Continuing discoveries of new and surprising mechanisms of gene regulation suggest that our understanding of this complex and ubiquitous biological process remains incomplete. Emerging examples illustrate that many and perhaps all genes are regulated at multiple steps including transcription, posttranscriptional processing, nuclear export and localization, stability, and translation of mature mRNA molecules. Translation itself is regulated by a diverse collection of mechanisms that act not only at the initiation step but also during elongation and termination and even after termination.
Among the various cis elements in mRNAs (43) that participate in regulating translation are AUG codons within transcript leaders (upstream AUGs [uAUGs]) and, in some cases, associated upstream open reading frames (uORFs). Based on a 1987 survey, less than 10% of eukaryotic mRNAs contain AUG codons within their transcript leader regions (often erroneously referred to as 5′ untranslated regions). However, uAUGs are conspicuously common in certain classes of genes, including two-thirds of oncogenes and many other genes involved in the control of cellular growth and differentiation (29, 31, 42). Despite the wealth of sequence data being generated by large-scale sequencing projects, extracting an up-to-date, comprehensive, and accurate estimate of the number of genes with uORFs is a formidable task. Only a minority of database entries are based on careful mRNA mapping data with annotations that identify the precise start of the transcript leader. Moreover, the use of alternative transcriptional start sites, alternative RNA processing, and alternative initiation codons complicates the determination of what exactly constitutes the transcript leader. Nonetheless, it is clear that uAUGs are not uncommon in genes with critical cellular roles, and identifying when and how they function is necessary if we are to achieve a comprehensive understanding of the interesting genes that contain these elements and of eukaryotic gene regulation in general.
Some of the general principles by which uORFs participate in translational control are beginning to be understood. In this article, we first review these principles, which include the process of recognition of uORFs, regulation of reinitiation at downstream cistrons after translation of uORFs, and regulatory effects of peptides encoded by uORFs. We then illustrate how these principles are applied by reviewing several specific examples where the roles of uORFs in translational control have been well characterized.
AFTER TRANSLATING A UORF, WHAT ARE THE OPTIONS FOR A RIBOSOME?
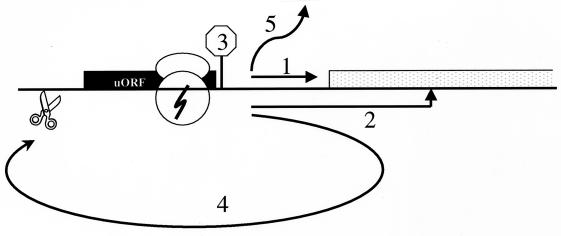
If a uORF is recognized and translated by a scanning ribosome (factors influencing uORF recognition are discussed below), multiple alternative fates appear to be available to the ribosome (Fig. 1). One option is for the ribosome to remain associated with the mRNA, continue scanning, and reinitiate further downstream, at either a proximal or distal AUG codon (Fig. 1, options 1 and 2). Ribosomes can, and often do, translate a uORF and then reinitiate downstream with high efficiency, such that the presence of the uORF appears not to affect gene expression (7, 41). The potential of a ribosome to reinitiate further downstream, as well as the site at which it reinitiates, can vary depending on both trans-acting factors and the structure of the mRNA. We have some understanding of how these elements cooperate to determine where and when a ribosome reinitiates (discussed below), but our knowledge is far from complete.
FIG. 1.
Alternative fates available to a ribosome after translating a uORF. See the text for a detailed discussion.
With some mRNAs, another option for the ribosome is to stall during either the elongation or termination phase of uORF translation, creating a blockade to additional ribosome scanning (Fig. 1, option 3). In the known cases, ribosome stalling is mediated by the structure of the peptide encoded by the uORF. The peptide sequences that have been characterized thus far do not share any recognizable consensus sequences; moreover, they exhibit significant mechanistic differences, suggesting that the different peptides interact with distinct sites in the translation machinery. Nonetheless, they appear to have the common outcome of perturbing the normal sequence of events needed for efficient translation termination, and they thereby create a ribosomal roadblock at the uORF that can impede translation of the downstream cistron.
In addition to influencing the action of ribosomes during and after termination, uORFs may affect gene expression by altering mRNA stability (Fig. 1, option 4). The similarity in the cistronic organization of a uORF-containing mRNA to that of an mRNA containing a 5′-proximal nonsense mutation has suggested the potential of a uORF-bearing mRNA to trigger the nonsense-mediated decay pathway (53). Studies with yeast and mammalian cells have revealed that among the set of trans-acting factors that participate in this regulatory mechanism, there are factors involved in translation termination or that associate with known termination factors (12). Thus, a model is emerging of a multicomponent complex that acts at translation termination to regulate the fate of the ribosome and the mRNA. At this point, the possibility of a uORF triggering nonsense-mediated decay as part of a naturally occurring regulatory mechanism remains hypothetical, although it is interesting that the yeast CPA1 transcript, which contains a uORF, is degraded via the nonsense-mediated decay pathway (54).
UORF RECOGNITION BY SCANNING RIBOSOMES
For a uORF to function as a regulatory element, its initiation codon must be recognized, at least at certain times, by the scanning 40S ribosomal subunit and associated initiation factors. When uORF recognition is regulated by a so-called leaky-scanning mode of regulation, ribosomes either ignore the upstream AUG codon and scan past it or recognize it and initiate translation, depending on the conditions. The parameters that determine the efficiency with which the ribosome preinitiation complex recognizes an AUG codon are the same for uORFs as for the ORFs that encode the major gene products. The effects of the nucleotides immediately flanking the AUG codon, the proximity of the AUG codon to the cap site, and the presence of nearby secondary structure in the mRNA all can affect AUG codon recognition (7, 32, 52, 64, 65). In most examples of regulatory uORFs described thus far, the initiation codon is a conventional AUG triplet. However, a GUG codon appears to initiate a uORF in the Fli-1 mRNA (56), illustrating the need to remain cognizant of the potential for non-AUG codons to serve as uORF initiation sites. cis-acting elements such as internal ribosomal entry sites or sequences that promote ribosomal shunting may enable ribosomes to circumvent the uAUG codon altogether and thus evade the regulatory effects of the uORF (21, 47, 68).
