Abstract
The Chinese tree shrew (Tupaia belangeri chinensis) is emerging as an important experimental animal in multiple fields of biomedical research. Comprehensive reference genome annotation for both mRNA and long non-coding RNA (lncRNA) is crucial for developing animal models using this species. In the current study, we collected a total of 234 high-quality RNA sequencing (RNA-seq) datasets and two long-read isoform sequencing (ISO-seq) datasets and improved the annotation of our previously assembled high-quality chromosome-level tree shrew genome. We obtained a total of 3 514 newly annotated coding genes and 50 576 lncRNA genes. We also characterized the tissue-specific expression patterns and alternative splicing patterns of mRNAs and lncRNAs and mapped the orthologous relationships among 11 mammalian species using the current annotated genome. We identified 144 tree shrew-specific gene families, including interleukin 6 (IL6) and STT3 oligosaccharyltransferase complex catalytic subunit B (STT3B), which underwent significant changes in size. Comparison of the overall expression patterns in tissues and pathways across four species (human, rhesus monkey, tree shrew, and mouse) indicated that tree shrews are more similar to primates than to mice at the tissue-transcriptome level. Notably, the newly annotated purine rich element binding protein A (PURA) gene and the STT3B gene family showed dysregulation upon viral infection. The updated version of the tree shrew genome annotation (KIZ version 3: TS_3.0) is available at http://www.treeshrewdb.org and provides an essential reference for basic and biomedical studies using tree shrew animal models.
Keywords: Tree shrew, Genome annotation, Transcriptome, Gene family, Virus infection
INTRODUCTION
Suitable animal models are essential for expanding our knowledge regarding fundamental biological questions and for developing new drugs, vaccines, and therapeutics (McGonigle & Ruggeri, 2014; Robinson et al., 2019; Yao et al., 2015). An ideal animal model should possess many features, including high genetic similarity to humans, similar pathobiology and symptoms, efficacy with drug prediction and response, low cost, and low restriction (Bennett & Panicker, 2016; McGonigle & Ruggeri, 2014; Robinson et al., 2019; Yao et al., 2015). The Chinese tree shrew (Tupaia belangeri chinensis) is a small rat-sized (100–150 g) mammal with a short reproductive cycle (~6 weeks) (Yao, 2017; Zheng et al., 2014), and is widely distributed in Southeast Asia and South and Southwest China. In the past few decades, tree shrews have been widely used in a variety of biomedical studies, including research on viral infections (Amako et al., 2010; Li et al., 2018; Xu et al., 2007, 2020c), cancer (Ge et al., 2016; Lu et al., 2021), myopia (He et al., 2014; Levy et al., 2018; Phillips et al., 2000), visual cortex function (Fitzpatrick, 1996; Lee et al., 2016; Petry & Bickford, 2019), and neuroscience (Dimanico et al., 2021; Fan et al., 2018; Ni et al., 2018; Savier et al., 2021; Wei et al., 2017). Our research on tree shrews began with the genetic dissection of the Chinese tree shrew genome (Fan et al., 2013). We also aimed to promote the use of this animal in basic and biomedical research by continuing to update relevant genome information (Fan et al., 2014, 2019). Moreover, we developed two immortalized tree shrew cell lines for resource sharing (Gu et al., 2019b; Zhang et al., 2020b) and established the first genetic manipulation of tree shrews using spermatogonial stem cells to successfully generate transgenic offspring (Li et al., 2017). Compared with commonly used animal models such as rodents, tree shrews are phylogenetically closer to primates (Fan et al., 2013, 2019), and can more accurately mimic the physiological and pathological conditions of humans.
Accurate genome assembly and annotation are crucial for understanding tree shrew biology and for developing disease models using this animal. Indeed, creating an animal model of human disease using a tree shrew genome-based approach (Yao, 2017) is dependent on high-quality annotations of the tree shrew genome. Many attempts have been made to decipher the tree shrew genome in great detail and accuracy (Fan et al., 2013, 2019; Sanada et al., 2019). We successfully assembled the first high-quality genome of the Chinese tree shrew (KIZ version 1: TS_1.0) using high depth (~79X) short-read sequencing technology (Fan et al., 2013) and the first chromosome-level tree shrew genome (KIZ version 2: TS_2.0) using single-molecule real-time (SMRT) sequencing technology (Fan et al., 2019). The release of two versions of the tree shrew genome at www.treeshrewdb.org (Fan et al., 2014, 2019) has undoubtedly enhanced our knowledge on the usage of this species. Recently, Sanada et al. (2019) assembled a tree shrew genome using short reads for coding sequence (CDS) annotation. However, despite efforts to improve the annotation of the tree shrew genome, our understanding of the coding and non-coding genes of the tree shrew remains incomplete and unlikely to meet the growing needs of the research field.
RNA sequencing (RNA-seq) technology provides accurate and massive amounts of information regarding the direct transcription status of a genome (Cardoso-Moreira et al., 2019; Stark et al., 2019). The emergence of third-generation sequencing, which features long sequence reads that can cover the full-length of most transcripts (Gordon et al., 2016; Sharon et al., 2013), has greatly improved the accuracy of transcript structure annotation. Previous studies using next-generation sequencing (NGS) based on RNA-seq and long-read isoform sequencing (ISO-seq) have revealed the complexity and characteristics of eukaryotic transcriptomes (Chen et al., 2017). Using ortholog and de novo annotations (Garber et al., 2011; Yandell & Ence, 2012), transcriptome sequencing has been widely used to annotate the genomes of plants (Purugganan & Jackson, 2021; Wang et al., 2019), model animals (Ji et al., 2020; Nudelman et al., 2018; Zhang et al., 2020a), and livestock (Beiki et al., 2019; Foissac et al., 2019). In this study, we aimed to provide a more comprehensive tree shrew genome annotation using a wide range of transcriptome sequencing data. We collected high-quality RNA-seq datasets of tree shrews from publicly available sources (Supplementary Table S1), as well as two ISO-seq datasets and 139 RNA-seq datasets newly generated in this study. These transcriptome datasets included expression data of tree shrew cells and tissues under different conditions, including viral infection (Sanada et al., 2019; Yan et al., 2012), normal tissue (Fan et al., 2013; Han et al., 2020), and pathological tissue (Li et al., 2017; Lin et al., 2014; Tu et al., 2019; Wu et al., 2016b; Zhang et al., 2020b). Using a stringent pipeline, we obtained a total of 53 298 newly annotated coding transcripts and 115 562 newly annotated non-coding transcripts and produced a relatively complete and reliable tree shrew genome annotation (KIZ version 3: TS_3.0). Based on this comprehensive annotation, we further explored the spatial expression and alternative splicing patterns of the tree shrew transcripts and characterized the orthologous relationships among tree shrews and other species. We also compared expression similarity across species and provided a landscape of the innate immune response in tree shrews upon viral infection.
MATERIALS AND METHODS
Animals and tissue collection
Nine adult Chinese tree shrews were purchased from the Experimental Animal Center of the Kunming Institute of Zoology, Chinese Academy of Sciences. Animals were anesthetized with pentobarbital and intracardially perfused with phosphate-buffered saline (PBS). Eight tissues (small intestine, liver, heart, kidney, spleen, ovary, brain, and testis) from four animals were collected and snap-frozen in liquid nitrogen. The remaining animals were used for the isolation of tree shrew primary renal cells (TSPRCs) for viral infection assays. All animal experiments were approved by the Institutional Review Board of the Kunming Institute of Zoology, Chinese Academy of Sciences.
ISO-seq for tree shrew tissues
Tissues from two adult Chinese tree shrews were used for ISO-seq (Supplementary Table S2). RNA extraction, library construction, and sequencing were performed by Annoroad Gene Technology (China). In brief, total RNA from each sample was isolated using a NEBNext® UltraTM RNA Library Prep Kit for Illumina® (Catalog # 7530; New England Biolabs Inc., USA) and processed following the manufacturer’s protocols. RNA degradation and contamination were monitored by 1% agarose gels and RNA purity was checked using a NanoPhotometer® spectrophotometer (IMPLEN, USA). RNA integrity was assessed using a Qubit® RNA Assay Kit with a Qubit® 2.0 Fluorometer (Life Technologies, USA) and an RNA Nano 6000 Assay Kit with the Bioanalyzer 2100 system (Agilent Technologies, USA). Equal amounts of RNA from each tissue of the two tree shrews were pooled as one mixed RNA sample for ISO-seq. Two ISO-seq libraries (<4 kb and >4 kb) were prepared according to the Isoform Sequencing protocol (ISO-seqTM) using a Clontech SMARTer PCR cDNA Synthesis Kit and the BluePippinTM Size Selection System (Sage Science, USA) protocol as described by PacBio (Menlo Park, USA). SMRT sequencing was performed on the Pacific Bioscience Sequel System using two SMRT cells.
The ISO-seq data were processed using IsoSeq v3.4.0 (https://github.com/PacificBiosciences/IsoSeq). Only sequence reads containing both 5' and 3' adaptors were retained to cover the entire transcript. We used LoRDEC (Salmela & Rivals, 2014) to correct errors in the SMRT reads by referring to the RNA-seq data. Subsequently, the corrected SMRT reads were aligned to the tree shrew reference genome TS_2.0 (Fan et al., 2019) using GMAP (Wu et al., 2016a) to locate the position of the predicted genes on the pseudochromosomes.
Compilation of publicly available tree shrew RNA-seq datasets
To ensure a robust and complete annotation of the tree shrew genome, we obtained all publicly available RNA-seq datasets of tree shrews from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), DNA Data Bank of Japan (DDBJ, https://www.ddbj.nig.ac.jp/index-e.html), and China National Center for Bioinformation (BIGD, https://bigd.big.ac.cn/). These transcriptome sequencing datasets were originally obtained by sequencing normal and pathological tissues or cells and represent a wide spectrum of biological and pathological conditions (Supplementary Table S1).
We used the following strategy for quality control (QC) of the RNA-seq data and filtered those data that did not meet requirements. Briefly, raw sequencing reads were processed by Trimmomatic (v0.38) (Bolger et al., 2014) to trim adaptor and low-quality sequences, with the parameters “LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36”. After read filtering, the quality of the clean reads was assessed by FastQC (https://sourceforge.net/projects/fastqc.mirror/). Datasets that passed QC (Q30>20) were aligned to the chromosome-level tree shrew genome (KIZ version 2: TS_2.0; https://www.treeshrewdb.org/download) using STAR (v2.6.0c) (Dobin et al., 2013). We discarded those datasets that failed to pass QC, i.e., mapping ratio to genome below 75%. After QC, a total of 91 publicly available RNA-seq datasets were retained for analysis (Supplementary Table S1).
