Abstract
Cells are constantly challenged by internal or external genotoxic assaults, which may induce a high frequency of DNA lesions, leading to genome instability. Accumulation of damaged DNA is severe or even lethal to cells and can result in abnormal proliferation that can cause cancer in multicellular organisms, aging or cell death. Eukaryotic cells have evolved a comprehensive defence system termed the DNA damage response (DDR) to monitor and remove lesions in their DNA. The DDR has been extensively studied in the budding yeast Saccharomyces cerevisiae. Emerging evidence indicates that DDR genes in the pathogenic fungus Candida albicans show functional consistency with their orthologs in S. cerevisiae, but may act through distinct mechanisms. In particular, the DDR in C. albicans appears critical for resisting DNA damage stress induced by reactive oxygen species (ROS) produced from immune cells, and this plays a vital role in pathogenicity. Therefore, DDR genes could be considered as potential targets for clinical therapies. This review summarizes the identified DNA damage checkpoint and repair genes in C. albicans based on their orthologs in S. cerevisiae, and discusses their contribution to pathogenicity in C. albicans.
Keywords: DNA damage response, DNA damage checkpoint, DNA damage repair, Candida albicans, Pathogenicity
1. Introduction
Humans are constantly exposed to fungi capable of causing a variety of diseases. Numerous medically important fungi, especially Candida species, including Candida albicans, Candida tropicalis, C. dubliniensis and the highly pathogenic fungus Candida auris, have been identified to cause mild infections, severe cutaneous infections, or life-threatening systemic infections [1]. Among them, C. albicans is the most frequently isolated agent of candidiasis, accounting for 64% of Candida infections [2], [3]. C. albicans is skilled at adapting to various environments since it can switch between either yeast or hyphae forms as well as white, grey or opaque forms, each exhibiting distinct pathogenicities [1], [4]. In addition to morphology switching, other virulence factors, such as adhesins and invasive enzymes, also contribute to the pathogenicity of C. albicans [1].
The invasion and spreading of C. albicans cells within the human body are challenged by the immune system. In particular, macrophages and neutrophils can recognize and phagocytose invading pathogens and release reactive oxygen species (ROS) or other active substances to attack the invaders’ proteins or DNA. This protective mechanism represents a key part of the first line of defence against such pathogens as C. albicans [5], [6], [7]. However, eukaryotic cells have evolved a set of responses, termed the DNA damage response (DDR), to fight against DNA damage stresses and maintain the stability of the genome. This predicts that the DDR system in Candida cells will be critical for the pathogen’s resistance to the attack from the host immune system and its survival and proliferation inside the host.
2. DNA damage response
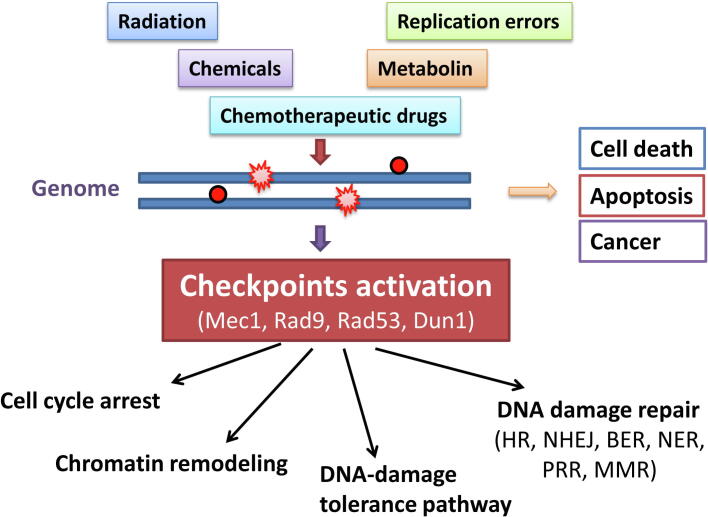
The integrity of the genome is critical for the transfer of genetic information between generations [8]. During growth and development, cells may be challenged by both extrinsic factors, such as radiation, chemicals, chemotherapeutic drugs, and intrinsic factors, such as replication errors, reactive oxygen species [9]. As a result, these stresses introduce modified bases, mismatches, intra- or inter-strand crosslinks, and single- or double-strand breaks (DSBs) to the genome [10], [11]. The accumulation of DNA lesions may result in direct cell death, apoptosis or can trigger the aberrant proliferation that characterizes cancer in mammals. To defend against such DNA-damaging events, cells have evolved a set of conserved mechanisms, termed the DNA damage response (DDR), to sense and repair damaged DNA, and thus ensure the fidelity of genetic information transfer [12]. In general, this DDR includes cell cycle checkpoints, chromatin remodeling, DNA repair and DNA-damage tolerance pathways (Fig. 1).
Fig. 1.
Schematic diagram of DNA damage response. External and internal stresses induce DNA lesions. The damaged DNA may activate the DNA damage response signaling pathway, where DNA damage checkpoints play a central role in arresting the cell cycle and mediating the DNA repair process. The unsuccessful repairing of DNA lesions may cause apoptosis, cell death or cancer. Homologous recombination (HR); Non-homologous end joining (NHEJ); Base excision repair (BER); Nucleotide excision repair (NER); Postreplication repair (PRR); Mismatch repair (MMR).
Checkpoints function in coordinating cell cycle progression and DNA repair and thus play central roles in DDR. The checkpoints have been extensively studied in the budding yeast Saccharomyces cerevisiae due to its relatively simple genetic framework compared to mammals [13], [14], [15], [16]. Several sensor proteins function to receive the signal of damaged DNA and transfer the damage signal to effector kinase. Among the identified sensor proteins, the checkpoint kinase ScRad53 is a widely studied G2/M checkpoint in S. cerevisiae. In the presence of DNA damage, ScRad53 can be activated through a set of regulatory phosphorylations by the upstream damage sensor kinases ScMec1 or ScTel1 [17], [18], [19]. Adaptor proteins ScRad9 or ScMrc1 are indispensable in mediating the recruitment of ScRad53 to DNA lesions [17], [18], [19]. Once the checkpoint is activated, it mediates a series of cellular processes, including cell cycle arrest, inhibition of origin firing, protection and restart of stalled replication forks, as well as DNA damage repair [13]. Generally, different types of DNA lesions rely on specific repair pathways. After DNA damage is corrected, the checkpoint kinase needs to be deactivated to permit resumption of the normal cell cycle.
The genetic framework of C. albicans is close to that of S. cerevisiae, and thus comparative genomics between these two fungi can help to illuminate many points. Such points include the evolutionary processes that led to the pathogenicity of C. albicans in contrast to the nonpathogenic yeast S. cerevisiae. This review summarizes current observations on DNA damage checkpoint and repair genes in C. albicans, mainly structured around the findings in S. cerevisiae.
3. DNA damage checkpoints in C. albicans
Activation of DNA damage checkpoint is the core event of DDR, which further regulates downstream cell cycle arrests, histone modifications, transcriptional changes and post-translational modifications of proteins to facilitate the repair process. In S. cerevisiae, this activation involves two highly conserved apical protein kinases ScMec1 (ATM) and ScTel1 (ATR), initiating evolutionarily conserved signal transduction cascades. Generally, ScMec1 functions to sense a wide range of DNA lesions that induces the generation of ssDNA (single-stranded DNA), whereas ScTel1 is mainly activated by DSBs [20].
For checkpoint signal initiation, ScMec1 associates with ScDdc2, which recognizes and binds RPA-coated ssDNA in DNA lesions [21]. Moreover, activation of ScMec1 relies on other factors, including the 9–1-1 complex (ScDcd1-ScMec3-ScRad17), the replication factor ScDpb11 and the DNA helicase ScDna2 [22]. Subsequently, activated ScMec1, with the participation of adaptors ScRad9 or ScMrc1, phosphorylates and activates a series of effector kinases, including ScRad53, ScChk1 and downstream ScDun1 [13]. In addition, ScMec1 (or ScTel1) also phosphorylates ScSae2, which interacts with ScRad53 and ScDun1, contributing to DNA repair and genome maintenance [23]. Generally, ScMec1 is considered as the principal sensor kinase since mec1 mutants confer comprehensive genotoxic defects, including sensitivity to hydroxyurea (HU) and methyl methane sulfonate (MMS), persistent replication fork stalling followed by irreversible fork collapse, DSB formation and cell death [14], [24], [25]. In particular, ScMec1 was found to coordinate checkpoint signaling and HR-mediated DNA repair through regulating the ScSgs1-ScTop3-ScRmi1 (STR) complex [26]. Intriguingly, unlike ScMec1 in S. cerevisiae, this checkpoint kinase is not essential in C. albicans. These differences in gene essentiality also happen to other genes, which may arise during evolutionary progress. Since C. albicans cells inhabit mammals, they have more chances, compared to the budding yeast, to be stressed by the host immune system and evolve substituted DDR signaling pathways. However, CaMec1 in C. albicans plays a similarly critical role in maintaining genome stability. The Camec1 deletion mutants show strong sensitivity to various genotoxic stresses [27]. Deleting CaMEC1 increases cell size, suggesting cell division defects that may support, at least partially, a checkpoint-related function [28]. In addition, CaMec1 was found to be involved in phosphorylation of CaRfa2, an ssDNA-binding protein, which provides evidence for the role in checkpoint activation [29].
ScTel1 acts partially redundant with ScMec1 in mediating cell cycle checkpoints to initiate DNA repair, particularly DSB repair in S. cerevisiae [19], [30], [31]. ScTel1 may participate, together with the MRX (ScMre11-ScRad50-ScXrs2) complex, in ScMec1-dependent DSB-induced checkpoint activation by increasing the efficiency of ssDNA accumulation at the ends of DSBs [32]. The ScTel1–ScMre11 complex triggers the activation of ScRad53 and its interaction with ScRad9 in mitotic cells [33]. Moreover, ScTel1 may also activate the checkpoint response to DSBs independently of ScMec1 [32]. In C. albicans, the function of CaTel1 remains uncharacterized, but sequence analysis suggests it contains a similar N terminus TAN domain to that found in ScTel1, implying DNA damage repair-related functions. However, unlike ScTel1, sequence analysis also shows that there is no PI3Kc domain at the C terminus of CaTel1 in C. albicans, suggesting a potential functional diversity of Tel1 in these two yeast species.
