Abstract
Aims
Flavonoids and related compounds, such as quercetin‐based antiviral drug Gene‐Eden‐VIR/Novirin, inhibit the protease of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). The alkylated chalcones isolated from Angelica keiskei inhibit SARS‐CoV proteases. In this study, we aimed to compare the anti‐SARS CoV‐2 activities of both newly synthesized chalcone derivatives and these two drugs.
Methods
Determination of the potent antiviral activity of newly synthesized chalcone derivatives against SARS‐CoV‐2 by calculating the RT‐PCR cycling threshold (Ct ) values.
Results
Antiviral activities of the compounds varied because of being dose dependent. Compound 6, 7, 9, and 16 were highly effective against SARS‐CoV‐2 at the concentration of 1.60 µg/mL. Structure‐based virtual screening was carried out against the most important druggable SARS‐CoV‐2 targets, viral RNA‐dependent RNA polymerase, to identify putative inhibitors that could facilitate the development of potential anti‐coronavirus disease‐2019 drug candidates.
Conclusions
Computational analyses identified eight compounds inhibiting each target, with binding affinity scores ranging from −4.370 to −2.748 kcal/mol along with their toxicological, ADME, and drug‐like properties.
What’s known
Chalcone derivatives are the building blocks of pharmaceutical raw materials that have been used for a long time to treat various diseases.
Alkylated chalcones isolated from Angelica keiskei have been reported to inhibit SARS‐CoV proteases.
No potent drug or molecule has been found in the world that is specifically effective against SARS CoV‐2.
What’s new
In this study, various modifications have been made to the chalcone structure to design new and active compounds against SARS CoV‐2 and to reach target molecules.
It has been shown that eight new chalcone derivatives synthesized for the first time have very significant antiviral activity against SARS‐CoV‐2.
These molecules have been shown to be very promising syntheses in the treatment of Covid‐19 disease.
1. INTRODUCTION
Coronavirus disease‐2019 (COVID‐19) is a highly contagious disease caused by a novel virus identified as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). To date, there is no specific effective antiviral drug against SARS‐CoV‐2. The pandemic has adversely affected the global healthcare system. Treatment options for COVID‐19 can be categorized into four types: general treatment, coronavirus‐specific treatment, antiviral treatment, and other methods. 1 , 2 General treatment includes nutritional supplements, immune boosters, and some alternative medications in folk medicine. 3 Various treatment strategies based on the use of available drugs have been tried against SARS‐CoV‐2 globally, but no effective treatment protocol has been found. There are many drugs, such as chloroquine/hydroxychloroquine, favipiravir, ribavirin, remdesivir, or combinations of these drugs, which are found to be effective against SARS‐CoV‐2. 4 Although some studies initially reported that one or more of these drugs were effective against SARS‐CoV‐2 in some countries, later some scientific studies reported that these drugs did not have the expected level of effectiveness against SARS‐CoV‐2. 4 , 5
Previous treatments targeting SARS‐CoV and Middle East respiratory syndrome coronavirus may accelerate the development of the COVID‐19 treatment because of their structural and genomic similarities. Antiviral effective therapies act by arresting viral replication stages and the inactivation or inhibition of the enzymes or steps involved in these stages. One of the best characterized and conserved drug targets of coronaviruses is integrase, the main protease proteins. Inhibiting the activity of this enzyme would block viral replication, for which protease inhibitors are used. One of the most important targets that can inhibit viral replication against SARS‐CoV‐2 is the inactivation of the RNA‐dependent RNA polymerase (RdRp) enzyme. Inactivation of this enzyme prevents virus replication. Today, some of the available protease and RNA polymerase enzyme inhibitor drugs are being redesigned to treat SARS‐CoV‐2. 6 , 7 , 8 , 9
In cases where vaccines against SARS‐CoV‐2 may be insufficient, humanity's most important weapon would be an effective antiviral drug. For this reason, it is crucial to produce an effective antiviral drug against this highly contagious and deadly virus. Small modifications to the inhibitory ligand can significantly induce conformational changes in the molecules by changing the protein's binding modes and interactions to the active site and thus their activity.
In this context, some compounds that inhibit viral RNA polymerases in the literature were examined to find a scaffold, which would be active against COVID‐19. The scaffold was determined to show the best activity against COVID‐19. Various modifications were made to chalcone structure to design a newer, more active compound and reach the target molecules. A target molecule that could be effective against COVID‐19 was designed.
Chalcone derivatives are the building blocks of pharmaceutical raw materials that have long been used to treat various diseases, and medicinal plants are structural analogs of the flavonoids they carry. Flavonoids and related compounds, such as quercetin‐based antiviral drug Gene‐Eden‐VIR/Novirin, inhibit the protease of SARS‐CoV‐2. 10 Chalcones also exhibit inhibitory activity against tobacco mosaic virus, 11 HIV, 12 herpes simplex virus, 13 and dengue virus. 14 It also exhibits antioxidant, 15 antimicrobial, 16 antimycobacterial, 17 and antileishmanial 18 properties. Antiviral properties of chalcones have been determined in studies on plant viruses and human rhinoviruses. 19 Studies on hydroxy or methoxy‐containing chalcones have confirmed that these chalcone derivatives show antiviral properties depending on the positions of the OH/OMe groups and the nature of the chalcone skeleton. 20 Additionally, the alkylated chalcones isolated from Angelica keiskei inhibit SARS‐CoV proteases. 21
The current study aimed to determine the inhibitory activities of new eight chalcone derivatives against SARS‐CoV‐2 by calculating the SARS‐CoV‐2 RT‐PCR Cycling threshold (Ct ) values. Moreover, the toxicological, ADME, and drug‐like properties of the compounds were also determined by in silico method.
2. MATERIALS AND METHODS
2.1. Chemistry experimental procedure
2.1.1. General
All reagents used were commercially available unless otherwise specified, and all solvents were distilled before use. Melting points were measured with Gallenkamp (UK) melting point devices. IR Spectra: PerkinElmer Spectrum One FT‐IR spectrometer (PerkinElmer Life and Analytical Sciences, Beaconsfiled, UK). 1H‐ and 13C‐NMR Spectra: Varian 400 (Oxford Instruments, England) and Bruker 400 (Bruker Biospin AG, Fallanden, Switzerland) spectrometers. Elemental analysis results were obtained on a Leco CHNS‐932 instrument.
2.2. General procedure for preparation of compounds (6‐9)
To a solution of flouro/triflouro‐substituted acetophenone derivatives (2‐5) (1 mmol) in THF: H2O (5:1, 10 mL) 2,4,6‐trimethoxybenzaldehyde (1) (1,5 mmol) and LiOH.H2O (10 mmol) were added, and the resultant mixture was sequentially stirred overnight at room temperature. The solvent was evaporated in vacuum. HCl solution of 2 mol/L (5 mL) was added, and the crude material was extracted with ethyl acetate (EtOAc) (25 mL × 3). The combined extracts were dried over Na2SO4. The solvent was removed in vacuum and the remaining residue was purified via column chromatography over silicagel using gradient elution with EtOAc and hexanes to yield compound 6‐9.
2.2.1. (E)‐1‐(2,5‐Difluorophenyl)‐3‐(2,4,6‐trimethoxyphenyl)prop‐2‐en‐1‐one (6)
The above procedure was followed with 2,5‐difluoroacetophenone (2) to yield 6 as a yellow solid (61% yield). Rf (EtOAc/Hexanes 20:80) = 0.46; mp = 121‐123°C; IR (KBr, cm−1) νmax 2939, 2836, 1558, 1204; 1H NMR (400 MHz, CDCl3) δ 8.20 (d, J = 16.0 Hz, 1H), 7.70 (d, J = 16.0 Hz, 1H), 7.51‐7.43 (m, 1H), 7.19‐7.04 (m, 2H), 6.11 (s, 2H), 3.88 (s, 6H), 3.85 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 189.3, 163.7, 161.9, 158.6 (C‐5ʺ, d, J C,F = 243.5 Hz), 156.9 (C‐2ʺ, d, J C,F = 247.2 Hz), 137.3, 129.4 (C‐1ʺ, dd, J C,F = 16.2, 6.7 Hz), 124.8, 119.4 (C‐4ʺ, dd, J C,F = 24.5, 8.9 Hz), 117.7 (C‐3ʺ, dd, J C,F = 26.3, 7.8 Hz), 116.9 (C‐6ʺ, dd, J C,F = 24.4, 4.2 Hz), 106.3, 90.5, 55.8, 55.4; Anal. calcd for C18H16F2O4: C, 64.67; H, 4.82; Found: C, 64.71; H, 4.72.
