Abstract
The development of medical imaging artificial intelligence (AI) systems for evaluating COVID‐19 patients has demonstrated potential for improving clinical decision making and assessing patient outcomes during the recent COVID‐19 pandemic. These have been applied to many medical imaging tasks, including disease diagnosis and patient prognosis, as well as augmented other clinical measurements to better inform treatment decisions. Because these systems are used in life‐or‐death decisions, clinical implementation relies on user trust in the AI output. This has caused many developers to utilize explainability techniques in an attempt to help a user understand when an AI algorithm is likely to succeed as well as which cases may be problematic for automatic assessment, thus increasing the potential for rapid clinical translation. AI application to COVID‐19 has been marred with controversy recently. This review discusses several aspects of explainable and interpretable AI as it pertains to the evaluation of COVID‐19 disease and it can restore trust in AI application to this disease. This includes the identification of common tasks that are relevant to explainable medical imaging AI, an overview of several modern approaches for producing explainable output as appropriate for a given imaging scenario, a discussion of how to evaluate explainable AI, and recommendations for best practices in explainable/interpretable AI implementation. This review will allow developers of AI systems for COVID‐19 to quickly understand the basics of several explainable AI techniques and assist in the selection of an approach that is both appropriate and effective for a given scenario.
Keywords: AI, COVID‐19, deep learning, explainability, interpretability
1. INTRODUCTION
Over the past decade, machine intelligent techniques that attempt to emulate human information processing and decision making have experienced a strong emergence in medicine. 1 , 2 , 3 , 4 These techniques span a variety of medical tasks, including computer‐aided diagnosis (CAD) systems and drug discovery, providing augmented information to improve patient management and clinical outcomes. 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 However, a key obstacle in applying artificial intelligence (AI) systems is the lack of transparency in technology contributing to critical decisions. 11 , 12 , 13 Because of the perception of AI methods as “black box” algorithms, which require little or no explicit human intervention, it can be difficult to ethically justify their use in high‐stake decisions, especially because this type of technique lends little indication of when it is likely to fail. 11 , 12 , 13 , 14 , 15 Thus, the investigation of methods that can explain why an AI system provided a particular prediction is critically important.
In medical imaging, machine learning (ML) is typically applied to improve medical image assessment and workflow, including CAD, automatic image segmentation, and image review scheduling (e.g., triaging high‐priority images that require immediate attention). 3 , 4 , 5 , 6 , 7 , 8 , 9 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 The choice in ML method is dictated by the imaging task, which then influences which interpretability techniques may be appropriate. In this paper, we review several approaches to providing interpretable radiological AI systems.
The terms “explainability” and “interpretability” have been increasingly discussed in the AI community, particularly as they pertain to AI performance and ethics, and have raised several important questions. 27 , 28 , 29 Will radiologists more heavily weigh AI output with improved interpretability? Can the incorporation of explainable techniques also benefit model performance? Who is responsible when inappropriate decisions are made based on AI output? These and similar questions have instigated several attempts to define “explainability” and “interpretability” in AI; however, many definitions have considerable overlap or clash. 27 , 28 , 29 In our review, “explainability” refers to techniques applied by a developer to explain and improve the AI system, while “interpretability” refers to understanding algorithm output for end‐user implementation. Questions portraying the intended meaning of each term are given in Figure 1.
FIGURE 1.
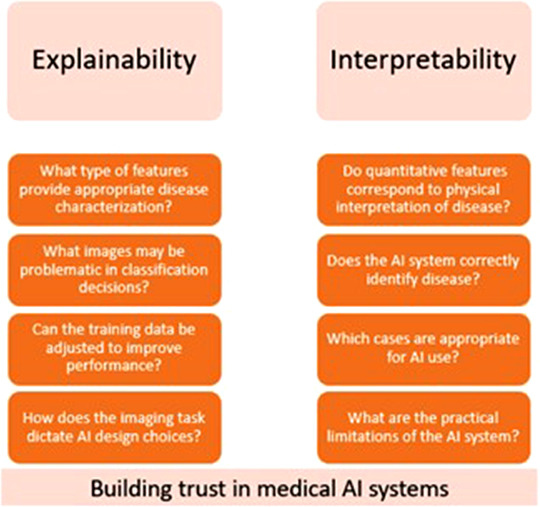
Examples of questions regarding “explainability” and “interpretability” as defined in this review. While the two imply similar meaning, the intended audience and implementation of the model output is different between the two
Several groups have provided excellent surveys of explainable AI and visualization. 11 , 28 , 30 , 31 , 32 , 33 , 34 However, such reviews tended to focus on more general problems in both medical and nonmedical disciplines (e.g., nonimage assessments), whereas our review prioritizes rapid, seamless implementation and discussion from a practical perspective for both radiology developer and end‐user standpoints. In Section 2, we define several relevant medical imaging tasks, then proceed in Section 3 with a brief discussion of general methods. In Sections 4, 5, 6, we categorize techniques into three groups, providing brief discussion of their function, advantages, and disadvantages, as well as example applications from developer and end‐user perspectives. Finally, we discuss appropriate metrics for explainable AI evaluation in Section 7, applications to problems specific to COVID‐19 in Section 8, and provide recommendations for use in Section 9.
2. MEDICAL TASKS OF INTEREST
The most common task in medical imaging AI is disease detection and diagnosis, which include methods that predict the presence or absence of a condition, the classification between different subtypes of a condition, and/or the localization of a condition within the image. 7 , 35 , 36 While these tasks are often performed using separate models, they can be treated similarly. For example, does a “normal” image class contain only images with no underlying disease, or does this classification imply the possibility of presence of other abnormalities? Is there a region of the image where localization would be nonsensical, such as disease identification outside of the body? These distinctions have important implications for understanding the true performance of an AI system and should be clearly defined. Examples of detection/diagnosis tasks include screening CXR images for the presence of COVID‐19 and identifying regions in an image that are indicative of COVID‐19. Note that detection within an image can include a localization task, that is, the AI determines that the image contains an abnormality, then locates the abnormality, and finally classifies it. Thus, detection indicates potential disease and diagnosis works to classify the detection as disease or not.
Disease prognosis is often considered similarly to disease detection and diagnosis, but with a different medical interpretation. Prognosis tasks include severity or subtype classifications or direct classifications into prognostic subgroups that are predictive of future patient developments, including disease progression, response to therapy, and mortality. Often, the “normal” subclass is absent from prognostic AI systems with the assumption of prior disease diagnosis. Further, the form of disease severity quantification can vary depending on the disease in question (e.g., lung opacity and size of tumor), so the AI technique and interpretability/explainability approach must be chosen appropriately for the given task as well as be understandable by the intended audience. Prognostic evaluations often have life or death implications, thus the explainability of such decisions is a necessity for aiding the physician and patient in making informed treatment decisions.
3. OVERVIEW OF EXPLAINABILITY/INTERPRETABILITY APPROACHES
In general, a tradeoff exists between the complexity/depth of an AI system and its interpretability, with classical, shallow algorithms, such as decision trees, providing more explainable output with a potentially reduced performance. 34 , 37 , 38 Figure 2 depicts this phenomenon for several commonly used algorithms. It is important to note that finding the optimal operating point between system performance, which will improve patient management, and system interpretability, which will lead to more frequent implementation and trust in radiological practice, is critical.
FIGURE 2.
