Ectatosticta spiders from the Qinghai-Tibet Plateau were studied. Multilocus molecular and morphological data identified 16 putative species, including seven new species: Ectatosticta wenshu Lin & S. Li sp. nov., Ectatosticta baima Lin & S. Li sp. nov., Ectatosticta helii Lin & S. Li sp. nov., Ectatosticta shaseng Lin & S. Li sp. nov., Ectatosticta puxian Lin & S. Li sp. nov., Ectatosticta qingshi Lin & S. Li sp. nov., and Ectatosticta baixiang Lin & S. Li sp. nov. This increase in the number of Ectatosticta species from a single species in 2008 to 16 in the current study highlights the Linnean shortfall in China. The previously known distribution of Ectatosticta spiders was from one locality in Shaanxi and is now expanded to the east and south of the Qinghai-Tibet Plateau.
Taxonomists have described 1438769 species since binomial nomenclature began in 1753 (Mora et al., 2011), with an average of 5 576 species described each year for the past 258 years. Recent technological advances have accelerated the discovery of new taxa. However, the predicted total number of terrestrial species is 8 750 000 and marine species is 2 210 000 (Mora et al., 2011). It would take more than 1 000 years to document the remaining predicted species on the planet, even if we were able to describe 10 000 new species each year using advanced technologies. Clearly, knowledge of species diversity remains inadequate as most species are still not formally described (i.e., Linnean shortfall) and because geographical distributions of most species are poorly understood and usually contain many gaps (i.e., Wallacean shortfall) (Lomolino, 2004; Whittaker et al., 2005).
Spiders of the genus Ectatosticta Simon, 1892 (family Hypochilidae Marx, 1888) are an exemplary example of these shortfalls in China. Only two genera of hypochilids are known worldwide, i.e., Hypochilus Marx, 1888, which is endemic to the US (from California to New Mexico and the Appalachian Mountains), and Ectatosticta, which is endemic to China. Although 10 species of Hypochilus are now recognized in the US, the number of hypochilid species in China was limited to a single species, Ectatosticta davidi Simon, 1889, for 120 years due to Linnean and Wallacean shortfalls, with a second species not discovered until 2009—Ectatosticta deltshevi Platnick & Jäger, 2009. These two species are known from the northern Qinghai-Tibet Plateau. However, recent discoveries ( Lin & Li, 2020, 2021, Wang et al., 2021) have confirmed considerable Ectatosticta diversity in the southern Qinghai-Tibet Plateau, with higher species richness and wider distribution than that of Hypochilus in the USA. Thus, we conducted an in-depth study ofEctatosticta to challenge the Wallacean and Linnean shortfalls.
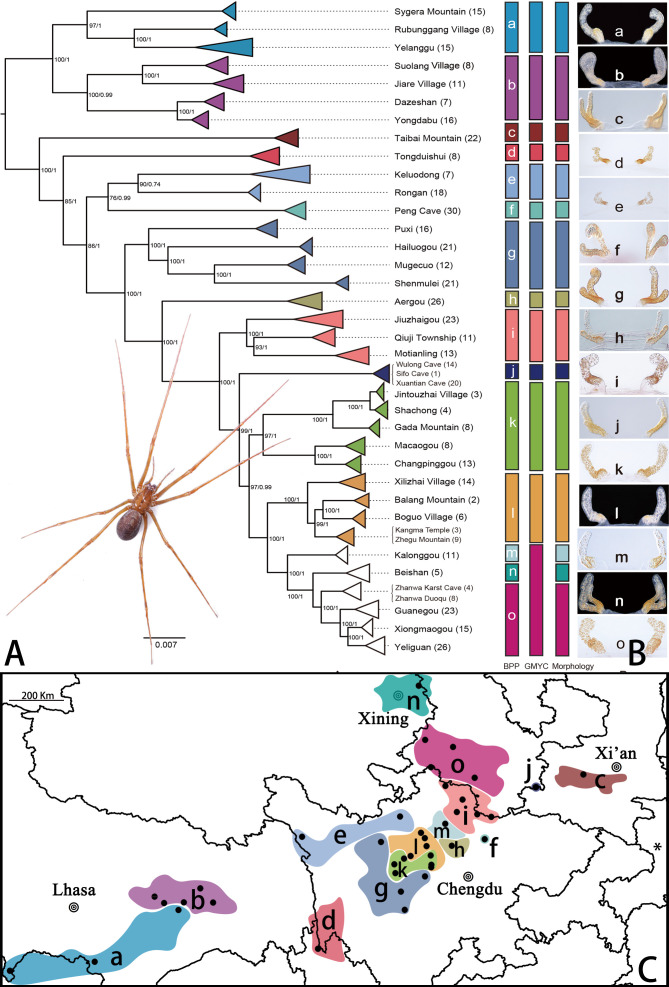
In total, 505 Ectatosticta specimens from 40 localities on the Qinghai-Tibet Plateau were studied, and seven genetic makers were analyzed to produce a robust phylogeny. Based on multilocus molecular and morphological data, we identified 16 putative species, i.e., Ectatosticta deltshevi Platnick & Jäger, 2009, Ectatosticta wenshu Lin & S. Li sp. nov., Ectatosticta davidi (Simon, 1889), Ectatosticta yukuni Lin & S. Li, 2021, Ectatosticta rulai Lin & S. Li, 2021, Ectatosticta baima Lin & S. Li sp. nov., Ectatosticta helii Lin & S. Li sp. nov., Ectatosticta shaseng Lin & S. Li sp. nov., Ectatosticta wukong Lin & S. Li, 2020, Ectatosticta puxian Lin & S. Li sp. nov., Ectatosticta bajie Lin & S. Li, 2021, Ectatosticta xuanzang Lin & S. Li, 2020, Ectatosticta baixiang Lin & S. Li sp. nov., Ectatosticta dapeng Lin & S. Li, 2021, Ectatosticta qingshi Lin & S. Li sp. nov., andEctatosticta shennongjiaensis Wang, Zhao, Irfan & Zhang, 2021 ( Figure 1). New synonyms are proposed based on molecular analysis and morphological evidence, including Ectatosticta nyingchiensis Wang et al., 2021 syn. nov. as a junior synonym of Ectatosticta dapeng;Ectatosticta pingwuensis Wang et al., 2021 syn. nov. and Ectatosticta songpanensis Wang et al., 2021 syn. nov. as junior synonyms of Ectatosticta rulai. For detailed morphological descriptions, diagnoses, illustrations, and identification key of all Ectatosticta species, please see the online Supplementary Material. The actual distribution of Ectatosticta spiders suggests that they are a psychrophilic species, mainly distributed east and south of the Qinghai-Tibet Plateau at an altitude of 2 000–4 000 m a.s.l. with strict temperature and humidity requirements.
