Abstract
Background and purpose
Treatment planning of radiotherapy for locally advanced breast cancer patients can be a time consuming process. Artificial intelligence based treatment planning could be used as a tool to speed up this process and maintain plan quality consistency. The purpose of this study was to create treatment plans for locally advanced breast cancer patients using a Convolutional Neural Network (CNN).
Materials and methods
Data of 60 patients treated for left-sided breast cancer was used with a training, validation and test split of 36/12/12, respectively. The in-house built CNN model was a hierarchically densely connected U-net (HD U-net). The inputs for the HD U-net were 2D distance maps of the relevant regions of interest. Dose predictions, generated by the HD U-net, were used for a mimicking algorithm in order to create clinically deliverable plans.
Results
Dose predictions were generated by the HD U-net and mimicked using a commercial treatment planning system. The predicted plans fulfilling all clinical goals while showing small (≤0.5 Gy) statistically significant differences (p < 0.05) in the doses compared to the manual plans. The mimicked plans show statistically significant differences in the average doses for the heart and lung of ≤0.5 Gy and a reduced D2% of all PTVs. In total, ten of the twelve mimicked plans were clinically acceptable.
Conclusions
We created a CNN model which can generate clinically acceptable plans for left-sided locally advanced breast cancer patients. This model shows great potential to speed up the treatment planning process while maintaining consistent plan quality.
Keywords: Locally advanced breast cancer, Convolutional neural networks, Dose prediction, Dose mimicking, Machine learning
1. Introduction
Worldwide, breast cancer is the most common type of cancer with over 2.2 million incidences in 2020, with a predicted increase to more than 2.4 million incidences by 2025 [1]. Postoperative radiotherapy of the breast is part of the breast-conserving treatment, while radiation of the regional lymph nodes is indicated in some patients with lymph node metastases with the aim of preventing a locoregional recurrence and improving survival [2], [3]. Treatment planning of these patients can be time-consuming and this time and also the quality of the plan may depend on the experience of the treatment planner [4], [5].
In order to decrease treatment planning time and inter-treatment planner variations, different methods are used to automate the treatment planning process for breast cancer patients. There are several studies focusing on specific hard-coded algorithms which can automate the planning process [6], [7], [8]. Other types of studies are focusing on methods that are using previously obtained treatment plans in order to predict dose volume histograms (DVHs) [9], dose distributions [10], [11] or even generate clinically deliverable plans [12]. Many of these studies are using artificial intelligence (AI) involving convolutional neural networks (CNNs) to generate these predictions.
All aforementioned studies showed possible solutions for automating the (breast) planning process, but most CNN based studies focus only on the DVH or dose prediction itself without converting the solution into a clinically deliverable plan and performing an evaluation on the clinical acceptability of these plans. During an earlier study from our institution this challenge was tackled by creating a CNN model for the dose prediction of left-sided node negative breast cancer patients and combining it with a dose mimicking algorithm to transform this prediction into a clinically deliverable plan [13].
However, this model was not optimized for locally advanced breast cancer patients. Besides that, its 2D nature resulted in less spatial information during the prediction phase and mimicking lead to higher average and maximum doses to the regions of interest (ROIs) compared to the clinical dose. Therefore, the purpose of this study was to create clinically deliverable plans for left sided locally advanced breast cancer patients using a new CNN model in combination with a dose mimicking algorithm.
