Abstract
Background and Aims.
Brain imaging-derived structural correlates of alcohol involvement have largely been speculated to arise as a consequence of alcohol exposure. However, they may also reflect predispositional risk.
Methods.
In substance naïve children of European ancestry who completed the baseline session of the Adolescent Brain Cognitive Development (ABCD) Study (n=3,013), mixed-effects models estimated whether polygenic risk scores (PRS) for Problematic Alcohol Use (PAU-PRS) and Drinks Per Week (DPW-PRS) are associated with magnetic resonance imaging-derived brain structure phenotypes (i.e., total and regional: cortical thickness, surface area and volume; subcortical volume; white matter volume, fractional anisotropy, mean diffusivity). Follow-up analyses evaluated whether any identified regions were also associated with polygenic risk among substance naïve children of African ancestry (n=898).
Results:
After adjustment for multiple testing correction, polygenic risk for problematic alcohol use was associated with lower volume of the left frontal pole and greater cortical thickness of the right supramarginal gyrus (|βs|>0.009; ps<0.001; psfdr<0.046; r2s < 0.004). PAU PRS and DPW PRS showed nominally significant associations with a host of other regional brain structure phenotypes (e.g., insula surface area and volume). None of these regions showed any, even nominal association among children of African ancestry.
Conclusions:
Genomic liability to alcohol involvement may manifest as variability in brain structure during middle childhood prior to alcohol use initiation. Broadly, alcohol-related variability in brain morphometry may partially reflect predisposing genomic influence. Larger discovery GWASs and target samples of diverse ancestries are needed to determine whether observed associations may generalize across ancestral origins.
Introduction
Excessive alcohol use is an escalating international health problem that accounts for over 5% of global deaths and disease burden (1). Alcohol use and use disorders have been reliably associated with magnetic resonance imaging (MRI)-derived brain structure phenotypes, particularly among regions and pathways that feature prominently in executive function, incentive salience, and negative emotionality (2,3). For instance, the largest mega-analysis of Alcohol Use Disorder in adults (n cases = 898, n controls = 292) found lower volume and cortical thickness of subcortical (n=11; e.g., hippocampus, amygdala, nucleus accumbens, putamen) and cortical regions (n=27; e.g., insula, superior frontal gyrus; orbitofrontal cortex), respectively (2). Consistent with evidence from non-human animal models, it is widely speculated that these structural reductions arise as a consequence of chronic alcohol exposure and contribute to the development of alcohol-related comorbidities (e.g., risk taking, Alzheimer’s Disease) (4,5). However, emerging data challenging this common interpretation suggest that these brain structure correlates may, at least partially, reflect genetically conferred predisposing risk factors for alcohol involvement (6,7). For example, studies of substance naïve children of parents with AUD have observed similar associations with reduced gray matter metrics (8). However, these studies have also reported apparently paradoxical findings that non-exposed children of parents with AUD are characterized by increased gray matter structure in the precentral gyrus, the inferior and caudal frontal gyrus, the temporal partial junction, and the interior-temporal gyrus (2,8).
Building upon twin and family studies documenting the moderate heritability of alcohol use and use disorders (30–50%) (9,10), large-scale genome-wide association studies (GWASs) have begun to reliably characterize the polygenic architecture of alcohol involvement (11,12). Polygenic risk scores (PRS) that effectively capture this polygenic liability can be combined with neuroimaging data to investigate whether individual differences in neural phenotypes may be partially attributable to common underlying genomic vulnerability and/or arise following substance exposure, use, and/or problematic use. However, given the ubiquity of lifetime alcohol use, efforts to disentangle such predispositional effects from neurotoxic consequences of chronic exposure have been limited.
Here, among 3,013 substance-naïvea children (age=9.92±0.62 years, 47% Female; 100% genomically-confirmed European ancestry; Supplemental Table 1) who completed the baseline session of the ongoing Adolescent Brain Cognitive DevelopmentSM Study (ABCD Study®; data release 2.0.1), we test whether PRS for alcohol use and problematic use (including alcohol use disorder) are associated with brain structure phenotypes. To this end, we generated PRS using the largest available GWASs of alcohol use (i.e., alcohol drinks/week [DPW-PRS]; training n=537,349) and problematic use (i.e., alcohol use disorder/problem use [PAU-PRS]; training n=352,365) and estimated their association with total and region variability in cortical thickness, cortical surface area, cortical volume, subcortical volume, as well as white matter volume and integrity, among substance naïve children. Due to divergent findings among adults with AUD and children at familial risk for AUD (2,8), we tested all regions with correction for multiple testing. We tested whether any observed associations were present in substance naïve children of African ancestry (n=997), with polygenic risk scores generated using summary statistics generated from individuals of African ancestry (n= 62,447). Finally, we expected that variability in brain structure may indirectly link alcohol involvement PRS to behavioral risk factors (i.e., cognition and externalizing problems) believed to play an etiologic role in the development of substance use disorder according to stage-based theories of addiction (13).
