Abstract
Molecular analysis of chromosomal DNA from 193 multidrug-resistant (MDR) Salmonella enterica serovar Typhi isolates from 1990 to 1995 from Pakistan, Kuwait, Malaysia, Bangladesh, and India produced a total of five major different pulsed-field gel electrophoresis (PFGE) patterns. Even within a particular country MDR S. enterica serovar Typhi DNA was found to be in different PFGE groups. Similar self-transferable 98-MDa plasmids belonging to either incompatibility group incHI1 or incHI1/FIIA were implicated in the MDR phenotype in S. enterica serovar Typhi isolates from all the locations except Quetta, Pakistan, where the majority were of incFIA. A total of five different PFGE genotypes with six different plasmids, based on incompatibility and restriction endonuclease analysis groups, were found among these MDR S. enterica serovar Typhi isolates.
It is estimated that each year there are over 20 million cases of typhoid fever, which result in 700,000 deaths worldwide (25). In developed countries, the incidence of cases and death has been greatly decreased by a combination of improved sanitation and hygiene, vaccines, and effective antimicrobial chemotherapy. The first two are difficult if not impossible to implement in many developing countries, and, unfortunately, the effectiveness of antimicrobial chemotherapy is also being eroded by the emergence of antibiotic resistance (5, 16, 27). In developing countries, the antibiotics most readily available for treatment of typhoid are chloramphenicol, ampicillin, and co-trimoxazole. Plasmid-encoded chloramphenicol resistance emerged first in the early 1970s (2), followed by large epidemics in Central America (20). Although slightly less effective than chloramphenicol, ampicillin was used both for therapy and for elimination of the carrier state (22). Again, plasmid-encoded resistance soon developed (1, 20). Finally, co-trimoxazole was introduced in 1980, and plasmid-encoded resistance to trimethoprim and sulfonamides was observed shortly afterwards (7). The first cases of typhoid due to Salmonella enterica serovar Typhi carrying plasmid-encoded resistance to chloramphenicol, ampicillin, and co-trimoxazole were reported from southeast Asia (14). Cases in the area increased exponentially, and it is possible to define an epidemic zone for multidrug-resistant (MDR) S. enterica serovar Typhi which encompasses China, southeast Asia, and the Indian subcontinent (16). There is also a potential epidemic zone in the Middle East (Kuwait, Oman, and Saudi Arabia) due to importation of MDR S. enterica serovar Typhi by migrant workers from the epidemic zone. Interestingly MDR S. enterica serovar Typhi is rarely, if at all, reported from Africa or South and Central America, where some of the early epidemics of chloramphenicol-resistant typhoid were first reported (2, 20). We investigated the antimicrobial susceptibility and the genetic basis of resistance of MDR S. enterica serovar Typhi isolates from various countries in Asia and the Middle East. Pulsed-field gel electrophoresis (PFGE) was used to study the genotypic relationship among the isolates.
MATERIALS AND METHODS
Bacterial isolates.
A total of 193 MDR S. enterica serovar Typhi isolates obtained from 1990 to 1995 were studied; 149 isolates were from patients from Rawalpindi and Quetta, Pakistan, 30 were from Bangladesh, 6 were from Kuwait, 6 were from Malaysia, and 2 were from India. The identity of S. enterica serovar Typhi isolates was confirmed using biochemical tests on Analytab Products strips (bioMerieux, Basingstoke, United Kingdom), and they were serotyped using agglutinating antisera (Murex Diagnostics, Dartford, United Kingdom). Isolates were stored at −70°C on Protect beads (Technical Service Consultants Ltd., Heywood, United Kingdom) until analyzed.
Antimicrobial susceptibility testing.
