioides species propagules inhabit a complex environment in the soil with several amoeboid predators that can impose selective pressure, selecting for virulence traits (Albuquerque et al. 2019). A hypothesis has been raised in recent years whereby biodiversity loss may increase pathogen transmission and disease incidence, especially if it reduces predation and competition on reservoir hosts, thereby increasing their density (Keesing et al. 2010).
Our data shows that Paracoccidioides is a heterothallic fungus with a single mating-type locus that produces two alleles, MAT1-1 and MAT1-2, in agreement with a previous report (Torres et al. 2010). The initial stages of a sexual cycle in Paracoccidioides have been observed under laboratory conditions (Torres et al. 2010, Teixeira et al. 2013), and along with the recombination events reported in genomic studies (Muñoz et al. 2016) could support the hypothesis of a sexual cycle leading to diversification in these pathogens (Teixeira et al. 2013). After evolutionary divergence, genetic hybridization may be mainly observed when (i) the ranges of closely related species overlap, or (ii) one species is uncommon, and individuals have to find mates from a closely related species. In the first scenario, this can lead to two species being genetically more related when in parapatry or sympatry than in regions where they are in allopatry (Palme et al. 2004, Behm et al. 2010, McKinnon et al. 2010). On the other hand, asymmetric introgression may occur when one species exists at a low density (Choleva et al. 2014). Unbalanced gene flow may also be affected by sex-biased dispersal or philopatry. Therefore, it is tempting to hypothesize that this phenomenon could orchestrate genetic hybridization among members of the P. brasiliensis complex, as our results reveal a mixed-mode of reproduction in Paracoccidioides that occurs in sympatry. A mating-type idiomorph-biased distribution was not found to be a significant feature in P. brasiliensis s. str., P. venezuelensis, and P. lutzii, but in P. americana and P. restrepiensis. Skewed MAT loci distribution could result from the scarcity of sexual reproduction, strong selection for pleiotropic effects of a mating-type allele (Nieuwenhuis & James 2016), or even a phenomenon of small populations (Valero et al. 2018). This paradoxical reproduction system has been observed in Sporothrix (Teixeira et al. 2015, de Carvalho et al. 2021b), Histoplasma (Rodrigues et al. 2020a), and Cryptococcus (Nielsen et al. 2005), with species prevalently clonal along with recombinant molecular siblings coexisting in the same geographical range.
Geographical trends observed for P. brasiliensis s. str. revealed a widely distributed species throughout Latin American, present in Argentina, Brazil, Bolivia, Guadeloupe Island, Paraguay, Peru, Uruguay, and Venezuela. Our results corroborate the distribution reported previously by other authors (Matute et al. 2006, Teixeira et al. 2009, Theodoro et al. 2012), including Bolivia and Guadeloupe Island areas for lineage S1. P. americana geographical distribution agrees to the countries described in the literature for this genetic group, i.e., Argentina, Brazil, Uruguay, and Venezuela (Theodoro et al. 2012, Roberto et al. 2016).
Initially, it was thought that P. restrepiensis was restricted to Colombia, but this phylogenetic species has already been found in Brazil and Venezuela (Roberto et al. 2016, Cocio et al. 2020a, Mattos et al. 2021). Although most isolates originated from Colombia, we found a single isolate occurring in Uruguay. P. venezuelensis was the last group recognized in the P. brasiliensis complex, and until recently, it was thought to be exclusive to Venezuela (Teixeira et al. 2014b, Turissini et al. 2017). However, phylogenetic analysis detected one strain from São Paulo, Brazil, characterizing a new location for this species.
Finally, we confirm that P. lutzii has its epicentre in Central-west Brazil with an overwhelming occurrence in Mato Grosso state (Gegembauer et al. 2014, Hahn et al. 2014, Teixeira et al. 2014b, Hahn et al. 2019, Rodrigues et al. 2020b). The distribution patterns in Paracoccidioides species have a notable impact on the serological diagnosis of PCM (Rodrigues et al. 2020b). Therefore, our data draws attention to the urgent need to expand the offer of serological diagnostic tests using antigenic preparations from P. lutzii (Gegembauer et al. 2014, Queiroz Junior et al. 2014, Maifrede et al. 2021) or the availability of PCR assays for the detection of P. lutzii DNA (Pinheiro et al. 2021), which is occurring in areas beyond the known endemic range in Brazil.
