Highlights
-
•
Fabry is an X-linked lysosomal storage disease with deficiency in α-galactosidase.
-
•
This deficiency results in the accumulation of glycosphinogolipids.
-
•
Diagnosis is often made by analysis of globotriaosylceramide in fluids and tissues.
-
•
Fabry is often misdiagnosed in female patients due to residual enzyme activity.
-
•
Lipidomics by LC-HRMS enables identification of new biomarkers in Fabry.
Abbreviations: α-GAL A, α-Galactosidase A; IPA, 2-Propanol; CAN, Acetonitrile; CDH, Cerebrodihexoside; Chcl3, Chloroform; Cnvs, Copy Number Variants; ERT, Enzyme Replacement Therapy; EIC, Extracted Ion Chromatogram; Ga2, Galabiosylceramide; GLA, Glactosidase Alpha; Gb3, Globotriaosylceramide; Lyso-Gb3, Globotriaosylsphingosine; LC/MS, Liquid Chromatography-Mass Spectrometry; LSD, Lysosomal Storage Disorder; Meoh, Methanol; ND, Not Detected; OCT, Optimal Cutting Temperature; SRM, Selected Reaction Monitoring; SECIM, Southeast Center for Integrated Metabolomics; MS/MS, Tandem Mass Spectrometry; UHPLC-HRMS, Ultra-High Pressure Liquid Chromatography-High-Resolution Mass Spectrometry
Abstract
Introduction
Lipidomics analysis or lipid profiling is a system-based analysis of all lipids in a sample to provide a comprehensive understanding of lipids within a biological system. In the last few years, lipidomics has made it possible to better understand the metabolic processes associated with several rare disorders and proved to be a powerful tool for their clinical investigation. Fabry disease is a rare X-linked lysosomal storage disorder (LSD) caused by a deficiency in α-galactosidase A (α-GAL A). This deficiency results in the progressive accumulation of glycosphingolipids, mostly globotriaosylceramide (Gb3), globotriaosylsphingosine (lyso-Gb3), as well as galabiosylceramide (Ga2) and their isoforms/analogs in the vascular endothelium, nerves, cardiomyocytes, renal glomerular podocytes, and biological fluids.
Objectives
The primary objective of this study was to evaluate lipidomic signatures in renal biopsies to help understand variations in Fabry disease markers that could be used in future diagnostic tests.
Methods
Lipidomic analysis was performed by ultra-high pressure liquid chromatography-high-resolution mass spectrometry (UHPLC-HRMS) on kidney biopsies that were left over after clinical pathology analysis to diagnose Fabry disease.
Results
We employed UHPLC-HRMS lipidomics analysis on the renal biopsy of a patient suspicious for Fabry disease. Our result confirmed α-GAL A enzyme activity declined in this patient since a Ga2-related lipid biomarker was substantially higher in the patient's renal tissue biopsy compared with two controls. This suggests this patient has a type of LSD that could be non-classical Fabry disease.
Conclusion
This study shows that lipidomics analysis is a valuable tool for rare disorder diagnosis, which can be conducted on leftover tissue samples without disrupting normal patient care.
Introduction
Lysosomal storage disorders (LSDs) are a heterogeneous group of about 50 inherited disorders in humans that result in an accumulation of undegraded substrates originating from a disturbed catabolic pathway that generally occurs in lysosomes [1], [2], [3]. One of these disorders is Fabry disease (OMIM 301500), which is a multisystemic X-linked LSD caused by glactosidase alpha (GLA) gene mutations leading to decreased, or absence of, α-galactosidase A (α-GAL A, EC 3.2.1.22) activity, resulting in the accumulation of glycosphingolipids, predominantly globotriaosylceramide (Gb3), in biological fluids and multiple organs and tissues, such as the walls of small blood vessels, unmyelinated nerves, heart, and kidney [4], [5], [6]. Although Fabry disease is an X-linked disease that occurs earlier in males and is clinically more severe in affected males, many heterozygous females are affected as well [7]. The spectrum of disease in heterozygous female patients is broad and ranges from asymptomatic to mild or severe [8]. Fabry patients may survive into adulthood, but the life-long progression of this disease results in patients suffering from permanent and intense pain. Signs and symptoms of Fabry disease can overlap with other common conditions, such as renal, cerebral, and cardiovascular disease; consequently, diagnosis can be complicated and challenging, and patients are often misdiagnosed or belatedly diagnosed [9]. Treatment to prevent and/or cure Fabry disease has not been realized. Still, enzyme replacement therapy (ERT) delays Fabry's progression by intravenously injecting recombinant α-GAL A into the bloodstream to reduce excess lipid deposits [10], [11], [12]. ERT therapy has been beneficial for improving patients' quality of life, slowing disease progression, and stabilizing renal function and cardiac size [13]. Besides ERT, pharmacological chaperone therapy, which uses oral small molecules to boost endogenous enzyme activity and Gb3 degradation, has been used to slow the progression of Fabry’s disease [14].
Gb3 has been identified as a Fabry disease biomarker that can be used to diagnose, screen, and monitor Fabry patients. Recently, the deacylated form of Gb3, globotriaosylsphingosine (lyso-Gb3), has also been considered as another potential diagnostic biomarker for Fabry disease [15], [16], [17]. Due to the complexity of diagnosing patients with residual α-GAL A enzyme activity, numerous attempts have been made to discover more potent Fabry biomarkers. New studies have revealed various isoforms and analogues of Gb3 and lyso-Gb3 that can also be used as Fabry biomarkers. As described by Auray-Blais et al., a multivariate statistical analysis performed on a subset of male urinary samples revealed seven novel Fabry lysoGb3 analogue biomarkers, all having modified sphingosine moieties (−C2H4, − C2H4 + O, − H2, − H2 + O, + O, + H2O2,+ H2O3) [18]. These seven biomarkers were present in all of the Fabry cohort study participants, but not in the control group. In another study performed by Manwaring et al., five new potential Gb3-related biomarkers in untreated male Fabry disease patients were evaluated and identified using a mass spectrometric metabolomics approach [19]. Three of these novel biomarkers corresponded to Gb3, which has an extra double bond on the sphingosine base with C16:0, C18:0, and C22:1 fatty acyl chains. The next two biomarkers corresponded to a mixture of two structural isomers, the first with a d16:1 sphingosine base and a C16:0 fatty acyl chain and the second with a d18:1 sphingosine base and a C14:0 fatty acyl chain [19]. Besides Gb3 and lyso-Gb3 and their isoforms/analogues that can be potential Fabry biomarkers, Galabiosylceramide (Ga2), also known as digalactosylceramide or cerebrodihexoside (CDH), which is a component of cellular membranes, was also found to have a very high concentration in biological fluids of Fabry patients. As described by Boutin et al., a metabolomic approach was used to identify and study Ga2-related isoforms/analogs of Fabry disease biomarkers. These biomarkers were significantly higher in urine samples collected from untreated Fabry males than healthy controls and can, therefore, be considered as additional potential biomarkers of Fabry disease [20].