Experimentally, the recognition that a uORF is involved in controlling expression is often based on detecting altered expression or function of the protein encoded by the downstream cistron after mutating the uAUG codon. However, additional studies are needed to distinguish whether the effect of such a mutation is due to elimination of the initiation function of the AUG triplet or results from an alternative function of one or more of the A, U, and G nucleotides. For example, the third uAUG codon in the Rous sarcoma virus transcript leader appears to serve as a translation initiation site (16), but these nucleotides also influence viral RNA packaging, independent of their role in translation initiation (2). In most well-characterized examples of regulatory uORFs, mutation of the uAUG codon alters protein expression without affecting mRNA abundance. If removal of a uAUG alters mRNA levels, then the AUG nucleotides may be altering transcription or RNA stability, independent of translation of the uORF. However, another possibility is that the uAUG codon does function as an initiation codon and that translation of the uORF alters RNA stability, for example by triggering the nonsense-mediated decay pathway.
Although the efficiency of initiation at an upstream AUG codon can often modulate the regulatory consequences of an associated uORF, other steps may be at least as influential. As noted above, AUG codons that are recognized efficiently may have little or no impact on downstream translation. On the other hand, only a small fraction (∼10%) of ribosomes that load on the cytomegalovirus (CMV) gpUL4 mRNA initiate at the second uAUG codon yet the associated uORF (uORF2) reduces downstream translation by ∼10-fold (7). These results support a model in which the few ribosomes that translate the uORF stall on the mRNA and block many subsequent ribosomes that would otherwise have reached the downstream AUG codon by leaky scanning. Thus, initiation rates at upstream AUG codons do not fully explain the regulatory activities of all uORFs.
REINITIATING AFTER TRANSLATION OF THE UORF
At present, very little is known about the fate of a ribosome after completing termination of a uORF. The two options are for the ribosome to remain associated with the mRNA (Fig. 1, options 1 and 2) or for it to dissociate (Fig. 1, option 5). In bacteria, an essential protein known as ribosome recycling factor catalyzes the dissociation of the ribosome from the mRNA after termination (25), but no analogous activity has yet been detected in the cytosol of eukaryotes. In some eukaryotic uORFs, ribosomes probably remain associated with the mRNA at a significant frequency, since reinitiation does occur at downstream AUG codons.
Multiple features of an mRNA influence the efficiency with which ribosomes reinitiate downstream translation after translation of a uORF. For example, increasing the intercistronic spacing up to approximately 50 to 80 nucleotides reduces or eliminates the inhibitory effects of uORFs in some cases (11, 30), implying enhanced reinitiation. However, studies with several systems reveal that the sequence of the intercistronic region, not just its length, can affect reinitiation. Replacement of the intercistronic region in the maize Lc gene with a sequence of similar length increases translational expression 15-fold in transgenic plants (61). In the GCN4 gene, the 10 nucleotides immediately downstream from the uORFs are critical in determining whether ribosomes will be able to reinitiate (18). Substitution of a single nucleotide in the 7-nucleotide intercistronic region in the CCAAT/enhancer binding protein alpha (C/EBPα) mRNA dramatically increases reinitiation (36). The mechanisms by which the intercistronic sequences influence reinitiation are unknown. For Lc, the abundance of termination codons was postulated to disrupt reinitiation (61), but this hypothesis has not been tested. Introduction of hairpin structure downstream from the YAP2 uORF may promote ribosome release, leading to destabilization of the mRNA (60). In addition to the intercistronic region, sequences upstream from and within the uORF and even the particular downstream cistron all can affect reinitiation efficiency, demonstrating that the control of reinitiation is very complex (18–20).
A working model proposes that after having translated a uORF, recharging of the ribosome with initiation factors is the limiting step required for reinitiation (24). This model would account for the observed enhancement of downstream translation upon lengthening some intercistronic spacers, since the ribosome would have additional time available to reacquire reinitiation factors prior to encountering the downstream AUG codon. Structures in the intercistronic sequence might impede ribosomal transit after it completes translation of the uORF and thereby paradoxically stimulate downstream translation by providing more time for recharging. As well, the abundance of the factors needed for reinitiation would be expected to influence the rate of ribosome recharging, as is the case for eIF2-GTP and Met-tRNA in the GCN4 system (24). Finally, this model is consistent with the observation that lengthening a uORF diminishes reinitiation efficiency (39). If factors that are necessary for reinitiation are shed from the ribosome as it translates longer uORFs, the time needed for ribosome recharging would be extended. Although the results obtained with the GCN4 system are consistent with a recharging model, our understanding of the molecular details of the recharging mechanism is incomplete. The factors present in the reinitiation complex (other than eIF2-GTP and Met-tRNA) and the way their recruitment to the ribosome is controlled are unknown. In fact, no data are available that would indicate whether a ribosome resumes scanning as a 40S subunit or as a complete 80S ribosome.
NASCENT PEPTIDES MEDIATE REGULATION BY SOME UORFS
Several uORFs affect downstream translation by mechanisms that depend on the amino acid sequence of the encoded peptide. Experimentally, sequence-dependent uORFs are characterized by a change in downstream translation resulting from missense but not synonymous mutations of codons within the uORF. Caution is needed in interpreting the effects of missense mutations since the nucleotide alterations might affect transcription or mRNA processing and thereby alter expression by a translation-independent pathway.