Conclusion
Our study illustrates the need to improve genetic surveillance in endemic areas for Paracoccidioides species to ensure that the results of molecular epidemiological studies are accurate. AFLP analysis identifies P. brasiliensis s. str. and P. lutzii as the most diverse species in the genus. In contrast, markedly low genetic diversity was noted for P. americana, P. restrepiensis, and P. venezuelensis. This straightforward typing method will enable the cost-effective analysis of more Paracoccidioides isolates to improve our understanding of the eco-epidemiology trends in PCM infections, help progress toward a consensus taxonomy, assess species boundaries, and explore the significance of genetic diversity in Paracoccidioides species in the clinical scenario.
Acknowledgments
The authors acknowledge the financial support granted by the São Paulo Research Foundation (FAPESP 2017/27265-5 and FAPESP 2018/21460-3), National Council for Scientific and Technological Development (CNPq 429594/2018-6), and Coordination for the Improvement of Higher Education Personnel (CAPES 88887.177846/2018-00). TNR was a fellow of FAPESP (2013/05405-9). AMR is a CNPq Research Productivity Fellow (CNPq 304902/2020-9). The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript. MAB was funded by the Wellcome Trust (#206194). MCF was funded by the UK Medical Research Council and Wellcome Trust and is a Fellow in the CIFAR ‘Fungal Kingdoms’ program. For the purpose of Open Access, the author has applied a CC-BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.simyco.2021.100131.
Contributor Information
Z.P. de Camargo, Email: zpcamargo1@gmail.com.
A.M. Rodrigues, Email: amrodrigues.amr@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Albuquerque P., Nicola A.M., Magnabosco D.A.G., et al. A hidden battle in the dirt: Soil amoebae interactions with Paracoccidioides spp. PLoS Neglected Tropical Diseases. 2019;13 doi: 10.1371/journal.pntd.0007742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida F.A., Neves F.F., Mora D.J., et al. Paracoccidioidomycosis in Brazilian patients with and without human immunodeficiency virus infection. American Journal of Tropical Medicine and Hygiene. 2017;96:368–372. doi: 10.4269/ajtmh.16-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida Fd. Estudos comparativos do granuloma coccidióidico nos Estados Unidos e no Brasil. Novo gênero para o parasito brasileiro. Anais da Faculdade de Medicina da Universidade de São Paulo. 1930;5:125–141. [Google Scholar]
- Altman D.G. Chapman and Hall; London: 1991. Practical statistics for medical research; p. 624. [Google Scholar]
- Arnaud-Haond S., Alberto F., Teixeira S., et al. Assessing genetic diversity in clonal organisms: Low diversity or low resolution? Combining power and cost efficiency in selecting markers. Journal of Heredity. 2005;96:434–440. doi: 10.1093/jhered/esi043. [DOI] [PubMed] [Google Scholar]
- Babicki S., Arndt D., Marcu A., et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Research. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm J.E., Ives A.R., Boughman J.W. Breakdown in postmating isolation and the collapse of a species pair through hybridization. The American Naturalist. 2010;175:11–26. doi: 10.1086/648559. [DOI] [PubMed] [Google Scholar]
- Beuchle R., Grecchi R.C., Shimabukuro Y.E., et al. Land cover changes in the Brazilian Cerrado and Caatinga biomes from 1990 to 2010 based on a systematic remote sensing sampling approach. Applied Geography. 2015;58:116–127. [Google Scholar]
- Bocca A.L., Amaral A.C., Teixeira M.M., et al. Paracoccidioidomycosis: eco-epidemiology, taxonomy and clinical and therapeutic issues. Future Microbiology. 2013;8:1177–1191. doi: 10.2217/fmb.13.68. [DOI] [PubMed] [Google Scholar]
