Abstract
Arginine methylation is a prevalent posttranslational modification which is deposited by a family of protein arginine methyltransferases (PRMTs), and is found in three different forms in mammalian cells: monomethylarginine (MMA), asymmetric dimethylarginine (ADMA) and symmetric dimethylarginine (SDMA). Pan-methylarginine antibodies are critical for identifying proteins that are methylated on arginine residues, and are also used for evaluating signaling pathways that modulate this methyltransferase activity. Although good pan-MMA, -ADMA and -SDMA antibodies have been developed over the years, there is still room for improvement. Here we use a novel antigen approach, which involves the separation of short methylated motifs with inert polyethylene glycol (PEG) linkers, to generate a set of pan antibodies to the full range of methylarginine marks. Using these antibodies, we observed substrate scavenging by PRMT1, when PRMT5 activity is blocked. Specifically, we find that the splicing factor SmD1 displays increased ADMA levels upon PRMT5 inhibitor treatment. Furthermore, when the catalysis of both SDMA and ADMA is blocked with small molecule inhibitors, we demonstrate that SmD1 and SMN no longer interact. This could partially explain the synergistic effect of PRMT5 and type I PRMT inhibition on RNA splicing and cancer cell growth.
Keywords: arginine methylation, MMA, ADMA, SDMA, SmD1
1. Introduction
Posttranslational modifications (PTMs) are a major regulator of protein function. PTMs can subtly alter protein structure, either by causing local or distant changes to protein folds, or by creating a docking site for effector proteins if the PTM occurs in an unstructured region of a protein. There are over 200 different types of PTMs [1] and examples of the most common PTMs include: 1) the phosphorylation of serine (pS), threonine (pT) and tyrosine (pY) residues; 2) the acetylation of lysine residues; 3) the modification of lysine by ubiquitin and ubiquitin-like proteins; and 4) the methylation of lysine and arginine residues. More than 5% of the human proteome is dedicated to enzymes that mark proteins with PTMs [2], which can be further broken down to about 1% of the proteome focused on the regulation of phosphorylation, and another 1% dedicated to the regulation of methylation [3]. Most PTMs are revisable, so the enzymes that regulate these modifications include families of proteins that both write and erase these marks.
A central theme, when studying PTMs, is the development of antibodies that recognize these different marks. These antibodies are used to track the deposition and removal of specific marks during signal transduction, and also to identify novel substrates in combination with mass spectrometric approaches. For the latter application, it is ideal to have antibodies that are pan (recognize all modified substrates) or semi-pan (recognize a subset of modified substrates) for a particular PTM. Antibodies that recognize a broad spectrum of substrates were first developed for tyrosine phosphorylated protein detection, and include monoclonals like 4G10 and P-Y20 [4, 5]. The development of broadly reactive phosphoserine and phosphothreonine specific antibodies has been more difficult, although the development of kinase consensus motif antibodies for AKT and PKC sites has been successful [6]. Currently, there are a number of commercial sources for pan phosphoserine/ threonine antibodies, as well as antibodies that recognize all three phospho-states – pY, pS and pT. Similarly, acetyllysine-specific antibodies have also been successfully developed. Early work, before the turn of the century, with histone H4K12ac-specific antibodies investigated X-chromosome inactivation [7], and an antibody to acetylated tubulin was used to map the site of acetylation to K40 [8]. Pan-acetyllysine antibodies have also been developed [9], and they are available from commercial sources.
The focus of this study is the development of pan methylarginine-specific antibodies. There are three types are arginine methylation – monomethylarginine (MMA), symmetric dimethylarginine (SDMA) and asymmetric dimethylarginine (ADMA) [10]. These three methylarginine marks are deposited by a family of nine related protein arginine methyltransferases (PRMTs), as well as a tenth mitochondrial enzyme called NDUFAF7 [11], which is not highly related to the PRMT family. The arginine methyltransferases fall into three categories according to their catalytic activities: PRMT1, 2, 3, 6, 8 and CARM1 (also called PRMT4) are type I PRMTs that deposit the ADMA mark. PRMT5, PRMT9 and NDUFAF7 are type II enzymes that deposit the SDMA mark [12]. PRMT7 is type III PRMT which is primarily responsible for depositing the MMA mark [13]. Arginine methylation is a very abundant PTM, with approximately 0.5% of all arginine residues methylated in human proteome, and the relative ratios of the three different types of arginine methylation are roughly as follows: 1500 / 3 / 2 / 1 for arginine / ADMA / SDMA / MMA [14]. Most of the PRMTs methylate a very similar motif, which is glycine and arginine rich (GAR motifs). Using GAR motifs with different methylated states as antigens, it is possible to generate polycolnal antibodies that recognize many PRMT substrates. Although we refer to these antibodies as pan-methylarginine antibodies, they will likely not recognize all PRMT substrates, because they will not identify proteins that harbor methylation site outside of GAR motifs. These non-GAR motif methylation sites are not very usual for the dominant enzymes like PRMT1 and PRMT5, but are very common for CARM1. Indeed, pioneering work from the Richard lab, generated the first pan-methylarginine specific antibodies that recognized SDMA marks (SYM10 & 11) and ADMA marks (ASYM24 & 25), and these antibodies were used, in conjunction with mass spectrometry, to identify a slew of novel substrates for PRMT1 and PRMT5 [15]. Subsequently, in collaboration with Cell Signaling Technology (CST), we developed a panel of rabbit monoclonal antibodies against MMA, SDMA and ADMA marks [16]. The advantage of having rabbit monoclonal antibodies for the different methylarginine marks is that the hybridomas serve as a renewable resource of the antibodies. The disadvantage is that you will lose a large number of immune-reactive bands when you select a rabbit with good reactivity and then generate monoclonal antibodies. To alleviate some of this loss, different monoclonal antibodies to the same mark can be pooled, and this is what CST did. These CST antibodies have been used by us to demonstrate cross-talk between the different types of arginine methylation [14], and by others to mine the proteome for more PRMT substrates [17, 18].