2. Materials and methods
2.1. Patient database
The patient database consisted of 60 patients treated for locally advanced (stage III) left-sided breast cancer including patients treated for lymph nodes level 1 and 2 (30) and level 1, 2, 3 and 4 (30). All patients were treated with step-and-shoot IMRT to a total dose of 40.05 Gy in 15 fractions (15 × 2.67 Gy). Two tangential IMRT beams were used with at least one open segment per direction which combined deliver at least 200 MU to be robust to small breast shape changes. An additional four IMRT beams were added with gantry angles of 30°, 330°, 290° and 190° (for right sided plans). These angles could be adjusted depending on patient anatomy. A total of 20 IMRT segments per plan was allowed. In cases where the D98% clinical goal for the PTVp was not fulfilled after mimicking, the dose was scaled to exactly fulfil this clinical goal. The mean age was 61 years with a range of 45–86 years and all patients were treated between 2018 and 2020 in our institution. Ethical approval was granted by the local ethics committee. From each patient, the relevant ROIs and a manually created treatment plan were used for training purposes. The target volumes were delineated according to the ESTRO guidelines [14]. The manual treatment plans were optimized in such a way that all clinical goals were fulfilled including as much as possible reduction of the dose to the organs at risk (OARs) with a focus on the mean heart and lung doses. An overview of the clinical goals and the relevant ROIs is shown in Table 1. All the ROIs and plans were checked by experienced planners and approved by radiation oncologists to ensure optimal plan quality. The TPS used in this study was RayStation 9B (RaySearch Laboratories AB, Sweden).
Table 1.
Overview of all clinical goals used for the optimized plans in this study. Percentages are indicating the relative volume of the ROI and goals are set with a prescription dose of 40.05 Gy. The ROI names follow the nomenclature according to AAPM TG 263 [15]. The clinical goals are based on the consensus statement of the Dutch society for radiation Oncology [18].
| ROI | Description | Goal |
|---|---|---|
| PTVp | PTV of the whole left breast cropped with 5 mm from external | D98% ≥ 38.0 Gy |
| D2% ≤ 42.8 Gy | ||
| 39.6 Gy ≤ Dmean ≤ 40.4 Gy | ||
| PTVn1n2 | PTV of lymph nodes level 1&2 cropped with 5 mm from external | D98% ≥ 38.0 Gy |
| D2% ≤ 42.8 Gy | ||
| PTVn3n4 | PTV of lymph nodes level 3&4 cropped with 5 mm from external | D98% ≥ 38.0 Gy |
| D2% ≤ 42.8 Gy | ||
| Heart | Heart ROI | Dmean ≤ 2.5 Gy |
| Lungs | Lungs ROI | Dmean ≤ 6.0 Gy |
| V5Gy ≤ 50% | ||
| External-PTV | Full patient body, without all PTVs | V42.85y ≤ 10 cm3 |
| Humerus-PRV10 | Humerus ROI with uniform expansion of 10 mm | V38Gy ≤ 2 cm3 |
| Breast_CL | Contralateral breast | Dmean ≤ 1 Gy |
| Thyroid | Thyroid ROI | V30Gy ≤ 50% |
| Esophagus | Esophagus ROI | V30Gy ≤ 5% |
For training the CNN model, the dataset was split into a training, validation and test set (36/12/12). This split was balanced in such a way that the two different patient groups used in this study were divided equally among the different subsets (lymph nodes level 1 and 2 and level 1, 2, 3 and 4). Furthermore, a 4-fold cross validation method was used to create different splits in the training and validation data during the training phase.
2.2. Deep learning model
The deep learning network used within this study was an enhanced 2D hierarchically, densely connected (HD) CNN. The input channels for this model consisted of all relevant ROI contours, which were also used during the manual planning process. These ROIs were represented by 2D transversal slices of the patient. In total, five different ROI contours were used as an input: PTV (combined PTVp, PTVn1n2 and PTVn3n4), Heart, Lungs, Humerus_PRV10 and External-PTV. All other ROIs were not used, because they were not used during the manual planning process so they should not influence the plan outcome. The five ROI contours were represented by Euclidean distance maps, which are matrices of voxels containing distance information about the ROI. These Euclidean distance maps were used to keep proximity information about the ROIs in the craniocaudal direction, thereby preventing the need for a full 3D model. Mathematically, these Euclidean distance maps can be written as:
where is the distance map value of voxel , the Euclidean distance between voxel and the boundary of the ROI, , and represents the ROI itself. Figure 1 shows a visualization of the Euclidean distance map transformation. The batch size during the training phase was 24, by selecting 8 slices per 3 different patients. The slice selection was done through Gaussian sampling to select the more important central slices containing the PTVs. The Gaussian scheme had a standard deviation that was equal to one-third of the distance from the central slice to the end slice as previously used by Bakx et al. [13].
