Abstract
Circular RNAs (circRNAs) are a class of single-stranded covalently closed non-coding RNAs without a 5′ cap structure or 3′ terminal poly (A) tail, which are expressed in a variety of tissues and cells with conserved, stable and specific characteristics. Glioblastoma (GBM) is the most aggressive and lethal tumor in the central nervous system, characterized by high recurrence and mortality rates. The specific expression of circRNAs in GBM has demonstrated their potential to become new biomarkers for the development of GBM. The specific expression of circRNAs in GBM has shown their potential as new biomarkers for GBM cell proliferation, apoptosis, migration and invasion, which provides new ideas for GBM treatment. In this paper, we will review the biological properties and functions of circRNAs and their biological roles and clinical applications in GBM.
Keywords: circRNAs, non-coding RNAs, biomarker, glioblastoma, miRNAs sponge
Introduction
Glioblastoma (GBM) is one of the most malignant primary brain tumors in adults, characterized by an expansile and infiltrative growth pattern (Jackson et al., 2019; Tan et al., 2020; McKinnon et al., 2021). According to the World Health Organization (WHO) classification for central nervous system (CNS) tumors, GBM is classified as the highest grade IV (Broekman et al., 2018; Caragher et al., 2018). Currently, the standard of therapy for GBM is surgical resection with maximum safety followed by concurrent radiotherapy and adjuvant chemotherapy (Alifieris and Trafalis, 2015; Karachi et al., 2018; Choi et al., 2019; Geraldo et al., 2019). However, the efficacy of this regimen is limited, and the median survival of patients after treatment is only 15 months (Touat et al., 2017; Lim et al., 2018; Balca-Silva et al., 2019). To better treat GBM patients and improve their survival time and quality of life remains a huge challenge. Therefore, the study of mechanisms regulating the malignant progression of GBM and the explore for early GBM biomarkers are important for the early diagnosis, treatment and prognosis of GBM.
Circular RNAs (circRNAs) are covalently contiguous closed loops without 5′ and 3′ ends, and are structurally more stable than linear RNAs and less susceptible to degradation by nucleic acid exonucleases (Jakobi and Dieterich, 2019; Huang and Zhu, 2021). Initially circRNAs were thought to be products of missplicing or intermediates escaping from the lasso structure of introns (Chen and Huang, 2018; Tsitsipatis and Gorospe, 2021). With the widespread use of transcriptome sequencing technologies, numerous studies have identified circRNAs as a class of endogenous, numerous molecules that are stably present in mammalian cells with certain organizational, temporal, and disease properties and are no longer considered a class of RNA molecules with no role in the human body (Han et al., 2018; Wu J. et al., 2021; Choudhary et al., 2021). CircRNAs present in mammalian cells, there are over 400 circRNAs in normal humans whose abnormal expression can induce tumorigenesis (Ebbesen et al., 2017; Zhou et al., 2020; Zhang et al., 2021a; Shao et al., 2021).
Studies have showed that circRNAs are involved in the occurrence and development of GBM due to their highly stable ring structure, high abundance in cancer tissues and relative tissue specificity, and their altered expression is expected to become a new marker for early diagnosis and prognostic assessment of GBM or a new target for effective treatment. This review summarized the research progress of circRNAs in GBM in recent years, including the mechanism of circRNAs occurrence, function and application research in GBM.
Biogenesis and Classification of Circular RNAs
CircRNAs were first identified in RNA viruses in 1976, and in 1979, Hsu et al. discovered a ring-like molecule with covalently linked 3 and 5′ ends in Hela cells by electron microscopy (Hsu and Coca-Prados, 1979). Because of its special structure, it was often ignored as an abnormal shear by-product. It was not until 1993 that the existence of this structurally unique closed-loop noncoding RNA was confirmed in humans (Farooqi et al., 2021; Wang X. et al., 2021; Lauretti et al., 2021; Sempere et al., 2021). In recent years, with the widespread application of transcriptomic gene sequencing and biophysical techniques, the biological functions of circRNAs and their roles in the development of human diseases are gradually being better understood with the help of high-throughput sequencing technologies. The circRNAs are mainly formed by processing protein-coding genes by RNA polymerase II(Ashwal-Fluss et al., 2014; Li et al., 2019; Ali et al., 2021; Sinha et al., 2021). Meanwhile circRNAs biosynthesis is mediated by RNA binding proteins, intron pair driven and lasso driven, and thus has an important role in regulating adjacent splice sites and promoting circular biosynthesis (Wu et al., 2021a; van Zonneveld et al., 2021; Zhao et al., 2021). Currently circRNAs have also been shown to have many characteristics. Diversity and abundance, circRNAs are widely found in eukaryotic cells and are very diverse (Glazar et al., 2014; Liu et al., 2020; Han et al., 2021). Stability, unlike linear RNA, circRNAs is a single-stranded, covalent closed-loop structure without a 5′ cap structure and a 3′ terminal ploy(A) tail. This structure may protect it from degradation by RNA exonuclease (RNAser) and thus has higher stability than linear RNA (Suzuki and Tsukahara, 2014; Di Timoteo et al., 2020; Wang et al., 2021h). Conservative, circRNAs is highly conserved across species (AbouHaidar et al., 2014; Mao et al., 2021; Meyer et al., 2021). Specificity, mainly in terms of cell type specificity and tissue specificity (Shang et al., 2019; Gokool et al., 2020; Huang and Zhu, 2021).
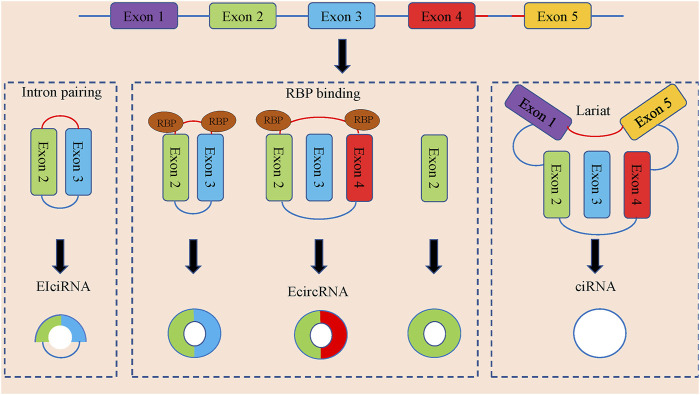
CircRNAs can be divided into three categories according to the composition of splicing (Figure 1). Exonic circRNAs (E-circRNAs) are composed of backward-sheared exons, intron circRNAs (ciRNAs) are composed of introns only, exon-intron circRNAs (eiciRNAs). Contains both exons and introns. Jeck et al., (Jeck et al., 2013), proposed two different exon cyclization modes, lariat-driven cyclization mode and intron pairing-driven cyclization mode.
FIGURE 1.
The formation and classification of circRNAs. CircRNAs are commonly divided into EIciRNAs, EcircRNAs and ciRNAs in accordance with their components, which were derived from exons and introns, and both of them in pre-mRNAs, respectively. Latant ecircRNAs can be generated from one pre-mRNA via alternative splicing. The red segment and blue segment between Exon 4 and Exon 5 represented a 7-nt GU-rich motif near the 5′ splice site and an 11-nt C-rich motif at the branchpoint site, respectively, which promoted the generation of ciRNAs.
Biological Functions of Circular RNAs
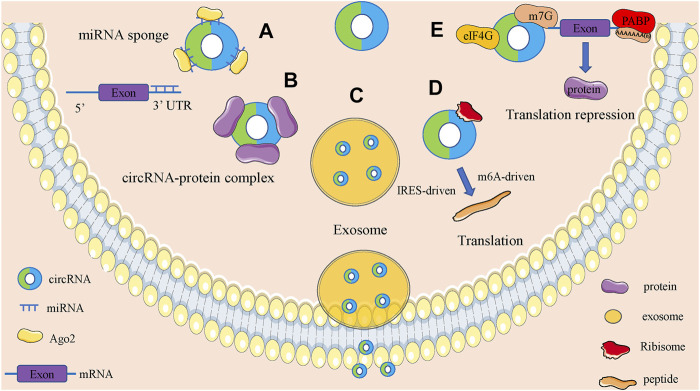
CircRNAs are functionally diverse and are often found to function as microRNAs (miRNAs) sponges because they are rich in miRNAs binding sites. In addition, circRNAs also have roles in regulating parent gene expression, regulating parent gene selective splicing, translating protein functions and participating in intercellular communication by entering exosomes (Figure 2).
FIGURE 2.
Biological function of circRNAs. (A). CircRNAs can act as miRNA sponges to regulate the expression of downstream genes. (B). CircRNAs can bind to RNA binding protein and regulate parental gene expression. (C). Some circRNAs can be carried by exosomes and involved in the process of cell-cell communication. (D). CircRNAs can encode proteins based on IRES-driven and m6A-driven models. (E). CircRNAs can bind with proteins to establish circRNA-protein complexes and alter the functions of some proteins.
MicroRNAs Sponge
Through miRNAs response elements, non-coding RNAs and coding RNAs form a large-scale regulatory network in the transcriptome. MiRNAs are negative regulators of gene expression, reducing the stability of target genes or limiting their translational function (Salmena et al., 2011; Su and Lv, 2020; Shen et al., 2021). CircRNAs are rich in miRNAs binding sites and competitively repress transcriptional regulation of miRNAs, a new class of highly expressed and stable ceRNAs (Shi et al., 2013). The cyclic RNA hsa_circ_0043280 can reduce PAQR3 levels by competitively absorbing miR-203a-3p and blocking miR-203a-3p, and can function as a tumor suppressor to inhibit tumor growth and metastasis in cervical cancer (Zhang et al., 2021b). Hsa_circ_0006349 promotes MKP1 expression through uptake of miR-98, which enhances proliferation and glycolysis of non-small-cell lung cancer (NSCLC) cells and promotes malignant progression of tumors (Qin et al., 2021). Circ-PPP1CB is downregulated in bladder cancer and negatively correlates with clinical stage and histological grade. Circ-PPP1CB regulates cell growth, metastasis and epithelial mesenchymal transition (EMT) by interacting with the miR-1307-3p/SMG1 axis (Wang F. et al., 2021). Circ-EYA3 is elevated in pancreatic ductal adenocarcinoma (PDAC) tissues and cells, and higher levels of circ-EYA3 are significantly associated with poorer prognosis in PDAC patients. Circ-EYA3 can enhance c-Myc expression by acting as an endogenous miR-1294 sponge, which in turn promotes ATP synthesis to increase energy production and promote malignant progression of PDAC (Rong et al., 2021a).
Interaction with RNA-Binding Proteins
CircRNAs can alter splicing patterns or RNA stability by binding to RNA binding protein (RBP) (Huang A. et al., 2020; Zhou et al., 2020; He A. T. et al., 2021; Li J. et al., 2021). Muscleblind (MBL) is a muscleblind-like 1 (mbnll) MBL promotes the production of circmbl, which has a specific MBL binding site, and circmbl has a strong direct interaction with MBL protein (Kristensen et al., 2019). Circ-Mbl regulates MBL protein levels and reduces its own mrna production by promoting circmbl production when MBL is in excess. Circmbl can also eliminate excess MBL by binding to MBL (Ashwal-Fluss et al., 2014). Circrna can facilitate the interaction between DNA, RNA, and RBP to perform biological functions by binding to related proteins (Qi et al., 2021). Circ-RNF13 prolongs the half-life of SUMO2 by binding to the 3′ untranslated regions (3′-UTR) of SUMO2 gene, which leads to sumoylation of GLUT1 and ubiquitination to regulate the AMPK-mtor pathway, ultimately promoting proliferation and metastasis of nasopharyngeal carcinoma (NPC) (Mo et al., 2021). Circ-RHOBTB3 expression is reduced in colorectal cancer tissues, and lower circ-RHOBTB3 levels are significantly associated with advanced clinical stage and greater risk of metastasis. Circ-RHOBTB3 binds to hur to promote β-Trcp1-mediated hur ubiquitination, which in turn inhibits the invasive effects of CRC (Chen J. et al., 2021).
