Abstract
Both B cells and T cells are involved in an effective immune response to SARS‐CoV‐2, the disease‐causing virus of COVID‐19. While B cells—with the indispensable help of CD4+ T cells—are essential to generate neutralizing antibodies, T cells on their own have been recognized as another major player in effective anti‐SARS‐CoV‐2 immunity. In this report, we provide insights into the characteristics of individual HLA‐A*02:01‐ and HLA‐A*24:02‐restricted SARS‐CoV‐2‐reactive TCRs, isolated from convalescent COVID‐19 patients. We observed that SARS‐CoV‐2‐reactive T‐cell populations were clearly detectable in convalescent samples and that TCRs isolated from these T cell clones were highly functional upon ectopic re‐expression. The SARS‐CoV‐2‐reactive TCRs described in this report mediated potent TCR signaling in reporter assays with low nanomolar EC50 values. We further demonstrate that these SARS‐CoV‐2‐reactive TCRs conferred powerful T‐cell effector function to primary CD8+ T cells as evident by a robust anti‐SARS‐CoV‐2 IFN‐γ response and in vitro cytotoxicity. We also provide an example of a long‐lasting anti‐SARS‐CoV‐2 memory response by reisolation of one of the retrieved TCRs 5 months after initial sampling. Taken together, these findings contribute to a better understanding of anti‐SARS‐CoV‐2 T‐cell immunity and may contribute to paving the way toward immunotherapeutics approaches targeting SARS‐CoV‐2.
Keywords: COVID‐19, SARS‐CoV‐2, single‐cell sequencing, T‐cell receptor
T cell receptors (TCRs) directed against previously identified SARS‐CoV‐2 target epitopes were isolated from convalescent COVID‐19 patients by single‐cell sequencing. Upon ectopic re‐expression, these SARS‐CoV‐2 TCRs conferred potent effector functions such as cytotoxicity and Interferon‐gamma production.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) causes coronavirus disease 2019 (COVID‐19), with a wide range of clinical symptoms ranging from asymptomatic or mild cases to severe cases with increased mortality in high‐risk patients [1, 2, 3, 4].
Similar to other viral infections, the immune response to COVID‐19 encompasses both B cell‐mediated humoral responses through antibodies as well T‐cell activity [5]. In the early days of the pandemic, investigations have focused on the role of the humoral immune response, where neutralizing antibodies have emerged as key to viral clearance. After an acute antibody response, however, antibody levels cease quickly in COVID‐19 convalescent patients and a humoral response is not necessarily detected in all affected individuals [6, 7, 8]. In contrast, a growing body of evidence combined from prior coronaviruses, SARS‐CoV‐1 and Middle East respiratory syndrome coronavirus (MERS‐CoV), and the recent SARS‐CoV‐2 suggests that T‐cell responses are important for both early viral clearance as well as conferring protection through memory T cells for extended time periods [9, 10, 11, 12, 13, 14]. On the flip side, a strong but dysregulated cellular immune response may be detrimental to the course of the infection [15, 16]. Thus, studying the T‐cell immune response against SARS‐CoV‐2, including the underlying mechanisms, contributes to a better understanding of COVID‐19.
Since the description of the first SARS‐CoV‐2 infections, a lot of progress has been made in the understanding of such T‐cell responses. Many SARS‐CoV‐2‐derived T‐cell epitopes, presented by infected cells through the HLA system, have recently been identified and characterized [17, 18, 19, 20]. In contrast to detecting T‐cell activation after stimulation with large pools of overlapping peptides or protein fractions, this has enabled the tracking of individual T‐cell responses against single SARS‐CoV‐2‐derived peptides, for example, in acutely infected or convalescent individuals over time [21, 22, 23]. It is now also possible to differentiate between responses against SARS‐CoV‐2‐specific epitopes and epitopes that show cross‐reactivity with other human common cold coronaviruses [24].
While single‐cell analysis of SARS‐CoV‐2‐reactive T cells has progressed significantly, information about the TCRs driving these responses is still limited. Understanding which TCRs are involved in T‐cell responses to SARS‐CoV‐2 is necessary to delineate the nature of such a T‐cell response. It is, for instance, still a matter of debate to what extent heterologous immunity–provided by cross‐reactivity of memory T cells generated as part of an immune reaction to previous viral infections–is beneficial, prohibitive, or irrelevant. More importantly, qualitative assessment of TCRs derived from a T‐cell response unique to SARS‐CoV‐2 (induced in the course of natural infection or by vaccination) by means of TCR re‐expression is required to truly understand such T‐cell responses on a clonal level. First results have already shown tracking of different clones at multiple time points during infections and allowed comparison of larger numbers of patients, identifying diverse TCR repertoires with partly shared binding motifs [25, 26, 27, 28]. So far though, data linking individual TCRs reactive for SARS‐CoV‐2‐specific epitopes with their functional properties are still scarce [29].
Here, we report on single‐cell TCR isolation specific for HLA‐A*02:01‐ and HLA‐A*24:02 SARS‐CoV‐2‐derived T‐cell epitopes (derived from the nucleocapsid protein, ORF3 and ORF1) from PBMCs of convalescent patients after PCR‐confirmed SARS‐CoV‐2 infection. We further provide re‐expression data on the identified TCRs showing excellent functionality of the majority of retrieved TCRs as assessed by reporter assays, cytokine production, and in vitro cytotoxicity assays.
Results
SARS‐CoV‐2 tetramer‐positive cells are detectable ex vivo in COVID‐19 convalescent individuals
Previously, we characterized several SARS‐CoV‐2 epitopes using IFN‐γ ELISPOT assays with PBMCs of convalescent individuals (SCD) and prepandemic healthy donors (PRE) [17]. From this panel of epitopes, we selected two HLA‐A*02:01‐ and two HLA‐A*24:02‐restricted SARS‐CoV‐2‐specific T‐cell epitopes (Table 1) that were specifically recognized by T cells of the SCD group but not the PRE group (Fig. 1A). To test whether we could identify CD8+ T‐cell clones recognizing one of the four SARS‐CoV‐2‐derived T‐cell epitopes directly ex vivo, HLA‐A*02:01+ and/or HLA‐A*24:02+ SCD group samples (n = 11) collected between 31 and 56 days after positive PCR test and a variable degree of antibody response (Supporting information Table S1) were probed with fluorescently labeled peptide‐HLA‐tetramers and analyzed using flow cytometry. PRE group samples acquired from separate individuals (n = 6) served as controls. Using a sensitive combinatorial staining approach (Supporting information Fig. S1) [30, 31], we were able to identify 2D tetramer‐positive cells for all SARS‐CoV‐2 T‐cell epitopes in at least one of the SCD group samples (Fig. 1B upper row, Supporting information Fig. S2), indicative of target‐specific CD8+ T cells. In contrast, tetramer staining of PRE group samples revealed only sporadic events which did not make up distinct 2D tetramer populations (Fig. 1B lower row, Supporting information Fig. S2). Overall, we observed homogeneously absent or very low frequency of tetramer‐positive cells in PRE group samples, but significantly higher frequencies in many of the SCD group samples with matching HLA alleles for the respective targets, but not in HLA‐unmatched samples (Fig. 1C). These results followed a similar pattern as previously reported for T‐cell responses to SARS‐CoV‐2‐specific epitopes [17]. Of note, tetramer‐positive populations could also be detected in samples that did not test positive for antispike and/or antinucleocapsid antibodies (Supporting information Fig. S3).
Table 1.
SARS‐CoV‐2‐derived HLA class I T‐cell epitopes
| Peptide ID | Sequence | HLA restriction | Protein | Protein class | Position |
|---|---|---|---|---|---|
| A02_P03 | ALSKGVHFV | A*02 | ORF3 | accessory | 72‐80 |
| A02_P09 | LLLLDRLNQL | A*02 | ORF9 nuc | structural | 221‐230 |
| A24_P01 | VYIGDPAQL | A*24 | ORF1 | non‐structural | 5721‐5729 |
| A24_P03 | VYFLQSINF | A*24 | ORF3 | accessory | 112‐120 |
Selected SARS‐CoV‐2‐specific T‐cell epitopes and their origins. Nuc, nucleocapsid protein.
Figure 1.
