Abstract
As low-field MRI technology is being disseminated into clinical settings around the world, it is important to assess the image quality required to properly diagnose and treat a given disease and evaluate the role of machine learning algorithms, such as deep learning, in the enhancement of lower quality images. In this post hoc analysis of an ongoing randomized clinical trial, we assessed the diagnostic utility of reduced-quality and deep learning enhanced images for hydrocephalus treatment planning. CT images of post-infectious infant hydrocephalus were degraded in terms of spatial resolution, noise, and contrast between brain and CSF and enhanced using deep learning algorithms. Both degraded and enhanced images were presented to three experienced pediatric neurosurgeons accustomed to working in low- to middle-income countries (LMIC) for assessment of clinical utility in treatment planning for hydrocephalus. In addition, enhanced images were presented alongside their ground-truth CT counterparts in order to assess whether reconstruction errors caused by the deep learning enhancement routine were acceptable to the evaluators. Results indicate that image resolution and contrast-to-noise ratio between brain and CSF predict the likelihood of an image being characterized as useful for hydrocephalus treatment planning. Deep learning enhancement substantially increases contrast-to-noise ratio improving the apparent likelihood of the image being useful; however, deep learning enhancement introduces structural errors which create a substantial risk of misleading clinical interpretation. We find that images with lower quality than is customarily acceptable can be useful for hydrocephalus treatment planning. Moreover, low quality images may be preferable to images enhanced with deep learning, since they do not introduce the risk of misleading information which could misguide treatment decisions. These findings advocate for new standards in assessing acceptable image quality for clinical use.
Keywords: Low field MRI, Image quality, Deep learning, Risk assessment, Hydrocephalus treatment planning
1. Introduction
With an estimated 400,000 new cases worldwide each year, childhood hydrocephalus is the most common pediatric condition requiring neurosurgery globally (Dewan et al., 2018). Over 90% of cases occur in low- and middle-income countries (LMIC) (Dewan et al., 2018). In sub-Saharan Africa, approximately 180,000 infants per year are affected (Warf, 2013). Hydrocephalus is characterized by a build up of intracranial cerebrospinal fluid (CSF) that, in infants, causes the head to enlarge. These infants need surgical treatment to survive requiring intracranial imaging for planning. In planning surgery it is important to know where the CSF is in relation to brain, and how many compartments are loculated where fluid is trapped. An imaging technology capable of showing contrast between brain and CSF at an appropriate resolution is required. We have previously suggested that a voxel size approaching 100 (e.g 3 × 3 × 10 mm3) could be sufficient for planning treatment (Obungoloch et al., 2018).
The brain is an organ where the soft tissue and fluid encased within the skull have limited alternatives for imaging. Ultrasound is only effective within the first year of life before skull fusion closes the acoustical windows of the fontanels. The ionizing radiation associated with CT poses exceptional risks to infants (Frush et al., 2003, Brenner and Eric, 2007); however, in sub-Saharan Africa CT is more prevalent than MRI (World Health Organization, 2011) due to its lower cost. Although MRI is the gold standard for pediatric neuro-imaging, the high cost, strict siting requirements, and demanding maintenance schedule render high-field cryogenic systems infeasible for most of the developing world (Gatrad et al., 2007, Klein, 2015, Malkin, 2007, World Health Organization, 2011).
According to a 2014 baseline country survey on medical devices conducted by the World Health Organization, Uganda has 0.45 CT machines per million people and only 0.08 MRI machines per million people. By comparison, a high income country such as the Netherlands, has 12 CT and 12 MRI machines per million people (roughly 27 times more CT/million and 150 times more MRI/million people) (World Health Organization, 2011). Placed in the context of new hydrocephalus cases per year, with rates at least 10 times more per year in Africa than in Europe (Dewan et al., 2018), the clinical need for globally sustainable diagnostic imaging devices is clear. Low-field MRI devices have been recently developed that are feasible for the developing world and show diagnostic promise for the treatment and management of illnesses such as hydrocephalus (Obungoloch et al., 2018, O’Reilly et al., 2020, Sheth et al., 2020, Cooley et al., 2021).
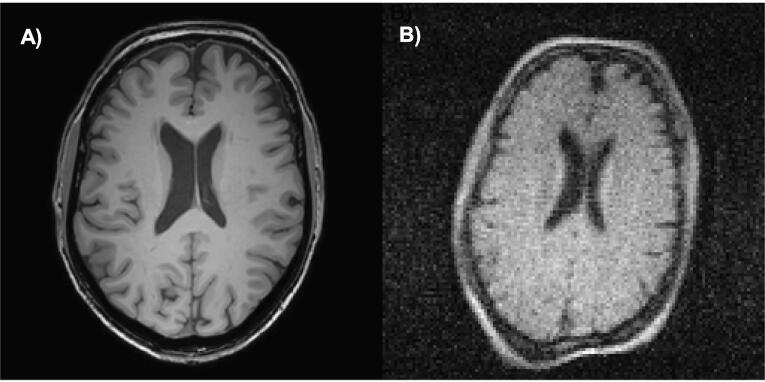
The quality of an MRI image ultimately depends on the signal-to-noise ratio (SNR) per voxel. Higher field strength systems (>1.5 Tesla) can produce increased signal-to-noise and pushing voxel size as low as hundreds of micrometers (Rutland et al., 2020). Low-field systems (<0.1 Tesla) inherently suffer from low signal-to-noise placing limits on achievable voxel size and including more baseline noise than most clinicians are accustomed to. Fig. 1 demonstrates the difference in brain image quality between a high-field (Fig. 1A) and a low-field (Fig. 1B) MRI system.