Involved in Intercellular Communication Through Exosomes
The main function of exosomes is to mediate intercellular communication through their contents under physiological and pathological conditions (Wang J. et al., 2021; Chen Q. et al., 2021; Shahzad et al., 2021). In addition, exosomes play a role in coagulation, antigen presentation, immune regulation, and viral replication. Exosome contents, such as proteins, mRNAs, and miRNAs, have been shown to act in receptor cells, thereby activating multiple signaling pathways (Kalluri and LeBleu, 2020; Wen et al., 2020; Rong et al., 2021b; Wang et al., 2021e; Reese and Dhayat, 2021; Tian et al., 2021). Tumor cells can secrete more exosomes than normal cells with some variation in contents, and tumor cell-derived exosomes can provide a suitable microenvironment for tumor development, such as cell proliferation, angiogenesis and metastasis, drug resistance and formation of pre-metastatic microenvironment (Li and Wang, 2017; Yan and Chen, 2020). It has been shown that circRNAs can be encapsulated into exosomes and thus participate in tumorigenesis and progression. Exosome-derived hsa_circ_0000337 accelerates Chemoresistance resistance in esophageal cancer cells by regulating the miR-377-3p/JAK2 axis (Zang et al., 2021). Plasma exosomes from colorectal cancer (CRC) patients are enriched in circ-133. Exosomes circ-133 from hypoxic cells are transmitted to normoxic cells and promote CRC metastasis by acting on the miR-133a/GEF-H1/rhoa axis (Yang H. et al., 2020).
Peptide Translation
Normally circRNAs cannot be flipped, but with deeper research, it was found that exon sequences of some circRNAs can be translated into proteins (Chen and Shan, 2021; Dodbele et al., 2021). Some circRNAs contain internal ribosome entry site (IRES) sequences and can bind directly to ribosomes and can be translated in eukaryotic cells (Abe et al., 2015; He L. et al., 2021). The 40S subunit of eukaryotic ribosomes binds to circRNAs and can directly initiate translation (Wang and Wang, 2015). CircRNAs can also be efficiently translated in an E. Coli cell-free translation system with an open reading flame (ORF) (Thompson, 2012). It has also been shown that eukaryotic endogenous circRNAs can drive protein translation through m6a methylation (Yang et al., 2017; Dai et al., 2020).
Regulation of Gene Expression
CircRNAs can interact with RNA to participate in post-transcriptional regulation. CircRNAs are formed with a balance between competitive complementary pairing between introns and linear RNAs, which affects mrna expression and translation (Shao et al., 2021). The ORF-containing circRNAs produced by COL6A3 encodes a novel 198-aa functional peptide, and hsa_circ_0006401–198-aa promotes the stability of the host gene COL6A3 mrna, thereby facilitating CRC proliferation and translocation (Zhang et al., 2021c). Circ-PTEN can promote CRC proliferation and translocation by acting as a miR-155 Circ-PTEN can increase the expression of its host gene PTEN by acting as a sponge for miR-155 and miR-330-3p, which in turn regulates the PI3K/AKT signaling pathway (Wang et al., 2021i). Regulation of parent genes through RNA polymerase II and epigenetic modifications. Some intron derived circRNAs are mainly localized in the nucleus and can interact with RNA polymerase II to promote transcription of their own coding genes (Li et al., 2015). CircRNAs can also regulate parent gene expression through epigenetic modifications. Recently, some circRNAs were also found to have m6a modifications, which affect the stability of the parent gene (Zhou et al., 2017). Thus, circRNAs can be used to regulate the transcription of disease-related parent genes, which in turn affect the expression of the parent gene and its target genes, providing new ideas for the treatment of corresponding diseases.
Biological Function and Molecular Mechanism of Circular RNAs in Glioblastoma
Studies have shown that circRNAs have important roles in a variety of tumors, and they can be involved in tumorigenesis and progression through many different mechanisms and are closely associated with the clinical features of tumors (Goodall and Wickramasinghe, 2020). Here, we briefly summarize the circRNAs involved in GBM tumorigenesis and progression and analyze their correlation with the clinical features of GBM.
Circular RNAs are associated with Proliferation in Glioblastoma
CircRNAs can regulate the cell proliferation ability of GBM by regulating gene expression or downstream signaling pathways (Table 1). A total of 417 aberrantly expressed circRNAs were found in GBM tissues compared with adjacent normal brains by second-generation sequencing, with hsa_circ_0008344 being the most differentially expressed. Overexpression of hsa_circ_0008344 significantly promoted proliferation, colony formation and decreased apoptosis in GBM cells (Zhou et al., 2018). Eukaryotic initiation factor 4A3 (EIF4A3) bound to MMP9 mrna transcripts induced circ-MMP9 cyclization and promoted circ-MMP9 expression in GBM. MMP9 promotes the proliferative capacity of GBM cells by targeting miR-124 to regulate the expression of CDK4 and AURKA (Wang et al., 2018b). Hsa_circ_0074027 expression is significantly upregulated in GBM and is associated with clinical features. Hsa_circ_0074027 promotes IL17RD expression through sponge binding of miR-518a-5p, which in turn promotes the proliferative capacity of cells (Qian et al., 2019). Circ-PITX1 enhances MAP3K2 expression by binding miR-379-5p as a competitive endogenous RNA (ceRNA) sponge, which promotes cell proliferation and inhibits apoptosis in GBM(Lv et al., 2019). In addition, Cao et al.,. Also found that circ-PITX1 was significantly overexpressed in GBM tissues and cells, and knockdown of circpitx1 inhibited cell proliferation and tumor growth. Circ-PITX1/miR-584-5p/KPNB1 axis may be a potential therapeutic target for GBM (Cao et al., 2021). Hsa_circ_0001801 upregulates HMGB3 expression in GBM through sponge binding of miR-628-5p, thereby promoting cell proliferation (Chen et al., 2019). Zhu et al., Found that circentpd7 (circbase ID:hsa_circ_0019421) was upregulated in GBM tissues, and knockdown of circentpd7 significantly inhibited GBM cell motility and proliferation (Zhu et al., 2020). Hsa_circ-0043278-miR-638/-HOXA9 regulatory axis has an important role in GBM progression by regulating miR-638/- HOXA9. Hsa_circ-0043278-miR-638/- HOXA9 regulatory axis has an important role in GBM progression and can be involved in GBM tumorigenesis and progression by regulating cell proliferation (Wu et al., 2020). Circ-ABCC3 acts as a sponge for miR-770-5p, which targets SOX2, and knockdown of Circ-ABCC3 significantly inhibits tumor growth in vivo (Zhang and Xu, 2021). Wang et al., Found that hsa_circ_0006168 may promote tumor growth in GBM by acting as a competitive endogenous RNA for miR-628-5p and regulating the IGF1R/Ras/Erk pathway (Wang T. et al., 2021). Circ_0001588 may promote the proliferative capacity of GBM cells by regulating the miR-211-5p/YY1 signaling pathway (Wang Q. et al., 2021).
TABLE 1.
The proliferation-related circular RNAs in GBM.
| circRNAs | Expression | Mechanism | Biological function | Ref.PMID |
|---|---|---|---|---|
| hsa_circ_0008344 | Up | — | Promote cell proliferation, colony formation and inhibit cell apoptotic rate | 29687495 |
| circ-MMP9 | Up | miR-124/CDK4 | Promote cell proliferation | 30470262 |
| hsa_circ_0029426 | Up | miR-197 | Promote cell proliferation and inhibit cell apoptosis | 30548670 |
| hsa_circ_0074027 | Up | miR-518a-5p/IL17RD | Promote cell proliferation, colony formation and inhibit cell apoptotic rate | 30738578 |
| hsa_circ_0067934 | Up | PI3K-AKT | Promote cell proliferation | 31081099 |
| circ-PITX1 | Up | miR-379–5p/MAP3K2 | Promote cell proliferation and inhibit cell apoptosis | 31493405 |
| circ-FOXO3 | Up | miR-138-5p/miR-432-5p/NFAT5 | Promote cell proliferation | 31504797 |
| hsa_circ_0001801 | Up | miR-628-5p/HMGB3 | Promote cell proliferation | 31858556 |
| circ-EPB41L5 | Up | miR-19a/EPB41L5 | Promote cell proliferation and colony formation | 31905344 |
| circ-ENTPD7 | Up | miR-101-3p/ROS1 | Promote cell proliferation | 32308563 |
| hsa_circ_0043278 | Up | miR-638/HOXA9 | Promote cell proliferation | 33154193 |
| circ-SMO | Up | SMO-193aa | Promote cell proliferation | 33446260 |
| circ-SKA3 | Up | miR-1 | Promote cell proliferation | 33500664 |
| circ-PARP4 | Up | miR-125a-5p | Promote cell proliferation | 33520365 |
| circ-PITX1 | Up | miR-584-5p/KPNB1 | Promote cell proliferation | 33763840 |
| circ-ABCC3 | Up | miR-770-5p/SOX2 | Promote cell proliferation and inhibit cell apoptosis | 33811842 |
| hsa_circ_0006168 | Up | miR-628-5p/IGF1R | Promote cell proliferation, colony formation and inhibit cell apoptotic rate | 34024251 |
| hsa_circ_0001588 | Up | miR-211-5p/YY1 | Promote cell proliferation | 34105224 |
| circ-FLN1 | Up | miR-199-3p | Promote cell proliferation | 34498720 |
| circ-SERPINE2 | Up | miR-361-3p/miR-324-5p/BCL2 | Promote cell proliferation, colony formation and inhibit cell apoptotic rate | 34553034 |
| — | — | — | ||
| circ-LGMN | Up | miR-127-3p/LGMN | Promote cell proliferation | 34582975 |
| circ-NF1 | Up | miR-340 | Promote cell proliferation | 34589042 |
| circ-NT5E | Down | miR-422a | Inhibit cell proliferation | 29967262 |
| hsa_circ_0001946 | Down | miR-671-5p/CDR1 | Inhibit cell proliferation and promote cell apoptosis | 30663767 |
| circ-MTO1 | Down | miR-92/WWOX | Inhibit cell proliferation | 31456594 |
| circ-AKT3 | Down | AKT3-174aa/PI3K/AKT | Inhibit cell proliferation | 31470874 |
| hsa_circ_01844 | Down | — | Inhibit cell proliferation and promote cell apoptosis | 32804726 |
| circ-CDR1as | Down | p53/MDM2 | Inhibit cell proliferation | 32894144 |
Moreover, circRNAs can also be involved in regulating cell proliferation by binding multiple miRNAs to regulate the expression of downstream genes. Found that circ-FOXO3 can bind both miR-138-5p and miR-432-5p to regulate NFAT5 expression and thus promote cell proliferation (Zhang S. et al., 2019). Li et al., Found that the circ-SERPINE2-miR-361-3p/miR-324-5p/BCL2 signaling pathway plays an important role in the tumor growth process of GBM (Li D. et al., 2021). Besides, hsa_circ_0008344 (Zhou et al., 2018), hsa_circ_0029426 (Zhang G. et al., 2019), hsa_circ_0067934 (Xin et al., 2019), circ-SKA3 (Zhou M. et al., 2021), circ-PARP4 (Zhou J. et al., 2021), circ-FLN1 (Sun et al., 2021) and circ-NF1(Liu L. et al., 2021) were also shown to significantly promote the proliferative capacity and tumor growth of GBM cells, but the specific molecular mechanisms remain to be further explored.
Hsa_circ_0001946 inhibits cell proliferation and promotes apoptosis by binding to miR-671-5p and regulating CDR1 expression (Li and Diao, 2019). Circ-MTO1 inhibits tumor growth of GBM through miR-92/WWOX regulatory axis (Zhang X. et al., 2019). Circ-NT5E inhibits cell proliferation of GBM by binding miR-422a (Wang et al., 2018a). Circ-CDR1as regulates the malignant growth of GBM by modulating the p53/MDM2 signaling pathway (Lou et al., 2020).