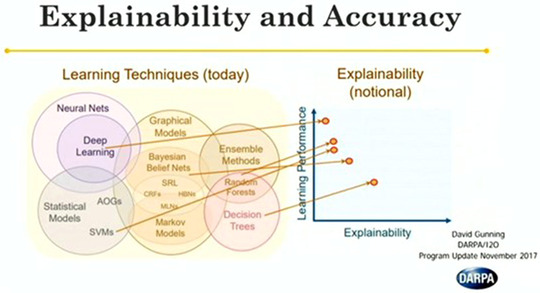
Portrayal of the tradeoff between learning performance, which is often associated with the number of learned parameters, and explainability. Note that deep networks are among the most common techniques for ML‐based medical image evaluation, but also have generally low interpretability. There has been a strong push in recent years to develop techniques for general explanation of neural network predictions. Image acquired from Gunning (publicly available presentation with open distribution) 37
4. FEATURE ANALYSIS FOR EXPLAINABLE AND INTERPRETABLE AI
4.1. Feature distribution analysis
Feature extraction has been a mainstay in ML for decades. Early on, features were handcrafted functions (i.e., human‐engineered features) that attempted to identify intuitive information for computer vision tasks, such as shape, intensity, and morphology, while recent studies utilize automatically identified deep learning features either solely or in fusion with handcrafted features. 7 , 39 , 40 , 41 , 42 Feature distribution analysis can provide information for improving both performance and understanding of AI systems. For example, plotting data in feature space can enable the visualization of class distributions and decision boundaries. However, this is infeasible for cases with high‐dimensional feature spaces of most modern deep learning networks. Thus, several techniques have been developed for feature dimensional reduction (DR), including principal component analysis (PCA), t‐distributed stochastic neighbor embedding (t‐SNE), and uniform manifold approximation and projection (UMAP), among others. 43 , 44 , 45 Each DR technique provides different advantages and disadvantages for feature distribution analysis, some of which are addressed below. Note one critical tradeoff for data visualization and explainability: as discussed by McInnes et al., DR methods tend to prioritize the maintenance of either local or global structural trends in feature space. 45 In general, local structure preservation has proven more effective for visualization purposes, but the loss of global structure is potentially detrimental for other aspects of AI techniques, such as classification performance. 45 Potentially, a balance between the two will be most effective.
PCA is one of the oldest techniques for feature DR and visualization and is still widely used in many fields, including medical imaging. 43 Briefly, PCA identifies orthogonal bases that provide uncorrelated features ranked in order of highest to lowest by degree of variation. Plotting data in the feature space with the first two/three principal components allows for visualization that, in some cases, can account for a large degree of total variance within the dataset. Thus, class distributions can be visualized in this simple, deterministic approach. There are considerable weaknesses to PCA though; it is likely that the first few principal components (which would likely be those chosen for low‐dimensional visualization) only account for a moderate percentage of the total variance within a dataset. Additionally, PCA prioritizes global structure preservation in a linear fashion over local preservation; while this may be more useful for ML system performance, it is likely suboptimal for explainable/interpretable output.
Alternatively, t‐SNE is a DR approach specifically developed for data visualization using information theory techniques. 44 This technique optimizes the Kullbeck–Leibler divergence between joint probabilities in the high‐dimensional feature space and the low‐dimensional embeddings used for visualization, allowing for the identification of local neighborhoods within a dataset. Since its inception in 2002, t‐SNE has been widely implemented in medical imaging feature visualization. Compared to PCA, t‐SNE has several advantages. It allows for nonlinear transformation and preservation of local structure, which may be beneficial for visualization as previously mentioned. Several variants of t‐SNE have been developed to overcome its major flaws, including run‐time improvements and parametric t‐SNE to allow for transform application to unseen data and variants that can effectively scale to large amounts of data. 46 , 47 From a purely interpretive/explanatory perspective, the transformation performed by PCA has clearly identifiable geometric explanations, whereas t‐SNE does not.
UMAP is a promising approach to DR for visualization that potentially improves upon t‐SNE through scalability to large amounts of data, flexibility of embedding dimensionality, and potentially superior preservation of global structure while maintaining visualization quality of local structures compared to t‐SNE. A relatively recent publication, the original paper on UMAP directly compares the algorithm to t‐SNE and LargeVis, 44 , 48 another commonly used visualization technique. Essentially, UMAP constructs local fuzzy simplicial sets followed by an optimized spectral embedding. Because of the recency of UMAP development, there are still several questions that are under investigation, including the impact of hyperparameter selection and the preservation of global and local structures in the embedded space. While investigations into these and other topics are ongoing, the current indication is that UMAP is generally superior to other modern feature analysis techniques, particularly for feature visualization.
4.2. Visualization of feature space
There are several critical problems that must be addressed before AI systems can be applied to medical imaging problems both from the developer and end‐user perspectives. For example, understanding which unseen cases will be difficult for automatic assessment is critical; a developer can adjust the model accordingly based on failed experimental cases and an end‐user is more likely to trust the algorithm if there is prior knowledge of which cases may be problematic. Feature distribution analysis can provide insight into developing solutions to these problems; thus, the discussion below will provide examples of how these algorithms can be applied to improve developer understanding and build end‐user trust for problems related to COVID‐19 assessment.
4.2.1. Developer perspective
As a developer, the use of feature analysis is dependent on the imaging task. Thus, this section focuses on disease detection and prognosis evaluations from a developer's standpoint.
The difficulty of determining presence or absence of disease is largely dependent on the disease in question. For example, it may be easier to detect a broken bone in a right arm radiograph, where it is unlikely that other abnormalities will be visible in the image, than to detect the presence of COVID‐19 pneumonia on a thoracic CT scan, which could present several other confounding presentations (e.g., other viral pneumonias). Visualizing feature space can identify those cases that are difficult to accurately classify. Consider a detection/diagnosis problem in which a binary classification must be made between radiographs of patients with COVID‐19 and those without COVID‐19. Using feature DR techniques, all data can be plotted in a visualizable space (e.g., t‐SNE/UMAP space) and colored to represent the ground truth disease present within each image. This will allow the developer to identify if there are specific cases, such as those that also present lung cancers or other viral pneumonias, that will potentially be confusing during classification and overlap with the disease in question, that is, COVID‐19. The developer can then make better choices in algorithm selection or model adjustment accordingly in order to attempt to correct the overlap between confusing cases, for example, by including more difficult cases in the training set or using bootstrapping.
Alternatively, these techniques could also be applied to predict patient prognosis. Cohen et al. present this concept by showing how UMAP embeddings can change between patients who are healthy, patients who are unlikely to recover from disease, and patients who are in indeterminate. 49 However, as Cohen discusses and we present in Figure 3, there are case embeddings of patients who recovered that appear within the general distribution of “embeddings of no return.” 49 This overlap is highly undesirable for clinical implementation. An end‐user may identify that an unseen case lies within this distribution and incorporate that information into their assessment of survival likelihood; however, if there is a significant number of cases with good outcomes overlapping with this distribution, then including such information could be misleading and lead to patient mismanagement and poorer outcomes. Thus, adjusting the model to minimize overlap is critical.
FIGURE 3.
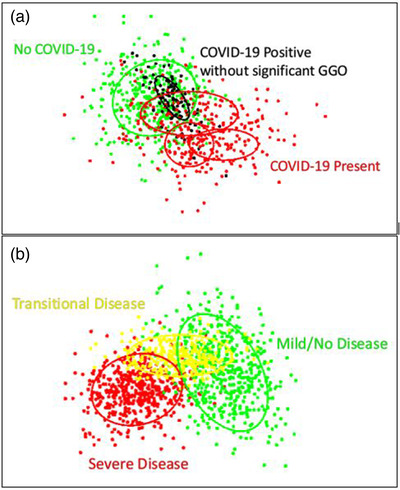
(a) Example of embeddings for a diagnosis task. Red points show positive embeddings, while green and black points show negative embeddings. The ovals depict distributions estimated from the points, with the different ovals referencing different presentations of disease. For example, presentations of COVID‐19 on CT images include ground glass opacities, crazy paving, and architectural distortions. The black oval indicates a positive presentation that significantly overlaps with the negative class distribution, indicating that these cases are problematic and may need a larger prevalence in the training set. (b) Example of embeddings for prognostic clinical evaluation. Green, yellow, and red points refer to healthy/mild, intermediate, and severe disease stages with corresponding distributions depicted through ovals. Clinically, this could be used to help identify in which stage a patient lies and appropriately guide clinical decisions
4.2.2. End‐user perspective
From an end‐user's perspective, the key goals of AI systems include providing useful information and added comprehension that will enhance diagnostic performance and/or reduce reading time. One aspect of this is enabling user trust in the model, which can be conveyed through visualization of dimensionally reduced features.