2.2.2. (E)‐1‐(3,5‐Difluorophenyl)‐3‐(2,4,6‐trimethoxyphenyl)prop‐2‐en‐1‐one (7)
The above procedure was followed with 3,5‐difluoroacetophenone (3) to yield 7 as a yellow solid (65% yield). Rf (EtOAc/Hexanes 20:80) = 0.5; mp = 173‐175°C; IR (KBr, cm−1) νmax 3084, 2942, 1563, 1121; 1H NMR (400 MHz, CDCl3) δ 8.29 (d, J = 16.0 Hz, 1H), 7.75 (d, J = 16.0 Hz, 1H), 7.57‐7.45 (m, 2H), 7.03‐6.93 (m, 1H), 6.14 (s, 2H), 3.92 (s, 6H), 3.87 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 189.4, 163.7, 162.9 (C‐3ʺ, dd, J C,F = 236.1, 12.0 Hz), 162.0, 142.5 (C‐1ʺ), 137.6, 120.6, 111.3 (C‐2ʺ, dd, J C,F = 18.7, 6.9 Hz), 107.0 (C‐4ʺ), 106.4, 90.6, 55.9, 55.4; Anal. calcd for C18H16F2O4: C, 64.67; H, 4.82; Found: C, 64.79; H, 4.75.
2.2.3. (E)‐1‐(2‐Fluoro‐5‐(trifluoromethyl)phenyl)‐3‐(2,4,6‐trimethoxyphenyl)prop‐2‐en‐1‐one (8)
The above procedure was followed with 2‐fluoro‐5‐trifluoromethylacetophenone (4) to yield 8 as a yellow solid (70% yield). Rf (EtOAc/Hexanes 20:80) = 0.5; mp = 143‐145°C; IR (KBr, cm−1) νmax 2945, 2313, 1566, 1333; 1H NMR (400 MHz, CDCl3) δ 8.22 (d, J = 15.6 Hz, 1H), 8.11‐8.04 (m, 1H), 7.79‐7.65 (m, 2H),7.32‐7.23 (m, 1H), 6.14 (s, 2H), 3.91 (s, 6H), 3.89 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 189.4, 163.8, 162.0, 159.7 (C‐2ʺ, J C,F = 304.7 Hz), 137.8, 129.7, 128.7, 127.5, 126.8, 124.8 (C‐4ʺ, J C,F = 6.1 Hz), 122.2, 117.2 (C‐3ʺ, J C,F = 24.5 Hz), 106.2, 90.5, 55.9, 55.4; Anal. calcd for C19H16F4O4: C, 59.38; H, 4.20; Found: C, 59.49; H, 4.22.
2.2.4. (E)‐1‐(3,5‐Bis(trifluoromethyl)phenyl)‐3‐(2,4,6‐trimethoxyphenyl)prop‐2‐en‐1‐one (9)
The above procedure was followed with 3,5‐bistrifluoromethylacetophenone (5) to yield 9 as a yellow solid (73% yield). Rf (EtOAc/Hexanes 20:80) = 0.6; mp = 193‐195°C; IR (KBr, cm−1) νmax 2994, 2302, 1561, 1278; 1H NMR (400 MHz, CDCl3) δ 8.43 (bs, 2H), 8.34 (d, J = 15.6 Hz, 1H), 8.05 (bs, 1H), 7.83 (d, J = 15.6 Hz, 1H), 6.17 (s, 2H), 3.95 (s, 6H), 3.90 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 189.4, 164.0, 162.1, 140.9, 138.8, 132.0, 131.7, 128.5, 125.1, 120.3, 106.2, 90.6, 55.9, 55.5; Anal. calcd for C20H16F6O4: C, 55.31; H, 3.71; Found: C, 55.33; H, 3.75.
2.3. Synthesis procedure for 1‐(3‐bromo‐2‐hydroxy‐4,6‐dimethoxyphenyl)ethan‐1‐one (11)
To a solution of ammonium cerium (IV) nitrate (1 eq.) and LiBr (1 eq.) in CH3CN (2.5 mL/1 mmol of substrate) was added 2‐hydroxy‐4,6‐dimethoxyacetophenone (10) (1 eq.). The mixture was stirred for 6 hours at room temperature. After 6 hours, the reaction was monitored by TLC and stopped with H2O (25 mL), extracted with DCM (3 × 25 mL). The combined extracts were dried over Na2SO4. The solvent was removed under reduced pressure to afford 1‐(3‐bromo‐2‐hydroxy‐4,6‐dimethoxyphenyl)ethan‐1‐one (11) as a light pink solid (91%). The 1H NMR and 13C NMR spectra are in agreement with the reported data. 22
2.4. General procedure for preparation of Compounds (13‐16)
To a solution of 1‐(3‐bromo‐2‐hydroxy‐4,6‐dimethoxyphenyl)ethan‐1‐one (11) (1 eq.) in MeOH (20 mL/1.1 mmol of substrate) benzaldehyde derivatives (2‐4, and 12) (1,6 eq.) and 50% aqueous KOH solution (7 mL/1.1 mmol of substrate) were added sequentially stirred for 15 hours at room temperature. After 15 hours solvent was evaporated. NH4Cl solution (20 mL/1.1 mmol of substrate) was added, and crude product was extracted with DCM (25 mL/1.1 mmol of substrate × 3). The combined extracts were dried over Na2SO4. The solvent was removed in vacuum.
2.4.1. (E)‐1‐(3‐Bromo‐2‐hydroxy‐4,6‐dimethoxyphenyl)‐3‐(2,5‐difluorophenyl)prop‐2‐en‐1‐one (13)
The above procedure was followed with 2,5‐difluorobenzaldehyde (2) to yield 13 as a yellow solid (61% yield). Rf (EtOAc/Hexanes 20:80) = 0.43; mp = 219‐221°C; IR (KBr, cm−1) νmax 3076, 1631, 1585, 720; 1H NMR (400 MHz, DMSO) δ 13.83 (bs, 1H), 7.89 (d, J = 15.8 Hz, 1H), 7.71‐7.59 (d, 2H), 7.40‐7.27 (m, 2H), 6.42 (s, 1H), 4.00 (s, 6H); 13C NMR (100 MHz, DMSO, 55°C) δ 192.6, 162.6, 161.6, 161.3 (C‐2ʹ, d, J C,F = 259.3 Hz), 160.2 (C‐5ʹ, d, J C,F = 270.6 Hz), 157.7, 133.8, 131.5, 124.5 (C‐1ʹ, dd, J C,F = 19.9, 8.2 Hz), 119.1 (C‐3ʹ, dd, J C,F = 24.7, 8.8 Hz), 118.3 (C‐4ʹ, dd, J C,F = 25.0, 9.0 Hz), 115.9 (C‐6ʹ, d, J C,F = 25.0 Hz), 107.3, 91.2, 89.7, 57.4, 57.2; Anal. calcd for C17H13BrF2O4: C, 51.15; H, 3.28; Found: C, 51.24; H, 3.31.
2.4.2. (E)‐1‐(3‐Bromo‐2‐hydroxy‐4,6‐dimethoxyphenyl)‐3‐(3,5‐difluorophenyl)prop‐2‐en‐1‐one (14)
The above procedure was followed with 3,5‐difluorobenzaldehyde (3) to yield 14 as a yellow solid (70% yield). Rf (EtOAc/Hexanes 20:80) = 0.5; mp = 221‐223°C; IR (KBr, cm−1) νmax 3061, 1631, 1585, 720; 1H NMR (400 MHz, DMSO) δ 13.87 (bs, 1H), 7.82 (d, J = 15.6 Hz, 1H), 7.62 (d, J = 15.6 Hz, 1H), 7.55‐7.49 (m, 2H), 7.36‐7.28 (m, 1H), 6.41 (s, 1H), 3.98 (d, 6H); 13C NMR (100 MHz, DMSO, 55°C) δ 193.0, 162.8, 162.7, 163.3 (C‐3ʹ, dd, J C,F = 244.6, 13.4 Hz), 161.6, 140.6, 139.2, 130.6, 112. 1 (C‐2ʹ, d, J C,F = 25.7), 107.4, 106.2 (C‐4ʹ), 91.1, 89.7, 57.6, 57.5; Anal. calcd for C17H13BrF2O4: C, 51.15; H, 3.28; Found: C, 51.25; H, 3.40.
2.4.3. (E)‐1‐(3‐Bromo‐2‐hydroxy‐4,6‐dimethoxyphenyl)‐3‐(2‐fluoro‐5‐(trifluoromethyl)phenyl)prop‐2‐en‐1‐one (15)
The above procedure was followed with 2‐fluoro‐5‐trifluoromethylbenzaldehyde (4) to yield 15 as a yellow solid (45% yield). Rf (EtOAc/Hexanes 20:80) = 0.56; mp = 206‐208°C; IR (KBr, cm−1) νmax 3332, 1630, 1524, 726; 1H NMR (400 MHz, DMSO) δ 8.14 (s, 1H), 7.91 (d, J = 16 Hz, 1H), 7.87‐7.77 (m, 1H), 7.65 (d, J = 16.0 Hz, 1H), 7.59‐7.47 (m, 1H), 6.33 (s, 1H), 3.97 (s, 3H), 3.95 (s, 3H); 13C NMR (100 MHz, DMSO, 55°C) δ 192.5, 162.4, 163.1 (C‐2ʹ, d, J C,F = 256 Hz), 162.0, 161.9, 132.9, 132.5, 129.3, 128.5, 127.8, 126.5, 124.4 (C‐1ʹ, d, J C,F = 12.1 Hz), 117.9 (C‐3ʹ, d, J C,F = 23.6 Hz), 108.1, 91.96, 88.9, 57.3, 57.1; Anal. calcd for C18H13BrF4O4: C, 48.13; H, 2.92; Found: C, 48.17; H, 3.05.