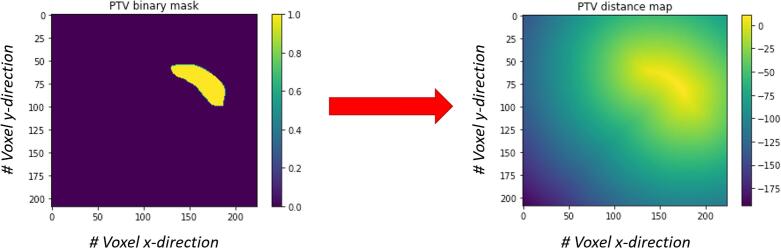
Figure 1.
Visualization of the Euclidean distance map transformation. This example shows one slice of the primary PTV (PTVp) as a binary mask (left) and the distance map transformation (right). Voxels within the PTVp have positive values, voxels outside the PTVp have negative values.
The CNN model was based on a hierarchically, densely connected U-net architecture (HD U-net) which has been used earlier for head and neck cancer patients by Nguyen et al. [16]. A schematic representation of the network is shown in Figure 2. All model parameters were initially a combination of the parameters used by Bakx et al. [13] and the HD U-net from Nguyen et al. [16]. Different models with varying parameters were tested to find an optimal working model for our particular patient group [17], [18]. Eventually, the model represented in Figure 2 was compiled with a mean squared error (MSE) loss function and an Adam optimizer with a learning rate of 0.0001. The model was trained for 800 epochs using a 4-fold cross validation. Model construction and training was done by using Keras API with the TensorFlow backend (v2.2.0) in Python 3.6. Training was done on a NVIDIA V100 GPU with 16 GB RAM and took approximately 4 h. More details about the construction of the model itself can be found in the Supplementary material.
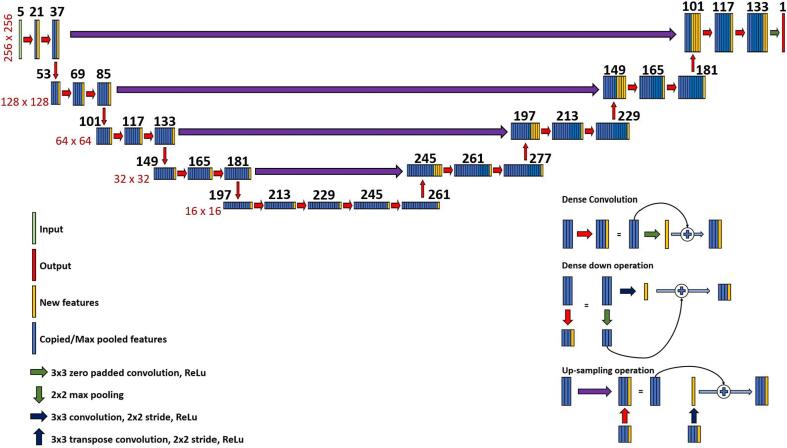
Figure 2.
Schematic representation of the used deep learning architecture: a HD U-net. The black numbers above the feature maps indicate the number of features in that particular layer. The red numbers alongside the feature maps indicate the dimension of the layers in that row.
2.3. Dose mimicking
The voxel-wise dose prediction predicted by the HD U-net, was used as input for the mimicking algorithm in RayStation. The mimicking algorithm used the dose prediction and pre-set ROI goals as inputs. The outcome of this iterative mimicking algorithm is a clinically deliverable plan which best reproduces the predicted dose.