Examining the prognosis example given from the developer perspective can also provide insight into how to build trust for the end‐user. One option for providing interpretable information is identifying the similarities between the test case and training data. This could include highlighting the k‐nearest training set neighbors of the test case with color coding to identify the neighbor classes, or a heatmap/distribution overlay highlighting class distributions as in Figure 3. Each of these would inspire trust in model output if, for example, all neighbors belong to the same class. In some cases, the test case can be visualized in the embedded space if the DR transformation can be reapplied (e.g., PCA and UMAP), which could also assist in interpretability. For problems with more concrete decision, such as detection and diagnosis, visualizing other aspects of the feature space, such as the decision boundary, could also increase trust.
5. INFLUENTIAL REGION IDENTIFICATION
While image classification algorithms have achieved high performance for many tasks, many have posed questions about what in the image caused a classification decision to be made. Because of this, several groups have developed methods that attempt to identify the region(s) of input images, which were influential in the classification decision; ideally, these methods highlight the diseased tissue within an image. In this section, we review current methods of influential region visualization and discuss how these can be used by developers to improve models and develop trust for the end‐user.
5.1. Region proposal
Generally, region proposal refers to techniques that attempt to determine which regions of an image are likely to be of interest. These techniques have been developed and investigated over the past 25 years, including approaches that predate the current ML boom, culminating with state‐of‐the‐art approaches, such as Mask R‐CNN that integrate region proposal and object detection. 50 However, these techniques can be computationally expensive, so methods with a lower memory footprint may be useful if there is minimal drop in performance.
While there are several region proposal approaches that do not utilize deep learning, such as classical sliding window techniques, selective search rises to the top due to fast runtime and high recall. 51 Essentially, selective search first provides semantic oversegmentations following a graph‐based method, 52 followed by region segmentation through hierarchical similarity measures comparing size, shape, color, and texture. Bounding boxes are generated based on similar regions, which can then be used for downstream tasks, such as image classification or for interpretable output. While selective search is effective and more easily understandable than deep network‐based algorithms, the runtime is generally longer (although this may be practically irrelevant in clinical practice) and performance fails to match that of deep networks.
More recently, state‐of‐the‐art region proposals have been produced through a class of deep networks called region proposal networks (RPNs). 53 , 54 , 55 , 56 RPNs evaluate an input image to produce a set of potentially important regions by applying a sliding window to the output of a convolutional layer in a deep network. A sliding window technique moves multi‐scale shift invariant anchor windows of varying input size and shape across all points within the image and classify if an anchor centered at a given location identifies a region of interest. As is often the case for deep networks, RPNs provide improved performance and runtime at the expense of decreased interpretability of why a given bounding box was selected/classified. However, the use of RPNs as an interpretability tool reduces the need for understanding the network output as the purpose of the RPN is to provide guidance to the radiologist, not to provide a computer classification based on the RPN output.
5.2. Heatmap visualization
Compared to region proposals, heatmaps can convey a more complete representation of the influential regions of an input image and provide a heightened sense of interpretability. Heatmaps serve as a powerful explanatory tool that are typically produced from some component of a deep network (nondeep learning image heatmaps exist but will not be discussed here). However, because the method of production can change the appearance and intended meaning of the heatmap, they should be used cautiously and with clear intent for end‐user interpretation. They could unnecessarily increase reading time or cause confusion if used inappropriately, thus, heatmap utility should be assessed on a per task basis before clinical implementation. Note that several methods discussed below have been successfully applied to a broad range of fields, but this discussion will be restricted to application in medical imaging.
There are multiple components of deep networks that can be utilized in heatmap production. The simplest and most generally applicable approach is the use of saliency maps, which refer to a set of techniques that aim at identifying the regions of an input image that are influential in the final classification decision. 57 These techniques are varied, including the use of deconvolutional networks 58 and backpropagation gradients at different layers of the network, 57 culminating in guided backpropagation, a common approach to saliency map production. 59 Guided backpropagation algorithms serve as the basis for many explainable/interpretable AI techniques, providing fast heatmaps with a relatively fast runtime.
While saliency maps are determined from the intermediate layers of the network, a more recent approach to heatmap production stems from the final layers of a convolutional neural network (CNN) instead. Class activation mapping (CAM) algorithms derive a weighting factor from the final layers of a CNN immediately prior to classification; in the original variant of CAM, this weighting factor was simply the learned model weight applied to a global pooling layer for classification. 60 More advanced techniques, such as Gradient‐weighted CAM (Grad‐CAM), improve upon this approach by calculating weight factors based on the gradient information from the final convolutional layer rather than the global pooling layer. 61 This allows for improved versatility in architecture applicability and improved performance for coarse heatmap production. Further, Grad‐CAM can be combined with the guided backpropagation algorithm to produce more finely resolved heatmaps, which may be desirable depending on the typical size and shape of a desired anatomical region of interest.
The final approach to heatmap production discussed here is attention gating, a technique commonly used to improve network performance that may be used to provide interpretable output. 62 , 63 , 64 , 65 Essentially, activation gates attempt to enhance AI system performance by emphasizing potentially important regions and suppressing background/irrelevant signal. This is often accomplished by producing learned weighting maps and performing element‐wise multiplication between an attention map and a corresponding input signal (e.g., an intermediate CNN layer). While CAM approaches utilize characteristics of the network with no influence on the classification performance, activation gates are a learned component of the network that can have a significant impact on network output. Many recent efforts have attempted to implement the self‐attention mechanisms used in vision transformer architectures for computer vision techniques, which can provide both global and local attention. 66
5.3. Developer perspective
In contrast to feature space visualization, the visualization of influential image regions provides a more specific, detailed evaluation of classification models and their potential misuses can be identified through influential region identification. Here, three studies are briefly discussed, which could inform future model production and improvement based on the use of heatmaps.
Hu investigated the potential of soft tissue image inclusion, acquired either through dual energy subtraction or through other postprocessing techniques, for automatic COVID‐19 diagnosis on chest radiographs. 67 The study trained three classification models for standard radiographs, soft tissue images, and a variant which fused features from the two image types, but found no notable improvement in classification, which was supported by little difference in Grad‐CAM heatmaps from the different classifiers (Figure 4). Similarly, Oh et al. utilized a novel Grad‐CAM‐based method to visualize classifications of lung patches between normal, COVID‐19 pneumonia, and multiple other lung diseases, including other viral pneumonias. 68 The visualizations demonstrate that there is little to no visual signature contributed from non‐COVID‐19 diseases to the heatmap output, indicating that the model appropriately specifies COVID‐19.
FIGURE 4.