Fig. 1.
A comparison of the image quality between a high-field (3T) and a low-field (0.05 T) image of the brain of the same volunteer taken at the Leiden University Medical Center. A) A 256 × 256 3D T1 weighted TFE with Field of View: 200 × 175 × 156 mm, Resolution: 1.15 × 1.15 × 1.2 mm, TR/TE/TI = 9.8 ms/4.6 ms/1050 ms, ETL = 166, scan duration: 3 min 13 s; B) A 128 × 128 image at 0.05 T with Field of view: 256 × 256 × 200mm, Resolution: 2 × 2 × 4 mm, TR/TE = 400 ms/15 ms, echo train length = 6, scan duration: 7 min 7 s.
The adoption of low-field MRI into clinical practice depends largely on a longstanding and recently growing body of evidence that higher image quality does not always lead to better diagnostic accuracy or better patient outcome (Jhaveri, 2015). In clinical practice there exists a threshold of image quality for specific pathologies, above which no further outcome-based value can be observed (Durand et al., 2013). It has been demonstrated that 0.5 Tesla MRI can be as diagnostically accurate as 1.5 Tesla MRI for a variety of diseases including central nervous system pathologies (Jack et al., 1990), hepatic lesions (Steinberg et al., 1990), and multiple-sclerosis (Lee et al., 1995). It has also been shown that a 0.064 Tesla MRI can have comparable diagnostic accuracy to a 1.5 Tesla MRI for neoplasms and white matter disease (Orrison et al., 1991). Although the threshold of image quality required to plan effective hydrocephalus treatment has not been previously explored, we hypothesized that the level of resolution, tissue contrast, and SNR provided by CT or high-field MRI substantially exceeds this threshold.
Various machine learning-based methods have previously been used to perform super-resolution enhancement of low-quality MRI images. Interpolation based methods (Lehmann et al., 1999) are simple to implement but lack prior information often resulting in blurring. Model-based methods (Manjón et al., 2010, Manjón et al., 2010, Shi et al., 2015) explore the stochastic mechanism in the MRI generating process and model it with prior information; nevertheless, the design of a suitable regularization for the model can be difficult. Learning-based methods have the advantage of modeling and learning the mapping of low-quality images to high-quality images from data alone (Alexander et al., 2014, Yang et al., 2010, Wang et al., 2014, Jia et al., 2017). Recently, deep learning has shown impressive performance in the field of super-resolution of MRI (Chen et al., 2018, Pham et al., 2017, Zhu et al., 2018, Cherukuri et al., 2019, Cherukuri et al., 2017).
In the present work, we assess the diagnostic utility of reduced-quality and deep learning enhanced images for hydrocephalus treatment planning. We focus on the most common form of infant hydrocephalus in sub-Saharan Africa – postinfectious (Paulson et al., 2020). This form of hydrocephalus is uncommon outside of LMIC (Dewan et al., 2018), and the only abundant high-resolution comparative images are from CT. We developed an image utility assessment which was completed by three senior neurosurgeons with extensive experience in the treatment and management of hydrocephalus in low-resource settings (Kulkarni et al., 2017, Paulson et al., 2020, Schiff et al., 2021). Qualitative and quantitative measures of image utility are used to classify images revealing the quality threshold for treatment planning of hydrocephalus in terms of resolution, noise, and contrast between brain and CSF. We further evaluate how machine learning can lead to misleading modifications during the enhancement of low-resolution imagery.
2. Methods
Three experienced pediatric neurosurgeons accustomed to working in LMIC, with particular experience in interpretation of postinfectious hydrocephalus imagery of African infants, were chosen as participants in the image utility assessment. CT images were acquired from a repository of 90 patients enrolled in an ongoing randomized clinical trial (median age of 3.1 months, 39% female Kulkarni et al., 2017) and treated at the CURE Children’s Hospital of Uganda for post-infectious hydrocephalus. The center-most image slice from each patient was chosen for the assessment as either a test image (10 randomly selected, Fig. S5) or a learning library image (remaining 80). The images are 512 × 512 with 0.4 mm resolution (20.48 cm field of view). Each slice is 5 mm thick.
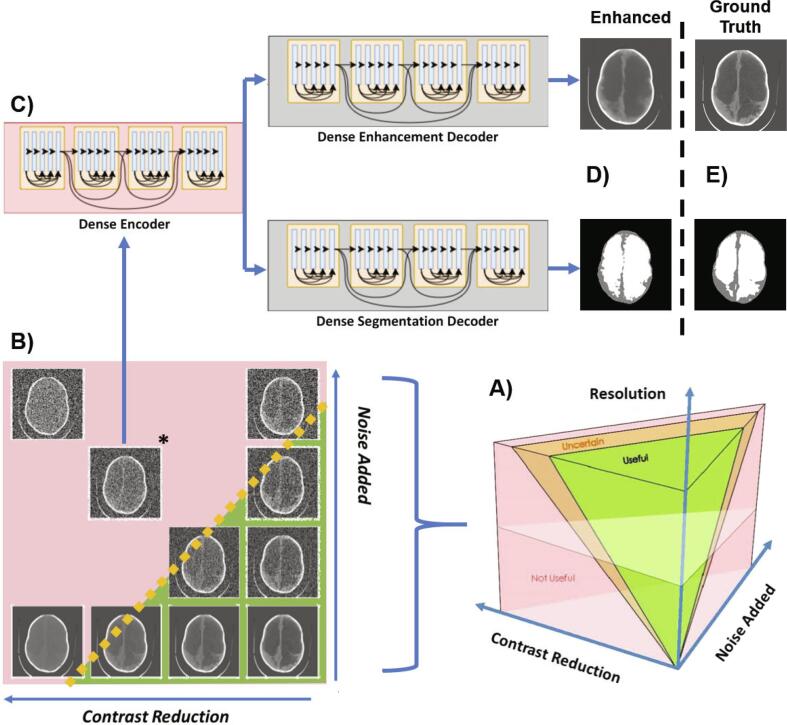
The 10 test images were degraded in terms of resolution, noise, and contrast between brain and CSF. Since the field of view remained constant for all images, resolution was adjusted by reducing the matrix size of the image. Because of this relationship, we adopt the term ”resolution” to describe changes in image matrix size for the present work. An image parameter space, as shown in Fig. 2A-B, was constructed consisting of the variables: 1) resolution (32 × 32, 64 × 64, 128 × 128, 512 × 512); 2) contrast reduction (20 levels between 0 and 1), 3) and noise added (20 levels between 0 and 1) resulting in 1,600 possible parameter combinations.