- Borst A., Theelen B., Reinders E., et al. Use of amplified fragment length polymorphism analysis to identify medically important Candida spp., including C. dubliniensis. Journal of Clinical Microbiology. 2003;41:1357–1362. doi: 10.1128/JCM.41.4.1357-1362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., White R.L., Skolnick M., et al. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Brummer E., Castaneda E., Restrepo A. Paracoccidioidomycosis: an update. Clinical Microbiology Reviews. 1993;6:89–117. doi: 10.1128/cmr.6.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D., Moulton V. Neighbor-Net: An Agglomerative Method for the Construction of Phylogenetic Networks. Molecular Biology and Evolution. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Carrero L.L., Nino-Vega G., Teixeira M.M., et al. New Paracoccidioides brasiliensis isolate reveals unexpected genomic variability in this human pathogen. Fungal Genetics and Biology. 2008;45:605–612. doi: 10.1016/j.fgb.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Choleva L., Musilova Z., Kohoutova-Sediva A., et al. Distinguishing between incomplete lineage sorting and genomic introgressions: Complete fixation of allospecific mitochondrial DNA in a sexually reproducing fish (Cobitis; Teleostei), despite clonal reproduction of hybrids. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0080641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocio T.A., Nascimento E., Kress M.R., et al. Characterization of a Paracoccidioides spp. strain from southeastern Brazil genotyped as Paracoccidioides restrepiensis (PS3) and review of this phylogenetic species. Genetics and Molecular Biology. 2020;43 doi: 10.1590/1678-4685-GMB-2019-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocio T.A., Nascimento E., von Zeska Kress M.R., et al. Phylogenetic species of Paracoccidioides spp. isolated from clinical and environmental samples in a hyperendemic area of paracoccidioidomycosis in Southeastern Brazil. Journal of Fungi. 2020;6:132. doi: 10.3390/jof6030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida S.M., Roza T.H., Salvador G.L.O., et al. Autopsy and biopsy study of paracoccidioidomycosis and neuroparacoccidioidomycosis with and without HIV co-infection. Mycoses. 2018;61:237–244. doi: 10.1111/myc.12737. [DOI] [PubMed] [Google Scholar]
- de Carvalho J.A., Beale M.A., Hagen F., et al. Trends in the molecular epidemiology and population genetics of emerging Sporothrix species. Studies in Mycology. 2021;100:100129. doi: 10.1016/j.simyco.2021.100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho J.A., Hagen F., Fisher M.C., et al. Genome-wide mapping using new AFLP markers to explore intraspecific variation among pathogenic Sporothrix species. PLoS Neglected Tropical Diseases. 2020;14 doi: 10.1371/journal.pntd.0008330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho J.A., Pinheiro B.G., Hagen F., et al. A new duplex PCR assay for the rapid screening of mating-type idiomorphs of pathogenic Sporothrix species. Fungal Biology. 2021;125:834–843. doi: 10.1016/j.funbio.2021.05.005. [DOI] [PubMed] [Google Scholar]
- de Groot T., Puts Y., Berrio I., et al. Development of Candida auris short tandem repeat typing and its application to a global collection of isolates. MBio. 2020;11 doi: 10.1128/mBio.02971-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Macedo P.M., Teixeira MdM., Barker B.M., et al. Clinical features and genetic background of the sympatric species Paracoccidioides brasiliensis and Paracoccidioides americana. PLoS Neglected Tropical Diseases. 2019;13 doi: 10.1371/journal.pntd.0007309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vienne D.M., Giraud T., Martin O.C. A congruence index for testing topological similarity between trees. Bioinformatics. 2007;23:3119–3124. doi: 10.1093/bioinformatics/btm500. [DOI] [PubMed] [Google Scholar]
- Del Negro G., Garcia N., Rodrigues E., et al. Note on Paracoccidioides tenuis Moore 1938 a possible synonym for Paracoccidioides brasiliensis. Revista Iberoamericana de Micología. 1993;10:69–71. [Google Scholar]