ScRad53, the main checkpoint effector, is classified as an essential element in DDR in S. cerevisiae [34]. In C. albicans, CaRad53 was first identified by the Wang Lab in 2007 [35]. CaRad53 can be phosphorylated by the treatment of either HU or MMS [35], [36]. Similar to the pattern of ScRad53, the phosphorylated CaRad53 in C. albicans shows a slower gel migration, supplying an effective tool to study its phosphorylation status [35], [36]. Nevertheless, according to current data, CaRAD53 is not essential in C. albicans, either the BWP17, SN148 or CEC3194 backgrounds [35], [37], [38]. Deleting CaRAD53 does, however, cause severe sensitivity to various genotoxic stresses, including MMS and HU [35]. Loss of function of CaRad53, either through deletion or point mutations, causes defects in G2/M arrest, suggesting functional similarity to ScRad53 [35]. Raphaël Loll‐Krippleber et al. further reported that CaRad53 maintains heterozygosity in C. albicans, and its deletion results in frequent aneuploidies, due to break‐induced replication/mitotic cross‐over or chromosome loss [38]. Site‐directed point mutation on CaRad53 reveals that its kinase activity and N‐terminal phosphorylation sites are crucial for its function in the resistance to genotoxic stress [38]. Therefore, CaRad53 shows critical roles in regulating the cell cycle and maintaining genome integrity in C. albicans.
Checkpoint kinase ScChk1 plays an additional role to ScRad53 in coordinating a DNA damage signal to cell cycle arrest [39]. ScChk1 shows ScMec1-dependent phosphorylation in response to DNA damage [14]. Several studies reveal that ScChk1 functions to arrest the cell cycle by regulating the stability of ScPds1 [40], [41]. However, there is no ortholog of ScCHK1 in either C. albicans, C. dubliniensis or C. parapsilosis. Although there is a protein in C. albicans named CaChk1, it is not an ortholog of ScChk1, showing only 2.8% sequence identity with ScChk1, and has a completely different function from ScChk1. The possibility could be that C. albicans cells lost the ortholog of ScCHK1 during the evolutionary progress, and other checkpoint kinases take the function of ScChk1 in response to DNA damage stress.
Adaptors CaMrc1 and CaRad9 are required for activating CaRad53 in C. albicans, but may function in response to different genotoxic stresses [35]. Deleting either CaRAD9 or CaMRC1 generates sensitivity to MMS and HU, but Carad9 mutant cells were more sensitive to MMS, whereas Camrc1 mutant cells were more sensitive to HU [42]. Consistently, deleting CaRAD9 blocks the filamentous growth induced by MMS or UV light specifically, but not that induced by HU or aphidicolin [35]. In contrast, CaMRC1 deletion impairs DNA synthesis and causes cell elongation even in the absence of external genotoxic agents, suggesting it has different roles from CaRad9 in DDR [35]. Moreover, phosphorylation of CaRad53 was significantly impaired in response to MMS in strains deleted for CaRAD9, while phosphorylation of CaRad53 was nearly blocked in response to HU treatment in cells with deletions of CaMRC1 [42]. In particular, CaRad9 and CaMrc1 mediate the phosphorylation of CaRad53 through different residues. Specifically, CaRad9 mediates the phosphorylation of CaRad53 in response to MMS at T7, S9, T24 of the TQ domain and S350 residues [42]. In contrast, CaMrc1 mediates the phosphorylation of Rad53 mainly at different residues in response to HU [42]. These observations support that adaptors CaRad9 and CaMrc1 mediate the phosphorylation of the checkpoint kinase CaRad53 in response to DNA damage or DNA replication stress via different signaling pathways.
ScDun1 is a downstream kinase of ScRad53 and regulates a wide range of cellular responses to DNA damage, including dNTP concentrations, by inhibiting the ScCrt1 repressor of RNR genes [22]. In C. albicans, CaDun1 also plays crucial roles in response to genotoxic stresses, suggesting a potential signaling pathway in the DDR [35]. However, CaDUN1 in C. albicans is not significantly overexpressed with the treatment of HU, whereas ScDUN1 is strongly upregulated in S. cerevisiae, showing a difference in response to genotoxic stress [43]. Thus, the specific function and working model of CaDun1 in C. albicans remain to be uncovered.
In general, candidate orthologs of checkpoint-related genes in budding yeast have been identified in C. albicans. The essentiality of the checkpoint elements is quite different from their orthologs in S. cerevisiae, yet they show similar functionality (Table 1). Furthermore, the signal transduction pattern of checkpoint elements in C. albicans remains to be clarified, which may be uncovered by protein interaction assays, genetic interaction assays or RNAseq assays in the future.
Table1.
Typical DNA damage checkpoint and repair genes in S. cerevisiae and C. albicans.
| Functional Group | S. cerevisiae | C. albicans | Name Description | Function Description |
|---|---|---|---|---|
| Checkpoint activation | ||||
| Sensors | DDC2/LCD1 | LCD1 | Lethal, Checkpoint-defective | Interacts physically with Mec1 and contributes to its activation; uncharacterized in C. albicans. |
| DDC1 | DDC1 | DNA Damage Checkpoint | 9–1-1 complex for Mec1 activation; uncharacterized in C. albicans. | |
| RAD17 | RAD17 | RADiation sensitive | 9–1-1 complex for Mec1 activation; uncharacterized in C. albicans. | |
| MEC3 | MEC3 | Mitosis Entry Checkpoint | 9–1-1 complex for Mec1 activation; uncharacterized in C. albicans. | |
| RAD24 | RAD24 | RADiation senstive | Loads Rad17-Mec3-Ddc1 onto DNA; uncharacterized in C. albicans. | |
| DPB11 | DPB11 | DNA Polymerase B (II) | DNA replication initiation protein; prevents accumulation of chromatin bridges by stimulating the Mec1 kinase and suppressing homologous recombination; uncharacterized in C. albicans. | |
| DNA2 | DNA2 | DNA synthesis defective | Tripartite DNA replication factor; involved in DNA repair/processing of meiotic DSBs; uncharacterized in C. albicans. | |
| MRE11 | MRE11 | Meiotic REcombination | Nuclease subunit of the MRX complex with Rad50 and Xrs2; complex functions in repair of DSBs and in telomere stability. | |
| RAD50 | RAD50 | RADiation sensitive | Subunit of MRX complex with Mre11 and Xrs2; complex is involved in processing DSBs. | |
| XRS2 | NA | X-Ray Sensitive | FHA domain-containing component of the Mre11 complex; no ortholog in C. albicans. | |
4. DNA damage checkpoint related protein phosphatases in C. albicans
The timely deactivation of checkpoint kinases, especially ScRad53, is critical for the essential resumption of cell cycle progression during recovery from or adaption to DNA damage. Ser/Thr phosphatases are assumed to play crucial roles during this progress since many checkpoint kinases are Ser/Thr kinases [38]. Up to now, phosphatase ScPph3, ScPtc2, ScPtc3 as well as ScGlc7, have been identified as necessary for the dephosphorylation of checkpoint kinase ScRad53 in S. cerevisiae [44].
4.1. Protein phosphatase 4 (PP4) complex
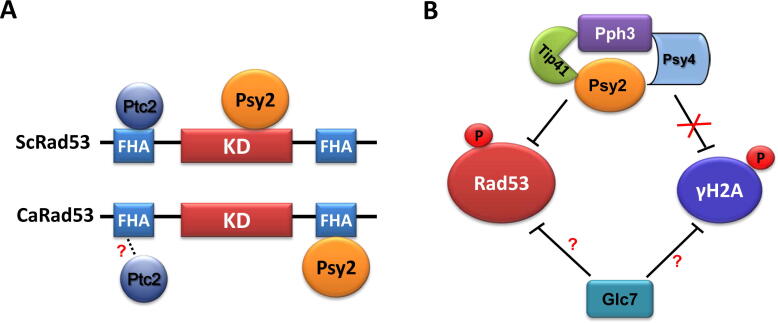
The protein phosphatase 4 (PP4) complex consists of the catalytic subunit ScPph3 and the regulatory subunits ScPsy2 and ScPsy4 in S. cerevisiae and is indispensable for the dephosphorylation of Rad53 and γH2A. Similar to the mechanism by which ScPph3 dephosphorylates checkpoint kinase ScRad53, CaPph3 and its regulator CaPsy2 are essential for the dephosphorylation of CaRad53 in C. albicans [42]. The single gene deletion or the double gene deletion of CaPPH3 and CaPSY2 both cause strong genotoxic sensitivities and persistent phosphorylation status of CaRad53 during the recovery from genotoxic stress [36]. A protein interaction assay revealed that CaPsy2 associates with both CaPph3 and CaRad53 in C. albicans, but the specific mechanism is quite different from the pattern in S. cerevisiae; ScPys2 interacts with ScPph3 in S. cerevisiae, while CaPsy2 shows no direct interaction with CaPph3 according to current data in C. albicans [36], [45]. Instead, we found CaTip41 may act as an adaptor protein for CaPph3 in C. albicans, since it interacts with both CaPph3 and CaPsy2, and contributes to deactivating CaRad53 [46]. In addition, ScPsy2 interacts with the kinase domain (KD) of ScRad53; but CaPsy2 interacts with the FHA2 domain (C terminus) of CaRad53 via its N terminus sequence in C. albicans. In consistent, blocking the interaction between CaPsy2 and CaRad53 through a point mutation of the Y33 residue at the N-terminus of CaPsy2, inhibits the dephosphorylation of CaRad53 and causes strong sensitivity to genotoxic stress in C. albicans (Fig. 2) [36]. This finding suggests a model where the direct interaction between CaPsy2 and CaRad53 is crucial for the dephosphorylation of CaRad53 in C. albicans.
Fig. 2.