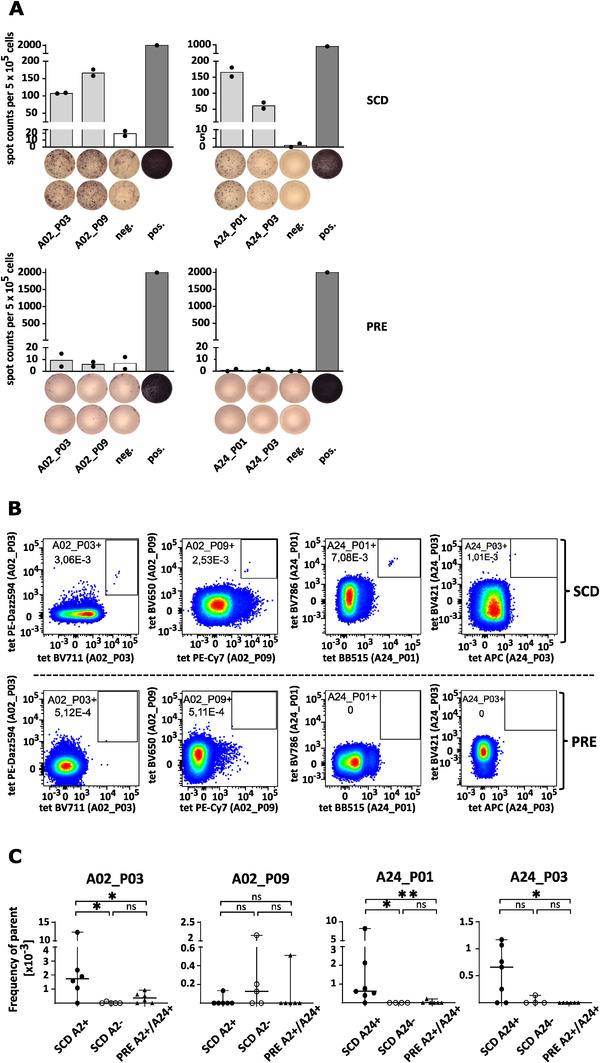
SARS‐CoV‐2 tetramer‐positive cells enriched in convalescent samples. (A) Representative IFN‐γ ELISPOT assays of peptide‐specific T cells from HLA‐matched SARS‐CoV‐2 convalescent donors (SCD‐12 and SCD‐13) (SCD, upper row) and unexposed prepandemic healthy donors (PRE, lower row) after 12‐d in vitro stimulation with SARS‐CoV‐2‐derived HLA‐A*02‐ and A*24‐binding peptides. T‐cell responses were considered positive when the mean spot counts in ELISPOT assays were at least threefold higher than the negative control. ELISPOT data are presented as a scatter dot plot with mean. Neg. = negative control using an irrelevant HLA‐matched peptide (HLA‐A*02 YLLPAIVHI DDX5_HUMAN148‐156, HLA‐A*24 AYVHMVTHF BI1_HUMAN45‐53). Pos. = positive control (PHA). The experiment was conducted once with technical duplicates. (B) Exemplary 2D tetramer FACS plots for convalescent (SCD, upper panel) or prepandemic healthy donor control samples (PRE, lower panel). An equal number of CD8+ T cells as parent population is shown for all samples. The depicted numbers in the gates indicate the respective frequency of parent. Axis are scaled logarithmically. The respective target antigen is indicated on the axis in brackets. For an example of the full gating strategy, please refer to Supporting information Fig. S1. (C) Scatter plot showing the cumulative frequencies of tetramer‐positives cells in convalescent samples (subdivided into HLA/target matched and unmatched) compared to prepandemic controls. Statistical significance was determined with the Mann Whitney test using GraphPad Prism (*p < 0.05, **p < 0.01, ns, not significant). The experiment was conducted once, n (SCD) = 11, n (PRE) = 6.
TCR isolation from SARS‐CoV‐2 convalescent PBMC samples
Having established that there are clearly discernible SARS‐CoV‐2 tetramer‐positive populations detectable in SCD group samples, we used FACS to isolate SARS‐CoV‐2 tetramer‐positive T cells for subsequent TCR sequence retrieval by single‐cells next generation sequencing (10× Genomics + Illumina platform). Out of 11 tested SCD group samples, 9 (82%) were positive for at least one of the analyzed T‐cell epitopes, largely matching the HLA allele phenotype of the donor (Table 2). In addition, single‐cell sorting followed by 5’RACE in combination with Sanger sequencing was used as a complementary approach to retrieve TCR sequences. Combining these efforts, we were able to identify 16 unique TCRs (Table 3).
Table 2.
Tetramer‐detected responses against SARS‐CoV‐2 epitopes in convalescent donors
| Donor ID | A02_P03 | A02_P09 | A24_P01 | A24_P03 | HLA Type | |
|---|---|---|---|---|---|---|
| HLA‐A*02 positive | SCD‐11 | ++ | ‐ | ‐ | ‐ | A*02, A*29, B*27, B*51, C*02, C*15 |
| SCD‐09 | ++ | ‐ | ‐ | ‐ | A*02, A*03, B*14, B*51, C*08, C*15 | |
| SCD‐05 | ‐ | ‐ | ‐ | ‐ | A*02, A*03, B*13, B*15, C*06, C*12 | |
| SCD‐06 | +++ | ‐ | ‐ | ‐ | A*02, B*40, B*44, C*03, C*05 | |
| HLA‐A*24 positive | SCD‐01 | ‐ | ‐ | + | ‐ | A*24, A*30, B*13, B*35, C*04, C*06 |
| SCD‐02 | ‐ | ++ | ++ | ++ | A*24, A*25, B*18, B*51, C*12 | |
| SCD‐08 | ‐ | ‐ | + | + | A*11, A*24, B*18, B*35, C*04, C*07 | |
| SCD‐07 | ‐ | ‐ | ‐ | ‐ | A*24, A*68, B*41, B*47, C*06, C*17 | |
| SCD‐04 | ‐ | ‐ | +++ | ‐ | A*11, A*24, B*15, B*51, C*07, C*15 | |
| HLA‐A*02/24 positive | SCD‐10 | ++ | ‐ | + | + | A*02, A*24, B*07, B*40, C*03, C*07 |
| SCD‐03 | ++ | ‐ | ‐ | ++ | A*02, A*24, B*27, B*40, C*02, C*03 |
Detected tetramer responses against SARS‐CoV‐2‐specific T cell epitopes in PBMC samples from convalescent donors. Donors grouped by HLA type.“+” represent frequency of tetramer‐positive cells of all CD8+ T cells: +++ = % tet+ > 0.005%, ++ = % tet+ > 0.001%, + = % tet+ > 0.0005%
Table 3.
Properties of SARS‐CoV‐2‐reactive TCRs
| TCR ID | Vα | Jα | Cα | Vβ | Jβ | Cβ | CDR3α (aa) | CDR3β (aa) |
|---|---|---|---|---|---|---|---|---|
| SCV‐001 | TRAV1‐1 | TRAJ22 | TRAC | TRBV27 | TRBJ2‐4 | TRBC2 | CAAPISSGSARQLTF | CASSLPGAGHIQYF |
| SCV‐002 | TRAV12‐1 | TRAJ24 | TRAC | TRBV12‐5 | TRBJ2‐1 | TRBC2 | CVVNDSWGKLQF | CASRASGGYNEQFF |
| SCV‐003 | TRAV12‐1 | TRAJ44 | TRAC | TRBV9 | TRBJ2‐1 | TRBC2 | CVVNPTGTASKLTF | CASSIAGGLYEQFF |
| SCV‐004 | TRAV12‐1 | TRAJ44 | TRAC | TRBV9 | TRBJ2‐7 | TRBC2 | CVVNPTGTASKLTF | CASSVAGGLYEQYF |
| SCV‐005 | TRAV12‐1 | TRAJ44 | TRAC | TRBV9 | TRBJ2‐7 | TRBC2 | CVVNIDGTASKLTF | CASSVAGGLYEQYF |
| SCV‐006 | TRAV17 | TRAJ57 | TRAC | TRBV7‐9 | TRBJ2‐7 | TRBC2 | CATDRGLVF | CASSLPGNYEQYF |
| SCV‐007 | TRAV19 | TRAJ33 | TRAC | TRBV9 | TRBJ2‐7 | TRBC2 | CALRKYSNYQLIW | CASSAGTSGSGSTYEQYF |
| SCV‐008 | TRAV19 | TRAJ40 | TRAC | TRBV30 | TRBJ2‐1 | TRBC2 | CALSEAPSGTYKYIF | CAWSALFPSGRFGNEQFF |
| SCV‐009 | TRAV29/DV5 | TRAJ44 | TRAC | TRBV20‐1 | TRBJ2‐1 | TRBC2 | CAATFTGTASKLTF | CSARGGSAEASTDTQYF |
| SCV‐010 | TRAV29/DV5 | TRAJ50 | TRAC | TRBV10‐1 | TRBJ2‐2 | TRBC2 | CVFKRSRKVIF | CASSLAGSTGELFF |
| SCV‐011 | TRAV29/DV5 | TRAJ50 | TRAC | TRBV24‐1 | TRBJ2‐3 | TRBC2 | CVFKRSRKVIF | CATVHRGFTDTQYF |
| SCV‐012 | TRAV35 | TRAJ41 | TRAC | TRBV19 | TRBJ2‐2 | TRBC2 | CAGQLGSGYALNF | CASSPQTSGSSFAGELFF |
| SCV‐013 | TRAV6 | TRAJ15 | TRAC | TRBV14 | TRBJ1‐2 | TRBC1 | CALGWTALIF | CASSPRAGTPNYGYTF |
| SCV‐014 | TRAV8‐6 | TRAJ56 | TRAC | TRBV5‐6 | TRBJ1‐5 | TRBC1 | CAVSTNTGANSKLTF | CASSFRGLNQPQHF |
| SCV‐015 | TRAV12‐2*02 | TRAJ48*01 | TRAC | TRBV27*01 | TRBJ2‐1*01 | TRBC2 | CAVDHFGNEKLTF | CASSFLSGANLYNEQFF |
| SCV‐016 | TRAV27*01 | TRAJ40*01 | TRAC | TRBV4‐1*01 | TRBJ1‐5*01 | TRBC1 | CAGGWVLTTSGTYKYIF | CASAWSGVGNQPQHF |
Summary of the retrieved αβ TCRs, including the respectively used gene segments and the complementarity‐determining regions (CDRs) for each α‐ and β‐chain, respectively.