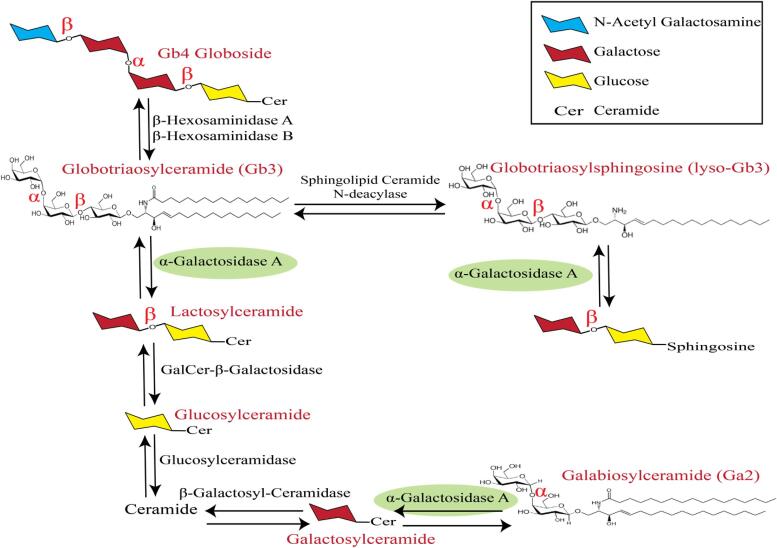
Currently, measuring α-GAL A enzyme activity in plasma and genetic testing are the most common Fabry disease screening methods in clinical labs [21], [22], [23], [24]. The heterogeneity between patients, even those having the same Fabry mutation, has been a considerable issue for Fabry patients’ diagnosis, especially for patients with some residual enzyme activity and patients with unknown mutations [25]. In these cases, analyzing Gb3, lyso-Gb3, andGa2 and their isoforms and analogues can significantly improve Fabry patient diagnosis and screening. Further, demonstrating an elevated level of Gb3 and other related biomarkers in a disease relevant organ like the kidney is a more specific approach to determining whether α-GAL A enzyme deficiency is clinically significant [26]. Fig. 1 shows the metabolic and catabolic pathway of α-GAL A enzyme activity, describing why decreased activity or absence of this enzyme can lead to accumulation of Gb3, lyso-Gb3, and Ga2.
Fig. 1.
Metabolic and catabolic pathway of α-GAL A enzyme activity. As described in this figure, α-GAL A is able to break down α bonds in Glycosphingolipid species, therefore, decreased level or absence of this enzyme can lead to the accumulation of Gb3, lyso-Gb3, and Ga2.
The study of the cellular, tissue or plasma lipidome using methods and principles of analytical chemistry is known as lipidomics [27], [28], [29]. Lipidomics provides a powerful tool for the advancement of lipid biomarkers to study disease states. Primarily, this approach enables studying cellular metabolism by quantifying the differences of individual lipid classes, subclasses, and molecular species that indicate metabolic variations [30]. Liquid chromatography-mass spectrometry (LC/MS), which integrates excellent separation efficiency, high sensitivity, and specificity, is the most essential and mainstream method for lipidomics research [31], [32]. Lipidomics has already proven useful for clinical diagnosis of rare diseases and has allowed a deeper understanding of patients’ associated lipidomic processes while proving to be a powerful tool for the clinical investigation of rare diseases [33], [34], [35]. In the same way, LC/MS-based lipidomics analysis is rapidly becoming the preferred technique to profile novel biomarkers in Fabry's disease [17], [36], [37]. In this study, by employing ultra-high pressure liquid chromatography-high-resolution mass spectrometry (UHPLC-HRMS), we analyzed the lipidomic profile of a female patient with features of Fabry disease using her leftover renal tissue biopsy with results compared to two control samples. This patient showed a normal level of α-GAL A enzyme activity. Sequence analysis of the entire GLA gene using whole genome sequencing revealed no reportable sequence or copy number variants (CNVs). To better diagnose this patient, we presented targeted profiling of all Fabry lipid biomarkers mentioned previously, including Gb3, lyso-Gb3, Ga2, and their isoforms and analogues that can be considered as Fabry biomarkers.
Materials and methods
Chemicals and reagents
LC-MS grade water with 0.1% formic acid, acetonitrile (ACN), 2-propanol (IPA), and all other analytical-grade solvents, including ammonium acetate, methanol (MeOH), ammonium formate, and chloroform (CHCl3) were purchased from Thermo Fisher Scientific (Waltham, MA). SPLASH® LipidoMix® Internal Standard Mix and all lipid injection standards were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL).
Patient information and biopsy details
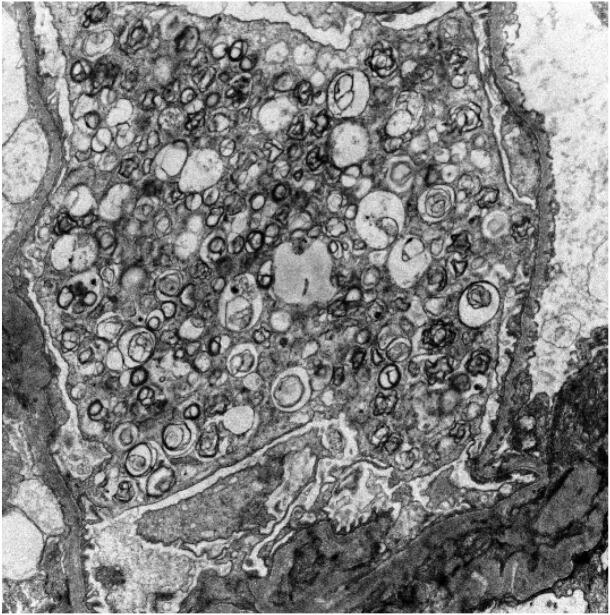
The patient was a 57–year old female presenting with proteinuria. She had a history of breast cancer (1991 and 2012 treated with radiation and chemotherapy). Genetic testing revealed no mutations in the BRCA1, BRCA2 or p53 genes. Symptoms included tinnitus, hypohidrosis, abdominal pain episodes and a peripheral neuropathy, which was thought to be secondary to chemotherapy. There was no family history of renal disease or any known genetic disease. Medications included aspirin, atorvastatin, gabapentin, levothyroxine, metformin and valsartan. The patient had a history of diabetes. Proteinuria, documented since 2013, was believed secondary to diabetes. However, the patient’s proteinuria (elevated microalbumin/creatinine of 2501, a ratio of microalbumin (mcg/L) to creatinine (mg/L) less than 30 is considered normal; a ratio of 30 to 300 denotes microalbuminuria, and values over 300 are macroalbuminuria [38], [39]) seemed more severe than that associated with her diabetes (the HbA1c value is 7). A renal biopsy revealed minimal interstitial fibrosis and tubular atrophy (<10%), no significant acute tubulointerstitial changes, focal mild arterial hyalinosis and normocellular glomeruli with podocytes displaying foamy cytoplasm. Immunofluorescence showed tubular cytoplasmic protein droplets staining for albumin, but no glomerular staining for immunoglobulin or complement components. Electron microscopy revealed abundant lamellated lipid inclusions (myeloid bodies) in the cytoplasm of podocytes (Fig. 2). No electron dense deposits or tubuloreticular inclusions were identified. There was no known exposure of the patient to hydroxychloroqine, chloroquine, amiodarone, gentamicin, or silicone; agents that have been associated with similar podocyte lamellated inclusions [40], [41], [42]. The diagnosis was ultrastructural findings consistent with lysosomal storage disease, favoring Fabry disease. The α-GAL A enzyme levels were within normal limits at 6.03 µmol/L/hr (normal > 1.10 µmol/L/hr). Lyso-Gb3 was 1.59 ng/mL, which was outside the reference range (normal < 1.11 ng/mL [6], [43]). Analysis of the entire GLA gene using whole genome sequencing did not identify any reportable sequence or copy number variants.