In order to better investigate the role of arginine methylation, we used a novel antigen design to develop a panel of antibodies that specifically recognize MMA, ADMA and SDMA substrates (Figure 1). We have previously shown that with PRMT1 loss, or small molecule inhibition of Type I PRMTs (MS023), as expected ADMA levels are dramatically reduced, and we also showed that under the same conditions, MMA and SDMA levels unexpectedly go up, suggesting that PRMT5 and PRMT1 target the same substrates for different types of methylation [14]. Using our new antibody set, we showed that the opposite is also true; when PRMT5 is inhibited (using EPZ015666) there are substrates that display reduced SDMA levels and increased ADMA levels. In particular, the splicing factor SmD1 displays a remarkable increase of ADMA levels under EPZ015666 treatment. Furthermore, we demonstrated that combined EPZ015666 and MS023 treatment synergistically inhibited the interaction between SmD1 and SMN. The disrupted interaction between SmD1 and SMN by combined PRMT5 and type I PRMT inhibition might be partially responsible for the reported synergistic effect of these two inhibitors on RNA splicing [17].
Figure 1.
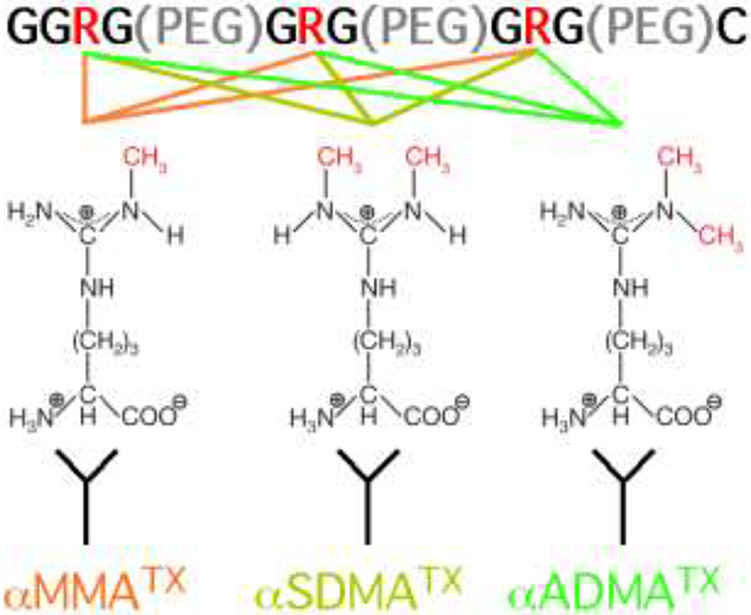
A scematic representing the use of a PEG linked antigen to develop MMA, SDMA and ADMA antibodies. All the antigens have same amino acid sequence but harbor different modification at the arginine(R) sites: one methyl group was added on the nitrogen atom of each arginine site for MMA antigen, two methyl groups were added on the same nitrogen atom of each arginine site for ADMA antigen, and two methyl groups were added on different nitrogen atoms of each arginine site for SDMA antigen. The three short GAR motifs present in the antigen are seperated by PEG linkers.
2. Materials and Methods
2.1. Materials
The antigens for the antibody production were generated by New England Peptide (NEP). The same antigen structure was used to generate all three types of antibody (MMA, ADMA, SDMA), namely N2H-GGRG(dPEG4)GRG(dPEG4)GRG(dPEG4)C-amid. All the R residues in the peptide were modified with either MMA, SDMA or ADMA, depending on the antibody project. The cysteine residue at the C-terminal end of the peptides was used for attachment to KLH. The MMA, ADMA, and SDMA antibodies were produced in rabbits by NEP. The antibodies used in this study were not affinity purified or negativel depleted the unmodified peptide backbone or PEG. Other antibodies used were anti-β-actin (Sigma, #A1978), anti-MMA (CST, #8711), anti-ADMA (CST, #13522), anti-SDMA (CST, #13222), anti-SmD1 (Santa Cruz, #sc-166650), and anti-SMN (BD Biosciences, #610646).