2.4.4. (E)‐1‐(3‐Bromo‐2‐hydroxy‐4,6‐dimethoxyphenyl)‐3‐(2,4,5‐trifluorophenyl)prop‐2‐en‐1‐one (16)
The above procedure was followed with 2,4,5‐trifluorobenzaldehyde (12) to yield 16 as a yellow solid (60% yield). Rf (EtOAc/Hexanes 20:80) = 0.46; mp = 209‐211°C; IR (KBr, cm−1) νmax 3022, 1638, 1316, 869; 1H NMR (400 MHz, DMSO) δ 13.9 (bs, 1H), 8.01‐7.96 (m, 1H), 7.86 (d, J = 16 Hz, 1H), 7.71‐7.63 (m, 1H), 7.60 (d, J = 16 Hz, 1H), 6.40 (s, 1H), 3.97 (6H); 13C NMR (100 MHz, DMSO, 55°C) δ 192.7, 162.7, 162.8, 161.9, 157.2 (C‐2ʹ, ddd, J C,F = 249.0, 11.1, 2.4 Hz), 151.0 (C‐5ʹ, dd, J C,F = 238.4, 13.7 Hz), 147.2 (C‐4ʹ, ddd, J C,F = 257.2, 12.1, 3.0 Hz), 133.4, 131.1, 120.3 (C‐1ʹ), 117.9 (C‐6ʹ, dd, J C,F = 20.2, 3.5 Hz), 107.6 (C‐3ʹ, dd, J C,F = 28.9, 21.5 Hz), 107.2, 91.1, 89.7, 57.6, 57.4; Anal. calcd for C17H12BrF3O4: C, 48.94; H, 2.90; Found: C, 49.01; H, 2.95.
2.5. Biological part
2.5.1. Viral propagation
The SARS‐CoV‐2 Hatay strain (SARS‐CoV‐2, Hatay, 2020), isolated by Professor Dr Nizami Duran, was used in the study. The SARS‐CoV‐2 strain was propagated in the Vero E6 cell line (African green monkey kidney cells), obtained from the American Type Culture Collection (ATCC C1008). After 7 days of incubation at 37°C in an atmosphere of 5% CO2, the supernatant was harvested. All necessary permissions have been obtained for the present study.
Vero E6 cell line was used for viral isolation. Cell cultures were maintained with 10% fetal calf serum in RPMI‐1640 (Sigma‐Aldrich, USA) medium containing 10 mM HEPES, 4 IM glutamine, and 100 IU/mL penicillin/streptomycin. Incubation of cells was carried out at 37°C in an incubator with 95% air and 5% CO2. Cell density was adjusted to 1 × 105 cells/mL. Experiments were performed by adding cell maintenance medium to 10% of the culture dish in culture dishes. Incubation of cell cultures continued for 96 hours. Cells were removed from the culture vessels with 0.25% trypsinization solution and collected in 50 mL centrifuge tubes by centrifugation at 300 g for 10 minutes at the end of incubation. Cell number and viability were determined by hemocytometer with 1% trypan blue dye prepared in 0.9% NaCl.
Viral growth was confirmed both by determining the presence of cytopathological effect (CPE) and by the real‐time PCR method. The formation of CPE was detected on the 4th day in the Vero cell line.
2.5.2. Proliferation experiments
DMSO was used to dissolve the chemical compounds at a concentration nontoxic to Vero cells. Proliferation experiments in cell culture were performed on 24‐well flat‐bottom microplates. The wells were prepared with RPMI‐1640 medium containing 10% fetal calf serum at a density of 1 × 105 cells/mL.
2.5.3. Titration of the virus
Stock viral strains (maintained at −80°C) were rapidly dissolved in 37°C water (in a bain‐marie) and were re‐activated in cell lines (Vero E6 cell line). The cells were then exploded by “freezing and thawing.” Cells were collected from the culture dish and centrifuged at 1200 g for 20 minutes, and the supernatant virus was collected as a stock solution. Like in our previous study in which TCID50 of the virus was calculated; in the present study, three different titers of SARS‐CoV‐2 (1, 10, and 100 TCID50) were chosen to be studied. 23
2.6. Activity studies
2.6.1. 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide method
This method is based on the principle of the cleavage of the 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) tetrazolium ring in active mitochondria. In this method, cultured live cells can be detected colorimetrically and quantitatively. The MTT method, first described by Mosmann and later developed, is a practical method used to determine cell viability. 24
3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide is a substance that is actively absorbed into cells and is reduced to colored water‐insoluble form as a result of its activity in mitochondria. The MTT reduction feature of cells is considered a measure of cell viability. The density of the dye obtained as a result of MTT analysis correlates with the number of live cells. This method was used to detect the presence of living cells to demonstrate the effectiveness of the chemical compounds at different concentrations against SARS‐CoV‐2. As viral replication would decrease the live‐cell ratio, a detected decrease in cell viability was indicated as increased viral reproduction.
In the MTT assay, the decrease in cell viability was associated with increased viral replication, whereas high cell viability was considered as an indicator of antiviral activity. For this purpose, seawater at different concentrations was added to the SARS‐CoV‐2 inoculated cell lines and was incubated for 96 hours in an incubator with 5% carbon dioxide at 37°C. MTT, 10 µL, was added to each well, and the plates were incubated for 4 hours under the same conditions. Absorbance measurements were made at 570 nm on a spectrophotometer.
2.7. Real‐time PCR
Severe acute respiratory syndrome coronavirus 2 replication in cell culture was verified by the real‐time PCR method. Compared with the control group using the real‐time PCR method, the presence or increase of viral replication was described as an increased number of viral copies. Viral RNA extraction was performed using a commercial kit under the commercial kit procedure. In the studies, the SYBR‐Green RT‐qPCR method was performed in accordance with the QuantiTect SYBR‐Green PCR kit (Qiagen, Germany) test procedure. The PCR cycle starts with an initial denaturation at 95°C for 15 minutes, followed by three steps of 45 cycles of 94°C for 15 seconds, annealing at 60°C for 30 seconds and elongation at 72°C for 30 seconds, and a final extension temperature of 72°C for 10 minutes. The Human β‐actin gene (QuantiTect Primer Assays; Qiagen) was used as the reference gene. All reactions were tested in triplicate at different intervals. 25
2.8. Cycle threshold value
In isolation, “Ct ” values provide a relative measure of the viral quantity in the sample. Different known quantities of standards were included in the same study and tested in parallel with viral cultures. Thus, the “Ct ” value could potentially provide some measure of viral copies. 26 , 27 An 8‐ to 10‐fold dilutions series was prepared from the culture filtrate for SARS‐CoV‐2. Five replicates of each dilution were tested in parallel. All replicates were detected at dilutions of 10−1 to 10−5. 28
2.9. Molecular docking studies
Molecular modeling studies were conducted using Maestro 11.8 (Schrödinger, LLC, New York, NY, USA) software. All ligands were drawn using the 2D Skether module of the Maestro 11.8. Possible ionization states and tautomers of the ligands were determined by LigPrep (Schrödinger, LLC). Then, all ligands were optimized using the conjugate gradient method with force field parameters MacroModel and OPLS3 (Schrödinger, LLC). 29 After the proteins (PDB ID: 6M71, 6YB7, and 6LZG) were downloaded from the RCSB Protein Database (www.rcsb.org), 30 this version of Protein Preparation Wizard was used for docking studies (Epik, Prime, Impact: Schrödinger, LLC). 31 In this process, unwanted structures in proteins were removed, hydrogen atoms and missing side‐chain atoms were added, partial charges and bonds were assigned, ionization states, hydrogen, and disulfide bonds were formed. Active regions of proteins were determined for docking, and grid maps of proteins were created using Maestro's receptor grid generation panel. Ligands were docked on these maps 100 times in extra precision mode using Glide (Schrödinger, LLC) software. The poses obtained from docking were examined, and appropriate docking scores were determined, as given in Table 1. 32
TABLE 1.
Docking scores in kcal/mol for molecules in target proteins
| Compound | 6M71 | 6YB7 | 6LGZ |
|---|---|---|---|
| 6 | −4.370 | −3.953 | −4.127 |
| 7 | −3.921 | −3.674 | −3.543 |
| 8 | −3.328 | −3.555 | −3.876 |
| 9 | −3.855 | −3.427 | −3.467 |
| 13 | −3.266 | −2.748 | −2.974 |
| 14 | −3.462 | −2.810 | −2.772 |
| 15 | −3.342 | −2.773 | −3.666 |
| 16 | −3.768 | −3.370 | −3.405 |
2.9.1. Statistical analysis
All data were evaluated by the statistical tests (ie χ2, paired t‐test, and independent sample t‐test) using Statistical Package for Social Sciences (SPSS for Windows. 18.0, Chicago, IL, USA) software. P‐values < .05 were considered to be statistically significant.