RNA-seq of tree shrew tissues and cells with or without viral infection
To enhance the credibility of the genome annotation, we generated new RNA-seq data from tissues and/or cells of nine tree shrews (Supplementary Table S2) for further analysis with the publicly available RNA-seq data.
To explore the changes in gene expression in tree shrew cells in response to viral infection, we performed RNA-seq of virus-infected TSPRCs. We used the same procedure to isolate and culture TSPRCs and challenge cells with or without virus as described in our previous studies (Xu et al., 2016, 2020a; Yu et al., 2014). Briefly, TSPRCs were infected with a DNA virus (herpes simplex virus type 1, HSV-1; multiplicity of infection (MOI)=10) and RNA viruses (Sendai virus (SeV, 20 hemagglutinating units/mL), encephalomyocarditis virus (EMCV, MOI=2), and Newcastle disease virus (NDV, MOI=1)), respectively, for the indicated times before harvesting for RNA-seq (Supplementary Table S3). RNA-seq of tree shrew tissues and infected cells was performed by Annoroad Gene Technology (China). Approximately 1 μg of total RNA from each sample was used to construct the RNA-seq libraries (500–1 000 bp) with a NEBNext® UltraTM RNA Library Prep Kit for Illumina®. The quality of each library was assessed using the Agilent Bioanalyzer 2100 system. Libraries were sequenced on the Illumina NovaSeq platform and 150 bp paired-end reads were generated. We followed the same QC procedures as described above for processing the publicly available RNA-seq data. We also included transcriptome datasets of lung tissues from influenza A virus (IAV)-infected tree shrews and of hepatitis C virus (HCV)-infected tree shrew primary hepatocytes in the current analyses (authors’ unpublished data).
Evaluation of coding ability of transcripts and annotation of coding genes
We assembled RNA transcripts based on the RNA-seq reads from publicly available datasets (Supplementary Table S1) and the new data generated in this study (Supplementary Tables S2, S3) using StringTie (v2.1.1) in a reference-guided manner (-G) (Pertea et al., 2015). The assembled RNA-seq transcript models were merged with the SMRT models by StringTie merge (--merge option) to obtain a transcript model covering all transcriptome datasets. To ensure the credibility of the transcripts, we only retained transcripts with high-confidence expression levels (fragments per kilobase per million (FPKM)>0.5 in at least one sample) during the merging of the RNA-seq and SMRT transcripts. We used Gffcompare (https://ccb.jhu.edu/software/stringtie/gffcompare.shtml) (Pertea & Pertea, 2020) to compare the assembled transcript models with the transcript models of the reference genome TS_2.0 (reference model). We treated those transcripts with no matches to the reference models (class code “=”) as newly identified transcripts.
We evaluated the coding potential of the newly identified transcripts by incorporating the results predicted using the following approaches to achieve a more reliable annotation: (1) The Coding Potential Assessment Tool (CPAT) (Wang et al., 2013) was applied to evaluate the transcript coding ability using a logistic regression model. The hexamer frequency table was built using “make_hexamer_tab.py” script, and the logit model was built using “make_logitModel.py” script. (2) We used the Coding Potential Calculator 2 (CPC2) (Kang et al., 2017; Kong et al., 2007) to evaluate the coding ability of the transcripts employing a novel discriminative model based on four sequence-intrinsic features. (3) We used TransDecoder (https://github.com/TransDecoder/) to predict the high-confidence open reading frames (ORF) of each transcript. (4) Pfam (El-Gebali et al., 2019), which contains a comprehensive archive of protein domains, and UniProtKB/Swiss-Prot (Boutet et al., 2007), which contains a comprehensive archive of protein sequences from multiple species, were used to identify potentially translated ORFs. Except for CPAT, we ran all other programs with their default parameters and integrated the prediction results with the following procedures. First, we integrated the prediction results from both CPAT and CPC and only transcripts that met the coding cut-off of both approaches (CPAT, coding potential>0.4; CPC, designated as “coding”) were subjected to further analyses. Second, the ORF of each transcript was predicted using TransDecoder (https://github.com/TransDecoder/) and only transcripts with at least one high-confidence ORF were retained. Third, we scanned the potentially translated ORFs against the Pfam (http://rfam.xfam.org/) (El-Gebali et al., 2019) and UniProtKB/Swiss-Prot databases (Boutet et al., 2007). Those transcripts with at least one predicted Pfam domain or high protein sequence identity (E-value>1e-5) with at least one known protein were defined as coding transcripts. We selected the longest transcript from each gene locus as the representative transcript of the gene. We BLASTed the representative transcripts against the UniProtKB/Swiss-Prot and UniProtKB/Trembl databases (Boutet et al., 2007) using blastall (https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download). The best hit gene name of each gene locus was designated as the name of the newly annotated gene. For multiple gene loci with the same best hit, we modified the gene name by adding “LI (like)+number” to avoid gene name redundancy. We used eggnog-mapper (Huerta-Cepas et al., 2017) to BLAST the translated ORFs for each transcript against the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Kanehisa et al., 2017) and Gene Ontology (GO) database (http://geneontology.org/) for gene function annotation. The best KEGG pathway and GO term hits were designated for each gene. Fourth, lncRNAs were identified and annotated from the transcripts using the following criteria: (1) transcripts are >200 nucleotides (nt) long and meet the non-coding cut-offs of CPAT (coding potential<0.4) and CPC (designated as “noncoding”); (2) transcripts have a predicted ORF<100 nt; and (3) transcripts have a low similarity (E-value>1e-5) with the tRNA family in the Rfam database ( El-Gebali et al., 2019) and UniProtKB/Swiss-Pro database (Boutet et al., 2007). We defined those transcripts showing inconsistent prediction of coding RNAs and lncRNAs based on the above approaches as biased transcripts.
After coding potential evaluation and gene annotation, we merged the TS_2.0 genome annotation file (Fan et al., 2019) with the newly annotated transcripts to generate the TS_3.0 genome annotation. We verified the accuracy of the TS_3.0 transcripts by comparing the annotated transcripts with those characterized by molecular cloning. In total, 30 transcripts reported in our previous studies (Gu et al., 2019a; Luo et al., 2018; Yao et al., 2019; Yu et al., 2014, 2016) (Supplementary Table S4) were selected for comparison using blastall (https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download). An E-value<1e-5 was used as a cutoff to define whether a transcript obtained by molecular cloning was included in the TS_3.0 genome annotation.
Benchmarking universal single-copy orthologs (BUSCO) analysis
We used BUSCO (Seppey et al., 2019) to evaluate the completeness of the TS_3.0 genome annotation using the Mammalia and Eukaryota BUSCO datasets (Simão et al., 2015; Waterhouse et al., 2018). To ensure that the sequences of each species originated from the genome annotation, we selected protein-coding genes according to the information provided in the gene transfer format (GTF) file for each species, and the longest transcript of each gene was selected for consideration. We compared the TS_3.0 annotation with the previous tree shrew genome annotations (Supplementary Table S6). We used Gffread (Pertea & Pertea, 2020) to extract the sequences from the reference genome according to the annotation information. BUSCO evaluation was run in transcriptome mode (-m tran).
Alternative splicing prediction, differential gene expression, and dimension reduction analyses
We used Kallisto (Bray et al., 2016) to quantify the expression level of each transcript. Briefly, the clean reads obtained from each RNA-seq dataset were mapped to the transcriptome constructed using the TS_3.0 annotation. We used SUPPA2 (Trincado et al., 2018) to predict alternative splicing events, including skipped exons (SE), alternative 5' splice sites (A5), alternative 3' splice sites (A3), mutually exclusive exons (MXE), and retained introns (RI). The splicing level of each gene in each RNA-seq dataset was quantified using the Percent Spliced-In (PSI) index. The PSI values of each gene were calculated based on the transcript model and expression level (transcripts per million, TPM) of all transcripts for the gene (Trincado et al., 2018).
To characterize tissue-specific gene/transcript expression levels and alternative splicing events, we identified specifically expressed genes by applying the Wilcoxon rank-sum test and Dunn test in the R (http://www.R-project.org/) package Seurat (Butler et al., 2018). P-values were adjusted (Padjust) by the Benjamini-Hochberg (BH) method. Genes/transcripts with a Padjust<0.05 were defined as differentially/specifically expressed in a given condition. Using Seurat (Butler et al., 2018), we performed uniform manifold approximation and projection (UMAP) (McInnes et al., 2020) for each tree shrew tissue based on TPM of mRNA and lncRNA at the gene and transcript levels, respectively. Those genes/transcripts expressed in all samples of a given tissue and with a gene/transcript |log2 fold-change|>0.5 when compared with other tissues were regarded as tissue-specific genes/transcripts.
We used the R package DESeq2 (Love et al., 2014) to identify differentially expressed genes (DEGs) under virus infection conditions. The Padjust values were calculated using the BH method, as described above. Genes were identified as dysregulated upon viral infection if Padjust<0.05 and |log2 fold-change|>1 were met. KEGG and GO enrichment analyses were performed using the R package ClusterProfiler (Yu et al., 2012), with P-values adjusted by the BH method. A pathway with a Padjust<0.05 was defined as significantly enriched.
Gene family analyses
We obtained protein sequences of multiple mammals from the Ensembl database (https://asia.ensembl.org/index.html), including Homo sapiens (GRCh38.p13), Pan troglodytes (Pan_tro_3.0), Gorilla gorilla gorilla (gorGor4), Macaca mulatta (Mmul_10), Rattus norvegicus (Rnor_6.0), Mus musculus (GRCm39), Sus scrofa (Sscrofa11.1), Bos taurus (ARS-UCD1.2), Canis lupus familiaris (CamFam3.1), and Oryctolagus cuniculus (OryCun2.0) (Supplementary Table S5). The orthologous relationships among species were calculated using OrthoFinder (Emms & Kelly, 2019). We used CAFÉ (De Bie et al., 2006; Mendes et al., 2020) to detect gene family size changes, including expansion and contraction, based on the orthogroups and phylogenetic tree constructed by OrthoFinder (Emms & Kelly, 2019). The phylogenetic tree was constructed using all protein-coding genes of the genomes.
Two gene families showed expansion in this study, i.e., STT3 oligosaccharyltransferase complex catalytic subunit B (STT3B) and subunit A (STT3A) and the interleukin 6 (IL6) gene family, which were featured for their potential roles in viral infection. We constructed gene trees of these gene families using the maximum-likelihood (ML) method (K2+G model) with 1 000 bootstraps. Trees were based on protein sequence alignment and constructed using MEGA (Kumar et al., 2018).