Phosphatases involved in the dephosphorylation of checkpoint kinase Rad53. (A) In S. cerevisiae, ScPsy2 and ScPtc2 are required for the dephosphorylation of ScRad53; ScPsy2 interacts with the kinase domain (KD) and ScPtc2 interacts with the FHA (N terminus) domain. In C. albicans, Psy2 interacts with the FHA (C terminus) domain, but Ptc2 shows no clear interaction with Rad53. (B) Pph3 and Psy2 form a complex and play a dominant role in the dephosphorylation of Rad53 in C. albicans. Psy4 and Tip41 act as adaptors for Pph3 and Psy2. In particular, Tip41 plays an important role in the dephosphorylation of Rad53 during the recovery from DNA damage stress, while Psy4 seems dispensable for the dephosphorylation of Rad53. Unlike the pattern in S. cerevisiae, the Pph3-Psy2-Psy4 complex is not involved in the dephosphorylation of H2A. Additionally, Glc7 regulates the dephosphorylation of Rad53 both in S. cerevisiae and C. albicans, but the direct interaction between Rad53 and Glc7 is unclear. Moreover, Glc7 is involved in the dephosphorylation of γH2A in S. cerevisiae, but the role in C. albicans remains to be established. The red question mark means uncovered interaction according to current data. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Furthermore, the ScPph3-ScPsy2-ScPsy4 complex takes part in regulating the phosphorylation status of γH2AX in S. cerevisiae, whose phosphorylation at the carboxy-terminal SQE motif to create γH2AX-containing nucleosomes is the earliest mark of DSBs [47]. In contrast, CaPph3 and CaPsy2 do not show direct roles in dephosphorylating γH2AX, even though CaPph3, CaPsy2 and CaPsy4 can form a complex in C. albicans [48]. Therefore, the regulating functions on γH2AX by Pph3-Psy2-Psy4 complex appear different in these two yeast species.
4.2. Type 2C protein phosphatase (PP2C)
Functional differences in DDR are also observed in Ptc2, a type 2C protein phosphatase. In S. cerevisiae, ScPtc2 interacts with the FHA1 domain (N terminus) of ScRad53 and contributes to ScRad53 dephosphorylation together with ScPtc3 (Fig. 2-A) [49]. In C. albicans, the identified CaPtc2 shows strong sequence identity with both ScPtc2 and ScPtc3. No specific ortholog of ScPtc3 was found in this pathogen, suggesting a potential dual function of CaPtc2 in C. albicans that was distributed to the two orthologs in S. cerevisiae after the whole-genome duplications (WGD) [50]. Deletion of CaPTC2 causes moderate sensitivity to genotoxic stresses [51]. In addition, CaPtc2 offers no direct interaction with CaRad53 and no significant roles in the dephosphorylation of CaRad53 [36], [51], [52]. Therefore, CaPtc2 may not play a dominant role in the dephosphorylation of checkpoint kinase CaRad53 in C. albicans.
4.3. Protein phosphatase 1 (PP1)
In S. cerevisiae. ScGlc7, a protein phosphatase of the PP1 family, is necessary for the dephosphorylation of ScRad53 during the recovery from replication fork stalling caused by HU stress [53]. In addition, ScGlc7 is involved in the dephosphorylation of γH2AX at S129 residue and is required for the recovery from replication fork stalling caused by dNTP depletion [53]. In C. albicans, CaSds22 mediates CaRad53 dephosphorylation probably through inhibitory binding to CaGlc7 [54]. However, no direct interaction between Glc7 and Rad53 was reported in either of these two yeast species, suggesting an indirect role of Glc7 in regulating Rad53. We also can not preclude the transient interaction between Glc7 and Rad53; therefore, combined protein interaction assays may be adopted to further check the interaction between Glc7 and Rad53.
Compared to Rad53, dephosphorylation of other checkpoint kinases remains less studied in both S. cerevisiae and C. albicans. In general, protein phosphatases function differentially during the response to the genotoxic stress in these two yeast species. Here, we propose a working model of checkpoint-related phosphatases in C. albicans, according to the signaling pathway in S. cerevisiae (Fig. 2-B).
5. DNA damage repair pathways in C. albicans
5.1. Homologous recombination (HR)
In the presence of DNA damage, specific repair pathways are utilized to remove different DNA lesions. External stress like MMS may induce DNA alkylation, yielding single-strand breaks (SSBs) and also DSBs, resulting in lethality. Fixing DSBs relies on homologous recombination (HR) and also non-homologous end joining (NHEJ). HR in S. cerevisiae depends on phosphorylation controlled by ScMec1 [13]. Established HR repair proteins are ScRad51, ScRad52, ScRad54, ScRad55, ScRad57, ScRad59, ScSlx4, ScRad50, ScMre11, ScMms4, ScMus81, ScSgs1, ScDna2, ScMms22 and ScMms1[55].
ScRad51 plays a central role in this process, and its binding to ssDNA is a key step in HR [55]. In addition, ScRad52 acts to attenuate resection of the ends of DSB and stimulate strand exchange by facilitating ScRad51 binding to single-stranded DNA [56], [57]. In C. albicans, CaRAD51 appears to be involved in HR since its deletion causes increased sensitivity to compounds that cause DSBs [58]. Furthermore, deletion of CaRAD52 results in strong sensitivity to genotoxic stress and increased loss of heterozygosity in C. albicans [59], [60]. In S. cerevisiae, ScRad52 plays a more prominent role than ScRad51 in HR, where ScRad59 functions cooperatively with ScRad52 in inverted-repeat recombination by a strand-annealing mechanism in a ScRad51-independent pathway [61]. In C. albicans, CaRAD52 is epistatic to CaRAD51 for growth rate, colony morphology, viability and filamentation, suggesting the more prominent role of CaRAD52 than CaRAD51 in the repair of DSBs in C. albicans [58], [62]. Additionally, ScRad51 functions cooperatively with ScRad54, a member of the SWI/SNF family of DNA translocases, to mediate HR, where ScRad54 acts as a motor protein to translocate along dsDNA and performs several important functions in HR [63]. In C. albicans, deletion of CaRAD54 creates hypersensitivity to MMS and menadione, and causes an aberrant cell and nuclear morphology [64], but a potential cooperation with Rad51 in HR remains to be established.
In S. cerevisiae, ScRad50, ScMre11 and ScXrs2 take part in the early steps of DSB repair, which is required for both HR and NHEJ. In C. albicans, both CaRAD50 and CaMRE11 genes were identified, and their deletions cause dramatic sensitivity to UV-induced DNA damage stress and oxidative stress. As well, deleting CaRAD50 and CaMRE11 both increase genome instability [65]. But strikingly, CaXRS2 gene is missing from the genome of C. albicans, suggesting different DDR pathways in these two yeast species. These observations indicate that the DNA damage repair related functions of CaRad50 and CaMre11 are also active in C. albicans.
ScSLX4 encodes an endonuclease and acts as a scaffold with ScSlx1 in genome maintenance, supplying a role in HR [66], [67], [68]. ScSlx4 works together with PP4 phosphatase to down-regulate ScRad53 activity to allow HR machinery to repair DNA lesions [69]. In C. albicans, the Caslx4 mutant exhibits increased sensitivity to the DNA damaging agent MMS but not to the DNA replication inhibitor HU, thus showing a DNA damage related role [70]. Similarly, CaSLX4 expression is activated in a checkpoint kinase CaRad53 dependent manner during MMS-induced DNA damage, implying a checkpoint-related regulating mechanism in C. albicans [70]. In addition, ScMms22-ScMms1 complex is involved in HR and mainly functions as S-phase specific recombination-promoting factor [71]. In C. albicans, CaMMS22 is essential for the HR induced by camptothecin, while CaMMS1 is missing from the genome according to current data [72]. Moreover, the ScRad55-ScRad57 complex shows unique nonredundant functions in recombination, and mutations in any one of these components can lead to recombination defects. In C. albicans, CaRAD57 is critical for responding to MMS, HU, or ionizing radiation (IR) damage but is only essential for camptothecin-incuded damage repair in the absence of CaMMS22 [72]. However, CaRAD55 is also missing from the genome of C. albicans, according to current data. These differences in MMS1 and RAD55 may supply another evidence for the idea that DDR in C. albicans does not completely follow the pattern in S. cerevisiae. A list of the potential HR-involved genes in both S. cerevisiae and C. albicans is found in Table 1.
5.2. Non-homologous end joining (NHEJ)
NHEJ provides a relatively simple bypass strategy for repairing DSBs, allowing the free ends to be ligated without the participation of homology. In C. albicans, CaLIG4 is an orthologue to both yeast DNL4/LIG4 and human ligase IV, which are involved in NHEJ to correct DSBs [73]. CaLIG4 can complement the defect of Sclig4 in NHEJ, suggesting an NHEJ related role of CaLig4 in C. albicans [74]. However, CaLIG4 is not essential for DNA replication or for repairing DNA damage induced by ionizing radiation or UV light in C. albicans, suggesting that these lesions may be repaired primarily by HR [73], [75]. Moreover, in S. cerevisiae, ScRtt109 functions in the NHEJ pathway through the interaction with ScVps75 [76]; and in C. albicans, deletion of CaRTT109 generates strong sensitivity to DNA damage stress and elevated transcription of DDR genes [77], but the specifics of the role in NHEJ remains to be established. In addition, ScYku80, a subunit of the telomeric Ku complex, and ScDoa1, a Trp-Asp (WD)-repeat protein involved in ubiquitin-mediated protein degradation, seem to play roles in NHEJ, since their orthologs in S. cerevisiae promote NHEJ process [78]. Nevertheless, their detailed roles in NHEJ are unknown in C. albicans up to now.
5.3. Base excision repair (BER)
The base excision repair (BER) pathway repairs the damage that creates minor disturbances in the DNA helix [37]. This process is initiated by a series of glycosylases, including ScUng1, ScMag1, ScOgg1, ScNtg1, ScOgg2 and ScNtg2 in S. cerevisiae (Table 1). They recognize damaged or abnormal bases and cleave the glycosylic bond linking the base to the sugar-phosphate backbone [79]. Moreover, apurinic/apyrimidinic (AP) endonucleases, ScApn2, ScApn1, and other additional factors, ScRad27, ScPol30, ScPol4, ScPol3, ScPol2, ScCdc9, seem necessary for the BER pathway (Table 1) [80]. In C. albicans, the BER genes CaNTG1, CaAPN1 and CaOGG1 have been characterized, and deletion mutants of these genes show no change in susceptibility to DNA-damaging agents, yet ScAPN1 shows essential roles in response to MMS, UV light and gamma rays [81]. The possibility could be that BER and other DNA repair pathways have overlapping functions in correcting DNA lesions in C. albicans.