3. RESULTS
The synthesis of the target molecules (6‐9) was accomplished in one step using the Claisen–Schmidt condensation reaction, as illustrated in Figure 1. The corresponding chalcones were obtained in high purity and good yields by treating 2,4,6‐trimethoxy benzaldehyde (1) with fluoro and trifluoromethyl‐substituted acetophenone derivatives (2‐5) in an aqueous alkali medium. LiOH was the preferred alkaline base in the series. The reason for that nucleophilic aromatic substitution product is also obtained in reactions carried out in methanol and in a strongly basic medium, especially if the aromatic benzene ring contains fluorine. As a result of the fluorine atom's substitution by the methoxy group, methoxy chalcone structures arise as a by‐product. To prevent the result from affecting the reaction efficiency, LiOH was used as an alkaline base. Tetrahydrofuran:water mixture (THF:H2O) was determined as the reaction solvent. 33
FIGURE 1.
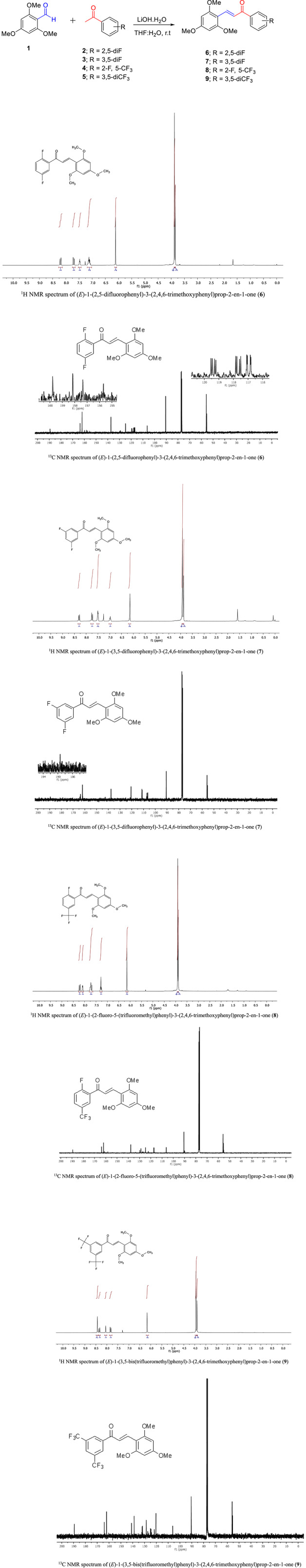
Preparation of fluoro/trifluoromethyl‐substituted trimethoxy chalcones (6‐9)
The synthesis of the second group of compounds (13‐16) was accomplished in two steps using bromination of compound 10 followed by the Claisen–Schmidt condensation reaction, as shown in Figure 2. By treating 2,4,6‐trimethoxyacetophenone (10) with LiBr and ammonium cerium (IV) nitrate in CH3CN, the corresponding compound 11 was obtained in high yield. Subsequently, target compounds (13‐16) were obtained in high purity and good yield by the Claisen–Schmidt reaction of compound 11 with the corresponding benzaldehydes in an aqueous alkali medium (2‐4, and 12).
FIGURE 2.
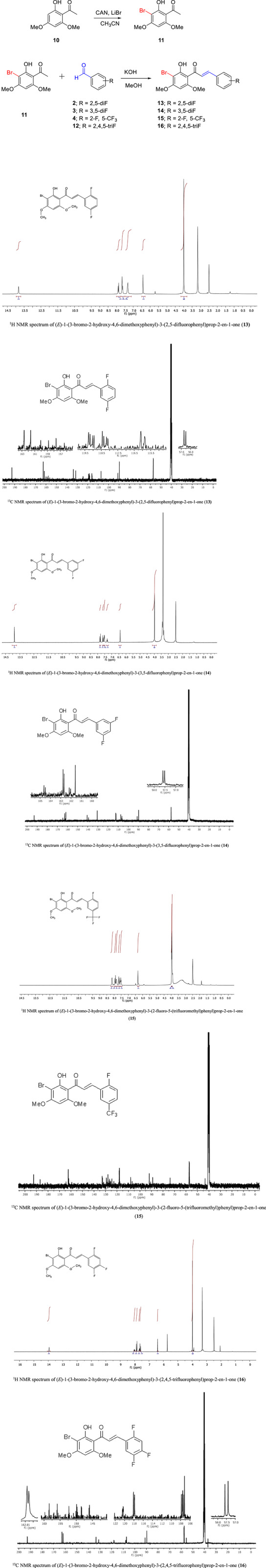
Preparation of bromo/fluoro/trifluoromethyl‐substituted chalcones (13‐16)
Antiviral effects of all newly synthesized compounds against SARS‐CoV‐2 were evaluated in comparison with the virus control group. The highest non‐toxic concentrations of the compounds on Vero cells were selected for antiviral efficacy studies. The antiviral effects of three different concentrations of chalcone derivatives have been investigated.
In the study, viral growth in the Vero E6 cell line was detected by CPE in the cells from the 4th day of incubation, as shown in Figure 3. Activity studies in cell culture were performed after 96 hours of incubation. The characteristic CPE of SARS‐CoV‐2 was not detected in cell cultures in which viral reproduction was inhibited. Morphological analysis was performed by recording the CPE rate in the cells, especially the presence/absence of CPE.
FIGURE 3.
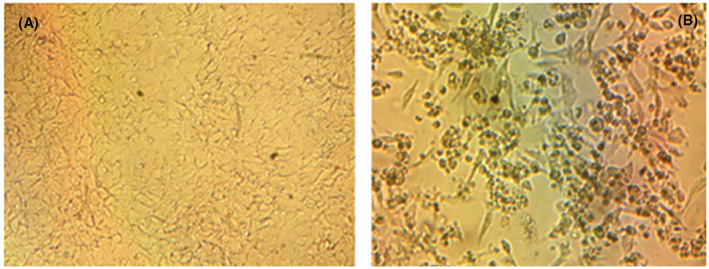
Cytopathological effects of the SARS‐CoV‐2 in Vero E6 cells compared to the control group. A, Control group, (B) cytopathological changes caused by SARS‐CoV‐2. A, Image of Vero cells at the end of the 96th hours of incubation (RPMI‐1640 medium contained 1% fetal calf serum). B, Picture of Vero cells infected with SARS CoV‐2 at the end of 96th hours of incubation (RPMI‐1640 medium contained 100 TCID50 virus and 1% fetal calf serum). A, B, Under of inverted microscope at 100‐fold magnification. SARA‐CoV‐2, severe acute respiratory syndrome coronavirus 2
The “Ct ” values give a relative measure of the viral copy number in the sample compared with the curves using the standards. Different known quantities of standards were included in the same study and tested in parallel with viral cultures. Therefore, the “Ct ” value was evaluated as the potential viral (SARS‐CoV‐2) copy number. In this study, the inhibitory activities of new chalcone derivatives against SARS‐CoV‐2 were determined by calculating the SARS‐CoV‐2 RT‐PCR “Ct ” values. 26 , 27
It has been reported that low Ct values (high viral load) can be considered an indicator of severe disease and high contagiousness. In contrast, high Ct values (low viral load) indicate a decrease in both the risk of infection and contagiousness. 34
As shown in Figure 4, the efficacy of compounds 6‐9, 14, and 16 on SARS‐CoV‐2 at three different concentrations (0.40, 0.80, and 1.60 µg/mL) was compared with the virus control group in terms of “Ct ” values. When the efficacy of the newly synthesized compounds was compared with the virus control group (no drugs or chemicals included), the compounds have been found to exhibit significant antiviral activity against SARS CoV‐2. It was determined that the antiviral activities of the newly synthesized compounds were statistically significantly higher than the virus control group.
FIGURE 4.
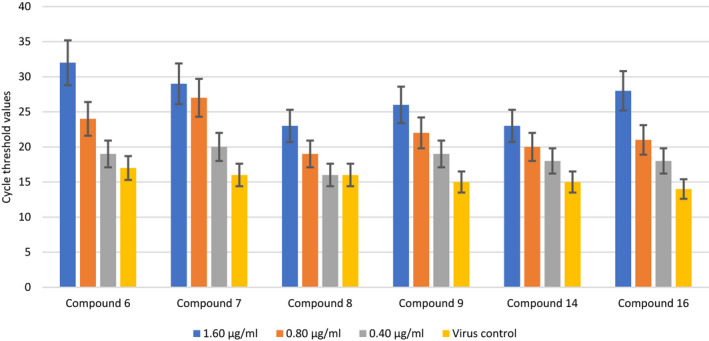
The effect of compounds 6, 7, 8, 9, 14, and 16 at three different concentrations on severe acute respiratory syndrome coronavirus 2 replication. P values: C6 (1,60) & VC: P < .01; C6 (0,80) & VC: P < .05; C6 (0,40) & VC: P > .05; C7 (1,60) & VC: P < .01; C7 (0,80) & VC: P < .01; C7 (0,40) & VC: P > .05; C8 (1,60) & VC: P < .05; C8 (0,80) & VC: P > .05; C8 (0,40) & VC: P > .05; C9 (1,60) & VC: P < .05; C9 (0,80) & VC: P < .05; C9 (0,40) & VC: P < .05; C14 (1,60) & VC: P < .01; C14 (0,80) & VC: P > .05; C14 (0,40) & VC: P > .05; C16 (1,60) & VC: P < .01; C16 (0,80) & VC: P < .05; C16 (0,40) & VC: P < .05
The antiviral activities of compounds 6‐9, 14, and 16 at high concentrations (1.6 and 0.8 μg/mL) were statistically significantly higher compared with the virus control group. No statistically significant difference was found when the antiviral activity of compound 8 at a concentration of 0.4 µg/mL was compared with the virus control group. The antiviral activity of compound 8 was lower than that of the other compounds.