In recent years, circRNAs have been found to be involved in disease processes by encoding peptides. Found that circ-SMO encodes a peptide of length 193aa SMO-199aa. Knockdown deprivation of SMO-193aa in GBM stem cells significantly attenuated Hedgehog signaling and inhibited self-renewal, proliferation in vitro and tumorigenicity in vivo (Wu X. et al., 2021). Xia et al., Found that circ-AKT3 encodes a novel 174 aa protein, AKT3-174aa, and overexpression of AKT3-174aa significantly reduced cell proliferation, radioresistance and tumorigenicity of GBM cells in vivo, while overexpression of circ-AKT3 suppressed the malignant phenotype of GBM(Xia et al., 2019).
Circular RNAs are associated with Invasion and Metastasis in Glioblastoma
Invasion and metastasis of tumor cells are the main cause of death in most patients with malignant tumors (Asif et al., 2021). This ability to invade and metastasize allows tumor cells to leave their primary location within tissues, enter lymphatic vessels and blood vessels, and colonize distant organs with the blood circulation. Metastasis of tumor cells is a complex, dynamic process that occurs through cytoskeletal remodeling to form leading edge protrusions, thereby generating mechanical forces that retract and separate the cell tails from the extracellular matrix. CircRNAs have vital roles in the invasion and metastasis of GBM. Hsa_circ_0008344 (Zhou et al., 2018), circ-MMP9(Wang et al., 2018b), hsa_circ_0029426 (Zhang G. et al., 2019), hsa_circ_0074027 (Qian et al., 2019), hsa_circ_0067934 (Xin et al., 2019), circ-FOXO3 (Zhang S. et al., 2019), hsa_circ_0001801 (Chen et al., 2019), circ-EPB41L5 (Lv et al., 2020), circ-ENTPD7 (Zhu et al., 2020), hsa_circ_0043278 (Wu et al., 2020), circ-PARP4 (Zhou J. et al., 2021), circ-SMARCA5 (Barbagallo et al., 2021), circ-PITX1 (Cao et al., 2021), circ-ABCC3(Zhang and Xu, 2021), hsa_circ_0006168 (Wang T. et al., 2021), hsa_circ_0001588 (Wang Q. et al., 2021), circ-MELK (Zhou F. et al., 2021), circ-FLN1 (Sun et al., 2021), circ-LGMN(Chen B. et al., 2021), circ-NT5E (Wang et al., 2018a), hsa_circ_0001946 (Li and Diao, 2019) and circ-MTO1 (Zhang X. et al., 2019) are involved in the regulation of GBM invasion and metastasis (Table 2).
TABLE 2.
The migration and invasion-related circular RNAs in GBM.
| circRNAs | Expression | Mechanism | Biological function | Ref.PMID |
|---|---|---|---|---|
| hsa_circ_0008344 | Up | — | Promote cell migration and invasion | 29687495 |
| circ-MMP9 | Up | miR-124/CDK4 | Promote cell migration and invasion | 30470262 |
| hsa_circ_0029426 | Up | miR-197 | Promote cell migration and invasion | 30548670 |
| hsa_circ_0074027 | Up | miR-518a-5p/IL17RD | Promote cell migration and invasion | 30738578 |
| hsa_circ_0067934 | Up | PI3K-AKT | Promote cell migration and invasion | 31081099 |
| circ-FOXO3 | Up | miR-138-5p/miR-432-5p/NFAT5 | Promote cell migration and invasion | 31504797 |
| hsa_circ_0001801 | Up | miR-628-5p/HMGB3 | Promote cell migration and invasion | 31858556 |
| circ-EPB41L5 | Up | miR-19a/EPB41L5 | Promote cell migration and invasion | 31905344 |
| circ-ENTPD7 | Up | miR-101-3p/ROS1 | Promote cell migration and invasion | 32308563 |
| hsa_circ_0043278 | Up | miR-638/HOXA9 | Promote cell migration and invasion | 33154193 |
| circ-PARP4 | Up | miR-125a-5p | Promote cell migration and invasion | 33520365 |
| circ-SMARCA5 | Up | — | Promote cell migration and invasion | 33562358 |
| circ-PITX1 | Up | miR-584-5p/KPNB1 | Promote cell migration and invasion | 33763840 |
| circ-ABCC3 | Up | miR-770-5p/SOX2 | Promote cell migration and invasion | 33811842 |
| hsa_circ_0006168 | Up | miR-628-5p/IGF1R | Promote cell migration and invasion | 34024251 |
| hsa_circ_0001588 | Up | miR-211-5p/YY1 | Promote cell migration and invasion | 34105224 |
| circ-MELK | Up | miR-593/EphB2 | Promote cell migration and invasion | 34168916 |
| circ-FLN1 | Up | miR-199-3p | Promote cell migration and invasion | 34498720 |
| circ-LGMN | Up | miR-127-3p/LGMN | Promote cell migration and invasion | 34582975 |
| circ-NT5E | Down | miR-422a | Inhibit cell migration and invasion | 29967262 |
| hsa_circ_0001946 | Down | miR-671-5p/CDR1 | Inhibit cell migration and invasion | 30663767 |
| circ-MTO1 | Down | miR-92/WWOX | Inhibit cell migration and invasion | 31456594 |
Epithelial-mesenchymal transition (EMT) is mainly involved in embryogenesis, organogenesis, and tissue healing in humans, but also in tumorigenesis and metastasis, promoting tumor cell invasion and motility by altering intercellular interactions and cell-matrix interactions (Inoue et al., 2015; Wu N. et al., 2021; Satcher and Zhang, 2021; Xiong et al., 2021). About 90% of tumor patient deaths result from tumor invasion and metastasis, which suggests that regulation of the EMT process is important for tumor prevention and treatment. E⁃cadherin expression is suppressed upon EMT activation, resulting in the loss of the typical polygonal cobblestone morphology of epithelial cells, while cells acquire a spindle-shaped mesenchymal morphology and express markers associated with the mesenchymal cell state, particularly N-cadherin, wave proteins and fibronectin (Cristofanilli and Mendelsohn, 2006; Cai et al., 2021; Zhang N. et al., 2021), therefore, the activation status of EMT can be assessed by changes in the expression of E⁃cadherin and N-cadherin. It has been shown that growth factors such as epidermal growth factor (EGF), transcription factors such as Snail, Slug, Twist, E⁃box binding zinc finger protein (ZEB) and signaling pathways such as TGF, Wnt, Notch and Hedgehog can mediate the EMT. Mucin (MUC) acts as an inducer to activate various signaling pathways that also contribute to EMT (Ponnusamy et al., 2013). We found that among the circRNAs involved in GBM, circ-MMP9(Wang et al., 2018b), hsa_circ_0067934 (Xin et al., 2019), hsa_circ_0001801 (Chen et al., 2019), circ-PARP4 (Zhou J. et al., 2021), circ-PITX1 (Cao et al., 2021), hsa_circ_0006168 (Wang T. et al., 2021), hsa_circ_0001588 (Wang Q. et al., 2021) and circ-MELK (Zhou F. et al., 2021) can regulate GBM invasion and metastasis by modulating the EMT process. Hsa_circ_0067934 (Xin et al., 2019), circ-EPB41L5 (Lv et al., 2020) and circ-ABCC3(Zhang and Xu, 2021) can participate in the invasion and metastasis of GBM by regulating the PI3K/Akt/mtor signaling pathway.
Circular RNAs are associated with angiogenesis in Glioblastoma
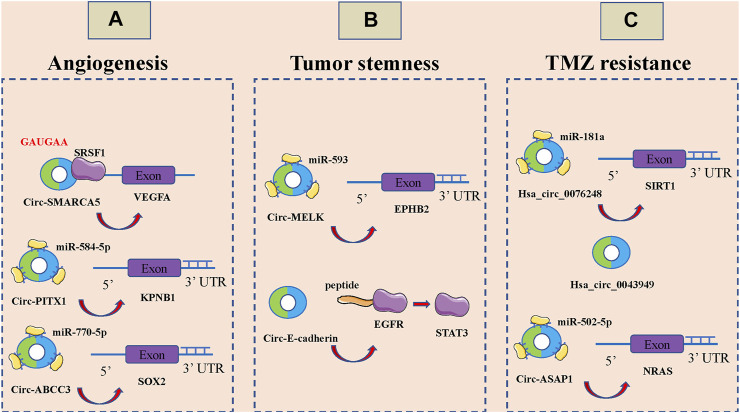
Tumor development, invasion and metastasis are highly dependent on neovascularization (Goncalves et al., 2021; Rimini and Casadei-Gardini, 2021). Under physiological conditions, angiogenesis is intricately and precisely regulated by multiple molecules and mechanisms that allow the formation of highly tissue-specific, structured and hierarchical vascular networks to sustain the physiological processes of embryonic development, growth and tissue repair (Lai et al., 2021; Martin and Gurevich, 2021; Narasimhan et al., 2021). However, tumor cells are able to release large amounts of vascular endothelial growth factor (VEGF) and inhibit the secretion of angiogenesis inhibitory factor, which unbalances the regulatory mechanism of angiogenesis, resulting in rapid, uncontrolled proliferation of tumor neovascularization and a large, abnormally disordered replenishment network (Liu Y. et al., 2021; Uemura et al., 2021; Zhu et al., 2021). CircRNAs can also be involved in the malignant invasion and metastasis of GBM by affecting angiogenesis (Figure 3A). Barbagallo et al., Found that circ-SMARCA5 regulates VEGFA mrna Splicing and angiogenesis through binding to SRSF1, which in turn leads to malignant progression of GBM(Barbagallo et al., 2019). In addition, they demonstrated that the GAUGAA motif is a key sequence for binding of circ-SMARCA5 to SRSF1(Barbagallo et al., 2021). Cao et al., The circ-PITX1/miR-584-5p/KPNB1 regulatory axis was found to be an important molecular mechanism mediating GBM angiogenesis (Cao et al., 2021). Circ-ABCC3 regulates GBM angiogenesis and tumor malignancy progression through PI3K/AKT signaling pathway and miR-770-5p/SOX2 axis (Zhang and Xu, 2021).
FIGURE 3.
CircRNAs associated with the angiogenesis, tumor stemness and temozolomide (TMZ) resistance of glioblastoma (GBM). (A). CircRNAs associated with the angiogenesis of GBM, including circ-SMARCA5, circ-PITX1 and circ-ABCC3. (B). CircRNAs associated with the tumor stemness of GBM, including circ-MELK and circ-E-cadherin. (C): CircRNAs associated with the temozolomide (TMZ) resistance of glioblastoma GBM, including hsa_circ_0076248, hsa_circ_0043949 and circ-ASAP1.
Circular RNAs are associated with Tumor Stemness in Glioblastoma
Stem cells are widely involved in body growth and development and organ formation, and have the ability of self-renewal, infinite proliferation and multidirectional differentiation (Jing et al., 2021; Mehraj et al., 2021; Mirzadeh Azad et al., 2021; Otero-Albiol and Carnero, 2021). The strong self-renewal ability, inherent high proliferative capacity and multidirectional differentiation together constitute the basic characteristics of malignant stem cells, among which the self-renewal ability is closely related to tumorigenesis and malignancy (Ferragut et al., 2021; Mehraj et al., 2021; Yue et al., 2021). Over the past three to 4 decades, numerous studies have noted a potential link between stem cell systems and certain tumors, and a small proportion of tumor-initiating cells with stem cell properties, also known as tumor stem cells, have been identified in a variety of organs (Cermeno and Garcia, 2016; Hass et al., 2020; Pan et al., 2021; Ryskalin et al., 2021). Tumor stem cells of glioma tissue origin have the capacity for self-renewal, homotransplantation into tumors, and differentiation into neurons and glial cells; it is now thought that they may be derived from genetically mutated neural stem cells, transiently expanded cells, neural progenitor cells, and even highly differentiated astrocytes and oligodendrocytes in normal brain tissue. It has been found that circRNAs also play an important role in GBM cell stemness (Figure 3B). Zhou et al., Found that circ-MELK expression was significantly increased in GBM tissues and that circ-MELK could regulate GBMEMT progression and glioma stem cells (GSCs) maintenance by binding to miR-593 to promote ephb2 expression (Zhou F. et al., 2021). Gao et al., Demonstrated that circ-E-cadherin encodes a 14 amino acid peptide that binds to the CR2 structural domain of EGFR and activates EGFR-STAT3 signalling, thereby maintaining the tumorigenicity of glioma stem cells (Gao et al., 2021).