Methods
Participants
The Adolescent Brain and Cognitive Development (ABCD) Study is an ongoing multi-site longitudinal study of child health and development. The ABCD Study is led by the National Institute on Drug Abuse and measured children on a range of cognitive, behavioral, personality, and biological measures. These measures total in the several thousands and so a complete description of the is beyond the scope of this work (see the NIMH data archive: https://nda.nih.gov/data_dictionary.html?source=ABCD%2BRelease%2B3.0&submission=ALL, for more on the ABCD study see: Volkow et al. 2018 (14)). Our total sample at baseline includes 11,875 children, including 2,108 twins and 30 triplets, ages 8.9–11 were recruited from 22b sites across the United States, to complete the ABCD Study baseline assessment. We restricted our sample to participants of genomically-confirmed non-Hispanic European ancestry (n=4,737) who self-reported no exposure to substances and screened negative for substances according to toxicology, leaving a final analytic sample of 3,434 children (mean age =9.92 years, std age = 0.62, 47 % Female, n=3,013 with no missing data). Our African American sample consisted of 997 individuals (9.90 years of age, std age = 0.60, 49% Female) with 898 remaining after exclusion for substance exposure.
Measures
Genotyping, quality control and ancestry estimation
The Rutgers University Cell and DNA repository genotyped saliva samples on the Smokescreen array (15). Genotyped calls were aligned to GRCh37 (hg19), and all individuals self-reporting ancestral origins (i.e., self-reported race) other than European or African American were excluded because these were the only ancestral populations in which GWAS summary statistics are available. Ancestrally homogenous samples were used due to differing genomic LD structure across ancestries and evidence that GWAS-derived summary statistics from one ancestral group cannot be meaningfully applied across ancestries (16,17). While novel techniques are being developed to leverage GWAS summary statistics across ancestries, these currently require availability of GWAS summary statistics from each ancestry. We separated individuals into self-reported ancestries prior to QC and conducted QC separately in the self-reported European and African ancestries subsamples. Following QC, genomic data were then used to exclude genomic ancestral outliers (described below).
The following preprocessing steps were conducted with the Ricopili pipeline (18): Single Nucleotide Polymorphisms (SNPs) with call rates ≥ 0.95 and MAF ≥ 1% were retained. Individuals with high rates of missingness (>5%) and autosomal heterozygosity deviation (FHET) outside of ± 2 SD were removed. After sample QC, SNPs were further filtered to call rate ≥ 0.98 and Hardy-Weinberg p-values > 1E-6 (founders only), which yielded 372,342 SNPs. In order to reconcile mismatches, sex checks were conducted with follow-up. Individuals whose data passed the first phase of QC were then checked for relatedness--both known and cryptic--and Mendelian errors were resolved. Next, using data from unrelated individuals (pi-hat ≤ 0.20) and an LD pruned set of common (MAF>0.05) and non-palindromic SNPs (and excluding MHC and chromosome 8 inversion region), principal components analysis (PCA) was performed in EIGENSTRAT using the European and African 1000 Genomes Project phase 3 data. yielding a sample of 4,737 of European Americans and 1232 African individuals. Due to the sensitivity of the PRS approach to admixture, we took a conservative approach and performed stringent exclusion for ancestral outliers, consistent with the Psychiatric Genomics Consortium’s Ricopili pipeline (13). After selection, a final ancestrally-informative PCA was conducted, and the first 20 PCs were projected from founders to other relatives. Imputation to 1000 Genomes and Haplotype Reference Consortium (HRC) data for Europeans and the Consortium on Asthma among African-ancestry Populations in the Americas (CAAPA) for African Americans was conducted using strictly QCed SNPs. Data were converted to hard-call genotypes using Plink, and only SNPs with imputation r2 scores ≥ 0.3 were used to create polygenic scores.
Polygenic Risk Scores
Polygenic Risk Scores (PRS) were generated using the PRS-cs software package (19). The PRS-cs auto approach calculates PRS by assuming a general distribution of effect sizes across the genome, and then reweighting Single Nucleotide Polymorphisms (SNPs) based on this assumption, their effect size in the original GWAS, and their linkage disequilibrium (LD) patterns from 1000 genomes phase 3 to create weights for every SNP that are then summed for a final score. We chose PRS-cs so that all common variation across the genome could be leveraged given the polygenicity of substance use phenotypes. Our models were trained using two well-powered GWAS: 1) Drinks per week (DPW, original GWAS N=537,349) from the GWAS & Sequencing Consortium of Alcohol and Nicotine use (GSCAN) (20), and 2) a problematic alcohol use (PAU) meta-analysis of GWASs (12)of: A) alcohol use disorder (specifically, alcohol dependence from the Psychiatric Genomics Consortium (11), B) ICD codes for alcohol dependence (ICD-9: 303.* or ICD-10: F10.2*) and alcohol abuse (ICD-9: 305.* or ICD-10: F10.1*) from the Million Veteran Program(12,21), and C) the problem subscale of the Alcohol Use Disorders Identification Test (AUDIT-P) (22) (Original GWAS N=352,391) from the UK Biobank. We chose to use both DPW-PRS and PAU-PRS because of past work that shows genetic separability in alcohol use and use disorder (11).