Fig. 2.
Schematic of study. In A) the image parameter space describing all possible combinations of noise, contrast between brain and CSF, and image resolution are visualized. There is likely to be a region of parameter combinations yielding images which are useful for hydrocephalus treatment planning (green volume), a region of parameter combinations that are not useful (red volume), and a region of uncertainty in between (orange volume). In B) we show a single plane from image parameter space in which all images have 512 × 512 resolution. The lower right corner has maximum contrast between brain and CSF and least noise considered in this study and the upper left corner has the lowest contrast and most noise. In C) the starred image from panel B) is chosen to be enhanced with a single encoder dual decoder (SEDD) architecture following the DenseNet network described in (Guo et al., 2019, Cherukuri et al., 2019). The output of such enhancement is seen in the upper panel of D) with corresponding segmentation in the lower panel of D). The ground truth version of the enhancement and segmentation from the original image without degradation or enhancement is shown in E) and called “ground truth”.
Resolution was down-sampled from the 512 × 512 image using bi-linear interpolation. The averaging between pixels in bi-linear interpolation can be considered an approximation of a partial volume effect.
Contrast between brain and CSF was reduced using histogram compression (Fig. S6), an algorithm developed specifically for this purpose. In histogram compression the histogram of gray-scale values for brain and CSF are iteratively compressed into a smaller gray-scale bandwidth to simulate loss in tissue contrast.
Gaussian noise with mean equal to variance was added according to known noise characteristics of CT images (Diwakar and Kumar, 2018). Since lower resolution images are more sensitive to noise, the noise added was scaled by clinical inspection for each resolution so that both useful and not useful images would be represented. The noise variance added was scaled by resolution as follows and normalized to the maximum value: from 0 to 0.001 (32 × 32), 0 to 0.01 (64 × 64), 0 to 0.05 (128 × 128, and 0 to 0.13 (512 × 512).
In Cherukuri et al., 2019, Cherukuri et al., 2017, deep learning networks took advantage of low-rank structural prior information to enhance low quality images. Building on this work, we developed a deep learning network capable of simultaneously enhancing and segmenting CT images of infant hydrocephalus that have been artificially degraded. Following the DenseNet network described in Guo et al. (2019), a single encoder dual decoder (SEDD) architecture was used to enhance CT images that have reduced quality. Deep learning networks, as shown in Fig. 2C-E, were trained for two resolutions (64 × 64 and 128 × 128) at seven locations in parameter space using library images. With noise added as the x-coordinate and contrast reduction as the y-coordinate, networks were trained for both resolutions at: 1) (0.3,0.3), 2) (0.6,0.3), 3) (0.3,0.6), 4) (0.6,0.6), 5) (0.9,0.6), 6) (0.6,0.9), 7) (0.9,0.9) (Fig. S7). The least degraded network is network 1. The networks were built by degrading the 80 library images at each of the 14 network locations and training with the original non-degraded image as ground truth. After training, the 10 test images were degraded at the network locations and enhanced generating 140 deep learning enhanced images.
From the 1,600 parameter combinations applied to the 10 test images, 420 cases were randomly presented to the panel of experts along with all 140 deep learning enhanced images. The image utility assessment was divided into two parts. In Part 1, the images were shown in 140 panels of 4 images each, as shown in Fig. 3A. In each of the 140 panels, one image location was randomly selected for an enhanced image and the other three were degraded images. The expert was not told that there would be enhanced images. In each panel, the expert was asked to select which, if any, of the 4 images are clinically useful for planning hydrocephalus treatment (see Supplementary Methods for full instructions). The data from the three experts were combined by addition of scores for each image in order to be classified as useful, uncertain, or not useful. If all three experts agreed that an image was useful, this image received a 3 (i.e. Useful). If all experts agreed that an image was not useful this image received a 0 (i.e. Not Useful). Uncertain images received a score of either 1 or 2.
Fig. 3.
The figure shows results from Part 1 of the Assessment. In A) we show an example panel from Part 1 of the assessment. The lower left image is an enhanced image and all other images are degraded. The experts must indicate which (if any) is useful. The left panel of B) shows raw classification data from Part 1 for 64 × 64 images. Solid lines are lines of constant contrast-to-noise ratio (CNR). Dashed lines show lines of constant usefulness likelihood from the multivariate logistic regression. The right panel of B) shows the receiver operating characteristic curves. In C) we show the univariate logistic regression models for each resolution with CNR as the predictor. The diamond and circle datapoints show the calculated CNR values for the low-field and high-field MRI images shown in Fig. 1, respectively. Resolution for these images lie between the 128 × 128 and 512 × 512 curves, which overlap for the CNR values reported. The bottom four panels of C) show the raw classification data for each resolution.