Circular RNAs are associated with Temozolomide Resistance in Glioblastoma
TMZ is an alkylating agent with nearly 100% oral bioavailability and easily crosses the blood-brain barrier, and is currently the first-line chemotherapeutic agent for the treatment of GBM(Al-Toubah et al., 2021; Li F. et al., 2021; Soni et al., 2021). The breakdown products of TMZ can cause DNA methylation after entering tumor cells, which can interfere with cellular DNA replication and cause DNA damage to inhibit the proliferation of tumor cells (Tomar et al., 2021; Winkler, 2021). However, there is a strong DNA damage repair system and complex damage repair mechanisms in GBM cells, which are important in mediating the development of resistance to TMZ in GBM (Trillo Aliaga et al., 2021; Zhang X.-N. et al., 2021). It has been shown that circRNAs are also involved in TMZ resistance of GBM (Figure 3C). Lei et al., Hsa_circ_0076248 was found to be involved in the malignant progression of glioma by binding miR-181a to promote SIRT1 expression, and upregulation of hsa_circ_0076248 significantly inhibited temozolomide chemotherapy sensitivity (Lei et al., 2019). Zhao et al., The expression profiles of circRNAs in three pairs of secondary temozolomide-resistant GBM and the corresponding primary GBM tissues were examined by microarray. The high expression of hsa_circ_0043949 was found to be closely associated with the resistance of TMZ (Zhao et al., 2020). Wei et al., Inhibition of circ-ASAP1 was found to be effective in restoring the sensitivity of TMZ-resistant xenografts to TMZ treatment in vivo, possibly by regulating NRAS expression through binding to miR-502-5p (Wei et al., 2021).
Relationships Between Circular RNAs Levels and Clinicopathologic Characteristics in Glioblastoma
Studies have shown that the expression levels of circRNAs significantly correlate with many clinicopathological features of GBM, including tumor size, grading, differentiation and staging, and tumor recurrence. Found that the expression of hsa_circ_0029426 was correlated with tumor size and WHO classification (Zhang G. et al., 2019). The expression of hsa_circ_0074027 was found to be closely associated with larger tumor size and higher WHO grade (Qian et al., 2019). It was found that high levels of circ-ENTPD7 correlated with advanced GBM classification and tumor size (Zhu et al., 2020). Sun et al., Demonstrated that the expression level of circ-FLNA correlated significantly with the presence of necrosis in MRI scans (Sun et al., 2021) (34498720). Liu et al., A multivariate Cox regression analysis revealed that circnf1 expression was an independent prognostic factor for GBM patients (Liu L. et al., 2021). Lv et al., The analysis found that the expression of circ-EPB41L5 correlated with age, number of lesions, necrotic changes, recurrence and survival in GBM patients (Lv et al., 2020). Wang et al., found that hsa_circ_0006168 expression significantly correlated with WHO classification, T-stage, N-stage and M-stage (Wang T. et al., 2021).
Circular RNAs as Prognostic Biomarkers for Glioblastoma
The expression levels of circRNAs were found to be used to predict the prognosis of tumor patients. To further analyze the prognostic value of circRNAs in GBM, we evaluated the association of circRNAs expression levels with the overall survival (OS) rate of GBM patients. Twelve upregulated circRNAs were reported to predict poorer OS in GBM patients (Zhang G. et al., 2019; Qian et al., 2019; Xin et al., 2019; Zhu et al., 2020; Chen B. et al., 2021; Li D. et al., 2021; Liu L. et al., 2021; Wang Q. et al., 2021; Wu X. et al., 2021; Zhou M. et al., 2021; Sun et al., 2021), and three downregulated circRNAs predicted poorer OS in GBM patients (Zhang X. et al., 2019; Lv et al., 2020; Wang T. et al., 2021). It was shown that lower expression of has_circ_0067934 was associated with longer disease-free survival (DFS) (Xin et al., 2019). Higher expression of circ-EPB41L5 was associated with longer progress-free survival (PFS) (Lv et al., 2020). Kaplan-Meier analysis showed that GBM patients with low circ-ASAP1 expression showed better OS after TMZ treatment compared to GBM patients with high circ-ASAP1 expression (Wei et al., 2021) (Table 3).
TABLE 3.
Utility of circRNAs for clinical of GBM.
| circRNAs | Expression | Clinical Sample | Diagnostic | Utility Prognostic | Predictive | Ref.PMID |
|---|---|---|---|---|---|---|
| has_circ_0029426 | Up | Tissues | — | √ | √ | 30548670 |
| has_circ_0074027 | Up | Tissues | — | — | √ | 30738578 |
| has_circ_0067934 | Up | Tissues | — | √ | — | 31081099 |
| circ-ENTPD7 | Up | Tissues | — | √ | √ | 32308563 |
| circ-SMO | Up | Tissues | — | √ | — | 33446260 |
| circSKA3 | Up | Tissues | — | √ | — | 33500664 |
| hsa_circ_0001588 | Up | Tissues | — | √ | — | 34105224 |
| circ-FLNA | Up | Tissues | — | √ | √ | 34498720 |
| circ-SERPINE2 | Up | Tissues | — | √ | — | 34553034 |
| circ-LGMN | Up | Tissues | — | √ | — | 34582975 |
| circ-NF1 | Up | Tissues | — | √ | √ | 34589042 |
| circ-ASAP1 | Up | Tissues | — | √ | — | 32,926,734 |
| circ-MTO1 | Down | Tissues | — | √ | — | 31456594 |
| circ-EPB41L5 | Down | Tissues | — | √ | √ | 31905344 |
| hsa_circ_0006168 | Down | Tissues | — | — | √ | 34024251 |
Conclusion and Future Prospects
CircRNAs are highly stable, richly expressed, and functionally diverse, and have begun to attract the attention of researchers in recent years, but research on circRNAs is currently in its infancy (Ducoli and Detmar, 2021; Winkle et al., 2021). Nevertheless, the almost complete sequence overlap between circRNAs and linear RNAs makes the accurate assessment of the expression and function of circRNAs still challenging (Yang X. et al., 2020; Jusic et al., 2020). For example, if exonic circRNAs are formed by reverse splicing, how does the spliceosome specifically recognize the exons of circRNAs, but not those of linear RNAs. Recent studies have found that m6a-modified circRNAs are usually derived from exons that are not methylated in mRNAs, and circRNAs from methylated mrna exons are less stable, and it is still unclear whether m6a modification affects the stability of circRNAs. CircRNAs are degraded, and the loop structure may confer different properties to their corresponding linear RNAs. The functional implications of circRNAs have only been tentatively explored, probably due to the limitations of the research tools.
Recent studies on circRNAs have focused on their miRNAs sponge function, researchers have verified the binding sites of circRNAs and miRNAs by luciferase reporter system, pull-down experiments using biotin-labeled probes to capture miRNAs, Ago2 immunoprecipitation can also further investigate miRNAs regulatory targets, co-localization of circRNAs and miRNAs in cells can be verified using FISH technique, and these mature experimental techniques now demonstrate the miRNAs sponge function of circRNAs, making the whole cerna network more complete and complex.
Nowadays, there are more and more researches on the mechanism of circRNAs generation. In addition to RBP can bind to circRNAs to reduce the regulation of RBP on target genes and affect tumorigenesis (Huang C.-K. et al., 2020; Li et al., 2020; Yang J. et al., 2021; Song et al., 2021), some RBP can also regulate the generation of circRNAs, which also has an important role in tumor development (2016; Li H. et al., 2021; Liu Z. et al., 2021; Wang et al., 2021d; Wang and Lei, 2021; Yang T. et al., 2021). The translation function of circRNAs is also becoming a hot topic, and the translation initiation mechanism of circRNAs mainly includes cap-dependent, IRES-dependent, m6a-dependent and small ORF-dependent translation initiation (He L. et al., 2021; Wang X. et al., 2021), and circRNAs encode proteins with the function of suppressing tumor activity and protecting proteins from degradation (Chen C.-K. et al., 2021; Wang et al., 2021d; Ma et al., 2021; Qi et al., 2021). In summary, the interactions between circRNAs and proteins and its translation function are of great significance for the study of tumors, and more in-depth studies should be conducted in this area in the future.
CircRNAs are highly stable and widely expressed in a variety of tissues and body fluids, and thus can be used as potential diagnostic and prognostic biomarkers. Studies have shown that circRNAs are abundant and stable in exosomes, and some circRNAs are more highly expressed in blood than in tissues (D'Ambrosi et al., 2021; Reese and Dhayat, 2021; Tian et al., 2021; van Zonneveld et al., 2021). The high expression and stability of circRNAs in body fluids would be more beneficial for their clinical applications (Bu et al., 2021; Fontemaggi et al., 2021; Wang et al., 2021h; Zhao et al., 2021). However, despite the identification of thousands of tissue- and disease-specific circRNAs by RNA-seq (Kaushik et al., 2021; Tian-Zhao et al., 2021), the understanding of the mechanism of circRNAs generation and biological functions is limited at this stage. Further research is needed to explore the circRNAs associated with GBM, with the aim of being able to be used in combination with traditional biological diagnostic indicators for clinical adjuvant screening of GBM at an early stage, thus indirectly improving the survival rate of GBM patients and achieving early detection and treatment. It is believed that as more and more GBM-related and structurally diverse circRNAs are discovered, the elucidation of complex molecular regulatory mechanisms of GBM and the application of circRNAs-based GBM diagnosis and treatment will have a broad prospect.
In summary, circRNAs are important regulators of GBM genesis and can act as endogenous RNAs or miRNAs sponges competitively repressing miRNAs, thereby altering target gene expression and participating in the development of GBM. Although the exact role of circRNAs in GBM genesis and prognosis is unknown, it is speculated that aberrant expression of circRNAs and their biased distribution in tumors may be common, with different circRNAs expressed up- or down-regulated in GBM cells and tissues, acting through different mechanisms of action.
Acknowledgments
We thank the generous support by Liaoning Cancer Hospital and Institute (Shenyang).
Data Availability Statement
The data in the current study are available from the corresponding authors on reasonable request.
Author Contributions
Original draft preparation, allocation, revision, supplement and edition: XG and HP. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CircRNAs, circular RNAs; GBM, glioblastoma; CNS, central nervous system; Exonic circRNAs, E-circRNAs; ciRNAs, intron circRNAs; eiciRNAs, exon-intron circRNAs; miRNAs, microRNAs; NSCLC, non-small-cell lung cancer; EMT, epithelial mesenchymal transition; PDAC, pancreatic ductal adenocarcinoma; RBP, RNA binding protein; 3′-UTR, 3′ untranslated regions; NPC, nasopharyngeal carcinoma; CRC, colorectal cancer; IRES, internal ribosome entry site; ORF, open reading flame; EIF4A3, Eukaryotic initiation factor 4A3; ceRNA, competitive endogenous RNA; EDF, epidermal growth factor; E⁃box binding zinc finger protein, ZEB; MUc, Mucin; VEGF, vascular endothelial growth factor; GSCs, glioma stem cells; TMZ, temozolomide; OS, overall survival; DFS, disease-free survival; PFS, progress-free survival.
