Although it first appeared almost two years ago, the COVID-19 pandemic continues to have an impact on a global scale, in part due to newly emerging SARS-CoV-2 variants such as Delta and Lambda. The B.1.621 variant, first identified in Colombia in January 2021, was classified as a variant of interest (VOI) and designated as Mu by the World Health Organization (WHO) in August 2021. However, its infectivity and resistance to neutralizing antibodies remain largely unknown. Here, in comparison to Delta, the Mu variant showed an unexpectedly enhanced immune resistance to inactivated vaccine-elicited antibodies. Nevertheless, Mu demonstrated less infectivity than Delta, implying a biological trade-off between viral transmission and immune escape. This study strongly calls for urgent evaluation of the protective efficacy of current COVID-19 vaccines against the Mu variant.
Variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are of concern regarding control of the global COVID-19 pandemic (Wang et al., 2021). The SARS-CoV-2 B.1.621 variant was first identified in Colombia in January 2021. Considering its epidemiological prevalence, the WHO defined B.1.621 (named Mu) as a VOI on 30 August 2021. As of September 2021, the WHO has classified four variants of concern (VOC), i.e., Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2), and two VOI, i.e., Lambda (C.37) and Mu (B.1.621) (Supplementary Figure S1A).
At the beginning of January 2021, the Mu variant was sporadically documented and its prevalence among sequenced COVID-19 cases was below 0.1% (https://unric.org/en/covid-19-what-is-the-mu-variant/). However, certain epidemiological aspects of Mu have subsequently worsened, and this variant is now responsible for 39% and 13% of infections in Colombia and Ecuador, respectively. As of 15 September 2021, the Mu variant has been detected in more than 20 countries (Supplementary Figure S1B, C), with over 5 000 sequences, including sub-lineage B.1.621.1, designated as Mu, predominantly in the USA (2 513), Colombia (1 042), Spain (518), and Mexico (379) (Supplementary Figure S1D and Table S1). In early September, three people carrying the Mu variant were identified in Hong Kong, suggesting that this new coronavirus variant can be transmitted to broader regions. As the fifth VOI, the epidemiological prevalence of the Mu variant should be of global concern.
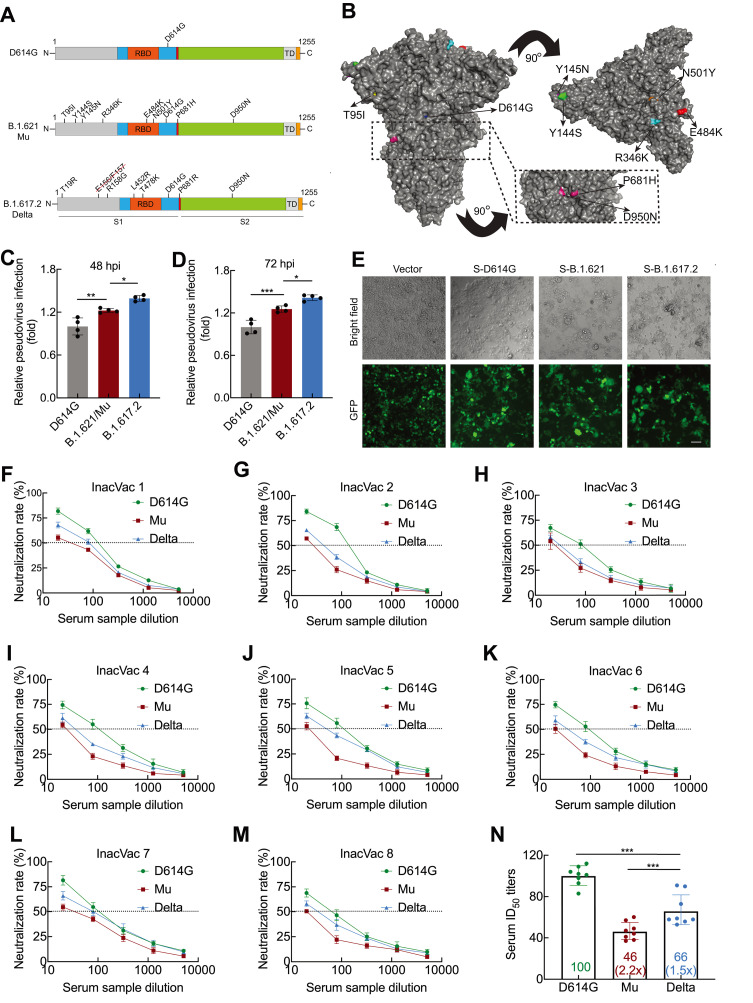
The Mu variant spike protein carries eight mutations, including T95I, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, and D950N (Figure 1A). Several mutations of concern have been identified in VOCs: e.g., E484K in Beta and Gamma, N501Y in Alpha and Beta, P681H in Alpha, and D950N in Delta. Based on spike mutations, the Mu variant is distinguishable from the Delta variant (Figure 1B), with the latter possessing strong transmission capacity and currently accounting for most COVID-19 cases worldwide. Thus, we examined the effect of the spike mutations on infectivity of the Mu variant using lentiviral pseudotyped SARS-CoV-2 with luciferase reporter. As expected, in HEK293T-ACE2 cells (which express human ACE2), the Delta variant showed substantial 1.393- and 1.415-fold increases in viral transduction at 48 (Figure 1C) and 72 h (Figure 1D) post-infection, respectively, compared to the spike D614G variant (Supplementary