5.4. Nucleotide excision repair (NER)
The nucleotide excision repair (NER) pathway represents a key repair system to remove distorted DNA regions induced by UV light. In S. cerevisiae, the damage binding factors ScRad14, RPA, the ScRad4–ScRad23 and the ScRad7–ScRad16 complex bind to UV damaged DNA; DNA helicases ScRad3 and ScRad25/ScSsl2 create a bubble structure; two endonucleases, the ScRad1–ScRad10 complex and ScRad2 may incise the damaged DNA strand on the 5′- and 3′-side of the lesion [82]. In C. albicans, CaRad23 and CaRad4 have been identified to have redundant roles in UV induced DDR, with CaRAD4 epistatic to CaRAD23 [83]. CaRAD3 in C. albicans seems essential, and when expressed in yeast has a helicase activity on a duplex DNA substrate and complements the defects in both NER pathway and transcription [84]. Furthermore, C. albicans versions of NER genes CaRAD2 and CaRAD10 were identified, but the level of UV sensitivity of the C. albicans Carad2 and Carad10 mutants was higher than that of the Scrad2 and Scrad10 mutants [81]. This difference could be caused by different levels of exposure to the external environment, where S. cerevisiae cells have more chance to be stressed by UV light, compared to C. albicans cells.
5.5. Postreplication repair (PRR)
The DNA postreplication repair (PRR) pathway fills DNA damage-induced single-stranded gaps without removing the replication-blocking lesions [85]. In S. cerevisiae, ScRad6 forms a heterodimer with ScRad18, which is central to translesion synthesis [86], [87], [88]. PRR can be further divided into two pathways: translesion DNA synthesis mode, that involves ScRev3, ScRev7, ScRev1 and error-free mode, which includes ScUbc13, ScMms2, ScRad5, Polδ-PCNA (Proliferating Cell Nuclear Antigen) [85]. In C. albicans, the identified CaRAD6 gene can complement the defect of an Scrad6 null mutation in UV sensitivity [37], [89]. In addition, CaRad6 depletion causes UV sensitivity in C. albicans, showing a conservative DNA damage-related role [37], [89]. We have also characterized the phenotype of deleting CaRAD18, a putative transcription factor with a zinc finger DNA-binding motif, which causes increased sensitivity to the genotoxic stresses induced by MMS and HU [37]. Moreover, ScRad6-ScRad18 complex regulates monoubiquitination at the Lysine (K) 164 residue of PCNA, which can be recruited to DNA lesions in S. cerevisiae [90]; in contrast, the detailed mechanism by which CaRad6-CaRad18 regulates PPR remains unclear in C. albicans.
5.6. Mismatch repair (MMR)
The DNA mismatch repair (MMR) pathway functions to correct mismatches generated during DNA replication and recombination. In S. cerevisiae, the ScMsh2–ScMsh6 complex is responsible for the repair of mispaired bases, while the ScMsh2–ScMsh3 complex is primarily responsible for the repair of larger insertion/deletion mispairs [91], [92]. Other proteins, including ScMlh1-ScPms1, ScMlh1–ScMlh3, DNA polymerase δ, RPA, PCNA, RFC, ScExo1, ScRad27 and the DNA polymerase δ and ε associated exonucleases, are implicated in this process [91]. In C. albicans, MMR genes have not been extensively characterized; only CaMSH2 and CaPMS1 mutants have been constructed, but they cause no apparent genotoxic phenotypes [65].
6. DNA damage response allows pathogenesis in C. albicans
6.1. DNA damage response contributes to morphogenesis
Morphology of C. albicans cells is the key pathogenic character and can be affected by damage response. In cases of DNA damage, the checkpoints may be activated to arrest the cell cycle and further induce polarization/extension of Candida cells, generating a form of filamentous-like growth. For instance, genotoxic stresses like HU and MMS induce filamentous growth of Candida cells, accompanied by the activation of checkpoint CaRad53 [36], [38]. In addition, the DDR deficiency in C. albicans caused by deleting the DNA damage repair genes CaRAD52, CaMMS22 and CaMMS21, results in a change in cell morphology, creating an increased ratio of elongated and connected cells [59], [93]. However, this checkpoint-induced filamentous growth is distinct from classical hyphal forms, and the pathogenicity of this kind of filamentous form remains to be established.
6.2. DNA damage response contributes to biofilm formation
Biofilm formation is also a critical virulence factor for C. albicans cells and is closely associated with morphogenesis. Since many DDR genes influence morphogenesis, they may show critical roles in biofilm formation. For instance, CaDDR48 is a stress-associated gene involved in DDR, and its transcription is upregulated in biofilm formation. In consistent, deletion of CaDDR48 decreases biofilm formation in C. albicans [94]. Moreover, deletion of NER genes, CaRAD4 or CaRAD23, significantly increases the biofilm formation ability in C. albicans [83]. Since deleting CaRAD4 or CaRAD23 does not influence morphology or ture hyphae formation, the increased biofilm formation may attribute to altered transcription of several cell wall genes. Therefore, the potential regulation on cell wall structure or biofilm formation by DDR deserves further studying.
6.3. Positive roles of DNA damage response in pathogenicity
DDR genes may contribute to pathogenicity, which is consistent with their roles in defending against the DNA damage stress generated by host immune cells. For instance, deleting CaRTT109 increases sensitivity to H2O2 and other DNA damage stressing agents in C. albicans [77]. Moreover, CaRTT109 deletion results in significantly less pathogenicity in the murine model and increased susceptibility to killing by macrophages that can be suppressed by the NADPH oxidase (NOX) inhibitor diphenylene-iodonium chloride (DPI) [77]. This is a piece of direct evidence that the DNA damage response in the fungal pathogen is necessary for fighting against oxidative DNA damage stress. Similarly, deletion of CaRAD52 confers strong filamentous growth, but the filamentous growth does not increase the pathogenicity, with the mutant showing decreased virulence in a murine model [95]. In addition, CaLIG4, a typical NHEJ gene in C. albicans, also plays a positive role in regulating pathogenicity [73]. Additionally, CaRFX2 is a DNA damage responsive gene that regulates the expression of CaRAD6 and CaDDR48 in C. albicans, and the CaRFX2 deletion significantly reduces virulence in a murine model [96]. Those DDR genes, like CaRTT109, CaRAD52, CaLIG4, show indispensable roles in pathogenicity and cell morphogenesis in C. albicans.
Some other DDR genes, with no significant roles in morphogenesis, show clear roles in pathogenicity. Recently, we reported that NER protein CaRad23 is needed for pathogenicity in a murine model but plays no significant role in morphogenesis [83]. The mechanism may be the regulation by CaRad23 on virulence-related genes. But whether the regulation of pathogenicity mediated by CaRad23 is related to NER or other DDR pathways remains to be established.
6.4. Negative roles of DNA damage response in pathogenicity
The DDR-related genes can also show negative roles in pathogenicity. Depleting CaPph3, one of the phosphatases involved in deactivating CaRad53, increases fungal pathogenicity in the murine model [36]. A similar consequence also happens to the deletion of or even point mutation in CaPSY2 [36]. One possibility could be the increased true hyphae formation caused by deleting CaPPH3. Interestingly, the double gene deletion of CaPPH3 and CaPTC2 shows decreased pathogenicity, although the CaPTC2 single gene deletion shows no significant role in pathogenicity [52]. In budding yeast, ScPph3 acts redundantly with ScPtc2 and ScPtc3 in DSB repair [97]. Kim et al. showed that the ScPPH3 ScPTC2 ScPTC3 triple gene depletion sensitizes cells to several genotoxic drugs such as MMS, HU and cisplatin [97]. Similarly, in C. albicans, double deletion of both CaPPH3 and CaPTC2 results in dramatic genotoxic sensitivity, which may reflect the cells’ poor survival ability in the host. Therefore, multiple functions of DDR genes in pathogenicity may be observed according to the different genetic backgrounds in C. albicans.
In addition, CaTOP2, which encodes DNA topoisomerase II, also plays a negative role in pathogenicity [98]. The underlying mechanism could be the high capacity to produce true hyphae and the increased phospholipase as well as proteinase activities in the mutant [98]. By contrast, deletion of CaTOP1, encoding DNA topoisomerase I, results in reduced pathogenicity [99]. This difference in pathogenicity may suggest that CaTop1 and CaTop2 regulate virulence independent of the activity of topoisomerase.
In general, whether the negative roles of DDR genes in regulating the pathogenicity is a direct or independent role from DDR signaling pathways remains unclear. Since the deletion of PP4 genes and CaTOP2 promotes hyphae growth, it is possible that DDR genes function in pathogenicity, at least partially, via affecting morphogenesis.
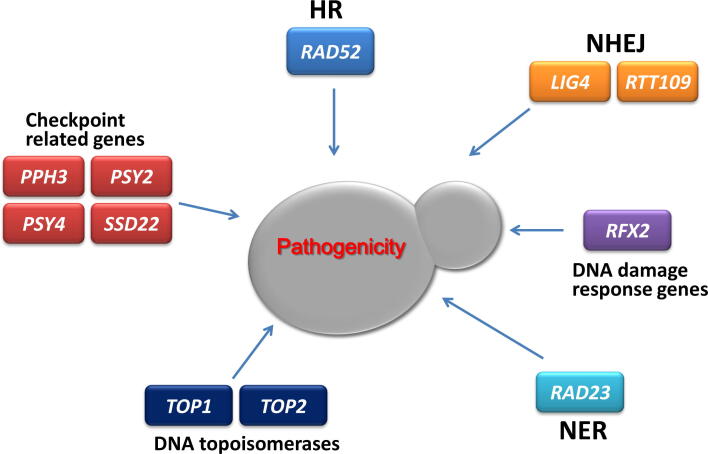
Taken together, many DNA damage response genes are indispensable for pathogenicity (Fig. 3). Some DDR genes, like CaRTT109, are needed for alleviating cellular DNA damage stress; some, like CaPPH3, are linked to morphogenesis and the remaining genes have, up to now, unclear involvement. The pathogenicity in the host is a comprehensive effect that depends on the direct and indirect effects of DDR genes. Therefore, the specific mechanisms by which DDR genes control pathogenicity need to be uncovered through the model cell and animal assays.
Fig. 3.
DNA damage response genes that are responsible for the pathogenicity of C. albicans cells. Typical genes involved in different DDR pathways are selected. RAD52, LIG4, RTT109, RFX2, RAD23 and TOP2 play positive roles in pathogenicity. TOP1, PPH3, PSY2 and PSY4 play negative roles in pathogenicity. SDS22 overexpression attenuates pathogenicity.