References
- Abe N., Matsumoto K., Nishihara M., Nakano Y., Shibata A., Maruyama H., et al. (2015). Rolling Circle Translation of Circular RNA in Living Human Cells. Sci. Rep. 5, 16435. 10.1038/srep16435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- AbouHaidar M. G., Venkataraman S., Golshani A., Liu B., Ahmad T. (2014). Novel Coding, Translation, and Gene Expression of a Replicating Covalently Closed Circular RNA of 220 Nt. Proc. Natl. Acad. Sci. USA 111 (40), 14542–14547. 10.1073/pnas.1402814111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Toubah T., Pelle E., Valone T., Haider M., Strosberg J. R. (2021). Efficacy and Toxicity Analysis of Capecitabine and Temozolomide in Neuroendocrine Neoplasms. J. Natl. Compr. Canc Netw., 1–8. 10.6004/jnccn.2021.7017 [DOI] [PubMed] [Google Scholar]
- Ali S. A., Peffers M. J., Ormseth M. J., Jurisica I., Kapoor M. (2021). The Non-coding RNA Interactome in Joint Health and Disease. Nat. Rev. Rheumatol. 17, 692–705. 10.1038/s41584-021-00687-y [DOI] [PubMed] [Google Scholar]
- Alifieris C., Trafalis D. T. (2015). Glioblastoma Multiforme: Pathogenesis and Treatment. Pharmacol. Ther. 152, 63–82. 10.1016/j.pharmthera.2015.05.005 [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M., et al. (2014). circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 56 (1), 55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Asif P. J., Longobardi C., Hahne M., Medema J. P. (2021). The Role of Cancer-Associated Fibroblasts in Cancer Invasion and Metastasis. Cancers 13 (18), 4720. 10.3390/cancers13184720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balça-Silva J., Matias D., Carmo A. d., Sarmento-Ribeiro A. B., Lopes M. C., Moura-Neto V. (2019). Cellular and Molecular Mechanisms of Glioblastoma Malignancy: Implications in Resistance and Therapeutic Strategies. Semin. Cancer Biol. 58, 130–141. 10.1016/j.semcancer.2018.09.007 [DOI] [PubMed] [Google Scholar]
- Barbagallo D., Caponnetto A., Barbagallo C., Battaglia R., Mirabella F., Brex D., et al. (2021). The GAUGAA Motif Is Responsible for the Binding between circSMARCA5 and SRSF1 and Related Downstream Effects on Glioblastoma Multiforme Cell Migration and Angiogenic Potential. Ijms 22 (4), 1678. 10.3390/ijms22041678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbagallo D., Caponnetto A., Brex D., Mirabella F., Barbagallo C., Lauretta G., et al. (2019). CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme through the Binding of SRSF1. Cancers 11 (2), 194. 10.3390/cancers11020194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekman M. L., Maas S. L. N., Abels E. R., Mempel T. R., Krichevsky A. M., Breakefield X. O. (2018). Multidimensional Communication in the Microenvirons of Glioblastoma. Nat. Rev. Neurol. 14 (8), 482–495. 10.1038/s41582-018-0025-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu T., Qiao Z., Wang W., Yang X., Zhou J., Chen L., et al. (2021). Diagnostic Biomarker Hsa_circ_0126218 and Functioning Prediction in Peripheral Blood Monocular Cells of Female Patients with Major Depressive Disorder. Front. Cell Dev. Biol. 9, 651803. 10.3389/fcell.2021.651803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Cui Y., Yang J., Wang S. (2021). Epithelial-mesenchymal Transition: When Tumor Cells Meet Myeloid-Derived Suppressor Cells. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1876 (1), 188564. 10.1016/j.bbcan.2021.188564 [DOI] [PubMed] [Google Scholar]
- Cao Y., Wang F., Chen Y., Wang Y., Song H., Long J. (2021). CircPITX1 Regulates Proliferation, Angiogenesis, Migration, Invasion, and Cell Cycle of Human Glioblastoma Cells by Targeting miR-584-5p/KPNB1 Axis. J. Mol. Neurosci. 71 (8), 1683–1695. 10.1007/s12031-021-01820-y [DOI] [PubMed] [Google Scholar]
- Caragher S. P., Hall R. R., Ahsan R., Ahmed A. U. (2018). Monoamines in Glioblastoma: Complex Biology with Therapeutic Potential. Neuro Oncol. 20 (8), 1014–1025. 10.1093/neuonc/nox210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermeño E. A., García A. J. (2016). Tumor-Initiating Cells: Emerging Biophysical Methods of Isolation. Curr. Stem Cell Rep 2 (1), 21–32. 10.1007/s40778-016-0036-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Huang S. (2018). Circular RNA: An Emerging Non-coding RNA as a Regulator and Biomarker in Cancer. Cancer Lett. 418, 41–50. 10.1016/j.canlet.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Chen B., Wang M., Huang R., Liao K., Wang T., Yang R., et al. (2021a). Circular RNA circLGMN Facilitates Glioblastoma Progression by Targeting miR-127-3p/LGMN axis. Cancer Lett. 522, 225–237. 10.1016/j.canlet.2021.09.030 [DOI] [PubMed] [Google Scholar]
- Chen C.-K., Cheng R., Demeter J., Chen J., Weingarten-Gabbay S., Jiang L., et al. (2021b). Structured Elements Drive Extensive Circular RNA Translation. Mol. Cell 81, 4300–4318. 10.1016/j.molcel.2021.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wu Y., Luo X., Jin D., Zhou W., Ju Z., et al. (2021c). Circular RNA circRHOBTB3 Represses Metastasis by Regulating the HuR-Mediated mRNA Stability of PTBP1 in Colorectal Cancer. Theranostics 11 (15), 7507–7526. 10.7150/thno.59546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Shan G. (2021). CircRNA in Cancer: Fundamental Mechanism and Clinical Potential. Cancer Lett. 505, 49–57. 10.1016/j.canlet.2021.02.004 [DOI] [PubMed] [Google Scholar]
- Chen Q., Li Y., Liu Y., Xu W., Zhu X. (2021d). Exosomal Non-coding RNAs-Mediated Crosstalk in the Tumor Microenvironment. Front. Cell Dev. Biol. 9, 646864. 10.3389/fcell.2021.646864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. L., Jiang L., Wang J. S., Liao C. X. (2019). Circ-0001801 Contributes to Cell Proliferation, Migration, Invasion and Epithelial to Mesenchymal Transition (EMT) in Glioblastoma by Regulating miR-628-5p/HMGB3 axis. Eur. Rev. Med. Pharmacol. Sci. 23 (24), 10874–10885. 10.26355/eurrev_201912_19791 [DOI] [PubMed] [Google Scholar]
- Choi B. D., Maus M. V., June C. H., Sampson J. H. (2019). Immunotherapy for Glioblastoma: Adoptive T-Cell Strategies. Clin. Cancer Res. 25 (7), 2042–2048. 10.1158/1078-0432.CCR-18-1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary A., Madbhagat P., Sreepadmanabh M., Bhardwaj V., Chande A. (2021). Circular RNA as an Additional Player in the Conflicts between the Host and the Virus. Front. Immunol. 12, 602006. 10.3389/fimmu.2021.602006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofanilli M., Mendelsohn J. (2006). Circulating Tumor Cells in Breast Cancer: Advanced Tools for "tailored" Therapy? Proc. Natl. Acad. Sci. 103 (46), 17073–17074. 10.1073/pnas.0608651103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai F., Wu Y., Lu Y., An C., Zheng X., Dai L., et al. (2020). Crosstalk between RNA m6A Modification and Non-coding RNA Contributes to Cancer Growth and Progression. Mol. Ther. - Nucleic Acids 22, 62–71. 10.1016/j.omtn.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosi S., Visser A., Antunes-Ferreira M., Poutsma A., Giannoukakos S., Sol N., et al. (2021). The Analysis of Platelet-Derived circRNA Repertoire as Potential Diagnostic Biomarker for Non-small Cell Lung Cancer. Cancers 13 (18), 4644. 10.3390/cancers13184644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Timoteo G., Rossi F., Bozzoni I. (2020). Circular RNAs in Cell Differentiation and Development. Development 147 (16). 10.1242/dev.182725 [DOI] [PubMed] [Google Scholar]
- Dodbele S., Mutlu N., Wilusz J. E. (2021). Best Practices to Ensure Robust Investigation of Circular RNAs: Pitfalls and Tips. EMBO Rep. 22 (3), e52072. 10.15252/embr.202052072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducoli L., Detmar M. (2021). Beyond PROX1: Transcriptional, Epigenetic, and Noncoding RNA Regulation of Lymphatic Identity and Function. Developmental Cell 56 (4), 406–426. 10.1016/j.devcel.2021.01.018 [DOI] [PubMed] [Google Scholar]
- Ebbesen K. K., Hansen T. B., Kjems J. (2017). Insights into Circular RNA Biology. RNA Biol. 14 (8), 1035–1045. 10.1080/15476286.2016.1271524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqi A. A., Attar R., Yulaevna I. M., Berardi R. (2021). Interaction of Long Non-coding RNAs and Circular RNAs with microRNAs for the Regulation of Immunological Responses in Human Cancers. Semin. Cell Developmental Biol. S1084-9521 (21), 00138-5. 10.1016/j.semcdb.2021.05.029 [DOI] [PubMed] [Google Scholar]
- Ferragut F., Vachetta V. S., Troncoso M. F., Rabinovich G. A., Elola M. T. (2021). ALCAM/CD166: A Pleiotropic Mediator of Cell Adhesion, Stemness and Cancer Progression. Cytokine Growth Factor. Rev. 61, 27–37. 10.1016/j.cytogfr.2021.07.001 [DOI] [PubMed] [Google Scholar]
- Fontemaggi G., Turco C., Esposito G., Di Agostino S. (2021). New Molecular Mechanisms and Clinical Impact of circRNAs in Human Cancer. Cancers 13 (13), 3154. 10.3390/cancers13133154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Xia X., Li F., Zhang M., Zhou H., Wu X., et al. (2021). Circular RNA-Encoded Oncogenic E-Cadherin Variant Promotes Glioblastoma Tumorigenicity through Activation of EGFR-STAT3 Signalling. Nat. Cell Biol 23 (3), 278–291. 10.1038/s41556-021-00639-4 [DOI] [PubMed] [Google Scholar]
- Geraldo L. H. M., Garcia C., da Fonseca A. C. C., Dubois L. G. F., de Sampaio e Spohr T. C. L., Matias D., et al. (2019). Glioblastoma Therapy in the Age of Molecular Medicine. Trends Cancer 5 (1), 46–65. 10.1016/j.trecan.2018.11.002 [DOI] [PubMed] [Google Scholar]
- Glažar P., Papavasileiou P., Rajewsky N. (2014). circBase: a Database for Circular RNAs. RNA 20 (11), 1666–1670. 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokool A., Loy C. T., Halliday G. M., Voineagu I. (2020). Circular RNAs: The Brain Transcriptome Comes Full Circle. Trends Neurosciences 43 (10), 752–766. 10.1016/j.tins.2020.07.007 [DOI] [PubMed] [Google Scholar]
- Gonçalves R. C., Banfi A., Oliveira M. B., Mano J. F. (2021). Strategies for Re-vascularization and Promotion of Angiogenesis in Trauma and Disease. Biomaterials 269, 120628. 10.1016/j.biomaterials.2020.120628 [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Wickramasinghe V. O. (2020). RNA in Cancer. Nat. Rev. Cancer 21, 22–36. 10.1038/s41568-020-00306-0 [DOI] [PubMed] [Google Scholar]