With this cautionary note, the peptide products of several prokaryotic and eukaryotic uORFs seem to play active roles in translational control mechanisms. In certain bacterial antibiotic resistance genes, the nascent peptide product of a short uORF binds to and interferes with the structure and activity of the ribosomal peptidyltransferase center (37). When the antibiotic effector is present, ribosomes stall during elongation through the uORF in a manner that depends on the amino acid sequence of the encoded peptide. These observations suggest that the nascent peptide, in the presence of the coregulatory antibiotic, inhibits peptidyltransferase activity and causes ribosomal stalling. As a result, the mRNA is believed to assume an altered structure, which exposes a Shine-Dalgarno sequence and thereby enables translation of the downstream ORF. Although eukaryotic genes are unlikely to employ an identical mechanism, these observations illustrate how a short nascent peptide can influence the function of the ribosome that synthesized it. Another prokaryotic example of a sequence-dependent uORF is in the tryptophanase operon of Escherichia coli, in which a 24-residue peptide seems to act in cis to influence ribosome release at the termination codon of the uORF (28).
At present, six eukaryotic mRNAs have been found in which translation is repressed by sequence-dependent uORFs (Table 1). Two of these, from the fungal CPA1 and arg-2 genes, encode the carbamoylphosphate synthetase involved in arginine biosynthesis in Saccharomyces cerevisiae and Neurospora crassa, respectively. Two other mRNAs encode receptors for extracellular signals, RAR-β2 and β2-adrenergic receptor. S-Adenosylmethionine decarboxylase (AdoMetDC) is an enzyme involved in polyamine biosynthesis in mammals and shows at least two modes of translational control involving the uORF in its mRNA. The mRNA from the CMV gpUL4 gene (formerly called gp48) encodes a virion glycoprotein of unknown function. Five of these uORFs range from 19 to 25 codons, while the AdoMetDC uORF is only 6 codons. In each case, missense mutations release the influence of the uORF on translation of the downstream cistron without affecting mRNA levels. The appearance of these uORF-regulated mRNAs in diverse biological systems, including fungi, viruses, and mammalian cells, suggests that this regulatory strategy is widespread among biological systems. However, the small number of examples and the differences among the functions of these genes and in the sequences of the uORFs have frustrated attempts to identify other examples.
TABLE 1.
Eukaryotic sequence-dependent uORFsa
| Gene | uORF peptide | Reference(s) |
|---|---|---|
| Mammals | ||
| AdoMedDC | MAGDIS | 23, 41 |
| β2-Adrenergic receptor | MKLPGVRPRPAAPRRRCTR | 46 |
| RAR-β2 | MIRGWEKDQQPTCQKRGRV | 50 |
| Virus | ||
| CMV UL4 | MQPLVLSAKKLSSLLTCKYIPP | 1, 13 |
| Fungi | ||
| CPA1 | MFSLSNSQYTCQDYISDHIWKTSHH | 65 |
| arg-2 | MNGRPSVFTSQDYLSDHLWRALNA | 64 |
Positions are indicated in which changes either singly (bold) or in combination (underlined) reduce the inhibitory effects of the uORF.
Several lines of evidence suggest that at least some of these uORF peptide products act by interfering with translation termination. The most direct evidence for this hypothesis is derived from mapping of ribosome stall sites using primer extension inhibition (or toeprint) assays. Both in cell-free translation systems and in infected cell extracts, ribosomes stall when the termination codon of the gpUL4 wild-type uORF is within or very close to the A site in the ribosome (8). No such stalling occurs with uORF mutants that fail to inhibit downstream translation. Removing the termination codons of the gpUL4 and AdoMetDC uORFs eliminates their inhibitory effects (7, 23, 41, 57), implying that these nascent peptides act on only on terminating ribosomes and not on elongating ones. Finally, in cell-free translation assays, the wild-type gpUL4 uORF nascent peptide remains linked to the tRNA responsible for decoding the final sense codon of the uORF while missense peptides from mutant uORFs are efficiently hydrolyzed under these conditions (9, 10). These data suggest that the nascent peptide product of the uORF interferes with a step during translation termination. The persistence of the linkage of the nascent peptide to the tRNA indicates that the affected step is prior to completion of peptidyl-tRNA hydrolysis. For example, the entry or activity of a peptide release factor or a termination-specific activity of the peptidyltransferase center could be perturbed by the presence of the uORF peptide. It should be noted that the nucleotide sequence surrounding a termination site affects the rate of termination in some instances (59); however, differences in the termination codon or its adjacent nucleotides have never been found to influence inhibitory mechanisms involving sequence-dependent uORFs (13, 23, 41).
The uORFs in the arg-2 and CPA-1 mRNAs act through a mechanism with striking similarities yet also with significant differences compared to that used by gpUL4 and AdoMetDC. These fungal uORF-encoded peptides apparently can act on either terminating or elongating ribosomes (see below), suggesting that the target of their mechanism might be the peptidyltransferase center or another site that participates in both reactions. In fact, the gpUL4 and AdoMetDC uORFs could also inhibit the peptidyltransferase center but function only during termination because the rate of termination is low enough to allow the nascent peptide to associate with a factor in a way that does not occur during elongation.