Figure 1.
Phylogenetic tree and distribution of Ectatosticta species
A: Phylogeny of 505 specimens inferred from IQTree and MrBayes using seven genetic markers. Support for each branch is shown at the nodes, with bootstrap percentages given first, followed by posterior probabilities. Different species delimitation methods are shown on the right. Photo by Ye-Jie Lin. B: Genitalia of females (a: Ectatosticta xuanzang; b: Ectatosticta dapeng; c: Ectatosticta davidi; d: Ectatosticta baixiang sp. nov.; e: Ectatosticta qingshi sp. nov.; f: Ectatosticta heliisp. nov.; g: Ectatosticta bajie; h: Ectatosticta shaseng sp. nov.; i: Ectatosticta rulai; j: Ectatosticta yukuni; k: Ectatosticta puxian sp. nov.; l: Ectatosticta wukong; m: Ectatosticta baima sp. nov.; n: Ectatosticta deltshevi; o: Ectatosticta wenshusp. nov.). C: Distribution of Ectatosticta spiders in China. Dark spots represent locations of samples and correspond to Supplemnetary Table S1. The asterisk represents the species Ectatosticta shennongjiaensis.
Our study used a multi-gene approach, including non-protein-coding and protein-coding mitochondrial and nuclear genes. The seven markers included three mitochondrial gene fragments (COX1, COX2, and COX3), two nuclear protein-coding loci (ANKRD50 and MOGS), and two nuclear rDNA genes (18S and 28S). Phylogenetic reconstruction was performed using IQTree v1.5.5 (Nguyen et al., 2015) for fast maximum-likelihood (ML) analysis and MrBayes 3.1.2 (Ronquist & Huelsenbeck 2003) for Bayesian inference (BI) analysis. We used two different molecular methods combined with morphological data to detect putative cryptic species: i.e., generalized mixed Yule-coalescent (GMYC) and Bayesian multispecies coalescent approaches with Bayesian Phylogenetics and Phylogeography (BPP) software. The GMYC model was used to identify genealogical clusters that may also correspond to cryptic species lineages. ML was used to fit the GMYC model to an ultrametric tree to identify a threshold time (T) that corresponded to Yule-coalescent transition. The BPP method was used to detect signals of species divergence for putative cryptic species suggested by the phylogeny (Yang, 2015). The sequence alignments were analyzed under the multispecies coalescent model (MSC) with a reversible-jump Markov chain Monte Carlo (rjMCMC) algorithm. Each analysis was run for 200 000 iterations with a sampling frequency of 5 and burn-in of 10 000 iterations.
The phylogenetic tree was generally well resolved, with both ML and BI analyses producing an identical topology at the species level (Figure 1). The Tibet species were basal to the other species. BPP analysis identified 15 provisional species for the dataset, which were relatively consistent with morphology. GMYC analysis recognized 13 provisional species. The differences in the BPP and GMYC results focused on individuals mainly distributed in Gansu, Beishan in Qinghai, and Kalong in Sichuan. The GMYC analysis recognized three lineages as one species, whereas BPP analysis and morphological evidence supported their status as separate species. Specimens from Beishan were described as Ectatosticta deltshevi, and this location is geographically distant from Ectatosticta baima sp. nov. in Kalong and Ectatosticta wenshu sp. nov. in Gansu. Furthermore, Ectatosticta baima sp. nov. and Ectatosticta wenshu sp. nov. were easily distinguished from Ectatosticta deltshevi based on copulatory organs.
