Abstract
Objectives
Neutrophil/lymphocyte ratio (NLR), platelet/lymphocyte ratio (PLR), monocyte/lymphocyte ratio (MLR), eosinophil/lymphocyte ratio (ELR), and C‐reactive protein (CRP)/lymphocyte ratio (CLR) are well‐established inflammatory indices. This study aimed to examine whether NLR, PLR, MLR, ELR and CLR could differentiate coronavirus disease 2019 (COVID‐19) patients with pneumonia from those of without.
Methods
We retrospectively examined the laboratory parameters including CRP, D‐dimer, procalcitonin and complete blood count of 306 COVID‐19 patients (pneumonic = 152 and non‐pneumonic = 154). NLR, PLR, MLR, ELR and CLR values of each patient were calculated. The ability of these indices to distinguish COVID‐19 patients with and without pneumonia was determined by receiver operating characteristic (ROC) analysis.
Results
NLR, PLR and CLR values were higher while ELR value was lower in pneumonic COVID‐19 patients compared with patients with non‐pneumonic COVID‐19 infection. MLR value was similar in the two groups. NLR, PLR and CLR were positively correlated with CRP and procalcitonin. ELR was negatively correlated with CRP. The ROC analysis revealed that the optimal cut‐off value of CLR for discriminating COVID‐19 patients with pneumonia from those without pneumonia was 1.14 and the area under curve (AUC) for CLR was 0.731 (sensitivity = 81.5% and specificity = 55.6%), which was markedly higher than the AUCs of NLR (0.622), PLR (0.585) and ELR (0.613). However, no statistical differences were observed between AUC values of NLR, PLR and ELR (P > .05).
Conclusion
Our findings showed that NLR, PLR, ELR and CLR indices can be used in differentiating COVID‐19 patients with or without pneumonia. Among them, the CLR index was the best predictor of pneumonia in COVID‐19 patients.
What's known
COVID‐19 can cause life‐threatening pneumonia.
Hyperinflammation plays a crucial role in COVID‐19 lung damage.
NLR, PLR, MLR, ELR and CLR are well‐established inflammatory indices.
Early diagnosis and treatment are crucial to reduce mortality in COVID‐19 patients with pneumonia.
What's new
NLR, PLR and CLR were higher and ELR was lower in pneumonic COVID‐19 patients compared with non‐pneumonic COVID‐19 patients.
There was a positive correlation between NLR, PLR, CLR values and CRP and procalcitonin levels. ELR value was negatively correlated with CRP.
CLR was more efficient than NLR, PLR and ELR in differentiating COVID‐19 patients with or without pneumonia.
1. INTRODUCTION
The coronavirus disease 2019 (COVID‐19) infection caused by a novel coronavirus, firstly emerged in Hubei Province, China, spread across all continents of the world in a short time. This respiratory tract infection possess a spectrum of symptoms ranging from mild to life‐threatening consequences such as acute respiratory distress syndrome (ARDS) and multiple organ failure. 1 , 2 COVID‐19 patients are classified according to clinical presentation as mild, moderate, severe and critical. The main criteria for classification of patients are involvement of lung and severity of pneumonia. 3 COVID‐19 pneumonia is a common cause of hospital admission and death. Computed tomography (CT) images play a key role in determining the presence and severity of pneumonia in COVID‐19 patients. 4 Unfortunately, CT scanning may not be available in all emergency departments. 5 Besides, inside the CT suites are high‐risk areas for nosocomial COVID‐19 transmission. 6 For this reason, there is need for fast, cheap and widely accessible biomarkers to simplify the diagnostic process and administer timely treatment of COVID‐19 pneumonia.
In recent studies, the neutrophil/lymphocyte ratio (NLR), platelet/lymphocyte ratio (PLR), monocyte/lymphocyte ratio (MLR) and C‐reactive protein (CRP)/lymphocyte ratio (CLR), which are novel inflammatory markers, have been considered as useful indicators for diagnosis and prognosis of various infectious diseases, including COVID‐19 infection. 7 , 8 , 9 , 10 , 11 , 12 Eosinophil/lymphocyte ratio (ELR) is another marker of inflammation and can be easily calculated by the ratio of eosinophil count to lymphocyte count. 13 However, the diagnostic values of these indices for COVID‐19 patients with pneumonia have not yet been investigated. Herein, we aimed to investigate the utility of NLR, PLR, MLR, CLR and ELR to distinguish COVID‐19 patients with pneumonia from those without pneumonia.