SARS‐CoV‐2‐reactive TCRs are responsive at a nanomolar antigen dose
To characterize and validate retrieved TCRs, we ectopically re‐expressed the TCRs and set up T‐cell activation assays. All TCRs were strongly expressed after electroporation according to anti‐CD3 antibody staining in comparison to mock control (Supporting information Fig. S4). TCR‐reconstituted αβ‐TCR‐deficient CD8+ Jurkat reporter cells and peptide pulsed parental‐ (T2) or HLA‐A*24:02‐expressing T2 cells (T2‐A24) were used in coculture assays as effector and target cells, respectively. An initial screening was conducted using an intermediate peptide dose (100 nM) of target peptide or irrelevant control peptide. Luciferase activity, expressed by an NFAT‐inducible reporter construct by the Jurkat reporter cells, was measured after 6 h of coculture. T‐cell activation was determined as fold change of the ontarget response over activation against an irrelevant peptide control (Fig. 2A). Based on previous experience using a similar type of assay [32], a threefold cut‐off was selected to define TCRs as being responsive. Based on this cut‐off, 12 of the 16 (75%) tested TCRs were responsive against one SARS‐CoV‐2‐specific epitope: two TCRs (SCV‐012 and SCV‐015) responded to A2_P03, seven TCRs responded to A24_P01 (SCV‐001, ‐002, ‐003, ‐004, ‐005, ‐007, ‐010), three TCRs were reactive to A24_P03 (SCV‐006, ‐014, ‐016), and four TCRs showed no‐ or a subthreshold response (Fig. 2A). None of the tested TCRs showed reactivity to A2_P09. To assess the potency of the selected TCRs more quantitatively, EC50 values of the 12 reactive SARS‐CoV‐2 TCRs were determined, using peptide titrations on T2 or T2‐A24 cells with A02_P03, A24_P01, or A24_P03. The well‐described affinity‐enhanced 1G4_a95LY TCR, which recognizes the cancer‐testis antigen Ny‐ESO‐1 with high affinity, was used as a positive control [33, 34]. All 12 selected SARS‐CoV‐2‐reactive TCRs showed a robust response, with 11 TCRs showing strong reactivity and EC50 values in the subnanomolar range, comparable to the 1G4 positive control (Fig. 2B and Table 4).
Figure 2.
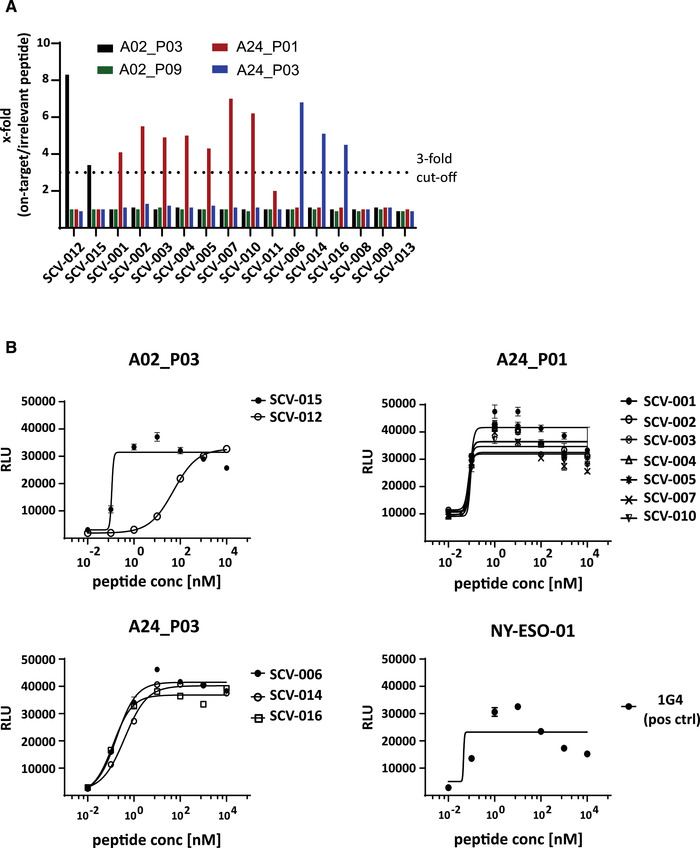
Signaling of SARS‐CoV‐2 TCRs and EC50 determination. The identified unique SARS‐CoV‐2 TCRs were electroporated into KO‐Jurkat cells expressing a NFAT‐luciferase reporter to determine reactivity and potency. TCR‐reconstituted KO‐Jurkat cells were used as effector cells and cocultured with T2 cells, pulsed with target peptide or irrelevant control peptide. (A) Bar graph summarizing the NFAT luciferase reporter response against the four target peptides, shown as fold change compared to the irrelevant peptide control. The dotted line indicates the threefold threshold. TCRs are grouped by their target reactivity. Experiment was conducted once with triplicates. (B) EC50 dose‐response curves, grouped by target epitope. Dots mark the relative light units (RLU) values measured for each individual peptide concentration (10 pM to 10 μM). Curves show the nonlinear curve fit (4PL sigmoidal) calculated with GraphPad Prism. Data shown are representative of two independently conducted experiments (n = 2) with technical triplicates.
Table 4.
Potency of SARS‐CoV‐2‐reactive TCRs in NFAT reporter assay
| TCR ID | Target | EC50 value (nM) |
|---|---|---|
| SCV‐012 | A02_P03 | 50.34 |
| SCV‐015 | A02_P03 | 0.12 |
| SCV‐001 | A24_P01 | 0.09 |
| SCV‐002 | A24_P01 | 0.08 |
| SCV‐003 | A24_P01 | 0.08 |
| SCV‐004 | A24_P01 | 0.09 |
| SCV‐005 | A24_P01 | 0.08 |
| SCV‐007 | A24_P01 | 0.07 |
| SCV‐010 | A24_P01 | 0.08 |
| SCV‐011 | A24_P01 | 0.09 |
| SCV‐006 | A24_P03 | 0.18 |
| SCV‐014 | A24_P03 | 0.39 |
| SCV‐016 | A24_P03 | 0.13 |
| 1G4 | NY‐ESO‐01 | 0.05 |
Summary of EC50 values for tested SARS‐CoV‐2‐reactive TCRs and 1G4 control TCR.
SARS‐CoV‐2‐reactive TCRs confer potent functionality including cytotoxicity
The selected SARS‐CoV‐2‐reactive TCRs were subsequently expressed in primary T cells to test for more complex T‐cell functionality using in vitro cytotoxicity assays. Primary CD8+ T cells were electroporated with the SARS‐CoV‐2‐responsive TCRs and used as effector cells. TCR surface expression was detectable for most TCRs by flow cytometry if matching anti‐Vbeta antibodies were available (Supporting information Fig. S5). T2 or T2‐A24 target cells were pulsed with 100 nM target peptide and plate at a fixed cells number per well. Electroporated effectors were used at titrated E/T ratios of 1:1 to 1:32. Cytotoxicity was determined using LDH release as a read‐out. We observed target peptide‐specific killing which correlated well with the titrated E/T ratio (Fig. 3A, dark blue to light blue). Alike the EC50 results, for most of the tested TCRs the level of cytotoxicity was similar to that mediated by the 1G4 TCR positive control (Fig. 3A). Another important feature of a productive T‐cell response is the secretion of cytokines, among which IFN‐γ plays a critical role. We, therefore, probed the collected supernatant for IFN‐γ with an ELISA (Fig. 3B). In accordance with cytotoxicity, we measured pronounced IFN‐γ secretion, although IFN‐γ production did not match the amplitude of cytotoxicity for all TCRs (Fig. 3A and 3B). In contrast, both killing and cytokine expression in the irrelevant peptide controls were very low for all TCRs (Supporting information Fig. S6).