Brain Structure.
Magnetic resonance imaging (MRI) acquisition and processing have been previously described, and are detailed in the Supplement (23,24). We examined regional volume, thickness and surface area of 34 bilateral cortex region in each hemispheres defined in the Desikan-Killany atlas as well as volumes of 12 bilateral subcortical Freesurfer segmentations (25). Fractional anisotropy and mean diffusivity was estimated for 36 white matter tracks defined by AtlasTrack (23). We also evaluated global estimates of each modality (e.g., total cortical thickness).
Putative Behavioral Risk Factors
Trait-related vulnerability to elevated externalizing problems as well as reduced cognition have been speculated to contribute to vulnerability to stage-based addiction transitions (26). We used the full scale IQ estimation generated from the National Institutes of Health Toolbox Cognitive Battery (27) and externalizing problems as assessed on The Child Behavior Checklist (CBCL) (28) to test whether alcohol involvement PRS and related brain structure correlates are associated with variability in cognition and externalizing.
Statistical Analysis
Associations between alcohol involvement PRS (i.e., PAU-PRS and DPW-PRS) and brain structure phenotypes were estimated using mixed-effects models in the lme4 (29) package in R. Family ID and scanner MRI serial number were included as random effects to account for non-independence of measurement associated with relatedness and scanner/site. We controlled for the first 20 ancestry principal components, mean of each modality (separately when predicting regions of that modality, e.g., mean thickness predicting regional thickness), prenatal exposure to alcohol (i.e., retrospective report of maternal use of alcohol during pregnancy prior to [no/yes] or after [no/yes] maternal knowledge of the pregnancy (30)), age, sex, age by sex, socioeconomic status proxies (i.e., caregiver education and combined household income), genotyping batch as fixed effects in each model. We ran a test of association separately for each brain region within each modality (e.g., cortical thickness) with each PRS. False-discovery rate (FDR) correction within modality (e.g., cortical thickness) was used to adjust for multiple testing. (Gray matter cortex = 34 regions, white matter = 36 tracts, subcortex = 26 regions). Results surviving FDR correction are discussed here; we make all results (significant and non-significant) available in the Supplement Results by region and PRS. Each brain region that was significantly associated with PRS was then tested for association with full-scale IQ, and externalizing problems.
Data Availability Statement
Summary statistics and data are publicly available through various sources. The Million Veterans Project summary statistics were accessed via dbGaP (phs001672.v1.p1) as part of #24806: Neurobiological bases of psychiatric traits. The authors thank Million Veteran Program (MVP) staff, researchers, and volunteers, who have contributed to MVP, and especially participants who previously served their country in the military and now generously agreed to enroll in the study. (See https://www.research.va.gov/mvp/ for more details).
The citation for MVP is Gaziano, J.M. et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016).
This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by the Veterans Administration (VA) Cooperative Studies Program (CSP) award #G002. Data for this study were provided by the Adolescent Brain Cognitive Development (ABCD) study, which was funded by awards U01DA041022, U01DA041025, U01DA041028, U01DA041048, U01DA041089, U01DA041093, U01DA041106, U01DA041117, U01DA041120, U01DA041134, U01DA041148, U01DA041156, U01DA041174, U24DA041123, and U24DA041147 from the NIH and additional federal partners (https://abcdstudy.org/federal-partners.html). All data used here in are publicly available through the GSCAN consortium, dbGAP, and the ABCD consortium.
Results
PAU-PRS was associated with reduced left frontal pole gray matter volume (standardized β= −0.054, r2=0.003, pfdr=0.031; Figure 1; Table 1). PAU-PRS was also associated with greater right supramarginal gyrus cortical thickness (standardized β=0.009, r2=0.001, pfdr=0.045; Figure 1; Table 1). Cognition as well as externalizing problems were not associated with frontal pole gray matter volume or supramarginal gyrus cortical thickness (All p > .21; Supplemental Table 4).
Figure 1.