In Part 2, the experts were shown enhanced images in a side-by-side comparison with their corresponding 512 × 512 non-degraded versions as seen in Fig. 4A. The experts were asked to assess whether the spatial errors in the enhanced version were acceptable or would alter treatment decisions (see Supplementary Methods for full instructions). The data from Part 2 were also combined by addition of scores. Part 2 enhanced images receiving a 3 were classified as useful (i.e. useful in both Part1 and Part2), those receiving a 1 or 2 were classified as uncertain, and those receiving a 0 were classified as misleading (i.e. useful in Part 1, but shown to have unacceptable error in Part 2).
Fig. 4.
The figure shows results from Part 2 of the assessment. A) An example panel from Part 2 of the assessment. The left column of images are ground truth and the right column are the enhanced versions. B) shows the usefulness likelihood curves based on image CNR. The triangles show the average CNR for each network location before enhancement and the circles show the average CNR for each network after enhancement. C) shows the predicted usefulness likelihood of the enhanced images based on CNR after enhancement, the actual Part 1 classification of the enhanced images, and the Part 2 re-classification of the enhanced images after comparison with ground truth. In D) we compare the usefulness likelihood of the degraded images with the risk of a misleading result if the image is enhanced for 128 × 128 images. The left vertical axis shows the usefulness likelihood of the degraded image and the right vertical axis shows the risk of a misleading result if the corresponding degraded image were enhanced. In D) we also show an example degraded image on the left with CNR = 1, the enhanced version of this image on the right with CNR` = 8 after enhancement and corresponding high likelihood of misleading results after enhancement. Finally, E) shows the ground truth version of the example image in D) for comparison.
In addition, an analysis of inter-rater reliability was performed using a variation of Cohen’s Kappa, as described in Byrt et al. (1993), which accounts for the existence of prevalence in the data and bias between evaluators (see Supplementary Methods). For this analysis, the data was divided into three parts: 1) classification of Part1 degraded images, 2) classification of Part 1 enhanced images, and 3) classification of Part 2 enhanced images. The Kappa statistic of Byrt et al. (1993) was calculated for all possible pairings of evaluators and conclusions regarding agreement were drawn based on the interpretation of Kappa values as suggested in Byrt et al., 1993, Hallgren, 2012.
Univariate and multivariate logistic regression was used to investigate the ability of contrast, noise, and contrast-to-noise ratio to predict image classification. A deviance statistic was used to assess goodness of fit of the logistic regression models. The deviance of the model is a chi-squared statistic which assesses the difference between the maximum log likelihood of the chosen model and that of the null model (i.e. the average probability of a classification at a given resolution being useful).
3. Results
3.1. Part 1: What makes an image useful?
We first characterize the relationship between resolution, contrast, noise and usefulness. The inter-rater reliability for the classification of degraded images shows fair agreement between evaluators 1 and 2 (K = 0.33), substantial agreement between evaluators 2 and 3 (K = 0.94), and fair agreement between evaluators 1 and 3 (K = 0.36). For all three evaluators, there was a high prevalence for classifying Part 1 enhanced images as being useful (see Supplementary results). As such, inter-rater reliability calculations for this data are not informative and all evaluators are in near perfect agreement. In Fig. 3A we show several degraded images, of which the lower left is enhanced by deep learning. The left panel of Fig. 3B shows how the contrast and noise of each image relates to the image classification determinations at 64 × 64 resolution (see Fig. S10 for full dataset results). The solid contour lines in Fig. 3B show lines of constant contrast-to-noise ratio between brain and CSF averaged from the full dataset of images. In comparison, the dotted lines show constant usefulness likelihood based upon a multivariate logistic regression model with contrast and noise as predictors. For images with each of the four resolutions considered, the multivariate logistic regression model provided a significant fit with p-values less than 0.01 (p32 × 32 = 7e-6, p64 × 64 = 4e-27, p128 × 128 = 2e-17, p512 × 512 = 8e-32). Note that there is qualitative agreement between the average contrast-to-noise contours and the lines of constant likelihood that the image is useful. On the right of Fig. 3B receiver operating characteristic curves demonstrate that average contrast-to-noise and likelihood are both comparably effective classifiers of image utility with areas under their curves > 0.85 (curves for full dataset in Fig. S11).
Since average contrast-to-noise appeared to be an effective classifier, Fig. 3C shows that individual image contrast-to-noise alone is a significant predictor of usefullness likelihood, stratified by resolution. The grey circle shows the usefullness likelihood of the 256 × 256 brain image from the 3 Tesla system in Fig. 1A based on its contrast-to-noise ratio (CNR = 13). The grey diamond shows the same for the 128 × 128 brain image from the 0.05 Tesla system in Fig. 1B (CNR = 4). Though the image generated by the 3T system has twice the resolution and 3 times the CNR, both share a predicted usefulness likelihood of 1. For each resolution, the raw classification data from Part 1 can be seen in the four inset panels of Fig. 3C. The solid lines show the logistic regression model and the dashed lines show the 95% confidence intervals around the fit.