S. enterica serovar Typhi isolates were tested for susceptibility to antimicrobials by a controlled disk diffusion technique according to the guidelines provided by the National Committee for Clinical Laboratory Standards (18) on diagnostic sensitivity testing (DST) agar (Oxoid Ltd., Basingstoke, United Kingdom) plates containing 5% lysed horse blood. The antibiotic discs (all from Oxoid) contained ampicillin (10 μg), tetracycline (30 μg), co-trimoxazole (1.25 μg), chloramphenicol (30 μg), streptomycin (30 μg), gentamicin (10 μg), amoxicillin (20 μg)-clavulanic acid (10 μg), ofloxacin (10 μg), ciprofloxacin (10 μg), and nalidixic acid (10 μg). MICs of these antibiotics were determined using the E-test strips (AB BIODISK, Solna, Sweden) following the manufacturer's instructions. Adjusted inocula of bacteria (ca. 106 CFU/ml [ca. 0.5 MacFarland standard]) were used on DST agar plates. Escherichia coli ATCC 25922 (the MICs for which are known) was used as a control for potency of antibiotics.
Beta-lactamase study.
Preparations of protein extracts from S. enterica serovar Typhi isolates were made using the method of Corkill et al. (6) and were analyzed by isoelectric focusing on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (Pharmacia, St. Albans, United Kingdom). Ampholytes with pIs in the range of 3.5 to 9.5 were used to generate the pH gradient, and proteins of known pIs were included as controls, as were extracts from bacteria expressing TEM-1 and TEM-2.
PFGE of macrorestricted genomic DNA.
Chromosomal DNA from S. enterica serovar Typhi isolates was prepared in agarose plugs as described by Thong et al. (26). DNA in agarose plugs was digested using 25 U of XbaI or 20 U of SpeI (Life Technologies, Paisley, United Kingdom). PFGE of agarose plug inserts was then performed on a CHEF-DR II system (Bio-Rad Laboratories, Richmond, Calif.) on a horizontal 1% agarose gel for 22 h at 120 V, with a pulse time of 1 to 40 s at 14°C. A lambda DNA digest consisting of a ladder (ca. 22 fragments) of increasing size from 48 kb to approximately 1,000 kb was included as a DNA size standard. The gel was stained with ethidium bromide and photographed on a UV transilluminator (UVP Inc., San Gabriel, Calif.). The restriction endonuclease (RE) digest patterns were compared, and their similarities were scored by the method of Tenover et al. (24). By these criteria isolates that gave PFGE banding patterns that were indistinguishable were assumed to be from a single outbreak strain. Isolates that gave banding patterns showing differences in fewer than four bands were assumed to be closely related, as they may represent isolates differing by a single genetic event. In addition, isolates with differences in four to six bands may also be part of an outbreak. However, isolates showing a difference in more than seven bands of their banding patterns may represent more than three genetic events, in which case they were considered epidemiologically unrelated.
Conjugation of plasmids and incompatibility grouping.
Conjugation experiments were carried out in broth using the method of Walia et al. (28) with E. coli K-12 (Nalr Lac+) as the recipient. All S. enterica serovar Typhi isolates were susceptible to nalidixic acid. Transconjugants were selected on MacConkey agar (Oxoid) supplemented with nalidixic acid (32 mg/l each) and ampicillin or chloramphenicol (32 mg/l each). Plasmid DNA was then extracted from the transconjugants using an alkaline lysis method (3). Plasmids were electrophoresed on horizontal 0.8% agarose gels and stained with 0.05% ethidium bromide (Sigma, Poole, United Kingdom). DNA bands were then visualized using a UV transilluminator (UVP).
Incompatibility grouping was performed on plasmids extracted from E. coli K-12 transconjugants using DNA-DNA hybridization (15) on nitrocellulose membranes (Sigma). A total of eight rep probes (incFII, incFI, incB/O, incHI1, incK, incN, incP, and incW) labeled with biotinylated UTP by nick translation were employed as described previously (13). Probe-target hybridization was detected using a streptavidin-alkaline phosphatase conjugate and nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate for visualization (Sigma).
Restriction endonuclease analysis (REA).
Plasmid DNA was extracted from MDR S. enterica serovar Typhi transconjugants as described previously (4) and subsequently was digested using HindIII and EcoRI (Life Technologies) according to the manufacturer's instructions. Restriction fragments were resolved by agarose gel electrophoresis. HindIII digests of lambda DNA were used as size standards.