Tissue expression pattern and pathway gene similarity across species
We retrieved expression data from five tissues (liver, brain, kidney, testis, and heart) of mice (https://www.ebi.ac.uk/arrayexpress/E-MTAB-6798), rhesus macaques (https://www.ebi.ac.uk/arrayexpress/E-MTAB-6813), and humans (https://www.ebi.ac.uk/arrayexpress/E-MTAB-6814) from ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) (Cardoso-Moreira et al., 2019). Principal component analysis of gene expression levels (TPM of each gene) for each tissue was constructed using the R package FactoMineR based on 11059 one-to-one orthologous genes and 3291 highly variable genes (defined by a coefficient of variation>0.1 TPM). Pathway gene information was retrieved from the KEGG database (Kanehisa et al., 2017). Protein sequence identities were calculated by BLASTing the tree shrew genes against human homologs and mouse genes against human homologs, respectively. Comparisons of protein sequence identity between mice and humans and between tree shrews and humans were performed using the Wilcoxon rank-sum test adopted in the R package. Here, P<0.05 was considered statistically significant.
RESULTS
Identification of high-quality tree shrew transcripts
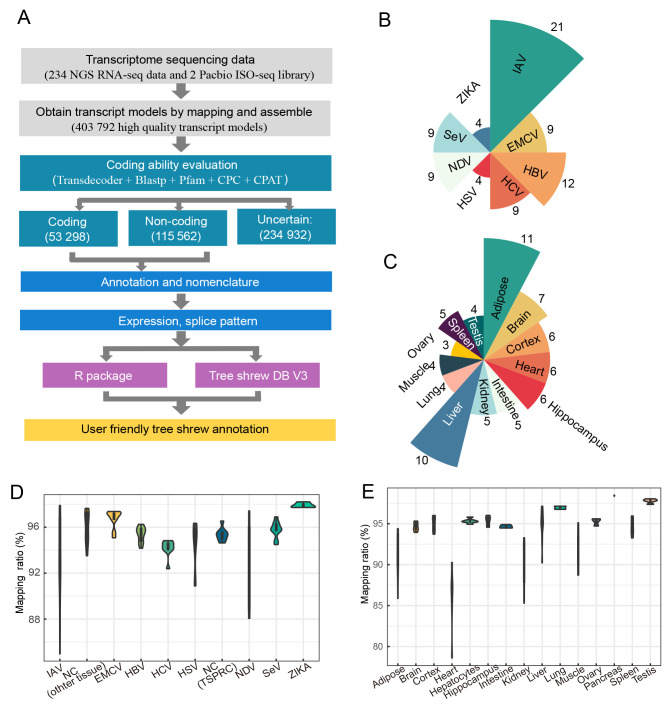
To refine and annotate the chromosome-level tree shrew reference genome TS_2.0 (Fan et al., 2019), we adopted a stringent pipeline to integrate the related RNA-seq and SMRT datasets (Figure 1A). We collected and curated tree shrew transcriptome data across different tissues and cells with different viral infections. A total of 234 tree shrew transcriptome datasets were used in this study (Supplementary Tables S1–S3). These datasets covered a wide range of biological and pathological conditions, including cells infected with different viruses (Figure 1B) and normal and pathological tissue expression (Figure 1C). We used these datasets of diverse conditions to ensure that we captured the diversity and quantity of the transcripts, especially those with low abundancy in normal conditions. After QC, we discarded four samples with a mapping ratio below 75% from further analysis. The remaining datasets had a mean mapping ratio of 93.26% (Figure 1C, D), reflecting relatively high completeness of the reference genome and high quality of the RNA-seq datasets used. Based on the transcriptome datasets, we assembled a transcript model with FPKM>0.5 for each sample using StringTie (Pertea et al., 2015) and obtained 423 965 transcripts for the tree shrew (hereafter referred as RNA-seq transcripts).
Figure 1.
Reference-guided transcriptome assembly of tree shrew TS_3.0 genome annotation
A: Integrative pipeline for tree shrew genome annotation using publicly available and newly generated transcriptome datasets. B, C: Number of RNA-seq datasets of virus-infected tissues/cells (B) and normal tissues (C) analyzed in this study. D, E: Mapping ratio of RNA-seq data of virus-infected tissues/cells (D) and normal tissues (E) relative to reference tree shrew genome TS_2.0 (Fan et al., 2019). Sample information in (D) is listed in Supplementary Table S1.
To verify the accuracy of the tree shrew RNA-seq transcripts, we constructed two ISO-seq full-length transcriptome libraries based on pooled RNA samples from eight tissues of two tree shrews. After QC and error correction, we obtained 36381 non-redundant non-chimeric full-length transcripts located at 12366 loci (hereafter referred as ISO-seq transcripts). The mean length of the ISO-seq transcripts was 2371 nt and the longest ISO-seq transcript (death effector domain containing [DEDD] gene) was 9417 nt. A total of 10968 transcripts were matched between the RNA-seq and ISO-seq transcripts, accounting for 30.15% of the ISO-seq transcripts. Nearly all ISO-seq transcripts were captured by the RNA-seq transcripts, and only 0.9% (1317/146347) of exons and 2.6% (319/12366) of loci captured by ISO-seq were missed by the RNA-seq transcripts. We combined the RNA-seq and ISO-seq transcripts and obtained a total of 403792 transcripts located in 98142 loci in the tree shrew genome. The compiled tree shrew transcripts were deposited in the Tree shrew Database (http://www.treeshrewdb.org/download.html).
Expanded list of tree shrew coding and long non-coding transcripts
Both lncRNAs and mRNAs share similar biogenesis pathways and are involved in multiple biological processes (Jiang et al., 2019; Quinn & Chang, 2016), but exert their functions in different manners (Dahariya et al., 2019; Melé et al., 2017). We categorized transcripts into high-confidence coding transcripts (mRNAs), lncRNA transcripts, and biased transcripts. Among the 403792 newly obtained transcripts, 115562 (>200 nt) were located in 56401 loci and were predicted to be lncRNA transcripts. Among these lncRNA transcripts, 50576 were antisense, 60118 were intergenic, and 4868 were bidirectional. We predicted 53298 coding transcripts located in 19242 loci. In addition, 234932 transcripts located in 93426 loci had inconsistent prediction regarding the characteristics of coding and lncRNA transcripts.
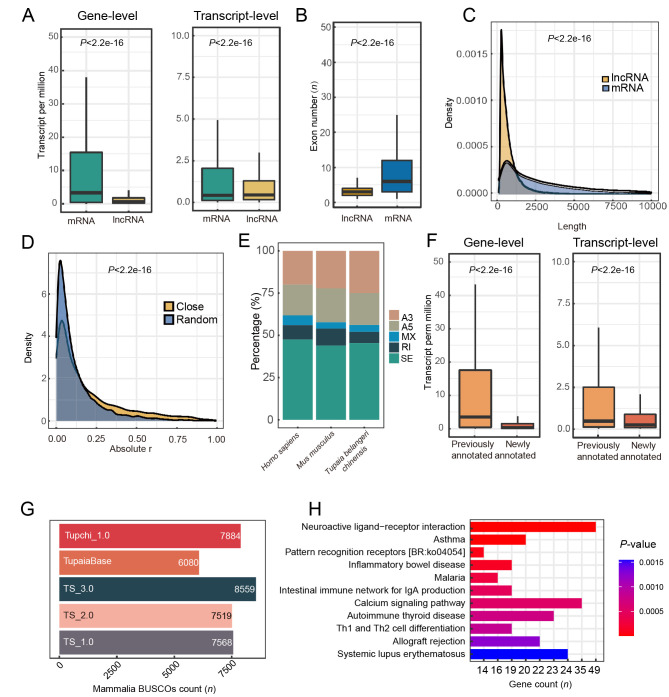
Overall, the expression levels of tree shrew lncRNAs were significantly lower than the expression levels of mRNAs at the gene and transcript levels (Figure 2A), consistent with previous reports on the human transcriptome (Iyer et al., 2015; Jiang et al., 2019). The exon number per lncRNA transcript was also significantly lower than that of mRNA in the tree shrew (Figure 2B), as observed in humans (Jiang et al., 2019). Moreover, the length of lncRNA was significantly shorter than that of mRNA in the tree shrew (Figure 2C). It has been reported that lncRNAs can regulate mRNA expression in a cis-regulatory manner (Jiang et al., 2019; Ørom et al., 2010; Ponjavic et al., 2009). Here, we calculated the expression correlation (Pearson correlation coefficient) between 10 000 mRNAs and their closest lncRNAs in the same genomic region across all RNA-seq datasets and compared the expression correlation between 10 000 randomly selected pairs of mRNAs and lncRNAs. Results showed that the expression correlation between closely located mRNA-lncRNA pairs was significantly stronger (Wilcoxon tests, P<2.2e-16) than that between randomly chosen pairs (Figure 2D). This provides additional evidence for the good accuracy of the lncRNA and mRNA annotations in the tree shrew genome.
Figure 2.
Characteristics of tree shrew TS_3.0 transcripts
A: Expression level of mRNAs is greater than that of lncRNAs at gene and transcript levels. B: mRNA transcripts have a higher number of exons than lncRNA transcripts. C: Average length of mRNA transcripts is longer than that of lncRNA transcripts. Density plot was drawn based on kernel density and statistical analysis was performed with Wilcoxon rank-sum test. D: Tree shrew lncRNAs exert cis-regulatory function on expression of proximal mRNAs. Close, mRNA and lncRNA pairs neighboring each other on pseudochromosome of tree shrew. Random, randomly selected mRNA and lncRNA pairs distant from each other in the genome. E: Percentages of alternative splicing events in reference genome annotations of tree shrew (TS_3.0), mouse (GRCm39), and human (GRCh38.p13). SE, skipped exon; A5, alternative 5’ splice site; A3, alternative 3’ splice site; MXE, mutually exclusive exons; RI, retained intron. F: Expression level of newly annotated coding genes is lower than that of previously annotated genes at gene and transcript levels. G: BUSCO evaluation of different tree shrew genome annotations showing that current version (TS_3.0) is superior. H: Pathway enrichment analysis of newly annotated genes showing enrichment in 11 pathways (Padjust<0.05). Values in A, B and F are presented as a boxplot, and statistical analyses were performed by Wilcoxon rank-sum test.
We further compared 30 transcripts obtained by molecular cloning in our previous studies (Gu et al., 2019a; Luo et al., 2018; Yao et al., 2019; Yu et al., 2014, 2016) (Supplementary Table S4) with those predicted in the TS_3.0 genome annotation. All 30 transcripts showed very good alignment with the currently annotated transcripts (blastall, E-value<1e-5), and 47 additional transcripts were identified in these gene loci according to the TS_3.0 genome annotation, suggesting high accuracy and completeness of the transcript annotation. For instance, we observed all tree shrewTLR gene family members (Yu et al., 2016) and six IL7 transcripts (Yu et al., 2014) reported in our previous studies in TS_3.0. The four IL7 transcripts showed a complete sequence match with the corresponding transcripts in TS_3.0 (Supplementary Figure S1). Among the five alternative splicing events in the tree shrew transcriptome, SE was the most common type of alternative splicing event in the TS_3.0 transcripts (Figure 2E). This pattern is consistent with that of humans and mice (Figure 2E).