Figure S2A, B). This increase in infectivity of the Delta variant is consistent with previous research (Mlcochova et al., 2021). Compared to the D614G variant, the Mu variant also displayed significant 1.223- and 1.255-fold increases in viral transduction at 48 (Figure 1C) and 72 h (Figure 1D) post-infection, respectively. These results suggest that the spike protein mutations facilitate viral entry of the Mu variant into ACE2-expressing cells. Nevertheless, viral transduction of the Mu variant was still lower than that of the Delta variant (Figure 1C, D). As the coronavirus spike protein is characterized by inducing fusion between cells, we next examined the effect of the Mu spike protein mutations on cell-cell fusion. Compared to D614G, a pronounced fusion between cells was observed in the Delta spike protein. In comparison to Delta, the Mu spike protein caused moderate cell-cell fusion (Figure 1E). Collectively, these results suggest that Mu is likely to be less transmissible than Delta, although more evidence is needed.
Figure 1.
Infectivity and neutralizing activity of immune serum for Mu variant
A: Diagram of SARS-CoV-2 spike protein from D614G, B.1.621/Mu, and B.1.617.2/Delta variants. D614G variant pseudovirus (containing D614G spike mutation); B.1.621/Mu variant pseudovirus (containing T95I, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, and D950N spike mutations); B.1.617.2/Delta variant pseudovirus (containing E156 and F157 deletions and T19R, R158G, L452R, T478K, D614G, P681R, and D950N spike mutations). B: Surface representation of SARS-CoV-2 B.1.621/Mu spike trimer (PDB: 6ZGE). Black dashed box indicates location of P681H and D950N. C, D: Infectivity of D614G, B.1.621/Mu, and B.1.617.2/Delta variant pseudoviruses assessed in HEK293T-ACE2 cells. Cells were inoculated with equivalent doses of each pseudotyped virus. At 12 h post-inoculation, supernatants were replaced with fresh culture medium. At 48 (C) and 72 h (D), cells were lysed and analyzed for firefly luciferase activity. Data are mean±standard deviation (SD). n=4. *: P<0.05,**: P<0.01, and***: P<0.001 were calculated by one-way analysis of variance (ANOVA) followed by Tukey’spost hoc test. E: Quantitative cell-cell fusion assay. HEK193T cells expressing SARS-CoV-2 spike variants D614G, B.1.621/Mu, and B.1.617.2/Delta were mixed with ACE2-expressing target HEK293T cells (ratio 1:1), and cell-cell fusion was analyzed by measuring presence of syncytia by fluorescence microscopy. F–N: Neutralizing activity of inactivated vaccine serum to D614G, B.1.621/Mu, and B.1.617.2/Delta variants. Pseudotypes were incubated with different serum dilutions for 60 min at 37 °C, and then incubated with HEK293T-ACE2 cells. At 72 h, cells were lysed and analyzed for firefly luciferase activity. Assay of each serum sample was performed in triplicate to determine 50% neutralization titer, as reflected by black dashed line (F–M). Each data point in N represents 50% neutralization titer obtained with a serum sample against indicated pseudovirus. Bar graphs indicate geometric mean titers (GMTs) with 95% confidence. Numbers within box indicate average fold-change in neutralization resistance of indicated spike variants compared to D614G variant in each serum sample (N). n=8. ***: P<0.001 was calculated by one-way ANOVA followed by Tukey’spost hoc test.