This method was implemented by RaySearch and improved throughout the years [19]. The main purpose of the mimicking algorithm was to approximate the predicted dose distribution. The pre-set ROI goals were similar to the used clinical goals in Table 1 and were equal for all patients. The beam energy and gantry angles were copied from the manual plans of the corresponding patients. The outcome of this mimicking algorithm was a treatment plan in the form of actual machine parameters and a dose distribution, which made the mimicked plan clinically deliverable. The mimicking was done in a research version of RayStation (v8.99). RayStation versions 9 and 8.99 were used for clinical and mimicked dose respectively. The two versions have a type b dose calculation algorithm. For both versions, the same clinical and mimicked dose grid (3 mm3) was used. More details about the mimick settings are given in the supplementary material.
2.4. Model evaluation
The evaluation of the model has been performed based on the clinical goals in Table 1. However, not all clinical goals in Table 1 were considered to be equally relevant. The reason for this was that some of the clinical goals were very specific for our own institution and are not commonly used in other studies or institutes. The most relevant clinical goals were set to be; all PTV goals, the average heart dose, the average lung dose, and the external-PTV clinical goal. An evaluation of all clinical goals from Table 1 was added in Table S1 of the Supplementary material. Besides an evaluation on the clinical goals, the actual obtained DVH values were evaluated as well. A comparison has been made between the manually optimized doses, the predicted doses and the mimicked doses. To find significant differences between the predicted and mimicked doses compared to the manually optimized doses, a Wilcoxon signed rank test was used to determine the p-values. A p-value of <0.05 was considered to be significant.
3. Results
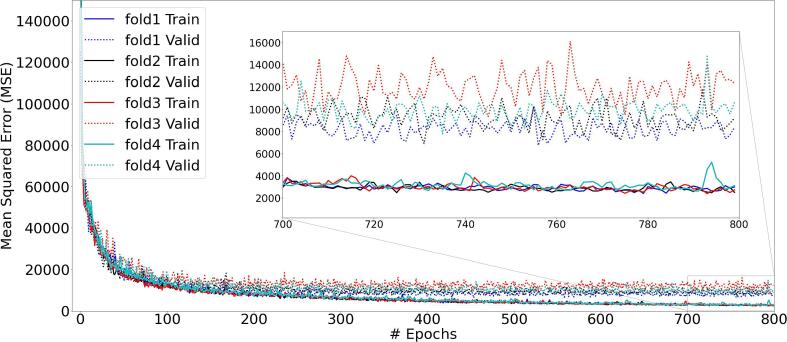
The training and validation losses are converging adequately and there is no sign of overfitting as the results all folds stabilize after a few hundred epochs (Figure 3). The differences in the training losses between the different folds were very small, compared to the differences in the validation losses. Fold 1 was chosen for further testing of the model, because it had the lowest validation loss of all folds.
Figure 3.
Training and validation loss of the HD U-net model for all different folds. All folds show a convergence in the losses while no sign of overfitting is shown.
An evaluation on the DVH values of the relevant clinical goals was performed using 12 test patients. For illustration purposes, the visualization of the evaluated plans (manual, predicted and mimicked) for one test patient is shown in Figure 4.
Figure 4.
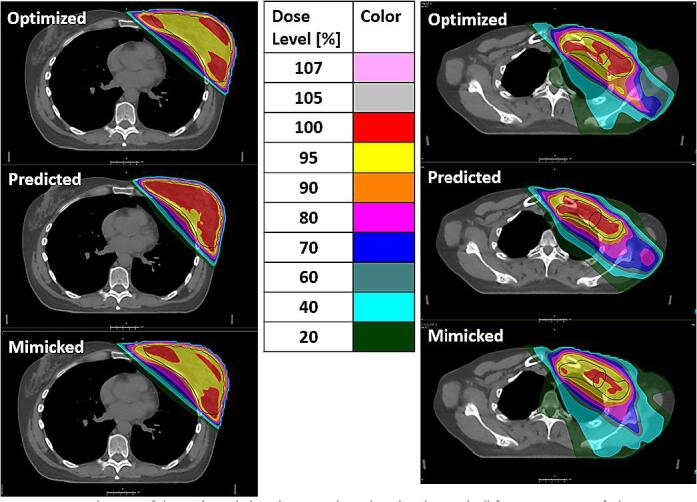
Visualization of the evaluated plans (manual, predicted and mimicked) for one test patient. The colors indicate the relative dose with respect to the prescription dose of 40.05 Gy. Left: PTVp, right: lymph node regions (PTVn1n2 and PTVn3n4). Black contours are indicating the target volumes.