Compared to the TS_2.0 tree shrew genome (Table 1), we found 6 126 coding transcripts (including 207 single-exon transcripts) located in 3 514 loci, none of which had been previously annotated and thus represented newly annotated genes. We profiled the gene expression patterns of these newly annotated genes across all RNA-seq datasets and found that the expression levels of the genes were significantly lower than those of the previously annotated ones (Figure 2F). The low abundancies of these newly annotated genes may be the reason for missing annotation in our previous studies (Fan et al., 2013, 2019). The newly annotated genes were enriched (BH adjusted, Padjust<0.05) in immune-related KEGG pathways, such as “Pattern recognition receptors” and “Inflammatory bowel disease (ko05321)” (Figure 2H), partly due to the bias of the RNA-seq datasets of tree shrew cells with viral infection. Combined with the previously annotated genes (Fan et al., 2019), 27 082 coding genes were finally annotated in the tree shrew genome.
Table 1. Comparisons of five tree shrew genome annotations.
| Parameters | TupChi_1.0 (NCBI) | TupaiaBase | TS_1.0 | TS_2.0 | TS_3.0 |
| Tree shrew genome annotations TS_1.0 (Fan et al., 2013), TS_2.0 (Fan et al., 2019), and TS_3.0 (this study) were established in our studies. Tupchi_1.0, NCBI tree shrew annotation (https://www.ncbi.nlm.nih.gov/assembly/GCF_000334495.1/). TupaiaBase was reported by (Sanada et al., 2019). BUSCO: Benchmarking with Universal Single-Copy Orthologs. –: Not available. | |||||
| Coding genes | |||||
| Total number of coding genes | 23 527 | 19 230 | 22 121 | 23 568 | 27 082 |
| Transcript per coding gene | 1.59 | 1 | 1 | 1 | 2.17 |
| Annotated coding genes | 23 537 | 12 612 | 20 225 | 20 811 | 25 127 |
| Average mRNA length | 48 104 | 33 712 | 40 114 | 41 239 | |
| Average CDS length | 1 682 | 1 419 | 1 404 | 1 527 | 1 684 |
| Average exon number | 8.34 | 7.68 | 7.54 | 8.86 | 9.32 |
| Average exon length | 229 | 185 | 186 | 172 | 181 |
| Average intron length | 6 003 | 3 411 | 4937 | 4907 | 4 863 |
| Complete BUSCOs
(Eukaryota 255 genes) |
216
(84.7%) |
195
(76.5%) |
221 (86.7%) | 235
(92.2%) |
250
(98.0%) |
| Complete BUSCOs
(Mammalia 9 224 genes) |
7 884
(85.5%) |
6 080
(65.9%) |
7 568
(82.0%) |
7 519
(81.5%) |
8 559
(92.8%) |
| Non-coding genes | |||||
| Total number of lncRNA genes | 3 718 | – | – | – | 56 401 |
| Transcripts per lncRNA gene | 5 179 | – | – | – | 2.05 |
| Average lncRNA length | 914 | – | – | – | 823 |
| Average exon number | 3.54 | – | – | – | 3.06 |
| Average intron length | 17 614 | – | – | – | 4 658 |
We further compared the gene, transcript, and lncRNA numbers between TS_3.0 and the previously reported versions of tree shrew genome annotation and found remarkable improvement (Table 1). Evaluation of gene completeness of the TS_3.0 annotation relative to the TS_2.0 annotation by BUSCO (Figure 2G; Supplementary Table S6) showed that the ratio of complete BUSCOs increased from 92.16% to 98.04% for Eukaryota BUSCOs (255 genes) and from 81.52% to 92.80% for Mammalia BUSCOs (9 224 genes). Compared with the NCBI TupChi_1.0 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000334495.1) and TupaiaBase (Sanada et al., 2019), the current TS_3.0 annotation showed better completeness and better quality (Table 1).
Tissue expression and alternative splicing profiles of TS_3.0
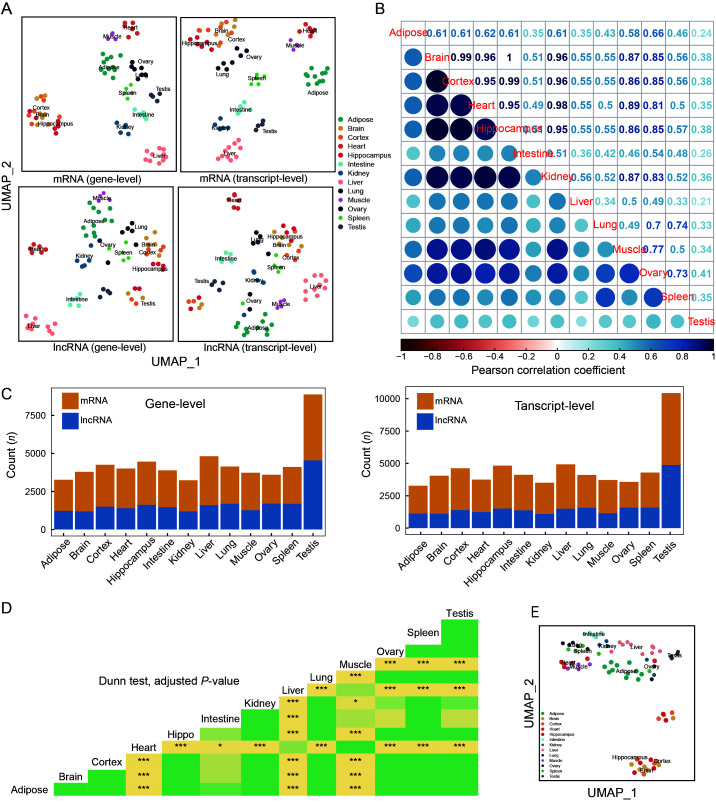
To characterize the tissue-specific expression and alternative splicing patterns of each gene, we analyzed the transcriptome datasets from each tissue using UMAP and calculated the expression correlations. Both mRNAs and lncRNAs showed clear tissue-specificity in the context of UMAP (Figure 3A). The correlation matrix showed that the testis had the most unique tissue expression pattern compared with other tissues (Figure 3B). Notably, the testis possessed the most unique mRNAs and lncRNAs at the gene and transcript level (Figure 3C), consistent with the patterns reported in humans (Djureinovic et al., 2014). Many of the tree shrew testis-specific genes were involved in spermatogenesis (Supplementary Figure S2), as also reported in rats (Ji et al., 2020).
Figure 3.
Tissue expression and alternative splicing profiles of tree shrew TS_3.0 transcripts
A: Tissue expression profiles of mRNAs and lncRNAs annotated in TS_3.0 at gene level (left panel) and transcript level (right panel). UMAP_1, UMAP dimension 1; UMAP_2, UMAP dimension 2. Detailed information on RNA-seq datasets of 13 tree shrew tissues is listed in Supplementary Tables S1, S2. B: Expression correlation matrix among different tree shrew tissues based on expression levels of mRNAs and lncRNAs. C: Tissue-specific expression patterns of mRNAs and lncRNAs at gene level (left) and transcript level (right). D: Comparison of PSI across 13 tree shrew tissues. P-value was calculated based on Dunn test and adjusted by Benjamini-Hochberg method. *: Padjust<0.05;***: Padjust<0.0005. Different colors indicate differentPadjust values in triangle map. E: UMAP was constructed based on PSI of each gene in 13 tree shrew tissues.
We also characterized the specificity and intensity of RNA alternative splicing. Alternative splicing intensity of genes differed significantly across the 13 tree shrew tissues under study (Kruskal-Wallis rank-sum test, P<2.2e-16) (Figure 3D). Brain-related tissues (including the brain, hippocampus, and cortex) showed the highest PSI, whereas heart tissue had the lowest PSI. Furthermore, UMAP using PSI showed that the alternative splicing pattern for each gene also presented tissue specificity (Figure 3E). Collectively, we found that alternative splicing intensity and gene specificity were meticulously regulated across different tree shrew tissues (Figure 3E), which may account for the different functions of the respective tissues and organs.
Orthologous relationships of tree shrew genes with other mammals
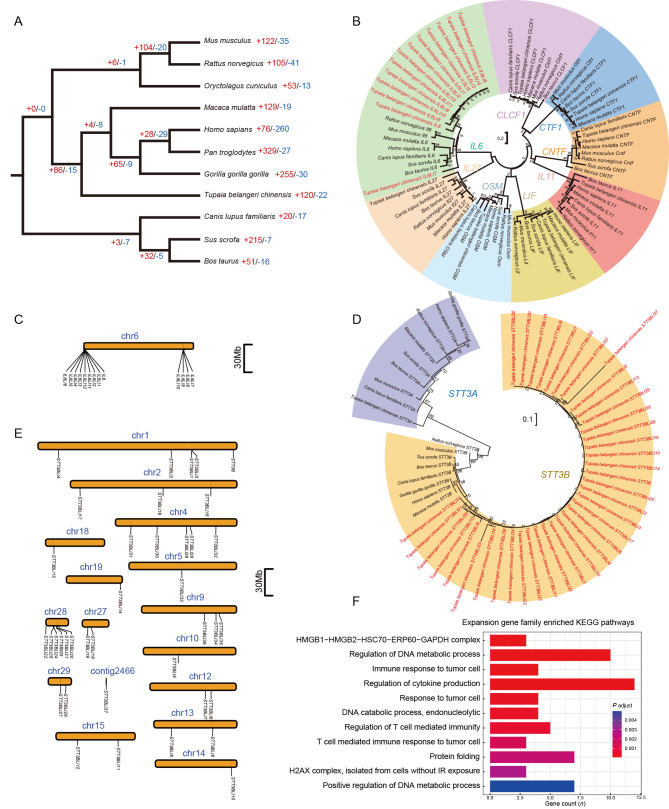
We identified a total of 272 814 genes in the 11 mammals under study (Supplementary Table S5). Among them, 95.1% (259 313 genes) could be assigned to an orthogroup (i.e., a set of genes from multiple species descended from a single gene from the last common ancestor of that set of species) (Emms & Kelly, 2019). In total, 25 249 tree shrew genes could be assigned to 12 485 orthogroups. Among these orthogroups, 191 were paralogs and appeared to be tree shrew specific. We identified 17 299 orthogroups (including 14 549 one-to-one orthologs) shared between humans and tree shrews (Supplementary Table S7), which is a substantial improvement compared with the 12 840 one-to-one orthologs in TS_2.0 (Fan et al., 2019). Based on the current comprehensive orthologous relationships among species, we constructed a phylogenetic tree using the STAG algorithm (Emms & Kelly, 2019). We confirmed that the tree shrew is phylogenetically closer to primates than to rodents (Figure 4A), as described in our previous study based on 2 117 single-copy one-to-one orthologs (Fan et al., 2013).