Fig. 2.
Electron micrograph showing numerous myeloid bodies within podocyte cytoplasm.
The University of Florida IRB has issued the following case report guidance: A case report for IRB purposes is a retrospective analysis of one, two or three clinical cases. A case report is a medical activity and does not have to be reviewed by the UF IRB. If no HIPPA identifiers are in the data/manuscript, authors do not need to obtain a signed privacy authorization from the patient.
Sample preparation
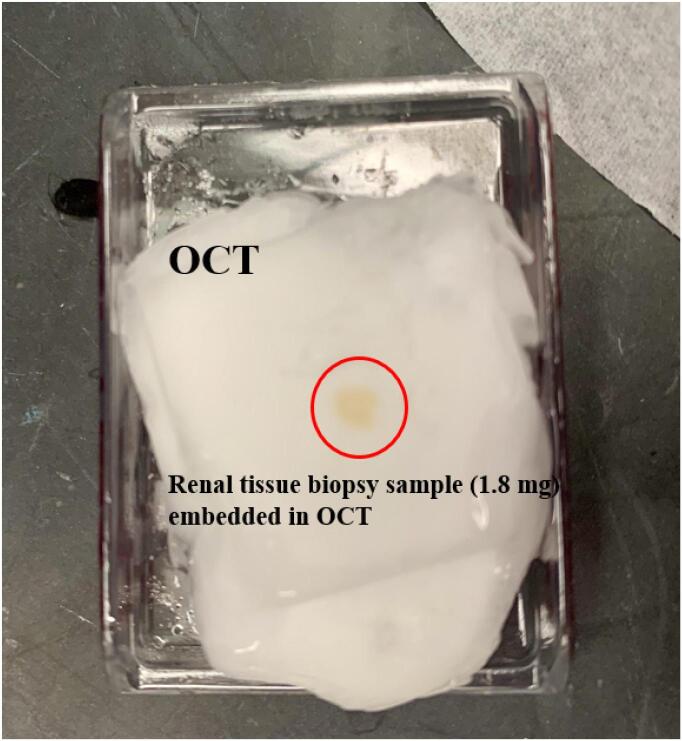
Frozen renal tissue biopsy samples from the patient and two controls, in which there was no diagnostic abnormality, were received as leftover specimens after the renal biopsy evaluations and diagnosis. Two control kidney specimens from similar age patients with no diagnostic abnormality were also used. The samples were received embedded in Optimal Cutting Temperature (OCT) compound as part of routine renal biopsy handling (Fig. 3). The OCT compound (Sakura Finetek USA, Inc, Torrance, CA) is routinely used to provide a solid matrix to encapsulate tissues for consistent frozen sectioning in a cryostat. The biopsy specimen is routinely placed in a mold with OCT, frozen and situated in a cryostat at −20C for sectioning. Removal of the frozen tissue from OCT was necessary for UHPLC-HRMS lipidomic analyses. First, as much of the OCT as possible was cut away using scalpel and forceps. Next, the remaining OCT was allowed to thaw at room temperature for about 10 min, the melted OCT was blotted away, and the tissue biopsies were transferred to microcentrifuge tubes and kept on ice and out of UV light at all steps wherever possible to preserve sample quality. Once the tissue biopsies were weighed, 50 µL of cold 5 mM ammonium acetate in water was added to each sample, and the whole tissue was homogenized with a probe sonicator that was set on level 8 (Fisher Scientific, Model 100). IPA, water, and methanol sequentially were used to wash the probe sonicator between samples. The samples were then incubated on ice for 30 min. A revised version of the Folch extraction method was used to extract lipids from the tissue biopsies [44]. First, the samples were centrifuged at 20,000 × g for 10 min at 4 °C to pellet the tissue debris; the supernatants were transferred into a clean glass centrifuge tube with a screw cap. Next, 5 μL of SPLASH® LipidoMix® Internal Standard Mix (containing a set of 14 stable isotope internal standards) was added to the samples followed by vortex mixing. All the vortex-mixing steps in this study were less than 30 s. Afterward, a total of 200 μL of ice-cold methanol and 400 μL of ice-cold chloroform were added to each sample. The samples were incubated at 4 °C for 20 min with vortex mixing every 5 min. Then, 100 μL of LC-MS grade water was added, the samples were vortex mixed and again incubated at 4 °C for 10 min with vortex mixing every 5 min. Next, the aqueous and organic layers were separated by centrifuging at a speed of 3260 × g at 4 °C for 10 min, and 400 μL of the organic layer (bottom layer) containing the lipid content of the samples was transferred using a glass pipette to a new glass centrifuge tube and cooled on ice. The aqueous layer (top layer) was re-extracted by adding 200 μL of ice-cold 2/1 (v/v) chloroform/methanol, followed by vortex mixing and incubating on ice for 10 min. The samples were centrifuged at 3260 × g and 4 °C for 10 min, followed by the removal of another 200 μL sample from the second organic layer, which was combined with the first organic layer. Finally, lipid extracts were dried under nitrogen gas at 30 °C using an Organomation Associates MultiVap (Berlin, MA, USA) and then reconstituted in 50 μL of IPA and vortex mixed. Afterwards, the samples were centrifuged at 3260 × g at 4 °C for 10 min. The entire reconstituted sample was transferred to a labeled glass LC vial with a fused insert and 2 µL of Lipidomics injection standard (1000 ppm of: lysophosphatidylcholine 19:0 (LPC(19:0)), phosphatidylcholine 19:0/19:0 (PC(19:0/19:0)), phosphatidylglycerol 17:0/17:0 (PG(17:0/17:0)), phosphatidylethanolamine 17:0/17:0) (PE(17:0/17:0)), phosphatidylserine 17:0/17:0 (PS(17:0/17:0)), and triacylglycerol 17:0/17:0/17:0 (TG(17:0/17:0/17:0))) was spiked into the samples, and vortex mixed to ensure homogeneity and make sure there were no air bubbles. Additionally, extraction blanks (a 25 μL water sample that experienced identical extraction steps as the samples, in order to assess variation of the extraction procedures) and reconstitution blanks were prepared for quality control purposes.