2.2. PRMT inhibitor treatment
MS023 was purchased from Sigma (#SML1555, 1mM in DMSO as stock). TP064 was purchased from Tocris (#6008, 10mM in DMSO as stock). EPZ015666 was purchased from Sigma (#SML1421, 5mM in DMSO as stock). All the inhibitors were used at 1:1000 dilution for 48h for cell treatment, and DMSO was used as the untreated control.
2.3. Co-immunoprecipitation and western blotting
293T cells were harvested and total cell extracts were prepared in mild lysis buffer (10mM Tris/Cl pH 7.5; 150mM NaCl; 0.5mM EDTA; 0.5% NP-40). SMN antibody was incubated with Protein A/G beads (Thermo Scientific) for 2h at 4°C, washed three times with lysis buffer, followed by incubation with the cell lysates for 1h at 4°C. Then the beads were washed three times with lysis buffer and boiled in loading buffer to elute the bounded proteins. The cell extracts or immunoprecipitated samples were separated by SDS-PAGE and transferred onto PVDF membranes. Blots were blocked in PBS containing 5% non-fat dry milk, then incubated with the appropriate primary antibody in the blocking buffer overnight at 4°C. The blots were then washed with PBST (PBS with 0.05% tween-20) and probed with an HRP-labelled secondary antibody for 1h at room temperature. After washing three times with PBST, the membranes were incubated with ECL reagent and the signal was detected on X-ray film.
2.4. LC-MS/MS (liquid chromatography-tandem mass spectrometry)
293T cells were treated with DMSO or EPZ015666 for 48h, then were harvested and the cell extracts were prepared in mild lysis buffer. ADMA antibody was incubated with Protein A/G beads, washed, followed by incubation with the cell lysates at 4°C for 2h. The bounded beads were washed and boiled in SDS loading buffer, and analyzed using SDS-PAGE followed by silver staining. After comparison with DMSO-treated samples, the differentially expressed band within 10~15kD in EPZ015666-treated sample was cut from the gel and the protein was identified with LC-MS/MS on a Thermo Ultimate 3000 RSLCnano UPLC in-line with an Orbitrap Fusion at the Proteomics Facility at UT Austin. Proteins were identified with Proteome Discoverer 2.2 (Thermo) using the Sequest HT search engine, with 10 ppm mass tolerance for the MS at 0.6 Da for the MS/MS. Identifications were validated with Scaffold 4.1 (Proteome Software) using protein threshold of 1% FDR for 2 peptides at peptide threshold of 0.1% FDR.
2.5. RNA interference
For small interfering(siRNA)-mediated knockdown of SmD1, cells were transfected with 200 pmol of either targeting or control siRNA using Lipofectamine 2000 (Invitrogen) for 48h, and the cells were harvested for western blot analysis. siSmD1-1 (hs.Ri.SNRPD1.13.1), siSmD1-2(hs.Ri.SNRPD1.13.2) and siNC (Negative Control DsiRNA, 51-01-14-03) were purchased from IDT, and used according to manufacturer’s instructions.
3. Results and discussion
3.1. Generation and evaluation of new MMA, ADMA and SDMA antibodies
The majority of PRMTs prefer to methylate arginine residues within glycine- and arginine-rich (GAR) motifs [19, 20]. In order to develop a panel of antibodies that recognize MMA, ADMA, and SDMA marks, we designed immunogens that harbored short GAR motifs (GGRG or GRG) that were linked together. As a linker/spacer, we used polyethylene glycol (dPEG4), which is highly flexible and displays reduced immunogenicity. This approach allows us to obtain antibodies that interact preferentially with the focused small methylated epitopes. We chose the same strategy for the design and synthesis of all three immunogens. A daisy-chain of PEG-linked short amino acid motifs has been used previously on a handful of occasions, including the generation of antibodies that recognize γ-carboxylated glutamic acid [21], and arginine-derived advanced glycation end-products [22, 23].
Upon completion of synthesis, the immunogen peptides were conjugated to keyhole limpet hemocyanin (KLH), which is a widely employed carrier protein for effective immune response and antibody production. The conjugate was then used to immunize rabbits. Serum samples were collected three times from the same rabbit. The “Pre-bleed” serum sample was collected before immunization, and the “Bleed 1” serum sample was collected one month later after the first immunization. The rabbit was then immunized again, which we call a “boost”; the serum sample (Bleed 2) was collected 10 days after the “boost”. The processes of immunization and serum sample collection are the same for all of our antibody development and two rabbits were used for each of the three (MMA, SDMA & ADMA) antibody development projects.