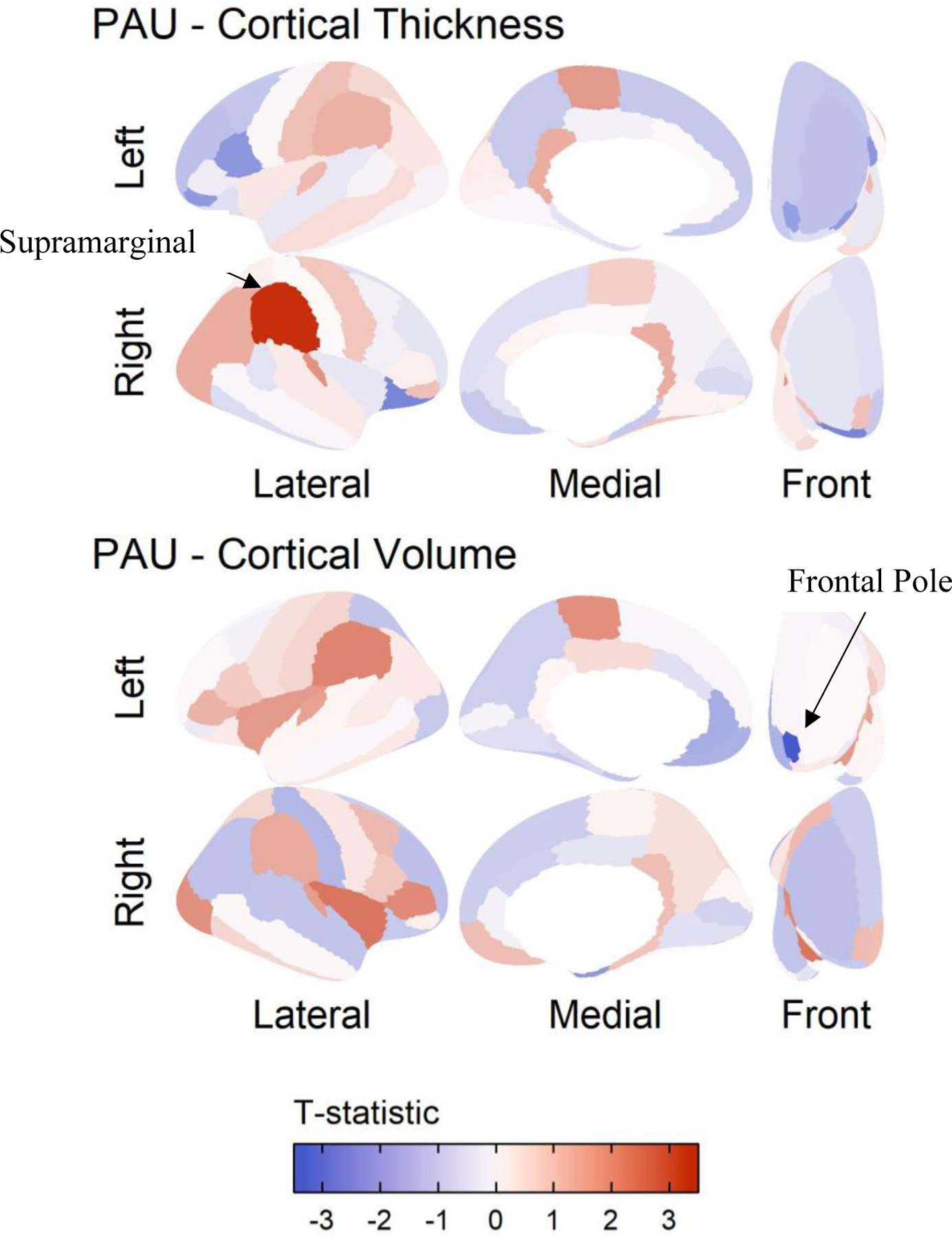
Results of polygenic risk prediction for Problematic Alcohol Use (PAU) plotted on the cortex. The results are shown for those modalities that had significant regions in the Desikan atlas after controlling for mean of the modality, prenatal alcohol exposure, age, sex, age by sex, first 20 principal components, SES, and accounting for family and site as random nested effects. Red indicates areas where higher PAU polygenic risk predicted more of that modality. In the top panel, we focus on supramarginal gyrus cortical thickness prediction by PRS, as this association was significant after multiple testing correction. In the bottom panel, we focus on volume of the temporal pole, as this was significantly associated with PAU PRS after multiple testing correction.
Table 1.