2. MATERIALS AND METHODS
This retrospective single‐center study was performed with 306 adult COVID‐19 patients (aged ≥18 years) admitted to the Sanliurfa Training and Research Hospital, Turkey between 1 April and 30 July 2020. All COVID‐19 cases had positive PCR test results in collected nasopharyngeal swab samples to detect SARS‐CoV‐2. The COVID‐19 patients were divided into two groups: pneumonic group (n = 152) and non‐pneumonic group (n = 154) according to clinical symptoms, CT findings, and laboratory results. All patients were admitted to the general wards of the hospital. Patients with rheumatological disease, malignancy, haematological disorder, thyroid disease, chronic renal failure, chronic liver disease, cerebrovascular disease, allergic disorders, pregnancy, immunosuppressive medication and blood transfusions were excluded from the study. This study was approved by the Harran University Ethics Committee (Protocol number: HRU/20/15/09).
Basic demographic information, comorbidities and laboratory data of the patients on admission were retrieved from the hospital database. Serum urea, creatinine, CRP and procalcitonin levels as well as plasma D‐dimer level were measured according to standard methods using Cobas 6000 analyzer (Roche, Germany). Prothrombin time and international normalized ratio were detected with Sysmex CS‐2000i analyzer (Sysmex, Japan). Complete blood counts (White blood cell, neutrophil, lymphocyte, monocyte, eosinophil and platelet counts) were measured in EDTA‐anticoagulated blood samples using Sysmex XN‐1000 analyzer (Sysmex). Afterwards, NLR, PLR, MLR, ELR and CLR values were calculated for each of the patients.
2.1. Statistical analysis
SPSS version 21.0 (SPSS Inc) and MedCalc version 16.8.4 (MedCalc Software) were used for the statistical analysis. Normality of data was assessed using Kolmogorov‐Smirnov test. Variables were compared using Student's t test, Mann‐Whitney U‐test or chi‐square test as appropriate. The relationship between inflammatory markers and NLR, PLR, ELR and CLR was determined by Spearman test. The diagnostic potential of derived blood lymphocyte parameters (NLR, PLR, ELR and CLR) for COVID‐19 patients with pneumonia was determined by ROC analysis. The area under curve (AUC) values of the NLR, PLR, ELR and CLR were compared using the z test. P < .05 were considered significant.
3. RESULTS
Table 1 shows the baseline characteristics of COVID‐19 patients classified by pneumonia status. Compared with patients with non‐pneumonic COVID‐19 infection, pneumonic COVID‐19 patients had higher levels of CRP, D‐dimer, NLR, PLR and CLR; and lower levels of lymphocyte, monocyte, eosinophil and ELR. Gender ratio, mean age, incidences of comorbidities, procalcitonin and MLR values were similar between the two groups.
TABLE 1.
Demographic characteristics and laboratory results of COVID‐19 patients on admission
| Non‐pneumonic (n = 154) | Pneumonic (n = 152) | P value | |
|---|---|---|---|
| Age, years | 38.1 ± 13.7 | 39.4 ± 11.5 | .370 |
| Gender, male/female | 76/78 | 71/81 | .644 |
| Comorbidities | |||
| Diabetes mellitus, n (%) | 13 (8.4) | 17 (11.2) | .420 |
| Hypertension, n (%) | 24 (15.6) | 20 (13.2) | .545 |
| Cardiovascular disease, n (%) | 9 (5.8) | 6 (3.9) | .442 |
| Hyperlipidemia, n (%) | 8 (5.2) | 9 (5.9) | .782 |
| Asthma/COPD, n (%) | 7 (4.5) | 10 (6.6) | .437 |
| Laboratory examinations | |||
| WBC, ×103/µL | 5.64 (2.59‐11.56) | 5.48 (2.59‐13.08) | .530 |
| Neutrophil, ×103/µL | 3.05 (0.88‐8.67) | 3.25 (1.48‐8.85) | .081 |