The newly developed antibodies were then subjected to Western blot (WB) analysis. To help evaluate the specificity of this antibody set, 293T cells were treated with different PRMT inhibitors for 48 hours, and total cell lysates were then harvested for WB analysis. The following PRMT inhibitors were used: 1) EPZ015666 which is a very selective PRMT5 inhibitor [24]; 2) MS032 which is a type I PRMT inhibitor that blocks the activity of PRMT1, 2, 3, 4, 6, and 8 [25]; and 3) TP064 which is a CARM1-specific inhibitor [26].
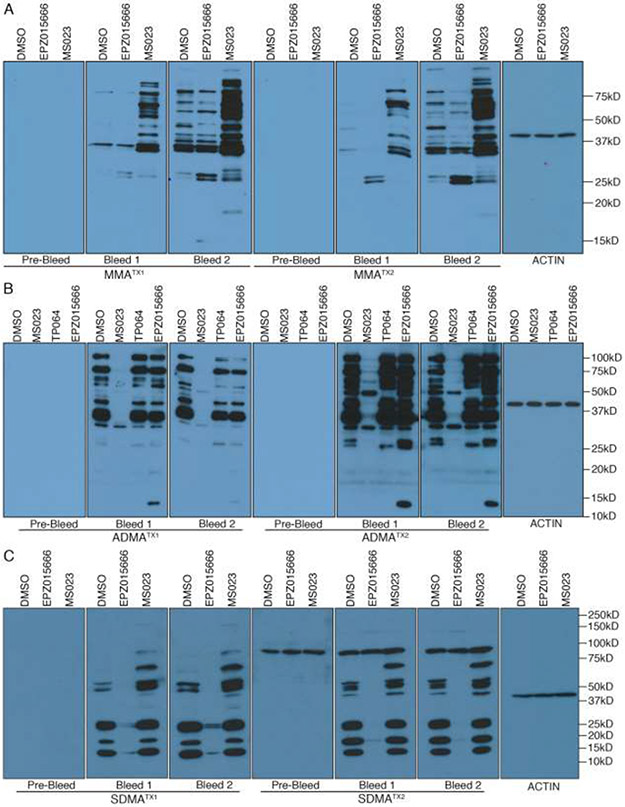
For both MMATX1 and MMATX2 antibodies, no signal was detected on the WB membrane when it was incubated with pre-bleed sera. However, when bleeds 1 and 2 were used, we saw multiple bands in the DMSO-treated lane, for both MMATX1 and MMATX2 antibodies (Figure 2A). Furthermore, both MMA antibodies display an increase in immuno-reactivity when cells are pre-treated with the PRMT5 (a few bands increase) or Type 1 inhibitors (many bands increase). This phenomenon is called substrate scavenging and was reported by us a number of years ago [14], and confirmed by others more recently [17, 18, 27]. These data indicated that both MMATX1 and MMATX2 are high specificity antibodies.
Figure 2.
Evaluation of newly developed MMA, ADMA and SDMA antibodies by WB. 293T was treated with different PRMT inhibitors for 48 hours, whole cell lysates were harvested and analyzed with MMA (A), ADMA (B) or SDMA (C) antibodies. For each antibody project, the pre-bleed sera was also tested. All the antibodies were used at 1:2000 dilution. The PRMT inhibitors used include MS023 (Type I PRMT inhibitor), EPZ015666 (PRMT5 inhibitor), and TP064 (CARM1 inhibitor). For each antibody type (MMA, SDMA & ADMA) two rabbits were immunized with the antigen depicted in Figure 1, and the individual rabbits were referred to as TX1 and TX2.
A similar approach was taken to evaluate the the ADMATX1 and ADMATX2 antibodies. Again, we saw no bands when WB membranes were incubated with pre-bleed, while strong signals were observed with the two bleeds for both rabbits (Figure 2B). Moreover, a dramatic loss of bands was seen when cells were treated with MS023. These data indicated that the ADMATX1 and ADMATX2 antibodies are also highly specific and can recognize many different substrates for the Type I PRMTs. Interestingly, ADMATX2 antibody recognizes a 50kD protein in the presence of MS023-treatment, and this band is lost in the adjacent lane with TP064-treatment, suggesting that ADMATX2 can recognize at least one CARM1 substrate. Also, the WB results show that ADMATX2 antibody can detect a strong 13kD band when cells are treated with the PRMT5 inhibitor (Figure 2B), which suggests “scavenging” by PRMT1 when the deposition of SDMA marks is blocked. Scavenging in this direction is not as common as the broad impact that Type I inhibitors has on methylarginine type switching that results in an obvious elevation of MMA levels (Figure 2A), but it has been reported in quantitative mass spectrometry studies performed by the Bonaldi group [17, 18].