Nominally and FDR significant results for the PAU and DPW PRS Prediction
| PAU | ||||
|---|---|---|---|---|
| Modality | Region | Beta | STE | Nominal P-value |
| Area | Right Insula | 0.027 | 0.014 | 0.046 |
| Area | Right Pars Triangularis | 0.035 | 0.016 | 0.033 |
| Thickness | Left Pars Orbitalis | −0.031 | 0.016 | 0.049 |
| Thickness | Left Pars Opercularis | −0.028 | 0.014 | 0.043 |
| Thickness | Right Lateral Orbital Frontal Gyrus | −0.035 | 0.015 | 0.017 |
| Thickness | Right Supramarginal Gyrus | 0.032 | 0.010 | 0.001* |
| Volume | Left Frontal Pole | −0.057 | 0.017 | 0.001* |
| fractional anisotropy | right anterior thalamic radiations | −0.024 | 0.012 | 0.046 |
| fractional anisotropy | left anterior thalamic radiations | −0.025 | 0.012 | 0.030 |
| White Matter volume | Corpus Callosum | −0.010 | 0.005 | 0.045 |
| White Matter volume | Right Parahippocampal cingulum | −0.030 | 0.014 | 0.041 |
| White Matter volume | Forceps Minor | −0.023 | 0.009 | 0.014 |
| DPW | ||||
| Area | Right Insula | 0.031 | 0.014 | 0.023 |
| Area | Left Superior Parietal Gyrus | −0.033 | 0.013 | 0.013 |
| Thickness | Right Inferior Parietal Gyrus | 0.024 | 0.010 | 0.012 |
| Thickness | Right Pericalcarine Cortex | −0.034 | 0.016 | 0.039 |
| Thickness | Right Temporal Pole | −0.033 | 0.017 | 0.048 |
| Volume | Right Entorhinal Cortex | −0.047 | 0.018 | 0.008 |
| Volume | Left Insula | 0.029 | 0.013 | 0.024 |
| Volume | Right Insula | 0.027 | 0.013 | 0.039 |
| Volume | Left Superior Parietal Gyrus | −0.029 | 0.014 | 0.044 |
| Volume | Left Frontal Pole | −0.034 | 0.017 | 0.049 |
| Volume | Left Pallidum | −0.030 | 0.015 | 0.049 |
| Mean Diffusivity | Right Parahippocampal Cingulum | −0.033 | 0.016 | 0.041 |
| Mean Diffusivity | Right Inferior Longitudinal Fasiculus | 0.029 | 0.013 | 0.029 |
Note. Nominal significant results shown by region across all modalities.
STE= Standard Error.
Area was significant after FDR correction.
No brain regions were discovered for typical alcohol use after correction for multiple comparisons (DPW-PRS, Supplemental Results). Several nominally significant associations arose in analyses for both PAU and DPW (Table 1). Notably, DPW-PRS showed a nominal association with left frontal pole volume (Table 1). Given that typical and problematic alcohol are genetically correlated (SNP-rg=0.78) (12), such consistency is unsurprising. A regression with PAU-PRS and DPW-PRS entered simultaneously show that this finding is driven primarily by PAU-PRS (Table 2).
Table 2.
Problematic Alcohol Use (PAU) and Drinks Per Week (DPW) PRS Prediction of TopOutcomes in Multiple Linear Model, Accounting for the Other PRS
| Region | PAU Beta | PAU P-value | PAU CI | DPW Beta | DPW P-Value | DPW CI |
|---|---|---|---|---|---|---|
| Frontal Pole | −0.0522 | 0.0060 | ±0.0371 | −0.0129 | 0.0191 | ±0.9786 |
| Supramarginal Gyrus | 0.0331 | 0.0025 | ±0.0355 | −0.0034 | 0.7575 | ±0.0213 |
Note. Both the PAU and DPW Polygenic Risk Scores (PRS) were entered as covariates in the same linear mixed effects model to test whether prediction of each outcome was specific to polygenic risk for each alcohol behavior. The standardized beta weights, p-values and confidence intervals around the beta were drawn from the mixed effects regression model. Bold values are those significant in the multiple regression.
Finally, PAU-PRS was not significantly associated with frontal pole gray matter volume or supramarginal gyrus cortical thickness among individuals of African ancestry and no FDR-corrected significant associations emerged for any other brain structure variable examined (n=898 in ABCD and n=62,447 for original GWAS of individuals of African ancestry; Online Methods; Supplemental Table 3).c
Discussion
Here, among 3,013 substance-naïve children, we find evidence that alcohol-related differences in brain structure may, at least partially, reflect genetically-conferred pre-dispositional risk factors that precede alcohol exposure. Broadly, these findings, combined with evidence from humans and non-human animal models, challenge widespread speculation that brain-based associations with alcohol are solely attributable to the neurotoxic effects of alcohol (6,31).
We discovered two main associations between PAU-PRS and brain morphometry. First, PAU-PRS were associated with lower frontal pole volume; this finding aligns with evidence that adolescents who drink heavily have lower volume here (32). Unlike past studies showing reduced volume is associated with increased impulsive behaviors (33), we found no evidence that a proxy behavior of externalizing psychopathology is associated with frontal pole volume. This may be because of differences in measurement between trait impulsivity and trait externalizing.