Ribosome stalling at a uORF probably inhibits downstream translation because the resulting blockade prevents the stalled ribosome, as well as ribosomes that subsequently load onto the mRNA, from reaching the downstream cistron. For example, the half time of ribosomes remaining at the gpUL4 uORF termination codon is ∼10 min in cell-free translation extracts (10). If ribosomes load onto the mRNA at a rate of ∼10 per min (45), one stalled ribosome would be expected to block ∼100 ribosomes, resulting in a ∼100-fold inhibition of downstream translation. However, the uAUG codon is surrounded by a suboptimal context of nucleotide, resulting in an average of ∼10 scanning 40S subunits bypassing the uAUG and translating the downstream ORF after the stalled ribosome vacates the termination site (7). These calculations predict that the uORF should reduce translation 10-fold, a number consistent with experimental observations (7, 57).
EXAMPLES OF GENES REGULATED BY UORFS
In the preceding paragraphs, we have discussed the various parameters that govern the influence of uORFs on translation of a downstream cistron. These parameters are modulated in diverse ways, depending on the particular mRNA, to achieve translational control. The available mechanisms depend in part on whether a particular mRNA contains one or multiple uORFs. Regulation of an mRNA with a single uORF must depend on properties inherent either in the uORF (or its encoded peptide) or in the interaction of the uORF with other structures in the mRNA, usually within the leader. Here we discuss three well-characterized examples of regulation through single uORFs: AdoMetDC, arg-2, and C/EBP. With mRNAs containing multiple uORFs, the repertoire of mechanisms expands to include possible regulatory interactions among uORFs. In the group of genes with multiple uORFs, yeast GCN4 stands out as the best understood, but we also discuss an instructive example from mammalian organisms, oncogene mdm2.
Mammalian AdoMetDC.
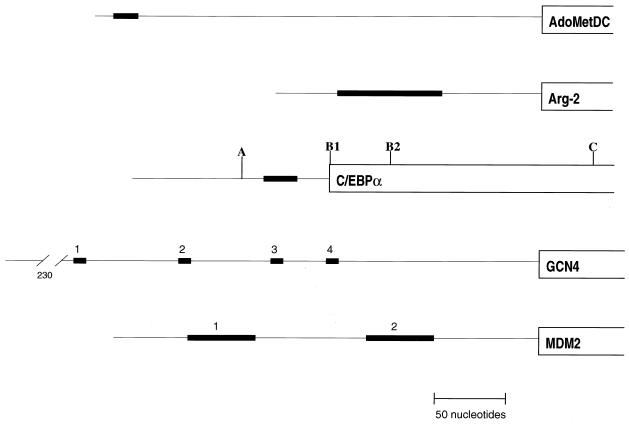
AdoMetDC catalyzes a key regulated step in the biosynthesis of the polyamines spermidine and spermine. The rate of translation of AdoMetDC mRNA depends on cell type (22), cellular polyamine content (51, 66), and, for T lymphocytes, growth status (40). This mRNA contains a single uORF that is a key regulatory element both in feedback control by polyamine levels and in cell-specific regulation, albeit by different mechanisms (23, 51, 52). The uORF is located approximately 14 nucleotides from the 5′ cap (Fig. 2) (22), and its regulatory effects are sequence dependent, with a strict specificity that extends from mammalian cells to yeasts (41).
FIG. 2.
Structures of the leaders of selected mRNAs. uORFs contained in these mRNAs are depicted by the thick horizontal lines.
In resting normal T cells and in T-cell lines with normal cellular levels of polyamines, ribosome loading on the AdoMetDC mRNA is suppressed to a point where the mRNA is largely associated with single ribosomes, whereas in many cell lines of nonlymphoid origin, molecules of this mRNA can contain 5 to 10 ribosomes (22). This cell-specific translation of AdoMetDC depends not only on the amino acid sequence of the uORF-encoded peptide but also on the close proximity of the uORF initiation codon to the cap in the wild-type mRNA (52). Extending the distance between the cap and the initiation codon from 14 to 47 nucleotides enhances recognition of the uORF in nonlymphoid cells to a level similar to that in T cells, with concomitant suppression of translation. It has been suggested that this cell-type-specific difference in recognition of an initiation codon close to the 5′ cap could arise from differences in the level and/or activity of a translation initiation factor (52).
In cells depleted of endogenous polyamines, AdoMetDC mRNA becomes more highly loaded with ribosomes (66). In contrast to cell-type-specific regulation, this feedback regulation of AdoMetDC translation by endogenous polyamine levels is independent of the position of the uORF in the mRNA (51). The uORF appears to be the sole element in the AdoMetDC mRNA necessary for polyamine regulation, since introduction of the wild-type uORF sequence into an unrelated leader confers polyamine regulation on translation of the heterologous gene (51). As in cell-specific control of AdoMetDC translation, regulation by endogenous polyamines requires a specific amino acid sequence at the carboxy-terminal end of the uORF peptide. These results have led to a model where intracellular polyamines modulate the interaction of the peptide product of the uORF with its target, leading to regulated arrest of the translating ribosome at termination (51). Consistent with this prediction, the in vitro rate of translation of the peptide from the wild-type uORF is much more sensitive to spermidine concentration than is translation from altered uORFs (49), which may result from spermidine-mediated stalling at the termination codon (55).
Neurospora arg-2.