There was also a dose‐dependent change in the antiviral efficacy of the compounds. While the highest efficacy was determined at a concentration of 1.60 µg/mL, it was evinced that antiviral activity decreased at a concentration of 0.8 and 0.4 µg/mL. Comparing the antiviral activities of the compounds at different concentrations, compound 6 > 7 > 16 > 9 > 14 > 8 at 1.60 μg/mL; compound 7 > 6 > 9 > 16 > 14 > 8 at 0.80 μg/mL; compound 7 > 6 = 9 > 16 = 14 > 8 at 0.40 μg/mL ranking has been determined. After evaluating the results, it was found that the antiviral activities of 6, 7, 9 and 16 of the first group compounds were remarkable. Hence it was noted that compounds 6, 7, 9, 14, and 16 exhibited statistically significantly higher antiviral activities than compound 8 against SARS‐CoV‐2 at all concentrations.
The antiviral effects of the second group of compounds (13 and 15) at three different concentrations (2.0, 1.0, 0.5 μg/mL) have been examined. As shown in Figure 5, there was no significant difference between the 2.0 and 1.0 µg/mL concentrations of compound 13 against SARS‐CoV‐2. Compound 13 was found to exhibit a dose‐dependent antiviral activity. However, at a concentration of 0.5 µg/mL, antiviral efficacy was not different from the virus control group. Also, the antiviral efficacy of compound 15 was statistically significantly higher than the virus control group at all three tested concentrations. It was determined that the antiviral efficacy of compound 15 at a concentration of 2 µg/mL against SARS‐CoV‐2 was not significantly different from that of a concentration of 1 µg/mL. It was also noted that the compound exhibited a dose‐dependent antiviral activity like other compounds.
FIGURE 5.
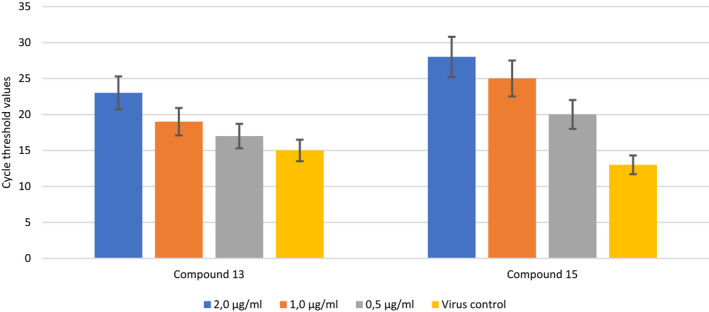
The effect of compounds 13 and 15 at three different concentrations on severe acute respiratory syndrome coronavirus 2 replication. P values: C13 (2,0) & VC: P < .05; C13 (1,0) & VC: P > .05; C13 (0,5) & VC: P > .05; C15 (2,0) & VC: P < .01; C15 (1,0) & VC: P < .01; C15 (0,5) & VC: P < .05
In this study, eight new chalcone derivative compounds, expected to be effective against SARS‐CoV2, have been synthesized. Molecular docking studies examine the binding of these compounds to target proteins and their interactions with essential residues in the proteins’ active regions at the atomic level. Also, the structure–activity relationships (SAR) in this group of compounds were analyzed by comparing docking scores with in vitro results, as given in Table 1.
The primary targets of drugs developed against SARS‐CoV2 are specified as Mpro, RdRp, and spike proteins in the literature. In silico studies have investigated the SARS‐CoV2 inhibition potential of newly synthesized compounds using the target proteins. 35 In this context, 6M71 PDB‐encoded protein (SARS‐CoV‐2 RdRp in complex with cofactors) was used for RdRp, 6YB7 PDB‐encoded protein (SARS‐CoV‐2 main protease with unliganded active sites) for Mpro, and 6LZG PDB‐encoded protein (Structure of novel coronavirus spike receptor‐binding domain complexed with its receptor angiotensin‐converting enzyme 2 [ACE2]) was used for spike protein. The interactions of synthesized compounds and reference molecules with target proteins are given in Table 2.
TABLE 2.
Interactions of compounds with active sites of the target proteins
| Entry | Interactions Type | 6M71 | 6YB7 | 6LGZ |
|---|---|---|---|---|
| 6 | H‐bound | Ser814 | Lys5 | Arg403, Gly496 |
| Hydrophobic | Ala762, Leu758, Cys813, Cys799, Trp800, Trp617, Tyr619 | Leu286, Tyr126 | Tyr495, Tyr505, Phe497, Tyr453, Ala387, Pro389, Phe390 | |
| Charged (−) | Asp618, Asp761, Asp760, Glu811 | Glu288, Asp289, Glu290, Asp197 | Asp38, Glu37, Glu35, Glu406 | |
| Charged (+) | Lys798 | Arg131, Lys137 | Lys353, Arg393 | |
| Polar | Ser139, Gln127 | Ser494, Gln493, His34, Asn33, Gln389 | ||
| 7 | H‐bound | Ser814 | Lys5 | Arg403 |
| Hydrophobic | Ala762, Cys813, Cys622, Tyr619, Trp800, Trp617, Tyr619 | Leu286, Cys128, Ala129 | Tyr495, Tyr505, Phe497, Tyr453, Ala387, Pro389, Phe390 | |
| Charged (−) | Asp618, Asp761, Asp760, Glu811 | Glu288, Asp289, Glu290, Asp197 | Asp38, Glu37, Glu35, Glu406 | |
| Charged (+) | Lys798 | Arg131, Lys137 | Arg393 | |
| Polar | His810 | Ser284 | Ser494, Gln493, His34, Asn33, Gln389 | |
| 8 | H‐bound | |||
| Hydrophobic | Ala762, Phe812, Cys813, Pro620 Trp800, Trp617, Tyr619, Val763 | Leu286, Leu287, Cys128 | Tyr505, Ala387, Ala386, Phe356, Val503 | |
| Charged (−) | Asp618, Asp761, Asp760, Glu811 | Glu288, Asp289, Glu290 | Asp405 | |
| Charged (+) | Lys798, Lys621 | Lys137, Lys5 | Arg403, Arg393 | |
| Polar | Gln127, Ser284, Thr199 | His34, Thr324 | ||
| 9 | H‐bound | |||
| Hydrophobic | Ala762, Leu758, Cys813, Phe812 Trp800, Trp617, Tyr619, Val763 Cys622, Pro620 | Leu286, Tyr126, Leu287, Tyr237, Tyr239 | Tyr505, Ala387, Ala387, Tyr453 | |
| Charged (−) | Asp618, Asp761, Asp760, Glu811 | Glu288, Asp289, Glu290, Asp197 | Glu37, Glu35, Glu406 | |
| Charged (+) | Lys798, Lys621 | Arg131, Lys137, Lys5 | Arg403, Arg408, Lys353, Lys417 | |
| Polar | Asn238, Thr199, Thr198 | His34 | ||
| 13 | H‐bound | Trp800 | ||
| Hydrophobic | Cys813, Cys799, Phe812, Trp800, Trp617, Ala797 | Leu286, Leu287, Tyr239 | Tyr505, Ala387, Tyr453, Tyr495 | |
| Charged (−) | Asp618, Asp761, Glu811 | Glu288, Asp289, Glu290, | Glu37, Asp38, Glu406, Asp405 | |
| Charged (+) | Lys798 | Arg131, Lys137 | Arg403, Lys353, Lys414 | |
| Polar | Ser814, His810 | Ser284, Thr199 | His34, Gln409 | |
| 14 | H‐bound | |||
| Hydrophobic | Cys799, Tyr619, Phe812, Trp800, Trp617, Ala797 | Leu286, Leu287, Tyr239 | Tyr505, Ala387, Tyr453, Tyr495 | |
| Charged (−) | Asp618, Asp761, Glu796, Glu811 | Glu288, Asp289, Glu290 | Glu37, Asp38, Glu406, Asp405 | |
| Charged (+) | Lys798 | Arg131, Lys137 | Arg403, Arg393, Lys417 | |
| Polar | Ser284, Thr199 | His34, Asn33, Ser494 | ||
| 16 | H‐bound | Ser814 | Asp289 | Gly496, Arg403 |
| Hydrophobic | Leu758, Phe812, Cys813, Trp800, Trp617, Tyr619 | Leu286, Leu287, Thr126, Cys128 | Tyr505, Ala387, Tyr453, Tyr495 | |
| Charged (−) | Asp618, Asp761, Asp760, Glu811 | Glu288, Glu290 | Glu37, Asp38, Glu406 | |
| Charged (+) | Lys798 | Arg131, Lys137, Lys5 | Arg403, Lys353, Arg393 | |
| Polar | Ser139, Gln127 | His34, Asn33, Ser494 | ||
| 15 | H‐bound | Ser814 | Lys5 | Arg403 |
| Hydrophobic | Cys813, Cys799, Trp800, Phe812, Ala797 | Leu286, Leu287, Ala285, Leu282 | Tyr505, Ala387, Ala386, Phe356 | |
| Charged (−) | Asp618, Asp761, Glu811 | Glu288, Asp289, Glu290 | Glu37, Asp405, Glu406 | |
| Charged (+) | Lys798 | Lys137, Arg131 | Arg403, Lys353, Arg393, Arg408, Lys417 | |
| Polar | His810 | Thr199 | His34, Gln409 |
The replication mechanism of SARS‐CoV‐2 is mainly governed by RdRp, the nsps 12, 8, and 7 complexes. 36 This 6M71 PDB‐encoded RdRp protein is mainly extracted from SARS‐CoV2. 37 It has been reported that Ser759, Asp760, Asp761, Asp618, residues located in the catalytic region of the protein, are required for replication. 9 , 37 The docking scores of the compounds varied between −4,340 and −3,266 kcal/mol. As a result of docking studies, compound 6 made a hydrogen bond with Ser814; a charged (negative) interaction with Asp760, Asp761, Asp618, and its binding affinity was determined at −4,370 kcal/mol, as shown in Figure 6. It has been reported that quercetin is a potential inhibitor of the 6M71‐encoded protein. In the docking study in extra precision mode, the docking score of quercetin was calculated as −6,108 kcal/mol. 38
FIGURE 6.