- Han B., Chao J., Yao H. (2018). Circular RNA and its Mechanisms in Disease: From the Bench to the Clinic. Pharmacol. Ther. 187, 31–44. 10.1016/j.pharmthera.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Han Y., Zhang H., Bian C., Chen C., Tu S., Guo J., et al. (2021). Circular RNA Expression: Its Potential Regulation and Function in Abdominal Aortic Aneurysms. Oxidative Med. Cell Longevity 2021, 1–21. 10.1155/2021/9934951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass R., von der Ohe J., Ungefroren H. (2020). Impact of the Tumor Microenvironment on Tumor Heterogeneity and Consequences for Cancer Cell Plasticity and Stemness. Cancers 12 (12), 3716. 10.3390/cancers12123716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A. T., Liu J., Li F., Yang B. B. (2021a). Targeting Circular RNAs as a Therapeutic Approach: Current Strategies and Challenges. Sig Transduct Target. Ther. 6 (1), 185. 10.1038/s41392-021-00569-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Man C., Xiang S., Yao L., Wang X., Fan Y. (2021b). Circular RNAs' Cap-independent Translation Protein and its Roles in Carcinomas. Mol. Cancer 20 (1), 119. 10.1186/s12943-021-01417-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M.-T., Coca-Prados M. (1979). Electron Microscopic Evidence for the Circular Form of RNA in the Cytoplasm of Eukaryotic Cells. Nature 280 (5720), 339–340. 10.1038/280339a0 [DOI] [PubMed] [Google Scholar]
- Huang A., Zheng H., Wu Z., Chen M., Huang Y. (2020a). Circular RNA-Protein Interactions: Functions, Mechanisms, and Identification. Theranostics 10 (8), 3503–3517. 10.7150/thno.42174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.-K., Kafert-Kasting S., Thum T. (2020b). Preclinical and Clinical Development of Noncoding RNA Therapeutics for Cardiovascular Disease. Circ. Res. 126 (5), 663–678. 10.1161/CIRCRESAHA.119.315856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Zhu Q. (2021). Mechanisms Regulating Abnormal Circular RNA Biogenesis in Cancer. Cancers 13 (16), 4185. 10.3390/cancers13164185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Umezawa A., Takenaka T., Suzuki H., Okada H. (2015). The Contribution of Epithelial-Mesenchymal Transition to Renal Fibrosis Differs Among Kidney Disease Models. Kidney Int. 87 (1), 233–238. 10.1038/ki.2014.235 [DOI] [PubMed] [Google Scholar]
- Jackson C. M., Choi J., Lim M. (2019). Mechanisms of Immunotherapy Resistance: Lessons from Glioblastoma. Nat. Immunol. 20 (9), 1100–1109. 10.1038/s41590-019-0433-y [DOI] [PubMed] [Google Scholar]
- Jakobi T., Dieterich C. (2019). Computational Approaches for Circular RNA Analysis. WIREs RNA 10 (3), e1528. 10.1002/wrna.1528 [DOI] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 19 (2), 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing N., Gao W.-Q., Fang Y.-X. (2021). Regulation of Formation, Stemness and Therapeutic Resistance of Cancer Stem Cells. Front. Cell Dev. Biol. 9, 641498. 10.3389/fcell.2021.641498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusic A., Devaux Y., Devaux Y. (2020). Mitochondrial Noncoding RNA-Regulatory Network in Cardiovascular Disease. Basic Res. Cardiol. 115 (3), 23. 10.1007/s00395-020-0783-5 [DOI] [PubMed] [Google Scholar]
- Kalluri R., LeBleu V. S. (2020). The Biology , Function , and Biomedical Applications of Exosomes. Science 367 (6478). 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachi A., Dastmalchi F., Mitchell D. A., Rahman M. (2018). Temozolomide for Immunomodulation in the Treatment of Glioblastoma. Neuro Oncol. 20 (12), 1566–1572. 10.1093/neuonc/noy072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik A. C., Wu Q., Lin L., Li H., Zhao L., Wen Z., et al. (2021). Exosomal ncRNAs Profiling of Mycobacterial Infection Identified miRNA-185-5p as a Novel Biomarker for Tuberculosis. Brief Bioinform 22 (6), bbab210. 10.1093/bib/bbab210 [DOI] [PubMed] [Google Scholar]
- Kristensen L. S., Andersen M. S., Stagsted L. V. W., Ebbesen K. K., Hansen T. B., Kjems J. (2019). The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- Lai V., Neshat S. Y., Rakoski A., Pitingolo J., Doloff J. C. (2021). Drug Delivery Strategies in Maximizing Anti-angiogenesis and Anti-tumor Immunity. Adv. Drug Deliv. Rev., 113920. 10.1016/j.addr.2021.113920 [DOI] [PubMed] [Google Scholar]
- Lauretti E., Dabrowski K., Praticò D. (2021). The Neurobiology of Non-coding RNAs and Alzheimer's Disease Pathogenesis: Pathways, Mechanisms and Translational Opportunities. Ageing Res. Rev. 71, 101425. 10.1016/j.arr.2021.101425 [DOI] [PubMed] [Google Scholar]
- Lei B., Huang Y., Zhou Z., Zhao Y., Thapa A. J., Li W., et al. (2019). Circular RNA Hsa_circ_0076248 Promotes Oncogenesis of Glioma by Sponging miR‐181a to Modulate SIRT1 Expression. J. Cell Biochem 120 (4), 6698–6708. 10.1002/jcb.27966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Li L., Chen X., Yang W., Cao Y. (2021a). Circular RNA SERPINE2 Promotes Development of Glioblastoma by Regulating the miR-361-3p/miR-324-5p/BCL2 Signaling Pathway. Mol. Ther. - Oncolytics 22, 483–494. 10.1016/j.omto.2021.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Chen S., Yu J., Gao Z., Sun Z., Yi Y., et al. (2021b). Interplay of M 6 A and Histone Modifications Contributes to Temozolomide Resistance in Glioblastoma. Clin. Translational Med. 11 (9), e553. 10.1002/ctm2.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Deng Z., Yang H., Pan X., Wei Z., Shen H.-B., et al. (2021c). circRNA-binding Protein Site Prediction Based on Multi-View Deep Learning, Subspace Learning and Multi-View Classifier. Brief Bioinform, bbab394. 10.1093/bib/bbab394 [DOI] [PubMed] [Google Scholar]
- Li H. M., Ma X. L., Li H. G. (2019). Intriguing Circles: Conflicts and Controversies in Circular RNA Research. WIREs RNA 10 (5), e1538. 10.1002/wrna.1538 [DOI] [PubMed] [Google Scholar]
- Li J., Wei M., Liu X., Xiao S., Cai Y., Li F., et al. (2021d). The Progress, Prospects, and Challenges of the Use of Non-coding RNA for Diabetic Wounds. Mol. Ther. - Nucleic Acids 24, 554–578. 10.1016/j.omtn.2021.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Diao H. (2019). Circular RNA Circ_0001946 Acts as a Competing Endogenous RNA to Inhibit Glioblastoma Progression by Modulating miR‐671‐5p and CDR1. J. Cell Physiol 234 (8), 13807–13819. 10.1002/jcp.28061 [DOI] [PubMed] [Google Scholar]
- Li X., Wang X. (2017). The Emerging Roles and Therapeutic Potential of Exosomes in Epithelial Ovarian Cancer. Mol. Cancer 16 (1), 92. 10.1186/s12943-017-0659-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ge Y.-z., Xu L., Jia R. (2020). Circular RNA ITCH: A Novel Tumor Suppressor in Multiple Cancers. Life Sci. 254, 117176. 10.1016/j.lfs.2019.117176 [DOI] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., et al. (2015). Exon-intron Circular RNAs Regulate Transcription in the Nucleus. Nat. Struct. Mol. Biol. 22 (3), 256–264. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Lim M., Xia Y., Bettegowda C., Weller M. (2018). Current State of Immunotherapy for Glioblastoma. Nat. Rev. Clin. Oncol. 15 (7), 422–442. 10.1038/s41571-018-0003-5 [DOI] [PubMed] [Google Scholar]
- Liu C., Wu Y., Ma J. (2020). Interaction of Non-coding RNAs and Hippo Signaling: Implications for Tumorigenesis. Cancer Lett. 493, 207–216. 10.1016/j.canlet.2020.08.012 [DOI] [PubMed] [Google Scholar]
- Liu L., Jia L., Shao J., Liu H., Wu Q., Wu X. (2021a). Circular RNA circNF1 siRNA Silencing Inhibits Glioblastoma Cell Proliferation by Promoting the Maturation of miR-340. Front. Neurol. 12, 658076. 10.3389/fneur.2021.658076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Huang N., Liao S., Rothzerg E., Yao F., Li Y., et al. (2021b). Current Research Progress in Targeted Anti‐angiogenesis Therapy for Osteosarcoma. Cell Prolif 54 (9), e13102. 10.1111/cpr.13102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wang T., She Y., Wu K., Gu S., Li L., et al. (2021c). N6-methyladenosine-modified circIGF2BP3 Inhibits CD8+ T-Cell Responses to Facilitate Tumor Immune Evasion by Promoting the Deubiquitination of PD-L1 in Non-small Cell Lung Cancer. Mol. Cancer 20 (1), 105. 10.1186/s12943-021-01398-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J., Hao Y., Lin K., Lyu Y., Chen M., Wang H., et al. (2020). Circular RNA CDR1as Disrupts the p53/MDM2 Complex to Inhibit Gliomagenesis. Mol. Cancer 19 (1), 138. 10.1186/s12943-020-01253-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv T., Miao Y., Xu T., Sun W., Sang Y., Jia F., et al. (2020). Circ-EPB41L5 Regulates the Host Gene EPB41L5 via Sponging miR-19a to Repress Glioblastoma Tumorigenesis. Aging 12 (1), 318–339. 10.18632/aging.102617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X., Wang M., Qiang J., Guo S. (2019). Circular RNA Circ-PITX1 Promotes the Progression of Glioblastoma by Acting as a Competing Endogenous RNA to Regulate miR-379-5p/MAP3K2 axis. Eur. J. Pharmacol. 863, 172643. 10.1016/j.ejphar.2019.172643 [DOI] [PubMed] [Google Scholar]
- Ma J., Du W. W., Zeng K., Wu N., Fang L., Lyu J., et al. (2021). An Antisense Circular RNA circSCRIB Enhances Cancer Progression by Suppressing Parental Gene Splicing and Translation. Mol. Ther. 29 (9), 2754–2768. 10.1016/j.ymthe.2021.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Cao Y., Guo Z., Wang L., Xiang C. (2021). Biological Roles and Therapeutic Potential of Circular RNAs in Osteoarthritis. Mol. Ther. - Nucleic Acids 24, 856–867. 10.1016/j.omtn.2021.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P., Gurevich D. B. (2021). Macrophage Regulation of Angiogenesis in Health and Disease. Semin. Cell Developmental Biol. 119, 101–110. 10.1016/j.semcdb.2021.06.010 [DOI] [PubMed] [Google Scholar]
- McKinnon C., Nandhabalan M., Murray S. A., Plaha P. (2021). Glioblastoma: Clinical Presentation, Diagnosis, and Management. BMJ 374, n1560. 10.1136/bmj.n1560 [DOI] [PubMed] [Google Scholar]
- Mehraj U., Ganai R. A., Macha M. A., Hamid A., Zargar M. A., Bhat A. A., et al. (2021). The Tumor Microenvironment as Driver of Stemness and Therapeutic Resistance in Breast Cancer: New Challenges and Therapeutic Opportunities. Cell Oncol. 10.1007/s13402-021-00634-9 [DOI] [PubMed] [Google Scholar]
- Meyer T., Sand M., Schmitz L., Stockfleth E. (2021). The Role of Circular RNAs in Keratinocyte Carcinomas. Cancers 13 (16), 4240. 10.3390/cancers13164240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Azad F., Polignano I. L., Proserpio V., Oliviero S. (2021). Long Noncoding RNAs in Human Stemness and Differentiation. Trends Cell Biol. 31 (7), 542–555. 10.1016/j.tcb.2021.02.002 [DOI] [PubMed] [Google Scholar]
- Mo Y., Wang Y., Zhang S., Xiong F., Yan Q., Jiang X., et al. (2021). Circular RNA circRNF13 Inhibits Proliferation and Metastasis of Nasopharyngeal Carcinoma via SUMO2. Mol. Cancer 20 (1), 112. 10.1186/s12943-021-01409-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan B., Narasimhan H., Lorente-Ros M., Romeo F. J., Bhatia K., Aronow W. S. (2021). Therapeutic Angiogenesis in Coronary Artery Disease: a Review of Mechanisms and Current Approaches. Expert Opin. Investig. Drugs 30 (9), 947–963. 10.1080/13543784.2021.1964471 [DOI] [PubMed] [Google Scholar]