Second, we found that PAU-PRS are associated with thicker supramarginal gyrus cortex. Greater supramarginal gyrus thickness has been found in substance-naïve children of alcohol dependent individuals (8); however, the supramarginal gyrus is thinner among those with alcohol dependence (2). It is possible that some brain structure correlates of alcohol involvement may be developmentally dependent. Indeed, throughout childhood and adolescence neuronal pruning appears to support long term planning, working memory performance, language, and attention (34). It has been postulated that greater indices of gray matter among those at familial risk for alcohol problems may reflect a developmental delay in neural pruning that places adolescents at risk for substance use, which, in turn, accelerates neuronal pruning in later development (35). Given that the supramarginal gyrus is thicker among those at elevated risk for problematic alcohol use and this region supports cognition and language (36), this is plausible. However, supramarginal gyrus cortical thickness was not significantly correlated with cognition (Supplemental Table 4).
Notably, some of the nominal associations identified in our study (Table 1) showed a similar directional pattern wherein among substance naïve children, associations with PRS were positive (e.g., greater insula volume and area, inferior and superior parietal cortex cortical thickness and volume) that have observed to be negatively associated with alcohol dependence in a large meta-analysis (2). It is possible that brain regions exerting the most dominant influence may show fluid changes across development, and that genomic variability as well as alcohol exposure (37) may drive developmentally dependent differences.
It is important to consider the limitations of this study. First, while the study of alcohol and substance naïve children allowed us to preclude that associations may be attributable to alcohol exposure, we were unable to test whether brain structure correlates of genomic risk are associated with onsets of alcohol use and other drinking milestones. As the longitudinal ABCD study continues to collect data, it will be particularly interesting to examine how the interplay between polygenic risk and exposure influences brain structure trajectories, and vice versa (i.e., how malleability of brain development modulates escalation or desistance in alcohol use). Second, PRS typically have low cross-trait (i.e., from GWAS phenotype to a related phenotype) predictive utility. While our findings suggest that variability in brain structure may represent predispositional biomarkers for alcohol involvement, the small effect sizes (i.e., maximum R2=0.003) and resource intensive nature of neuroimaging limit their clinical utility. Further, these small effects combined with multiple testing burden may have contributed to false negatives; as such null effects should not be interpreted to suggest that an association observed in adults may solely reflect neurotoxicity. Third, as the discovery GWAS were of individuals of predominantly European descent, we restricted our ABCD sample to children of similar ancestral background. Our analyses in the smaller African-ancestry sample (N = 898) of ABCD revealed no significant results (Supplemental Table 3), likely due to low power of the discovery GWAS of matched ancestry (N = 62,447 vs. 352,365 for the European-ancestry GWAS) as well as the smaller ABCD sample. This highlights the need for more and larger non-European discovery GWAS.
Limitations notwithstanding, our study provides initial evidence that alcohol-related brain structure correlates may represent genomic risk factors for alcohol involvement. This does not preclude the possibility of neurotoxic effects. However, these findings suggest that genomic vulnerability to alcohol involvement may be conveyed through brain alterations that emerge during middle childhood prior to alcohol exposure.
Supplementary Material
Acknowledgments
ASH receives support from DA007261-17. ECJ receives support from AA027435-02. AA receives support from MH109532 and K02DA032573. DAAB receives support from MH018951. Dr. Bogdan (AG052564, AA027827, DA046224). ASH, ECJ, AA, and RB developed the research questions. ASH conducted analyses, with help from ECJ, DAAB, and SEP. ASH and RB drafted the manuscript. ECJ, AA, DAAB, and SEP provided critical revision of the manuscript for important intellectual content. ASH had full access to all data in the study and take responsibility for the integrity of the data and accuracy of the data analyses. Conflict of interest disclosures: No disclosures were reported.
Footnotes
The authors have no declarations of interest to declare
Children self-reported no substance use (including substances other than alcohol or tobacco) and screened negative according to hair toxicology (see Supplementary Information for a table of exclusions). All participants were required to have non-missing data on all variables used in analyses.
Cornell University was an original collection site that collected data from 34 participants, before being moved to Yale University. ABCD documentation reports 21 data collection sites and does not list Cornell; our analyses nested data based on 22 data collection sites, including the original Cornell site.
GWAS summary statistics for Drinks Per Week are not currently available for those of African ancestry.