Figure 3.
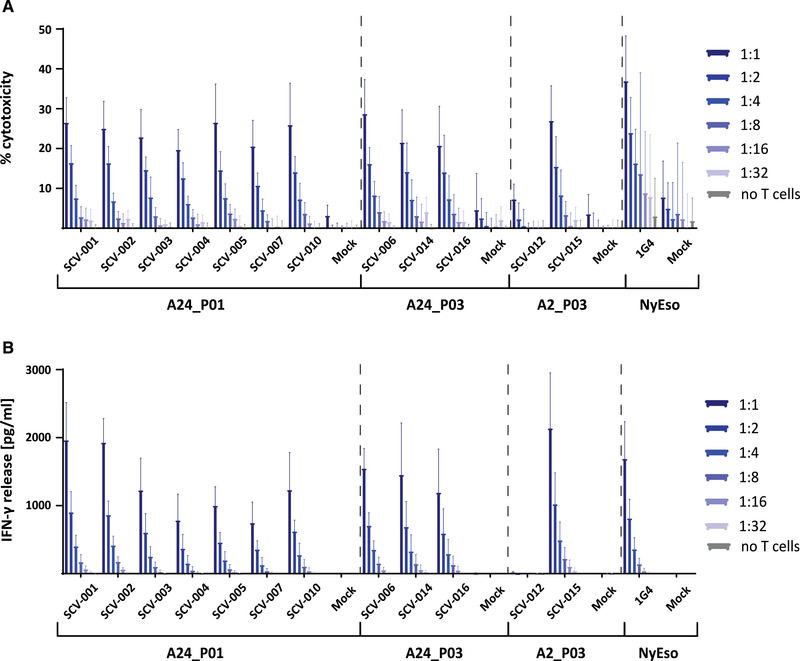
SARS‐CoV‐2‐reactive TCRs confer potent effector function to primary CD8+ T cells. SARS‐CoV‐2‐reactive TCRs passing the threefold cut‐off criterium were electroporated into primary CD8+ T cells. T2‐A2 and T2‐A24 cells were pulsed with 100 nM of the respective target peptide and plated at a fixed number of 30 × 103 cells/well (96‐well U‐bottom plate) as target cells. Electroporated CD8+ T cells were added in at titrated effector/target (E/T) ratios of 1:1, 1:2, 1:4, 1:8, 1:16, and 1:32 (30 × 103 ‐ ∼1 × 103). The experiment was conducted three times (n = 3, 5 donors total). Error bars represent standard deviation. (A) Bar graph summarizing the cytotoxicity shown as % cytotoxicity. Triton‐lysed cells were used as maximum lysis (100% killing) reference. Dark blue to light blue indicates the titrated E/T ratio. (B) Bar graph summarizing the IFN‐γ response as measured by ELISA (in pg/mL). Dark blue to light blue indicates the titrated E/T ratio.
SARS‐CoV‐2‐reactive T‐cell clones persist for months after infection
The characterized 12 unique SARS‐CoV‐2‐directed TCRs were isolated from a total of nine convalescent donor samples. The persistence of anti‐SARS‐CoV‐2 immunity is still a matter of ongoing discussion. We, therefore, wanted to assess whether we could reisolate one or more of these TCRs months after the initial TCR isolation. Two of the donors (SCD‐03 and SCD‐06) were available for follow‐up blood donation 5 months after the first donation. PBMCs were prestimulated in vitro for 12 days with the peptides A02_P03 and A24_P03, as previously described [17]. Samples were subsequently stained with A02_P03‐ and A24_P03‐tetramers (Fig. 4A). Tetramer‐positive cells (SCD‐03: A2_P03+ and A24_P03+, SCD‐06: A2_P03+) were FACS‐sorted and further processed for single‐cell TCR identification.
Figure 4.
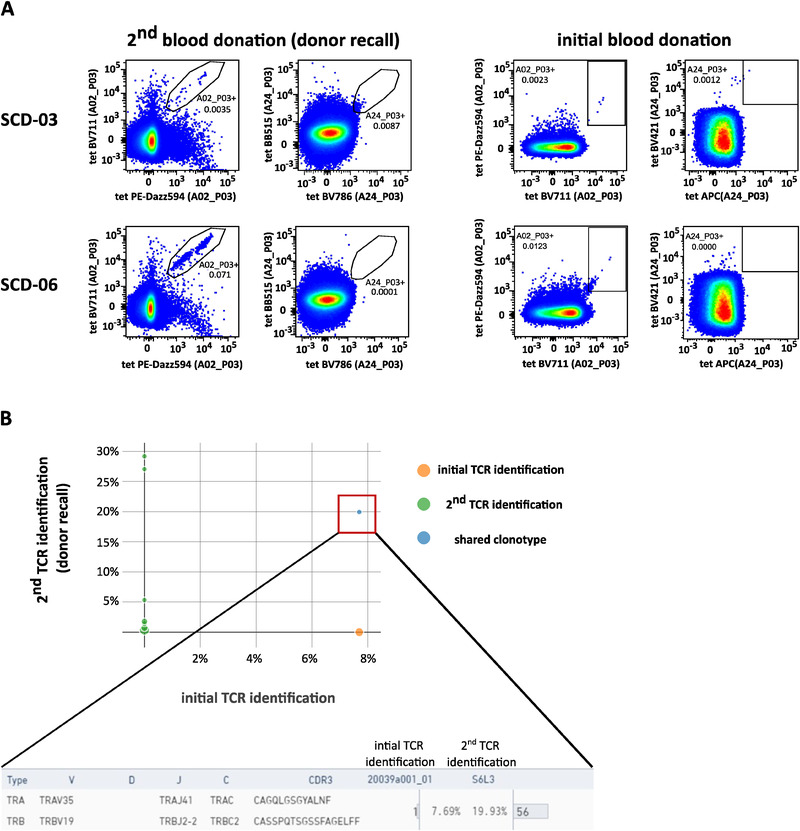
SARS‐CoV‐2‐reactive T‐cell clones may be persistent. PBMCs from two SARS‐CoV‐2 convalescent donors (SCD‐03 and SCD‐06) isolated 5 months after initial blood donation were stimulated in vitro for 12 days with the target peptides A02_P03 and A24_P03 and then stained with tetramers specific for these targets. The experiment was conducted once (n = 2). (A) FACS plots showing the 2D tetramer staining for SARS‐CoV‐2 epitopes A02_P03 and A24_P03. Upper row shows donor SCD‐03, lower row shows donor SCD‐06. For comparison, the plots on the right show the respective tetramer staining from the initial donation of the same donors. The depicted numbers in the gates indicate the respective frequency of parent. Axis are scaled logarithmically. The respective target antigen is indicated on the axis in brackets. Note that the plots shown on the right are also part of Supporting information Fig. S1. (B) The tetramer‐positive cells (shown above) were FACS‐sorted and subjected to single‐cell TCR sequencing using the 10× Chromium system. The diagram shows a V(D)J loop browser visualization of the initial TCR identification data set (x‐axis) compared to the second TCR identification (y‐axis). The blue dot indicates a shared clonotype, which was identified as SCV‐012.
We were able to isolate a total of 45 distinct TCRs from all these populations (data not shown). One of these TCRs was identical to one of the initially identified SARS‐CoV‐2‐reactive TCRs, namely SCV‐012 (Fig. 4B). This demonstrates that SARS‐CoV‐2‐reactive T‐cell clones may indeed persist for an extended period of time.
Discussion
In this study, we provide the first insights into the functional avidity of individual SARS‐CoV‐2‐reactive TCRs derived from CD8+ T cells reactive to SARS‐CoV‐2‐specific epitopes at the single‐cell level. There is a growing body of evidence suggesting that T‐cell responses are of high importance in the immune response in COVID‐19 infections and its efficient clearance [35]. Recently, it has been reported that impaired T‐cell activity, for example, measured by decreased IFN‐γ secretion, may be associated with more severe disease progression [36, 37, 38]. This argues for a need to identify factors contributing to constrained responses in severe as compared to mild courses of COVID‐19. While multiple variables may influence the observed phenomenon, we suggest that detailed characterization of the TCRs at the core of these responses will be a key to better understand and monitor SARS‐CoV‐2 infection and the associated immune reaction.