3.2. Part 2: Is reconstruction error acceptable?
Next we investigate the effect of deep learning enhancement on image classification. The inter-rater reliability for the classification of enhanced images in Part 2 shows slight agreement between evaluators 1 and 2 (K = 0.15), fair agreement between evaluators 2 and 3 (K = 0.24), and moderate agreement between evaluators 1 and 3 (K = 0.48). Fig. 4A shows a side by side comparison of ground truth (left column) with corresponding enhanced images (right column). Note the subtle errors in brain and CSF locations in the top right image and the more substantial errors in the lower right image. Regardless of these spatial errors, CNR is significantly increased by the enhancement network, as shown in the plot in Fig. 4B where average CNR of test images at each network location are shown before and after enhancement using the logistic models developed in Part 1. These data predict very high usefulness likelihood for enhanced images based on increased CNR. The table in Fig. 4C shows that while the Part 1 classification of enhanced images does closely follow the prediction of high usefulness likelihood, re-classification of enhanced images in Part 2 reveals that many enhanced images contain errors that are not clinically acceptable. We use an additional classification of Misleading for these images (i.e. images that were deemed Useful in Part 1, but had unacceptable errors in Part 2).
Since the logistic models developed in Part 1 do not describe the Part 2 classification, a new logistic regression model was constructed for Part 2 with pre-enhancement noise and contrast of images as predictors. Only contrast showed significance (Figs. S13 and S14) so noise was removed from the model. In order to compare the usefulness likelihood of a degraded image (Part 1) with the risk of misleading errors in an enhanced image (Part 2), an additional logistic regression model with CNR as the predictor was computed based on Part 2 classification (Fig. 4D). Risk of misleading results is calculated to be 1 minus the usefulness likelihood of the enhanced images based on a univariate logistic regression with CNR prior to enhancement as the predictor. As CNR increases, a 128 × 128 image is more likely to be useful in its degraded state (left vertical axis) and less likely to be misleading if enhanced (right vertical axis). Note that there exists no CNR value for which there is low usefulness likelihood of the degraded image and low risk of generating a misleading image through enhancement.
4. Discussion
4.1. Utility of Low CNR Images
The image quality threshold required for treatment planning of hydrocephalus is significantly lower than the quality typically provided by CT or high-field MRI imaging systems. The results in Fig. 3B-C can be viewed in several different ways. CNR is a comparison between the signal-to-noise ratio of two regions of interest. This implies that the true limiting factor of image quality is per voxel signal-to-noise, for which high-field MRI has an inherent advantage over low-field MRI. However, Fig. 3B-C suggests that there are options for using low CNR or low resolution images that may be advantageous. For a high-field system imaging infant hydrocephalus, a short scan time is desirable, in which case resolution and signal-to-noise can be traded for a faster scan. Alternatively, in the resource limited setting of an LMIC, a low-field MRI system has the potential to provide equivalent diagnostic information at a significant reduction in cost and complexity. The trade-off for this low cost and complexity is lower signal-to-noise and interpretability. It is the interpretability that sets the threshold for the lower bound of signal-to-noise.
The usefulness likelihood for the 3T (CNR = 13) and 0.05 T (CNR = 4) MRI images without deep learning enhancement featured in Fig. 1 are indicated in Fig. 3C. Although the visual quality of the two images is strikingly different, they are predicted to have the same utility for hydrocephalus treatment planning.
To put this in the context of global sustainability, the acquisition cost of the 0.05 T system used for producing the image in Fig. 1B is less than $20,000 USD. A 3T system costs at least an additional $2.8 million USD (excluding siting, maintenance, and consumables) and it can provide over three times the CNR (Fig. 1A). However, for the cost of a single 3T system, 150 low-field MRI systems could be placed throughout the region, providing increased access to the hydrocephalus patient population without compromise in diagnostic utility.
In addition to being a substantial global health need for children’s medicine, hydrocephalus is also an exceptionally straightforward technical challenge for low-field MRI systems. In the vast majority of hydrocephalic children, there is no need to differentiate contrast within the brain parenchyma for diagnosis, triage, monitoring, or treatment planning. For MRI the signal strength from the water-based CSF is the strongest signal within the head. Although our results support substantial utility from images with reduced quality in hydrocephalus management, more complex diagnostic and treatment decision-making in other diseases will pose additional challenges to such technologies.
4.2. Enhanced Images: Benefit or risk?
Image enhancement appears to perform exceptionally well based on Part 1 data, as shown in Fig. 4A where even the worst network locations are more than 85% likely to be rendered useful. However, data from Part 2 reveals that enhancement yields images that appear useful, but in fact would mislead treatment decisions due to unacceptable errors in brain and CSF location. Subtle features in the configuration of the CSF spaces, such as increased rounding of brain ventricles, are important signs of increased intracranial pressure suggesting that surgery might be required to improve CSF diversion through a shunt or endoscopic fenestration. If features such as these are a product of the enhancement network and not indicative of the true condition of the disease, clinicians may be led to make poor treatment decisions.
The key difference in using a degraded image versus its enhanced counterpart in a clinical setting is the source of risk. A degraded image is either useful or it is not - the risk of using it to diagnose or treat disease rests with the judgement of the clinician. Enhanced images in this study yield useful looking images 99% of the time, however 75% of these images are shown to have uncertain utility or to be misleading after comparison with ground truth. The risk of enhancement arises from the black box of the deep learning network. Furthermore, as shown in Fig. 4D, there is never a CNR for which there is low risk of producing a misleading image and low usefulness likelihood of the degraded image without enhancement. For example, a 128 × 128 degraded image with modest CNR yielding 75% usefulness likelihood still has a 14% chance of producing a misleading image through enhancement. Enhancing highly degraded images can improve the usefulness likelihood, but with substantially increased risk of misleading results. We find no scenario in which enhancement is safely beneficial. Note also that the CNR of the 0.05 T system studied had a very high useful likelihood and would not have required enhancement. Yet acceptance of such unenhanced images as shown in Fig. 1B would constitute a cultural shift in current standards of diagnostic acceptability.