Reference Cited
- 1.World Health Organization. Global status report on alcohol and health 2018. Geneva: World Health Organization; 2018. Licence: CC BY-NC-SA 3.0 IGO. Poznyak V, Rekve D, editors. 2018. 478 p. [Google Scholar]
- 2.Mackey S, Allgaier N, Chaarani B, Spechler P, Orr C, Bunn J, et al. Mega-Analysis of Gray Matter Volume in Substance Dependence: General and Substance-Specific Regional Effects. Am J Psychiatry [Internet]. 2019. February [cited 2019 Dec 15];176(2):119–28. Available from: http://ajp.psychiatryonline.org/doi/10.1176/appi.ajp.2018.17040415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hampton WH, Hanik IM, Olson IR. Substance abuse and white matter: Findings, limitations, and future of diffusion tensor imaging research. Vol. 197, Drug and Alcohol Dependence. Elsevier Ireland Ltd; 2019. p. 288–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crews FT. Alcohol-related neurodegeneration and recovery: mechanisms from animal models. Alcohol Res Health [Internet]. 2008. [cited 2019 Dec 15];31(4):377–88. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23584011 [PMC free article] [PubMed] [Google Scholar]
- 5.Yu R, Deochand C, Krotow A, Le’ao R, Tong M, Agarwal AR, et al. Tobacco Smoke-Induced Brain White Matter Myelin Dysfunction: Potential Co-Factor Role of Smoking in Neurodegeneration. J Alzheimer’s Dis. 2016. January 1;50(1):133–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baranger DAA, Demers CH, Elsayed NM, Knodt AR, Radtke SR, Desmarais A, et al. Convergent Evidence for Predispositional Effects of Brain Gray Matter Volume on Alcohol Consumption. Biol Psychiatry. 2019. September; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robert GH, Luo Q, Yu T, Chu C, Ing A, Jia T, et al. Association of Gray Matter and Personality Development with Increased Drunkenness Frequency during Adolescence. JAMA Psychiatry. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holla B, Bharath RD, Venkatasubramanian G, Benegal V. Altered brain cortical maturation is found in adolescents with a family history of alcoholism. Addict Biol. 2019. July 1;24(4):835–45. [DOI] [PubMed] [Google Scholar]
- 9.Verhulst B, Neale MC, Kendler KS. The heritability of alcohol use disorders: A meta-analysis of twin and adoption studies. Psychol Med. 2015. April 15;45(5):1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walters GD. THE HERITABILITY OF ALCOHOL ABUSE AND DEPENDENCE: A META-ANALYSIS OF BEHAVIOR GENETIC RESEARCH. Am J Drug Alcohol Abuse [Internet]. 2002. January 5 [cited 2020 Jan 28];28(3):557–84. Available from: http://www.tandfonline.com/doi/full/10.1081/ADA-120006742 [DOI] [PubMed] [Google Scholar]
- 11.Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018. December 1;21(12):1656–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou H, Sealock JM, Sanchez-Roige S, Clarke T-K, Levey DF, Cheng Z, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci [Internet]. 2020. May 25 [cited 2020 May 30];1–10. Available from: http://www.nature.com/articles/s41593-020-0643-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volkow ND, Koob GF, McLellan AT. Neurobiologic Advances from the Brain Disease Model of Addiction. Longo DL, editor. N Engl J Med [Internet]. 2016. January 28 [cited 2020 Jun 1];374(4):363–71. Available from: http://www.nejm.org/doi/10.1056/NEJMra1511480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volkow ND, Koob GF, Croyle RT, Bianchi DW, Gordon JA, Koroshetz WJ, et al. The conception of the ABCD study: From substance use to a broad NIH collaboration. Vol. 32, Developmental Cognitive Neuroscience. Elsevier Ltd; 2018. p. 4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baurley JW, Edlund CK, Pardamean CI, Conti DV., Bergen AW. Smokescreen: A targeted genotyping array for addiction research. BMC Genomics. 2016;17(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogdan R, Baranger DAA, Agrawal A. Polygenic Risk Scores in Clinical Psychology: Bridging Genomic Risk to Individual Differences. Annu Rev Clin Psychol. 2018. May 7;14(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet [Internet]. 2017. April 6 [cited 2020 Dec 13];100(4):635–49. Available from: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam M, Awasthi S, Watson HJ, Goldstein J, Panagiotaropoulou G, Trubetskoy V, et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. bioRxiv. 2019. April 11;587196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge T, Chen CY, Ni Y, Feng YCA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019. Dec 1;10(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Vol. 51, Nature Genetics. Nature Publishing Group; 2019. p. 237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun [Internet]. 2019. December 1 [cited 2021 Jan 7];10(1). Available from: https://pubmed.ncbi.nlm.nih.gov/30940813/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, Adams MJ, Howard DM, et al. Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts. Am J Psychiatry [Internet]. 2019. February 19 [cited 2019 Feb 22];176(2):107–18. Available from: http://ajp.psychiatryonline.org/doi/10.1176/appi.ajp.2018.18040369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagler DJ, Hatton SN, Cornejo MD, Makowski C, Fair DA, Dick AS, et al. Image processing and analysis methods for the Adolescent Brain Cognitive Development Study. Neuroimage. 2019. November 15;202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casey BJ, Cannonier T, Conley MI, Cohen AO, Barch DM, Heitzeg MM, et al. The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites. Dev Cogn Neurosci [Internet]. 2018. August 1 [cited 2019 Mar 27];32:43–54. Available from: https://www.sciencedirect.com/science/article/pii/S1878929317301214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage [Internet]. 2006. July 1 [cited 2020 Aug 26];31(3):968–80. Available from: https://pubmed.ncbi.nlm.nih.gov/16530430/ [DOI] [PubMed] [Google Scholar]