We demonstrate here that CD8+ T cells recognizing SARS‐CoV‐2‐specific epitopes are significantly enriched in PBMCs from convalescent donors compared to prepandemic controls but that the overall frequencies detected were relatively low. This is in line with a previous study showing that the frequencies of tetramer‐detectable SARS‐CoV‐2−specific CD8+ T cells were low compared to other viral infections [39]. While the typical contraction phase after infection may contribute to these low—compared to prepandemic controls nonetheless elevated—frequencies observed here, atypical low starting frequencies of target‐specific CD8+ T cells even during the acute phase as described by Habel et al. may also explain such low frequencies and may be a contributing factor to exacerbated disease progression in some COVID‐19 patients.
Despite the low abundance of below 0.001% (of total), we were able to isolate specific T cells and multiple TCRs directly ex vivo from a restricted amount of starting material, comprising as few as 20 × 106 cells per donor. This is important, as direct ex vivo analysis is well suited to reflect the in vivo T‐cell response caused by SARS‐CoV‐2 infection with limited risk to introduce bias. The fact that SARS‐CoV‐2‐reactive T‐cell clones are detectable even when an antibody response is not or no longer detectable (Supporting information Fig. S3) underscores the high sensitivity of tetramer‐based detection.
Most identified TCRs were able to elicit strong functional avidity in in vitro experiments, evident by both high NFAT signaling in the presence of target cells loaded with low amounts of peptide antigen as well as pronounced IFN‐γ secretion and cytotoxicity. These responses were comparable to the 1G4 reference TCR, which is in line with its reported binding affinity compared to other TCRs against viral antigens [40]. This suggests that, in general, the immune system is capable of mounting strong T‐cell responses against the tested SARS‐CoV‐2 epitopes.
We envision that applying TCR‐focused strategies in further studies covering more alleles and larger sample sizes—allowing the stratification into moderate and severe cases—will help to better understand the T‐cell response against SARS‐CoV‐2 and how such responses differ in both scenarios. A combination of functional analysis as demonstrated here with ongoing NGS efforts aimed at deciphering the TCR repertoire involved in COVID‐19 could also prove to be useful [25, 28]. Identifying patterns linking the potency of individual TCRs to shared CDR3 motifs may facilitate the use of deep sequencing as an elegant solution to provide meaningful analysis at the population level as well. Analyzing T‐cell responses may become one diagnostic tool for severe SARS‐CoV‐2 infections in the future. Furthermore, it is conceivable that the SARS‐CoV‐2‐reactive TCRs described here might be of interest for immunotherapies aiming at redirecting autologous T cells toward SARS‐CoV‐2. Since none of these TCRs are directed against the SARS‐CoV‐2 spike protein, this may become relevant in severe cases of SARS‐CoV‐2 where virus variants have evaded protection through existing vaccines by antigen escape.
When studying T‐cell responses against SARS‐CoV‐2, it seems important to distinguish between responses to SARS‐CoV‐2‐specific epitopes and T‐cell epitopes that share similarities with other coronaviruses. As we and others have previously shown, cross‐reactive T cells can also be detected in a large percentage of unexposed individuals [17, 41]. It is unclear, however, how strong such pre‐existing responses contribute to the immune response during infection with SARS‐CoV‐2. While some reports show significant overlap between immunodominant epitopes in healthy prepandemic and in infected individuals, others have found a lesser degree of reactivity for such epitopes in samples from similar groups [26, 42, 43]. In this report, we demonstrate that SARS‐CoV‐2‐specific epitopes relevant during infection can elicit strong T‐cell reactions. This observation is also in accordance with a recently published dataset of two TCRs against the SARS‐CoV‐2 – specific HLA‐A*11:01 epitope ORF3a78‐80 KTFPPTEPK, which demonstrated good reactivity expressed in CD8+ and CD4+ T cells [29]. Thus, our data emphasize the importance of the analyzed T‐cell epitopes and the TCRs recognizing them. Beyond the basic understanding of SARS‐CoV‐2 immunity, knowledge on individual SARS‐CoV‐2‐reactive TCRs may be useful during immunomonitoring of vaccine‐induced T‐cell responses.
Materials and methods
Flow cytometry antibodies
CD8 APC‐Cy7 (#301016, BioLegend), CD3 PE‐Cy7 (#300420, BioLegend), CD3 PerCPCy5.5 (#300430, BioLegend), CD4 PE (#317410, BioLegend), eBioscience™ Fixable Viability Dye eFluor™ 506 (#65‐0866‐18, Thermo), streptavidin Brilliant Blue 515 (#564453, BD), streptavidin Brilliant Violet 421 (#405225, BioLegend), streptavidin Brilliant Violet 711 (#563262, BD), streptavidin Brilliant Violet 786 (#563858, BD), streptavidin APC (#405243, BioLegend), streptavidin PE‐Dazz594 (#405248, BioLegend).
Peptides
Synthetic peptides were provided by EMC Microcollections GmbH and INTAVIS Peptide Services GmbH & Co. KG.
Cell samples
Blood samples from convalescent volunteers after SARS‐CoV‐2 infection were collected at the University Hospital Tübingen from April to May 2020 (SCD, n = 11). The collection of separate unexposed individuals (PRE, n = 6) includes samples of healthy blood donors (blood donations acquired through DRK Institut für Transfusionsmedizin und Immunologie, Mannheim and Department of Transfusion Medicine, University Hospital Tübingen) that were never exposed to SARS‐CoV‐2, as the PBMCs of these donors were isolated and asserted (Immatics Biotechnologies GmbH, Tübingen and Department of Immunology, Tübingen) before the SARS‐CoV‐2 pandemic (April 2016 to August 2019). Informed consent was obtained in accordance with the Declaration of Helsinki protocol. The study was approved by and performed according to the guidelines of the local ethics committees (179/2020/BO2, MC 288/2015). SARS‐CoV‐2 infection was confirmed by PCR test after nasopharyngeal swab. SARS‐CoV‐2 convalescent donor recruitment was performed by online and paper‐based calls. Sample collection for SARS‐CoV‐2 convalescents was performed approximately 3–7 weeks after the end of symptoms and/or negative virus smear. PBMCs were isolated by density gradient centrifugation and stored at −80°C until further use. HLA typing was carried out by Immatics Biotechnologies GmbH and the Department of Hematology and Oncology at the University Hospital Tübingen. SARS‐CoV‐2 convalescent donor characteristics are provided in Supporting information Table S1.
In vitro 12‐day prestimulation and IFN‐γ enzyme‐linked immunospot (ELISPOT) assay
PBMCs were pulsed with peptides (1 μg/mL per peptide) and cultured for 12 days adding 20 U/mL interleukin‐2 (Novartis) on days 3, 5, and 7. Peptide‐stimulated PBMCs were analyzed after 12 days prestimulation by enzyme‐linked immunospot (ELISPOT) assay in duplicates. A total of 2–8 × 105 cells per well were incubated with 1 μg/mL of single peptides in 96‐well plates coated with anti‐IFN‐γ (clone 1‐D1K, 2 μg/mL, MabTech). PHA (Sigma‐Aldrich) served as a positive control, irrelevant HLA‐matched control peptides as negative control (HLA‐A*02 YLLPAIVHI DDX5_HUMAN148‐156, HLA‐A*24 AYVHMVTHF BI1_HUMAN45‐53). After 24 h incubation, spots were revealed with anti‐IFN‐γ biotinylated detection antibody (clone 7‐B6‐1, 0.3 μg/mL, MabTech), ExtrAvidin‐alkaline phosphatase (1:1000 dilution, Sigma‐Aldrich) and BCIP/NBT (5‐bromo‐4‐chloro‐3‐indolyl‐phosphate/nitro‐blue tetrazolium chloride, Sigma‐Aldrich). Spots were counted using an ImmunoSpot S5 analyser (CTL) and T‐cell responses were considered positive when the mean spot count was at least threefold higher than the mean spot count of the negative control.
Generation of peptide‐exchanged HLA complexes using UV‐mediated ligand exchange
Recombinant HLA‐A*02:01 or HLA‐A*24:02 wild‐type heavy chain with C‐terminal BirA signal sequences and human β2m light chain were produced in E. coli as inclusion bodies and purified as previously described [44].
HLA complex refolding reactions with UV light‐cleavable peptides and peptide exchange reactions with UV light–cleavable peptides were performed as previously described with minor modifications [30]. In short, desired peptides were mixed with biotinylated UV light‐sensitive pHLAs at 100:1 molar ratio and subjected to at least 30 min of 366‐nm UV light (CAMAG). The success of peptide exchange reactions was determined before tetramerization using a β2m‐targeting ELISA.