7. DNA damage response in other fungal pathogens
DNA damage response in many other medical fungi also plays significant roles in regulating morphology, resistance to host cells and pathogenicity. For instance, deletion of DNA damage repair genes CgRTT107, CgSGS1 or chromatin organization genes CgRSC3, CgRTT109 in C. glabrata, results in decreased virulence in the murine model of disseminated candidiasis [100]. In addition, CgRAD53, the ortholog of ScRAD53 and CaRAD53, has been identified in C. glabrata. Constitutive expression of CgRAD53 supports the growth of C. glabrata cells in the presence of MMS, HU and cobalt chloride. CgRAD53 is also involved in biofilm formation and thus shows a potential correlation with virulence in C. glabrata [101]. Furthermore, it is well known that ScRad53 and CaRad53 are phosphorylated in response to genotoxic stresses, but strikingly in C. glabrata, exposure to DNA damage stresses does not induce CgRad53 phosphorylation, suggesting alterable signaling DDR pathways. Additionally, several key protectors of genome stability are upregulated in response to DNA damage stress in S. cerevisiae but downregulated in C. glabrata [102]. Therefore, the DNA damage response pathway could be different in these eukaryotic cells, which may contribute to rapidly generating genetic change, drug resistance and pathogenicity of C. glabrata . Finally, the C. auris cells usually do not produce hyphae, but the induction of DNA damage can cause filamentation growth, supplying a potential correlation to its pathogenicity [103].
A recent study in Cryptococcus neoformans reveals that perturbation of both CnRAD53 and CnCHK1, two main checkpoints, attenuates the virulence, probably by promoting phagosome maturation within the macrophage, reducing melanin production and increasing susceptibility to oxidative stresses [104]. This observation directly supports the idea that DNA damage checkpoint kinases are linked to pathogenesis. It also supplies the underlying mechanism by which checkpoints regulate pathogenicity during host infection. More recently, BER genes CnAPN1 and CnAPN2, were reported to modulate the DDR, melanin production, tolerance to drugs and also virulence of C. neoformans [105]. In addition, loss of CnPMS1, a mismatch repair gene in C. neoformans, increases mutation rates and reduces virulence [106]. These observations suggest that direct DNA damage response genes regulate the pathogenicity of C. neoformans.
In Aspergillus fumigates, several studies demonstrate that DDR or related pathways contribute to virulence and drug resistance. For instance, AfRtt109, a canonical histone acetyltransferase, functions in response to genotoxic agents but also regulates development as well as virulence [107]. Moreover, another study suggests that genetic instability caused by deleting AfAtmA and AfAtrA (ATM and ATR homologs in A. fumigates) can confer an adaptive advantage, mainly in the intensity of voriconazole resistance acquisition [108].
In the phytopathogenic fungus Ustilago maydis, during induction of the virulence program, the cell cycle is arrested on the plant surface and it is not resumed until the fungus enters the plant [109]. During the formation of the infective hypha in this phytopathogenic fungus, two DDR kinases, UnAtr1 and UmChk1, are required to induce a G2 cell cycle arrest, which in turn is essential to facilitate the virulence program [109], [110]. These observations suggest that DDR in U. maydis is associated with pathogenicity through regulating the cell cycle.
In general, current studies in these pathogenic fungi support the idea that DDR is closely related to pathogenicity, mainly through coordinating cell cycle progression. These findings may also help to understand the underlying mechanism by which DDR regulates pathogenicity in C. albicans.
8. Discussion
The phagocytosis of pathogens by host immune cells may stimulate the release of active molecules, including ROS or nitric oxide (NO) to damage the proteins, lipids and DNA as part of the pathogen destruction program. After the invading pathogens are engulfed by host immune cells, ROS stressing can induce various DNA lesions, including modified bases, apurinic/apyrimidinic (AP) sites, SSBs or DSBs inside the pathogen cells [111]. Several studies identify the crosstalk between the DDR and oxidative stress response. In humans, oxidative stresses, including ROS induce ATM activation, link to metabolic regulation and cancer progression [111], [112]. In addition, checkpoint activation through ATR can also be triggered by oxidative stress, but independent of ATM [111]. In S. cerevisiae, ScMec1 functions in sensing, responding to and affecting the redox state of the cell [111]. And in C. albicans, the potential function of CaMec1 in oxidative stress is unknown. Instead, the checkpoint kinase CaRad53 is found to be activated in response to oxidative stress in a CaTrx1 dependent manner [5]. These observations suggest the close relationship between DDR and oxidative stress. In addition, DNA damage repair genes, such as CaAPN1, CaSRS2, CaPOL30, CaRAD57, CaRDH54 and several oxidative stress response genes in C. albicans cells are induced during the incubation with macrophages [113]. More immediately, several DDR genes, like CaRTT109, are essential for the pathogenicity in dependent of its role in fighting against ROS [77]. Taken together, the DDR in response to oxidative stress seems crucial for the resistance to host immune cells. On the other hand, DDR genes are involved in damaging host cells. A large-scale analysis of interactions between C. albicans cells with macrophages reveals that several DNA damage response genes, such as CaRAD3 and CaRAD26 are required to induce macrophage pyroptosis [114]. In general, the DDR genes seem indispensable for the pathogenicity of C. albicans cells, either for colonization in or escaping from host immune cells. But the specific mechanism for different DDR genes in pathogenicity remains to be determined.
Up to now, numerous DDR genes have been identified in C. albicans. The majority of them act following the rules of their orthologs in S. cerevisiae, but the specific mechanism can be inconsistent, especially for the checkpoint protein phosphatases. This difference may arise during the evolutionary process, probably due to the different external living environments. In contrast to the DDR-related function, some other points of DDR genes may need to be further explored in the future. In particular, the detailed genetic interaction relationships between different DDR genes, which are critical for understanding the DDR signaling pathway, remain less examined in C. albicans. The main reason could be that C. albicans cells are diploid; to get visual phenotypes, two copies of a single gene should be deleted with two sequential rounds of transformation, leading to low genetic manipulation efficiency. Recently, CRISPR/Cas9-mediated gene editing has been developed to enable facile knockout in Candida species [115], [116]. As a consequence, the large-scale genetic interaction assay may be accomplished in the future, which may help understand the specific regulating mechanism of DDR in C. albicans. Furthermore, the potential involvement of DDR genes in pathogenicity remains poorly understood. The difficulty could be that it is time- and fund-consuming to perform classical virulence assay through murine models. Nowadays, several alternative model organisms like Galleria mellonella or Caenorhabditis elegans [117], [118], which show significant advantages, can be used to test pathogenicity. Finaly, some computational systems, like the C. albicans coexpression network (CalCEN) seem useful for identifying virulence factors, which could be used for studying the importance of DDR genes [119].
Taken together, DDR signaling pathways and genes in C. albicans show significant roles in fighting against DNA damage stress, including the internal ROS stress induced by the host immune system. Therefore, they may supply the potential value for clinical therapy.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Dr Malcolm Whiteway for his constructive comments and careful edits on the manuscript. This work was supported by the National Natural Science Foundation of China to J.F. (No. 82072261), the Priority Academic Program Development of Jiangsu Higher Education Institutions, China.
References
- 1.Mayer F.L., Wilson D., Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4(2):119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papon N., Courdavault V., Clastre M., Bennett R.J., Heitman J. Emerging and emerged pathogenic Candida species: beyond the Candida albicans paradigm. PLoS Pathog. 2013;9(9):e1003550. doi: 10.1371/journal.ppat.100355010.1371/journal.ppat.1003550.g00110.1371/journal.ppat.1003550.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown G.D., Denning D.W., Gow N.A.R., Levitz S.M., Netea M.G., White T.C. Hidden killers: human fungal infections. Sci Transl Med. 2012;4(165) doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 4.Jacobsen I.D., Hube B. Candida albicans morphology: still in focus. Expert Rev Anti Infect Ther. 2017;15(4):327–330. doi: 10.1080/14787210.2017.1290524. PubMed PMID: 28152317. [DOI] [PubMed] [Google Scholar]
