Abstract
The novel coronavirus disease 2019 (COVID‐19) remains a global health emergency, and understanding the interactions between the virus and host immune responses is crucial to preventing its lethal effects. The expansion of myeloid‐derived suppressor cells (MDSCs) in COVID‐19, thereby suppressing immune responses, has been described as responsible for the severity of the disease, but the correlation between MDSC subsets and COVID‐19 severity remains elusive. Therefore, we classified patients according to clinical and laboratory findings—aiming to investigate the relationship between MDSC subsets and laboratory findings such as high C‐reactive protein, ferritin and lactate dehydrogenase levels, which indicate the severity of the disease. Forty‐one patients with COVID‐19 (26 mild and 15 severe; mean age of 49.7 ± 15 years) and 26 healthy controls were included in this study. MDSCs were grouped into two major subsets—polymorphonuclear MDSCs (PMN‐MDSCs) and monocytic MDSCs—by flow cytometric immunophenotyping, and PMN‐MDSCs were defined as mature and immature, according to CD16 expressions, for the first time in COVID‐19. Total MDSCs, PMN‐MDSCs, mature PMN‐MDSCs and monocytic MDSCs were significantly higher in patients with COVID‐19 compared with the healthy controls (P < .05). Only PMN‐MDSCs and their immature PMN‐MDSC subsets were higher in the severe subgroup than in the mild subgroup. In addition, a significant correlation was found between C‐reactive protein, ferritin and lactate dehydrogenase levels and MDSCs in patients with COVID‐19. These findings suggest that MDSCs play a role in the pathogenesis of COVID‐19, while PMN‐MDSCs, especially immature PMN‐MDSCs, are associated with the severity of the disease.
Keywords: COVID‐19, MDSC subsets and acute‐phase reactants
1. INTRODUCTION
The novel coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), is a global pandemic that has caused millions of deaths. 1 , 2 The mortality rate of SARS‐CoV‐2 infection confirmed by polymerase chain reaction is reported to be about 2.7% in Turkey. 3 Risk factors affecting the severity of the disease are old age, male gender and comorbid conditions such as hypertension, coronary heart disease, diabetes, obesity, chronic lung disease and immunodeficiencies. 4 , 5 However, numerous studies have reported that the disease becomes severe or even leads to death in a certain proportion of middle‐aged patients without comorbidities who are exposed to SARS‐CoV‐2 infection. 6 Understanding immune responses that affect the severity of SARS‐CoV‐2 infection is vital to the diagnosis, treatment and management of the disease.
Prediction of severe COVID‐19 and treatment accordingly are vital to preventing mortality. Laboratory biomarkers such as lymphocyte count and C‐reactive protein (CRP), D‐dimer, ferritin, lactate dehydrogenase (LDH) and interleukin (IL) 6 serum levels are the factors used to determine the severity of the disease. 7 Lymphopenia and increased CRP, D‐dimer, ferritin and LDH serum levels have been shown to be responsible for severe inflammation and coagulation. 8
Myeloid‐derived suppressor cells (MDSCs) are a heterogeneous population of cells generated during various pathologies, ranging from cancer to obesity. Myeloid cells have emerged as a major contributor to protection against pathogens. Under physiological conditions, the cell‐signalling molecule granulocyte‐macrophage colony‐stimulating factor (GM‐CSF) drives myelopoiesis, and granulocyte colony‐stimulating factor (G‐CSF) and macrophage colony‐stimulating factor (M‐CSF) induce the differentiation of granulocytes and macrophages, respectively. In cancer and other pathological conditions, these factors are overproduced and favour the generation of MDSCs. 9 Thus, the accumulation of MDSCs takes place alongside the same differentiation pathways as for neutrophils and monocytes. 9 There are two subsets of MDSCs that differ morphologically, molecularly and functionally: polymorphonuclear MDSCs (PMN‐MDSCs), which are phenotypically and morphologically similar to neutrophils, and monocytic MDSCs (M‐MDSCs), which are phenotypically and morphologically similar to monocytes. 10 PMN‐MDSCs are a heterogeneous subset of MDSCs composed of mature and immature cells. Mature PMN‐MDSCs inhibit the proliferation and cytokine production of T cells, while immature PMN‐MDSCs suppress T cells by direct cytotoxicity. 11