The small subunit of the arginine-specific carbamoyl phosphate synthetase is encoded by the arg-2 gene of N. crassa and is negatively controlled at the translational level in response to the level of arginine in the culture medium. A 24-codon uORF is located in the leader of the arg-2 mRNA (Fig. 2), and similar uORFs are also found in the leaders of the corresponding mRNAs in S. cerevisiae and several other fungi (64). The arg-2 uORF is located approximately 40 nucleotides downstream of the closest transcriptional start site, and its presence is necessary for regulation by arginine both in vivo and in an isolated in vitro system. In the presence of high concentrations of arginine, the uORF causes ribosomes to arrest on the leader by a mechanism that is dependent on the structure of the encoded peptide (arginine attenuator peptide [AAP]) (38). This sequence-dependent ribosome stalling is reminiscent of that observed with gpUL4 (see the previous section) and of that suggested to take place with AdoMetDC, with an important distinction. AAP causes the ribosome to arrest even when the uORF termination codon of the arg-2 or yeast CPA1 gene is removed (14, 62, 63), demonstrating that in this case the uORF-encoded peptide can act on an elongating or terminating ribosome. The current model (64) suggests that at low arginine concentrations most ribosomes bypass the suboptimal uAUG codon by leaky scanning and translate the downstream cistron. At elevated concentrations of arginine, ribosomes translating the uORF stall in association with the AAP, creating a blockade to scanning ribosomes that subsequently load on the mRNA. In addition to the physical blockade by the arrested ribosome, it is possible that the prolonged time during which preinitiation complexes queue behind the AAP-stalled ribosome could augment recognition of the suboptimal uAUG codon.
Vertebrate C/EBPα and C/EBPβ.
C/EBPs are a family of transcription factors that regulate the expression of tissue-specific genes during differentiation of a variety of cell types. Several isoforms of C/EBPα and C/EBPβ exist that seem to arise from the use of different translation initiation codons (15, 35, 44). Truncated forms of the C/EBPs lack transcription activation domains but retain DNA binding activity and thus seem to act as antagonists of the full-length transcription factors.
C/EBPα and C/EBPβ mRNAs from several species contain a small, out-of-frame uORF (5) immediately upstream of the major translation start site (B1 in Fig. 2). Translation initiation at B1 appears to occur by combination of leaky scanning past the uORF AUG codon and reinitiation after uORF translation (6, 36). In contrast to its inhibitory effect on initiation at B1, the uORF enhances initiation at site C (Fig. 2), generating the truncated form of the transcription factor (5, 6). Apparently, translation of the uORF results in ribosomes bypassing sites B1 and B2 and thereby gaining access to site C.
The activities of two key translation initiation factors, eIF2 and eIF4E, are capable of regulating the expression of the various C/EBP proteins. Various treatments that reduce the phosphorylation of eIF2 enhance its activity and result in elevated production of the truncated C/EBP forms (6), possibly by increasing reinitiation efficiency. This is reminiscent of eIF2-regulated reinitiation in the yeast GCN4 gene (see below). These results suggest that the ratio of the C/EBP isoforms could be regulated by signal transduction pathways that alter the phosphorylation state of the α subunit of eIF2.
The activity of initiation factor eIF4E, the cap binding protein, is regulated by complex mechanisms involving both direct phosphorylation and inhibitory eIF4E binding proteins (4E-BPs). Overexpression of eIF4E enhances the expression of truncated C/EBP, while treatments that decrease phosphorylation of the 4E-BPs reduce the production of truncated forms (6). Since eIF4E stimulates upstream translation in bicistronic mRNAs (58), its influence on the expression of the C/EBP may result from enhanced recognition of the uORF.
The regulation of C/EBP isoform expression by the activities of these key initiation factors creates an important potential link between translational control through these uORFs and the establishment of cellular phenotype during differentiation. Roles for the signal transduction pathways regulating eIF2 and eIF4E in controlling cellular differentiation have yet to be directly demonstrated.
Yeast GCN4.
Translational control of the yeast GCN4 gene is the best understood example of regulatory interactions of multiple uORFs. The GCN4 gene encodes a transcription factor that activates the expression of approximately 50 genes of amino acid biosynthesis. During amino acid starvation, general protein synthesis is inhibited and translation of GCN4 mRNA is markedly enhanced (24).
This differential enhancement of GCN4 translation in response to amino acid starvation is mediated through the interaction of four small uORFs that contain two or three codons each (Fig. 2). Depending on which uORF is translated by ribosomes scanning the GCN4 leader, profoundly different effects can be generated on translation of a downstream open reading frame. The translational control observed with this gene is generated through the combined activities of the different uORFs (24). After translation of uORF1, ribosomes seem to be able to continue scanning and reinitiate at a downstream ORF with a relatively high efficiency of approximately 50%. In contrast, the downstream uORFs, especially uORF3 and uORF4, are much more inhibitory to translation of the major ORF. For example, uORF4 by itself in the leader inhibits translation of the GCN4 ORF by approximately 99%. It is not the particular codons within the uORFs that produce these uniquely different behaviors; rather, the nucleotide sequences located at and following the 3′ ends of the uORFs determine the efficiencies of reinitiation. uORF4 has a relatively GC-rich sequence surrounding its termination codon, while the same region of uORF1 is AU rich. The GC-rich 3′ region of uORF4 generates strong inhibitory activity when placed in conjunction with uORF1. Although no clear molecular mechanism has yet emerged, it has been suggested that this behavior reflects in some way the strength of the interaction of this region of the mRNA with ribosomes or some other component of the translational machinery.
The weak inhibitory activity of uORF1 works together with strong inhibition by the downstream uORFs to generate the observed regulation. Initiating ribosomes enter at the 5′ cap of the mRNA, translate uORF1, and resume scanning. It is thought that these ribosomes must then regain the ability to initiate as they scan down the leader. If this ability has been reacquired before reaching uORF3 or uORF4, one of these inhibitory uORFs is translated and the ribosome is rendered incapable of translating GCN4. In contrast, if the scanning ribosomes do not reacquire initiation capability until after scanning past the downstream uORFs, GCN4 can be successfully translated. Thus, uORF1 serves to derail initiation-competent ribosomes and forces them to reaccumulate required factors prior to subsequent initiation. The rate at which the ability to reinitiate is acquired is influenced by the availability of the eIF2–GTP–Met-tRNA ternary complex, which is controlled by several factors, most notably the phosphorylation state of the eIF2α (24, 48). Under starvation conditions, the high level of uncharged tRNAs activates the phosphorylation of eIF2α by the kinase product of the GCN2 gene. The resulting limitation in ternary-complex availability retards the acquisition of reinitiation activity by ribosomes that have translated uORF1 and thereby favors skipping the inhibitory uORFs and enhances the translation of GCN4.