- Desjardins C.A., Champion M.D., Holder J.W., et al. Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis. PLoS Genetics. 2011;7 doi: 10.1371/journal.pgen.1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice L.R. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- Duarte-Escalante E., Zúñiga G., Frías-De-León M.G., et al. AFLP analysis reveals high genetic diversity but low population structure in Coccidioides posadasii isolates from Mexico and Argentina. BMC Infectious Diseases. 2013;13:411. doi: 10.1186/1471-2334-13-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D.A., vonHoldt B.M. STRUCTURE harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Espírito-Santo M.M., Leite M.E., Silva J.O., et al. Understanding patterns of land-cover change in the Brazilian Cerrado from 2000 to 2015. Philosophical Transactions of the Royal Society B: Biological Sciences. 2016;371:20150435. doi: 10.1098/rstb.2015.0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Smouse P.E., Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava-Netto C. Contribuição para o estudo imunológico da blastomicose de Lutz (blastomicose sul-americana) Revista do Instituto Adolfo Lutz. 1961;21:99–194. [Google Scholar]
- Fava-Netto C., Vegas V.S., Sciannamea I.M., et al. The polysaccharidic antigen from Paracoccidioides brasiliensis. Study of the time of cultivation necessary for the preparation of the antigen. Revista do Instituto de Medicina Tropical de São Paulo. 1969;11:177–181. [PubMed] [Google Scholar]
- Feitosa L.S., Cisalpino P.S., dos Santos M.R., et al. Chromosomal polymorphism, syntenic relationships, and ploidy in the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genetics and Biology. 2003;39:60–69. doi: 10.1016/s1087-1845(03)00003-3. [DOI] [PubMed] [Google Scholar]
- Felix B., Roussel S., Pot B. In: Methods in Molecular Biology. Jordan K., Dalmasso M., editors. Humana Press; New York: 2015. Harmonization of PFGE profile analysis by using bioinformatics tools: example of the Listeria monocytogenes European Union Reference Laboratory network; pp. 9–28. [DOI] [PubMed] [Google Scholar]
- Ferguson R., Upton M.F. The isolation of Paracoccidioides brasiliensis from a case of South American blastomycosis. Journal of Bacteriology. 1947;53:376. [PubMed] [Google Scholar]
- Franco M., Montenegro M.R., Mendes R.P., et al. Paracoccidioidomycosis: a recently proposed classification of its clinical forms. Revista da Sociedade Brasileira de Medicina Tropical. 1987;20:129–132. doi: 10.1590/s0037-86821987000200012. [DOI] [PubMed] [Google Scholar]
- Garcia N.M., Del Negro G.M.B., Heins-Vaccari E.M., et al. Paracoccidioides brasiliensis, a new strain isolated from a fecal matter of a penguin (Pygoscelis adeliae) Revista do Instituto de Medicina Tropical de São Paulo. 1993;35:227–235. doi: 10.1590/s0036-46651993000300003. [DOI] [PubMed] [Google Scholar]
- Gegembauer G., Araujo L.M., Pereira E.F., et al. Serology of paracoccidioidomycosis due to Paracoccidioides lutzii. PLoS Neglected Tropical Diseases. 2014;8 doi: 10.1371/journal.pntd.0002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gezuele E. In: Proceedings IV Encuentro Internacional sobre Paracoccidioidomicosis. San-Blas G., editor. Instituto Venezolano de Investigaciones Científicas (IVIC); Caracas, Venezuela: 1989. Aislamiento de Paracoccidioides sp. de heces de pinguino de la Antartida. Caracas, Venezuela; pp. 10–14. [Google Scholar]
- Giacomazzi J., Baethgen L., Carneiro L.C., et al. The burden of serious human fungal infections in Brazil. Mycoses. 2016;59:145–150. doi: 10.1111/myc.12427. [DOI] [PubMed] [Google Scholar]
- Gonzalez Ochoa A., Esquivel E. Primer caso de granuloma paracoccidioide (blastomicosis sudamericana) en México. Revista Médica del Hospital General. 1950;13:159–167. [Google Scholar]
- Grecchi R.C., Gwyn Q.H.J., Bénié G.B., et al. Land use and land cover changes in the Brazilian Cerrado: A multidisciplinary approach to assess the impacts of agricultural expansion. Applied Geography. 2014;55:300–312. [Google Scholar]