| Lymphocyte, ×103/µL | 1.99 (0.48‐5.05) | 1.63 (0.50‐3.88) | .005 |
| Monocyte, ×103/µL | 0.59 (0.21‐1.50) | 0.48 (0.11‐1.62) | <.001 |
| Eosinophil, ×103/µL | 0.06 (0.0‐1.10) | 0.03 (0.0‐1.60) | <.001 |
| Platelet, ×103/µL | 242 (95‐422) | 230 (84‐590) | .333 |
| Urea, mg/dL | 29.2 ± 9.4 | 29.2 ± 9.2 | .974 |
| Creatinine, mg/dL | 0.88 ± 0.19 | 0.91 ± 0.21 | .330 |
| Prothrombin time, sn | 11.6 (9.6‐16) | 11.5 (10.1‐14.1) | .306 |
| INR | 1.0 (0.84‐1.35) | 1.0 (0.88‐1.17) | .762 |
| D‐dimer, µg/mL | 0.17 (0.15‐3.36) | 0.24 (0.11‐2.66) | .007 |
| CRP, mg/L | 2.03 (0.13‐39.71) | 6.24 (0.60‐202.6) | <.001 |
| Procalcitonin, ng/mL | 0.05 (0.02‐0.16) | 0.05 (0.02‐0.24) | .181 |
| NLR | 1.56 (0.39‐8.79) | 2.03 (0.59‐13.83) | <.001 |
| PLR | 126.9 (36.3‐650) | 145.2 (47.5‐478.2) | .010 |
| MLR | 0.28 (0.11‐1.73) | 0.29 (0.11‐1.13) | .830 |
| ELR | 0.03 (0.0‐0.76) | 0.02 (0.0‐1.09) | .001 |
| CLR | 0.97 (0.05‐21.32) | 4.14 (0.19‐181.18) | <.001 |
Abbreviations: CLR, C‐reactive protein/lymphocyte ratio; CRP, C‐reactive protein; ELR, Eosinophil/lymphocyte ratio; INR, International normalised ratio; MLR, Monocyte/lymphocyte ratio; NLR, Neutrophil/lymphocyte ratio; PLR, Platelet/lymphocyte ratio; WBC, White blood cell count.
In the correlation analysis of NLR, PLR, ELR and CLR indices with inflammatory biomarkers in the COVID‐19 patients, a positive correlation was observed between NLR, PLR, CLR values and CRP and procalcitonin. ELR value was negatively correlated with CRP (Table 2).
TABLE 2.
Correlation between derived blood lymphocyte parameters and inflammatory biomarkers in COVID‐19 patients
| NLR | PLR | ELR | CLR | |||||
|---|---|---|---|---|---|---|---|---|
| r | P value | r | P value | r | P value | r | P value | |
| CRP | 0.385 | <.001 | 0.245 | <.001 | −0.195 | 0.001 | 0.963 | <.001 |
| Procalcitonin | 0.238 | <.001 | 0.120 | .038 | −0.101 | 0.082 | 0.473 | <.001 |
Abbreviations: CLR, C‐reactive protein/lymphocyte ratio; CRP, C‐reactive protein; ELR, Eosinophil/lymphocyte ratio; MLR, Monocyte/lymphocyte ratio; NLR, Neutrophil/lymphocyte ratio; PLR, Platelet/lymphocyte ratio.
The AUC, cut‐off value, sensitivity and specificity of NLR, PLR, ELR and CLR are presented in Table 3. ROC curve analysis revealed that the cut‐off levels of NLR [AUC = 0.622 (0.565‐0.677), sensitivity = 62.5%, specificity = 60.4%], PLR [AUC = 0.585 (0.527‐0.641), sensitivity = 55.3%, specificity = 60.4%], ELR [AUC = 0.613 (0.556‐0.668), sensitivity = 48%, specificity = 72.7%] and CLR [AUC = 0.731 (0.677‐0.777), sensitivity = 81.5%, specificity = 55.6%] were 1.73, 139, 0.018 and 1.14, respectively. CLR index had significantly higher AUC value than NLR (z = 3.491, P = .001), PLR (z = 4.368, P = .000) and ELR (z = 2.876, P = .004) in distinguishing pneumonic COVID‐19 patients from patients with non‐pneumonic COVID‐19 infection. However, AUC values did not differ significantly among NLR, PLR and ELR (P > .05) (Figure 1).
TABLE 3.
ROC analysis results of the derived blood lymphocyte parameters
| AUC (95% CI) | Cut‐off level | Sensitivity (%) | Specificity (%) | P value | |
|---|---|---|---|---|---|
| NLR | 0.622 (0.565‐0.677) | 1.73 | 62.5 | 60.4 | <.001 |
| PLR | 0.585 (0.527‐0.641) | 139 | 55.3 | 60.4 | .009 |
| ELR | 0.613 (0.556‐0.668) | 0.018 | 48.0 | 72.7 | <.001 |
| CLR | 0.731 (0.677‐0.777) | 1.14 | 81.5 | 55.6 | <.001 |
Abbreviations: CLR, C‐reactive protein/lymphocyte ratio; ELR, Eosinophil/lymphocyte ratio; NLR, Neutrophil/lymphocyte ratio; PLR, Platelet/lymphocyte ratio.