MDSCs inhibit the differentiation of immature myeloid cells into dendritic cells and macrophages, preventing antigen presentation to T cells. 12 MDSCs suppress interferon gamma (IFN‐γ) secretion from T cells, as well as the antigen‐specific proliferation of T cells, by producing cytokines such as IL‐10 and tumour necrosis factor‐alpha (TNF‐α). MDSCs also contribute to the suppression of immune responses by inducing the differentiation of CD4+ T cells into regulatory T cells. 13 MDSCs perform their immunosuppression functions through reactive oxygen species (ROS), inducible nitric oxide synthase and arginase‐1 (Arg‐1) mediators. ROS, especially hydrogen peroxide, reduces the differentiation of immature myeloid cells into macrophages and dendritic cells and prevents T cell—major histocompatibility complex binding by catalysing the nitration of the T cell receptor. 14 Arg‐1 and inducible nitric oxide synthase nitrosylate the T cell receptor, resulting in the suppression or apoptosis of T cells. 15
However, viruses and cancer cells use the immunosuppressive properties of MDSCs to accumulate these cells at the infection site and in the tumour microenvironment, thereby suppressing the immune response. 13 , 16 In viral infections, IL‐6 produced by viral‐infected cells prevents the differentiation of MDSCs into macrophages and dendritic cells, while transforming growth factor‐beta (TGF‐β) produced by MDSCs is responsible for the accumulation of MDSCs at the inflammation site. 12 TNF‐α and IL‐1β produced by MDSCs promote the survival and accumulation of these cells at the site of infection. A significant direct correlation between TNF‐α, IL‐1β, IL‐6 and IL‐8 levels and PMN‐MDSCs has been reported in COVID‐19. 17 Also, plasma IL‐6 level was correlated with elevated CRP, fibrinogen, ferritin and LDH levels. Various studies have reported that CRP and LDH play a role in the development and suppressive functions of MDSCs. 18 , 19 CRP modulates the development and suppressive actions of MDSCs. 18 Also, high serum LDH levels indicate a more immunosuppressive MDSC‐enriched tumour microenvironment in cancer studies. 19
Several studies have reported that MDSCs in chronic viral diseases, including human immunodeficiency virus and hepatitis C virus, are associated with the pathogenesis of these diseases. 20 , 21 The expansion of MDSCs in patients with severe COVID‐19 has been reported in a few studies. 17 , 22 , 23 In this study, the role of MDSC subsets in the pathogenesis of COVID‐19 and their relationship with laboratory biomarkers that affect the severity of the disease, such as serum CRP, LDH and ferritin, were investigated. Mature and immature PMN‐MDSC subsets were evaluated for the first time in patients with COVID‐19.
2. MATERIALS AND METHODS
2.1. Study populations
This is a multicentre, prospective, case‐control study. Forty‐one patients diagnosed with COVID‐19 and confirmed with SARS‐CoV‐2 polymerase chain reaction testing between May and June 2020 and 26 healthy controls were included in this study (Figure 1). To prevent the test results from being affected by the treatment, biochemical and flow cytometric tests were performed within 24 hours on the blood taken at the time of the first admission before starting treatment. The patients’ demographic data and clinical features, such as age, gender, comorbidity, contact history, application symptoms, radiological and laboratory findings (including haemoglobin, leukocytes, lymphocytes and platelets), CRP, ferritin, LDH and other biochemical test results, as well as saturation measured with a pulse oximeter, were recorded at first admission. Arterial blood gas was measured in patients with SpO2 values below 90%. Patients using immunosuppressive drugs and patients with chronic renal disease and malignancy were excluded from this study. All patients with COVID‐19 were sampled at hospital admission, and written informed consent was obtained from all participants.
FIGURE 1.