Fluorescence activated cell sorting (FACS) of SARS‐CoV‐2 tetramer‐positive T cells
Cryopreserved PBMC samples were thawed and rested overnight. The next day, the cells were harvested, washed, and stained with tetramers to the SARS‐CoV‐2 targets A2_P03, A2_P09, A24_P01, and A24_P03. 2D tetramer staining was used to unambiguously identify T cells specific for one of these four targets. Following incubation for 30 min at 4°C, unbound tetramer was washed off and cells were resuspended in the cell surface antibody mix. Dead cells were excluded by the inclusion of a fixable viability dye. FSC‐A versus FSC‐H gating was used to exclude duplets. Viable CD3+CD4–CD8+ tetramer 2D+ cells were FACS‐sorted on a BD FACSAria™ Fusion Cell Sorter using the 70 μm nozzle. Cells were sorted into Protein LoBind® Tubes (Eppendorf).
SARS‐CoV‐2 IgG and IgA ELISA (EUROIMMUN)
The 96‐well SARS‐CoV‐2 IgG and IgA ELISA assay (EUROIMMUN, 2606A_A_DE_C03, as constituted on 22 April 2020) was performed on an automated BEP 2000 Advance® system (Siemens Healthcare Diagnostics GmbH) according to the manufacturer's instructions. The ELISA assay detects anti‐SARS‐CoV‐2 IgG and IgA directed against the S1 domain of the viral spike protein (including the immunologically relevant receptor binding domain) and relies on an assay‐specific calibrator to report a ratio of specimen absorbance to calibrator absorbance. The final interpretation of positivity is determined by ratio above a threshold value given by the manufacturer: positive (ratio ≥ 1.1), borderline (ratio 0.8‐1.0), or negative (ratio < 0.8). Quality control was performed following the manufacturer's instructions on each day of testing.
Elecsys® anti‐SARS‐CoV‐2 immunoassay (Roche Diagnostics GmbH)
The Elecsys® anti‐SARS‐CoV‐2 assay is an ECLIA (electrogenerated chemiluminescence immunoassay) designed by Roche Diagnostics GmbH and was used according to the manufacturer's instructions (V1.0, as constituted in May 2020). It is intended for the detection of high‐affinity antibodies (including IgG) directed against the nucleocapsid protein of SARS‐CoV‐2 in human serum. Readout was performed on the Cobas e411 analyser. Negative results were defined by a cut‐off index of <1.0. Quality control was performed following the manufacturer's instructions on each day of testing.
Single‐cell sequencing
FACS‐sorted, SARS‐CoV‐2 tetramer‐positive T cells were loaded onto the Chromium instrument (10× Genomics). TCRαβ‐ Illumina sequencing libraries were prepared using the Chromium Next GEM Single Cell 5ʹ Library and Gel Bead Kit version 1.1 (10× Genomics #1000165) and the Chromium Single Cell V(D)J Enrichment Kit, Human T Cell (10× Genomics #1000005) following the manufacturer's recommendations. The final TCRαβ‐Illumina sequencing libraries were sequenced on a MiSeq Micro (300 cycles, 4 Mio clusters) flow cell. After demultiplexing of the raw data, data were fed into the CellRanger software version 4.0 (10× Genomics) and the V(D)J Loupe browser version 3.0.0 (10× Genomics) was used for visualization and TCR sequence retrieval.
In vitro transcription and electroporation
For expression of desired T‐cell receptors in CD8+ T cells or Jurkat‐NFAT effector cells, α‐ and β‐chain encoding mRNA was produced by amplification of full length α‐ and β‐TCR sequences from DNA templates and an in vitro transcription reaction. Methylated pUC57 vector containing sequences for both α‐ and β‐chain of a TCR were procured from GenScript. For the constructs, variable domains of COVID‐specific TCRs were combined with murine TRAC and TRBC regions to prevent mispairing with endogenous TCRs [45, 46]. DNA templates were generated by PCR amplification (PrimeSTAR HS DNA Polyermase, Takara) using chain‐specific primers for either α or β followed by DNA precipitation. A T7 promoter and Kozak sequence were introduced into the amplified DNA fragment through the forward primer. In vitro transcription was performed using the mMESSAGE mMACHINE T7 KITs (Invitrogen, #AM1344) according to the manufacturer's instructions and frozen at −80°C until further use.
To express the respective TCRs in TCR‐KO Jurkats or primary CD8+ T cells, α‐ and β‐TCR mRNA was pooled in equal amounts and electroporated after mixing with the respective cell sample. The BTX ECM 830 Square Wave Electroporation System was used in the low voltage mode. Following electroporation, cells were cultured overnight in T‐cell medium containing 1 μg/mL DNase I (Roche # 4716728001).
αβTCR‐deficient Jurkat cells, expressing an NFAT‐luciferase reporter
The TCR‐KO Jurkat cell line was purchased from Promega. This cell line is based on the standard Jurkat cell line and has been manipulated to express a NFAT‐inducible luciferase reporter construct. The TCR‐KO line also constitutively expresses CD8. The disruption of the endogenous αβ TCR facilitates the expression of exogenously introduced TCRs of interest, here SARS‐CoV‐2‐ specific TCRs.
EC50 determination in TCR‐KO Jurkat cells
αβTCR‐deficient CD8+ Jurkat cells were electroporated with mRNA coding for SARS‐CoV‐2 TCRs. The next day, successful electroporation resulting in TCR surface expression was confirmed by flow cytometry. Standard T2 cells (T2‐A2) or T2 cells transduced to express HLA‐A*24:02 (T2‐A24) were pulsed with titrated amount of the respective target peptides (10 pM–10 μM) for 45 min at 37°C. Following incubation, T2 cells were washed with medium and adjusted to a density of 1 × 106/mL. Electroporated Jurkat cells were harvested, washed with medium, and adjusted to a density of 1 × 106/mL. Cells were plated to 96 U‐bottom wells (50 × 103 T2 cells + 50 × 103 CD8+ cells per well). After 6 h of coculture, cells were removed from the incubator and the Bio‐Glo‐NL reagent (Promega #J3082) was applied according to the manufacturer's recommendations. Luminescence was measured as relative light units (RLU) using a 0.5‐s integration time on a Synergy 2 microplate reader. EC50 values were determined by curve fitting using GraphPad Prism (v8.4.0) and a nonlinear regression model with a sigmoidal 4PL equation (Y = Bottom + (Top‐Bottom)/(1 + 10⋀((LogEC50‐X)*HillSlope).
In vitro cytotoxicity assay (CD8+)
CD8+ T cells were isolated from healthy donor PBMCs using MACS Miltenyi Biotech, # 130‐096‐495) and electroporated with mRNA coding for SARS‐CoV‐2‐reactive TCRs. The next day, successful electroporation resulting in TCR surface expression was confirmed by flow cytometry. Standard T2 cells (T2‐A2) or T2 cells transduced to express HLA‐A*24:02 (T2‐A24) were pulsed with the respective target peptides (100 nM) for 45 min at 37°C. Following incubation, T2 cells were washed with medium and adjusted to a density of 0.3 × 106/mL. Electroporated CD8+ T cells were harvested, washed with medium, and adjusted to a density of 0.3 × 106/mL (E/T ratio 1:1) and then serially diluted to 0.15 × 106/mL, 0.075 × 106/mL, etc. Finally, 100 μL of peptide‐pulsed T2 cells plus 100 μL of the respective CD8+ T‐cell dilution was plated onto 96‐well U‐bottom plates, resulting in a fixed number of T2 cells (30 × 103) and a titrated amount of electroporated CD8+ T cells (30 × 103 to ∼1 × 103), covering the E/T ratios of 1:1, 1:2, 1:4, 1:8, 1:16, and 1:32. Cells were cocultured for 18 h before flow cytometry analysis. Before the readout, plates were centrifuged, and supernatant transferred to a separate plate and frozen at −80°C for further analysis.
LDH release assay
LDH release during coculture of CD8+ T cells and peptide‐loaded target cells were determined using harvested supernatant and an LDH‐GloTM Cytotoxicity assay (Promega) according to manufacturer's instructions with minor modifications. In short, supernatant was diluted 1:50 in LDH Storage Buffer (200 mM Tris‐HCl [pH 7.3], 10% Glycerol, 1% BSA) and mixed with an equal volume of LDH Detection Mix freshly produced from LDH Detection Enzyme Mix and Reductase substrate. Samples were incubated in the dark at room temperature for 30 min. Afterward, luminescence was measured as RLU using a 0.5‐s integration time on a Synergy 2 microplate reader.
IFN‐γ ELISA
IFN‐γ ELISA readout to determine concentration in harvested supernatant was performed using a BioLegend ELISA MAX Deluxe IFN‐γ kit according to manufacturer's instructions with minor modifications. Washing steps were performed using an ELx405 plate washer. The IFN‐γ standard delivered with the kit was replaced with recombinant human IFN‐γ protein procured separately by Abcam. The standard concentration range was extended from 500 to 7.8 pg/mL in a 1:2 dilution series. Supernatant samples were diluted 1:12 in assay diluent before incubation and the HRP reaction stopped by the addition of 2N H2SO4. Absorbance at 450 and 570 nm was measured using a Synergy 2 microplate reader. Absorbance at 570 nm was subtracted from 450 nm and a standard curve was calculated for each plate individually based on IFN‐γ standard duplicates with a 4PL curve fit using the Gen5 software (BioTek).