Machine learning can generate attractive images from patterns with highly degraded information content. Philosophically, a learning library of other patient images enables utilization of information not present in the individual case undergoing enhancement. Such learned information brought to a new case image can be clinically misleading. This is a very different situation from machine learning faces or objects, or diagnosis classification from images, where there is only one correct match and the information required is already in the learning library. Hydrocephalus, as in so many other pathological conditions, tends to produce a unique structural pattern for each patient. For machine learning, automating the choice of a diagnosis is therefore very different from reconstructing an unknown unique architecture. This fundamental issue implies that while this study only employed one learning network architecture, this risk likely exists in other machine learning strategies and great care should be taken when employing these methods for anatomic reconstruction. A challenge for the machine learning community working with low-resolution and low-contrast images is to improve interpretation while minimizing risk of clinical errors.
4.3. Limitations
This study has limitations. Only three experts participated in the assessment. The single central slice from the image stack was chosen to demonstrate image quality and enhancement. Only image quality concerns inherent to low-field systems such as noise and contrast were considered, while distortions in the low-field image were not. Only one deep learning network architecture was employed and the number of training samples was relatively low (80). However, the size of available image archives is typically not large for diseases unique to LMIC such as post-infectious hydrocephalus in sub-Saharan Africa. A more complex machine learning strategy could incorporate a 3D array of connected slices for enhancement and clinical review.
Although motivation for this study stems from the advent of clinical low-field MRI as a tool for hydrocephalus treatment planning, the work was conducted with CT images. In high-resource settings where low-field MRI is being deployed (such as intensive care units), CT remains the high-resolution alternative of choice (Mazurek et al., 2021, Sheth et al., 2020). CT is the high-resolution modality most available in LMIC, and currently the only available repository of postinfectious hydrocephalus images where low-field MRI will soon be deployed. Note that we argue the potential benefits of low-field MRI using only one example image in Fig. 1B. This can be extended in the future as reliable low-field MR image repositories become available such as the new comparative repository in Adult stroke reported in (Mazurek et al., 2021). We anticipate that this quantifiable measure of CNR between brain and CSF will be generalizable to MRI at various field strengths as well as other CT studies of infant hydrocephalus treatment planning. Further evaluation will be necessary to determine whether CNR proves an important classifier for other conditions that may have more stringent image quality requirements.
5. Conclusion
The true value of a clinical medical image is in the treatment guiding information that it conveys to those providing care and in the patient outcomes that result, rather than its visual appeal. We have shown that lower quality images that are not customarily considered acceptable can be useful in planning hydrocephalus treatment. In addition, image resolution and contrast-to-noise ratio of brain and CSF predict the likelihood of a useful image for hydrocephalus treatment planning. Although deep learning can dramatically improve the visual quality of a highly degraded image, there is a substantial risk of misleading results, and algorithmic guidelines should be developed to avoid structural alterations which are potentially hazardous to clinical interpretation. At present, the most valuable low-resolution images may be less enhanced versions that maintain the structural details undistorted by excessive deep learning processing; indeed, emerging low-field MRI technologies are capable of producing useful images for hydrocephalus treatment planning without enhancement. Our findings advocate for new standards in assessing the cost-effectiveness of sustainable imaging technologies that can broaden global access to diagnostic imaging, and a reconsideration of acceptable image quality for clinical use.
CRediT authorship contribution statement
Joshua R. Harper: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Visualization. Venkateswararao Cherukuri: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Visualization. Tom O’Reilly: Conceptualization, Resources, Writing - review & editing, Visualization. Mingzhao Yu: Writing - original draft, Writing - review & editing. Edith Mbabazi-Kabachelor: Resources, Writing - review & editing. Ronald Mulando: Resources, Writing - review & editing. Kevin N. Sheth: Writing - review & editing, Conceptualization. Andrew G. Webb: Writing - review & editing, Conceptualization. Benjamin C. Warf: Writing - review & editing, Conceptualization, Resources. Abhaya V. Kulkarni: Writing - review & editing, Conceptualization, Resources. Vishal Monga: Writing - review & editing, Conceptualization, Methodology, Supervision. Steven J. Schiff: Writing - review & editing, Writing - original draft, Conceptualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Supported by US National Institutes of Health grant R01HD085853. ClinicalTrials.gov registration number NCT01936272.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.nicl.2021.102896.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alexander D.C., Zikic D., Zhang J., Zhang H., Criminisi A. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2014. Image quality transfer via random forest regression: applications in diffusion MRI; pp. 225–232. [DOI] [PubMed] [Google Scholar]
- Brenner D.J., Eric J. Hall. Computed tomography–an increasing source of radiation exposure. N. Engl. J. Med. 2007;357(22):2277–2284. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- Byrt T., Bishop J., Carlin J.B. Bias, prevalence and kappa. J. Clinical Epidemiol. 1993;46(5):423–429. doi: 10.1016/0895-4356(93)90018-v. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Y. Xie, Z. Zhou, F. Shi, A.G. Christodoulou, and D. Li (2018) ”Brain MRI super resolution using 3D deep densely connected neural networks,” in 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), IEEE, pp. 739–742.