Mammalian mdm2. Oncoprotein MDM2 forms part of a negative-feedback loop that regulates the activity of tumor suppressor p53 (17, 27). Since MDM2 binds to and antagonizes the activity of p53, overexpression of MDM2 leads to oncogenesis. Overexpression of MDM2 has been seen in a number of human tumors, particularly in those originating in soft tissues. Since elevated MDM2 levels are potentially detrimental, there is a clear biological necessity for tight regulation of expression of this protein, and indeed this is observed at both the transcriptional and translational levels. Transcription of the mdm2 gene is regulated by p53 through a binding site within the first intron (3, 26, 67). Thus, elevated p53 levels activate mdm2 expression, forming a negative-feedback loop. The first evidence for translational control of MDM2 protein expression came from the observation that overexpression of MDM2 in some human tumors resulted not from elevated levels of mdm2 mRNA but from enhanced translation of existing mRNA (34).
Transcription of the human mdm2 gene can arise from one of two promoters, yielding two alternatively spliced mRNAs that differ only in their leader sequences (26, 33, 67, 69). The long form of human mdm2 mRNA contains two uORFs, each of which contains 15 codons (Fig. 2). The short form lacks both uORFs and is much more efficiently loaded with ribosomes than is the long form (4, 33). The inefficient translation that is characteristic of the long form is transferred with the leader to a reporter gene. Mutational analysis suggested that the two uORFs acted synergistically to inhibit translational initiation. The mechanism of the apparent functional interaction between the two mdm2 uORFs is not understood, but this mechanism is of general interest, since multiple uORFs are often found in mammalian genes, particularly oncogenes (31).
CONCLUSIONS AND REFLECTIONS
uORFs clearly have potential to exert a major impact on gene expression, and some, but not all, serve as important regulatory elements. Although the number of well-characterized uORFs remains few and additional mechanisms will undoubtedly be discovered in the future, a few distinctive themes are beginning to emerge that provide a foundation for understanding the roles and mechanisms of regulation by uORFs. Initiation codon recognition, which has been well described in studies of conventional mRNAs that lack uORFs, is essential but of surprisingly low utility in predicting the effects of a uORF. Rather, the fate of ribosomes during and after termination of translation assumes special significance in considering mechanisms of uORF action.
Nascent peptide-dependent ribosomal stalling at termination appears to be widespread in biology. If it is of sufficiently long duration, such stalling will result in a reduction in translation of the downstream cistron. Exactly which biochemical step during the termination reaction is targeted by the nascent peptide is unknown. It seems most likely that the nascent peptide interacts with a protein or RNA in the ribosome that somehow prevents termination from proceeding efficiently. The diversity of uORF-encoded peptides suggests that perhaps multiple targets are available and that termination may be a particularly sensitive step in gene expression. The mechanisms by which corepressors, arginine and polyamines in the cases of arg-2 and AdoMetDC, modulate these interactions is unknown. Corepressors could interact directly with the peptide-target complex or could act indirectly through a less direct pathway.
The fate of a ribosome after having completed uORF translation appears to vary according to the particular gene. Current data hint that multiple determinants, including the RNA sequence in the vicinity and downstream of the uORF termination site, are quite important in determining the potential for ribosomal reinitiation downstream. Also, the availability of the eIF2–GTP–Met-tRNA ternary complex and perhaps other factors required for reinitiation plays a role. Still, many puzzles remain. For example, is the eIF4A/B helicase necessary for scanning the intercistronic region during reinitiation? We do not even know whether scanning of the intercistronic region is performed by the complete ribosome or the 40S subunit. A more complete understanding of the reinitiation process will be required before we can fully describe the mechanism of regulation by many uORFs.
Why are uORFs so frequent in genes with critical biological functions? Although production of a poorly translated mRNA seems inefficient, evolution has clearly tolerated and apparently exploited these elements for regulatory purposes. At present, it is unclear whether uORFs provide any unique opportunities for regulation that could not be supplied by other translational elements, such as structured transcript leaders or sites for repressor protein binding. At a minimum, uORFs provide alternative strategies for achieving the imposing goal of coordinating the expression of thousands of genes expressed in a cell. Future work will undoubtedly shed new light on the role of these intriguing regulatory elements in this overall scheme of gene regulation.
ACKNOWLEDGMENTS
We thank Lynn Law, Alexa Raney, and Matthew Sachs for helpful comments and suggestions.
The research from our laboratories was supported by research grants from the NIH (CA39053, CA71453, and AI26672).