Fig. 3.
Photo of a patient renal tissue biopsy sample embedded in OCT, showing that lipidomics analysis can be conducted on the leftover sample without disruption of normal patient care.
Data acquisition method
An AQUITY UPLC® BEH C18 column (1.7 μm × 2.1 mm × 50 mm) with corresponding VanGuard pre-column (Waters Corporation, Milford, MA, USA) kept at 50 °C was employed for chromatographic separation. A multi-solvent mixture of 90:8:2 IPA:ACN:water with 0.1% formic acid and 10 mM ammonium formate was used as mobile phase D and mobile phase C was 60:40 ACN:water with 0.1% formic acid and 10 mM ammonium formate. A stepwise gradient elution was performed as follows: 0–1 min 20% D, 1–3 min 20–30% D, 3–4 min 30–45% D, 4–6 min 45–60% D, 6–8 min 60–65% D, 8–10 min 65–65% D, 10–15 min 65–90% D, 15–17 min 90–98% D, 17–18 min 98–98% D, followed by 5.00 min column flush and re-equilibration. The entire runtime with equilibration was 23.00 min with a 0.5 mL/min flow rate and the injection volume was 3 μL for positive polarity and 5 μL for negative polarity.
The analyses were conducted on a Q Exactive™ Orbitrap™ Mass Spectrometer with heated electrospray ionization coupled with a Dionex Ultimate 3000 UHPLC system (Thermo Fisher Scientific) in both positive and negative ion modes using the following settings: spray voltage = 3500 V, aux gas = 5 (+mode) and 15 (- mode), sheath gas = 30 (+mode) and 25 (- mode), capillary temperature = 300 °C, sweep gas = 1 (+mode) and 0 (- mode), and S-lens RF level = 35). All samples were kept at 8 °C in the autosampler during the analysis. Mass spectra were acquired in full scan mode with 70,000 mass resolution from 200 to 2200 m/z. Triplicate full scan injections were conducted on every sample. The presence of Fabry disease biomarkers in the samples were screened using the exact mass, and later using tandem mass spectrometry (MS/MS) fragmentation analysis (35,000 mass resolution, 1.5 amu isolation, 20 collision energy) of all Fabry disease biomarkers found in the samples to provide secondary confirmation of identity.
Data processing
To monitor data quality and verify the reproducibility of the analysis, the performance of spiked injection and internal standards in all samples was validated and qualified as showing a relative standard deviation of less than 10%. Xcalibur v.4.1 software (Thermo Fisher Scientific) was used for data acquisition and peak integration analysis. All data were normalized using the lowest tissue biopsy weight (1.8 mg).
Results and discussion
To screen the patient's biopsy for Fabry disease, first, the most important biomarker, Gb3[(d18:1)(C16:0)], was analyzed using UHPLC-HRMS. Besides Gb3[(d18:1)(C16:0)], five new potential Gb3-related isoforms/analogues biomarkers were also checked (Gb3[(d18:2)(C16:0)], Gb3[(d18:2)(C22:1)], Gb3[(d18:2)(C18:0)], Gb3[(d16:0)(C16:0)], Gb3[(d18:1)(C14:0)]). As shown in Table 1, the Gb3 related biomarkers were not detected in the patient sample or the control samples. As is apparent from this result, the enzyme activity of α-GAL A in this patient is sufficient to break down globotriaosylceramides lipids.
Table 1.
Mean peak area of Gb3-Related Biomarker in patient renal sample injections compared to control sample injections (n = 3) in positive ionization mode (nd = “not detected”).
| Gb3 Related Biomarker | Expected Mass (m/z) | Patient | Control-1 | Control-2 |
|---|---|---|---|---|
| Gb3[(d18:1)(C16:0)] | 1024.6784 | nd | nd | nd |
| Gb3[(d18:2)(C16:0)] | 1022.6701 | nd | nd | nd |
| Gb3[(d18:2)(C22:1)] | 1104.7417 | nd | nd | nd |
| Gb3[(d18:2)(C18:0)] | 1051.7092 | nd | nd | nd |
| Gb3[(d18:1)(C14:0)] + Gb3[(d16:1)(C16:0)] | 996.6636 | nd | nd | nd |
To better evaluate the patient for possible Fabry disease, we further investigated the possibility of lyso-Gb3-related biomarker presence in the tissue biopsy. In addition to lyso-Gb3(d18:1), seven other potential lyso-Gb3-related isoforms/analogues biomarkers, which were previously confirmed having higher concentration in Fabry patients, were also checked [16]. Based on the results demonstrated in Table 2, none of lyso-Gb3-related potential biomarkers were identified in the patient sample; therefore, the globotriaosylsphingosine lipids levels were not elevated in the kidney tissue.
Table 2.