- Hagen F., Khayhan K., Theelen B., et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genetics and Biology. 2015;78:16–48. doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Hahn R.C., Rodrigues A.M., Della Terra P.P., et al. Clinical and epidemiological features of paracoccidioidomycosis due to Paracoccidioides lutzii. PLoS Neglected Tropical Diseases. 2019;13 doi: 10.1371/journal.pntd.0007437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn R.C., Rodrigues A.M., Fontes C.J., et al. Fatal Fungemia due to Paracoccidioides lutzii. The American Journal of Tropical Medicine and Hygiene. 2014;91:394–398. doi: 10.4269/ajtmh.13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming R.W. Error detecting and error correcting codes. The Bell System Technical Journal. 1950;29:147–160. [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Huson D.H., Kloepper T.H. Computing recombination networks from binary sequences. Bioinformatics. 2005;21:ii159–ii165. doi: 10.1093/bioinformatics/bti1126. [DOI] [PubMed] [Google Scholar]
- Jakobsson M., Rosenberg N.A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kasuga T., White T.J., Taylor J.W. Estimation of nucleotide substitution rates in Eurotiomycete fungi. Molecular Biology and Evolution. 2002;19:2318–2324. doi: 10.1093/oxfordjournals.molbev.a004056. [DOI] [PubMed] [Google Scholar]
- Keesing F., Belden L.K., Daszak P., et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen T. Springer Series in Information Sciences. 2001. Self-Organizing Maps; p. 502. [Google Scholar]
- Liu B.H. CRC press; Boca Raton: 1998. Statistical genomics: linkage, mapping, and QTL analysis. [Google Scholar]
- Lockhart S.R., Etienne K.A., Vallabhaneni S., et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clinical Infectious Diseases. 2017;64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücking R., Aime M.C., Robbertse B., et al. Fungal taxonomy and sequence-based nomenclature. Nature Microbiology. 2021;6:540–548. doi: 10.1038/s41564-021-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz A. Uma micose pseudococídica localizada na boca e observada no Brasil: contribuição ao conhecimento das hifoblastomicoses americanas. Brasil Médico. 1908;22:121–124. [Google Scholar]
- Lynch M., Milligan B.G. Analysis of population genetic structure with RAPD markers. Molecular Ecology. 1994;3:91–99. doi: 10.1111/j.1365-294x.1994.tb00109.x. [DOI] [PubMed] [Google Scholar]
- Macedo P.M., Almeida-Paes R., Almeida M.A., et al. Paracoccidioidomycosis due to Paracoccidioides brasiliensis S1 plus HIV co-infection. Memórias do Instituto Oswaldo Cruz. 2018;113:167–172. doi: 10.1590/0074-02760170310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maifrede S.B., Kruschewsky W.L.L., Patrício S.A., et al. Screening paracoccidioidomycosis by double immunodiffusion test in a referred diagnostic center in Brazilian southeastern: an accessible tool. Infection. 2021 doi: 10.1007/s15010-021-01704-8. [DOI] [PubMed] [Google Scholar]
- Mattos K., Cocio T.A., Chaves E.G.A., et al. An update on the occurrence of Paracoccidioides species in the Midwest region, Brazil: Molecular epidemiology, clinical aspects and serological profile of patients from Mato Grosso do Sul State. PLoS Neglected Tropical Diseases. 2021;15 doi: 10.1371/journal.pntd.0009317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute D.R., McEwen J.G., Puccia R., et al. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Molecular Biology and Evolution. 2006;23:65–73. doi: 10.1093/molbev/msj008. [DOI] [PubMed] [Google Scholar]
- McDonald B.A., Linde C. Pathogen population genetics, evolutionary potential, and durable resistance. Annual Review of Phytopathology. 2002;40:349–379. doi: 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- McKinnon G.E., Smith J.J., Potts B.M. Recurrent nuclear DNA introgression accompanies chloroplast DNA exchange between two eucalypt species. Molecular Ecology. 2010;19:1367–1380. doi: 10.1111/j.1365-294X.2010.04579.x. [DOI] [PubMed] [Google Scholar]
- Moore M. A new species of the Paracoccidioides Almeida (1930): P. cerebriformis Moore, (1935) Revista de Biologia e Higiene. 1935;6:148–162. [Google Scholar]