- Otero-Albiol D., Carnero A. (2021). Cellular Senescence or Stemness: Hypoxia Flips the coin. J. Exp. Clin. Cancer Res. 40 (1), 243. 10.1186/s13046-021-02035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G., Liu Y., Shang L., Zhou F., Yang S. (2021). EMT‐associated microRNAs and Their Roles in Cancer Stemness and Drug Resistance. Cancer Commun. 41 (3), 199–217. 10.1002/cac2.12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy M., Seshacharyulu P., Lakshmanan I., Vaz A., Chugh S., Batra S. (2013). Emerging Role of Mucins in Epithelial to Mesenchymal Transition. Ccdt 13 (9), 945–956. 10.2174/15680096113136660100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Han W., Chen D., Zhao J., Bai L., Huang F., et al. (2021). Engineering Circular RNA Regulators to Specifically Promote Circular RNA Production. Theranostics 11 (15), 7322–7336. 10.7150/thno.56990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L., Guan J., Wu Y., Wang Q. (2019). Upregulated Circular RNA Circ_0074027 Promotes Glioblastoma Cell Growth and Invasion by Regulating miR-518a-5p/IL17RD Signaling Pathway. Biochem. Biophysical Res. Commun. 510 (4), 515–519. 10.1016/j.bbrc.2019.01.140 [DOI] [PubMed] [Google Scholar]
- Qin C., Lu R., Yuan M., Zhao R., Zhou H., Fan X., et al. (2021). Circular RNA 0006349 Augments Glycolysis and Malignance of Non-small Cell Lung Cancer Cells through the microRNA-98/MKP1 Axis. Front. Cell Dev. Biol. 9, 690307. 10.3389/fcell.2021.690307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese M., Dhayat S. A. (2021). Small Extracellular Vesicle Non-coding RNAs in Pancreatic Cancer: Molecular Mechanisms and Clinical Implications. J. Hematol. Oncol. 14 (1), 141. 10.1186/s13045-021-01149-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimini M., Casadei-Gardini A. (2021). Angiogenesis in Biliary Tract Cancer: Targeting and Therapeutic Potential. Expert Opin. Investig. Drugs 30 (4), 411–418. 10.1080/13543784.2021.1881479 [DOI] [PubMed] [Google Scholar]
- Rong Z., Shi S., Tan Z., Xu J., Meng Q., Hua J., et al. (2021a). Circular RNA CircEYA3 Induces Energy Production to Promote Pancreatic Ductal Adenocarcinoma Progression through the miR-1294/c-Myc axis. Mol. Cancer 20 (1), 106. 10.1186/s12943-021-01400-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Z., Xu J., Shi S., Tan Z., Meng Q., Hua J., et al. (2021b). Circular RNA in Pancreatic Cancer: a Novel Avenue for the Roles of Diagnosis and Treatment. Theranostics 11 (6), 2755–2769. 10.7150/thno.56174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryskalin L., Biagioni F., Busceti C. L., Giambelluca M. A., Morelli L., Frati A., et al. (2021). The Role of Cellular Prion Protein in Promoting Stemness and Differentiation in Cancer. Cancers 13 (2), 170. 10.3390/cancers13020170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. P. (2011). A ceRNA Hypothesis: the Rosetta Stone of a Hidden RNA Language? Cell 146 (3), 353–358. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satcher R. L., Zhang X. H.-F. (2021). Evolving Cancer-Niche Interactions and Therapeutic Targets during Bone Metastasis. Nat. Rev. Cancer. 10.1038/s41568-021-00406-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere L. F., Powell K., Rana J., Brock A. A., Schmittgen T. D. (2021). Role of Non-coding RNAs in Tumor Progression and Metastasis in Pancreatic Cancer. Cancer Metastasis Rev. 40, 761–776. 10.1007/s10555-021-09995-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad U., Krumholtz S., Rutka J. T., Das S. (2021). Noncoding RNAs in Glioblastoma: Emerging Biological Concepts and Potential Therapeutic Implications. Cancers 13 (7), 1555. 10.3390/cancers13071555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Q., Yang Z., Jia R., Ge S. (2019). The Novel Roles of circRNAs in Human Cancer. Mol. Cancer 18 (1), 6. 10.1186/s12943-018-0934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao T., Pan Y.-h., Xiong X.-d. (2021). Circular RNA: an Important Player with Multiple Facets to Regulate its Parental Gene Expression. Mol. Ther. - Nucleic Acids 23, 369–376. 10.1016/j.omtn.2020.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Liu B., Xu J., Zhang B., Wang Y., Shi L., et al. (2021). Circular RNAs: Characteristics, Biogenesis, Mechanisms and Functions in Liver Cancer. J. Hematol. Oncol. 14 (1), 134. 10.1186/s13045-021-01145-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Sun M., Liu H., Yao Y., Song Y. (2013). Long Non-coding RNAs: a New Frontier in the Study of Human Diseases. Cancer Lett. 339 (2), 159–166. 10.1016/j.canlet.2013.06.013 [DOI] [PubMed] [Google Scholar]
- Sinha T., Panigrahi C., Das D., Panda A. (2021). Circular RNA Translation, a Path to Hidden Proteome. Wiley Interdiscip. Rev. RNA, e1685. 10.1002/wrna.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Zhang Y., Huang W., Shi J., Huang Q., Jiang M., et al. (2021). Circular RNA Cwc27 Contributes to Alzheimer's Disease Pathogenesis by Repressing Pur-α Activity. Cell Death Differ. 10.1038/s41418-021-00865-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni V., Adhikari M., Simonyan H., Lin L., Sherman J. H., Young C. N., et al. (2021). In Vitro and In Vivo Enhancement of Temozolomide Effect in Human Glioblastoma by Non-invasive Application of Cold Atmospheric Plasma. Cancers 13 (17), 4485. 10.3390/cancers13174485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q., Lv X. (2020). Revealing New Landscape of Cardiovascular Disease through Circular RNA-miRNA-mRNA axis. Genomics 112 (2), 1680–1685. 10.1016/j.ygeno.2019.10.006 [DOI] [PubMed] [Google Scholar]
- Sun Y., Ma G., Xiang H., Wang X., Wang H., Zhang Y., et al. (2021). circFLNA Promotes Glioblastoma Proliferation and Invasion by Negatively Regulating miR1993p Expression. Mol. Med. Rep. 24 (5). 10.3892/mmr.2021.12426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Tsukahara T. (2014). A View of Pre-mRNA Splicing from RNase R Resistant RNAs. Ijms 15 (6), 9331–9342. 10.3390/ijms15069331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A. C., Ashley D. M., López G. Y., Malinzak M., Friedman H. S., Khasraw M. (2020). Management of Glioblastoma: State of the Art and Future Directions. CA A. Cancer J. Clin. 70 (4), 299–312. 10.3322/caac.21613 [DOI] [PubMed] [Google Scholar]
- Thompson S. R. (2012). So You Want to Know if Your Message Has an IRES? WIREs RNA 3 (5), 697–705. 10.1002/wrna.1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Zhao Y., Zheng J., Jin S., Liu Z., Wang T. (2021). Circular RNA: A Potential Diagnostic, Prognostic, and Therapeutic Biomarker for Human Triple-Negative Breast Cancer. Mol. Ther. - Nucleic Acids 26, 63–80. 10.1016/j.omtn.2021.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian-Zhao D., Yang Y., Xing-Xuan W., Yu-Xin C., Xue-Lian W. (2021). Profiling of Circular RNAs and circTPCN/miR-634/mTOR Regulatory Pathway in Cervical Cancer. Genomics 113 (4), 2253–2263. 10.1016/j.ygeno.2021.05.026 [DOI] [PubMed] [Google Scholar]
- Tomar M. S., Kumar A., Srivastava C., Shrivastava A. (2021). Elucidating the Mechanisms of Temozolomide Resistance in Gliomas and the Strategies to Overcome the Resistance. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1876 (2), 188616. 10.1016/j.bbcan.2021.188616 [DOI] [PubMed] [Google Scholar]
- Touat M., Idbaih A., Sanson M., Ligon K. L. (2017). Glioblastoma Targeted Therapy: Updated Approaches from Recent Biological Insights. Ann. Oncol. 28 (7), 1457–1472. 10.1093/annonc/mdx106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trillo Aliaga P., Spada F., Peveri G., Bagnardi V., Fumagalli C., Laffi A., et al. (2021). Should Temozolomide Be Used on the Basis of O6-Methylguanine DNA Methyltransferase Status in Patients with Advanced Neuroendocrine Tumors? A Systematic Review and Meta-Analysis. Cancer Treat. Rev. 99, 102261. 10.1016/j.ctrv.2021.102261 [DOI] [PubMed] [Google Scholar]
- Tsitsipatis D., Gorospe M. (2021). Practical Guide for Circular RNA Analysis: Steps, Tips, and Resources. WIREs RNA 12 (1), e1633. 10.1002/wrna.1633 [DOI] [PubMed] [Google Scholar]
- Uemura A., Fruttiger M., D'Amore P. A., De Falco S., Joussen A. M., Sennlaub F., et al. (2021). VEGFR1 Signaling in Retinal Angiogenesis and Microinflammation. Prog. Retin. Eye Res. 84, 100954. 10.1016/j.preteyeres.2021.100954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zonneveld A. J., Kölling M., Bijkerk R., Lorenzen J. M. (2021). Circular RNAs in Kidney Disease and Cancer. Nat. Rev. Nephrol. 10.1038/s41581-021-00465-9 [DOI] [PubMed] [Google Scholar]
- Wang F., Zhang Y., Zhou X., Chen X., Xiang J., Fan M., et al. (2021a). Circular RNA CircPPP1CB Suppresses Tumorigenesis by Interacting with the MiR-1307-3p/SMG1 Axis in Human Bladder Cancer. Front. Cell Dev. Biol. 9, 704683. 10.3389/fcell.2021.704683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang Y., Liu L., Yang T., Song J. (2021b). Circular RNAs: New Biomarkers of Chemoresistance in Cancer. Cancer Biol. Med. 18, 421–436. 10.20892/j.issn.2095-3941.2020.0312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zheng D., Li Y., Zhang Y., Sui R., Chen Y., et al. (2021c). Circular RNA Circ_0001588 Sponges miR‐211‐5p to Facilitate the Progression of Glioblastoma via Up‐regulating YY1 Expression. J. Gene Med. 23 (10), e3371. 10.1002/jgm.3371 [DOI] [PubMed] [Google Scholar]
- Wang R., Zhang S., Chen X., Li N., Li J., Jia R., et al. (2018a). CircNT5E Acts as a Sponge of miR-422a to Promote Glioblastoma Tumorigenesis. Cancer Res. 78 (17), 4812–4825. 10.1158/0008-5472.CAN-18-0532 [DOI] [PubMed] [Google Scholar]
- Wang R., Zhang S., Chen X., Li N., Li J., Jia R., et al. (2018b). EIF4A3-induced Circular RNA MMP9 (circMMP9) Acts as a Sponge of miR-124 and Promotes Glioblastoma Multiforme Cell Tumorigenesis. Mol. Cancer 17 (1), 166. 10.1186/s12943-018-0911-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Latallo M. J., Zhang Z., Huang B., Bobrovnikov D. G., Dong D., et al. (2021d). Nuclear export and Translation of Circular Repeat-Containing Intronic RNA in C9ORF72-ALS/FTD. Nat. Commun. 12 (1), 4908. 10.1038/s41467-021-25082-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhang K., Tan S., Xin J., Yuan Q., Xu H., et al. (2021e). Circular RNAs in Body Fluids as Cancer Biomarkers: the New Frontier of Liquid Biopsies. Mol. Cancer 20 (1), 13. 10.1186/s12943-020-01298-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Mao P., Feng Y., Cui B., Zhang B., Chen C., et al. (2021f). Blocking Hsa_circ_0006168 Suppresses Cell Proliferation and Motility of Human Glioblastoma Cells by Regulating hsa_circ_0006168/miR-628-5p/IGF1R ceRNA axis. Cell Cycle 20 (12), 1181–1194. 10.1080/15384101.2021.1930357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ma R., Zhang X., Cui L., Ding Y., Shi W., et al. (2021g). Crosstalk between N6-Methyladenosine Modification and Circular RNAs: Current Understanding and Future Directions. Mol. Cancer 20 (1), 121. 10.1186/s12943-021-01415-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., He W., Ibrahim S. A., He Q., Jin J. (2021h). Circular RNAs: Novel Players in the Oxidative Stress-Mediated Pathologies, Biomarkers, and Therapeutic Targets. Oxidative Med. Cell Longevity 2021, 1–14. 10.1155/2021/6634601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Z. (2015). Efficient Backsplicing Produces Translatable Circular mRNAs. RNA 21 (2), 172–179. 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Z., Lu J., Zhang H. (2021i). Circular RNA Circ-PTEN Elevates PTEN Inhibiting the Proliferation of Non-small Cell Lung Cancer Cells. Hum. Cell 34 (4), 1174–1184. 10.1007/s13577-021-00526-y [DOI] [PubMed] [Google Scholar]