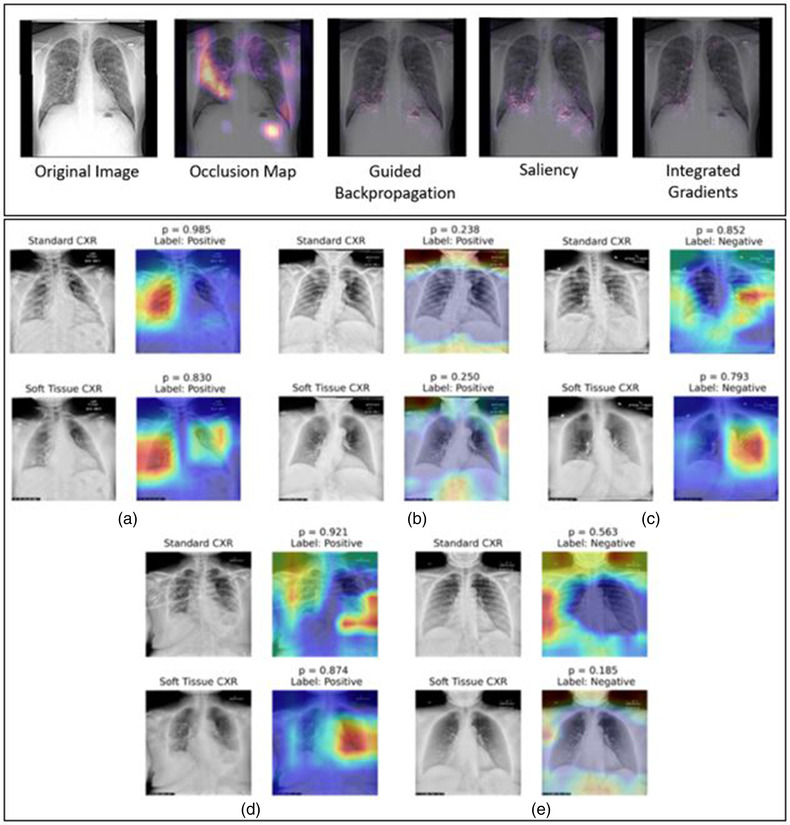
Example heatmaps obtained from a variety of techniques (top) 70 and from Grad‐CAM (bottom). 67 Each technique may provide different evaluations of influential regions, both in terms of relative importance and key locations. Further, note that Grad‐CAM may identify regions that are not important to a human observer. Regions outside the lungs were identified with relatively high influence for the network classification even though a radiologist likely would not use this information in COVID‐19 diagnosis. In the examples by Hu (bottom), some of the Grad‐CAM examples demonstrate reasonable heatmap localization (a–c), while others are less intuitive (d, e). Acquired with permission
Alternatively, Cohen utilized a deep learning model to predict the extent and opacity of COVID‐19 pneumonia in chest radiographs and incorporate saliency maps to evaluate model performance and provide interpretable output. 69 They also provided example heatmaps of cases that were successful in COVID‐19 region identification and of cases that were not. Interestingly, they provided one example that marks a region within the heart, not the lungs, as a success, claiming that this may indicate that the model uses the heart opacity to determine the relevant opacity of the COVID‐19 involvement. This result may suggest that certain preprocessing steps, such as those that remove the heart (e.g., via lung segmentation) prior to deep network analysis may be inappropriate for COVID‐19 severity assessments. This contradicts the study by Oh which utilized segmented images for input to the model, thus the influential region visualization indicated that one of these models may be suboptimal.
5.4. End‐user perspective
As a radiologist, the usability of influential region identification is relatively straightforward. The key questions to be asked are similar to those from feature space visualization: does the model and visualization improve radiologist performance? Is the reading speed significantly decreased and worth the longer reading time?
6. CONTRIBUTION AND IMPORTANCE OF IMAGE FEATURES
Feature importance refers to techniques that assign a score to input features based on how useful they are at predicting a target variable. Popular examples of feature importance scores include statistical correlation scores, coefficients calculated as part of linear models, decision trees, and permutation importance scores. Feature importance scores are the basis for dimensionality reduction and feature selection, which can improve the efficiency and effectiveness of a predictive model.
Feature selection methods reduce the number of features by eliminating features that present redundant information (i.e., features possessing zero variance between classes) or selecting relevant features. Doing so can lead to several benefits, such as improved accuracy, reduced overfitting risk, reduced training time, increased robustness and generalizability, as well as enhanced explainability of models. The optimal number of features to use is dependent on the amount of data available and by the complexity of the task. The feature reduction methods can be categorized by how they are coupled to the ML algorithms, as depicted in Figure 5. Filter methods select features on the basis of a calculated score by looking only at the intrinsic properties of the data, independent of the model. For example, one can filter out highly correlated features using a correlation matrix or use the t‐statistic and its multiclass variant ANOVA to calculate the p‐values for every feature. Wrapper methods, such as forward, backward, bidirectional, recursive feature elimination, and genetic algorithms, use the model's performance as the evaluation criteria when determining which features to include or exclude. This method can more accurately determine the optimal feature subset that contributes to the best model performance than filtering but is more computationally expensive. In embedded methods, the feature selection process is an integral part of the classification model. The performance value for every feature is given by an ML model based on how much each feature contributes to the model training. Embedded methods also have less computational costs compared to wrapper methods because the model is built only once to determine the feature scores, but they do require a parameter to specify the cutoff value of the feature scores.
FIGURE 5.
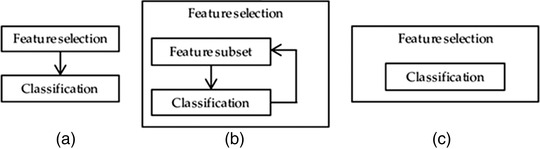
(a) Filter, (b) wrapper, and (c) embedded feature selection methods. Filter methods perform the feature selection independently of construction of the classification model. Wrapper methods iteratively select or eliminate a set of features using the prediction accuracy of the classification model. In embedded methods, the feature selection is an integral part of the classification model. 71 Obtained with permission under MDPI Open Access Policy
While feature selection is used to determine which features should be used in an ML model before or during training, feature importance methods attempt to explain which features are most influential to a model decision after the fact.
6.1. Shapley values
SHAP (SHapley Additive exPlanations) is among the most commonly cited approaches for explainability quantification. 72 This approach adapted from game theory is flexible to any type of deep learning model, including computer vision techniques. An estimated Shapley value is used to identify/explain which features lead to the prediction a model calculated as the contribution of a feature value to the difference between the actual prediction and the mean prediction. Features that cause highly scored predictions (e.g., positive predictions) are displayed in red (Figure 6), while lowly scored predictions are displayed in blue. One major advantage of Shapley values is that the difference between a given prediction and the average prediction is relatively distributed among the feature values.
FIGURE 6.
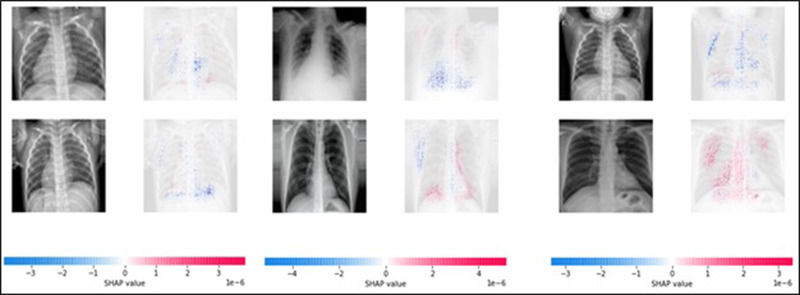
Shapley values acquired for classification of several example images. Note that this technique can identify both positively and negatively influential pixels. In general, these examples follow expectations with peripheral, lower lobe features providing generally more influence than other regions of the lung for COVID‐19 diagnosis
6.2. Local interpretable model‐agnostic explanation
Local interpretable model‐agnostic explanation (LIME) is also a general technique that can be used to explain any ML/black box classifier, including text from radiologist reports and medical images. 73 LIME samples input data used to train a classification model, slightly perturbs the training data, and evaluates the perturbed data with the classification model to evaluate how changes to input impact output. By repeating this process, LIME identifies how individual features lead to the prediction probabilities. A major drawback is that different kernels may be appropriate for each implementation, thus optimal use of LIME may be an iterative, time‐consuming process. Further, LIME may be detrimentally impacted by model bias, which may cause distrust for the end‐user.