FIGURE 1.
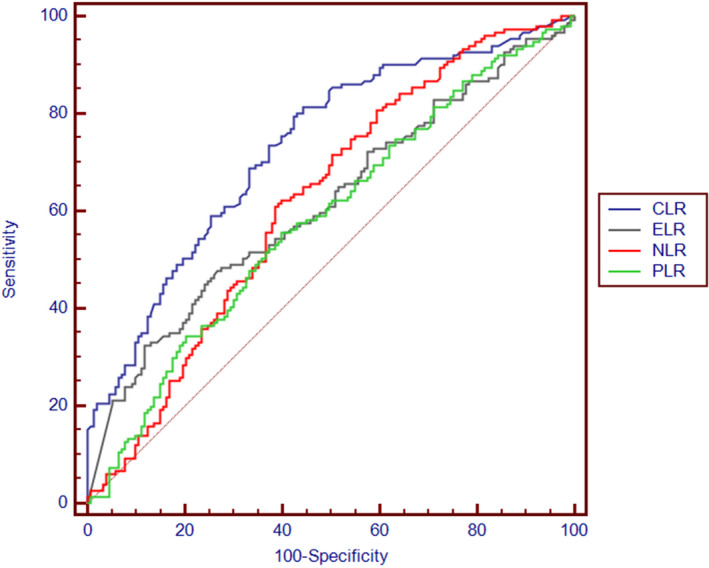
ROC curves of the derived blood lymphocyte parameters to predict COVID‐19 pneumonia
4. DISCUSSION
As far as we know, this is the first study that investigates the role of NLR, PLR, ELR and CLR indices as inflammatory biomarkers for differentiating COVID‐19 patients with and without pneumonia; and their correlation with CRP and procalcitonin. This study demonstrated that pneumonic COVID‐19 patients had significantly higher NLR, PLR and CLR values; and significantly lower ELR compared with the patients with non‐pneumonic COVID‐19 infection. There was a positive relationship between NLR, PLR, CLR and CRP and procalcitonin levels; and an inverse relationship between ELR and CRP levels. In addition, CLR was found to be more useful than other indices in identifying cases of COVID‐19 pneumonia.
An excessive and uncontrolled cytokine production plays an important role in the pathogenesis of COVID‐19 pneumonia. 14 The virus enters the alveolar cells via angiotensin converting enzyme 2 receptors 15 and triggers the release of inflammatory factors from the cells resulting in activation of macrophages in the alveolar tissue. 14 The inducing factors and chemokines released from macrophages cause the accumulation of mononuclear cells in the lung tissue. Extreme infiltration of inflammatory cells induces a cytokine storm leading to acute lung injury and ARDS, the severe consequences of COVID‐19 pneumonia. 14 , 16 Laboratory abnormalities such as decreased platelet, lymphocyte, monocyte and eosinophil counts; and increased neutrophil count and CRP level were reported in COVID‐19 patients. 17 , 18 , 19 , 20 WBC and its differential counts including lymphocytes, neutrophils, eosinophils and monocytes are associated with the inflammation and immune systems. 21 Platelets, which are anucleate blood cells produced from megakaryocytes in the bone marrow, play an important role in the host inflammatory and immune responses as well as regulation of hemostasis and thrombosis. 22 CRP, a positive acute phase protein, is one of the markers reflecting the systemic inflammatory response of body. 23 In the present study, we found that pneumonic COVID‐19 patients had lower lymphocyte, monocyte and eosinophil counts and higher neutrophil counts and CRP levels than non‐pneumonic group. Moreover, Li et al 24 reported that increased neutrophil percentage and CRP level and decreased lymphocyte and monocyte counts were closely related to the severity of COVID‐19 pneumonia. Xie et al 25 found that severe COVID‐19 patients had remarkably lower eosinophil counts compared with non‐severe COVID‐19 patients. Another study revealed that the increase in eosinophil count after admission may be a potential indicator of improvement in COVID‐19 patients. 26
Recently, NLR, PLR, MLR and CLR indices calculated from the abovementioned blood parameters have been extensively investigated for their role in assessing prognosis and severity of COVID‐19 infection. Yang et al 27 and Sun et al 28 examined some haematological indices in COVID‐19 patients and found that NLR, PLR and MLR values were significantly higher in severe patients than in non‐severe patients. Ding et al 29 found that NLR index positively correlated with the length of hospital stay and has a role in predicting the prognosis of disease for COVID‐19 patients. Another study reported an elevated NLR in non‐survivors than survivors and the magnitude of rising was correlated with severity of illness. 30 A recent meta‐analysis revealed that high NLR and low LCR (lymphocyte/CRP ratio) may be associated with poor prognosis in COVID‐19 patients. 