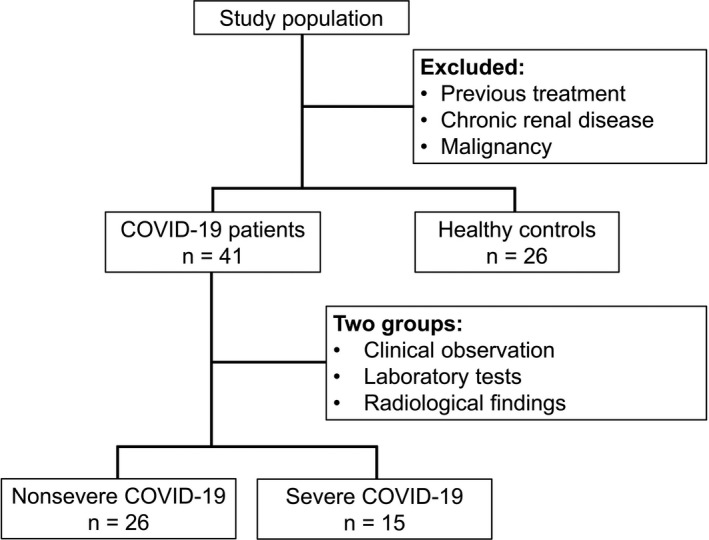
Flowchart of the study population
Patients with COVID‐19 were divided into two subgroups (mild and severe) according to the severity of the disease based on clinical observation and laboratory and radiological findings. 24 The mild subgroup included patients with no clinical symptoms or with mild clinical symptoms, normal or slightly increased infection parameters and unilateral radiological findings. The severe subgroup consisted of patients with clinical signs of respiratory failure, dyspnoea, tachypnoea (≥30/min), bilateral (>50%), multilobar, ground‐glass opacity and infiltration radiological findings, and laboratory findings that predict a poor prognosis (blood lymphocyte count <800/µl or CRP >10x the upper limit of the normal value or ferritin >500 ng/ml or D‐dimer >1000 ng/ml and so on). 24 , 25
Sepsis and systemic inflammatory response syndrome were diagnosed in patients with severe COVID‐19 according to the third international consensus definitions for sepsis and septic shock (Sepsis‐3). 26 , 27 Sepsis was defined as at least two or more of the following criteria in the presence of coinfection: fever or hypothermia (>38°C or <36°C, respectively), tachycardia (>90 min), tachypnoea (>20 min) or PaCO2 <32 mm Hg (4.3 kPa), white blood cell count >12 000/mm3 or <4000/mm3 or >10% immature bands, change in mental status, hypotension (systolic blood pressure <90 mm Hg or fallen by >40 from baseline, mean arterial pressure <70 mm Hg), hyperglycaemia (>140 mg/dL in someone without diabetes), significant oedema or positive fluid balance (>20 mL/kg/d), increased CRP (normal range >2 SD), procalcitonin (normal range >2 SD), lactate (> 2 mmol/L), creatinine (> 0.5 mg/dL), bilirubin (total bilirubin >4 mg/dL) and the international normalized ratio (INR) (>1.5 or activated partial thromboplastin time (aPTT) >60 seconds) and reduced platelet count (<100 000).
Clinical systemic inflammatory response syndrome criteria included tachypnoea, fever and lymphopenia, as well as increased inflammatory markers (ferritin and CRP) and proinflammatory cytokines in COVID‐19.
2.2. Flow cytometric analysis
Peripheral blood mononuclear cells were isolated by density gradient centrifugation at 2000 g for 30 minutes (without break) at room temperature using Histopaque 1077 (Sigma‐Aldrich, 10771) from fresh peripheral blood samples of patients with COVID‐19 on admission, and those of the control group were placed in ethylenediaminetetraacetic acid anticoagulated tubes (Becton Dickinson, 368856). The percentages of total MDSCs and their PMN‐MDSC and M‐MDSC subsets were determined using CD14‐PerCP Cy5.5 (clone HCD14), CD15‐FITC (clone SSEA‐1), CD16‐PE (clone 3G8), CD11b‐APC (clone ICRF44), CD66b‐PE Cy7 (clone G10F5) and HLA‐DR‐APC Cy7 (clone L243) monoclonal antibodies (BioLegend, San Diego, CA). MDSCs were analysed using FACSAria III flow cytometer (Becton Dickinson, CA, USA) with FACSDiva version 6.1.3 software package on at least 100,000 acquired events. Detailed information of reagents and consumables used in this study is provided in Table S1.