Data and statistical analysis
Analysis of flow cytometry data was done in FlowJo version 10.7. The DownSample plugin version 2.2 was used on the CD8+ population to generate equally sized parent populations for 2D tetramer plots used in Fig. 1. Statistical analysis was done using GraphPad Prism Version 8.4. The respective statistical analysis applied is stated in the corresponding figure legend. Fold change was calculated as the ratio of response to target peptide/response to irrelevant peptide. Statistical significance was indicated by asterisks, the respective significance level is specified in the corresponding figure legend. Inkscape 1.0.1 was used for visualization.
Funding
This work was supported by the Bundesministerium für Bildung und Forschung (BMBF, FKZ:01KI20130; J.W.), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, Grant WA 4608/1‐2; J.W.), the Deutsche Forschungsgemeinschaft under Germany's Excellence Strategy (Grant EXC2180 390900677; H.‐G.R., J.W.), the German Cancer Consortium (DKTK; H.‐G.R.), the Wilhelm Sander Stiftung (Grant 2016.177.2; J.W.), the José Carreras Leukämie‐Stiftung (Grant DJCLS 05 R/2017; J.W.). NGS sequencing methods were performed with the support of the DFG‐funded NGS Competence Center Tübingen (INST 37/1049‐1). Funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)–Project‐ID 286/2020B01–428994620.
Competing Interests
H.S. is CEO of Immatics N.V. . F.B., A.M. C.W., and D.M. are employees of Immatics Biotechnologies GmBH. H.‐G.R. is shareholder of Immatics N.V. and CureVac AG. A.N., T.B., H.‐G.R., and J.S.W. are listed as inventors on a patent related to peptides described in this manuscript secured under the numbers 20 169 047.6 (SARS‐CoV‐2 CD8+ und CD4+ T cell epitopes).
Author contributions
F.B., A.M., A.N., and J.S.W. conceived the study, designed experiments, and analysed data. F.B. and A.M. designed and performed UV exchange, FACS‐sorting and cellular experiments. A.N., T.B., J.S.H., S.H., and A.P. performed ELISPOT experiments and scoring of antibody responses. N.C. and S.A.K.F. performed 10× single‐cell sequencing experiments. C.W., D.M., J.S.W., H.S., and H.‐G.R. supervised the study. F.B. and A.M. wrote the manuscript, all authors reviewed the manuscript.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202149290
Abbreviations
- COVID‐19
coronavirus disease 2019
- ELISPOT
enzyme‐linked immunospot
- MERS‐CoV
Middle East respiratory syndrome coronavirus
- RLU
relative light units
- SARS‐CoV‐2
Severe acute respiratory syndrome coronavirus 2
Supporting information
Supporting information
Acknowledgments
We are indebted to Anna von Landenberg (A.v.L.) and Franziska Wetzel (F.W.) for expert technical support. A.v.L. and F.W. contributed to in vitro transcription of SARS‐CoV‐2–TCRs, Jurkat reporter assays, in vitro cytotoxicity assays and IFN‐γ–ELISA. A.v.L. performed 10× sample preparations for donor recall experiments. We thank Anne Kalweit (A.K.) for performing 5’RACE experiments and Martina Maas (M.M.) for performing ELISA for quality control of the UV exchange. We thank Janus Borner for bioinformatic analysis of the single‐cell sequencing data.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Mo, P. , Xing, Y. , Xiao, Y. , Deng, L. , Zhao, Q. , Wang, H. , Xiong, Y. , et al., Clinical characteristics of refractory COVID‐19 pneumonia in Wuhan, China. Clin. Infect. Dis. 2020. ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khan, S. , Siddique, R. , Shereen, M. A. , Ali, A. , Liu, J. , Bai, Q. , Bashir, N. , et al., Emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus 2: Biology and therapeutic options [Internet]. J. Clin. Microbiol. 2020. 58: e00187–e00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu, F. , Zhao, S. , Yu, B. , Chen, Y. M. , Wang, W. , Song, Z. G. , Hu, Y. , et al., A new coronavirus associated with human respiratory disease in China. Nature. 2020. 579: 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang, C. , Horby, P. W. , Hayden, F. G. and Gao, G. F. , A novel coronavirus outbreak of global health concern. Lancet North Am. Ed. 2020. 395: 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ni, L. , Ye, F. , Cheng, M. L. , Feng, Y. , Deng, Y. Q. , Zhao, H. , Wei, P. , et al., Detection of SARS‐CoV‐2‐specific humoral and cellular immunity in COVID‐19 convalescent individuals. Immunity. 2020. 52: 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang, A. T. , Garcia‐Carreras, B. , Hitchings, M. D. T. , Yang, B. , Katzelnick, L. C. , Rattigan, S. M. , Borgert, B. A. , et al., A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020. 11: 4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gallais, F. , Velay, A. , Nazon, C. , Wendling, M. ‐. J. , Partisani, M. , Sibilia, J. , Candon, S. , et al., Intrafamilial exposure to SARS‐CoV‐2 associated with cellular immune response without seroconversion, France. Emerg. Infect. Dis. 2021. 27: 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bilich, T. , Nelde, A. , Heitmann, J. S. , Maringer, Y. , Roerden, M. , Bauer, J. , Rieth, J. , et al., T cell and antibody kinetics delineate SARS‐CoV‐2 peptides mediating long‐term immune responses in COVID‐19 convalescent individuals. Sci. Transl. Med. 2021. 13: eabf7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao, J. , Zhao, J. and Perlman, S. , T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus‐infected mice. J. Virol. 2010. 84: 9318–9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li, C. K. , Wu, H. , Yan, H. , Ma, S. , Wang, L. , Zhang, M. , Tang, X. , et al., T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008. 181: 5490–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao, J. , Alshukairi, A. N. , Baharoon, S. A. , Ahmed, W. A. , Bokhari, A. A. , Nehdi, A. M. , Layqah, L. A. , et al., Recovery from the Middle East respiratory syndrome is associated with antibody and T cell responses. Sci. Immunol. 2017. 2: eaan5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Channappanavar, R. , Fett, C. , Zhao, J. , Meyerholz, D. K. and Perlman, S. , Virus‐specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014. 88: 11034–11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Altmann, D. M. and Boyton, R. J. , SARS‐CoV‐2 T cell immunity: Specificity, function, durability, and role in protection. Sci. Immunol. 2020. 5: eabd6160. [DOI] [PubMed] [Google Scholar]
- 14. Zuo, J. , Dowell, A. , Pearce, H. , Verma, K. , Hm, L. , Begum, J. , Aiano, F. , et al., Robust SARS‐CoV‐2‐specific T‐cell immunity is maintained at 6 months following primary infection. bioRxiv. 2020. 10.1101/2020.11.01.362319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuri‐Cervantes, L. , Pampena, M. B. , Meng, W. , Rosenfeld, A. M. , Ittner, C. A. G. , Weisman, A. R. , Agyekum, R. S. , et al., Comprehensive mapping of immune perturbations associated with severe COVID‐19. Sci. Immunol. 2020. 5: eabd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mathew, D. , Giles, J. R. , Baxter, A. E. , Oldridge, D. A. , Greenplate, A. R. , Wu, J. E. , Alanio, C. , et al., Deep immune profiling of COVID‐19 patients reveals distinct immunotypes with therapeutic implications. Science (80‐). 2020. 369: eabc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nelde, A. , Bilich, T. , Heitmann, J. S. , Maringer, Y. , Salih, H. R. , Roerden, M. , Lübke, M. , et al., SARS‐CoV‐2‐derived peptides define heterologous and COVID‐19‐induced T cell recognition. Nat. Immunol. 2020. 22: 74–85. [DOI] [PubMed] [Google Scholar]