The known number of species of Chinese spiders—5 252 species in 827 genera of 69 families—only accounts for about 5% of the entire Chinese spider fauna (Yao & Li, 2021). The increase in Ectatosticta species from a single species in 2008 to 16 in the current study highlights the Wallacean and Linnean shortfalls in China.
TYPE MATERIAL AND NOMENCLATURAL ACTS REGISTRATION
Type material in the paper is housed in the Institute of Zoology, Chinese Academy of Sciences (IZCAS) in Beijing, China. The electronic version of this article in portable document format represents a published work according to the International Commission on Zoological Nomenclature (ICZN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone (see Articles 8.5–8.6 of the Code). This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information can be viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org/.
Publication LSID:
urn:lsid:zoobank.org:pub:9523E776-C186-4A51-A86F-49085A6791AA
Ectatosticta baima Lin & S. Li, sp. nov. LSID:
urn:lsid:zoobank.org:act:85C4A158-8768-4CCB-A54C-24CA82229B7C
Ectatosticta baixiang Lin & S. Li, sp. nov. LSID:
urn:lsid:zoobank.org:act:6D52EA91-9028-4ACC-8E33-6A2061FC1A75
Ectatosticta helii Lin & S. Li, sp. nov. LSID:
urn:lsid:zoobank.org:act:532275A0-04E2-4C55-8655-AE60A5AD7E90
Ectatosticta puxian Lin & S. Li, sp. nov. LSID:
urn:lsid:zoobank.org:act:178BC945-4596-419C-975C-CE6B724D2B2B
Ectatosticta qingshi Lin & S. Li, sp. nov. LSID:
urn:lsid:zoobank.org:act:953D4F62-2ED2-446B-9152-555C799EA352
Ectatosticta shaseng Lin & S. Li, sp. nov. LSID:
urn:lsid:zoobank.org:act:59E7F931-512F-4767-9C5B-DC9559646192
Ectatosticta wenshu Lin & S. Li, sp. nov. LSID:
urn:lsid:zoobank.org:act:19972091-924A-4398-A4EA-2B8C193FA39D
SCIENTIFIC FIELD SURVEY PERMISSION INFORMATION
Permission for field surveys in Gansu, Sichuan, Qinghai, and Tibet was granted by the National Forestry and Grassland Administration, China.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
S.Q.L., X.Y.Y., and H.F.C. designed the study. Y.J.L. contributed to fieldwork and performed morphological species identification. J.N.L. acquired molecular data and completed descriptions. J.N.L. and S.Q.L. drafted and revised the manuscript. All authors read and approved the final version of the manuscript.
Funding Statement
This study was supported by the National Natural Science Foundation of China (32100363, 31970396)
Contributor Information
Xun-You Yan, Email: yanxunyou@163.com.
Shu-Qiang Li, Email: lisq@ioz.ac.cn.
Hai-Feng Chen, Email: chenhaifeng@lfnu.edu.cn.
References
- 1.Lin YJ, Li SQ Taxonomic studies on the genus Ectatosticta (Araneae, Hypochilidae) from China, with descriptions of two new species . ZooKeys. 2020;954(2):17–29. doi: 10.3897/zookeys.954.52254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin YJ, Li SQ Four new species of the genus Ectatosticta (Araneae, Hypochilidae) from China . Acta Arachnologica Sinica. 2021;30(1):1–8. [Google Scholar]
- 3.Lomolino MV. 2004. Conservation biogeography. In: Lomolino MV, Heaney LR. Frontiers of Biogeography: New Directions in the Geography of Nature. Sunderland, Massachusetts: Sinauer Associates, 293–296.
- 4.Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B How many species are there on Earth and in the Ocean. PLoS Biology. 2011;9(8):e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronquist F, Huelsenbeck JP MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 7.Wang LY, Zhao JX, Irfan M, Zhang ZS Review of the spider genus Ectatosticta Simon, 1892 (Araneae: Hypochilidae) with description of four new species from China . Zootaxa. 2021;5016(4):523–542. doi: 10.11646/zootaxa.5016.4.4. [DOI] [PubMed] [Google Scholar]
- 8.Whittaker RJ, Araújo MB, Jepson P, Ladle RJ, Watson JEM, Willis KJ Conservation biogeography: assessment and prospect. Diversity and Distributions. 2005;11(1):3–23. doi: 10.1111/j.1366-9516.2005.00143.x. [DOI] [Google Scholar]
- 9.Yang ZH The BPP program for species tree estimation and species delimitation. Current Zoology. 2015;61(5):854–865. doi: 10.1093/czoolo/61.5.854. [DOI] [Google Scholar]
- 10.Yao ZY, Li SQ Annual report of Chinese spider taxonomy in 2020. Biodiversity Science. 2021;29(8):1058–1063. doi: 10.17520/biods.2021140. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.