In the flow cytometric analysis, peripheral blood mononuclear cells and then the myeloid population, excluding lymphocytes, were gated. Total MDSCs were identified as HLA‐DR‐CD11b+. CD15+CD66b+ PMN‐MDSCs were gated on the HLA‐DR‐CD11b+ total MDSC population and subdivided into CD14‐CD16+ mature PMN‐MDSCs and CD14‐CD16‐ immature PMN‐MDSCs (Figure 2). M‐MDSCs were determined as CD14+ population on the CD15‐CD66b‐ gate (Figure 2). 28 , 29 The percentage of HLA‐DR‐CD11b+ total MDSCs, HLA‐DR‐CD11b+CD15+CD66b+CD14‐ PMN‐MDSCs (including CD16+ mature PMN‐MDSCs and CD16‐ immature PMN‐MDSCs) and HLA‐DR‐CD11b+CD15‐CD66b‐CD14+ M‐MDSC were given from the myeloid gate.
FIGURE 2.
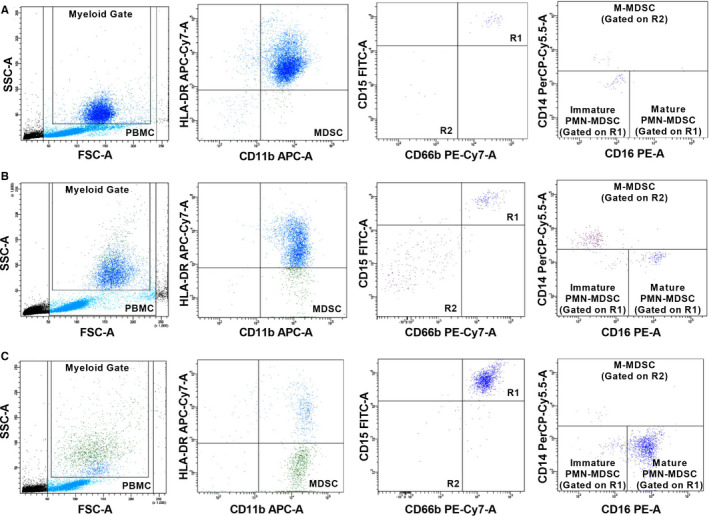
Flow cytometric analysis of MDSCs from (a) a control, (b) a patient in the mild subgroup and (c) a patient in the severe subgroup. Total MDSCs were identified as HLA‐DR‐CD11b+ on the myeloid gate. PMN‐MDSCs were determined as CD15+CD66b+ population (R1 gate) and classified as CD16+ mature and immature PMN‐MDSCs. M‐MDSCs were determined as CD14+ population in the CD15‐CD66b‐ population (R2 gate). M‐MDSCs, monocytic myeloid‐derived suppressor cells; MDSCs, Myeloid‐derived suppressor cells; PMN‐MDSCs, polymorphonuclear myeloid‐derived suppressor cells
2.3. Statistical analyses
Statistical analysis of the data was performed using IBM SPSS Statistics for Windows 21.0 statistical software package (IBM Corp, Armonk, NY, USA). The Kolmogorov‐Smirnov normality test was performed to verify the normal distribution, while the Levene test was used to evaluate the homogeneity of variances. For comparison of means between two groups, the Student t test was used for data with normal distribution, while the Mann‐Whitney U test was used for data without normal distribution. For comparison of more than two groups, one‐way analysis of variance was used for data with normal distribution, while the Kruskal‐Wallis test was used for data with non‐normal distribution. For correlation analysis, the Pearson correlation test was used to compare normally distributed data, whereas the Spearman correlation test was used for data that were not normally distributed. The results were expressed as mean ± standard deviation, and the significance level was accepted as P < .05.
3. RESULTS
3.1. Demographic characteristics and clinical features of the study population
The COVID‐19 group had 21 females and 20 males aged 24‐76 years (mean age = 49.7 ± 15 years). The control group comprised 13 females and 13 males aged 25‐82 years (mean age = 44.4 ± 17.6 years). There was no significant difference between the patient and control groups based on sex and age distribution.