7. EVALUATION OF EXPLAINABILITY AND INTERPRETABILITY
While the topic of explainable AI techniques has been of extreme importance and interest, the evaluation of these algorithms remains a difficult and as of yet relatively unexplored problem. For example, Adebayo and Arun attempt to characterize the trustworthiness of saliency mapping techniques in natural and medical images, respectively. 74 , 75 It is important to remember that the end‐goal of medical AI systems is for clinical use, and each end‐user may have different preferences or conditions that allow them, individually, to achieve their best performance. In this section, we discuss proposed approaches to explainable AI evaluation from both human and automatic evaluations. Importantly, we identify that there are two separate yet equally important aspects of evaluating explainable AI: (1) is clinical performance benefitted by the use of an explanation and (2) does the model explanation satisfactorily capture the cause of the AI prediction.
7.1. Human‐based explainable AI evaluation
The most straightforward approach for explainable AI evaluation is an observer study comparing radiologist (i.e., the end user) performance in reading images with explainable AI output and without explainable AI output (or with a different type of explainable output), while assessing reading time and performance accuracy. This approach most closely mimics clinical use, but human evaluators can be inconsistent and often fail to remain objective. This can be exacerbated by several factors, including personal preferences, hot topics in scientific literature, and desired outcome of a study, thus the removal of subjectivity from human‐based evaluations of explainable AI systems is of paramount importance.
Metrics for explainable AI assessment through human observation should be evaluated as in a multi‐reader multi‐case (MRMC) approach. 76 , 77 The use of multiple readers and a large set of diverse cases is important, as observer variability can strongly impact the actual performance of computer‐aided (AI‐aided) systems. 78 , 79 , 80 With that in mind, MRMC studies that utilize receiver operating characterisitic (ROC) and precision‐recall analyses with proper statistical characterization of variance are ideal for determining if an explanation benefits clinical performance. Other metrics include Cohen's κ, F1 score, and changes to accuracy and response time.
Understanding if an AI explanation adequately captures the root cause of a prediction (e.g., COVID‐19 positive or negative) is a more difficult problem that requires further exploration. One approach to this issue is proposed by Hase and Bansal through forward simulation and counterfactual simulation studies. 81 Forward simulations describe a scenario in which a reader, given some input, attempts to predict a model output, while counterfactual simulations involve predicting how a model output changes given a perturbation to the input. On the topic of explainable AI evaluation, this approach captures the question, “does the radiologist understand why a prediction was produced?” This kind of study can be approached in many ways. For example, Doshi‐Velez and Kim proposed to evaluate forward studies by providing an observer with model input and explanation; however, Hase and Bansal argue that a true objective evaluation of understanding requires that the explanation not be provided because this causes many cases to be trivial. 81 , 82 In reality, both approaches are useful and can provide valuable insight into how an approach can be improved and translated to the clinic. In these scenarios, the study aims to assess understanding, not performance, thus ROC analysis and precision‐recall analysis are inappropriate. Instead, evaluations should consider the rate at which an observer is able to appropriately predict a model's inference/change in inference upon input perturbation.
7.2. Automatic explainable AI evaluation
In general, there has been little exploration of automatic evaluations of computer‐based explainable AI output, as may be expected due to the goal use of human interpretation. It is insensible to consider the question “does explainable AI improve clinical performance” in this case, as the explanation is meant for radiologist interpretation rather than model performance. Thus, we only focus on the second question of understanding input perturbation effects. Currently, most attempts to produce automatic explanation evaluations utilize counterfactual simulations as identified above in combination with a feature importance metric. The features deemed important to a classification (ideally, those features/regions indicating COVID‐19 positivity) are removed from the input and the change in model output is evaluated. For example, the influential pixels of an input image may be identified by guided backpropagation, altered to reflect an unimportant region (e.g., patch of random noise and GAN‐based synthetic region adjustment), and the corresponding output should reflect a significantly reduced likelihood of positive classification. Notably, this approach can be used not only for counterfactual simulation, but also for important feature identification (Section 6.3). Studies have utilized a variety of metrics for this type of approach, with the area over the perturbation curve and switching point, which briefly evaluate the change in model output as a different amount of input features are removed from the classification step. Additionally, this change can be evaluated by assessing how the output traverses through feature space through feature removal and how this impacts class probability.
Finally, more standard semiautomatic approaches that are based on either manually or automatically produced ground truth may be appropriate depending on the explanatory task. For a simple case, consider a model trained to detect COVID‐19 with explanation provided through CAM. Any COVID‐19 presence, if visible, would likely be the most influential region as identified by a radiologist. If the model and explanation provide appropriate identification of this region, then the CAM heatmap would also highlight the region of COVID‐19 involvement. To evaluate if this is true, a threshold can be applied to the heatmap and compared to a radiologist's delineation of this region using a segmentation metric, such as the Dice coefficient or intersection over union. If the explanation significantly overlaps with the manual segmentation for a large, diverse image set, then the explanation may be deemed effective and understandable. This also allows for identification of those images for which the model explanation does not behave as expected by the radiologist and, in turn, can inform the developer and radiologist on which cases are most appropriate for AI system implementation. In this way, AI evaluation techniques that are standard in current literature can be applied to evaluate explanations if applied in an appropriate manner.
8. ADDRESSING COVID‐19 THROUGH EXPLAINABLE AI
To fully understand the clinical application of explainable AI for COVID‐19 assessments, we must first understand the data associated with COVID‐19. While reverse transcription polymerase chain reaction (RT‐PCR) tests are the most common tool for COVID‐19 detection, both radiography and CT can supplement RT‐PCR testing to improve detection accuracy and throughput. 83 However, no single radiography or CT finding is sufficient for COVID‐19 diagnosis, thus understanding the subtle differences between COVID‐19 and non‐COVID‐19 pneumonias makes differential diagnosis a difficult task and requires a complete view of all chest findings to provide high classification performance. 83 , 84 , 85 Explainable AI has the potential to fill this role and augment diagnosis tools for improved detection accuracy and differential diagnosis. 85 Further, standardized language for describing COVID‐19 and the construction of publicly available imaging datasets can satisfy data requirements and standardize model evaluation. 86 , 87 This includes large‐scale efforts, such as the NIH‐funded Medical Imaging and Data Resource Center, which is comprised of multi‐institutional, multi‐modal data acquired through the Radiological Society of North America, the American College of Radiology, and the American Association of Physicists in Medicine. Further, while this review has primarily focused on the use of radiography and chest CT, other modalities, including PET/CT, lung ultrasound, and MRI, may play a role in COVID‐19 patient management. 85
In many ways, the development of AI systems for COVID‐19 assessment is unchanged from development for other disease evaluations. In the current literature, the most common use of explainable AI for COVID‐19 evaluation is to ensure that the model correctly focuses on regions of interest in the input image that are indicative of disease presence, usually through heatmap visualization, as implemented by Mei, Bai, Wehbe, and Murphy with varying degrees of success. 88 , 89 , 90 , 91 In particular, Murphy and Wehbe demonstrate heatmaps for both positive and negative COVID‐19 cases, noting especially that the negative examples show low influence within the lungs. 90 , 91 Alternatively, the heatmaps shown by Bai correctly highlight COVID‐19 within lung segmentations but also identify regions with no content (e.g., outside the lung mask) as influential to the classification decision. 89 This finding provides limited understanding of model performance and should be further investigated prior to clinical implementation.