RESULTS
Antimicrobial susceptibility.
A total of 193 MDR S. enterica serovar Typhi isolates were examined. All the isolates were uniformly resistant to the five antimicrobial agents commonly available in developing countries, namely, ampicillin, co-trimoxazole, chloramphenicol, tetracycline, and streptomycin. However, they were all sensitive to nalidixic acid, cefuroxime, cefotaxime, ciprofloxacin, and ofloxacin. MICs of ampicillin, chloramphenicol, streptomycin, and tetracycline were all >256 mg/l, and the MICs of co-trimoxazole were 16 to 32 mg/l. MICs of nalidixic acid ranged between 2 and 4 mg/l, and amoxicillin-clavulanic acid MICs ranged between 0.25 and 10 mg/l. Resistance in each case was associated with self-transferable 98-MDa plasmids.
Beta-lactamases.
On isoelectric focusing all the MDR S. enterica serovar Typhi isolates produced beta-lactamases with a pI of 5.4 that cofocused with the control TEM-1 beta-lactamase. As these isolates did not show resistance to the extended-spectrum beta-lactams, no further analysis of their beta-lactamases was carried out.
Conjugation of resistance plasmids and incompatibility grouping.
As shown in Table 1 121 of the 193 (62.7%) MDR S. enterica serovar Typhi isolates from the various countries transferred resistance to three or more antimicrobials to E. coli K-12. For all the MDR S. enterica serovar Typhi isolates resistance was carried on 98-MDa plasmids. Apart from S. enterica serovar Typhi isolates from Quetta, Pakistan, which transferred three different resistance phenotypes, all the isolates transferred the same resistance phenotype in each case. The 98-MDa resistance plasmids fell into five different incompatibility groups (Table 2). Apart from resistance plasmids from Pakistan, which were in three different incompatibility groups, the plasmids were either of incHI1 or incHI1 cross-reacting with incFIIA (incHI1/FIIA). The distribution of the other plasmid incompatibility groups is shown in Table 2. Interestingly, the predominance of incFIA plasmids among the S. enterica serovar Typhi isolates from Pakistan contrasts with the general association between incHI1 plasmids and S. enterica serovar Typhi worldwide.
TABLE 1.
Patterns of antibiotic resistance transfer from MDR S. enterica serovar Typhi isolates using ampicillin and chloramphenicol for selection of transconjugant phenotypes
| Isolate origin (no.) | Phenotype of donora | No. of successful transfers | Phenotype transferred to E. coli K-12b |
|---|---|---|---|
| Rawalpindi (25) | Cm Ap Te Tm Su Sm | 25 | Cm Ap Te Tm Su Sm |
| Quetta (122) | Cm Ap Te Tm Su Sm | 72 | Cm Ap Te Tm Su Sm (62) |
| Cm Te Tm Sm (5) | |||
| Cm Ap Te Tm Sm (5) | |||
| Bangladesh (30) | Cm Ap Te Tm Su Sm | 10 | Cm Tm Ap |
| Kuwait (8) | Cm Ap Te Tm Su Sm | 6 | Cm Te Ap |
| Malaysia (6) | Cm Ap Te Tm Su Sm | 6 | Cm Te Ap |
| India (2) | Cm Ap Te Tm Su Sm | 2 | Cm Ap Te Tm |
Ap, ampicillin; Cm, chloramphenicol; Tm, trimethoprim; Su, sulfonamide; Te, tetracycline; Sm, streptomycin.
Numbers in parentheses are numbers of isolates with successful transfers.
TABLE 2.
Incompatibility groups of the transferable 98-MDa resistance plasmids from 121 E. coli transconjugants
| Source | No. of isolates with incompatibility groupa:
|
|||||
|---|---|---|---|---|---|---|
| FIA | FIIA | P | B/O | HI1 | HI1/FIIAb | |
| Rawalpindi | 25 | |||||
| Quetta | 69 | 2 | 1 | |||
| Bangladesh | 1 | 9 | ||||
| Kuwait | 5 | 1 | ||||
| India | 2 | |||||
| Malaysia | 1 | 5 | ||||
The prefix inc is omitted throughout.