Patients with COVID‐19 admitted to the hospital had the following symptoms: cough (68.3%), fever (46.3%), chills and shivering (34.1%), shortness of breath (34.1%), fatigue (24.4%), throat ache (9.8%), loss of taste and smell (9.8%) and diarrhoea (2.4%). Comorbidities in these patients were recorded as diabetes in 12 cases (29.3%) and hypertension in 12 cases (29.3%). Two patients with COVID‐19 had coinfections, and these patients had higher‐than‐normal procalcitonin, CRP and white blood cell values. Twenty‐seven patients (65.9%) had ground‐glass opacities compatible with pneumonia in chest tomography. Hospitalization days of the patients were 4‐32 days (mean = 10.4 ± 5.2 days). In the classification according to the severity of the disease, there were 26 patients in the mild subgroup and 15 patients in the severe subgroup. The mild subgroup consisted of 15 females and 11 males aged 24‐73 years (mean age = 45.1 ± 14.2 years), and the severe subgroup comprised 6 females and 9 males aged 37‐76 years (mean age = 57.7 ± 13.1 years). The oxygen saturation of the patients in the severe subgroup at admission was significantly lower than those in the mild subgroup (severe subgroup = 86.6 ± 9.1; mild subgroup = 93.6 ± 2.9, P = .011). Nine patients with severe COVID‐19 were treated with corticosteroids, and six were not taking corticosteroids. The mean was 15 ± 7.8 days in the patients who received corticosteroid treatment and 12 ± 3.7 days in the patients who did not receive corticosteroid treatment. There was no statistically significant difference in the mean hospitalization days between patients who received corticosteroid treatment and those who did not (P = .4). Intensive care treatment was required in three patients, and the duration of stay in the intensive care unit (ICU) was 1‐13 days. Regarding the patients in the ICU, all were receiving corticosteroid therapy, and two were undergoing convalescent plasma therapy. Two patients who had received convalescent therapy were survived; the other patient was died 2 days after intubation.
3.2. Laboratory findings
The laboratory findings of the patients with COVID‐19 and the mild and severe subgroups of patients are summarized in Table 1. CRP, LDH and procalcitonin levels were higher than the normal range in all the patients with COVID‐19. The levels of CRP, LDH, fibrinogen, ferritin and procalcitonin were significantly increased in the severe subgroup (P = .000, .005, .007, .040 and .013, respectively).
TABLE 1.
The laboratory findings of patients with COVID‐19 and comparison between mild and severe groups
| Mean ± SD |
All Patients (n = 41) |
Mild (n = 26) |
Severe (n = 15) |
P‐value Mild and Severe |
|---|---|---|---|---|
| Haematologic parameters | ||||
| White blood cell count (×109/L) | 5.7 ± 2.1 | 5.8 ± 2.3 | 5.4 ± 1.7 | .718** |
| Neutrophil (%) | 62.8 ± 12.9 | 60.6 ± 14.7 | 66.6 ± 8.2 | .104* |
| Neutrophil count (×109/L) | 3.8 ± 2.0 | 3.8 ± 2.2 | 3.9 ± 1.7 | .640** |
| Lymphocyte (%) | 26.7 ± 11.5 | 28.6 ± 12.9 | 23.5 ± 7.8 | .127* |
| Lymphocyte count (×109/L) | 1.3 ± 0.7 | 1.5 ± 0.7 | 1.1 ± 0.3 | .211** |
| Neutrophil‐to‐lymphocyte ratio | 4.1 ± 5.8 | 3.3 ± 2.5 | 5.5 ± 8.9 | .231** |
| Monocyte (%) | 9.1 ± 3.3 | 9.2 ± 3.8 | 9.0 ± 2.3 | .820** |
| Monocyte count (×109/L) | 0.5 ± 0.2 | 0.5 ± 0.2 | 0.5 ± 0.2 | .868** |
| Haemoglobin (g/L) | 13.6 ± 1.7 | 13.5 ± 1.8 | 14.1 ± 1.6 | .852* |
| Platelet count (×109/L) | 197.1 ± 64.2 | 204.2 ± 76.1 | 184.9 ± 34.4 | .583** |
| Acute‐phase reactants | ||||
| CRP (mg/L) | 31.2 ± 63.5 | 21.8 ± 69.3 | 47.6 ± 50 | .000** |
| D‐dimer (ng/mL) | 331.5 ± 261.0 | 278.6 ± 250.2 | 423.2 ± 262.0 | .059** |
| Albumin (g/dL) | 3.8 ± 0.4 | 3.9 ± 0.4 | 3.7 ± 0.4 | .472* |
| LDH (U/L) | 262.2 ± 95.8 | 231.2 ± 71.2 | 316.0 ± 110.8 | .005* |
| Troponin (ng/L) | 2.6 ± 3.1 | 2.1 ± 2.3 | 3.6 ± 4.0 | .242** |
| Fibrinogen (mg/dL) | 360.2 ± 139.6 | 331.9 ± 140.9 | 434.5 ± 111.6 | .007** |
| Ferritin (ng/mL) | 195.1 ± 263.8 | 137.6 ± 255.4 | 294.7 ± 255.9 | .040** |
| CPK (U/L) | 124.8 ± 133.4 | 106.0 ± 104.0 | 157.3 ± 172.5 | .478** |
| Procalcitonin (ng/mL) | 2.3 ± 13.3 | 0.3 ± 1.3 | 5.8 ± 21.9 | .013** |
Abbreviations: CPK, creatine phosphokinase; CRP, C‐reactive protein; LDH, lactate dehydrogenase.