Figure 4.
Orthologous relationships and gene family size changes among different species
A: Phylogenetic tree of 11 mammals using orthogroups. Numbers on tree branches refer to numbers of gene family expansion (+red) and contraction (-blue), respectively. B: Maximum-likelihood (ML) trees of IL6 gene family. Coding sequence of the longest transcript for each gene in each species was used to construct ML tree. Values on tree branches refer to support of 1 000 bootstraps. The tree shrewIL6 gene family had 13 copies, labeled in red in the tree. C: Locations of 13 tree shrew IL6 gene copies on pseudochromosome 6 (chr6). D: ML trees of STT3B gene family and STT3A. Tree shrew STT3B gene family had 39 copies. E: Locations of 39 tree shrew STT3B gene copies on 15 pseudochromosomes and one unplaced contig. Pseudochromosomes and unplaced contig were defined in reference tree shrew genome TS_2.0 (Fan et al., 2019). F: Pathway enrichment of genes from tree shrew-specific gene families with significant expansion.
Gene gain and loss are important evolutionary processes that allow organisms to adapt to their environment (Page, 1998). Here, we analyzed changes in gene family size across 11 mammal species, including tree shrews (Supplementary Table S8), to validate and refine the previously characterized gene expansion and contraction events. We identified 120 gene families showing rapid expansion and 22 gene families showing rapid contraction (Figure 4A). The gene family exhibiting the greatest expansion was long-interspersed element-1 (LINE-1) retrotransposable element ORF (LIRE1), with 180 LIRE1 genes identified in the TS_3.0 genome annotation. The gene family exhibiting the greatest contraction was immunoglobulin heavy variable 3–35 (Supplementary Table S9). The guanylate binding protein (GBP) gene family was found to have rapidly contracted, consistent with the findings of our previous study (Gu et al., 2019a). We found that IL6 (Supplementary Figure S3) and STT3B (Supplementary Figure S4) were significantly expanded, with 34 of 39 STT3B gene family members being newly annotated and four of 13 IL6 gene family members being refined in the TS_3.0 genome annotation, respectively. The IL6 family contains IL6, cardiotrophin like cytokine factor 1 (CLCF1), cardiotrophin 1 (CTF1), ciliary neurotrophic factor (CNTF), interleukin 11 (IL11), interleukin 27 (IL27), LIF interleukin 6 family cytokine (LIF), and oncostatin M (OSM) (Rose-John, 2018). The tree shrew contained all these IL6 family members, and 12 of the 13 IL6 copies in tree shrews were not detected in other species. We found that each member of the IL6 family was grouped into a single clade in the ML tree of the IL6 family genes, confirming the close relationship of the expanded copies of each IL6 gene family member (Figure 4B). IL6LI7 appeared to be the ancestral gene of the tree shrew IL6 gene copies. The tree shrew IL6 family members were all located on pseudochromosome 6 (Figure 4C), with consistent exon numbers for each family member (Supplementary Figure S5). This suggests that the IL6 gene copies were most likely generated from tandem duplication and segmental duplication. We constructed an ML tree for the tree shrew STT3B gene family members, together with those of the other mammals, and the STT3A paralog. The STT3A and STT3B copies showed a gene-specific clustering pattern (Figure 4D). In the clade for the expanded STT3B copies from the tree shrew, STT3BLI27 diverged first and appeared to be the ancestral gene of the tree shrew STT3B family. Intriguingly, all 39 copies of STT3B in the tree shrew were distributed on 15 pseudochromosomes and one unplaced contig (Figure 4E). Of note, the tree shrew STT3BLI27 had 16 exons, while the other copies of STT3B contained no more than four exons (Supplementary Figure S6), suggesting that expansion of the tree shrew STT3B was most likely caused by retrotransposon activity.
To further dissect the potential evolutionary roles of the tree shrew gene family size changes, we conducted enrichment analysis using the canonical genes of each rapidly changing gene family. Results showed that gene families that have undergone rapid size change were enriched in the “immune response to tumor cell”, “regulation of cytokine production”, and “regulation of DNA metabolic process” pathways (Figure 4F).
Expression similarity across different species
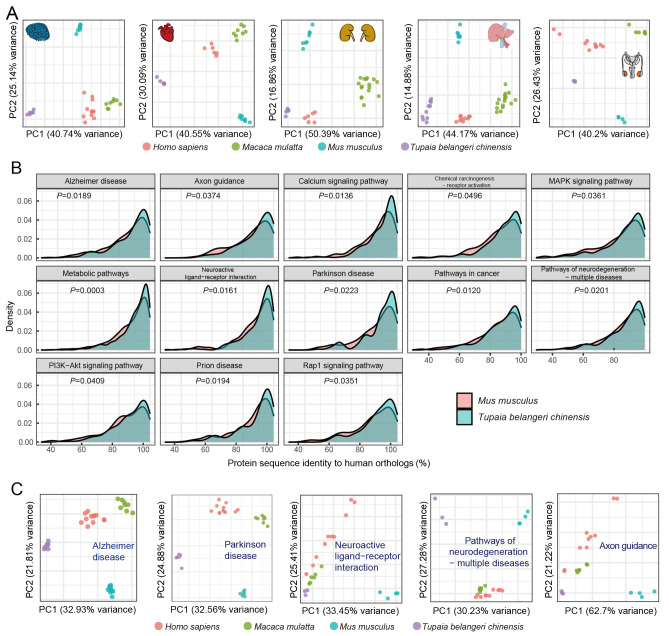
To study the mRNA expression patterns of tissues and related pathways across humans, rhesus monkeys, mice, and Chinese tree shrews, we retrieved tissue RNA-seq data of mice, monkeys, and humans (Cardoso-Moreira et al., 2019), and compared their clustering patterns via principal component (PC) analysis. The species clustering patterns based on expression data from brain, liver, testis, kidney, and heart tissues showed distant divergence of mice from primates and tree shrews in the second PC, whereas humans, monkeys, and tree shrews were mainly separated by the first PC (Figure 5A). However, these clustering patterns should be considered with caution as the first and second PCs only contributed to a proportion of expression variance.
Figure 5.
Expression similarities among different species
A: Tissue expression similarities among humans, rhesus monkeys, tree shrews, and mice. Expression patterns in five tree shrew tissues more closely resembled that of primates than that of mice. B: Comparisons of protein sequence identity of genes in KEGG pathways between tree shrews and humans and between mice and humans. C: Expression patterns of genes in brain-related pathways in tree shrews, rhesus monkeys, humans, and mice. Brain-related pathways included “Alzheimer’s disease”, “Parkinson disease”, “Neuroactive ligand-receptor interaction”, “Pathways of neurodegeneration-multiple diseases”, and “Axon guidance”.
We further determined the gene identity of tree shrews to humans using pathway analysis. For each human KEGG pathway, we compared protein sequence identities between mice and humans and between tree shrews and humans. Genes in 13 pathways showed greater protein sequence identity between tree shrews and humans than between mice and humans (Figure 5B). These 13 pathways included neuro-related pathways such as “Axon guidance”, “Parkinson disease”, and “Alzheimer disease”. Furthermore, proteins belonging to the “pathway in cancer” also showed higher identity between tree shrews and humans than between mice and humans (Figure 5B), suggesting that the tree shrew could be used to create valid cancer and neurodegenerative animal models. We also profiled the expression patterns of five pathways related to the brain (Figure 5C) and found that mice had a more distant clustering pattern than tree shrews with primates. Collectively, these results suggest that tree shrews are more genetically similar to primates than to mice at the transcriptomic level.
Changes in newly identified genes upon viral infection
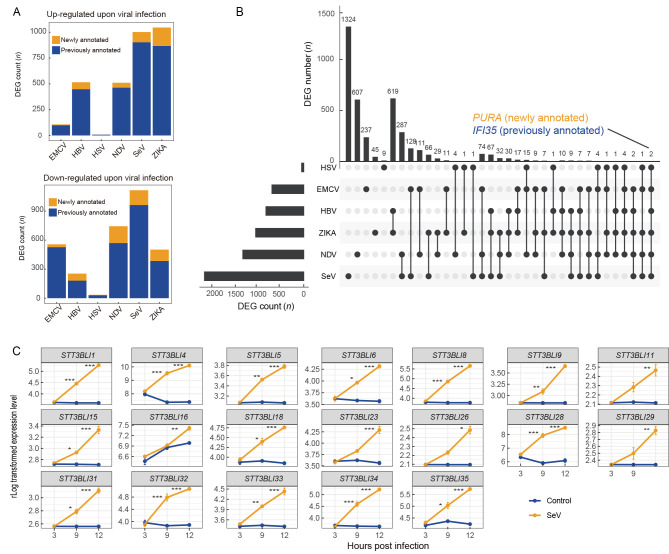
To profile the transcriptome patterns of host immune responses to viral infection using the TS_3.0 genome annotation, we focused on the differential expression of the newly identified genes upon viral infection. Results showed that the TS_3.0 genome annotation had better accuracy and resolution for gene identification in tree shrew cells with or without viral infection. Some of the newly annotated genes were significantly dysregulated upon infection with HBV (67 genes), SeV (99 genes), NDV (48 genes), EMCV (seven genes), and ZIKA (178 genes) (Figure 6A). Of the 3 779 DEGs identified from the RNA-seq datasets by comparing infected and uninfected cells, only purine rich element binding protein A (PURA) and interferon induced protein 35 (IFI35) were significantly dysregulated in cells with all virus infections. Both genes are reported to have a pro-viral effect in other species (Das et al., 2014; Gounder et al., 2018; Krachmarov et al., 1996). As PURA is a newly annotated gene in TS_3.0, more studies should be carried out to characterize its role in viral infections in tree shrews.
Figure 6.
Changes in expression of genes in virus-infected tree shrew tissues and cells
A: Changes in newly annotated genes upon viral infection. HBV, hepatitis B virus; NDV, Newcastle disease virus; EMCV, encephalomyocarditis virus; HSV-1, herpes simplex virus type 1; SeV Sendai virus; ZIKA, Zika virus. Genes were identified as differentially expressed genes (DEGs) upon virus infection if Padjust<0.05 and |log2 fold-change|>1. B: Plot of DEGs upon virus infection.PURA and IFI35 were dysregulated in cells and tissues infected with different viruses. Horizontal bar on left represents number of DEGs in each RNA-seq dataset. Dots and lines represent subsets of DEGs. Vertical histogram represents number of DEGs in each subset. C: Changes in expression of 19 gene copies of STT3B gene family upon SeV infection. Results are mean±standard deviation (SD). *: Padjust<0.05;**: Padjust<0.005;***, Padjust<0.0005.Padjust values were calculated using DESeq2.