HI1/FIA, plasmids hybridized with both the rep incHI1 and incFIA probes.
REA of resistance plasmids.
RE digestion of the 98-MDa plasmids from MDR S. enterica serovar Typhi with EcoRI or HindIII produced 7 to 12 fragments ranging in size from 2 to 10 kb (Table 3). Two isolates from Quetta, Pakistan, had distinct unique restriction digestion patterns. In order to improve resolution, the plasmid DNA was digested with HindIII and EcoRI together. There was better discrimination between strains following digestion with the two enzymes combined than with the individual enzymes. As expected more DNA fragments (15 to 18) were produced with the combined restriction enzyme digestion than with digestion by the individual enzymes, and their sizes varied from less than 2,000 to 10,000 bp (Table 3). With the combination of HindIII and EcoRI, REA patterns for plasmids from four of the five different countries (the exception being Pakistan [Rawalpindi and Quetta]) were similar (Table 3). REA of plasmids from Quetta using HindIII and EcoRI, either alone or in combination, gave two distinct patterns; these plasmids were clearly different from the plasmids from Rawalpindi, Pakistan, and from the other four countries.
TABLE 3.
REA patterns of 98-MDa plasmids and PFGE analysis of XbaI (5′-TCTAGA-3′)-digested chromosomal DNA
| Isolate origin (no.) | No. of REA fragments from digestion with:
|
REA pattern(s) of plasmids | PFGE pattern(s)a | ||
|---|---|---|---|---|---|
| HindIII | EcoRI | HindIII + EcoRI | |||
| Rawalpindi (25) | 9–10 | 9–10 | 15–18 | I | A1 (18), A2 (3), B1 (4) |
| Quetta (122) | 8–9 | 11–12 | 16–17 | III and IV | A1 (92), B1 (12), B2 (10), E1 (2), E2 (6) |
| Kuwait (8) | 7–8 | 8–9 | 15–18 | II | C3 (8) |
| Bangladesh (30) | 7–8 | 8–9 | 15–18 | II | C1 (30) |
| Malaysia (6) | 7–8 | 8–9 | 15–18 | II | D1 (1), D2 (2), E1 (3) |
| India (2) | 7–8 | 8–9 | 15–18 | II | C2 (2) |
The PFGE patterns were arbitrarily designated groups A to E. The pattern for each group differs by more than seven bands from those of the other groups. The subgroup patterns (e.g., C1, C2, C3) differed from each other by fewer than seven bands. Numbers in parentheses are numbers of isolates in the subgroup.
PFGE.
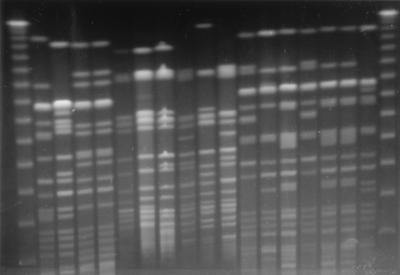
XbaI-digested DNA from S. enterica serovar Typhi isolates from the five different countries produced a total of five different patterns consisting of 13 to 22 fragments ranging in size from ca. 23 to 730 kb. Figure 1 shows the PFGE banding patterns of representative MDR S. enterica serovar Typhi isolates from various countries. The 30 MDR S. enterica serovar Typhi isolates from Bangladesh all produced the same PFGE fragment pattern (designated subgroup C1 in Table 3). This pattern was closely related to the PFGE patterns for the isolates from India (C2) and Kuwait (C3). The Malaysian isolates produced one pattern (D) that was distinct from all the other isolates and one (E) that was also found in eight isolates from Quetta, Pakistan. The MDR S. enterica serovar Typhi isolates from Rawalpindi, Pakistan, fell into two groups (A and B), and the majority of the isolates from Quetta, Pakistan, fell into these two groups, which were not detected among isolates from any of the other regions. As with the XbaI-digested DNA, the SpeI-digested DNA fragment pattern analysis resulted in similar types and a similar distribution of digestion patterns of S. enterica serovar Typhi isolates from the five countries and hence did not offer further discrimination among the S. enterica serovar Typhi isolates. However, SpeI digestion produced more fragments (18 to 24), ranging in size from ca. 20 to 750 kb.