Mean peak area of lyso-Gb3-related biomarker in patient sample injections compared to control sample injections (n = 3) in positive mode (nd = “not detected”).
| Lyso-Gb3 Related Biomarker | Expected Mass (m/z) | Patient | Control-1 | Control-2 |
|---|---|---|---|---|
| lyso-Gb3 | 786.4487 | nd | nd | nd |
| lyso-Gb3 (−C2H4) | 758.4174 | nd | nd | nd |
| lyso-Gb3 (−C2H4 + O) | 774.4123 | nd | nd | nd |
| lyso-Gb3 (−H2) | 784.4331 | nd | nd | nd |
| yso-Gb3 (−H2 + O) | 800.4280 | nd | nd | nd |
| lyso-Gb3 (+O) | 802.4436 | nd | nd | nd |
| lyso-Gb3 (H2O2) | 820.4542 | nd | nd | nd |
| lyso-Gb3 (H2O3) | 836.4491 | nd | nd | nd |
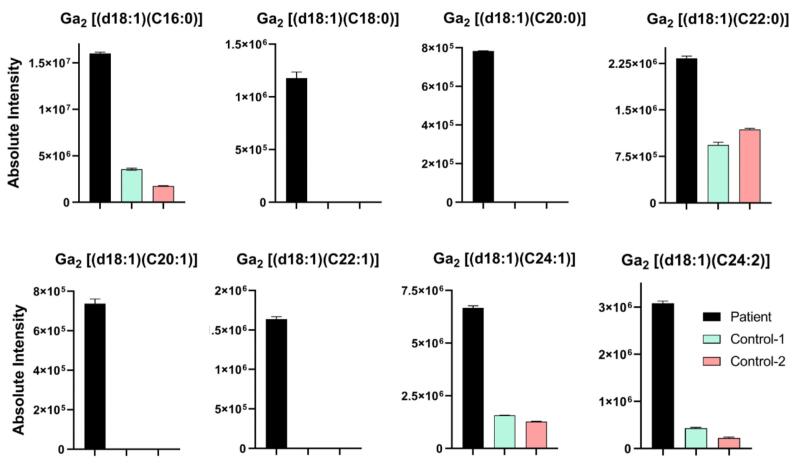
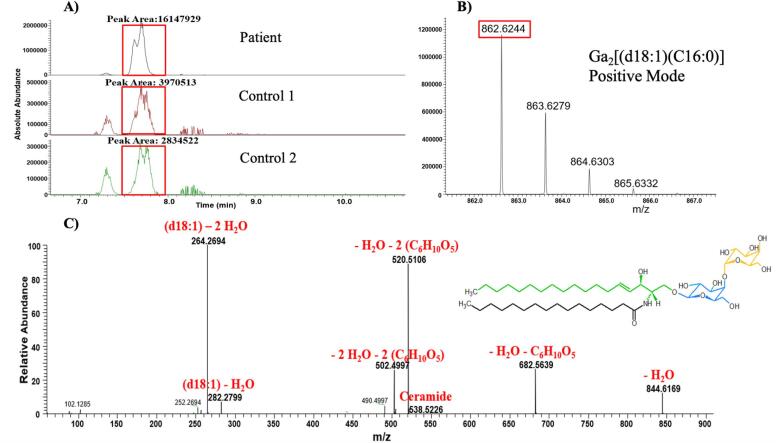
Finally, to obtain further information regarding Fabry disease biomarkers in this patient, all Ga2-related isoforms/analogs were then evaluated. Surprisingly, as depicted in Table 3 and Fig. 4, the Ga2-related lipid biomarker level was substantially higher in the patient’s renal tissue biopsy than in the two control samples. This additional investigation was possible because we analyzed the samples using an untargeted lipidomics approach and, thus, we were able to use the mass accuracy of the instrument to dig deeper into the lipids detected. These results confirm that this patient had some residual α-GAL A enzyme that had broken down Gb3 and lyso-Gb3 lipids, but was not sufficient to degrade all Ga2 lipids. The results validate that this patient appears to have a type of LSD, and since there was an observed disruption in α-GAL A enzyme activity in the patient’s renal biopsy, it could be non-classical Fabry disease. The representative extracted ion chromatograms (EICs) for Ga2[(d18:1)(C16:0)] in positive mode with associated mass spectra are shown in Fig. 5A and B to better present the intensity difference in the patient sample compared to control samples. Fig. 5C shows the corresponding MS/MS spectrum of Ga2[(d18:1)(C16:0)] that was obtained from the patient sample injection along with proposed structures of produced product ions. All product ions validate the presence of Ga2[(d18:1)(C16:0)].
Table 3.
Mean peak area of Ga2-related biomarker in patient sample injections compared to control sample injections (n = 3) in positive mode (nd = “not detected”). The “Absolute Intensity” columns in this table are derived from the integrated peak area in the chromatogram.
| Ga2 Related Biomarker | Expected Mass (m/z) | Measured Mass (m/z) | Mass Accuracy (ppm) | Absolute Intensity in Patient’s Sample | Absolute Intensity in Control-1′s Sample | Absolute Intensity in Control-2′s Sample | Patient/ Ave. Controls |
|---|---|---|---|---|---|---|---|
| Ga2 [(d18:1)(C16:0)] | 862.6256 | 862.6244 | −1.39 | 1.60E + 07 | 3.55E + 06 | 1.75E + 06 | 6.03 |
| Ga2 [(d18:1)(C18:0)] | 890.6569 | 890.6543 | −2.92 | 1.18E + 06 | nd | nd | – |
| Ga2 [(d18:1)(C20:0)] | 918.6882 | 918.6875 | −0.76 | 7.83E + 05 | nd | nd | – |
| Ga2 [(d18:1)(C22:0)] | 946.7195 | 946.7188 | −0.74 | 2.33E + 06 | 9.35E + 05 | 1.19E + 06 | 2.20 |
| Ga2 [(d18:1)(C20:1)] | 916.6725 | 916.6718 | −0.76 | 7.37E + 05 | nd | nd | – |
| Ga2 [(d18:1)(C22:1)] | 944.7038 | 944.7032 | −0.64 | 1.64E + 06 | nd | nd | – |
| Ga2 [(d18:1)(C24:1)] | 972.7351 | 972.7344 | −0.72 | 6.67E + 06 | 1.57E + 06 | 1.27E + 06 | 4.69 |
| Ga2 [(d18:1)(C24:2)] | 970.7195 | 970.7193 | −0.21 | 3.08E + 06 | 4.31E + 05 | 2.24E + 05 | 9.41 |
Fig. 4.
Bar plots showing Ga2-related Fabry biomarker intensities in the patient sample compared with the two control samples. These plots show Ga2 lipids had higher intensities in the patient samples, confirming disruption in α-GAL A enzyme activity.
Fig. 5.
A) Representative Extracted Ion Chromatogram (EIC) and B) mass spectrum of Ga2[(d18:1)(C16:0)] from the patient sample and two control samples based on UHPLC-HRMS, the mass spectrum is only from the top chromatogram panel and the spectrum is an average of multiple scans across the chromatographic peak. C) The corresponding MS/MS spectrum of Ga2[(d18:1)(C16:0)] obtained from the patient sample along with proposed structures of product ions, isolation width = 1.5 amu, collision energy = 20 V.