- Wang Z., Lei X. (2021). Prediction of RBP Binding Sites on circRNAs Using an LSTM-Based Deep Sequence Learning Architecture. Brief Bioinform 22 (6), bbab342. 10.1093/bib/bbab342 [DOI] [PubMed] [Google Scholar]
- Wei Y., Lu C., Zhou P., Zhao L., Lyu X., Yin J., et al. (2021). EIF4A3-induced Circular RNA ASAP1 Promotes Tumorigenesis and Temozolomide Resistance of Glioblastoma via NRAS/MEK1/ERK1-2 Signaling. Neuro Oncol. 23 (4), 611–624. 10.1093/neuonc/noaa214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen G., Zhou T., Gu W. (2020). The Potential of Using Blood Circular RNA as Liquid Biopsy Biomarker for Human Diseases. Protein Cell. 10.1007/s13238-020-00799-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkle M., El-Daly S. M., Fabbri M., Calin G. A. (2021). Noncoding RNA Therapeutics - Challenges and Potential Solutions. Nat. Rev. Drug Discov. 20 (8), 629–651. 10.1038/s41573-021-00219-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler F. (2021). Silencing Glioblastoma Networks to Make Temozolomide More Effective. Neuro Oncol. 23, 1807–1809. 10.1093/neuonc/noab186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Guo X., Wen Y., Huang S., Yuan X., Tang L., et al. (2021a). N6-Methyladenosine Modification Opens a New Chapter in Circular RNA Biology. Front. Cell Dev. Biol. 9, 709299. 10.3389/fcell.2021.709299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Li Z., Wang J., Geng L., Yue Y., Deng Z., et al. (2021b). Low Molecular Weight Fucoidan Attenuating Pulmonary Fibrosis by Relieving Inflammatory Reaction and Progression of Epithelial-Mesenchymal Transition. Carbohydr. Polym. 273, 118567. 10.1016/j.carbpol.2021.118567 [DOI] [PubMed] [Google Scholar]
- Wu X., Xiao S., Zhang M., Yang L., Zhong J., Li B., et al. (2021c). A Novel Protein Encoded by Circular SMO RNA Is Essential for Hedgehog Signaling Activation and Glioblastoma Tumorigenicity. Genome Biol. 22 (1), 33. 10.1186/s13059-020-02250-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Zheng M., Zhang Y., Xie M., Tian S., Ding T., et al. (2020). Hsa_circ_0043278 Functions as Competitive Endogenous RNA to Enhance Glioblastoma Multiforme Progression by Sponging miR-638. Aging 12 (21), 21114–21128. 10.18632/aging.103603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Li X., Li F., Wu X., Zhang M., Zhou H., et al. (2019). A Novel Tumor Suppressor Protein Encoded by Circular AKT3 RNA Inhibits Glioblastoma Tumorigenicity by Competing with Active Phosphoinositide-dependent Kinase-1. Mol. Cancer 18 (1), 131. 10.1186/s12943-019-1056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin J., Zhang X. Y., Sun D. K., Tian L. Q., Xu P. (2019). Up-regulated Circular RNA Hsa_circ_0067934 Contributes to Glioblastoma Progression through Activating PI3K-AKT Pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (8), 3447–3454. 10.26355/eurrev_201904_17709 [DOI] [PubMed] [Google Scholar]
- Xiong L., Ye X., Chen Z., Fu H., Li S., Xu P., et al. (2021). Advanced Maternal Age‐associated SIRT1 Deficiency Compromises Trophoblast Epithelial−Mesenchymal Transition through an Increase in Vimentin Acetylation. Aging Cell 20, e13491. 10.1111/acel.13491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Chen Y. G. (2020). Circular RNAs in Immune Response and Viral Infection. Trends Biochem. Sci. 45 (12), 1022–1034. 10.1016/j.tibs.2020.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Zhang H., Yang Y., Wang X., Deng T., Liu R., et al. (2020a). Hypoxia Induced Exosomal circRNA Promotes Metastasis of Colorectal Cancer via Targeting GEF-H1/RhoA axis. Theranostics 10 (18), 8211–8226. 10.7150/thno.44419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhang X., Cao J., Xu P., Chen Z., Wang S., et al. (2021a). Circular RNA UBE2Q2 Promotes Malignant Progression of Gastric Cancer by Regulating Signal Transducer and Activator of Transcription 3-mediated Autophagy and Glycolysis. Cell Death Dis 12 (10), 910. 10.1038/s41419-021-04216-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Shen P., Chen Q., Wu P., Yuan H., Ge W., et al. (2021b). FUS-induced circRHOBTB3 Facilitates Cell Proliferation via miR-600/NACC1 Mediated Autophagy Response in Pancreatic Ductal Adenocarcinoma. J. Exp. Clin. Cancer Res. 40 (1), 261. 10.1186/s13046-021-02063-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Liu M., Li M., Zhang S., Hiju H., Sun J., et al. (2020b). Epigenetic Modulations of Noncoding RNA: a Novel Dimension of Cancer Biology. Mol. Cancer 19 (1), 64. 10.1186/s12943-020-01159-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., et al. (2017). Extensive Translation of Circular RNAs Driven by N6-Methyladenosine. Cell Res 27 (5), 626–641. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J., Wu Y., Qiu L., Zhao R., Jiang M., Zhang H. (2021). LncRNAs Link Cancer Stemness to Therapy Resistance. Am. J. Cancer Res. 11 (4), 1051–1068. [PMC free article] [PubMed] [Google Scholar]
- Zang R., Qiu X., Song Y., Wang Y. (2021). Exosomes Mediated Transfer of Circ_0000337 Contributes to Cisplatin (CDDP) Resistance of Esophageal Cancer by Regulating JAK2 via miR-377-3p. Front. Cell Dev. Biol. 9, 673237. 10.3389/fcell.2021.673237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Ding R., Sun Y., Huo S. T., He A., Wen C., et al. (2021a). Circular RNA in Tumor Metastasis. Mol. Ther. - Nucleic Acids 23, 1243–1257. 10.1016/j.omtn.2021.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Liu P., Huang J., Liao Y., Pan C., Liu J., et al. (2021b). Circular RNA Hsa_circ_0043280 Inhibits Cervical Cancer Tumor Growth and Metastasis via miR-203a-3p/PAQR3 axis. Cell Death Dis 12 (10), 888. 10.1038/s41419-021-04193-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhou X., Geng X., Zhang Y., Wang J., Wang Y., et al. (2021c). Circular RNA Hsa_circ_0006401 Promotes Proliferation and Metastasis in Colorectal Carcinoma. Cell Death Dis 12 (5), 443. 10.1038/s41419-021-03714-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Sun W., Zhu L., Feng Y., Wu L., Li T. (2019a). Overexpressed Circ_0029426 in Glioblastoma Forecasts Unfavorable Prognosis and Promotes Cell Progression by Sponging miR‐197. J. Cell Biochem 120 (6), 10295–10302. 10.1002/jcb.28313 [DOI] [PubMed] [Google Scholar]
- Zhang H., Xu W. (2021). CircABCC3 Knockdown Inhibits Glioblastoma Cell Malignancy by Regulating miR-770-5p/SOX2 axis through PI3K/AKT Signaling Pathway. Brain Res. 1764, 147465. 10.1016/j.brainres.2021.147465 [DOI] [PubMed] [Google Scholar]
- Zhang N., Ng A. S., Cai S., Li Q., Yang L., Kerr D. (2021d). Novel Therapeutic Strategies: Targeting Epithelial-Mesenchymal Transition in Colorectal Cancer. Lancet Oncol. 22 (8), e358–e368. 10.1016/S1470-2045(21)00343-0 [DOI] [PubMed] [Google Scholar]
- Zhang S., Liao K., Miao Z., Wang Q., Miao Y., Guo Z., et al. (2019b). CircFOXO3 Promotes Glioblastoma Progression by Acting as a Competing Endogenous RNA for NFAT5. Neuro Oncol. 21 (10), 1284–1296. 10.1093/neuonc/noz128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-N., Yang K.-D., Chen C., He Z.-C., Wang Q.-H., Feng H., et al. (2021e). Pericytes Augment Glioblastoma Cell Resistance to Temozolomide through CCL5-CCR5 Paracrine Signaling. Cell Res 31 (10), 1072–1087. 10.1038/s41422-021-00528-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhong B., Zhang W., Wu J., Wang Y. (2019c). Circular RNA CircMTO1 Inhibits Proliferation of Glioblastoma Cells via miR-92/WWOX Signaling Pathway. Med. Sci. Monit. 25, 6454–6461. 10.12659/MSM.918676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li Z., Qin C., Li T., Wang Y., Cao H., et al. (2021). Mobius Strip in Pancreatic Cancer: Biogenesis, Function and Clinical Significance of Circular RNAs. Cell. Mol. Life Sci. 78 (17-18), 6201–6213. 10.1007/s00018-021-03908-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Gao Y., Guo R., Li H., Yang B. (2020). Microarray Expression Profiles and Bioinformatics Analysis of mRNAs, lncRNAs, and circRNAs in the Secondary Temozolomide-Resistant Glioblastoma. Invest. New Drugs 38 (5), 1227–1235. 10.1007/s10637-019-00884-3 [DOI] [PubMed] [Google Scholar]
- Zhou C., Molinie B., Daneshvar K., Pondick J. V., Wang J., Van Wittenberghe N., et al. (2017). Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-type-specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 20 (9), 2262–2276. 10.1016/j.celrep.2017.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Wang B., Wang H., Hu L., Zhang J., Yu T., et al. (2021a). circMELK Promotes Glioblastoma Multiforme Cell Tumorigenesis through the miR-593/EphB2 axis. Mol. Ther. - Nucleic Acids 25, 25–36. 10.1016/j.omtn.2021.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang H., Hong F., Hu S., Su X., Chen J., et al. (2021b). CircularRNA circPARP4 Promotes Glioblastoma Progression through Sponging miR-125a-5p and Regulating FUT4. Am. J. Cancer Res. 11 (1), 138–156. [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang H., Chu J., Huang Q., Li G., Yan Y., et al. (2018). Circular RNA Hsa_circ_0008344 Regulates Glioblastoma Cell Proliferation, Migration, Invasion, and Apoptosis. J. Clin. Lab. Anal. 32 (7), e22454. 10.1002/jcla.22454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Li H., Chen K. e., Ding W., Yang C., Wang X. (2021c). CircSKA3 Downregulates miR-1 through Methylation in Glioblastoma to Promote Cancer Cell Proliferation. Cmar 13, 509–514. 10.2147/CMAR.S279097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W.-Y., Cai Z.-R., Liu J., Wang D.-S., Ju H.-Q., Xu R.-H. (2020). Circular RNA: Metabolism, Functions and Interactions with Proteins. Mol. Cancer 19 (1), 172. 10.1186/s12943-020-01286-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Cheng C., Qin H., Wang H., Yu H. (2020). A Novel Circular RNA circENTPD7 Contributes to Glioblastoma Progression by Targeting ROS1. Cancer Cell Int 20, 118. 10.1186/s12935-020-01208-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Lama S., Tu L., Dusting G. J., Wang J.-H., Liu G.-S. (2021). TAK1 Signaling Is a Potential Therapeutic Target for Pathological Angiogenesis. Angiogenesis 24 (3), 453–470. 10.1007/s10456-021-09787-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data in the current study are available from the corresponding authors on reasonable request.