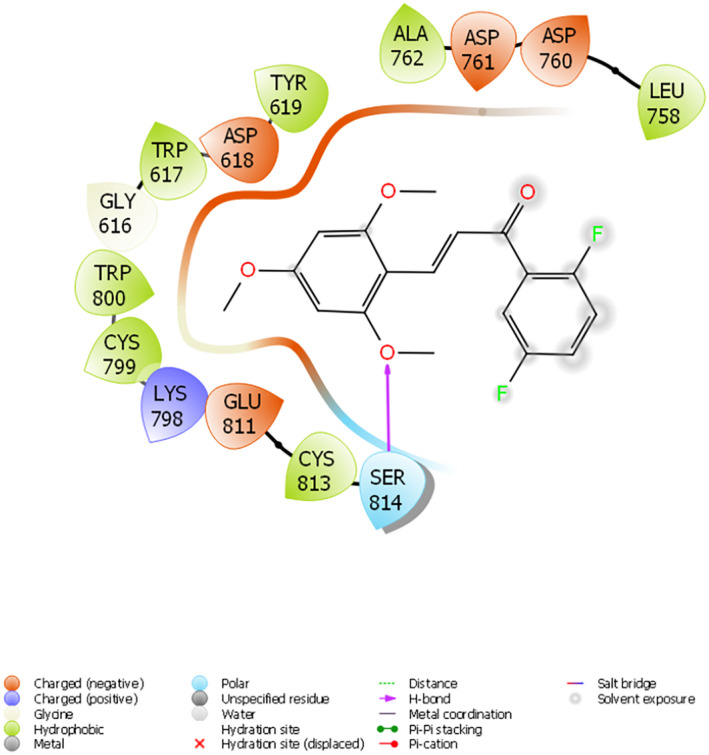
Interaction of compound 6 with the active site of 6M71
Viral RNAs were converted into various polyproteins with the effect of Mpro of SARS‐CoV2. Moreover, 6YB7 PDB‐encoded protein was used as a target protein in docking studies. The docking scores of the compounds ranged from −3,953 to −2,748 kcal/mol. It has been reported that Lys5, Ala285, Leu286, Glu288, Asp289, Glu290, located in the catalytic region of the protein, were responsible for the disruption of Mpro. 35 Compound 6 with the highest activity and docking score of −3,953 kcal/mol made hydrogen bond with Lys5; hydrophobic interaction with Leu286; and a charged (negative) interaction with lu288, Asp289, Glu290, as shown in Figure 7. Remdesivir and ritonavir are known to be a potential inhibitor of Mpro and interacts well with 6YB7‐encoded protein. Docking scores of remdesivir and ritonavir were calculated as −5,317 and 5.098 kcal/mol respectively in the docking study in extra precision mode. 39
FIGURE 7.
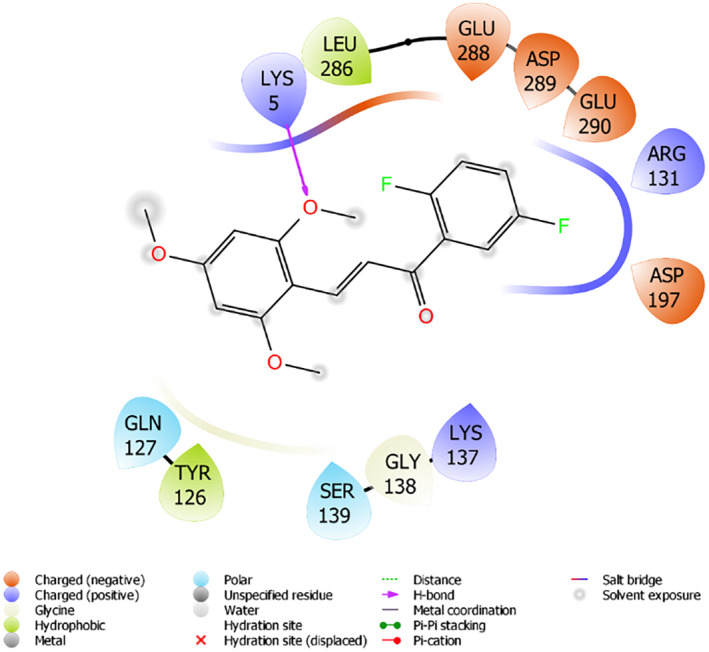
Interaction of compound 6 with the active site of 6YB7
The spike (S) protein, one of the target proteins for SARS‐CoV2, gives the virus its typical shape, contains a specific binding site that allows it to attach to the host cell. It attacks ACE‐2 receptors in the host cell. 6LZG PDB‐encoded protein was used as the target protein. The residues in the protein's catalytic region have been reported as Arg403, Tyr453, Gly496, Gln498, Asn501, Gly502, Tyr503, Tyr505. The docking scores of the compounds ranged from −4.127 to −2.772 kcal/mol. In addition to having a high docking score of −4.127 kcal/mol, compound 6 made a hydrogen bond with Arg403 and Gly496, as shown in Figure 6. It has been reported that lopinavir and kaempferol are a potential inhibitor of the 6LZG‐encoded protein and interacts with the spike protein. The docking scores of lopinavir and kaempferol were calculated as −5,708 kcal/mol and 5,266 kcal/mol respectively in the extra precision mode docking study. 40
Docking studies conducted for different target proteins are in harmony with biological activity studies. Compound 6 (Ct value: 32), the compound with the best activity against SARS‐CoV‐2, also has the highest docking scores in all target proteins. The most active compound concentration of compound 6 was 4.78 µM. These results show that the efficacy of compound 6 against SARS‐CoV‐2 is remarkable. Molecular docking studies also support these activity results.
Besides bioactivity studies and molecular docking studies, the physicochemical and ADMET properties of the compounds are critical in terms of drug‐likelihood. Many compounds with significant biological activity do not make it to clinical trials because of their unsuitable physicochemical and toxicological properties. These properties of the compounds are described in QikProp (Maestro, Schrodinger 11.8), Datawarrior v4.07.02, and pre‐ADMET software (https://preadmet.bmdrc.kr). 41 The compounds’ toxicological evaluation shows that none of them has the potential to be carcinogenic or mutagenic. Also, the compounds exhibit medium‐level ability to inhibit the hERG potassium channel and CYP450. For ADMET properties, it has been shown that the properties of all compounds are suitable. In addition, all of the synthesized compounds comply with Lipinski's rule of five. Details of the physicochemical and ADMET properties of the compounds are given in Tables 3 and 4, respectively.
TABLE 3.