In the case of COVID‐19, the problem is also slightly more nuanced than other diseases because of other confounding disease presentations that could be easily mistaken for COVID‐19, particularly those which present similar, nonspecific imaging findings to COVID‐19, such as other viral pneumonias. This is effectively demonstrated by Jin et al., who partitioned 11 356 CT scans into four disease categories, no pneumonia, influenza, community‐acquired pneumonia, and COVID‐19 pneumonia, and identify phenotypic errors that occurred for both human and AI readers. 92 Jin et al. utilized both Grad‐CAM and Guided Grad‐CAM to visual influential image regions and provide segmentations of diseased regions. 92 Similar to Bai's work, Grad‐CAM indicates that the model identifies regions both inside and outside the lungs as highly influential, while Guided Grad‐CAM improves heatmap visualization but does not capture all diseased lung tissue. 89 Further, they utilize t‐SNE to visualize feature embeddings of the different disease classes and identify image features that are problematic to the classification decision. 92 Importantly, there is overlap between embedded COVID‐19 and the no pneumonia classes. Thus, explainable AI systems that are translated to the clinic must include validation with explainable output tested on an independent set of cases, which have similar imaging findings to confirm model performance.
Another unique application of explainable AI for COVID‐19 evaluation is presented by Zhang et al. in which quantitative lesion features and clinical metadata are utilized to construct classifiers for predicting patient prognosis. 93 Zhang et al. utilize Shapley values to evaluate how individual features impact the risk classifier in terms of both the overall importance of each feature and whether each increased or decreased a prediction output. 93 Further, they evaluate different drug administrations and how the patient responded to treatment based on the lesions from which they extracted quantitative features. Because COVID‐19 is a relatively new disease, this type of analysis for understanding image content that is indicative of high risk, especially in conjunction with clinical metadata, is incredibly valuable and has implications in treatment decisions, such as how aggressively a patient should be treated and which drug should be used for the treatment. This is a clear example of how imaging paired with explainable AI can shape our understanding of COVID‐19 patient risk and be implemented clinically to assist with treatment decision making.
9. RECOMMENDATIONS
As AI systems are increasingly implemented worldwide, the inclusion of explanatory components becomes an increasingly important problem. The European Union has released guidelines for building trustworthy AI and will propose regulations on high‐risk AI use in 2021, thus, the incorporation of explanations should be a serious consideration of any systems produced for clinical use.
In many cases, the choice of explanation type may follow logically from the chosen AI approach. For example, if PCA or UMAP are already utilized in an AI pipeline, then there is little reason to not plot data in feature space to evaluate class distributions. Further, most convolutional neural network architectures are conducive to Grad‐CAM application, which has quickly become one of the most widely applied explainability techniques in medical imaging.
The question of interpretability technique then arises when the choice is not obvious based on a given AI pipeline. In this situation, one must consider several important points to provide an AI explanation. Is there an interpretable technique that can be directly applied to the current algorithm? Would an architecture alteration for incorporation of interpretable output affect model performance? What is the end‐goal of the explanation?
In general, this question is easier to answer for end‐user application than developmental investigation. End‐users value ease of use, clarity, and general performance improvement. In this regard, heatmaps are the most widely accepted approach and Grad‐CAM is the current primary standard. However, for nonclinical application, the choice is significantly more impacted by the question at hand. UMAP is the most flexible and generally appropriate technique for feature space visualization, while individual feature evaluations may contribute more easily interpretable information, such as understanding the physical implications of a given feature. In all, the choice in explanation technique should provide useful, actionable information for a developer and gain trust for end‐users.
Eventually, superior‐performing AI systems may not require interpretability output if the output is generalizable, exhibiting high performance, robust performance, and unbiased performance. Over time, the end user will trust the output, as clinicians do now with various medical tests, such as blood tests.
10. CONCLUSIONS
The recent COVID‐19 pandemic made clear that rapid clinical translation of AI systems may be critically important for assisting treatment‐related decisions and improving patient outcomes. This review has identified several approaches for providing explainable and interpretable AI output and discussed their advantages and disadvantages for key medical imaging scenarios related to COVID‐19 patient management. Further, this review provides recommendations for appropriate incorporation of these techniques to improve ML models and evaluate their performance.
CONFLICTS OF INTEREST AND DISCLOSURES
IEN has served as deputy editor of Medical Physics and discloses a relationship with Scientific Advisory Endectra, LLC. MLG is a stockholder and receives royalties, Hologic, Inc.; equity holder and co‐founder, Quantitative Insights, Inc (now Qlarity Imaging); shareholder, QView Medical, Inc.; royalties, General Electric Company, MEDIAN Technologies, Riverain Technologies, LLC, Mitsubishi, and Toshiba.
ACKNOWLEDGMENTS
This research is part of the Collaborative Research Project (CRP) #10 of MIDRC (The Medical Imaging Data Resource Center) and was made possible by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under contracts 75N92020C00008 and 75N92020C00021. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would also like to acknowledge Emily Townley for her administrative contributions to this effort.
Fuhrman JD, Gorre N, Hu Q, Li H, El Naqa I, Giger ML. A review of explainable and interpretable AI with applications in COVID‐19 imaging. Med. Phys. 2022;49:1–14. 10.1002/mp.15359
Senior author: Maryellen L. Giger m-giger@uchicago.edu
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this review article.