- 5.Dantas Ada S., Day A., Ikeh M., Kos I., Achan B., Quinn J. Oxidative stress responses in the human fungal pathogen. Candida albicans Biomol. 2015;5(1):142–165. doi: 10.3390/biom5010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poetsch A.R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput Struct Biotechnol J. 2020;18:207–219. doi: 10.1016/j.csbj.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z., Chen Y., Li B., Chen T., Tian S. Reactive oxygen species: A generalist in regulating development and pathogenicity of phytopathogenic fungi. Comput Struct Biotechnol J. 2020;18:3344–3349. doi: 10.1016/j.csbj.2020.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J.J., Lee S.Y., Miller K.M. Preserving genome integrity and function: the DNA damage response and histone modifications. Crit Rev Biochem Mol Biol. 2019;54(3):208–241. doi: 10.1080/10409238.2019.1620676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoeijmakers J.H. DNA damage, aging, and cancer. N Engl J Med. 2009;361(15):1475–1485. doi: 10.1056/NEJMra0804615. PubMed PMID: 19812404. [DOI] [PubMed] [Google Scholar]
- 10.Sancar A., Lindsey-Boltz L.A., Ünsal-Kaçmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73(1):39–85. doi: 10.1146/biochem.2004.73.issue-110.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 11.Roos W.P., Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12(9):440–450. doi: 10.1016/j.molmed.2006.07.007. PubMed PMID: 16899408. [DOI] [PubMed] [Google Scholar]
- 12.Zhou B.B., Elledge S.J. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408(6811):433–439. doi: 10.1038/35044005. PubMed PMID: 11100718. [DOI] [PubMed] [Google Scholar]
- 13.Lanz M.C., Dibitetto D., Smolka M.B. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 2019;38(18) doi: 10.15252/embj.2019101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cussiol J.R.R., Soares B.L., Oliveira F.M.B. From yeast to humans: Understanding the biology of DNA Damage Response (DDR) kinases. Genet Mol Biol. 2019;43(1 suppl 1) doi: 10.1590/1678-4685-GMB-2019-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper J.W., Elledge S.J. The DNA damage response: ten years after. Mol Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. PubMed PMID: 18082599. [DOI] [PubMed] [Google Scholar]
- 16.Longhese M.P., Foiani M., Muzi-Falconi M., Lucchini G., Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17(19):5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen J.B., Zhou Z., Siede W., Friedberg E.C., Elledge S.J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8(20):2401–2415. doi: 10.1101/gad.8.20.2401. PubMed PMID: 7958905. [DOI] [PubMed] [Google Scholar]
- 18.Lao JP, Ulrich KM, Johnson JR, Newton BW, Vashisht AA, Wohlschlegel JA, et al. The Yeast DNA Damage Checkpoint Kinase Rad53 Targets the Exoribonuclease, Xrn1. G3 (Bethesda). 2018;8(12):3931-44. doi: 10.1534/g3.118.200767. PubMed PMID: 30377154; PubMed Central PMCID: PMCPMC6288840. [DOI] [PMC free article] [PubMed]
- 19.Sanchez Y., Desany B.A., Jones W.J., Liu Q., Wang B., Elledge S.J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271(5247):357–360. doi: 10.1126/science.271.5247.357. PubMed PMID: 8553072. [DOI] [PubMed] [Google Scholar]
- 20.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn K., Lowndes N.F., Grenon M. Eukaryotic DNA damage checkpoint activation in response to double-strand breaks. Cell Mol Life Sci. 2012;69(9):1447–1473. doi: 10.1007/s00018-011-0875-3. PubMed PMID: 22083606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pardo B., Crabbe L., Pasero P. Signaling pathways of replication stress in yeast. FEMS Yeast Res. 2017;17(2) doi: 10.1093/femsyr/fow101. PubMed PMID: 27915243. [DOI] [PubMed] [Google Scholar]
- 23.Liang J., Suhandynata R.T., Zhou H. Phosphorylation of Sae2 Mediates Forkhead-associated (FHA) Domain-specific Interaction and Regulates Its DNA Repair Function. J Biol Chem. 2015;290(17):10751–10763. doi: 10.1074/jbc.M114.625293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato R., Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22(15):3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cha R.S., Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297(5581):602–606. doi: 10.1126/science.1071398. PubMed PMID: 12142538. [DOI] [PubMed] [Google Scholar]
- 26.Sanford E.J., Comstock W.J., Faça V.M., Vega S.C., Gnügge R., Symington L.S., et al. Phosphoproteomics reveals a distinctive Mec1/ATR signaling response upon DNA end hyper-resection. EMBO J. 2021;40(10) doi: 10.15252/embj.2020104566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legrand M., Chan C.L., Jauert P.A., Kirkpatrick D.T. The contribution of the S-phase checkpoint genes MEC1 and SGS1 to genome stability maintenance in Candida albicans. Fungal Genet Biol. 2011;48(8):823–830. doi: 10.1016/j.fgb.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sellam A., Chaillot J., Mallick J., Tebbji F., Richard Albert J., Cook M.A., et al. The p38/HOG stress-activated protein kinase network couples growth to division in Candida albicans. PLoS Genet. 2019;15(3):e1008052. doi: 10.1371/journal.pgen.1008052. PubMed PMID: 30921326; PubMed Central PMCID: PMCPMC6456229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J., Wang H., Wong A.-H., Zeng G., Huang Z., Wang Y., et al. Regulation of Rfa2 phosphorylation in response to genotoxic stress in Candida albicans. Mol Microbiol. 2014;94(1):141–155. doi: 10.1111/mmi.2014.94.issue-110.1111/mmi.12749. [DOI] [PubMed] [Google Scholar]
- 30.Cao J.P., Meyn M.S., Eckardt-Schupp F., Fritz E. TEL1 from Saccharomyces cerevisiae suppresses chromosome aberrations induced by ionizing radiation in ataxia-telangiectasia cells without affecting cell cycle checkpoints. Radiat Environ Biophys. 2001;40(4):309–315. doi: 10.1007/s00411-001-0125-4. PubMed PMID: 11820740. [DOI] [PubMed] [Google Scholar]
- 31.Lee M.S., Joo J.W., Choi H., Kang H.A., Kim K. Mec1 Modulates Interhomolog Crossover and Interplays with Tel1 at Post Double-Strand Break Stages. J Microbiol Biotechnol. 2020;30(3):469–475. doi: 10.4014/jmb.1909.09020. PubMed PMID: 31847509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mantiero D., Clerici M., Lucchini G., Longhese M.P. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 2007;8(4):380–387. doi: 10.1038/sj.embor.7400911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Usui T., Ogawa H., Petrini J.H. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell. 2001;7(6):1255–1266. doi: 10.1016/s1097-2765(01)00270-2. PubMed PMID: 11430828. [DOI] [PubMed] [Google Scholar]
- 34.Travesa A, Kuo D, de Bruin RA, Kalashnikova TI, Guaderrama M, Thai K, et al. DNA replication stress differentially regulates G1/S genes via Rad53-dependent inactivation of Nrm1. EMBO J. 2012;31(7):1811-22. doi: 10.1038/emboj.2012.28. PubMed PMID: 22333915; PubMed Central PMCID: PMCPMC3321207. [DOI] [PMC free article] [PubMed]
- 35.Shi Q.-M., Wang Y.-M., Zheng X.-D., Teck Ho Lee R., Wang Y., Pringle J. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol Biol Cell. 2007;18(3):815–826. doi: 10.1091/mbc.e06-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng J., Duan Y., Qin Y., Sun W., Zhuang Z., Zhu D., et al. The N-terminal pY33XL motif of CaPsy2 is critical for the function of protein phosphatase 4 in CaRad53 deactivation, DNA damage-induced filamentation and virulence in Candida albicans. Int J Med Microbiol. 2017;307(8):471–480. doi: 10.1016/j.ijmm.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Feng J., Islam A., Bean B., Feng J., Sparapani S., Shrivastava M., et al. Hof1 plays a checkpoint-related role in MMS-induced DNA damage response in Candida albicans. Mol Biol Cell. 2020;31(5):348–359. doi: 10.1091/mbc.E19-06-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loll-Krippleber R., d'Enfert C., Feri A., Diogo D., Perin A., Marcet-Houben M., et al. A study of the DNA damage checkpoint in Candida albicans: uncoupling of the functions of Rad53 in DNA repair, cell cycle regulation and genotoxic stress-induced polarized growth. Mol Microbiol. 2014;91(3):452–471. doi: 10.1111/mmi.2014.91.issue-310.1111/mmi.12471. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y., Vidanes G., Lin Y.C., Mori S., Siede W. Characterization of a Saccharomyces cerevisiae homologue of Schizosaccharomyces pombe Chk1 involved in DNA-damage-induced M-phase arrest. Mol Gen Genet. 2000;262(6):1132–1146. doi: 10.1007/pl00008656. PubMed PMID: 10660074. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez Y., Bachant J., Wang H., Hu F., Liu D., Tetzlaff M., et al. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286(5442):1166–1171. doi: 10.1126/science:286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y., Sanchez Y. Chk1 in the DNA damage response: conserved roles from yeasts to mammals. DNA Repair (Amst). 2004;3(8–9):1025–1032. doi: 10.1016/j.dnarep.2004.03.003. PubMed PMID: 15279789. [DOI] [PubMed] [Google Scholar]
- 42.Yao G., Wan J., Liu Q., Mu C., Wang Y., Sang J. Characterization of Pph3-mediated dephosphorylation of Rad53 during methyl methanesulfonate-induced DNA damage repair in Candida albicans. Biochem J. 2017;474(7):1293–1306. doi: 10.1042/BCJ20160889. PubMed PMID: 28183985. [DOI] [PubMed] [Google Scholar]
- 43.Bachewich C., Nantel A., Whiteway M. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol Microbiol. 2005;57(4):942–959. doi: 10.1111/j.1365-2958.2005.04727.x. PubMed PMID: 16091036. [DOI] [PubMed] [Google Scholar]
- 44.Travesa A., Duch A., Quintana D.G. Distinct phosphatases mediate the deactivation of the DNA damage checkpoint kinase Rad53. J Biol Chem. 2008;283(25):17123–17130. doi: 10.1074/jbc.M801402200. PubMed PMID: 18441009. [DOI] [PubMed] [Google Scholar]
- 45.O'Neill B.M., Szyjka S.J., Lis E.T., Bailey A.O., Yates J.R., Aparicio O.M., et al. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc Natl Acad Sci U S A. 2007;104(22):9290–9295. doi: 10.1073/pnas.0703252104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng J., Duan Y., Sun W., Qin Y., Zhuang Z., Zhu D., et al. CaTip41 regulates protein phosphatase 2A activity, CaRad53 deactivation and the recovery of DNA damage-induced filamentation to yeast form in Candida albicans. FEMS Yeast Res. 2016;16(2):fow009. doi: 10.1093/femsyr/fow009. [DOI] [PubMed] [Google Scholar]