The E484K mutation is considered to be responsible for reduced sensitivity to antibodies from both natural SARS-CoV-2 infection and vaccination (Harvey et al., 2021). The Mu variant shares three spike mutations, i.e., E484K, N501Y, and D614G, with the Beta variant, with the latter showing strong resistance to current vaccines (Planas et al., 2021). We assessed the effect of the Mu spike protein mutations on neutralization resistance by comparing neutralization efficacy against the D614G, Mu, and Delta spike pseudotyped viruses. Serum samples from eight individuals who had received two doses of an inactivated vaccine were collected (see Materials and Methods). The serum samples consistently showed pronounced neutralizing capacity against the D614G spike pseudotyped virus (Figure 1F–M). The average neutralizing potency of the serum was reduced 1.5-fold for the Delta variant (geometric mean titer (GMT): 66) and markedly reduced 2.2-fold for the Mu variant (GMT: 46) compared with activity against D614G (GMT: 100) (Figure 1N). These results suggest that the Mu variant has an unexpectedly prominent neutralizing resistance to inactivated vaccine-elicited antibodies.
Based on comparative analysis of the critical spike mutations, Mu is likely to share biological similarity with the Beta variant regarding infectivity and/or immune escape, with both jointly carrying E484K, N501Y, and D614G. Indeed, the enhanced immune escape of Mu relative to Delta was an unexpected observation in this study. Consistently, a recent preprint report (Uriu et al., 2021) showed that the Mu variant has comparable resistance to the Beta variant regarding mRNA vaccines and natural SARS-CoV-2 infection. Nevertheless, our study also showed that Mu has lower infectivity than Delta, implying a potential biological trade-off between viral transmission and immune escape. Considering the high immune resistance of Mu to inactivated and mRNA vaccines, which are commonly used worldwide, further global epidemiological monitoring of this variant is required. Thus, the above findings strongly call for emergent evaluation of the protective efficacy of current COVID-19 vaccines against the Mu variant.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
J.Z. conceived the research and wrote the manuscript. X.X., G.M., and X.L. performed the experiments and analyzed the data. J.B.H., X.L.F., Q.C.Z., and Z.H.D. coordinated the samples and revised the manuscript. All authors read and approved the final version of the manuscript.
We would like to acknowledge Professor Jing Sun from Guangzhou Medical University for the plasmid-expressing full-length spike protein and Professor Yong-Gang Yao from the Kunming Institute of Zoology, Chinese Academy of Sciences (CAS), for the pLenti-CMV-ACE2-flag plasmid. We also thank Ming-Hua Li from the Kunming National High-level Biosafety Research Center for Non-Human Primates, Kunming Institute of Zoology, for coordinating the serum samples.
ACKNOWLEDGMENTS
We would like to acknowledge Professor Jing Sun from Guangzhou Medical University for the plasmid-expressing full-length spike protein and Professor Yong-Gang Yao from the Kunming Institute of Zoology, Chinese Academy of Sciences (CAS), for the pLenti-CMV-ACE2-flag plasmid. We also thank Ming-Hua Li from the Kunming National High-level Biosafety Research Center for Non-Human Primates, Kunming Institute of Zoology, for coordinating the serum samples.
Funding Statement
This work was supported by start-up funding from the Kunming Institute of Zoology, Chinese Academy of Sciences to J.X.Z.
References
- 1.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al SARS-CoV-2 variants, spike mutations and immune escape. Nature Reviews Microbiology. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mlcochova P, Kemp S, Dhar MS, Papa G, Meng B, Ferreira IATM, et al SARS-CoV-2 B. 1.617. 2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 4.Uriu K, Kimura I, Shirakawa K, Takaori-Kondo A, Nakada TA, Kaneda A, et al. 2021. Ineffective neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine sera. bioRxiv, doi: 10.1101/2021.09.06.459005.
- 5.Wang CD, Wang ZF, Wang GY, Lau JYN, Zhang K, Li WM COVID-19 in early 2021: current status and looking forward. Signal Transduction and Targeted Therapy. 2021;6(1):114. doi: 10.1038/s41392-021-00527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.