- Moore M. Blastomycosis, coccidioidal granuloma and paracoccidioidal granuloma. Comparative study of North American, South American and European organisms and clinical types. Archives of Dermatology and Syphilology. 1938;38:163–190. [Google Scholar]
- Morais F.V., Barros T.F., Fukada M.K., et al. Polymorphism in the gene coding for the immunodominant antigen gp43 from the pathogenic fungus Paracoccidioides brasiliensis. Journal of Clinical Microbiology. 2000;38:3960. doi: 10.1128/jcm.38.11.3960-3966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J.F., Farrer R.A., Desjardins C.A., et al. Genome diversity, recombination, and virulence across the major lineages of Paracoccidioides. mSphere. 2016;1:e00213–e00216. doi: 10.1128/mSphere.00213-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J.F., Gallo J.E., Misas E., et al. Genome update of the dimorphic human pathogenic fungi causing paracoccidioidomycosis. PLoS Neglected Tropical Diseases. 2014;8:e3348. doi: 10.1371/journal.pntd.0003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafzadeh M.J., Sun J., Vicente V.A., et al. Molecular epidemiology of Fonsecaea species. Emerging Infectious Diseases. 2011;17:464–469. doi: 10.3201/1703.100555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath V.S., Senthil M., Hegde V.M., et al. Genetic diversity of Phytophthora colocasiae isolates in India based on AFLP analysis. 3 Biotech. 2013;3:297–305. doi: 10.1007/s13205-012-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery A.F., de Camargo Z.P., Rodrigues A.M., et al. Paracoccidioidomycosis due to P. lutzii: The importance of neutrophil/lymphocyte ratio in the symptomatic and asymptomatic phases in severe cases. Mycoses. 2021;64:874–881. doi: 10.1111/myc.13282. [DOI] [PubMed] [Google Scholar]
- Nery A.F., de Camargo Z.P., Rodrigues A.M., et al. Puzzling paracoccidioidomycosis: Factors associated with the severity of Paracoccidioides lutzii infections. International Journal of Infectious Diseases. 2021;107:284–290. doi: 10.1016/j.ijid.2021.05.002. [DOI] [PubMed] [Google Scholar]
- Nielsen K., Marra R.E., Hagen F., et al. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics. 2005;171:975–983. doi: 10.1534/genetics.105.045039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis B.P.S., James T.Y. The frequency of sex in fungi. Philosophical Transactions of the Royal Society B: Biological Sciences. 2016;371:20150540. doi: 10.1098/rstb.2015.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nino F.L. Seven new observations of paracoccidioidal granuloma in Argentina. Boletín Universidad de Buenos Aires. 1950;26:272–305. [PubMed] [Google Scholar]
- Palme A.E., Su Q., Palsson S., et al. Extensive sharing of chloroplast haplotypes among European birches indicates hybridization among Betula pendula, B. pubescens and B. nana. Molecular Ecology. 2004;13:167–178. doi: 10.1046/j.1365-294x.2003.02034.x. [DOI] [PubMed] [Google Scholar]
- Paris M., Bonnes B., Ficetola G.F., et al. Amplified fragment length homoplasy: in silico analysis for model and non-model species. BMC Genomics. 2010;11:287. doi: 10.1186/1471-2164-11-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R., Smouse P.E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R.O.D., Smouse P.E. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira E.F., Gegembauer G., Chang M.R., et al. Comparison of clinico-epidemiological and radiological features in paracoccidioidomycosis patients regarding serological classification using antigens from Paracoccidioides brasiliensis complex and Paracoccidioides lutzii. PLoS Neglected Tropical Diseases. 2020;14 doi: 10.1371/journal.pntd.0008485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro B.G., Hahn R.C., Camargo Z.P., et al. Molecular tools for detection and identification of Paracoccidioides species: Current status and future perspectives. Journal of Fungi. 2020;6:293. doi: 10.3390/jof6040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro B.G., Pôssa A.P., Della Terra P.P.D., et al. A new duplex PCR-assay for the detection and identification of Paracoccidioides species. Journal of Fungi. 2021;7:169. doi: 10.3390/jof7030169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell W., Morgante M., Andre C., et al. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding. 1996;2:225–238. [Google Scholar]