Next, the SDMA antibody set was also evaluated by WB analysis. We detected strong signals on the WB membranes that were incubated with both bleeds for the two rabbits (Figure 2C). Also, both antibodies (SDMATX1 and SDMATX2) displayed a decrease in signal when cells were treated with the PRMT5 inhibitor (EPZ015666). These data indicated that the SDMATX1 and SDMATX2 antibodies are also highly specific and can recognize many different substrates for PRMT5. It should be noted that all the SDMATX2 bleeds, including the pre-bleed, recognize a 100 kD band that is not impacted by any of the PRMT inhibitors. This represents a cross-reactive band that was present in the serum of this particular rabbit before it was immunized with the SDMA/PEG antigen. Thus, the SDMATX2 antibody, although useful for mass spectrometry studies and WB analysis, cannot be used for immunohistochemical approaches.
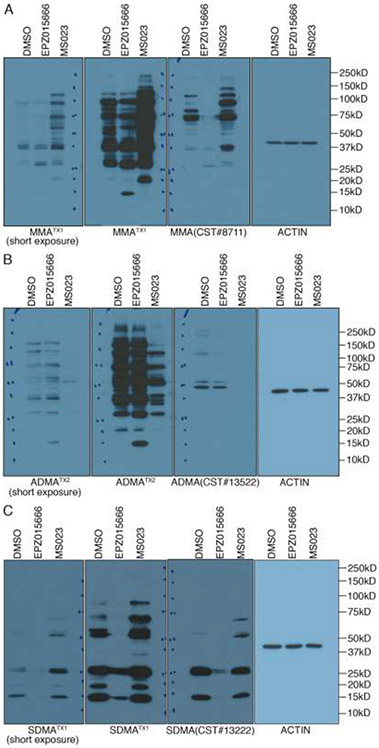
To expand the evaluation of the new developed antibodies, we compared our TX antibody series with the commercialized MMA, ADMA, and SDMA antibodies from CST. For WB analysis we used all antibodies at a 1:2000 dilution. The CST antibodies are largely weaker than the TX series, and we thus included a shorter exposure time for the TX antibodies to help with the comparison (Figure 3). In general, the TX antibodies recognize more methylated substrates than do the CST antibodies, and this is most obvious for the anti-ADMA antibody comparison (Figure 3B). The ability of our antibodies to recognize more arginine methylated substrates than the CST antibodies is likely due to the fact that CST has converted their rabbit polyclonal antibodies into rabbit monoclonal antibodies, which will clearly reduce the spectrum of PRMT substrates recognized.
Figure 3.
Comparative analysis of the TX antibodies (MMA, ADMA and SDMA) and a similar set of antibodies developed by CST. (A-C) 293T were treated with different PRMT inhibitors for 48 hours, whole cell lysates were harvested, and analyzed with the TX and CST antibodies. All the antibodies were used at 1:2000 dilution. In each case, a short exposure of the TX antibody is also shown.
3.2. SmD1 was identified as a SDMA-to-ADMA switching protein
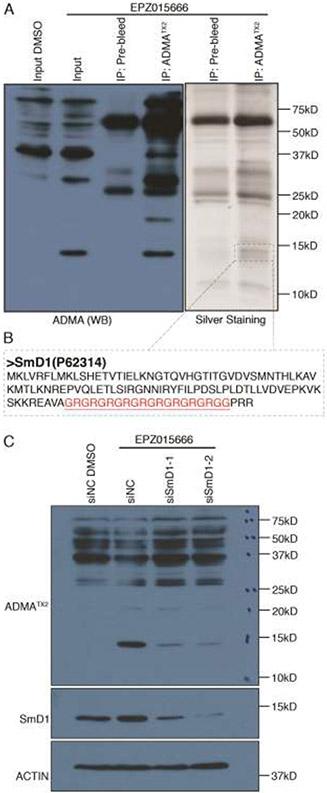
As we note above, the ADMATX2 antibody unexpectedly detected a new, very clear band between 10kD~15kD, upon EPZ015666 treatment (Figure 2B). We chose to identify this PRMT substrate to help us understand the potential consequence of this PRMT5 inhibitor-induced gain of ADMA. Importantly, this band can be immunoprecipitated (IPed) using the ADMATX2 antibody, but not with pre-bleed sera (an IgG control) (Figure 4A). We also ran a second gel in parallel with the same IPed sample and a silver stain could detect a candidate protein that only came down with the methyl-specific antibody. We next cut out this region of the gel (between 10~15kD) that included the specific low molecular weight protein detected with the ADMATX2 IP (Figure 4A), and subjected the proteins within this gel slice to tryptic digestion and subsequent mass spectrometry (MS) analysis. MS analysis identified several proteins in this gel slice, including SmD1 (Figure 4B). Indeed, when comparing the number of SmD1 normalized spectral counts that are IPed with the ADMATX2 antibody, after DMSO-treatment (15 normalized spectral counts) or EPZ-treatment (21 normalized spectral counts), we observed a 40% increase. To confirm the identity of this SDMA-to-ADMA switching protein, we performed SmD1 knock-down experiments using two independent siRNAs. WB analysis demonstrated that the knockdown was efficient with both siSmD1-1 and siSmD1-2, and the band we focused on was decreased when SmD1 was specifically knocked-down, which indicated that the switching band was indeed SmD1 (Figure 4C).