- Cherukuri V., Ssenyonga P., Warf B.C., Kulkarni A.V., Monga V., Schiff S.J. Learning based segmentation of CT brain images: application to postoperative hydrocephalic scans. IEEE Trans. Biomed. Eng. 2017;65(8):1871–1884. doi: 10.1109/TBME.2017.2783305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherukuri V., Guo T., Schiff S.J., Monga V. Deep MR Brain Image Super-Resolution Using Spatio-Structural Priors. IEEE Trans. Image Process. 2019;29:1368–1383. doi: 10.1109/TIP.2019.2942510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley C.Z., McDaniel P.C., Stockmann J.P., Srinivas S.A., Cauley S.F., Śliwiak M., Sappo C.R., Vaughn C.F., Guerin B., Rosen M.S., et al. A portable scanner for magnetic resonance imaging of the brain. Nature Biomed. Eng. 2021;5(3):229–239. doi: 10.1038/s41551-020-00641-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan M.C., Rattani A., Mekary R., Glancz L.J., Yunusa I., Baticulon R.E., Fieggen G., Wellons J.C., Park K.B., Warf B.C. Global hydrocephalus epidemiology and incidence: systematic review and meta-analysis. J. Neurosurg. 2018;130(4):1065–1079. doi: 10.3171/2017.10.JNS17439. [DOI] [PubMed] [Google Scholar]
- Diwakar M., Kumar M. A review on CT image noise and its denoising. Biomed. Signal Process. Control. 2018;42:73–88. [Google Scholar]
- Durand D.J., Carrino J.A., Fayad L.M., Huisman T.A., El-Sharkawy A.-M.M., Edelstein W.A. MRI pyschophysics: An experimental framework relating image quality to diagnostic performance metrics. J. Magn. Reson. Imaging. 2013;37(6):1402–1408. doi: 10.1002/jmri.23922. [DOI] [PubMed] [Google Scholar]
- Frush D.P., Donnelly L.F., Rosen N.S. Computed tomography and radiation risks: what pediatric health care providers should know. Pediatrics. 2003;112(4):951–957. doi: 10.1542/peds.112.4.951. [DOI] [PubMed] [Google Scholar]
- Gatrad A., Gatrad S., Gatrad A. Equipment donation to developing countries. Anaesthesia. 2007;62:90–95. doi: 10.1111/j.1365-2044.2007.05309.x. [DOI] [PubMed] [Google Scholar]
- Guo T., Cherukuri V., Monga V. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops. 2019. Dense123’color enhancement dehazing network. [Google Scholar]
- Guo T., Li X., Cherukuri V., Monga V. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops. 2019. Dense scene information estimation network for dehazing. [Google Scholar]
- Hallgren K.A. Computing inter-rater reliability for observational data: an overview and tutorial. Tutorials Quantitative Methods Psychol. 2012;8(1):23. doi: 10.20982/tqmp.08.1.p023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack J.C., Berquist T.H., Miller G.M., Forbes G.S., Gray J.E., Morin R.L., Ilstrup D.M. Field strength in neuro-MR imaging: a comparison of 0.5 T and 1.5 T. J. Computer Assisted Tomography. 1990;14(4):505–513. doi: 10.1097/00004728-199007000-00002. [DOI] [PubMed] [Google Scholar]
- Jhaveri K. Image quality versus outcomes. J. Magn. Reson. Imaging. 2015;41(4):866–869. doi: 10.1002/jmri.24622. [DOI] [PubMed] [Google Scholar]
- Jia Y., Gholipour A., He Z., Warfield S.K. A new sparse representation framework for reconstruction of an isotropic high spatial resolution MR volume from orthogonal anisotropic resolution scans. IEEE Trans. Med. Imaging. 2017;36(5):1182–1193. doi: 10.1109/TMI.2017.2656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H.-M. Springer; 2015. clinical low field strength magnetic resonance imaging: a practical guide to accessible MRI. [Google Scholar]
- Kulkarni A.V., Schiff S.J., Mbabazi-Kabachelor E., Mugamba J., Ssenyonga P., Donnelly R., Levenbach J., Monga V., Peterson M., MacDonald M., et al. Endoscopic treatment versus shunting for infant hydrocephalus in Uganda. N. Engl. J. Med. 2017;377(25):2456–2464. doi: 10.1056/NEJMoa1707568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Vellet A.D., Eliasziw M., Vidito L., Ebers G.C., Rice G.P., Hewett L., Dunlavy S. MR imaging field strength: prospective evaluation of the diagnostic accuracy of MR for diagnosis of multiple sclerosis at 0.5 and 1.5 T. Radiology. 1995;194(1):257–262. doi: 10.1148/radiology.194.1.7997564. [DOI] [PubMed] [Google Scholar]
- Lehmann T.M., Gonner C., Spitzer K. Survey: Interpolation methods in medical image processing. IEEE Trans. Med. Imaging. 1999;18(11):1049–1075. doi: 10.1109/42.816070. [DOI] [PubMed] [Google Scholar]
- Malkin R.A. “Design of health care technologies for the developing world, “ Annu. Rev. Biomed. Eng. 2007;9:567–587. doi: 10.1146/annurev.bioeng.9.060906.151913. [DOI] [PubMed] [Google Scholar]