- 26.Goldstein RZ, Volkow ND. Drug addiction and its underlying neurobiological basis: Neuroimaging evidence for the involvement of the frontal cortex. Vol. 159, American Journal of Psychiatry. 2002. p. 1642–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gershon RC, Wagster MV., Hendrie HC, Fox NA, Cook KF, Nowinski CJ. NIH toolbox for assessment of neurological and behavioral function. Neurology. 2013;80(11 Suppl 3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Achenbach TM, McConaughy SH, Howell CT. Child/adolescent behavioral and emotional problems: Implications of cross-informant correlations for situational specificity. Psychol Bull [Internet]. 1987. [cited 2016 Jun 2];101(2):213–32. Available from: http://doi.apa.org/getdoi.cfm?doi=10.1037/0033-2909.101.2.213 [PubMed] [Google Scholar]
- 29.Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015. October 1;67(1):1–48. [Google Scholar]
- 30.Paul SE, Hatoum AS, Fine JD, Johnson EC, Hansen I, Karcher NR, et al. Associations between Prenatal Cannabis Exposure and Childhood Outcomes: Results from the ABCD Study. JAMA Psychiatry [Internet]. 2021. January 1 [cited 2021 Mar 16];78(1):64–76. Available from: https://jamanetwork.com/journals/jamapsychiatry/fullarticle/2770964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siciliano CA, Noamany H, Chang CJ, Brown AR, Chen X, Leible D, et al. A cortical-brainstem circuit predicts and governs compulsive alcohol drinking. Science (80- ) [Internet]. 2019. November 22 [cited 2020 Aug 26];366(6468):1008–12. Available from: https://pubmed.ncbi.nlm.nih.gov/31754002/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heikkinen N, Niskanen E, Könönen M, Tolmunen T, Kekkonen V, Kivimäki P, et al. Alcohol consumption during adolescence is associated with reduced grey matter volumes. Addiction. 2017. April 1;112(4):604–13. [DOI] [PubMed] [Google Scholar]
- 33.Gröpper S, Spengler S, Stuke H, Gawron CK, Parnack J, Gutwinski S, et al. Behavioral impulsivity mediates the relationship between decreased frontal gray matter volume and harmful alcohol drinking: A voxel-based morphometry study. J Psychiatr Res. 2016. December 1;83:16–23. [DOI] [PubMed] [Google Scholar]
- 34.Selemon LD. A role for synaptic plasticity in the adolescent development of executive function. Vol. 3, Translational psychiatry. Nature Publishing Group; 2013. p. e238–e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winters KC, Arria A. Adolescent Brain Development and Drugs. Prev Res [Internet]. 2011. [cited 2020 Jun 19];18(2):21–4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22822298 [PMC free article] [PubMed] [Google Scholar]
- 36.Stoeckel C, Gough PM, Watkins KE, Devlin JT. Supramarginal gyrus involvement in visual word recognition. Cortex. 2009. October 1;45(9):1091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shnitko TA, Liu Z, Wang X, Grant KA, Kroenke CD. Chronic alcohol drinking slows brain development in adolescent and young adult nonhuman primates. eNeuro [Internet]. 2019. March 1 [cited 2020 Jul 15];6(2). Available from: /pmc/articles/PMC6464511/?report=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics and data are publicly available through various sources. The Million Veterans Project summary statistics were accessed via dbGaP (phs001672.v1.p1) as part of #24806: Neurobiological bases of psychiatric traits. The authors thank Million Veteran Program (MVP) staff, researchers, and volunteers, who have contributed to MVP, and especially participants who previously served their country in the military and now generously agreed to enroll in the study. (See https://www.research.va.gov/mvp/ for more details).
The citation for MVP is Gaziano, J.M. et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016).
This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by the Veterans Administration (VA) Cooperative Studies Program (CSP) award #G002. Data for this study were provided by the Adolescent Brain Cognitive Development (ABCD) study, which was funded by awards U01DA041022, U01DA041025, U01DA041028, U01DA041048, U01DA041089, U01DA041093, U01DA041106, U01DA041117, U01DA041120, U01DA041134, U01DA041148, U01DA041156, U01DA041174, U24DA041123, and U24DA041147 from the NIH and additional federal partners (https://abcdstudy.org/federal-partners.html). All data used here in are publicly available through the GSCAN consortium, dbGAP, and the ABCD consortium.