Figure 4.
SmD1 displays increased ADMA levels under EPZ015666 treatment. (A) 293T was treated with 5uM EPZ015666 for 48 hours, whole cell lysates were harvested and subjected to a co-IP assay using ADMATX2 antibody or pre-bleed serum. The samples were analyzed by Western blot and silver staining. The band that specifically occurred in ADMATX2 IP sample silver staining panel was cut out for MS analysis. (B) The sequence of the prime candidate (SmD1) is shown, and the GAR motif is highlighted in red. (C) 293T was transiently transfected with negative control siRNAs(siNC) or siSmD1, and then treated with 5uM EPZ015666 for 48 hours, whole cell lysates were harvested for western blot analysis.
3.3. Blocking both SDMA and ADMA prevents the SmD1/SMN interaction
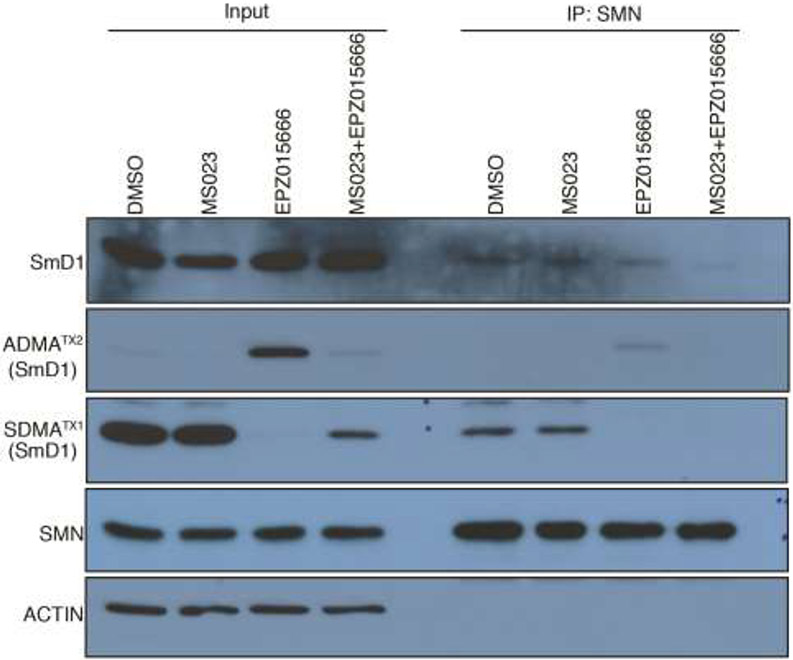
SmD1 is a small nuclear ribonucleoprotein that belongs to the snRNP core protein family. snRNPs are RNA-protein complexes that combine with pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein complex that orchestrates the splicing of pre-mRNAs. SMN plays an essential role in snRNP assembly and spliceosome formation, and a genetic defect mutation in the SMN1 gene results in a defect in snRNP dynamics and accounts for the motor neuron pathology observed in the genetic disorder spinal muscular atrophy (SMA) [28]. Through its Tudor domain, SMN binds Sm proteins in an arginine-methylation manner [29] and it recognizes both SDMA and ADMA modified splicing factors, albeit with a 3-4 fold preference for the SDMA mark [30, 31]. Thus, we are very interested in analyzing the interaction between SmD1 and SMN under PRMT5 or type I PRMT inhibition conditions, and also under combined PRMT5 and type I PRMT inhibition treatment. Even though SmD1 has been shown to interact with SMN through its SDMA marked GAR motif [32], we hypothesized that the ability of SmD1 to garner ADMA marks, in the absence of PRMT5 activity, will result in the partial rescue of the SmD1/SMN interaction. Indeed, our results showed that combined PRMT5 and type I PRMT inhibition had a more pronounced inhibitory effect on the SmD1/SMN interaction, as compared with either PRMT5 inhibition or type I PRMT inhibition alone (Figure 5). Thus, the reported synergistic effect of combined PRMT5 and type I PRMT inhibition on splicing and cell killing [17], might be due, in part, to the loss of the SmD1/SMN interaction.