11 In the present study, we determined the diagnostic values of NLR, PLR, CLR as well as ELR index for COVID‐19 pneumonia. In that regard, NLR, PLR, ELR and CLR indices and inflammatory markers were compared between pneumonic and non‐pneumonic COVID‐19 patients and ROC analysis of these indices were performed. We found that NLR, PLR and CLR values were higher and ELR values were lower in the pneumonic COVID‐19 patients compared with the non‐pneumonic patients and that there was a positive correlation between NLR, PLR, CLR indices and CRP and procalcitonin; and a negative correlation between ELR and CRP, suggesting these indices might be potential markers for diagnosis of COVID‐19 pneumonia. ROC analysis also showed that the AUC value of CLR (0.731) was higher than the NLR (0.622), PLR (0.585) and ELR (0.613) and there was no significant difference among NLR, PLR, and ELR, thus indicating that CLR was superior to NLR, PLR and ELR in identifying COVID‐19 pneumonia cases. Collectively, we speculated that these indices, particularly the CLR, could aid clinicians in early identification of patients with COVID‐19 pneumonia.
The major limitation of our study is that being conducted in a single center with retrospective design. Also, the time elapsed since the onset of symptoms, smoking habits and body mass index, which may affect laboratory indexes, could not be evaluated because of missing data.
5. CONCLUSION
Pneumonic COVID‐19 patients had significantly higher NLR, PLR and CLR; and significantly lower ELR compared with the non‐pneumonic patients. There was a positive correlation between NLR, PLR, CLR and CRP and procalcitonin levels; and an inverse correlation between ELR and CRP levels. Besides, ROC analysis indicated that the AUC of CLR was significantly greater than the AUCs of NLR, PLR and ELR, and therefore the predictive capacity of CLR for COVID‐19 pneumonia was better than the other three. Accordingly, CLR index could help clinicians to identify COVID‐19 patients with pneumonia.
ACKNOWLEDGEMENT
An oral presentation was made at the 9th Turkey EKMUD Scientific Platform held online from May 20 to 23, 2021.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest.
ETHICAL APPROVAL
This study was approved by the Harran University School of Medicine Ethics Committee Commission (Protocol number: HRU/20/15/09).
Damar Çakırca T, Torun A, Çakırca G, Portakal RD. Role of NLR, PLR, ELR and CLR in differentiating COVID‐19 patients with and without pneumonia. Int J Clin Pract. 2021;75:e14781. 10.1111/ijcp.14781
REFERENCES
- 1. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult in patients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID‐19) implicate special control measures. J Med Virol. 2020;92(6):568‐576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao W, Zhong Z, Xie X, Yu Q, Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID‐19) pneumonia: a multicenter study. Am J Roentgenol. 2020;214(5):1072‐1077. [DOI] [PubMed] [Google Scholar]
- 5. Poggiali E, Dacrema A, Bastoni D, et al. Can lung ultrasound help critical care clinicians in the early diagnosis of novel coronavirus (COVID‐19) pneumonia? Radiology. 2020;295(3):E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakajima K, Kato H, Yamashiro T, et al. COVID‐19 pneumonia: infection control protocol inside computed tomography suites. Jpn J Radiol. 2020;38(5):391‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Russell CD, Parajuli A, Gale HJ, et al. The utility of peripheral blood leucocyte ratios as biomarkers in infectious diseases: a systematic review and meta‐analysis. J Infect. 2019;78(5):339‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cunha BA, Connolly JJ, Irshad N. The clinical usefulness of lymphocyte:monocyte ratios in differentiating influenza from viral non‐influenza‐like illnesses in hospitalized adults during the 2015 influenza A (H3N2) epidemic: the uniqueness of HPIV‐3 mimicking influenza A. Eur J Clin Microbiol Infect Dis. 2016;35(1):155‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Curbelo J, Rajas O, Arnalich B, et al. Neutrophil count percentage and neutrophil‐lymphocyte ratio as prognostic markers in patients hospitalized for community‐acquired pneumonia. Arch Bronconeumol. 2019;55(9):472‐477. [DOI] [PubMed] [Google Scholar]