FIG. 1.
XbaI RE fragment patterns of representative S. enterica serovar Typhi isolates from various countries. Lanes 1 and 19 (numbering is from left to right), 48.5-kb ladder molecular size standard; lane 2, B1 from Bangladesh in PFGE group C1; lane 3, I1 from India in PFGE group C2; lanes 4 and 5, K1 and K2, respectively, from Kuwait in PFGE group C3; lanes 6, 7, 8, and 9, M1, M2, M3, and M4 from Malaysia in PFGE groups D1, D2, D2, and E1, respectively; lanes 10, 11, 12, 13, and 14, Q1, Q2, Q3, Q4, and Q5 from Quetta, Pakistan, in PFGE groups E2, E1, B1, B2, and A1, respectively; lanes 15, 16, 17, and 18, R1, R2, R3, and R4 from Rawalpindi, Pakistan, in PFGE groups B1, A1, A1, and A2, respectively.
DISCUSSION
In this study we have examined a total of 193 MDR isolates of S. enterica serovar Typhi from five different countries in Asia and the Middle East. Each was included because of resistance to chloramphenicol, ampicillin, and co-trimoxazole, and this resistance phenotype is predominant in many parts of the continent (9, 16, 19, 23, 27). The MICs of ampicillin, chloramphenicol, and co-trimoxazole among our MDR S. enterica serovar Typhi isolates are similar to those reported from India and Vietnam (23, 27). In each case ampicillin resistance was mediated by a β-lactamase with a pI of 5.4 that cofocused with TEM-1. This confirms and extends findings from Vellore, India, which demonstrated that all the MDR S. enterica serovar Typhi isolates examined expressed TEM-1 (23). All our MDR S. enterica serovar Typhi isolates were sensitive to cefuroxime and cefotaxime and, unlike those from Vietnam and Tajikistan (17, 27), all were sensitive to nalidixic acid, ciprofloxacin, and ofloxacin. This is of particular importance since the fluoroquinolones ciprofloxacin and ofloxacin are particularly valuable in the treatment of typhoid fever whether due to MDR S. enterica serovar Typhi or not (3, 5, 12).
In each case antimicrobial resistance was transferable to E. coli K-12 on ca. 98-MDa plasmids. In previous studies the large (90- to 110-MDa) self-transmissible plasmids from MDR S. enterica serovar Typhi have invariably been of incompatibility group IncHI1 (1, 10, 23). Although all of the tested plasmids from the MDR S. enterica serovar Typhi isolates from Bangladesh, Kuwait, India, and Rawalpindi, Pakistan, fell into this or a cross-reacting incHI1/FIIA group, none of the plasmids from the Quetta isolates did so. The majority of the Quetta plasmids were of incFIA but two were of incP and one was of IncB/O, a distribution which has not been reported previously. In addition one of five Malaysian plasmids was of incFIIA. RE digestion of the plasmids revealed a total of five different patterns. The plasmids from the MDR S. enterica serovar Typhi isolates from Kuwait, Bangladesh, Malaysia, and India all produced the same pattern regardless of their incompatibility groups and antibiotic resistances transferred. The isolates from Rawalpindi all produced the same plasmid RE pattern, which was different from all the others. Finally the isolates from Quetta carried plasmids that fell into two different RE patterns that were also unique. Those of incFIA produced one pattern, and the rest produced the other pattern. It is thus clear that the MDR phenotype in the isolates from our study is encoded on a variety of different plasmids, as determined by incompatibility group and REA. However, most of the diversity arises in the isolates from Pakistan.