- Manjón J.V., Coupé P., Buades A., Fonov V., Collins D.L., Robles M. Non-local MRI upsampling. Med. Image Anal. 2010;14(6):784–792. doi: 10.1016/j.media.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Manjón, J.V., P. Coupé, A. Buades, D.L. Collins, and M. Robles (2010) ”MRI superresolution using self-similarity and image priors,” International journal of biomedical imaging, 2010. [DOI] [PMC free article] [PubMed]
- Mazurek M., Cahn B., Yuen Mercy, Cahn Bradley, Yuen Matthew, Prabhat Anjali, Chavva Isha, Shah Jill, Crawford Anna, Welch E., Rothburg Jonathan, Sacolick Laura, Poole Michael, Wira Charles, Matouk Charles, Ward Adrienne, Timario Nona, Leasure Audrey, Beekman Rachel, Peng Teng, Witsch Jens, Antonios Joseph, Falcone Guido, Gobeske Kevin, Petersen Nils, Schindler Joseph, Sansing Lauren, Gilmore Emily, Hwang David, Kim Jennifer, Malhotra Ajay, Sze Gordon, Rosen Matthew, Kimberly Taylor, Sheth Kevin. Portable, bedside, low-field magnetic resonance imaging for evaluation of intracerebral hemorrhage. Nat. Commun. 2021 doi: 10.1038/s41467-021-25441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obungoloch J., Harper J.R., Consevage S., Savukov I.M., Neuberger T., Tadigadapa S., Schiff S.J. Design of a sustainable prepolarizing magnetic resonance imaging system for infant hydrocephalus. Magn. Reson. Mater. Phys., Biol. Med. 2018;31(5):665–676. doi: 10.1007/s10334-018-0683-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly T., Teeuwisse W.M., de Gans D., Koolstra K., Webb A.G. In vivo 3D brain and extremity MRI at 50 mT using a permanent magnet Halbach array. Magn. Reson. Med. 2020 doi: 10.1002/mrm.28396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrison W., Jr, Stimac G., Stevens E., LaMasters D., Espinosa M., Cobb L., Mettler F., Jr Comparison of CT, low-field-strength MR imaging, and high-field-strength MR imaging. Work in progress. Radiology. 1991;181(1):121–127. doi: 10.1148/radiology.181.1.1887020. [DOI] [PubMed] [Google Scholar]
- Paulson J.N., Williams B.L., Hehnly C., Mishra N., Sinnar S.A., Zhang L., Ssentongo P., Mbabazi-Kabachelor E., Wijetunge D.S., von Bredow B., et al. The Bacterial and Viral Complexity of Postinfectious Hydrocephalus in Uganda. Sci. Transl. Med. 2020;12(563) doi: 10.1126/scitranslmed.aba0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, C.-H., A. Ducournau, R. Fablet, and F. Rousseau (2017) ”Brain MRI super-resolution using deep 3D convolutional networks,” in 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), IEEE, pp. 197–200.
- Rutland J., Delman B., Gill C., Zhu C., Shrivastava R., Balchandani P. Emerging Use of Ultra-High-Field 7T MRI in the Study of Intracranial Vascularity: State of the Field and Future Directions. Am. J. Neuroradiol. 2020;41(1):2–9. doi: 10.3174/ajnr.A6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff, S., A. Kulkarni, K.E. Mbabazi, J. Mugamba, P. Ssenyonga, R. Donnelly, J. Levenbach, V. Monga, M. Peterson, V. Cherukuri, and B. Warf (2021) ”Brain growth after surgical treatment of infant post-infectious hydrocephalus in sub-Saharan Africa: two-year results of a randomized trial.” Journal of Neurosurgery Pediatrics. [DOI] [PMC free article] [PubMed]
- Sheth, K.N., M.H. Mazurek, M.M. Yuen, B.A. Cahn, J.T. Shah, A. Ward, J.A. Kim, E.J. Gilmore, G.J. Falcone, N. Petersen, et al. (2020) ”Assessment of Brain Injury Using Portable, Low-Field Magnetic Resonance Imaging at the Bedside of Critically Ill Patients,” JAMA neurology. [DOI] [PMC free article] [PubMed]
- Shi F., Cheng J., Wang L., Yap P.-T., Shen D. LRTV: MR image super-resolution with low-rank and total variation regularizations. IEEE Trans. Med. Imaging. 2015;34(12):2459–2466. doi: 10.1109/TMI.2015.2437894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg H., Alarcon J., Bernardino M. Focal hepatic lesions: comparative MR imaging at 0.5 and 1.5 T. Radiology. 1990;174(1):153–156. doi: 10.1148/radiology.174.1.2152980. [DOI] [PubMed] [Google Scholar]
- Wang Y.-H., Qiao J., Li J.-B., Fu P., Chu S.-C., Roddick J.F. Sparse representation-based MRI super-resolution reconstruction. Measurement. 2014;47:946–953. [Google Scholar]
- Warf B.C. Educate one to save a few. Educate a few to save many. World Neurosurg. 2013;79(2):S15–e15. doi: 10.1016/j.wneu.2010.09.021. [DOI] [PubMed] [Google Scholar]
- World Health Organization Baseline country survey on medical devices 2010. Tech. rep., World Health Organization. 2011 [Google Scholar]
- Yang J., Wright J., Huang T.S., Ma Y. Image super-resolution via sparse representation. IEEE Trans. Image Process. 2010;19(11):2861–2873. doi: 10.1109/TIP.2010.2050625. [DOI] [PubMed] [Google Scholar]
- Zhu B., Liu J.Z., Cauley S.F., Rosen B.R., Rosen M.S. Image reconstruction by domain-transform manifold learning. Nature. 2018;555(7697):487–492. doi: 10.1038/nature25988. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.