REFERENCES
- 1. Deo Rahul C. Machine learning in medicine. Circulation. 2015;132(20):1920‐1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leung MKK, Delong A, Alipanahi B, Frey BJ. Machine learning in genomic medicine: a review of computational problems and data sets. Proc IEEE. 2016;104(1):176‐197. [Google Scholar]
- 3. Fatima M, Pasha M. Survey of machine learning algorithms for disease diagnostic. J Intell Learn Syst Appl. 2017;09(01):1. [Google Scholar]
- 4. Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380(14):1347‐1358. [DOI] [PubMed] [Google Scholar]
- 5. Yanase J, Triantaphyllou E. A systematic survey of computer‐aided diagnosis in medicine: past and present developments. Expert Syst Appl. 2019;138:112821. [Google Scholar]
- 6. lin D, Vasilakos AV, Tang Y, Yao Y. Neural networks for computer‐aided diagnosis in medicine: a review. Neurocomputing. 2016;216:700‐708. [Google Scholar]
- 7. Giger ML. Machine learning in medical imaging. J Am Coll Radiol. 2018;15(3):512‐520. [DOI] [PubMed] [Google Scholar]
- 8. Yassin NIR, Omran S, El Houby EMF, Allam H. Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: a systematic review. Comput Methods Programs Biomed. 2018;156:25‐45. [DOI] [PubMed] [Google Scholar]
- 9. Sahiner B, Pezeshk A, Hadjiiski LM, et al. Deep learning in medical imaging and radiation therapy. Med Phys. 2019;46(1):e1‐e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vamathevan J, Clark D, Czodrowski P, et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18(6):463‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh A, Sengupta S, Lakshminarayanan V. Explainable deep learning models in medical image analysis. arXiv:200513799 [cs, eess]. 2020. Accessed May 11, 2021. http://arxiv.org/abs/2005.13799 [DOI] [PMC free article] [PubMed]
- 12. Holzinger A, Biemann C, Pattichis CS, Kell DB. What do we need to build explainable AI systems for the medical domain? arXiv:171209923 [cs, stat]. 2017. Accessed May 11, 2021. http://arxiv.org/abs/1712.09923
- 13. Shad R, Cunningham JP, Ashley EA, Langlotz CP, Hiesinger W. Medical imaging and machine learning. arXiv:210301938 [cs, eess]. 2021. Accessed May 11, 2021. http://arxiv.org/abs/2103.01938
- 14. Rudin C, Radin J. Why are we using black box models in AI when we don't need to? A lesson from an explainable AI competition. Harv Data Sci Rev. 2019;1(2). [Google Scholar]
- 15. Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019;1(5):206‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lynch CJ, Liston C. New machine‐learning technologies for computer‐aided diagnosis. Nat Med. 2018;24(9):1304‐1305. [DOI] [PubMed] [Google Scholar]
- 17. Lee H, Chen Y‐PP. Image based computer aided diagnosis system for cancer detection. Expert Syst Appl. 2015;42(12):5356‐5365. [Google Scholar]
- 18. Ahmad OF, Soares AS, Mazomenos E, et al. Artificial intelligence and computer‐aided diagnosis in colonoscopy: current evidence and future directions. Lancet Gastroenterol Hepatol. 2019;4(1):71‐80. [DOI] [PubMed] [Google Scholar]
- 19. McBee MP, Awan OA, Colucci AT, et al. Deep learning in radiology. Acad Radiol. 2018;25(11):1472‐1480. [DOI] [PubMed] [Google Scholar]
- 20. Mazurowski MA, Buda M, Saha A, Bashir MR. Deep learning in radiology: an overview of the concepts and a survey of the state of the art with focus on MRI. J Magn Reson Imaging. 2019;49(4):939‐954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song Y, Zheng S, Li L, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID‐19) with CT images. IEEE/ACM Trans Comput Biol Bioinform. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tajbakhsh N, Jeyaseelan L, Li Q, Chiang JN, Wu Z, Ding X. Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med Image Anal. 2020;63:101693. [DOI] [PubMed] [Google Scholar]
- 23. Minaee S, Boykov YY, Porikli F, Plaza AJ, Kehtarnavaz N, Terzopoulos D. Image segmentation using deep learning: a survey. IEEE Trans Pattern Anal Mach Intell. 2021. [DOI] [PubMed] [Google Scholar]
- 24. Yala A, Schuster T, Miles R, Barzilay R, Lehman C. A deep learning model to triage screening mammograms: a simulation study. Radiology. 2019;293(1):38‐46. [DOI] [PubMed] [Google Scholar]
- 25. Annarumma M, Withey SJ, Bakewell RJ, Pesce E, Goh V, Montana G. Automated triaging of adult chest radiographs with deep artificial neural networks. Radiology. 2019;291(1):196‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bien N, Rajpurkar P, Ball RL, et al. Deep‐learning‐assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. PLoS Med. 2018;15(11):e1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Escalante HJ, Escalera S, Guyon I, et al. Explainable and Interpretable Models in Computer Vision and Machine Learning. Springer International Publishing; 2018. [Google Scholar]
- 28. Tjoa E, Guan C. A survey on explainable artificial intelligence (XAI): toward medical XAI. IEEE Trans Neural Netw Learn Syst. 2020. [DOI] [PubMed] [Google Scholar]
- 29. Gunning D, Stefik M, Choi J, Miller T, Stumpf S, Yang G‐Z. XAI—explainable artificial intelligence. Sci Robot. 2019;4(37):eaay7120. [DOI] [PubMed] [Google Scholar]
- 30. Xu F, Uszkoreit H, Du Y, et al. A brief survey on history, research areas, approaches and challenges. In: Tang J, Kan M‐Y, Zhao D, Li S, Zan H, eds. Natural Language Processing and Chinese Computing. Springer International Publishing; 2019:563‐574. [Google Scholar]
- 31. Li X‐H, Cao CC, Shi Y, et al. A survey of data‐driven and knowledge‐aware eXplainable AI. IEEE Trans Knowl Data Eng. 2020. [Google Scholar]
- 32. Das A, Rad P. Opportunities and challenges in explainable artificial intelligence (XAI): a survey. arXiv:200611371 [cs]. 2020. Accessed May 12, 2021. http://arxiv.org/abs/2006.11371
- 33. Adadi A, Berrada M. Peeking inside the black‐box: a survey on explainable artificial intelligence (XAI). IEEE Access. 2018;6:52138‐52160. [Google Scholar]
- 34. Došilović FK, Brčić M, Hlupić N. Explainable artificial intelligence: a survey. In: 2018 41st International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO) . 2018:0210‐0215.
- 35. Petrick N, Sahiner B, Armato SG, et al. Evaluation of computer‐aided detection and diagnosis systems. Med Phys. 2013;40(8):087001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Giger ML. Computerized analysis of images in the detection and diagnosis of breast cancer. Semin Ultrasound CT MR. 2004;25(5):411‐418. [DOI] [PubMed] [Google Scholar]
- 37. Gunning D. Explainable Artificial Intelligence (XAI). Technical Report. Defense Advanced Research Projects Agency, DARPA/I20. 2017.
- 38. Wu L, Huang R, Tetko IV, Xia Z, Xu J, Tong W. Trade‐off predictivity and explainability for machine‐learning powered predictive toxicology: an in‐depth investigation with Tox21 data sets. Chem Res Toxicol. 2021;34(2):541‐549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nanni L, Ghidoni S, Brahnam S. Handcrafted vs. non‐handcrafted features for computer vision classification. Pattern Recognit. 2017;71:158‐172. [Google Scholar]
- 40. dos Santos CN, Xiang B, Zhou B. Classifying relations by ranking with convolutional neural networks. arXiv:150406580 [cs]. 2015. Accessed May 12, 2021. http://arxiv.org/abs/1504.06580
- 41. Hosny A, Aerts HJ, Mak RH. Handcrafted versus deep learning radiomics for prediction of cancer therapy response. Lancet Digit Health. 2019;1(3):e106‐e107. [DOI] [PubMed] [Google Scholar]
- 42. Zhang J, Xia Y, Xie Y, Fulham M, Feng DD. Classification of medical images in the biomedical literature by jointly using deep and handcrafted visual features. IEEE J Biomed Health Inform. 2018;22(5):1521‐1530. [DOI] [PubMed] [Google Scholar]
- 43. Pearson K. On lines and planes of closest fit to systems of points in space. Philos Mag. 1901;2(11):559‐572. [Google Scholar]
- 44. van der Maaten L, Hinton G. Visualizing data using t‐SNE. J Mach Learn Res. 2008;9(86):2579‐2605. [Google Scholar]
- 45. McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv:180203426 [cs, stat]. 2020. Accessed January 15, 2021. http://arxiv.org/abs/1802.03426
- 46. Van Der Maaten L. Accelerating t‐SNE using tree‐based algorithms. J Mach Learn Res. 2014;15(1):3221‐3245. [Google Scholar]
- 47. Van Der Maaten L. Learning a parametric embedding by preserving local structure. In: Proceedings of the 12th International Conference on Artificial Intelligence and Statistics. 2009:384‐391.
- 48. Tang J, Liu J, Zhang M, Mei Q. Visualizing large‐scale and high‐dimensional data. In: Proceedings of the 25th International Conference on World Wide Web . 2016:287‐297.
- 49. Cohen, JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M. COVID‐19 image data collection: prospective predictions are the future. 2021. https://arxiv.org/abs/2006.11988. arXiv:200611988 [cs, eess, q‐bio]. Accessed January 15, 2021.
- 50. He K, Gkioxari G, Dollár P, Girshick R. Mask R‐CNN. In: Proceedings of the IEEE International Conference on Computer Vision . 2017:2961‐2969.