- 47.Keogh M.C., Kim J.A., Downey M., Fillingham J., Chowdhury D., Harrison J.C., et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439(7075):497–501. doi: 10.1038/nature04384. PubMed PMID: 16299494. [DOI] [PubMed] [Google Scholar]
- 48.Sun L.L., Li W.J., Wang H.T., Chen J., Deng P., Wang Y., et al. Protein phosphatase Pph3 and its regulatory subunit Psy2 regulate Rad53 dephosphorylation and cell morphogenesis during recovery from DNA damage in Candida albicans. Eukaryot Cell. 2011;10(11):1565–1573. doi: 10.1128/EC.05042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leroy C., Lee S.E., Vaze M.B., Ochsenbien F., Guerois R., Haber J.E., et al. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol Cell. 2003;11(3):827–835. doi: 10.1016/S1097-2765(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 50.Gordon J.L., Byrne K.P., Wolfe K.H., Zhang J. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5(5):e1000485. doi: 10.1371/journal.pgen.1000485. PubMed PMID: 19436716; PubMed Central PMCID: PMCPMC2675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng J., Zhao J., Li J., Zhang L., Jiang L. Functional characterization of the PP2C phosphatase CaPtc2p in the human fungal pathogen Candida albicans. Yeast. 2010;27(9):753–764. doi: 10.1002/yea.1778. PubMed PMID: 20641018. [DOI] [PubMed] [Google Scholar]
- 52.Feng J., Shan A., Hu J., Cao Z., Lv R., Feng J. Genetic interaction between Ptc2 and protein phosphatase 4 (PP4) in the regulation of DNA damage response and virulence in Candida albicans. FEMS Yeast Res. 2019;19(8) doi: 10.1093/femsyr/foz075. PubMed PMID: 31644792. [DOI] [PubMed] [Google Scholar]
- 53.Bazzi M., Mantiero D., Trovesi C., Lucchini G., Longhese M.P. Dephosphorylation of gamma H2A by Glc7/protein phosphatase 1 promotes recovery from inhibition of DNA replication. Mol Cell Biol. 2010;30(1):131–145. doi: 10.1128/MCB.01000-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao G., Wan J., Mu C., Liu Q., Wang Y., Sang J. Sds22 participates in Glc7 mediated Rad53 dephosphorylation in MMS-induced DNA damage in Candida albicans. Fungal Genet Biol. 2016;93:50–61. doi: 10.1016/j.fgb.2016.06.003. PubMed PMID: 27328280. [DOI] [PubMed] [Google Scholar]
- 55.Aylon Y., Kupiec M. New insights into the mechanism of homologous recombination in yeast. Mutat Res. 2004;566(3):231–248. doi: 10.1016/j.mrrev.2003.10.001. PubMed PMID: 15082239. [DOI] [PubMed] [Google Scholar]
- 56.Lim G, Chang Y, Huh WK. Phosphoregulation of Rad51/Rad52 by CDK1 functions as a molecular switch for cell cycle-specific activation of homologous recombination. Sci Adv. 2020;6(6):eaay2669. doi: 10.1126/sciadv.aay2669. PubMed PMID: 32083180; PubMed Central PMCID: PMCPMC7007264. [DOI] [PMC free article] [PubMed]
- 57.Pohl T.J., Nickoloff J.A. Rad51-independent interchromosomal double-strand break repair by gene conversion requires Rad52 but not Rad55, Rad57, or Dmc1. Mol Cell Biol. 2008;28(3):897–906. doi: 10.1128/MCB.00524-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Prieto F., Gomez-Raja J., Andaluz E., Calderone R., Larriba G. Role of the homologous recombination genes RAD51 and RAD59 in the resistance of Candida albicans to UV light, radiomimetic and anti-tumor compounds and oxidizing agents. Fungal Genet Biol. 2010;47(5):433–445. doi: 10.1016/j.fgb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andaluz E., Ciudad T., Gomez-Raja J., Calderone R., Larriba G. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol Microbiol. 2006;59(5):1452–1472. doi: 10.1111/j.1365-2958.2005.05038.x. PubMed PMID: 16468988. [DOI] [PubMed] [Google Scholar]
- 60.Andaluz E, Bellido A, Gomez-Raja J, Selmecki A, Bouchonville K, Calderone R, et al. Rad52 function prevents chromosome loss and truncation in Candida albicans. Mol Microbiol. 2011;79(6):1462-82. doi: 10.1111/j.1365-2958.2011.07532.x. PubMed PMID: 21272099; PubMed Central PMCID: PMCPMC3564047. [DOI] [PMC free article] [PubMed]
- 61.Mott C., Symington L.S. RAD51-independent inverted-repeat recombination by a strand-annealing mechanism. DNA Repair (Amst). 2011;10(4):408–415. doi: 10.1016/j.dnarep.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bellido A., Andaluz E., Gomez-Raja J., Alvarez-Barrientos A., Larriba G. Genetic interactions among homologous recombination mutants in Candida albicans. Fungal Genet Biol. 2015;74:10–20. doi: 10.1016/j.fgb.2014.10.016. PubMed PMID: 25445312. [DOI] [PubMed] [Google Scholar]
- 63.Ceballos S.J., Heyer W.D. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. BBA. 2011;1809(9):509–523. doi: 10.1016/j.bbagrm.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoot S.J., Zheng X., Potenski C.J., White T.C., Klein H.L. The role of Candida albicans homologous recombination factors Rad54 and Rdh54 in DNA damage sensitivity. BMC Microbiol. 2011;11:214. doi: 10.1186/1471-2180-11-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Legrand M., Chan C.L., Jauert P.A., Kirkpatrick D.T. Role of DNA mismatch repair and double-strand break repair in genome stability and antifungal drug resistance in Candida albicans. Eukaryot Cell. 2007;6(12):2194–2205. doi: 10.1128/EC.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muñoz I.M., Hain K., Déclais A.-C., Gardiner M., Toh G.W., Sanchez-Pulido L., et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35(1):116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 67.Muñoz-Galván S., Tous C., Blanco M.G., Schwartz E.K., Ehmsen K.T., West S.C., et al. Distinct roles of Mus81, Yen1, Slx1-Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol Cell Biol. 2012;32(9):1592–1603. doi: 10.1128/MCB.00111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fricke W.M., Brill S.J. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17(14):1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jablonowski CM, Cussiol JR, Oberly S, Yimit A, Balint A, Kim T, et al. Termination of Replication Stress Signaling via Concerted Action of the Slx4 Scaffold and the PP4 Phosphatase. Genetics. 2015;201(3):937-49. doi: 10.1534/genetics.115.181479. PubMed PMID: 26362319; PubMed Central PMCID: PMCPMC4649662. [DOI] [PMC free article] [PubMed]
- 70.Wang Y., Wang N., Liu J., Zhang Y., Li X., Han Q. Homolog of Saccharomyces cerevisiae SLX4 is required for cell recovery from MMS-induced DNA damage in Candida albicans. FEMS Yeast Res. 2021;21(2) doi: 10.1093/femsyr/foab010. PubMed PMID: 33606011. [DOI] [PubMed] [Google Scholar]
- 71.Duro E., Vaisica J.A., Brown G.W., Rouse J. Budding yeast Mms22 and Mms1 regulate homologous recombination induced by replisome blockage. DNA Repair (Amst). 2008;7(5):811–818. doi: 10.1016/j.dnarep.2008.01.007. PubMed PMID: 18321796. [DOI] [PubMed] [Google Scholar]
- 72.Yan L, Xiong J, Lu H, Lv QZ, Ma QY, Cote P, et al. The Role of Mms22p in DNA Damage Response in Candida albicans. G3 (Bethesda). 2015;5(12):2567-78. doi: 10.1534/g3.115.021840. PubMed PMID: 26438292; PubMed Central PMCID: PMCPMC4683630. [DOI] [PMC free article] [PubMed]
- 73.Andaluz E., Calderone R., Reyes G., Larriba G. Phenotypic analysis and virulence of Candida albicans LIG4 mutants. Infect Immun. 2001;69(1):137–147. doi: 10.1128/IAI.69.01.137-147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andaluz E., Ciudad A., Rubio Coque J., Calderone R., Larriba G. Cell cycle regulation of a DNA ligase-encoding gene (CaLIG4) from Candida albicans. Yeast. 1999;15(12):1199–1210. doi: 10.1002/(SICI)1097-0061(19990915)15:12<1199::AID-YEA447>3.0.CO;2-S. PubMed PMID: 10487922. [DOI] [PubMed] [Google Scholar]
- 75.Andaluz E., Ciudad T., Larriba G. An evaluation of the role of LIG4 in genomic instability and adaptive mutagenesis in Candida albicans. FEMS Yeast Res. 2002;2(3):341–348. doi: 10.1016/S1567-1356(02)00094-6. PubMed PMID: 12702284. [DOI] [PubMed] [Google Scholar]
- 76.Jessulat M., Alamgir M., Salsali H., Greenblatt J., Xu J., Golshani A. Interacting proteins Rtt109 and Vps75 affect the efficiency of non-homologous end-joining in Saccharomyces cerevisiae. Arch Biochem Biophys. 2008;469(2):157–164. doi: 10.1016/j.abb.2007.11.001. PubMed PMID: 18036332. [DOI] [PubMed] [Google Scholar]
- 77.Lopes da Rosa J, Boyartchuk VL, Zhu LJ, Kaufman PD. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc Natl Acad Sci U S A. 2010;107(4):1594-9. doi: 10.1073/pnas.0912427107. PubMed PMID: 20080646; PubMed Central PMCID: PMCPMC2824404. [DOI] [PMC free article] [PubMed]
- 78.Kunze D., MacCallum D., Odds F.C., Hube B. Multiple functions of DOA1 in Candida albicans. Microbiology (Reading) 2007;153(Pt 4):1026–1041. doi: 10.1099/mic.0.2006/002741-0. PubMed PMID: 17379712. [DOI] [PubMed] [Google Scholar]
- 79.Kelley M.R., Kow Y.W., Wilson D.M., 3rd. Disparity between DNA base excision repair in yeast and mammals: translational implications. Cancer Res. 2003;63(3):549–554. PubMed PMID: 12566294. [PubMed] [Google Scholar]
- 80.Memisoglu A., Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451(1–2):39–51. doi: 10.1016/s0027-5107(00)00039-7. PubMed PMID: 10915864. [DOI] [PubMed] [Google Scholar]
- 81.Legrand M., Chan C.L., Jauert P.A., Kirkpatrick D.T. Analysis of base excision and nucleotide excision repair in Candida albicans. Microbiology (Reading) 2008;154(Pt 8):2446–2456. doi: 10.1099/mic.0.2008/017616-0. PubMed PMID: 18667577. [DOI] [PubMed] [Google Scholar]
- 82.Prakash S., Prakash L. Nucleotide excision repair in yeast. Mutat Res. 2000;451(1–2):13–24. doi: 10.1016/s0027-5107(00)00037-3. PubMed PMID: 10915862. [DOI] [PubMed] [Google Scholar]
- 83.Feng J, Yao S, Dong Y, Hu J, Whiteway M, Feng J. Nucleotide Excision Repair Protein Rad23 Regulates Cell Virulence Independent of Rad4 in Candida albicans. mSphere. 2020;5(1). doi: 10.1128/mSphere.00062-20. PubMed PMID: 32075883; PubMed Central PMCID: PMCPMC7031613. [DOI] [PMC free article] [PubMed]