Among the expanded gene family members, 19 of the 39 copies of the STT3B gene family in the tree shrew were up-regulated upon SeV infection. The oligosaccharyltransferase complex is known to be an essential host factor for dengue virus (DENV) replication (Lin et al., 2017). Considering that the 19 STT3B gene copies were not located on the same pseudochromosome, we speculated that they were up-regulated by the same transcription regulation system. The ancestral gene of the STT3B gene copies, STT3BLI27, was not dysregulated upon viral infection, suggesting that the 19 STT3B gene copies may have acquired the association with pro-viral function at a later stage of gene family expansion. However, further studies are required to confirm this speculation.
DISCUSSION
Comprehensive tree shrew genome annotation is crucial for developing animal models and for studying basic scientific questions (Yao, 2017). In this study, we annotated the Chinese tree shrew genome by integrating diverse RNA-seq datasets and newly generated ISO-seq datasets. We obtained a total of 27 082 coding genes (including 3 514 previously unannotated coding genes in TS_2.0 (Fan et al., 2019)) and 56 401 lncRNAs. Evaluation of the completeness of multiple tree shrew genome annotations using BUSCO (Seppey et al., 2019; Simão et al., 2015; Waterhouse et al., 2018) indicated that the current TS_3.0 annotation showed remarkable improvement in terms of completeness, which was achieved by incorporating diverse RNA-seq datasets that covered a wide range of biological and pathological conditions. The newly updated tree shrew TS_3.0 genome annotation can be downloaded from the Tree shrew Database (http://www.treeshrewdb.org/download.html).
Compared with the previous tree shrew genome annotation (Table 1), TS_3.0 provides a complete list of lncRNAs, which could help in the interpretation of the roles of lncRNAs in tree shrew biology and disease. The lack of lncRNA conservation among species is a considerable obstacle for functional annotation (Iyer et al., 2015). Here, we compared multiple characteristics between tree shrew mRNAs and lncRNAs and found smaller average exon number and shorter transcript length in lncRNAs than in mRNAs. We confirmed that the identified tree shrew lncRNAs may exert a cis-regulatory role on mRNA expression (Figure 3D). The overall characteristics of the tree shrew lncRNAs versus mRNAs resembled that of human lncRNAs versus mRNAs reported in previous studies (Iyer et al., 2015; Jiang et al., 2019; Ørom et al., 2010; Ponjavic et al., 2009).
Alternative splicing plays a key role in transcript processing and biological functions (Baralle & Giudice, 2017; Ule & Blencowe, 2019). By generating multiple transcripts from a particular gene, alternative splicing events can dramatically increase the diversity and complexity of the transcriptome, and can impact mRNA stability, localization, and translation (Baralle & Giudice, 2017; Ule & Blencowe, 2019). We previously showed that alternative splicing events in STING have played an important role in the innate immunity response of tree shrews against DNA and RNA viral infections (Xu et al., 2020a). Our updated annotation of tree shrew transcripts, especially from SMRT reads, provided an accurate model to characterize alternative splicing events in tree shrews. Using the TS_3.0 transcripts, we quantified and characterized the alternative splicing events and found a tissue-specific pattern of splicing intensity. The high occurrence of alternative splicing events in the tree shrew brain-related tissues is consistent with that found in humans (Rodriguez et al., 2020), suggesting that alternative splicing constitutes a straightforward strategy for enacting diverse functions such as tissue formation (Baralle & Giudice, 2017). It would be worth performing functional characterization of important genes that exhibit alternative splicing in different tree shrew tissues and/or in response to viral infection in the TS_3.0 genome annotation, as exemplified by the elegant functional assay for the STING isoform described in our recent study (Xu et al., 2020a).
The updated tree shrew TS_3.0 genome annotation could also provide insightful information regarding cross-species comparisons to initiate genome-based methods for creating animal models of human disease (Yao, 2017). We systematically characterized the orthologous relationships among experimental animals, including mice, monkeys, and tree shrews, using the newly updated tree shrew genome annotation. Orthologous comparison confirmed the closer relationship between primates and tree shrews than between primates and mice (Figure 4A), suggesting that tree shrews would be better model animals for biomedical research. We also compared the tissue expression patterns and related genes in particular pathways across four species, which again showed that tree shrews are closer to primates than to mice at the transcriptomic level (Figure 5C).
Gene expansion and contraction play key roles in environment adaptation (Yim et al., 2014). We re-appraised gene expansion and contraction events using the TS_3.0 transcripts and confirmed the gene families highlighted in our previous study (Fan et al., 2013). Among the 144 gene families that experienced size changes in the tree shrew, the IL6 and STT3B families may have particular biological implications. Notably, IL6 is thought to be actively involved in the cytokine storms observed in COVID-19 patients (Mehta et al., 2020; Vabret et al., 2020; Zhou et al., 2020) and therapy with the IL-6-receptor antagonist tocilizumab is considered a promising treatment for COVID-19 patients (Fu et al., 2020; Jones & Hunter, 2021). In SARS-CoV-2-infected tree shrews (Xu et al., 2020c; Zhao et al., 2020), different individuals demonstrated different susceptibility to SARS-CoV-2 and showed different viral loads after infection, though none of the infected tree shrews showed severe symptoms. Whether the expanded IL6 gene family played a role in this process is an interesting and important question. Cloning all 13 IL6 copies and characterizing the respective roles of each gene copy could help clarify why this gene family underwent expansion in the tree shrew. Among the 39 gene copies of the STT3B gene family, 19 were up-regulated upon SeV infection, whereas the other copies, including ancestral STT3BLI27, showed no such effect. The STT3B protein is a part of the oligosaccharyltransferase complex in humans (Lu et al., 2019), and is reported to play a pro-viral role in Dengue virus and HSV-1 infections (Lin et al., 2017; Lu et al., 2019). Expansion of the STT3B gene family may indicate a new immune response mechanism for tree shrews to counteract or facilitate these viral infections. However, more studies are required to characterize the function of the tree shrew STT3B gene family and to confirm the above speculation.
An important update of the TS_3.0 genome annotation was the inclusion of newly generated RNA-seq data from tree shrew cells and tissues challenged with different viruses. The inclusion of these datasets offers the chance to identify genes that are up-regulated or down-regulated upon viral infection for further study. Indeed, previously reported tree shrew genes that show altered expression upon viral infection (Gu et al., 2019a, 2021; Xu et al., 2016, 2020a, 2020b) could be confirmed. We identified several important targets showing a universal regulator effect, such as PURA and IFI35. The PURA gene encodes Pur-alpha, which has a repeated nucleic acid binding domain (Daniel & Johnson, 2018), and is reported to be regulated by transcription start sites Ⅰ and Ⅱ (Wortman et al., 2010). PURA is known to activate the John Cunningham virus in the glial cells of many acquired immunodeficiency syndrome patients (Krachmarov et al., 1996). In addition, IFI35 is an interferon-stimulated gene that negatively regulates RIG-I antiviral signals to support vesicular stomatitis viral replication (Das et al., 2014) and enhances H5N1 influenza disease symptoms (Gounder et al., 2018). We speculate that in vivo overexpression of both PURA and IFI35 may create tree shrew models more permissive to different viruses, including HCV and HBV, which have no feasible animal models at present.
In summary, we generated an improved tree shrew genome annotation using comprehensive RNA-seq and ISO-seq datasets. The updated version of the tree shrew genome annotation (TS_3.0) fixed some of the issues with previous versions, such as TS_1.0 (Fan et al., 2013) and TS_2.0 (Fan et al., 2019). Detailed annotation of the genes, gene families, and alternative splicing events in the tree shrew genome, as well as cross-comparison of expression patterns among different tissues and species, further illuminated the unique and common genetic features of tree shrews and provided further evidence of the considerable potential of tree shrews in biomedical research.
DATA AVAILABILITY
The TS_3.0 genome annotation data and newly generated RNA-seq and ISO-seq data are available from the Tree shrew Database (http://www.treeshrewdb.org/download/). Related data were also deposited in GSA (accession No. PRJCA006366).
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Y.G.Y. and M.S.Y. conceived and designed the experiments. L.B.L. provided living tree shrews and tissues. D.D.Y. and L.X. isolated tree shrew primary cells and performed viral infection and RNA extraction. Q.Y.Z. provided the transcriptome data of lung tissues from IAV-infected tree shrews. M.S.Y., J.Y.Z., M.X., and Y.F. collected transcriptome data and performed genome annotation and transcriptome analyses. M.S.Y. and Y.G.Y. wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding Statement
This study was supported by the National Natural Science Foundation of China (U1902215 to Y.G.Y. and 31970542 to Y.F.), Chinese Academy of Sciences (Light of West China Program xbzg-zdsys-201909 to Y.G.Y.), and Yunnan Province (202001AS070023 and 2018FB046 to D.D.Y. and 202002AA100007 to Y.G.Y.)
References
- 1.Amako Y, Tsukiyama-Kohara K, Katsume A, Hirata Y, Sekiguchi S, Tobita Y, et al. 2010. Pathogenesis of hepatitis C virus infection in Tupaia belangeri. Journal of Virology, 84(1): 303–311.