PFGE of macrorestricted chromosomal DNA from the MDR S. enterica serovar Typhi isolates produced five major patterns which differed from each other in more than seven bands. This was apparent when either XbaI or SpeI was used. Although until recently it has been suggested that S. enterica serovar Typhi represents a single clone with little interspecies divergence (21), molecular analyses such as that by PFGE are demonstrating much greater genetic heterogeneity among S. enterica serovar Typhi isolates (9, 23, 25, 26). It is possible that some of the apparent genomic diversity could result from homologous recombinations between rrn operons, as has been observed in epidemiologically linked outbreaks in Spain (8). It is unlikely that such events could account for all the diversity detected in the present study and by others, especially in those isolates that are not epidemiologically linked (8, 19, 21, 22). Previously PFGE analysis of S. enterica serovar Typhi isolates from Vellore, India, has demonstrated that although antibiotic-sensitive isolates showed great genomic variability, the MDR S. enterica serovar Typhi constituted a single genotype (23). Analysis of MDR S. enterica serovar Typhi isolates from visitors to Canada (10) and from Bangladesh (11) also demonstrated single genotypes. In the present study two different PFGE patterns were detected in both Pakistan and Malaysia. In contrast the isolates from India, Bangladesh, and Kuwait produced identical patterns. Thus a total of five different genotypes of MDR S. enterica serovar Typhi were detected. Similarly, Hampton et al. (9) were able to detect different genotypes among MDR S. enterica serovar Typhi isolates originating from Pakistan, India, Bangladesh, and Tajikistan. In conclusion, our study has demonstrated that there are at least five different genotypes of MDR S. enterica serovar Typhi in circulation in Asia. In addition it appears that there are at least six different plasmids encoding the MDR phenotype based on a combination of REA and incompatibility grouping. Clearly the MDR S. enterica serovar Typhi isolates that are circulating in Asia are not derived from a single clone of S. enterica serovar Typhi that has acquired one particular resistance plasmid.
ACKNOWLEDGMENT
We thank the Wellcome Trust for a grant to support this work.
REFERENCES
- 1.Anderson E S. Problems and implications of chloramphenicol resistance in the typhoid bacillus. J Hyg Camb. 1975;74:289–299. doi: 10.1017/s0022172400024360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson E S, Smith H R. Chloramphenicol resistance in the typhoid bacillus. Br Med J. 1972;3:329–331. doi: 10.1136/bmj.3.5822.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arshad A, Naseem S, Tasmin A. Enoxacin in the treatment of typhoid fever. Clin Ther. 1992;14:825–828. [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandra R, Srinivasan S, Nalini P, Rao R S. Multidrug resistant enteric fever. J Trop Med Hyg. 1992;95:284–287. [PubMed] [Google Scholar]
- 6.Corkill J E, Hart C A, McLennan A G, Aspinall S. Characterisation of a β-lactamase from Pseudomonas paucimobilis. J Gen Microbiol. 1991;137:1425–1429. doi: 10.1099/00221287-137-6-1425. [DOI] [PubMed] [Google Scholar]
- 7.Datta N, Richards H, Datta C. Salmonella typhi in-vivo acquires resistance to both chloramphenicol and cotrimoxazole. Lancet. 1981;i:1181–1183. doi: 10.1016/s0140-6736(81)92350-3. [DOI] [PubMed] [Google Scholar]
- 8.Echeita M A, Usera M A. Chromosomal rearrangements in Salmonella enterica serotype Typhi affecting molecular typing in outbreak investigations. J Clin Microbiol. 1998;36:2123–2126. doi: 10.1128/jcm.36.7.2123-2126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampton M D, Ward R L, Rowe B, Threlfall E J. Molecular fingerprinting of multidrug-resistant Salmonella enterica serotype Typhi. Emerg Infect Dis. 1998;4:317–320. doi: 10.3201/eid0402.980223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harnett N, McLeod S, AuYong Y, Wan J, Alexander S, Khakhria R, Krishnan C. Molecular characterization of multiresistant strains of Salmonella typhi from South Asia isolated in Ontario, Canada. Can J Microbiol. 1998;44:356–363. [PubMed] [Google Scholar]