Figure 5.
Combined MS023 and EPZ015666 treatment had synergistic inhibitory effect on the interaction between SmD1 and SMN. 293T was treated with PRMT inhibitors for 48 hours, whole cell lysates were harvest and subjected to Co-IP assay using anti-SMN antibody and analyzed by western blot.
4. Conclusion
In summary, we developed a panel of high-quality antibodies that recognizes MMA, ADMA and SDMA marks. We show that this panel of antibodies recognize many more substrates that do the commertially available methylarginine antibodies. Using the developed antibodies, we found a significant increase of ADMA levels of SmD1 under PRMT5 inhibition. Furthermore, combined PRMT5 and type I PRMT inhibition treatment had more pronounced inhibitory effects on the interaction between SmD1 and SMN. We propose that this finding may partially explain the synergistic effect on RNA splicing by combined PRMT5 and type I PRMT inhibition.
Highlights:
Using PEG linked antigens to develop pan-methylarginine antibodies
Successfully developed MMA, SDMA and ADMA antibody sets
SmD1 is a protein that substrate switches in the presence of PRMT5 inhibition
These antibodies can be used to evaluate target engagement of PRMT inhibitors
Acknowledgements
MTB is supported by an NIH grant (GM126421) and an NSF grant (1939814). We thank Michelle V. Gadush for assistance with the mass spec studies.
Footnotes
Disclosure
MTB is a co-founder of EpiCypher.
The other authors do not have any conflict to report
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Duan G, Walther D, The roles of post-translational modifications in the context of protein interaction networks, PLoS Comput Biol 11(2) (2015) e1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Minguez P, Parca L, Diella F, Mende DR, Kumar R, Helmer-Citterich M, Gavin AC, van Noort V, Bork P, Deciphering a global network of functionally associated post-translational modifications, Mol Syst Biol 8 (2012) 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Petrossian TC, Clarke SG, Uncovering the human methyltransferasome, Mol Cell Proteomics 10(1) (2011) M110 000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].White MF, Backer JM, Preparation and use of anti-phosphotyrosine antibodies to study structure and function of insulin receptor, Methods Enzymol 201 (1991) 65–79. [DOI] [PubMed] [Google Scholar]
- [5].Haque SJ, Wu Q, Kammer W, Friedrich K, Smith JM, Kerr IM, Stark GR, Williams BR, Receptor-associated constitutive protein tyrosine phosphatase activity controls the kinase function of JAK1, Proc Natl Acad Sci U S A 94(16) (1997) 8563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb MJ, Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs, J Biol Chem 277(42) (2002) 39379–87. [DOI] [PubMed] [Google Scholar]
- [7].Jeppesen P, Turner BM, The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression, Cell 74(2) (1993) 281–9. [DOI] [PubMed] [Google Scholar]
- [8].LeDizet M, Piperno G, Identification of an acetylation site of Chlamydomonas alpha-tubulin, Proc Natl Acad Sci U S A 84(16) (1987) 5720–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim SY, Sim CK, Zhang Q, Tang H, Brunmeir R, Pan H, Karnani N, Han W, Zhang K, Xu F, An Alternative Strategy for Pan-acetyl-lysine Antibody Generation, PLoS One 11(9) (2016) e0162528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bedford MT, Clarke SG, Protein arginine methylation in mammals: who, what, and why, Mol Cell 33(1) (2009) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rhein VF, Carroll J, Ding S, Fearnley IM, Walker JE, NDUFAF7 methylates arginine 85 in the NDUFS2 subunit of human complex I, J Biol Chem 288(46) (2013) 33016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yang Y, Hadjikyriacou A, Xia Z, Gayatri S, Kim D, Zurita-Lopez C, Kelly R, Guo A, Li W, Clarke SG, Bedford MT, PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145, Nat Commun 6 (2015) 6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG, Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues, J Biol Chem 287(11) (2012) 7859–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dhar S, Vemulapalli V, Patananan AN, Huang GL, Di Lorenzo A, Richard S, Comb MJ, Guo A, Clarke SG, Bedford MT, Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs, Sci Rep 3 (2013) 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Boisvert FM, Cote J, Boulanger MC, Richard S, A Proteomic Analysis of Arginine-methylated Protein Complexes, Mol Cell Proteomics 2(12) (2003) 1319–30. [DOI] [PubMed] [Google Scholar]