- Prevost A., Wilkinson M.J. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theoretical and Applied Genetics. 1999;98:107–112. [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz Junior L.P., de Camargo Z.P., Tadano T., et al. Serological and antigenic profiles of clinical isolates of Paracoccidioides spp. from Central Western Brazil. Mycoses. 2014;57:466–472. doi: 10.1111/myc.12183. [DOI] [PubMed] [Google Scholar]
- Restrepo A., Benard G., de Castro C.C., et al. Pulmonary paracoccidioidomycosis. Seminars in Respiratory and Critical Care Medicine. 2008;29:182–197. doi: 10.1055/s-2008-1063857. [DOI] [PubMed] [Google Scholar]
- Roberto T.N., Rodrigues A.M., Hahn R.C., et al. Identifying Paracoccidioides phylogenetic species by PCR-RFLP of the alpha-tubulin gene. Medical Mycology. 2016;54:240–247. doi: 10.1093/mmy/myv083. [DOI] [PubMed] [Google Scholar]
- Rodrigues A.M., Beale M.A., Hagen F., et al. The global epidemiology of emerging Histoplasma species in recent years. Studies in Mycology. 2020;97:100095. doi: 10.1016/j.simyco.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.M., de Hoog G.S., Zhang Y., et al. Emerging sporotrichosis is driven by clonal and recombinant Sporothrix species. Emerging Microbes & Infections. 2014;3:e32. doi: 10.1038/emi.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.M., Kubitschek-Barreira P.H., Pinheiro B.G., et al. Immunoproteomic analysis reveals novel candidate antigens for the diagnosis of paracoccidioidomycosis due to Paracoccidioides lutzii. Journal of Fungi. 2020;6:357. doi: 10.3390/jof6040357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombauts S., Van De Peer Y., Rouze P. AFLPinSilico, simulating AFLP fingerprints. Bioinformatics. 2003;19:776–777. doi: 10.1093/bioinformatics/btg090. [DOI] [PubMed] [Google Scholar]
- Salgado-Salazar C., Jones L.R., Restrepo Á., et al. The human fungal pathogen Paracoccidioides brasiliensis (Onygenales: Ajellomycetaceae) is a complex of two species: phylogenetic evidence from five mitochondrial markers. Cladistics. 2010;26:613–624. doi: 10.1111/j.1096-0031.2010.00307.x. [DOI] [PubMed] [Google Scholar]
- Salipante S.J., Hall B.G. Inadequacies of minimum spanning trees in molecular epidemiology. Journal of Clinical Microbiology. 2011;49:3568–3575. doi: 10.1128/JCM.00919-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelenz S., Hagen F., Rhodes J.L., et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrobial Resistance & Infection Control. 2016;5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober P., Boer C., Schwarte L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesthesia & Analgesia. 2018;126:1763–1768. doi: 10.1213/ANE.0000000000002864. [DOI] [PubMed] [Google Scholar]
- Shannon C.E. A mathematical theory of communication. The Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- Shikanai-Yasuda M.A., Mendes R.P., Colombo A.L., et al. Brazilian guidelines for the clinical management of paracoccidioidomycosis. Revista da Sociedade Brasileira de Medicina Tropical. 2017;50:715–740. doi: 10.1590/0037-8682-0230-2017. [DOI] [PubMed] [Google Scholar]
- Simoes R., Picoli M.C.A., Camara G., et al. Land use and cover maps for Mato Grosso State in Brazil from 2001 to 2017. Scientific Data. 2020;7:34. doi: 10.1038/s41597-020-0371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E.H. Measurement of Diversity. Nature. 1949;163 688–688. [Google Scholar]