Manual optimized plans and AI test results were for some DVH parameters significantly different (Table 2). The predicted plans showed a significant higher D98% for all PTVs while the D2% was significantly lower for all PTVs. Additionally, the average heart dose and the External-PTV clinical goal showed statistically significant higher values. The predicted plans fulfilled all clinical goals for all 12 test patients. For the mimicked plans, significant lower values for the D2% of all PTVs and higher average doses for the heart and lungs were observed. In total, 10 out of the 12 test patients fulfilled all relevant clinical goals. Full data of all test patients was added in the Supplementary material (Table S1).
Table 2.
Manual optimized planned and AI based test results. The bold values indicate a significant higher dose and the italic values indicate a significant lower dose compared to the optimized plans (p < 0,05, Wilcoxon signed rank test). All values are in Gy except for the External-PTV clinical goal, which is in cm3.
| ROI | n* | Clinical goals | Manual | Predicted | Mimicked |
|---|---|---|---|---|---|
| PTVp | 12 | D98% [Gy] ≥ 38.0 Gy | 38.2 ± 0.2 | 38.7 ± 0.2 | 38.2 ± 0.1 |
| 12 | D2% [Gy] ≤ 42.8 Gy | 41.5 ± 0.3 | 41.1 ± 0.1 | 41.2 ± 0.4 | |
| 12 | 39.6 Gy ≤ Dmean [Gy] ≤ 40.4 Gy | 40.1 ± 0.2 | 40.3 ± 0.1 | 40.0 ± 0.2** | |
| PTVn1n2 | 12 | D98% [Gy] ≥ 38.0 Gy | 38.3 ± 0.3 | 38.7 ± 0.2 | 38.4 ± 0.2 |
| 12 | D2% [Gy] ≤ 42.8 Gy | 41.4 ± 0.3 | 40.8 ± 0.1 | 40.8 ± 0.3 | |
| PTVn3n4 | 6 | D98% [Gy] ≥ 38.0 Gy | 38.3 ± 0.2 | 38.7 ± 0.1 | 38.3 ± 0.2 |
| 6 | D2% [Gy] ≤ 42.8 Gy | 41.6 ± 0.5 | 40.9 ± 0.0 | 40.8 ± 0.1 | |
| Heart | 12 | Dmean [Gy] ≤ 2.5 Gy | 1.3 ± 0.3 | 1.4 ± 0.3 | 1.3 ± 0.3 |
| Lungs | 12 | Dmean [Gy] ≤ 6.0 Gy | 4.3 ± 0.6 | 4.4 ± 0.5 | 4.7 ± 0.5 |
| External-PTV | 12 | V42.85Gy [cm3] ≤ 10.0 cm3 | 0.2 ± 0.5 | 0.9 ± 1.4 | 2.4 ± 5.8** |
*Number of patients included in the evaluation. **One patient in the test set did not fulfil this goal.
4. Discussion
A CNN model has been developed that is able to create dose predictions which fulfil all relevant clinical goals for locally advanced left-sided breast cancer patients. Next to that, it is established that the mimicking algorithm is able to create clinically acceptable plans in 10 out of the 12 cases with the use of these dose predictions. One of the treatment plans that failed one clinical goal had an average PTVp dose of 40.5 Gy which was only slightly higher than the maximum allowed average dose of 40.4 Gy.