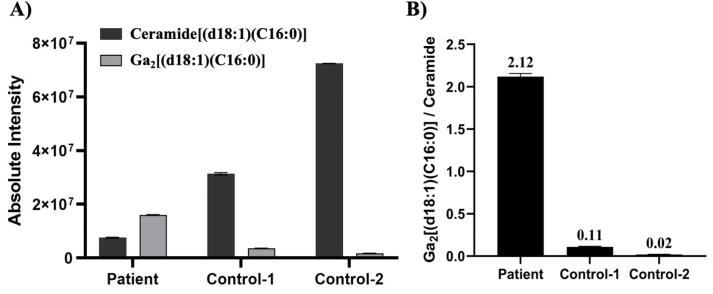
In this case study, we were aided by the ability to compare a suspected patient to two controls. However, in a clinical diagnostic setting, control subjects will likely not be available for comparison against a suspected Fabry patient. Thus, we must evaluate how a patient’s diagnosis could be made from a single sample injection. In such cases, the use of ratiometric metabolomics could help with the diagnosis. Ratiometric metabolomics has been used in clinical labs for screening of various disorders, including screening newborns for phenylketonuria based on the phenylalanine-to-tyrosine ratio in dried blood spots or in a research setting to test the immune system with the kynurenine-to-tryptophan ratio [45], [46]. This method has enabled the diagnosis phenylketonuria much earlier (as young as age < 24 h) and reliance on the assay with a reduced false-positive rate [47]. In the case of Fabry disease, we evaluated the ratio of Ga2[(d18:1)(C16:0)] to its corresponding ceramide (ceramide[(d18:1)(C16:0)], expected mass = 538.5199 m/z, measured mass = 538.5189 m/z, mass accuracy = -1.86 ppm) to better demonstrate the increased level of Ga2[(d18:1)(C16:0)] in the patient. Again, because we used HRMS for analysis, we were able to dig back into the already obtained data to evaluate the diagnostic potential of this ratio. As shown in Fig. 6, with the increasing level of Ga2[(d18:1)(C16:0)] in the patient, the corresponding ceramide[(d18:1)(C16:0)] level decreased and, as a result, the Ga2[(d18:1)(C16:0)]/Ceramide[(d18:1)(C16:0)] ratio increased more than 20 times in the patient sample compared to the two control samples. This result demonstrates that ratiometric metabolomics could be used to improve the diagnosis of rare diseases, but more patient samples will be needed to develop appropriate diagnostic reference ranges. We also assert that ratiometric metabolomics can be expanded to global metabolomic studies in which coverage of the metabolome or lipidome includes precursors and products of enzymatic reactions to generate broad ratiometric maps of metabolism. In clinical analysis, the availability of a reference standard is important and while Ga2[(d18:1)(C16:0)] is not available as a commercial standard, Ga2[(d18:1)(C17:0)] is available (Avanti Polar Lipids) and could serve as a reference standard.
Fig. 6.
A) Bar plot showing the absolute intensity of Ga2[(d18:1)(C16:0)] biomarker and ceramide[(d18:1)(C16:0)] in the patient and control samples. B) Bar plot showing the Ga2[(d18:1)(C16:0)]/Ceramide[(d18:1)(C16:0)] peak area ratio in the patient and control samples.
Conclusion
In this study, we demonstrated that lipidomic analysis offers the possibility to analyze the network of lipids and lipid biomarkers in patients with rare disorders and can contribute to the diagnostic evaluation when traditional anatomic and clinical pathology methods are unable to provide a definitive diagnosis. Moreover, the lipidomic analysis was performed on the affected kidney tissue, which is likely more clinically significant. Due to the rapid expansion of lipidomic analysis in the clinical laboratory and new perspectives it can provide through deeper annotation of lipids, lipidomics is expected to play a prominent role in the characterization of rare disorders. Specifically, it can aid in the early diagnosis of rare disorders, or help to better understand different manifestations of lipid disorders. We also showed the power of lipidomics data obtained from an HRMS platform to allow further evaluation of lipids without the need to re-inject samples, as would be needed with a fully targeted method. Finally, we were able to show the potential of lipid ratios to aid in disease diagnostics when control samples cannot be obtained. With additional samples, clinical reference ranges can be established to provide diagnostic results.
Finally, it is important to state that these samples were analyzed in a research mass spectrometry lab. While mass spectrometry is used routinely in clinical labs, adding a new assay for diagnostic testing requires additional validation that includes additional sensitivity and specificity testing, as well as comparison to already established methods for patient diagnosis. The assay we used was a non-quantitative assay and did not include an internal or reference standard for the target species. We used the ratiometric approach as a method for estimating enzyme activity. In clinical analysis, the availability of a reference standard is important for quality control and quantitation. While Ga2[(d18:1)(C16:0)] is not available as a commercial standard, Ga2[(d18:1)(C17:0)] is available (Avanti Polar Lipids, product 860701) and could serve as a reference standard since it would elute within a similar time frame and have a similar ionization efficiency. Although we used HRMS, this method could be transferred to a triple quadrupole where selected reaction monitoring (SRM) could be employed. However, when moving to a triple quadrupole, the method will lose the ability to identify other potential markers related to genetic disorders that could involve variants. This was one of the advantages of the method we employed versus of what is currently used for clinical diagnostics in Fabry disease. When those current markers failed to diagnose, we were able to expand to additional lipids that could be affected by variations in enzymatic activity. This type of approach could also be harnessed for other undiagnosed diseases to expand the ability to identify biomarkers to diagnose patients.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors gratefully acknowledge the scientific support and useful discussion of the Southeast Center for Integrated Metabolomics (SECIM) for which this work would not be possible.