Physicochemical properties
| Comp. | mol MW | SASA | FOSA | FISA | PISA | WPSA | Mol Vol | donorHB | accptHB | logP | PSA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 334.319 | 611.988 | 278.05 | 44.731 | 196.544 | 92.663 | 1052.629 | 0 | 4.25 | 4.347 | 46.299 |
| 6 | 334.319 | 618.356 | 290.337 | 39.884 | 203.367 | 84.769 | 1054.929 | 0 | 4.25 | 4.368 | 46.378 |
| 8 | 384.327 | 660.507 | 290.005 | 39.861 | 175.101 | 155.54 | 1136.17 | 0 | 4.25 | 4.042 | 46.338 |
| 9 | 434.334 | 674.253 | 274.353 | 45.126 | 132.87 | 221.904 | 1197.133 | 0 | 4.25 | 4.790 | 47.399 |
| 13 | 399.188 | 605.368 | 189.772 | 63.966 | 198.165 | 153.465 | 1037.028 | 0 | 3.25 | 3.744 | 55.99 |
| 14 | 399.188 | 607.615 | 193.461 | 64.012 | 187.997 | 162.145 | 1039.649 | 0 | 3.25 | 3.744 | 55.999 |
| 16 | 417.179 | 613.568 | 189.77 | 63.966 | 164.586 | 195.246 | 1052.298 | 0 | 3.25 | 3.845 | 55.989 |
| 15 | 449.196 | 651.589 | 201.216 | 65.711 | 159.479 | 225.183 | 1123.502 | 0 | 3.25 | 4.492 | 57.717 |
Determined by QikProp (Maestro, Schrodinger 11.8).
Abbreviations: accpt HB, estimated number of hydrogen bonds that would be accepted by the solute from water molecule in an aqueous solution; Donor HB, estimated number of hydrogen bonds that would be donated by the solute to water molecule in an aqueous solution; FISA, hydrophilic component of SASA; FOSA, hydrophobic component of SASA; LogP, predicted octanol/water partition coefficient; mol MW, molecular weight of molecule; Mol Vol, total solvent‐accessible volume in cubic angstroms using a probe with a 1.4 Å radius; PISA, Pi (carbon and attached hydrogen) component of SASA; PSA, polar surface area; SASA, total solvent accessible surface area in square angstroms using a probe with a 1.4 Å radius; WPSA, weakly polar component of the SASA (halogen, P and S). Total solvent‐accessible volume in cubic angstroms using a probe with a 1.4 Å radius.
TABLE 4.
Some predicted toxicological, ADME, and drug‐like properties
| Parameters | 7 | 6 | 8 | 9 | 13 | 14 | 16 | 15 |
|---|---|---|---|---|---|---|---|---|
| Toxicological | ||||||||
| Irritant a | None | None | None | None | None | None | None | None |
| Reproductive effects a | None | None | None | None | None | None | None | None |
| Carcinogenic a | None | None | None | None | None | None | None | None |
| Mutagenic a | None | None | None | None | None | None | None | None |
| hERG inhibition b | Medium risk | Medium risk | Medium risk | Medium risk | Medium risk | Medium risk | Medium risk | Medium risk |
| CYP450 inhibition b | 2C19, 2C9, 3A4 | 2C19, 2C9, 3A4 | 2C19, 2C9, 3A4 | 2C19,2C9, 3A4 | 2C19, 2C9, | 2C19, 2C9, | 2C19, 2C9, | 2C19, 2C9, |
| ADME | ||||||||
| Blood–brain barrier b | Low absorption | Low absorption | Middle absorption | Middle absorption | Middle absorption | Middle absorption | Middle absorption | High absorption |
| Human intestinal absorption b | Well absorbed | Well absorbed | Well absorbed | Well absorbed | Well absorbed | Well absorbed | Well absorbed | Well absorbed |
| Plasma protein binding b | Strongly bound | Strongly bound | Strongly bound | Strongly bound | Strongly bound | Strongly bound | Strongly bound | Strongly bound |
| Caco2 permeability b | Middle permeability | Middle permeability | Middle permeability | Middle permeability | Middle permeability | Middle permeability | Middle permeability | Middle permeability |
| Drug like | ||||||||
| Drug‐likeness score a | −1,155 | −1,155 | −7,005 | −7,005 | −2,911 | −2,911 | −2,911 | −8,761 |
| CMC‐like rule b | Qualified | Qualified | Qualified | Qualified | Qualified | Qualified | Qualified | Qualified |
| Lead‐like rule b | Violated | Violated | Violated | Violated | Violated | Violated | Violated | Violated |
| Rule of five b | Suitable | Suitable | Suitable | Suitable | Suitable | Suitable | Suitable | Suitable |
| WDI‐like rule b | Suitable | Suitable | Suitable | Out of %90 cutoff | Suitable | Suitable | Suitable | Out of %90 cutoff |
Determined by datawarrior v4.07.02.
Determined by pre‐admet (https://preadmet.bmdrc.kr).
4. DISCUSSION
Coronavirus disease‐2019 is a highly contagious viral disease caused by coronavirus SARS‐CoV‐2, belonging to the beta coronavirus family. 42 Currently, there is no effective specific antiviral treatment option for SARS‐CoV‐2. However, several studies for using existing antivirals in isolation or combination and developing new therapeutic drugs to treat COVID‐19 are underway. 43 More extensive studies are needed to determine the reliability and effectiveness of these new drug candidates and to guide clinical decisions. Today, COVID‐19 is a global health problem worldwide and is a serious and potential threat to human health. It is crucial to develop new protection strategies against SARS‐CoV‐2 and discover new active molecules against this virus. Studies are conducted on different structures; among them, one is chalcone scaffold. 44 , 45 Chalcones are bioactive scaffolds of great medicinal interest because of their various pharmacological and biological activities to provide a valuable scaffold for discovering new drug leads. The current study aimed to determine the inhibitory activities of new chalcone derivatives against SARS‐CoV‐2 by calculating the “Ct ” values of SARS‐CoV‐2 by RT‐PCR method.
In the study, the antiviral activities of new chalcone derivatives were evaluated by calculating their “Ct ” values compared with that of the virus control group. In addition, the morphological appearance of the cells in each group and the viability rates of the collected cells were also evaluated. Morphological evaluation by microscopy and cell viability can also be considered a verification of “Ct ” values. In cultures with a low “Ct ” value (high viral load), intense CPE was detected in cells, and low cell viability was interpreted as an indicator of high viral reproduction. In cultures with a high “Ct ” value (low viral load), the absence of CPE or the presence of minimal CPE in cells and high cell viability rates were interpreted as a low viral titer. Finally, in the experiments, it was indicated that compound 15 and all other compounds, especially the compounds 6, 7, and 9 showed remarkable antiviral activity against SARS‐CoV‐2. The SAR indicated that the flouro at differnt position of chalcone played a significant role on the antiviral activity. Multiple fluoro substituents (positions 2, 4 and 5) play an important role in the activity.
Unfortunately, currently, there is no effective drug against SARS‐CoV‐2. For most people, the disease may cause mild illness; however, it can cause severe morbidity and even mortality in some people. Although there is no clear explanation for this situation, it is reported to be closely associated with host immunity.
The present study found that the newly synthesized chalcone derivatives showed a significant level of antiviral activity. The results indicated that the antiviral activities of the synthesized compounds were dose dependent. Also, compound 6, 15, and 16 have been found to possess potent activity against SARS‐CoV‐2 at the concentrations of 1.60, 2.0, and 1.60 µg/mL, respectively.
5. COCNLUSIONS
Chalcone derivatives were also analyzed in silico for their pharmacokinetic properties and were validated as having drug‐like nature. This library was considered for tackling the modern‐day issue of SARS‐CoV‐2 and further tested against homology modeled Mpro, RdRp, and S proteins. Considering the potential efficacy of these newly synthesized compounds against SARS‐CoV‐2, we believe that these molecules may be the potentially effective drug candidates. However, further studies are needed to test the effectiveness of these molecules by in vivo tests.
DISCLOSURE
ND, MFP, DAA, MAA, EA, FC, ET, BA, SB and O.A declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
ND and OA: Planning and design of the study, DAA, MAA, BA and S.B: Syntheses and in‐silico studies, EA, FC and ET: In‐vitro activity studies.
Duran N, Polat MF, Aktas DA, et al. New chalcone derivatives as effective against SARS‐CoV‐2 agent. Int J Clin Pract. 2021;75:e14846. 10.1111/ijcp.14846
Nizami Duran, Serdar Burmaoglu and Oztekin Algul contributed equally.
Funding information
No sources of funding were used to assist in the preparation of this study.
Contributor Information
Nizami Duran, Email: nizamduran@hotmail.com.
Serdar Burmaoglu, Email: sboglu@atauni.edu.tr.