The Dutch society for radiation oncology recently reached a consensus about dose evaluation criteria for breast cancer radiotherapy. We based most of our criteria on this consensus statement where the average PTV dose should be within 1% of the prescribed dose. It was conceived as a variation that could be achieved in clinical practice based on a treatment planning benchmark. Currently, there is some discussion about the maximum average dose allowed and maybe the 1% might prove to be a little too stringent in clinical practice. Another treatment plan exceeded the allowed volume for the external clinical goal with 18.8 cm3 at a dose of 42.9 Gy instead of the maximum of 10 cm3. The reason for this can be sub-optimal mimick settings, as these were not optimized for the individual patient.
Although there are small differences in the OAR doses between the mimicked plans and the manual plans, it is unlikely that these differences in mean heart and lung dose of 0.1 to 0.4 Gy on average are clinically relevant. Darby et al. found an excess relative risk (ERR) for major coronary events of 7.4% per Gy on the mean heart dose [20], while Taylor et al. found an ERR for cardiac mortality of 4% per Gy on the mean heart dose and an ERR for RT induced lung cancer of 11% per Gy on the mean lung dose [21].
To our knowledge, this is the first CNN based model for locally advanced breast cancer patients which makes it hard to compare this study with other literature. Besides that, most CNN based breast planning models, in particular for node-negative patients, are only focusing on dose predictions and are not generating clinically applicable plans [9], [10], [11], [16]. This model was based on the HD U-net from Dan Nguyen et al., which was originally used for head and neck cancer patients. [16]. Their trained model was capable of accurately predicting dose distributions with the OAR max dose within 6.3% and the mean dose within 5.1% of the prescription dose. Our study showed that this type of CNN architecture is also able to successfully create dose predictions for a different treatment site and doing so with a 2D distance map based input instead of a full 3D input.
Uncertainty quantification in deep neural networks is still an open issue despite advances in this area. We believe that uncertainty quantification further details a model’s accuracy and facilitates a model’s generalizability. Different uncertainty quantification methods have been reported in diverse settings [22]. However, in this work we did not include uncertainty quantification. To the best of our knowledge, no study has assessed uncertainty for a HD U-net model for dose prediction. It requires a comprehensive literature review and testing the applicability of various uncertainty methods in a trial-and-error fashion, which is outside the scope of the current work presented. We plan to evaluate several methods, including probabilistic forecasting and prediction intervals, as they are the most widely used techniques in literature for uncertainty quantification of neural networks [22]. In general, Bayesian deep learning and Bayesian neural networks will be used to interpret model parameters. Some methods have been proposed for U-net models, including modifications in the original U-net model, specifically adding batch normalization and dropout after each convolutional layer. These methods will be comprehensively discussed in our future studies.
This study used data from 60 patients which is a relatively small cohort. This is due to the limited availability of suitable data from locally advanced breast cancer patients available within our institution. Patients treated before 2018 were considered to be unusable because of differences in clinical goals and ROI margins compared to current patients. Besides that, all patients were treated for left-sided locally advanced breast cancer, which means that cases that differ from this patient group (e.g., right-sided, node negative etc.) will likely be not suitable for this model. An extension of the database, in numbers and different patient groups, may result in a more accurate and widely applicable model. Besides that, the dose mimicking algorithm was only used with a focus on creating clinically deliverable plans, without trying to obtain optimal plans per individual patient. The vendor has already improved the mimick software in a newer software version, meaning that a future study may look into the optimization of new mimick settings for this particular model. This should result in creating mimicked dose distributions that are even better at sparing OARs. Finally, a more elaborate study that focuses on direct machine parameter optimization seems useful. Right now, our CNN model is able to create a voxel-wise dose prediction which is not directly clinically applicable without the use of dose mimicking and pre-set gantry angles. A model which predicts the actual machine parameters could replace the used two-step approach and directly predict a clinically applicable plan to decrease computation time and model complexity.