- 2.Baralle FE, Giudice J Alternative splicing as a regulator of development and tissue identity. Nature Reviews Molecular Cell Biology. 2017;18(7):437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beiki H, Liu H, Huang J, Manchanda N, Nonneman D, Smith TPL, et al Improved annotation of the domestic pig genome through integration of Iso-Seq and RNA-seq data. BMC Genomics. 2019;20(1):344. doi: 10.1186/s12864-019-5709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett AJ, Panicker S Broader impacts: international implications and integrative ethical consideration of policy decisions about US chimpanzee research. American Journal of Primatology. 2016;78(12):1282–1303. doi: 10.1002/ajp.22582. [DOI] [PubMed] [Google Scholar]
- 5.Bolger AM, Lohse M, Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A UniProtKB/Swiss-Prot. Methods in Molecular Biology. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 7.Bray NL, Pimentel H, Melsted P, Pachter L Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology. 2016;34(5):525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 8.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardoso-Moreira M, Halbert J, Valloton D, Velten B, Chen CY, Shao Y, et al Gene expression across mammalian organ development. Nature. 2019;571(7766):505–509. doi: 10.1038/s41586-019-1338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G, Shi TL, Shi LM Characterizing and annotating the genome using RNA-seq data. Science China Life Sciences. 2017;60(2):116–125. doi: 10.1007/s11427-015-0349-4. [DOI] [PubMed] [Google Scholar]
- 11.Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, Gutti RK Long non-coding RNA: classification, biogenesis and functions in blood cells. Molecular Immunology. 2019;112:82–92. doi: 10.1016/j.molimm.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Daniel DC, Johnson EM PURA, the gene encoding Pur-alpha, member of an ancient nucleic acid-binding protein family with mammalian neurological functions . Gene. 2018;643:133–143. doi: 10.1016/j.gene.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das A, Dinh PX, Panda D, Pattnaik AK Interferon-inducible protein IFI35 negatively regulates RIG-I antiviral signaling and supports vesicular stomatitis virus replication. Journal of Virology. 2014;88(6):3103–3113. doi: 10.1128/JVI.03202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Bie T, Cristianini N, Demuth JP, Hahn MW CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 15.Dimanico MM, Klaassen AL, Wang J, Kaeser M, Harvey M, Rasch B, et al Aspects of tree shrew consolidated sleep structure resemble human sleep. Communications Biology. 2021;4(1):722. doi: 10.1038/s42003-021-02234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djureinovic D, Fagerberg L, Hallström B, Danielsson A, Lindskog C, Uhlén M, et al The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Molecular Human Reproduction. 2014;20(6):476–488. doi: 10.1093/molehr/gau018. [DOI] [PubMed] [Google Scholar]
- 17.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al The Pfam protein families database in 2019. Nucleic Acids Research. 2019;47(D1):D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emms DM, Kelly S OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology. 2019;20(1):238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y, Huang ZY, Cao CC, Chen CS, Chen YX, Fan DD, et al Genome of the Chinese tree shrew. Nature Communications. 2013;4:1426. doi: 10.1038/ncomms2416. [DOI] [PubMed] [Google Scholar]
- 21.Fan Y, Luo RC, Su LY, Xiang Q, Yu DD, Xu L, et al Does the genetic feature of the Chinese tree shrew (Tupaia belangeri chinensis) support its potential as a viable model for Alzheimer's disease research? . Journal of Alzheimer's Disease. 2018;61(3):1015–1028. doi: 10.3233/JAD-170594. [DOI] [PubMed] [Google Scholar]
- 22.Fan Y, Ye MS, Zhang JY, Xu L, Yu DD, Gu TL, et al Chromosomal level assembly and population sequencing of the Chinese tree shrew genome. Zoological Research. 2019;40(6):506–521. doi: 10.24272/j.issn.2095-8137.2019.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan Y, Yu DD, Yao YG Tree shrew database (TreeshrewDB): a genomic knowledge base for the Chinese tree shrew. Scientific Reports. 2014;4:7145. doi: 10.1038/srep07145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzpatrick D The functional organization of local circuits in visual cortex: insights from the study of tree shrew striate cortex. Cerebral Cortex. 1996;6(3):329–341. doi: 10.1093/cercor/6.3.329. [DOI] [PubMed] [Google Scholar]
- 25.Foissac S, Djebali S, Munyard K, Vialaneix N, Rau A, Muret K, et al Multi-species annotation of transcriptome and chromatin structure in domesticated animals. BMC Biology. 2019;17(1):108. doi: 10.1186/s12915-019-0726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu BQ, Xu XL, Wei HM Why tocilizumab could be an effective treatment for severe COVID-19? Journal of Translational Medicine. 2020;18(1):164. doi: 10.1186/s12967-020-02339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garber M, Grabherr MG, Guttman M, Trapnell C Computational methods for transcriptome annotation and quantification using RNA-seq. Nature Methods. 2011;8(6):469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 28.Ge GZ, Xia HJ, He BL, Zhang HL, Liu WJ, Shao M, et al Generation and characterization of a breast carcinoma model by PyMT overexpression in mammary epithelial cells of tree shrew, an animal close to primates in evolution. International Journal of Cancer. 2016;138(3):642–651. doi: 10.1002/ijc.29814. [DOI] [PubMed] [Google Scholar]
- 29.Gordon D, Huddleston J, Chaisson MJP, Hill CM, Kronenberg ZN, Munson KM, et al Long-read sequence assembly of the gorilla genome. Science. 2016;352(6281):aae0344. doi: 10.1126/science.aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gounder AP, Yokoyama CC, Jarjour NN, Bricker TL, Edelson BT, Boon ACM Interferon induced protein 35 exacerbates H5N1 influenza disease through the expression of IL-12p40 homodimer. PLoS Pathogens. 2018;14(4):e1007001. doi: 10.1371/journal.ppat.1007001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu TL, Yu DD, Fan Y, Wu Y, Yao YL, Xu L, et al Molecular identification and antiviral function of the guanylate-binding protein (GBP) genes in the Chinese tree shrew (Tupaia belangeri chinesis) . Developmental & Comparative Immunology. 2019a;96:27–36. doi: 10.1016/j.dci.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Gu TL, Yu DD, Li Y, Xu L, Yao YL, Yao YG Establishment and characterization of an immortalized renal cell line of the Chinese tree shrew (Tupaia belangeri chinesis) . Applied Microbiology and Biotechnology. 2019b;103(5):2171–2180. doi: 10.1007/s00253-019-09615-3. [DOI] [PubMed] [Google Scholar]
- 33.Gu TL, Yu DD, Xu L, Yao YL, Zheng X, Yao YG Tupaia guanylate-binding protein 1 interacts with vesicular stomatitis virus phosphoprotein and represses primary transcription of the viral genome . Cytokine. 2021;138:155388. doi: 10.1016/j.cyto.2020.155388. [DOI] [PubMed] [Google Scholar]
- 34.Han YY, Wang WG, Jia J, Sun XM, Kuang DX, Tong PF, et al WGCNA analysis of the subcutaneous fat transcriptome in a novel tree shrew model. Experimental Biology and Medicine. 2020;245(11):945–955. doi: 10.1177/1535370220915180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He L, Frost MR, Siegwart JT Jr, Norton TT Gene expression signatures in tree shrew choroid during lens-induced myopia and recovery. Experimental Eye Research. 2014;123:56–71. doi: 10.1016/j.exer.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, et al Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Molecular Biology and Evolution. 2017;34(8):2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al The landscape of long noncoding RNAs in the human transcriptome. Nature Genetics. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji XJ, Li P, Fuscoe JC, Chen G, Xiao WZ, Shi LM, et al A comprehensive rat transcriptome built from large scale RNA-seq-based annotation. Nucleic Acids Research. 2020;48(15):8320–8331. doi: 10.1093/nar/gkaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang S, Cheng SJ, Ren LC, Wang Q, Kang YJ, Ding Y, et al An expanded landscape of human long noncoding RNA. Nucleic Acids Research. 2019;47(15):7842–7856. doi: 10.1093/nar/gkz621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones SA, Hunter CA Is IL-6 a key cytokine target for therapy in COVID-19? Nature Reviews Immunology. 2021;21(6):337–339. doi: 10.1038/s41577-021-00553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei LP, et al CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Research. 2017;45(W1):W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei LP, et al CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Research. 2007;35(W1):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krachmarov CP, Chepenik LG, Barr-Vagell S, Khalili K, Johnson EM Activation of the JC virus Tat-responsive transcriptional control element by association of the Tat protein of human immunodeficiency virus 1 with cellular protein Purα. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(24):14112–14117. doi: 10.1073/pnas.93.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S, Stecher G, Li M, Knyaz C, Tamura K MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee KS, Huang XY, Fitzpatrick D Topology of ON and OFF inputs in visual cortex enables an invariant columnar architecture. Nature. 2016;533(7601):90–94. doi: 10.1038/nature17941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levy AM, Fazio MA, Grytz R Experimental myopia increases and scleral crosslinking using genipin inhibits cyclic softening in the tree shrew sclera. Ophthalmic and Physiological Optics. 2018;38(3):246–256. doi: 10.1111/opo.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li CH, Yan LZ, Ban WZ, Tu Q, Wu Y, Wang L, et al Long-term propagation of tree shrew spermatogonial stem cells in culture and successful generation of transgenic offspring. Cell Research. 2017;27(2):241–252. doi: 10.1038/cr.2016.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li RF, Yuan B, Xia XS, Zhang S, Du QL, Yang CG, et al Tree shrew as a new animal model to study the pathogenesis of avian influenza (H9N2) virus infection. Emerging Microbes & Infections. 2018;7(1):166. doi: 10.1038/s41426-018-0167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin DL, Cherepanova NA, Bozzacco L, MacDonald MR, Gilmore R, Tai AW Dengue virus hijacks a noncanonical oxidoreductase function of a cellular oligosaccharyltransferase complex. mBio. 2017;8(4):e00939–e00917. doi: 10.1128/mBio.00939-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin JN, Chen GF, Gu L, Shen YF, Zheng MZ, Zheng WS, et al Phylogenetic affinity of tree shrews to Glires is attributed to fast evolution rate. Molecular Phylogenetics and Evolution. 2014;71:193–200. doi: 10.1016/j.ympev.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Love MI, Huber W, Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu H, Cherepanova NA, Gilmore R, Contessa JN, Lehrman MA Targeting STT3A-oligosaccharyltransferase with NGI-1 causes herpes simplex virus 1 dysfunction. The FASEB Journal. 2019;33(6):6801–6812. doi: 10.1096/fj.201802044RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu T, Peng HM, Zhong LP, Wu P, He J, Deng ZM, et al The tree shrew as a model for cancer research. Frontiers in Oncology. 2021;11:653236. doi: 10.3389/fonc.2021.653236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo MT, Fan Y, Mu D, Yao YG, Zheng YT Molecular cloning and characterization of APOBEC3 family in tree shrew . Gene. 2018;646:143–152. doi: 10.1016/j.gene.2017.12.060. [DOI] [PubMed] [Google Scholar]
- 56.McGonigle P, Ruggeri B Animal models of human disease: challenges in enabling translation. Biochemical Pharmacology. 2014;87(1):162–171. doi: 10.1016/j.bcp.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 57.McInnes L, Healy J, Melville J. 2020. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv: 1802.03426.