- 84.Seong K.M., Lee S.H., Kim H.D., Lee C.H., Youn H., Youn BuHyun, et al. Expression, purification, and characterization of putative Candida albicans Rad3, the product of orf19.7119. Biochemistry (Mosc) 2011;76(6):666–676. doi: 10.1134/S0006297911060071. [DOI] [PubMed] [Google Scholar]
- 85.Broomfield S., Hryciw T., Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res. 2001;486(3):167–184. doi: 10.1016/s0921-8777(01)00091-x. PubMed PMID: 11459630. [DOI] [PubMed] [Google Scholar]
- 86.Wu Z., He M.H., Zhang L.L., Liu J., Zhang Q.D., Zhou J.Q. Rad6-Bre1 mediated histone H2Bub1 protects uncapped telomeres from exonuclease Exo1 in Saccharomyces cerevisiae. DNA Repair (Amst) 2018;72:64–76. doi: 10.1016/j.dnarep.2018.09.007. PubMed PMID: 30254011. [DOI] [PubMed] [Google Scholar]
- 87.Bailly V., Lauder S., Prakash S., Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272(37):23360–23365. doi: 10.1074/jbc.272.37.23360. PubMed PMID: 9287349. [DOI] [PubMed] [Google Scholar]
- 88.Game J.C., Chernikova S.B. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair (Amst) 2009;8(4):470–482. doi: 10.1016/j.dnarep.2009.01.007. PubMed PMID: 19230796. [DOI] [PubMed] [Google Scholar]
- 89.Leng P., Sudbery P.E., Brown A.J. Rad6p represses yeast-hypha morphogenesis in the human fungal pathogen Candida albicans. Mol Microbiol. 2000;35(5):1264–1275. doi: 10.1046/j.1365-2958.2000.01801.x. PubMed PMID: 10712706. [DOI] [PubMed] [Google Scholar]
- 90.Lee KY, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol Cells. 2008;26(1):5-11. PubMed PMID: 18525240; PubMed Central PMCID: PMCPMC3518309. [PMC free article] [PubMed]
- 91.Kolodner R.D., Marsischky G.T. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9(1):89–96. doi: 10.1016/s0959-437x(99)80013-6. PubMed PMID: 10072354. [DOI] [PubMed] [Google Scholar]
- 92.Liu D., Keijzers G., Rasmussen L.J. DNA mismatch repair and its many roles in eukaryotic cells. Mutat Res Rev Mutat Res. 2017;773:174–187. doi: 10.1016/j.mrrev.2017.07.001. PubMed PMID: 28927527. [DOI] [PubMed] [Google Scholar]
- 93.Islam A, Tebbji F, Mallick J, Regan H, Dumeaux V, Omran RP, et al. Mms21: A Putative SUMO E3 Ligase in Candida albicans That Negatively Regulates Invasiveness and Filamentation, and Is Required for the Genotoxic and Cellular Stress Response. Genetics. 2019;211(2):579-95. doi: 10.1534/genetics.118.301769. PubMed PMID: 30530734; PubMed Central PMCID: PMCPMC6366906. [DOI] [PMC free article] [PubMed]
- 94.Cleary I.A., MacGregor N.B., Saville S.P., Thomas D.P. Investigating the function of Ddr48p in Candida albicans. Eukaryot Cell. 2012;11(6):718–724. doi: 10.1128/EC.00107-12. PubMed PMID: 22523369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chauhan N., Ciudad T., Rodriguez-Alejandre A., Larriba G., Calderone R., Andaluz E. Virulence and karyotype analyses of rad52 mutants of Candida albicans: regeneration of a truncated chromosome of a reintegrant strain (rad52/RAD52) in the host. Infect Immun. 2005;73(12):8069–8078. doi: 10.1128/IAI.73.12.8069-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hao B., Clancy C.J., Cheng S., Raman S.B., Iczkowski K.A., Nguyen M.H. Candida albicans RFX2 encodes a DNA binding protein involved in DNA damage responses, morphogenesis, and virulence. Eukaryot Cell. 2009;8(4):627–639. doi: 10.1128/EC.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim J.A., Hicks W.M., Li J., Tay S.Y., Haber J.E. Protein phosphatases pph3, ptc2, and ptc3 play redundant roles in DNA double-strand break repair by homologous recombination. Mol Cell Biol. 2011;31(3):507–516. doi: 10.1128/MCB.01168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zheng H., Yu Y.S. TOP2 gene is involved in the pathogenicity of Candida albicans. Mol Cell Biochem. 2012;364(1–2):45–52. doi: 10.1007/s11010-011-1203-9. PubMed PMID: 22203422. [DOI] [PubMed] [Google Scholar]
- 99.Jiang W., Gerhold D., Kmiec E.B., Hauser M., Becker J.M., Koltin Y. The topoisomerase I gene from Candida albicans. Microbiology (Reading). 1997;143(Pt 2):377–386. doi: 10.1099/00221287-143-2-377. PubMed PMID: 9043115. [DOI] [PubMed] [Google Scholar]
- 100.Rai M.N., Balusu S., Gorityala N., Dandu L., Kaur R., Cowen L.E. Functional genomic analysis of Candida glabrata-macrophage interaction: role of chromatin remodeling in virulence. PLoS Pathog. 2012;8(8):e1002863. doi: 10.1371/journal.ppat.1002863. PubMed PMID: 22916016; PubMed Central PMCID: PMCPMC3420920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gupta P., Meena R.C., Kumar N. Functional characterization of Candida glabrata ORF, CAGL0M02233g for its role in stress tolerance and virulence. Microb Pathog. 2020;149 doi: 10.1016/j.micpath.2020.104469. PubMed PMID: 32890635. [DOI] [PubMed] [Google Scholar]
- 102.Shor E, Garcia-Rubio R, DeGregorio L, Perlin DS. A Noncanonical DNA Damage Checkpoint Response in a Major Fungal Pathogen. mBio. 2020;11(6). doi: 10.1128/mBio.03044-20. PubMed PMID: 33323516; PubMed Central PMCID: PMCPMC7773997. [DOI] [PMC free article] [PubMed]
- 103.Bravo Ruiz G, Ross ZK, Gow NAR, Lorenz A. Pseudohyphal Growth of the Emerging Pathogen Candida auris Is Triggered by Genotoxic Stress through the S Phase Checkpoint. mSphere. 2020;5(2). doi: 10.1128/mSphere.00151-20. PubMed PMID: 32161147; PubMed Central PMCID: PMCPMC7067593. [DOI] [PMC free article] [PubMed]
- 104.Jung KW, Lee Y, Huh EY, Lee SC, Lim S, Bahn YS. Rad53- and Chk1-Dependent DNA Damage Response Pathways Cooperatively Promote Fungal Pathogenesis and Modulate Antifungal Drug Susceptibility. mBio. 2019;10(1). doi: 10.1128/mBio.01726-18. PubMed PMID: 30602579; PubMed Central PMCID: PMCPMC6315099. [DOI] [PMC free article] [PubMed]
- 105.Oliveira RKM, Hurtado FA, Gomes PH, Puglia LL, Ferreira FF, Ranjan K, et al. Base Excision Repair AP-Endonucleases-Like Genes Modulate DNA Damage Response and Virulence of the Human Pathogen Cryptococcus neoformans. J Fungi (Basel). 2021;7(2). doi: 10.3390/jof7020133. PubMed PMID: 33673204; PubMed Central PMCID: PMCPMC7917787. [DOI] [PMC free article] [PubMed]
- 106.Boyce KJ, Wang Y, Verma S, Shakya VPS, Xue C, Idnurm A. Mismatch Repair of DNA Replication Errors Contributes to Microevolution in the Pathogenic Fungus Cryptococcus neoformans. mBio. 2017;8(3). doi: 10.1128/mBio.00595-17. PubMed PMID: 28559486; PubMed Central PMCID: PMCPMC5449657. [DOI] [PMC free article] [PubMed]
- 107.Zhang Y., Fan J., Ye J., Lu L. The fungal-specific histone acetyltransferase Rtt109 regulates development, DNA damage response, and virulence in Aspergillus fumigatus. Mol Microbiol. 2021;115(6):1191–1206. doi: 10.1111/mmi.v115.610.1111/mmi.14665. [DOI] [PubMed] [Google Scholar]
- 108.Dos Reis TF, Silva LP, de Castro PA, Almeida de Lima PB, do Carmo RA, Marini MM, et al. The Influence of Genetic Stability on Aspergillus fumigatus Virulence and Azole Resistance. G3 (Bethesda). 2018;8(1):265-78. doi: 10.1534/g3.117.300265. PubMed PMID: 29150592; PubMed Central PMCID: PMCPMC5765354. [DOI] [PMC free article] [PubMed]
- 109.Mielnichuk N., Sgarlata C., Perez-Martin J. A role for the DNA-damage checkpoint kinase Chk1 in the virulence program of the fungus Ustilago maydis. J Cell Sci. 2009;122(Pt 22):4130–4140. doi: 10.1242/jcs.052233. PubMed PMID: 19861497. [DOI] [PubMed] [Google Scholar]
- 110.Tenorio-Gómez M., de Sena-Tomás C., Pérez-Martín J., Muzi-Falconi M. MRN- and 9–1-1-Independent Activation of the ATR-Chk1 Pathway during the Induction of the Virulence Program in the Phytopathogen Ustilago maydis. PLoS ONE. 2015;10(9):e0137192. doi: 10.1371/journal.pone.0137192. PubMed PMID: 26367864; PubMed Central PMCID: PMCPMC4573213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choi J.E., Chung W.H. Functional interplay between the oxidative stress response and DNA damage checkpoint signaling for genome maintenance in aerobic organisms. J Microbiol. 2020;58(2):81–91. doi: 10.1007/s12275-020-9520-x. PubMed PMID: 31875928. [DOI] [PubMed] [Google Scholar]
- 112.Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T. ATM activation by oxidative stress. Science. 2010;330(6003):517–521. doi: 10.1126/science.1192912. PubMed PMID: 20966255. [DOI] [PubMed] [Google Scholar]
- 113.Lorenz M.C., Bender J.A., Fink G.R. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3(5):1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O'Meara TR, Duah K, Guo CX, Maxson ME, Gaudet RG, Koselny K, et al. High-Throughput Screening Identifies Genes Required for Candida albicans Induction of Macrophage Pyroptosis. mBio. 2018;9(4). doi: 10.1128/mBio.01581-18. PubMed PMID: 30131363; PubMed Central PMCID: PMCPMC6106084. [DOI] [PMC free article] [PubMed]
- 115.Vyas V.K., Barrasa M.I., Fink G.R. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv. 2015;1(3) doi: 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Min K, Ichikawa Y, Woolford CA, Mitchell AP. Candida albicans Gene Deletion with a Transient CRISPR-Cas9 System. mSphere. 2016;1(3). doi: 10.1128/mSphere.00130-16. PubMed PMID: 27340698; PubMed Central PMCID: PMCPMC4911798. [DOI] [PMC free article] [PubMed]
- 117.Elkabti AB, Issi L, Rao RP. Caenorhabditis elegans as a Model Host to Monitor the Candida Infection Processes. J Fungi (Basel). 2018;4(4). doi: 10.3390/jof4040123. PubMed PMID: 30405043; PubMed Central PMCID: PMCPMC6309157. [DOI] [PMC free article] [PubMed]
- 118.Jacobsen I.D. Galleria mellonella as a model host to study virulence of Candida. Virulence. 2014;5(2):237–239. doi: 10.4161/viru.27434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O'Meara TR, O'Meara MJ. DeORFanizing Candida albicans Genes using Coexpression. mSphere. 2021;6(1). doi: 10.1128/mSphere.01245-20. PubMed PMID: 33472984; PubMed Central PMCID: PMCPMC7845621. [DOI] [PMC free article] [PubMed]