As a next step a clinical study will be performed to investigate the clinical implementation of AI based treatment planning. During this step focus will be on the time use of manually made versus automated treatment planning to investigate whether the latter already offers time benefits in daily clinical practice. Furthermore, this study will include a qualitative review by physicians, to better investigate clinical applicability.
In conclusion, we have developed a method for automatic treatment plan generation for locally advanced left-sided breast cancer patients using a combination of a CNN model with a dose mimicking algorithm. The resulting treatment plans showed that this method is able to create clinically acceptable treatment plans based on the patient's anatomy. Further improvements like an extension of the patient database, uncertainty quantification, a more optimized mimicking algorithm and a focus on direct machine parameter prediction could enhance the performance and applicability of this method. The current results show great potential for AI based locally advanced breast cancer treatment plan generation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Jorien van der Leer and Thérèse van Nunen for assisting in creating and checking the clinical treatment plans. Furthermore, we would like to thank Mats Holmström and Hanna Gruselius from RaySearch for their feedback and support and RaySearch for funding the PdEng position of Nienke Bakx. Lastly, we would like to thank Carola van Pul from Eindhoven University of Technology (TU/e) for providing capacity to train the model.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2021.11.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Global Cancer Observatory. https://gco.iarc.fr/ [accessed 13 January 2021].
- 2.Early Breast Cancer Trialists' Collaborative Group Effect of radiotherapy after breast-conserving surgery on 10-year recurrence and 15-year breast cancer death: meta-analysis of individual patient data for 10 801 women in 17 randomised trials. The Lancet. 2011;378(9804):1707–1716. doi: 10.1016/S0140-6736(11)61629-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGale P., Correa C., Cutter D., Duane F., Ewertz M., Gray R., et al. Effect of radiotherapy after mastectomy and axillary surgery on 10-year recurrence and 20-year breast cancer mortality: meta-analysis of individual patient data for 8135 women in 22 randomised trials. Lancet. 2014;383(9935):2127–2135. doi: 10.1016/S0140-6736(14)60488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hizam D.A., Jong W.L., Zin H.M., Ng K.H., Ung N.M. Evaluation of treatment plan quality for head and neck IMRT: a multicenter study. Med Dosim. 2021;46(3):310–317. doi: 10.1016/j.meddos.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Wang J., Hu W., Yang Z., Chen X., Wu Z., Yu X., et al. Is it possible for knowledge-based planning to improve intensity modulated radiation therapy plan quality for planners with different planning experiences in left-sided breast cancer patients? Rad Oncol. 2017;12(1) doi: 10.1186/s13014-017-0822-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo B., Shah C., Xia P. Automated planning of whole breast irradiation using hybrid IMRT improves efficiency and quality. J Appl Clin Med Phys. 2019;20(12):87–96. doi: 10.1002/acm2.v20.1210.1002/acm2.12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin T.-C., Lin C.-Y., Li K.-C., Ji J.-H., Liang J.-A., Shiau A.-C., et al. Automated hypofractionated IMRT treatment planning for early-stage breast Cancer. Rad Oncol. 2020;15(1) doi: 10.1186/s13014-020-1468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dragojević I., Hoisak J.D.P., Mansy G.J., Rahn D.A., Manger R.P. Assessing the performance of an automated breast treatment planning software. J Appl Clin Med Phys. 2021;22(4):115–120. doi: 10.1002/acm2.v22.410.1002/acm2.13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma M., Kovalchuk N., Buyyounouski M.K., Xing L., Yang Y. Dosimetric features-driven machine learning model for DVH prediction in VMAT treatment planning. Med Phys. 2019;46(2):857–867. doi: 10.1002/mp.2019.46.issue-210.1002/mp.13334. [DOI] [PubMed] [Google Scholar]