- Teixeira H., Rodríguez-Echeverría S., Nabais C. Genetic diversity and differentiation of Juniperus thurifera in Spain and Morocco as determined by SSR. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0088996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MdM., Cattana M.E., Matute D.R., et al. Genomic diversity of the human pathogen Paracoccidioides across the South American continent. Fungal Genetics and Biology. 2020;140:103395. doi: 10.1016/j.fgb.2020.103395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MdM., Rodrigues A.M., Tsui C.K.M., et al. Asexual propagation of a virulent clone complex in human and feline outbreak of sporotrichosis. Eukaryotic Cell. 2015;14:158–169. doi: 10.1128/EC.00153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MdM., Theodoro R.C., Derengowski LdS., et al. Molecular and morphological data support the existence of a sexual cycle in species of the genus Paracoccidioides. Eukaryotic Cell. 2013;12:380–389. doi: 10.1128/EC.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.M., Theodoro R.C., de Carvalho M.J.A., et al. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Molecular Phylogenetics and Evolution. 2009;52:273–283. doi: 10.1016/j.ympev.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Teixeira M.M., Theodoro R.C., Nino-Vega G., et al. Paracoccidioides species complex: Ecology, phylogeny, sexual reproduction, and virulence. PLoS Pathogens. 2014;10 doi: 10.1371/journal.ppat.1004397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.M., Theodoro R.C., Oliveira F.F., et al. Paracoccidioides lutziisp. nov.: biological and clinical implications. Medical Mycology. 2014;52:19–28. doi: 10.3109/13693786.2013.794311. [DOI] [PubMed] [Google Scholar]
- Tessier C., David J., This P., et al. Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theoretical and Applied Genetics. 1999;98:171–177. [Google Scholar]
- The R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2014. R: A Language and Environment for Statistical Computing, 2014.http://wwwR-projectorg/ [Google Scholar]
- Theodoro R.C., Bagagli E., Oliveira C. Phylogenetic analysis of PRP8 intein in Paracoccidioides brasiliensis species complex. Fungal Genetics and Biology. 2008;45:1284–1291. doi: 10.1016/j.fgb.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Theodoro R.C., Teixeira MdM., Felipe M.S.S., et al. Genus Paracoccidioides: Species recognition and biogeographic aspects. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0037694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres I., García A.M., Hernández O., et al. Presence and expression of the mating type locus in Paracoccidioides brasiliensis isolates. Fungal Genetics and Biology. 2010;47:373–380. doi: 10.1016/j.fgb.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Turissini D.A., Gomez O.M., Teixeira M.M., et al. Species boundaries in the human pathogen Paracoccidioides. Fungal Genetics and Biology. 2017;106:9–25. doi: 10.1016/j.fgb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero C., Gago S., Monteiro M.C., et al. African histoplasmosis: new clinical and microbiological insights. Medical Mycology. 2018;56:51–59. doi: 10.1093/mmy/myx020. [DOI] [PubMed] [Google Scholar]
- Varshney R.K., Chabane K., Hendre P.S., et al. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Science. 2007;173:638–649. [Google Scholar]
- Vauterin L., Vauterin P. In: Molecular Identification, Systematics, and Population Structure of Prokaryotes. Stackebrandt E., editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 2006. Integrated databasing and analysis; pp. 141–217. [Google Scholar]
- Vos P., Hogers R., Bleeker M., et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warris A., Klaassen C.H., Meis J.F., et al. Molecular epidemiology of Aspergillus fumigatus isolates recovered from water, air, and patients shows two clusters of genetically distinct strains. Journal of Clinical Microbiology. 2003;41:4101–4106. doi: 10.1128/JCM.41.9.4101-4106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S., et al. In: PCR Protocols: A Guide to Methods and Applications. Innis M., Gelfand D., Shinsky J., White T., editors. Academic Press; New York: 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; pp. 315–322. [Google Scholar]
- Wickham H. Use R! Springer; Cham: XVI: 2016. ggplot2: Elegant Graphics for Data Analysis; p. 260. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.