- 58.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ, et al COVID-19: consider cytokine storm syndromes and immunosuppression. The Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Melé M, Mattioli K, Mallard W, Shechner DM, Gerhardinger C, Rinn JL Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Research. 2017;27(1):27–37. doi: 10.1101/gr.214205.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendes FK, Vanderpool D, Fulton B, Hahn MW CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics. 2020:btaa1022. doi: 10.1093/bioinformatics/btaa1022. [DOI] [PubMed] [Google Scholar]
- 61.Ni RJ, Huang ZH, Luo PH, Ma XH, Li T, Zhou JN The tree shrew cerebellum atlas: systematic nomenclature, neurochemical characterization, and afferent projections. Journal of Comparative Neurology. 2018;526(17):2744–2775. doi: 10.1002/cne.24526. [DOI] [PubMed] [Google Scholar]
- 62.Nudelman G, Frasca A, Kent B, Sadler KC, Sealfon SC, Walsh MJ, et al High resolution annotation of zebrafish transcriptome using long-read sequencing. Genome Research. 2018;28(9):1415–1425. doi: 10.1101/gr.223586.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Page RD GeneTree: comparing gene and species phylogenies using reconciled trees. Bioinformatics. 1998;14(9):819–820. doi: 10.1093/bioinformatics/14.9.819. [DOI] [PubMed] [Google Scholar]
- 65.Pertea G, Pertea M GFF utilities: GffRead and GffCompare. F1000Research. 2020;9:304. doi: 10.12688/f1000research.23297.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petry HM, Bickford ME The second visual system of the tree shrew. Journal of Comparative Neurology. 2019;527(3):679–693. doi: 10.1002/cne.24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Phillips JR, Khalaj M, McBrien NA Induced myopia associated with increased scleral creep in chick and tree shrew eyes. Investigative Ophthalmology & Visual Science. 2000;41(8):2028–2034. [PubMed] [Google Scholar]
- 69.Ponjavic J, Oliver PL, Lunter G, Ponting CP Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genetics. 2009;5(8):e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Purugganan MD, Jackson SA Advancing crop genomics from lab to field. Nature Genetics. 2021;53(5):595–601. doi: 10.1038/s41588-021-00866-3. [DOI] [PubMed] [Google Scholar]
- 71.Quinn JJ, Chang HY Unique features of long non-coding RNA biogenesis and function. Nature Reviews Genetics. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 72.Robinson NB, Krieger K, Khan FM, Huffman W, Chang M, Naik A, et al The current state of animal models in research: a review. International Journal of Surgery. 2019;72:9–13. doi: 10.1016/j.ijsu.2019.10.015. [DOI] [PubMed] [Google Scholar]
- 73.Rodriguez JM, Pozo F, di Domenico T, Vazquez J, Tress ML An analysis of tissue-specific alternative splicing at the protein level. PLoS Computational Biology. 2020;16(10):e1008287. doi: 10.1371/journal.pcbi.1008287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rose-John S Interleukin-6 family cytokines. Cold Spring Harbor Perspectives in Biology. 2018;10(2):a028415. doi: 10.1101/cshperspect.a028415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salmela L, Rivals E LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30(24):3506–3514. doi: 10.1093/bioinformatics/btu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanada T, Tsukiyama-Kohara K, Shin IT, Yamamoto N, Kayesh MEH, Yamane D, et al Construction of complete Tupaia belangeri transcriptome database by whole-genome and comprehensive RNA sequencing . Scientific Reports. 2019;9(1):12372. doi: 10.1038/s41598-019-48867-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Savier E, Sedigh-Sarvestani M, Wimmer R, Fitzpatrick D A bright future for the tree shrew in neuroscience research: summary from the inaugural Tree Shrew Users Meeting. Zoological Research. 2021;42(4):478–481. doi: 10.24272/j.issn.2095-8137.2021.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seppey M, Manni M, Zdobnov EM BUSCO: assessing genome assembly and annotation completeness. Methods in Molecular Biology. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 79.Sharon D, Tilgner H, Grubert F, Snyder M A single-molecule long-read survey of the human transcriptome. Nature Biotechnology. 2013;31(11):1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 81.Stark R, Grzelak M, Hadfield J RNA sequencing: the teenage years. Nature Reviews Genetics. 2019;20(11):631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 82.Trincado JL, Entizne JC, Hysenaj G, Singh B, Skalic M, Elliott DJ, et al SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biology. 2018;19(1):40. doi: 10.1186/s13059-018-1417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tu Q, Yang D, Zhang XN, Jia XT, An SQ, Yan LZ, et al A novel pancreatic cancer model originated from transformation of acinar cells in adult tree shrew, a primate-like animal. Disease Models & Mechanisms. 2019;12(4):dmm038703. doi: 10.1242/dmm.038703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ule J, Blencowe BJ Alternative splicing regulatory networks: functions, mechanisms, and evolution. Molecular Cell. 2019;76(2):329–345. doi: 10.1016/j.molcel.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 85.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang GL, Park HJ, Dasari S, Wang SQ, Kocher JP, Li W CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Research. 2013;41(6):e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang K, Wang DH, Zheng XM, Qin A, Zhou J, Guo BY, et al Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton. Nature Communications. 2019;10(1):4714. doi: 10.1038/s41467-019-12575-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, et al BUSCO applications from quality assessments to gene prediction and phylogenomics. Molecular Biology and Evolution. 2018;35(3):543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wei S, Hua HR, Chen QQ, Zhang Y, Chen F, Li SQ, et al Dynamic changes in DNA demethylation in the tree shrew (Tupaia belangeri chinensis) brain during postnatal development and aging . Zoological Research. 2017;38(2):96–102. doi: 10.24272/j.issn.2095-8137.2017.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wortman MJ, Hanson LK, Martínez-Sobrido L, Campbell AE, Nance JA, García-Sastre A, et al Regulation of PURA gene transcription by three promoters generating distinctly spliced 5-prime leaders: a novel means of fine control over tissue specificity and viral signals . BMC Molecular Biology. 2010;11:81. doi: 10.1186/1471-2199-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu TD, Reeder J, Lawrence M, Becker G, Brauer MJ GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. Methods in Molecular Biology. 2016a;1418:283–334. doi: 10.1007/978-1-4939-3578-9_15. [DOI] [PubMed] [Google Scholar]
- 92.Wu XY, Xu HB, Zhang ZG, Chang Q, Liao SS, Zhang LQ, et al Transcriptome profiles using next-generation sequencing reveal liver changes in the early stage of diabetes in tree shrew (Tupaia belangeri chinensis) . Journal of Diabetes Research. 2016b;2016:6238526. doi: 10.1155/2016/6238526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu L, Yu DD, Fan Y, Peng L, Wu Y, Yao YG Loss of RIG-I leads to a functional replacement with MDA5 in the Chinese tree shrew. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(39):10950–10955. doi: 10.1073/pnas.1604939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu L, Yu DD, Ma YH, Yao YL, Luo RH, Feng XL, et al COVID-19-like symptoms observed in Chinese tree shrews infected with SARS-CoV-2. Zoological Research. 2020c;41(5):517–526. doi: 10.24272/j.issn.2095-8137.2020.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu L, Yu DD, Peng L, Wu Y, Fan Y, Gu TL, et al An alternative splicing of tupaia STING modulated anti-RNA virus responses by targeting MDA5-LGP2 and IRF3 . The Journal of Immunology. 2020a;204(12):3191–3204. doi: 10.4049/jimmunol.1901320. [DOI] [PubMed] [Google Scholar]
- 96.Xu L, Yu DD, Yao YL, Gu TL, Zheng X, Wu Y, et al Tupaia MAVS is a dual target during hepatitis c virus infection for innate immune evasion and viral replication via NF-κB . The Journal of Immunology. 2020b;205(8):2091–2099. doi: 10.4049/jimmunol.2000376. [DOI] [PubMed] [Google Scholar]
- 97.Xu XP, Chen HB, Cao XM, Ben KL Efficient infection of tree shrew (Tupaia belangeri) with hepatitis C virus grown in cell culture or from patient plasma . The Journal of General Virology. 2007;88(Pt9):2504–2512. doi: 10.1099/vir.0.82878-0. [DOI] [PubMed] [Google Scholar]
- 98.Yan H, Zhong GC, Xu GW, He WH, Jing ZY, Gao ZC, et al Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yandell M, Ence D A beginner's guide to eukaryotic genome annotation. Nature Reviews Genetics. 2012;13(5):329–342. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- 100.Yao YG Creating animal models, why not use the Chinese tree shrew (Tupaia belangeri chinensis)? . Zoological Research. 2017;38(3):118–126. doi: 10.24272/j.issn.2095-8137.2017.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yao YG, Chen YB, Liang B The 3rd symposium on animal models of primates - the application of non-human primates to basic research and translational medicine. Journal of Genetics and Genomics. 2015;42(6):339–341. doi: 10.1016/j.jgg.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 102.Yao YL, Yu DD, Xu L, Fan Y, Wu Y, Gu TL, et al Molecular characterization of the 2', 5'-oligoadenylate synthetase family in the Chinese tree shrew (Tupaia belangeri chinensis) . Cytokine. 2019;114:106–114. doi: 10.1016/j.cyto.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 103.Yim HS, Cho YS, Guang XM, Kang SG, Jeong JY, Cha SS, et al Minke whale genome and aquatic adaptation in cetaceans. Nature Genetics. 2014;46(1):88–92. doi: 10.1038/ng.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu DD, Wu Y, Xu L, Fan Y, Peng L, Xu M, et al Identification and characterization of toll-like receptors (TLRs) in the Chinese tree shrew (Tupaia belangeri chinensis) . Developmental & Comparative Immunology. 2016;60:127–138. doi: 10.1016/j.dci.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 105.Yu DD, Xu L, Liu XH, Fan Y, Lü LB, Yao YG Diverse interleukin-7 mRNA transcripts in Chinese tree shrew (Tupaia belangeri chinensis) . PLoS One. 2014;9(6):e99859. doi: 10.1371/journal.pone.0099859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu GC, Wang LG, Han YY, He QY clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS:A Journal of Integrative Biology. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang P, Chen JS, Li QY, Sheng LX, Gao YX, Lu BZ, et al Neuroprotectants attenuate hypobaric hypoxia-induced brain injuries in cynomolgus monkeys. Zoological Research. 2020a;41(1):3–19. doi: 10.24272/j.issn.2095-8137.2020.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang XM, Yu DD, Wu Y, Gu TL, Ma N, Dong SZ, et al Establishment and transcriptomic features of an immortalized hepatic cell line of the Chinese tree shrew. Applied Microbiology and Biotechnology. 2020b;104(20):8813–8823. doi: 10.1007/s00253-020-10855-x. [DOI] [PubMed] [Google Scholar]
- 109.Zhao Y, Wang JB, Kuang DX, Xu JW, Yang ML, Ma CX, et al Susceptibility of tree shrew to SARS-CoV-2 infection. Scientific Reports. 2020;10(1):16007. doi: 10.1038/s41598-020-72563-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zheng YT, Yao YG, Xu L. 2014. Basic Biology and Disease Models of Tree Shrews. Kunming: Yunnan Science and Technology Press, 1–475. (in Chinese)
- 111.Zhou YG, Fu BQ, Zheng XH, Wang DS, Zhao CC, Qi YJ, et al Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. National Science Review. 2020;7(6):998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
The TS_3.0 genome annotation data and newly generated RNA-seq and ISO-seq data are available from the Tree shrew Database (http://www.treeshrewdb.org/download/). Related data were also deposited in GSA (accession No. PRJCA006366).