- 10.Fan J., Xing L., Dong P., Wang J., Hu W., Yang Y. Data-driven dose calculation algorithm based on deep U-Net. Phys Med Biol. 2020;65(24) doi: 10.1088/1361-6560/abca05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn S.H., Kim EunSook, Kim C., Cheon W., Kim M., Lee S.B., et al. Deep learning method for prediction of patient-specific dose distribution in breast cancer. Rad Oncol. 2021;16(1) doi: 10.1186/s13014-021-01864-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan J., Wang J., Chen Z., Hu C., Zhang Z., Hu W. Automatic treatment planning based on three-dimensional dose distribution predicted from deep learning technique. Med Phys. 2019;46(1):370–381. doi: 10.1002/mp.2019.46.issue-110.1002/mp.13271. [DOI] [PubMed] [Google Scholar]
- 13.Bakx N., Bluemink H., Hagelaar E., van der Sangen M., Theuws J., Hurkmans C. Development and evaluation of radiotherapy deep learning dose prediction models for breast cancer. Phys Imag Rad Oncol. 2021;1(17):65–70. doi: 10.1016/j.phro.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strnad V., Hannoun-Levi J.-M., Guinot J.-L., Lössl K., Kauer-Dorner D., Resch A., et al. Recommendations from GEC ESTRO Breast Cancer Working Group (I): Target definition and target delineation for accelerated or boost Partial Breast Irradiation using multicatheter interstitial brachytherapy after breast conserving closed cavity surgery. Radiother Oncol. 2015;115(3):342–348. doi: 10.1016/j.radonc.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Mayo C.S., Moran J.M., Bosch W., Xiao Y., McNutt T., Popple R., et al. American Association of Physicists in Medicine Task Group 263: standardizing nomenclatures in radiation oncology. Int J Rad Oncol Biol Phys. 2018;100(4):1057–1066. doi: 10.1016/j.ijrobp.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen D., Jia X., Sher D., Lin M.-H., Iqbal Z., Liu H., et al. 3D radiotherapy dose prediction on head and neck cancer patients with a hierarchically densely connected U-net deep learning architecture. Phys Med Biol. 2019;64(6):065020. doi: 10.1088/1361-6560/ab039b. [DOI] [PubMed] [Google Scholar]
- 17.Yang X.S. Introduction to algorithms for data mining and machine learning. Academic Press. 2019;17:149–180. doi: 10.1016/C2018-0-02034-4. [DOI] [Google Scholar]
- 18.Raschka S. Model evaluation, model selection, and algorithm selection in machine learning. arXiv preprint arXiv:1811.12808. 2018. https://arxiv.org/abs/1811.12808.
- 19.Fredriksson A. Automated improvement of radiation therapy treatment plans by optimization under reference dose constraints. Phys Med Biol. 2012;57(23):7799–7811. doi: 10.1088/0031-9155/57/23/7799. [DOI] [PubMed] [Google Scholar]
- 20.Darby S.C., Ewertz M., McGale P., Bennet A.M., Blom-Goldman U., Brønnum D., et al. Risk of ischemic heart disease in women after radiotherapy for breast cancer. N Engl J Med. 2013;368(11):987–998. doi: 10.1056/NEJMoa1209825. [DOI] [PubMed] [Google Scholar]
- 21.Taylor C., Correa C., Duane F.K., Aznar M.C., Anderson S.J., Bergh J., et al. Estimating the risks of breast cancer radiotherapy: evidence from modern radiation doses to the lungs and heart and from previous randomized trials. J Clin Oncol. 2017;35(15):1641–1649. doi: 10.1200/JCO.2016.72.0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdar M., Pourpanah F., Hussain S., Rezazadegan D., Liu L.i., Ghavamzadeh M., et al. A review of uncertainty quantification in deep learning: Techniques, applications and challenges. Inf Fusion. 2021;76:243–297. doi: 10.1016/j.inffus.2021.05.008. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.