REFERENCES
- 1.Alderete J P, Jarrahian S, Geballe A P. Translational effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J Virol. 1999;73:8330–8337. doi: 10.1128/jvi.73.10.8330-8337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banks J D, Linial M L. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J Virol. 2000;74:456–464. doi: 10.1128/jvi.74.1.456-464.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barak Y, Gottlieb E, Juvengershon T, Oren M. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 4.Brown C Y, Mize G J, Pineda M, George D L, Morris D R. Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene. 1999;18:5631–5637. doi: 10.1038/sj.onc.1202949. [DOI] [PubMed] [Google Scholar]
- 5.Calkhoven C F, Bouwman P R J, Snippe L, Ab G. Translation start site multiplicity of the CCAAT enhancer binding protein alpha mRNA is dictated by a small 5′ open reading frame. Nucleic Acids Res. 1994;22:5540–5547. doi: 10.1093/nar/22.25.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calkhoven C F, Müller C, Leutz A. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J H, Geballe A P. Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J Virol. 1995;69:1030–1036. doi: 10.1128/jvi.69.2.1030-1036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao J H, Geballe A P. Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol. 1996;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J H, Geballe A P. Inhibition of nascent-peptide release at translation termination. Mol Cell Biol. 1996;16:7109–7114. doi: 10.1128/mcb.16.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao J H, Geballe A P. Ribosomal release without peptidyl tRNA hydrolysis at translation termination in a eukaryotic system. RNA. 1998;4:181–188. [PMC free article] [PubMed] [Google Scholar]
- 11.Child S J, Miller M K, Geballe A P. Translational control by an upstream open reading frame in the HER-2/neu transcript. J Biol Chem. 1999;274:24335–24341. doi: 10.1074/jbc.274.34.24335. [DOI] [PubMed] [Google Scholar]
- 12.Czaplinski K, Ruiz-Echevarria M J, Gonzalez C I, Peltz S M. Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. Bioessays. 1999;21:685–696. doi: 10.1002/(SICI)1521-1878(199908)21:8<685::AID-BIES8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Degnin C R, Schleiss M R, Cao J H, Geballe A P. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J Virol. 1993;67:5514–5521. doi: 10.1128/jvi.67.9.5514-5521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delbecq P, Werner M, Feller A, Filipkowski R K, Messenguy F, Pierard A. A segment of messenger RNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol Cell Biol. 1994;14:2378–2390. doi: 10.1128/mcb.14.4.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 16.Donze O, Damay P, Spahr P F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freedman D A, Levine A J. Regulation of the p53 protein by the MDM2 oncoprotein. Cancer Res. 1999;59:1–7. [PubMed] [Google Scholar]
- 18.Grant C M, Hinnebusch A G. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant C M, Miller P F, Hinnebusch A G. Requirements for intercistronic distance and level of eukaryotic initiation factor 2 activity in reinitiation on GCN4 mRNA vary with the downstream cistron. Mol Cell Biol. 1994;14:2616–2628. doi: 10.1128/mcb.14.4.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant C M, Miller P F, Hinnebusch A G. Sequences 5′ of the first upstream open reading frame in GCN4 mRNA are required for efficient translational reinitiation. Nucleic Acids Res. 1995;23:3980–3988. doi: 10.1093/nar/23.19.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemmings-Mieszczak M, Hohn T. A stable hairpin preceded by a short open reading frame promotes nonlinear ribosome migration on a synthetic mRNA leader. RNA. 1999;5:1149–1157. doi: 10.1017/s1355838299990325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill J R, Morris D R. Cell-specific translation of S-adenosylmethionine decarboxylase messenger RNA—Regulation by the 5′ transcript leader. J Biol Chem. 1992;267:21886–21893. [PubMed] [Google Scholar]
- 23.Hill J R, Morris D R. Cell-specific translational regulation of S-adenosylmethionine decarboxylase messenger RNA—dependence on translation and coding capacity of the cis-acting upstream open reading frame. J Biol Chem. 1993;268:726–731. [PubMed] [Google Scholar]
- 24.Hinnebusch A G. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 199–244. [Google Scholar]
- 25.Janosi L, Hara H, Zhang S, Kaji A. Ribosome recycling by ribosome recycling factor (RRF)—an important but overlooked step of protein biosynthesis. Adv Biophys. 1996;32:121–201. doi: 10.1016/0065-227x(96)84743-5. [DOI] [PubMed] [Google Scholar]
- 26.Juven T, Barak Y, Zauberman A, George D L, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- 27.Juvengershon T, Oren M. Mdm2: the ups and downs. Mol Med. 1999;5:71–83. [PMC free article] [PubMed] [Google Scholar]
- 28.Konan K V, Yanofsky C. Role of ribosome release in regulation of tna operon operon expression in Escherichia coli. J Bacteriol. 1999;181:1530–1536. doi: 10.1128/jb.181.5.1530-1536.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozak M. An analysis of vertebrate messenger RNA sequences—intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 33.Landers J E, Cassel S L, George D L. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res. 1997;57:3562–3568. [PubMed] [Google Scholar]
- 34.Landers J E, Haines D S, Strauss J F, George D L. Enhanced translation: a novel mechanism of mdm2 oncogene overexpression identified in human tumor cells. Oncogene. 1994;9:2745–2750. [PubMed] [Google Scholar]
- 35.Lin F T, MacDonald O A, Diehl A M, Lane M D. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: transcription activator lacking antimitotic activity. Proc Natl Acad Sci USA. 1993;90:9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lincoln A J, Monczak Y, Williams S C, Johnson P F. Inhibition of CCAAT/enhancer-binding protein alpha and beta translation by upstream open reading frames. J Biol Chem. 1998;273:9552–9560. doi: 10.1074/jbc.273.16.9552. [DOI] [PubMed] [Google Scholar]