Oztekin Algul, Email: oztekinalgul@mersin.edu.tr.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
REFERENCES
- 1. Lu H. Drug treatment options for the 2019‐new coronavirus (2019‐nCoV). Biosci Trends. 2020;14(1):69‐71. [DOI] [PubMed] [Google Scholar]
- 2. Simsek S, Unal YA. Antiviral treatment of covid‐19. Turk J Med Sci. 2020;50:611‐619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farooq SS, Ngaini Z. Natural and synthetic drugs as potential treatment for coronavirus disease 2019 (COVID‐2019). Chem Afr. 2020;4:1‐13. [Google Scholar]
- 4. Ayten O, Ozdemir C, Akturk U, Sen N. Potential treatment of COVID‐19. Eurasian J Pulmonol. 2020;22:35‐44. [Google Scholar]
- 5. Drożdżal S, Rosik J, Lechowicz K, et al. FDA approved drugs with pharmacotherapeutic potential for SARS‐CoV‐2 (COVID‐19) therapy. Drug Resist Updat. 2020;53:100719. doi: 10.1016/j.drup.2020.100719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280.e8. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID‐19 associated pneumonia in clinical studies. Biosci Trends. 2020;14(1):72‐73. [DOI] [PubMed] [Google Scholar]
- 8. Jean SS, Lee PI, Hsueh PR. Treatment options for COVID‐19: the reality and challenges. J Microbiol Immunol Infect. 2020;53(3):436‐443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yin W, Mao C, Luan X, et al. Structural basis for inhibition of the RNA‐dependent RNA polymerase from SARS‐CoV‐2 by remdesivir. Science. 2020;368(6498):1499‐1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Polansky H, Lori G. Coronavirus disease 2019 (COVID‐19): first indication of efficacy of Gene‐Eden‐VIR/Novirin in SARS‐CoV‐2 infection. Int J Antimicrob Agents. 2020;55(6):105971. doi: 10.1016/j.ijantimicag.2020.105971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang XU, Shijun SU, Chen M, et al. Novel chalcone derivatives containing a 1,2,4‐triazine moiety: design, synthesis, antibacterial and antiviral activities. RSC Adv. 2019;9:6011‐6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu JH, Wang XH, Yi YH, Lee KH. Anti‐AIDS agents 54. A potent anti‐HIV chalcone and flavonoids from genus Desmos. Bioorg Med Chem Lett. 2003;13:1813‐1815. [DOI] [PubMed] [Google Scholar]
- 13. Hassan ST, Masarčíková R, Berchová K. Bioactive natural products with anti‐herpes simplex virus properties. J Pharm Pharmacol. 2015;67(10):1325‐1336. [DOI] [PubMed] [Google Scholar]
- 14. Kiat TS, Pippen R, Yusof R, Ibrahim H, Khalid N, Rahman NA. Inhibitory activity of cyclohexenyl chalcone derivatives and flavonoids of fingerroot, Boesenbergia rotunda (L.), towards dengue‐2 virus NS3 protease. Bioorg Med Chem Lett. 2006;16(12):3337‐3340. [DOI] [PubMed] [Google Scholar]
- 15. Biradar JS, Sasidhar BS, Parveen R. Synthesis, antioxidant and DNA cleavage activities of novel indole derivatives. Eur J Med Chem. 2010;45:4074‐4078. [DOI] [PubMed] [Google Scholar]
- 16. Burmaoglu S, Algul O, Gobek A, et al. Design of potent fluoro‐substituted chalcones as antimicrobial agents. J Enzyme Inhib Med Chem. 2017;32(1):490‐495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chiaradia LD, Martins PG, Cordeiro MN, et al. Synthesis, biological evaluation, and molecular modeling of chalcone derivatives as potent inhibitors of Mycobacterium tuberculosis protein tyrosine phosphatases (PtpA and PtpB). J Med Chem. 2012;55(1):390‐402. [DOI] [PubMed] [Google Scholar]
- 18. de Mello MVP, Abrahim‐Vieira BA, Domingos TFS, et al. A comprehensive review of chalcone derivatives as antileishmanial agents. Eur J Med Chem. 2018;150:920‐929. [DOI] [PubMed] [Google Scholar]
- 19. Nowakowska Z. A review of anti‐infective and anti‐inflammatory chalcones. Eur J Med Chem. 2007;42:125‐137. [DOI] [PubMed] [Google Scholar]
- 20. Onyilagha JC, Malhotra B, Elder M, French CJ, Towers GHN. Comparativestudies of inhibitory activities of chalcones on tomato ringspot virus (ToRSV). J Indian Dent Assoc. 1997;19:133‐137. [Google Scholar]
- 21. Park JY, Ko JA, Kim DW, et al. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS‐CoV. J Enzyme Inhib Med Chem. 2016;31(1):23‐30. [DOI] [PubMed] [Google Scholar]
- 22. Cechinel‐Filho V, Vaz Z, Zunino L, Calixto J, Yunes R. Synthesis of xanthoxyline derivatives with antinociceptive and antioedematogenic activities. Eur J Med Chem. 1996;31(10):833‐839. [Google Scholar]
- 23. Allahverdiyev A, Duran N, Ozguven M, Koltas S. Antiviral activity of the volatile oils of Melissa officinalis L. against Herpes simplex virus type‐2. Phytomedicine. 2004;11:657‐661. [DOI] [PubMed] [Google Scholar]
- 24. Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1–2):55‐63. [DOI] [PubMed] [Google Scholar]
- 25. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keyaerts E, Vijgen L, Maes P, Duson G, Neyts J, Van Ranst M. Viral load quantitation of SARS‐coronavirus RNA using a one‐step real‐time RT‐PCR. Int J Infect Dis. 2006;10(1):32‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tom MR, Mina MJ. To interpret the SARS‐CoV‐2 test, consider the cycle threshold value. Clin Infect Dis. 2020;71(16):2252‐2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Buchan BW, Windham S, Balada‐Llasat JM, et al. Practical comparison of the biofire filmarray pneumonia panel to routine diagnostic methods and potential impact on antimicrobial stewardship in adult hospitalized patients with lower respiratory tract infections. J Clin Microbiol. 2020;58(7):e00135‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Algul O, Ersan RH, Alagoz MA, Duran N, Burmaoglu S. An efficient synthesis of novel di‐heterocyclic benzazole derivatives and evaluation of their antiproliferative activities. J Biomol Struct Dyn. 2020;8:1‐13. doi: 10.1080/07391102.2020.1803966 [DOI] [PubMed] [Google Scholar]
- 30. Alagoz MA. New molecule design with in‐silico methods for Covid‐19 treatment. Bioorg Med Chem Rep. 2020;3:32‐40. [Google Scholar]
- 31. Temirak A, Shaker YM, Ragab FA, et al. Synthesis, biological evaluation and docking studies of new 2‐furylbenzimidazoles as antiangiogenic agents. Eur J Med Chem. 2014;87:868‐880. [DOI] [PubMed] [Google Scholar]
- 32. Ersan RH, Alagoz MA, Dogen A, Duran N, Burmaoglu S, Algul O. Bisbenzoxazole derivatives: design, synthesis, in vitro antimicrobial, antiproliferative activity, and molecular docking studies. Polycycl Aromat Compd. 2020. doi: 10.1080/10406638.2020.1852589 [DOI] [Google Scholar]
- 33. Anil DA, Polat MF. Synthesis and characterization of some novel fluoro and methoxy substituted chalcone derivatives via nucleophilic aromatic substitution. Org Commun. 2020;13:1‐9. [Google Scholar]
- 34. Singanayagam A, Patel M, Charlett A, Lopez Bernal J, Saliba V. Gopal, duration of infectiousness and correlation with RT‐PCR cycle threshold values in cases of COVID‐19, England, January to May 2020. Eurosurveillance. 2020;25(32):2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vijayakumar BG, Ramesh D, Joji A, Prakasan J, Kannan T. In silico pharmacokinetic and molecular docking studies of natural flavonoids and synthetic indole chalcones against essential proteins of SARS‐CoV‐2. Eur J Pharmacol. 2020;886:173448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jácome R, Becerra A, Ponce de León S, Lazcano A. Structural analysis of monomeric RNA‐dependent polymerases: evolutionary and therapeutic implications. PLoS One. 2015;10(9):e0139001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao Y, Yan L, Huang Y, et al. Structure of the RNA‐dependent RNA polymerase from COVID‐19 virus. Science. 2020;368(6492):779‐782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Da Silva FM, Da Silva KP, De Oliveira LP, et al. Flavonoid glycosides and their putative human metabolites as potential inhibitors of the SARS‐CoV‐2 main protease (Mpro) and RNA‐dependent RNA polymerase (RdRp). Mem Inst Oswaldo Cruz. 2020;115:e200207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adegbola PI, Fadahunsi OS, Adegbola AE, Semire B. In silico studies of Potency and safety assessment of selected trial drugs for the treatment of COVID‐19. In Silico Pharm. 2021;45:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adejoro IA, Babatunde DD, Tolufashe GF. Molecular docking and dynamic simulations of some medicinal plants compounds against SARS‐CoV‐2: an in‐silico study. J Taibah Univ Sci. 2020;14:1563‐1570. [Google Scholar]
- 41. Ersan RH, Alagoz MA, Ertan‐Bolelli T, Duran N, Burmaoglu S, Algul O. Head‐to‐head bisbenzazole derivatives as antiproliferative agents: design, synthesis, in‐vitro activity, and SAR analysis. Mol Divers. 2020. doi: 10.1007/s11030-020-10115-0 [DOI] [PubMed] [Google Scholar]
- 42. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID‐19) outbreak‐an update on the status. Mil Med Res. 2020;7(1):11. doi: 10.1186/s40779-020-00240-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS‐CoV‐2 and discovery of its inhibitors. Nature. 2020;582(7811):289‐293. [DOI] [PubMed] [Google Scholar]
- 45. Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS‐CoV‐2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.