References
- 1.Vellodi A. Lysosomal storage disorders. Br. J. Haematol. 2005;128(4):413–431. doi: 10.1111/j.1365-2141.2004.05293.x. [DOI] [PubMed] [Google Scholar]
- 2.Futerman A.H., van Meer G. The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell Biol. 2004;5(7):554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 3.Platt F.M. Sphingolipid lysosomal storage disorders. Nature. 2014;510(7503):68–75. doi: 10.1038/nature13476. [DOI] [PubMed] [Google Scholar]
- 4.Gupta S., Ries M., Kotsopoulos S., Schiffmann R. The relationship of vascular glycolipid storage to clinical manifestations of Fabry disease: a cross-sectional study of a large cohort of clinically affected heterozygous women. Medicine (Baltimore) 2005;84(5):261–268. doi: 10.1097/01.md.0000178976.62537.6b. [DOI] [PubMed] [Google Scholar]
- 5.A. Mehta, M. Beck, G. Sunder-Plassmann. Fabry Disease: Perspectives from 5 Years of FOS. 2006. [PubMed]
- 6.Sakuraba H., Togawa T., Tsukimura T., Kato H. Plasma lyso-Gb3: a biomarker for monitoring fabry patients during enzyme replacement therapy. Clin. Exp. Nephrol. 2018;22(4):843–849. doi: 10.1007/s10157-017-1525-3. [DOI] [PubMed] [Google Scholar]
- 7.Deegan P.B., Baehner A.F., Barba Romero M.A., Hughes D.A., Kampmann C., Beck M., Investigators E.F. Natural history of Fabry disease in females in the Fabry Outcome Survey. J. Med. Genet. 2006;43(4):347–352. doi: 10.1136/jmg.2005.036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiffmann R., Hughes D.A., Linthorst G.E., Ortiz A., Svarstad E., Warnock D.G., West M.L., Wanner C., Bichet D.G., Christensen E.I., Correa-Rotter R., Elliott P.M., Feriozzi S., Fogo A.B., Germain D.P., Hollak C.E.M., Hopkin R.J., Johnson J., Kantola I., Kopp J.B., Kröner J., Linhart A., Martins A.M., Matern D., Mehta A.B., Mignani R., Najafian B., Narita I., Nicholls K., Obrador G.T., Oliveira J.P., Pisani A., Politei J., Ramaswami U., Ries M., Terryn W., Tøndel C., Torra R., Vujkovac B., Waldek S., Walter J. Screening, diagnosis, and management of patients with Fabry disease: conclusions from a “Kidney Disease: improving Global Outcomes” (KDIGO) Controversies Conference. Kidney Int. 2017;91(2):284–293. doi: 10.1016/j.kint.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Mehta A., Beck M., Eyskens F., Feliciani C., Kantola I., Ramaswami U., Rolfs A., Rivera A., Waldek S., Germain D.P. Fabry disease: a review of current management strategies. QJM. 2010;103(9):641–659. doi: 10.1093/qjmed/hcq117. [DOI] [PubMed] [Google Scholar]
- 10.Pisani A., Visciano B., Roux G.D., Sabbatini M., Porto C., Parenti G., Imbriaco M. Enzyme replacement therapy in patients with Fabry disease: state of the art and review of the literature. Mol. Genet. Metab. 2012;107(3):267–275. doi: 10.1016/j.ymgme.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Guffon N., Fouilhoux A. Clinical benefit in Fabry patients given enzyme replacement therapy–a case series. J. Inherit Metab. Dis. 2004;27(2):221–227. doi: 10.1023/B:BOLI.0000028726.11177.8b. [DOI] [PubMed] [Google Scholar]
- 12.Lenders M., Brand E. Effects of enzyme replacement therapy and antidrug antibodies in patients with Fabry disease. J. Am. Soc. Nephrol. 2018;29(9):2265–2278. doi: 10.1681/ASN.2018030329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weidemann F., Breunig F., Beer M., Sandstede J., Turschner O., Voelker W., Ertl G., Knoll A., Wanner C., Strotmann J.M. Improvement of cardiac function during enzyme replacement therapy in patients with Fabry disease: a prospective strain rate imaging study. Circulation. 2003;108(11):1299–1301. doi: 10.1161/01.CIR.0000091253.71282.04. [DOI] [PubMed] [Google Scholar]
- 14.McCafferty E.H., Scott L.J. Migalastat: a review in Fabry disease. Drugs. 2019;79(5):543–554. doi: 10.1007/s40265-019-01090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aerts J.M., Groener J.E., Kuiper S., Donker-Koopman W.E., Strijland A., Ottenhoff R., van Roomen C., Mirzaian M., Wijburg F.A., Linthorst G.E., Vedder A.C., Rombach S.M., Cox-Brinkman J., Somerharju P., Boot R.G., Hollak C.E., Brady R.O., Poorthuis B.J. Elevated globotriaosylsphingosine is a hallmark of Fabry disease. Proc. Natl. Acad. Sci. USA. 2008;105(8):2812–2817. doi: 10.1073/pnas.0712309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auray-Blais C., Ntwari A., Clarke J.T.R., Warnock D.G., Oliveira J.P., Young S.P., Millington D.S., Bichet D.G., Sirrs S., West M.L., Casey R., Hwu W.-L., Keutzer J.M., Zhang X.K., Gagnon R. How well does urinary lyso-Gb3 function as a biomarker in Fabry disease? Clin. Chim. Acta. 2010;411(23-24):1906–1914. doi: 10.1016/j.cca.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 17.Alharbi F.J., Baig S., Auray-Blais C., Boutin M., Ward D.G., Wheeldon N., Steed R., Dawson C., Hughes D., Geberhiwot T. Globotriaosylsphingosine (Lyso-Gb. J. Inherit. Metab. Dis. 2018;41(2):239–247. doi: 10.1007/s10545-017-0127-2. [DOI] [PubMed] [Google Scholar]
- 18.Auray-Blais C., Boutin M., Gagnon R., Dupont F.O., Lavoie P., Clarke J.T.R. Urinary globotriaosylsphingosine-related biomarkers for Fabry disease targeted by metabolomics. Anal. Chem. 2012;84(6):2745–2753. doi: 10.1021/ac203433e. [DOI] [PubMed] [Google Scholar]
- 19.Manwaring V., Boutin M., Auray-Blais C. A metabolomic study to identify new globotriaosylceramide-related biomarkers in the plasma of Fabry disease patients. Anal. Chem. 2013;85(19):9039–9048. doi: 10.1021/ac401542k. [DOI] [PubMed] [Google Scholar]
- 20.Boutin M., Auray-Blais C. Metabolomic discovery of novel urinary galabiosylceramide analogs as Fabry disease biomarkers. J. Am. Soc. Mass. Spectrom. 2015;26(3):499–510. doi: 10.1007/s13361-014-1060-3. [DOI] [PubMed] [Google Scholar]
- 21.Linthorst G.E., Bouwman M.G., Wijburg F.A., Aerts J.M.F.G., Poorthuis B.J.H.M., Hollak C.E.M. Screening for Fabry disease in high-risk populations: a systematic review. J. Med. Genet. 2010;47(4):217–222. doi: 10.1136/jmg.2009.072116. [DOI] [PubMed] [Google Scholar]
- 22.van der Tol L., Smid B.E., Poorthuis B.J.H.M., Biegstraaten M., Deprez R.H.L., Linthorst G.E., Hollak C.E.M. A systematic review on screening for Fabry disease: prevalence of individuals with genetic variants of unknown significance. J. Med. Genet. 2014;51(1):1–9. doi: 10.1136/jmedgenet-2013-101857. [DOI] [PubMed] [Google Scholar]