- 10. Peng J, Qi D, Yuan G, et al. Diagnostic value of peripheral hematologic markers for coronavirus disease 2019 (COVID‐19): a multicenter, cross‐sectional study. J Clin Lab Anal. 2020;34(10):e23475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lagunas‐Rangel FA. Neutrophil‐to‐lymphocyte ratio and lymphocyte‐to‐C‐reactive protein ratio in patients with severe coronavirus disease 2019 (COVID‐19): a meta‐analysis. J Med Virol. 2020;92(10):1733–1734. 10.1002/jmv.25819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qu R, Ling Y, Zhang YH, et al. Platelet‐to‐lymphocyte ratio is associated with prognosis in patients with coronavirus disease‐19. J Med Virol. 2020;92(9):1533‐1541. 10.1002/jmv.25767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen H, Chen S, Huang Z, et al. Relationship between blood parameters and Clonorchis sinensis infection: a retrospective single center study. Int Immunopharmacol. 2018;59:120‐126. [DOI] [PubMed] [Google Scholar]
- 14. Sun X, Wang T, Cai D, et al. Cytokine storm intervention in the early stages of COVID‐19 pneumonia. Cytokine Growth Factor Rev. 2020;53:38‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade‐long structural studies of SARS Coronavirus. J Virol. 2020;94(7):e00127‐e00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID‐19): a meta‐analysis. Clin Chem Lab Med. 2020;58(7):1021‐1028. [DOI] [PubMed] [Google Scholar]
- 18. Velavan TP, Meyer CG. Mild versus severe COVID‐19: laboratory markers. Int J Infect Dis. 2020;95:304‐307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kermali M, Khalsa RK, Pillai K, Ismail Z, Harky A. The role of biomarkers in diagnosis of COVID‐19—a systematic review. Life Sci. 2020;254:117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ponti G, Maccaferri M, Ruini C, Tomasi A, Ozben T. Biomarkers associated with COVID‐19 disease progression. Crit Rev Clin Lab Sci. 2020;57(6):389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leng SX, Xue QL, Tian J, Huang Y, Yeh SH, Fried LP. Associations of neutrophil and monocyte counts with frailty in community‐dwelling disabled older women: results from the Women's Health and Aging Studies I. Exp Gerontol. 2009;44(8):511‐516. [DOI] [PubMed] [Google Scholar]
- 22. van der Meijden PEJ, Heemskerk JWM. Platelet biology and functions: new concepts and clinical perspectives. Nat Rev Cardiol. 2019;16(3):166‐179. [DOI] [PubMed] [Google Scholar]
- 23. Yao Z, Zhang Y, Wu H. Regulation of C‐reactive protein conformation in inflammation. Inflamm Res. 2019;68:815‐823. [DOI] [PubMed] [Google Scholar]
- 24. Li K, Wu J, Wu F, et al. The clinical and chest CT features associated with severe and critical COVID‐19 pneumonia. Invest Radiol. 2020;55(6):327‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xie G, Ding F, Han L, Yin D, Lu H, Zhang M. The role of peripheral blood eosinophil counts in COVID‐19 patients. Allergy. 2021;76(2):471‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu F, Xu A, Zhang Y, et al. Patients of COVID‐19 may benefit from sustained Lopinavir‐combined regimen and the increase of Eosinophil may predict the outcome of COVID‐19 progression. Int J Infect Dis. 2020;95:183‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang AP, Liu JP, Tao WQ, Li HM. The diagnostic and predictive role of NLR, d‐NLR and PLR in COVID‐19 patients. Int Immunopharmacol. 2020;84:106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun S, Cai X, Wang H, et al. Abnormalities of peripheral blood system in patients with COVID‐19 in Wenzhou, China. Clin Chim Acta. 2020;507:174‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ding X, Yu Y, Lu B, et al. Dynamic profile and clinical implications of hematological parameters in hospitalized patients with coronavirus disease 2019. Clin Chem Lab Med. 2020;58(8):1365‐1371. [DOI] [PubMed] [Google Scholar]
- 30. Luo X, Zhou W, Yan X, et al. Prognostic value of C‐reactive protein in patients with COVID‐19. Clin Infect Dis. 2020;ciaa641. [DOI] [PMC free article] [PubMed] [Google Scholar]