- 11.Hermans P W M, Saha S K, van Leeuwen W J, Verbrugh H A, van Belkum A, Goessens W H F. Molecular typing of Salmonella typhi strains from Dhaka (Bangladesh) and development of DNA probes identifying plasmid-encoded multidrug-resistant isolates. J Clin Microbiol. 1996;34:1373–1379. doi: 10.1128/jcm.34.6.1373-1379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hien T T, Bethel D B, Hoa N T T, Wain J, Diep T S, Phi L T, Cuong B M, Duong N M, Thanh P T, Walsh A L, Day N P J, White N J. Short course of ofloxacin for treatment of multidrug-resistant typhoid. Clin Infect Dis. 1994;20:917–923. [PubMed] [Google Scholar]
- 13.Kariuki S, Gilks C, Corkill J, Kimari J, Benea A, Waiyaki P G, Hart C A. Multi-drug resistant non-typhi Salmonellae in Kenya. J Antimicrob Chemother. 1996;38:425–434. doi: 10.1093/jac/38.3.425. [DOI] [PubMed] [Google Scholar]
- 14.Ling J, Chau P Y. Plasmid mediating resistance to chloramphenicol, trimethoprim, and ampicillin in Salmonella typhi in the Southeast Asia region. J Infect Dis. 1984;149:652. doi: 10.1093/infdis/149.4.652. [DOI] [PubMed] [Google Scholar]
- 15.Meinkoth J, Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984;138:267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- 16.Mirza S H, Beeching N J, Hart C A. Multi-drug resistant typhoid: a global problem. J Med Microbiol. 1995;44:317–319. doi: 10.1099/00222615-44-5-317. [DOI] [PubMed] [Google Scholar]
- 17.Murdoch D A, Banatvaia N, Bone A, Shoismatulloev B I, Ward L R, Threlfall E J. Epidemic ciprofloxacin-resistant Salmonella typhi in Tajikistan. Lancet. 1998;351:339. doi: 10.1016/s0140-6736(05)78338-0. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. 4th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 19.Oh H M L, Masayu Z, Chew S K. Typhoid fever in hospitalized children in Singapore. J Infect. 1997;34:237–242. doi: 10.1016/s0163-4453(97)94283-3. [DOI] [PubMed] [Google Scholar]
- 20.Olarte J, Galindo E. Salmonella typhi resistant to chloramphenicol, ampicillin and other antimicrobial agents: strains isolated during an extensive typhoid fever epidemic in Mexico. Antimicrob Agents Chemother. 1973;4:597–601. doi: 10.1128/aac.4.6.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reeves M W, Evins G M, Heiba A A, Plikaytis B D, Farmer J J., III Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989;27:313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders W L. Treatment of typhoid fevers: a comparative trial of ampicillin and chloramphenicol. Br Med J. 1965;2:1226–1229. doi: 10.1136/bmj.2.5472.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shanahan P M A, Jesudason M V, Thomson C J, Amyes S G B. Molecular analysis of and identification of antibiotic resistance genes in clinical isolates of Salmonella typhi from India. J Clin Microbiol. 1998;36:1595–1600. doi: 10.1128/jcm.36.6.1595-1600.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thong K L, Cheong M Y, Puthucheary S, Koh C L, Pang T. Epidemiologic analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:1135–1141. doi: 10.1128/jcm.32.5.1135-1141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thong K L, Ngeow Y F, Altwegg P N, Pang T. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–1074. doi: 10.1128/jcm.33.5.1070-1074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wain J, Hoa N T, Chinh N T, Vinh H, Everett M J, Diep T S, Day N P, Solomon T, White N J, Piddock L J, Parry C M. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin Infect Dis. 1997;25:1404–1410. doi: 10.1086/516128. [DOI] [PubMed] [Google Scholar]
- 28.Walia S K, Madhavan T, Chagh T D, Sharma K B. Characterization of self-transmissible plasmids determining lactose fermentation and multiple antibiotic resistance in clinical strains of Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 1987;7:279–284. doi: 10.1016/0147-619x(87)90003-5. [DOI] [PubMed] [Google Scholar]