- 51. Uijlings JR, Van De Sande KE, Gevers T, Smeulders AW. Selective search for object recognition. Int J Comput Vision. 2013;104(2):154‐171. [Google Scholar]
- 52. Felzenszwalb PF, Huttenlocher DP. Efficient graph‐based image segmentation. Int J Comput Vision. 2004;59(2):167‐181. [Google Scholar]
- 53. Girshick R. Fast R‐CNN. In: Proceedings of the IEEE International Conference on Computer Vision . 2015:1440‐1448.
- 54. Ren S, He K, Girshick R, Sun J. Faster R‐CNN: towards real‐time object detection with region proposal networks. arXiv preprint arXiv:150601497. 2015. [DOI] [PubMed]
- 55. Redmon J, Divvala S, Girshick R, Farhadi A. You only look once: unified, real‐time object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition . 2016:779‐788.
- 56. Redmon J, Farhadi A. Yolov3: an incremental improvement. arXiv preprint arXiv:180402767. 2018.
- 57. Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv preprint arXiv:13126034. 2013.
- 58. Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. In: European Conference on Computer Vision . Springer; 2014:818‐833. [Google Scholar]
- 59. Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M. Striving for simplicity: the all convolutional net. arXiv preprint arXiv:14126806. 2014.
- 60. Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition . 2016:2921‐2929.
- 61. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad‐CAM: visual explanations from deep networks via gradient‐based localization. In: Proceedings of the IEEE International Conference on Computer Vision . 2017:618‐626.
- 62. Oktay O, Schlemper J, Folgoc LL, et al. Attention U‐net: learning where to look for the pancreas. arXiv preprint arXiv:180403999. 2018.
- 63. Schlemper J, Oktay O, Schaap M, et al. Attention gated networks: learning to leverage salient regions in medical images. Med Image Anal. 2019;53:197‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jetley S, Lord NA, Lee N, Torr PH. Learn to pay attention. arXiv preprint arXiv:180402391. 2018.
- 65. Woo S, Park J, Lee J‐Y, Kweon IS. CBAM: convolutional block attention module. In: Proceedings of the European Conference on Computer Vision (ECCV) . 2018:3‐19.
- 66. Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:201011929. 2020.
- 67. Hu Q, Drukker K, Giger ML. Role of standard and soft tissue chest radiography images in COVID‐19 diagnosis using deep learning. Medical Imaging 2021: Computer‐Aided Diagnosis. International Society for Optics and Photonics; 2021:1159704. [Google Scholar]
- 68. Oh Y, Park S, Ye JC. Deep learning COVID‐19 features on CXR using limited training data sets. IEEE Trans Med Imaging. 2020;39(8):2688‐2700. [DOI] [PubMed] [Google Scholar]
- 69. Cohen JP, Dao L, Roth K, et al. Predicting COVID‐19 pneumonia severity on chest X‐ray with deep learning. Cureus. 2020;12(7):e9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chatterjee S, Saad F, Sarasaen C, et al. Exploration of interpretability techniques for deep COVID‐19 classification using chest X‐ray images. arXiv:200602570 [cs, eess]. 2020. Accessed April 16, 2021. http://arxiv.org/abs/2006.02570 [DOI] [PMC free article] [PubMed]
- 71. Suppers A, van Gool AJ, Wessels HJ. Integrated chemometrics and statistics to drive successful proteomics biomarker discovery. Proteomes. 2018;6(2):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lundberg S, Lee S‐I. A unified approach to interpreting model predictions. arXiv preprint arXiv:170507874. 2017.
- 73. Ribeiro MT, Singh S, Guestrin C. “Why should i trust you?” Explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining . 2016:1135‐1144.
- 74. Adebayo J, Gilmer J, Muelly M, Goodfellow I, Hardt M, Kim B. Sanity checks for saliency maps. arXiv preprint arXiv:181003292. 2018.
- 75. Arun N, Gaw N, Singh P, et al. Assessing the (un) trustworthiness of saliency maps for localizing abnormalities in medical imaging. arXiv preprint arXiv:200802766. 2020. [DOI] [PMC free article] [PubMed]
- 76. Liu L, Dou Q, Chen H, Qin J, Heng P‐A. Multi‐task deep model with margin ranking loss for lung nodule analysis. IEEE Trans Med Imaging. 2019;39(3):718‐728. [DOI] [PubMed] [Google Scholar]
- 77. Dendumrongsup T, Plumb AA, Halligan S, Fanshawe TR, Altman DG, Mallett S. Multi‐reader multi‐case studies using the area under the receiver operator characteristic curve as a measure of diagnostic accuracy: systematic review with a focus on quality of data reporting. PLoS One. 2014;9(12):e116018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gallas BD, Brown DG. Reader studies for validation of CAD systems. Neural Netw. 2008;21(2):387‐397. [DOI] [PubMed] [Google Scholar]
- 79. Gallas BD, Chen W, Cole E, et al. Impact of prevalence and case distribution in lab‐based diagnostic imaging studies. J Med Imaging. 2019;6(1):015501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rodriguez‐Ruiz A, Lång K, Gubern‐Merida A, et al. Stand‐alone artificial intelligence for breast cancer detection in mammography: comparison with 101 radiologists. J Natl Cancer Inst. 2019;111(9):916‐922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hase P, Bansal M. Evaluating explainable AI: which algorithmic explanations help users predict model behavior? arXiv:200501831 [cs]. 2020. Accessed March 9, 2021. http://arxiv.org/abs/2005.01831
- 82. Doshi‐Velez F, Kim B. Towards a rigorous science of interpretable machine learning. arXiv preprint arXiv:170208608. 2017.
- 83. Cleverley J, Piper J, Jones MM. The role of chest radiography in confirming covid‐19 pneumonia. BMJ. 2020;370:m2426. [DOI] [PubMed] [Google Scholar]
- 84. Bai HX, Hsieh B, Xiong Z, et al. Performance of radiologists in differentiating COVID‐19 from non‐COVID‐19 viral pneumonia at chest CT. Radiology. 2020;296(2):E46‐E54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dong D, Tang Z, Wang S, et al. The role of imaging in the detection and management of COVID‐19: a review. IEEE Rev Biomed Eng. 2021;14:16‐29. [DOI] [PubMed] [Google Scholar]
- 86. Salehi S, Abedi A, Balakrishnan S, Gholamrezanezhad A. Coronavirus disease 2019 (COVID‐19) imaging reporting and data system (COVID‐RADS) and common lexicon: a proposal based on the imaging data of 37 studies. Eur Radiol. 2020;30(9):4930‐4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tsai EB, Simpson S, Lungren MP, et al. The RSNA International COVID‐19 Open Radiology Database (RICORD). Radiology. 2021;299(1):E204‐E213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mei X, Lee H‐C, Diao K, et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID‐19. Nat Med. 2020;26(8):1224‐1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bai HX, Wang R, Xiong Z, et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID‐19 from pneumonia of other origin at chest CT. Radiology. 2020;296(3):E156‐E165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wehbe RM, Sheng J, Dutta S, et al. DeepCOVID‐XR: an artificial intelligence algorithm to detect COVID‐19 on chest radiographs trained and tested on a large U.S. clinical data set. Radiology. 2021;299(1):E167‐E176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Murphy K, Smits H, Knoops AJG, et al. COVID‐19 on chest radiographs: a multireader evaluation of an artificial intelligence system. Radiology. 2020;296(3):E166‐E172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jin C, Chen W, Cao Y, et al. Development and evaluation of an artificial intelligence system for COVID‐19 diagnosis. Nat Commun. 2020;11(1):5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang K, Liu X, Shen J, et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID‐19 pneumonia using computed tomography. Cell. 2020;181(6):1423‐1433. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this review article.