- 23.Michaud M., Mauhin W., Belmatoug N., Garnotel R., Bedreddine N., Catros F., Ancellin S., Lidove O., Gaches F. When and how to diagnose Fabry disease in clinical pratice. Am. J. Med. Sci. 2020;360(6):641–649. doi: 10.1016/j.amjms.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Kok K., Zwiers K.C., Boot R.G., Overkleeft H.S., Aerts J.M.F.G., Artola M. Fabry disease: molecular basis, pathophysiology, diagnostics and potential therapeutic directions. Biomolecules. 2021;11(2):271. doi: 10.3390/biom11020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linthorst Gabor E., Poorthuis Ben J.H.M., Hollak Carla E.M. Enzyme activity for determination of presence of Fabry disease in women results in 40% false-negative results. J. Am. Coll. Cardiol. 2008;51(21):2082. doi: 10.1016/j.jacc.2008.02.050. [DOI] [PubMed] [Google Scholar]
- 26.Schiffmann R., Fuller M., Clarke L.A., Aerts J.M.F.G. Is it Fabry disease? Genet. Med. 2016;18(12):1181–1185. doi: 10.1038/gim.2016.55. [DOI] [PubMed] [Google Scholar]
- 27.Han X., Gross R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44(6):1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Lagarde M., Géloën A., Record M., Vance D., Spener F. Lipidomics is emerging. Biochim. Biophys. Acta. 2003;1634(3):61. doi: 10.1016/j.bbalip.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Lee S.H., Williams M.V., DuBois R.N., Blair I.A. Targeted lipidomics using electron capture atmospheric pressure chemical ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17(19):2168–2176. doi: 10.1002/rcm.1170. [DOI] [PubMed] [Google Scholar]
- 30.Han X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016;12(11):668–679. doi: 10.1038/nrendo.2016.98. [DOI] [PubMed] [Google Scholar]
- 31.Wang M., Wang C., Han X. Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-What, how and why? Mass Spectrom. Rev. 2017;36(6):693–714. doi: 10.1002/mas.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu T., Zhang J.-L. Mass-spectrometry-based lipidomics. J. Sep. Sci. 2018;41(1):351–372. doi: 10.1002/jssc.201700709. [DOI] [PubMed] [Google Scholar]
- 33.Gülbakan B., Özgül R.K., Yüzbaşıoğlu A., Kohl M., Deigner H.-P., Özgüç M. Discovery of biomarkers in rare diseases: innovative approaches by predictive and personalized medicine. EPMA J. 2016;7(1):24. doi: 10.1186/s13167-016-0074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb-Robertson B.-J., Stratton K.G., Kyle J.E., Kim Y.-M., Bramer L.M., Waters K.M., Koeller D.M., Metz T.O. Statistically driven metabolite and lipid profiling of patients from the undiagnosed diseases network. Anal. Chem. 2020;92(2):1796–1803. doi: 10.1021/acs.analchem.9b03522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin S.-Y., Fauman E.B., Petersen A.-K., Krumsiek J., Santos R., Huang J., Arnold M., Erte I., Forgetta V., Yang T.-P., Walter K., Menni C., Chen L., Vasquez L., Valdes A.M., Hyde C.L., Wang V., Ziemek D., Roberts P., Xi L., Grundberg E., Waldenberger M., Richards J.B., Mohney R.P., Milburn M.V., John S.L., Trimmer J., Theis F.J., Overington J.P., Suhre K., Brosnan M.J., Gieger C., Kastenmüller G., Spector T.D., Soranzo N. The multiple tissue human expression resource, C., an atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sueoka H., Ichihara J., Tsukimura T., Togawa T., Sakuraba H., Sommer C. Nano-LC-MS/MS for quantification of Lyso-Gb3 and its analogues reveals a useful biomarker for Fabry disease. PLoS One. 2015;10(5):e0127048. doi: 10.1371/journal.pone.0127048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You Q., Peng Q., Yu Z., Jin H., Zhang J., Sun W., Huang Y. Plasma lipidomic analysis of sphingolipids in patients with large artery atherosclerosis cerebrovascular disease and cerebral small vessel disease. Biosci. Rep. 2020;40(9) doi: 10.1042/BSR20201519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilde H.M., Banks D., Larsen C.L., Connors G., Wallace D., Lyon M.E. Evaluation of the Bayer microalbumin/creatinine urinalysis dipstick. Clin. Chim. Acta. 2008;393(2):110–113. doi: 10.1016/j.cca.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 39.Basi S., Fesler P., Mimran A., Lewis J.B. Microalbuminuria in type 2 diabetes and hypertension: a marker, treatment target, or innocent bystander? Diabetes Care. 2008;31(Supplement 2):S194–S201. doi: 10.2337/dc08-s249. [DOI] [PubMed] [Google Scholar]
- 40.Costa R.M., Martul E.V., Reboredo J.M., Cigarran S. Curvilinear bodies in hydroxychloroquine-induced renal phospholipidosis resembling Fabry disease. Clin. Kidney J. 2013;6(5):533–536. doi: 10.1093/ckj/sft089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Menezes Neves P.D.M., Machado J.R., Custódio F.B., Dos Reis Monteiro M.L.G., Iwamoto S., Freire M., Ferreira M.F., Dos Reis M.A. Ultrastructural deposits appearing as “zebra bodies” in renal biopsy: Fabry disease?- comparative case reports. BMC Nephrol. 2017;18(1):157. doi: 10.1186/s12882-017-0571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loh A.H., Cohen A.H. Drug-induced kidney disease–pathology and current concepts. Ann. Acad. Med. Singap. 2009;38(3):240–250. [PubMed] [Google Scholar]
- 43.Gold H., Mirzaian M., Dekker N., Joao Ferraz M., Lugtenburg J., Codée J.D., van der Marel G.A., Overkleeft H.S., Linthorst G.E., Groener J.E., Aerts J.M., Poorthuis B.J. Quantification of globotriaosylsphingosine in plasma and urine of fabry patients by stable isotope ultraperformance liquid chromatography-tandem mass spectrometry. Clin. Chem. 2013;59(3):547–556. doi: 10.1373/clinchem.2012.192138. [DOI] [PubMed] [Google Scholar]
- 44.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 45.Levy H.L. Newborn screening by tandem mass spectrometry: a new era. Clin. Chem. 1998;44(12):2401–2402. [PubMed] [Google Scholar]
- 46.Dharnidharka V.R., Al Khasawneh E., Gupta S., Shuster J.J., Theriaque D.W., Shahlaee A.H., Garrett T.J. Verification of association of elevated serum IDO enzyme activity with acute rejection and low CD4-ATP levels with infection. Transplantation. 2013;96(6):567–572. doi: 10.1097/TP.0b013e31829c7cec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chace D.H., Sherwin J.E., Hillman S.L., Lorey F., Cunningham G.C. Use of phenylalanine-to-tyrosine ratio determined by tandem mass spectrometry to improve newborn screening for phenylketonuria of early discharge specimens collected in the first 24 hours. Clin. Chem. 1998;44(12):2405–2409. [PubMed] [Google Scholar]