- 18. Oliveira, S. C. , de Magalhães, M. T. Q. and Homan, E. J. , Immunoinformatic analysis of SARS‐CoV‐2 nucleocapsid protein and identification of COVID‐19 vaccine targets. Front. Immunol. 2020. 11: 587615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prachar, M. , Justesen, S. , Steen‐Jensen, D. B. , Thorgrimsen, S. , Jurgons, E. , Winther, O. and Bagger, F. O. , Identification and validation of 174 COVID‐19 vaccine candidate epitopes reveals low performance of common epitope prediction tools. Sci. Rep. 2020. 10: 20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sekine, T. , Perez‐Potti, A. , Rivera‐Ballesteros, O. , Strålin, K. , Gorin, J. ‐. B. , Olsson, A. , Llewellyn‐Lacey, S. , et al., Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID‐19. Cell. 2020. 183: 158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chour, W. , Xu, A. , Ng, A. , Choi, J. , Xie, J. , Yuan, D. , Diana, C. , et al., Shared antigen‐specific CD8+ T cell responses against the SARS‐COV‐2 spike protein in HLA A*02:01 COVID‐19 participants. medRxiv. 2020. 10.1101/2020.05.04.20085779 [DOI] [Google Scholar]
- 22. Kared, H. , Redd, A. D. , Bloch, E. M. , Bonny, T. S. , Sumatoh, H. , Kairi, F. , Carbajo, D. , et al., CD8+ T cell responses in convalescent COVID‐19 individuals target epitopes from the entire SARS‐CoV‐2 proteome and show kinetics of early differentiation. bioRxiv. 2020. 10.1101/2020.10.08.330688 [DOI] [Google Scholar]
- 23. Saini, S. K. , Hersby, D. S. , Tamhane, T. , Povlsen, H. R. , Hernandez, S. P. A. , Nielsen, M. , Gao, A. O. , et al., SARS‐CoV‐2 genome‐wide mapping of CD8 T cell recognition reveals strong immunodominance and substantial CD8 T cell activation in COVID‐19 patients. bioRxiv. 2020. 10.1101/2020.10.19.344911 [DOI] [Google Scholar]
- 24. Schulien, I. , Kemming, J. , Oberhardt, V. , Wild, K. , Seidel, L. M. , Killmer, S. , Sagar, et al., Characterization of pre‐existing and induced SARS‐CoV‐2‐specific CD8+ T cells. Nat. Med. 2020. 27: 78–85. [DOI] [PubMed] [Google Scholar]
- 25. Snyder, T. M. , Gittelman, R. M. , Klinger, M. , May, D. H. , Osborne, E. J. , Taniguchi, R. , Zahid, H. J. , et al., Magnitude and dynamics of the T‐cell response to SARS‐CoV‐2 infection at both individual and population levels. medRxiv. 2020. 10.1101/2020.07.31.20165647 [DOI] [Google Scholar]
- 26. Ferretti, A. P. , Kula, T. , Wang, Y. , Nguyen, D. M. V. , Weinheimer, A. , Dunlap, G. S. , Xu, Q. , et al., Unbiased screens show CD8+ T cells of COVID‐19 patients recognize shared epitopes in SARS‐CoV‐2 that largely reside outside the spike protein. Immunity. 2020. 53: 1095–1107.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Minervina, A. A. , Komech, E. A. , Titov, A. , Koraichi, M. B. , Rosati, E. , Mamedov, I. Z. , Franke, A. , et al., Longitudinal high‐throughput TCR repertoire profiling reveals the dynamics of T cell memory formation after mild COVID‐19 infection. arXiv. 2020. 10.1101/2020.05.18.100545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shomuradova, A. S. , Vagida, M. S. , Sheetikov, S. A. , Zornikova, K V. , Kiryukhin, D. , Titov, A. , Peshkova, I. O. , et al., SARS‐CoV‐2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020. 53: 1245–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu, C. , Shen, M. , Han, X. , Chen, Q. , Li, L. , Chen, S. , Zhang, J. , et al., Identification of cross‐reactive CD8+ T cell receptors with high functional avidity to a SARS‐CoV‐2 immunodominant epitope and its natural mutant variants. bioRxiv. 2020. 10.1101/2020.11.02.364729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodenko, B. , Toebes, M. , Hadrup, S. R. , van Esch, W. J. E. , Molenaar, A. M. , Schumacher, T. N. M. , Ovaa, H. , et al., Generation of peptide‐MHC class I complexes through UV‐mediated ligand exchange. Nat. Protoc. 2006. 1: 1120–1132. [DOI] [PubMed] [Google Scholar]
- 31. Hadrup, S. R. , Toebes, M. , Rodenko, B. , Bakker, A. H. , Egan, D. A. , Ovaa, H. , Schumacher, T. N. M. , et al., High‐throughput T‐cell epitope discovery through MHC peptide exchange. Methods Mol. Biol. 2009. 524: 383–405. [DOI] [PubMed] [Google Scholar]
- 32. Moritz, A. A. , Anjanappa, R. , Wagner, C. , Bunk, S. , Hofmann, M. , Pszolla, G. , Saikia, A. , et al., High‐throughput peptide‐MHC complex generation and kinetic screenings of TCRs with peptide‐receptive HLA‐A*02:01 molecules. Sci. Immunol. 2019. 4: eaav0860. [DOI] [PubMed] [Google Scholar]
- 33. Chen, J.‐L. , Dunbar, P. R. , Gileadi, U. , Jäger, E. , Gnjatic, S. , Nagata, Y. , Stockert, E. , et al., Identification of NY‐ESO‐1 peptide analogues capable of improved stimulation of tumor‐reactive CTL. J. Immunol. 2000. 165: 948–955. [DOI] [PubMed] [Google Scholar]
- 34. Chen, J.‐L. , Stewart‐Jones, G. , Bossi, G. , Lissin, N. M. , Wooldridge, L. , Choi, E. M. L. , Held, G. , et al., Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J. Exp. Med. 2005. 201: 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sette, A. and Crotty, S. , Adaptive immunity to SARS‐CoV‐2 and COVID‐19. Cell. 2021. 184: 861–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tan, A. T. , Linster, M. , Tan, C. W. , Bert, L. , Chia, N. , Kunasegaran, W. N. , Zhuang, Y. , et al., Early induction of functional SARS‐CoV‐2 specific T cells associates with rapid viral clearance and mild disease in COVID‐19 patients. Cell. Rep. 2021. 34: 108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Le Bert, N. , Clapham, H. E. , Tan, A. T. , Chia, W. N. , Tham, C. Y. L. , Lim, J. M. , Kunasegaran, K. , et al., Highly functional virus‐specific cellular immune response in asymptomatic SARS‐CoV‐2 infection. bioRxiv. 2020. 10.1101/2020.11.25.399139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bacher, P. , Rosati, E. , Esser, D. , Martini, G. R. , Saggau, C. , Schiminsky, E. , Dargvainiene, J. , et al., Low avidity CD4+ T cell responses to SARS‐CoV‐2 in unexposed individuals and humans with severe COVID‐19. Immunity. 2020. 53: 1258–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Habel, J. R. , Nguyen, T. H. O. , van de Sandt, C. E. , Juno, J. A. , Chaurasia, P. , Wragg, K. , Koutsakos, M. , et al., Suboptimal SARS‐CoV‐2−specific CD8+ T cell response associated with the prominent HLA‐A*02:01 phenotype. Proc. Natl. Acad. Sci. USA. 2020. 117: 24384–24391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aleksic, M. , Liddy, N. , Molloy, P. E. , Pumphrey, N. , Vuidepot, A. , Chang, K. M. , Jakobsen, B. K. , et al., Different affinity windows for virus and cancer‐specific T‐cell receptors: Implications for therapeutic strategies. Eur. J. Immunol. 2012. 42: 3174–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grifoni, A. , Weiskopf, D. , Ramirez, S. I. , Mateus, J. , Dan, J. M. , Moderbacher, C. R. , Rawlings, S. A. , et al., Targets of T cell responses to SARS‐CoV‐2 coronavirus in humans with COVID‐19 disease and unexposed individuals. Cell. 2020. 181: 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lehmann, A. , Kirchenbaum, G. A. , Zhang, T. , Reche, P. A. and Lehmann, P. V. , Deconvoluting the T cell response to SARS‐CoV‐2: specificity versus chance‐ and cognate cross‐reactivity. bioRxiv. 2020. 10.1101/2020.11.29.402677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saini, S. K. , Stampe Hersby, D. , Tamhane, T. , Rus Povlsen, H. , Hernandez, S. P. A. , Nielsen, M. , Gang, A. O. , et al., SARS‐CoV‐2 genome‐wide mapping of CD8 T cell recognition reveals. bioRxiv. 2020. 10.1101/2020.10.19.344911 [DOI] [Google Scholar]
- 44. Garboczi, D. N. , Hung, D. T. and Wiley, D. C. , HLA‐A2‐peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. USA. 1992. 89: 3429–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haga‐Friedman, A. , Horovitz‐Fried, M. and Cohen, C. J. , Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J. Immunol. 2012. 188: 5538–5546. [DOI] [PubMed] [Google Scholar]
- 46. Jin, B. Y. , Campbell, T. E. , Draper, L. M. , Stevanović, S. , Weissbrich, B. , Yu, Z. , Restifo, N. P. , et al., Engineered T cells targeting E7 mediate regression of human papillomavirus cancers in a murine model. JCI insight. 2018. 3. e99488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