- 37.Lovett P S, Rogers E J. Ribosome regulation by the nascent peptide. Microbiol Rev. 1996;60:366–366. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo Z, Sachs M S. Role of an upstream open reading frame in mediating arginine-specific translational control in Neurospora crassa. J Bacteriol. 1996;178:2172–2177. doi: 10.1128/jb.178.8.2172-2177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luukkonen B G M, Tan W, Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mach M, White M W, Neubauer M, Degen J L, Morris D R. Isolation of a cDNA clone encoding S-adenosylmethionine decarboxylase. Expression of the gene in mitogen-activated lymphocytes. J Biol Chem. 1986;261:11697–11703. [PubMed] [Google Scholar]
- 41.Mize G J, Ruan H J, Low J J, Morris D R. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J Biol Chem. 1998;273:32500–32505. doi: 10.1074/jbc.273.49.32500. [DOI] [PubMed] [Google Scholar]
- 42.Morris D R. Growth control of translation in mammalian cells. Prog Nucleic Acid Res Mol Biol. 1995;51:339–363. doi: 10.1016/s0079-6603(08)60883-1. [DOI] [PubMed] [Google Scholar]
- 43.Morris D R. cis-acting mRNA structures in gene-specific translation control. In: Harford J B, Morris D R, editors. RNA metabolism and post-transcriptional gene regulation. New York, N.Y: Wiley-Liss, Inc.; 1997. pp. 165–180. [Google Scholar]
- 44.Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein messenger RNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmiter R D. Quantitation of parameters that determine the rate of ovalbumin synthesis. Cell. 1975;4:189–197. doi: 10.1016/0092-8674(75)90167-1. [DOI] [PubMed] [Google Scholar]
- 46.Parola A L, Kobilka B K. The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J Biol Chem. 1994;269:4497–4505. [PubMed] [Google Scholar]
- 47.Pelletier J, Flynn M E, Kaplan G, Racaniello V, Sonenberg N. Mutational analysis of upstream AUG codons of poliovirus RNA. J Virol. 1988;62:4486–4492. doi: 10.1128/jvi.62.12.4486-4492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu H, Hu C, Anderson J, Bjork G R, Sarker S, Hopper A K, Hinnebusch A G. Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2000;20:2505–2516. doi: 10.1128/mcb.20.7.2505-2516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raney A, Baron A C, Mize G J, Law G L, Morris D R. In vitro translation of the upstream open reading frame in the mammalian mRNA encoding S-adenosylmethionine decarboxylase. J Biol Chem. 2000;275:24444–24450. doi: 10.1074/jbc.M003364200. [DOI] [PubMed] [Google Scholar]
- 50.Reynolds K, Zimmer A M, Zimmer A. Regulation of RAR beta 2 mRNA expression: evidence for an inhibitory peptide encoded in the 5′-untranslated region. J Cell Biol. 1996;134:827–835. doi: 10.1083/jcb.134.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan H, Shantz L M, Pegg A E, Morris D R. The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J Biol Chem. 1996;271:29576–29582. doi: 10.1074/jbc.271.47.29576. [DOI] [PubMed] [Google Scholar]
- 52.Ruan H J, Hill J R, Fatemie-Nainie S, Morris D R. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA—influence of the structure of the 5′ transcript leader on regulation by the upstream open reading frame. J Biol Chem. 1994;269:17905–17910. [PubMed] [Google Scholar]
- 53.Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 54.Ruiz-Echevarria M J, Peltz S W. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–751. doi: 10.1016/s0092-8674(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 55.Ryabova L A, Hohn T. Ribosome shunting in the cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes Dev. 2000;14:817–829. [PMC free article] [PubMed] [Google Scholar]
- 56.Sarrazin S, Starck J, Gonnet C, Doubeikovski A, Melet F, Morle F. Negative and translation termination-dependent positive control of FLI-1 protein synthesis by conserved overlapping 5′ upstream open reading frames in Fli-1 mRNA. Mol Cell Biol. 2000;20:2959–2969. doi: 10.1128/mcb.20.9.2959-2969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schleiss M R, Degnin C R, Geballe A P. Translational control of human cytomegalovirus-gp48 expression. J Virol. 1991;65:6782–6789. doi: 10.1128/jvi.65.12.6782-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tahara S M, Dietlin T A, Dever T E, Merrick W C, Worrilow L M. Effect of eukaryotic initiation factor-4F on AUG selection in a bicistronic messenger RNA. J Biol Chem. 1991;266:3594–3601. [PubMed] [Google Scholar]
- 59.Tate W P, Mannering S A. Three, four or more: the translational stop signal at length. Mol Microbiol. 1996;21:213–219. doi: 10.1046/j.1365-2958.1996.6391352.x. [DOI] [PubMed] [Google Scholar]
- 60.Vilela C, Ramirez C V, Linz B, Rodriguespousada C, McCarthy J E G. Post-termination ribosome interactions with the 5′ UTR modulate yeast mRNA stability. EMBO J. 1999;18:3139–3152. doi: 10.1093/emboj/18.11.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Wessler S R. Inefficient reinitiation is responsible for upstream open reading frame-mediated translational repression of the maize R gene. Plant Cell. 1998;10:1733–1746. doi: 10.1105/tpc.10.10.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, Fang P, Sachs M S. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol Cell Biol. 1998;18:7528–7536. doi: 10.1128/mcb.18.12.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z, Gaba A, Sachs M S. A highly conserved mechanism of regulated ribosome stalling mediated by fungal arginine attenuator peptides that appears independent of the charging status of arginyl-tRNAs. J Biol Chem. 1999;274:37565–37574. doi: 10.1074/jbc.274.53.37565. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z, Sachs M S. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol Cell Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Werner M, Feller A, Messenguy F, Pierard A. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell. 1987;49:805–813. doi: 10.1016/0092-8674(87)90618-0. [DOI] [PubMed] [Google Scholar]
- 66.White M W, Degnin C, Hill J, Morris D R. Specific regulation by endogenous polyamines of translational initiation of S-adenosylmethionine decarboxylase messenger RNA in Swiss 3T3 fibroblasts. Biochem J. 1990;268:657–660. doi: 10.1042/bj2680657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 68.Yueh A, Schneider R J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- 69.Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23:2584–2592. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]