- [16].Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M, Kornhauser J, Jia X, Ren J, Beausoleil SA, Silva JC, Vemulapalli V, Bedford MT, Comb MJ, Immunoaffinity enrichment and mass spectrometry analysis of protein methylation, Molecular & cellular proteomics : MCP 13(1) (2014) 372–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fong JY, Pignata L, Goy PA, Kawabata KC, Lee SC, Koh CM, Musiani D, Massignani E, Kotini AG, Penson A, Wun CM, Shen Y, Schwarz M, Low DH, Rialdi A, Ki M, Wollmann H, Mzoughi S, Gay F, Thompson C, Hart T, Barbash O, Luciani GM, Szewczyk MM, Wouters BJ, Delwel R, Papapetrou EP, Barsyte-Lovejoy D, Arrowsmith CH, Minden MD, Jin J, Melnick A, Bonaldi T, Abdel-Wahab O, Guccione E, Therapeutic Targeting of RNA Splicing Catalysis through Inhibition of Protein Arginine Methylation, Cancer Cell 36(2) (2019) 194–209 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Musiani D, Bok J, Massignani E, Wu L, Tabaglio T, Ippolito MR, Cuomo A, Ozbek U, Zorgati H, Ghoshdastider U, Robinson RC, Guccione E, Bonaldi T, Proteomics profiling of arginine methylation defines PRMT5 substrate specificity, Sci Signal 12(575) (2019). [DOI] [PubMed] [Google Scholar]
- [19].Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S, PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins, J Biol Chem 276(35) (2001) 32971–6. [DOI] [PubMed] [Google Scholar]
- [20].Thandapani P, O'Connor TR, Bailey TL, Richard S, Defining the RGG/RG motif, Mol Cell 50(5) (2013) 613–23. [DOI] [PubMed] [Google Scholar]
- [21].Lacombe J, Rishavy MA, Berkner KL, Ferron M, VKOR paralog VKORC1L1 supports vitamin K-dependent protein carboxylation in vivo, JCI Insight 3(1) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang T, Streeter MD, Spiegel DA, Generation and characterization of antibodies against arginine-derived advanced glycation endproducts, Bioorg Med Chem Lett 25(21) (2015) 4881–4886. [DOI] [PubMed] [Google Scholar]
- [23].Streeter MD, Rowan S, Ray J, McDonald DM, Volkin J, Clark J, Taylor A, Spiegel DA, Generation and Characterization of Anti-Glucosepane Antibodies Enabling Direct Detection of Glucosepane in Retinal Tissue, ACS Chem Biol 15(10) (2020) 2655–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, Rioux N, Munchhof MJ, Jin L, Jacques SL, West KA, Lingaraj T, Stickland K, Ribich SA, Raimondi A, Scott MP, Waters NJ, Pollock RM, Smith JJ, Barbash O, Pappalardi M, Ho TF, Nurse K, Oza KP, Gallagher KT, Kruger R, Moyer MP, Copeland RA, Chesworth R, Duncan KW, A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models, Nat Chem Biol 11(6) (2015) 432–7. [DOI] [PubMed] [Google Scholar]
- [25].Eram MS, Shen Y, Szewczyk M, Wu H, Senisterra G, Li F, Butler KV, Kaniskan HU, Speed BA, Dela Sena C, Dong A, Zeng H, Schapira M, Brown PJ, Arrowsmith CH, Barsyte-Lovejoy D, Liu J, Vedadi M, Jin J, A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases, ACS Chem Biol 11(3) (2016) 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nakayama K, Szewczyk MM, Dela Sena C, Wu H, Dong A, Zeng H, Li F, de Freitas RF, Eram MS, Schapira M, Baba Y, Kunitomo M, Cary DR, Tawada M, Ohashi A, Imaeda Y, Saikatendu KS, Grimshaw CE, Vedadi M, Arrowsmith CH, Barsyte-Lovejoy D, Kiba A, Tomita D, Brown PJ, TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma, Oncotarget 9(26) (2018) 18480–18493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Noto PB, Sikorski TW, Zappacosta F, Wagner CD, Montes de Oca R, Szapacs ME, Annan RS, Liu Y, McHugh CF, Mohammad HP, Piccoli SP, Creasy CL, Identification of hnRNP-A1 as a pharmacodynamic biomarker of type I PRMT inhibition in blood and tumor tissues, Sci Rep 10(1) (2020) 22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Coady TH, Lorson CL, SMN in spinal muscular atrophy and snRNP biogenesis, Wiley Interdiscip Rev RNA 2(4) (2011) 546–64. [DOI] [PubMed] [Google Scholar]
- [29].Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R, Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein, Rna 7(11) (2001) 1531–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, Vedadi M, Min J, Pawson TJ, Blencowe BJ, Greenblatt JF, SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination, Nature 529(7584) (2016) 48–53. [DOI] [PubMed] [Google Scholar]
- [31].Liu K, Guo Y, Liu H, Bian C, Lam R, Liu Y, Mackenzie F, Rojas LA, Reinberg D, Bedford MT, Xu RM, Min J, Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30, PLoS One 7(2) (2012) e30375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G, SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets, Mol Cell 7(5) (2001) 1111–7. [DOI] [PubMed] [Google Scholar]