*Student t test, **Mann‐Whitney U test.
3.3. Flow cytometric analysis
The comparison of MDSCs and their subsets between the patients with COVID‐19 and the control group is summarized in Table 2. Total MDSCs were significantly increased in all the patients with COVID‐19 (15.3 ± 20.5% in patients and 4.0 ± 2.6% in controls, P =.001), especially in the severe subgroup (22.1 ± 25.1% in the severe subgroup vs 11.4 ± 16.7% in the mild subgroup, P = .001). When the patients were evaluated for lymphopenia, there was no significant difference in MDSCs between patients with lymphopenia and patients without lymphopenia.
TABLE 2.
Comparison of MDSCs and their subsets in patients with COVID‐19 and controls
| Mean ± SD (%) |
All Patients (n = 41) |
Control (n = 26) |
P‐value* |
Mild (n = 26) |
Severe (n = 15) |
P‐value** |
|---|---|---|---|---|---|---|
| Myeloid Gate | 36.1 ± 13.1 | 21.7 ± 5.4 | .001 | 34.6 ± 12.6 | 38.9 ± 14.0 | .001 a |
| Total MDSCs | 15.3 ± 20.5 | 4.0 ± 2.6 | .001 | 11.4 ± 16.7 | 22.1 ± 25.1 | .001 b |
| PMN‐MDSCs | 3.4 ± 2.9 | 1.8 ± 2.2 | .009 | 2.6 ± 2.9 | 5.1 ± 2.5 | .001 c |
| Mature PMN‐MDSCs | 0.7 ± 1.6 | 0.4 ± 1.0 | .003 | 0.7 ± 1.8 | 0.7 ± 1.1 | .004 d |
| Immature PMN‐MDSCs | 1.2 ± 1.9 | 0.7 ± 1.2 | .257 | 0.6 ± 1.2 | 2.4 ± 2.5 | .003 e |
| M‐MDSCs | 1.6 ± 1.6 | 0.8 ± 0.8 | .035 | 1.8 ± 1.7 | 1.3 ± 1.4 | .033 f |
Abbreviations: MDSCs, Myeloid‐derived suppressor cells; M‐MDSCs, monocytic myeloid‐derived suppressor cells; PMN‐MDSCs, polymorphonuclear myeloid‐derived suppressor cells.
*Differences between the patient and control groups for normally distributed data were evaluated using the independent sample t test, while the Mann‐Whitney U test was used to evaluate the data that were not normally distributed.
**ANOVA (Welch) and Kruskal‐Wallis tests were used to test differences between the control, mild and severe groups.
Control and mild groups, P = .001; control and severe groups, P = .001.
Control and mild groups, P = .008; control and severe groups, P = .001.
Control and severe groups, P = .001; mild and severe groups, P = .033.
Control and severe groups, P = .004.
Control and severe groups, P = .010; mild and severe groups, P = .004.
Control and mild groups, P = .031.
A comparison of the MDSC subsets revealed that PMN‐MDSCs and M‐MDSCs were significantly increased in patients with COVID‐19 (P = .009 and .035, respectively) (Figure 3). Increased PMN‐MDSCs were found in the severe subgroup compared to the mild subgroup (P = .033). While mature PMN‐MDSCs were increased in all patients, immature PMN‐MDSCs were increased only in the severe subgroup compared to the mild subgroup and the control group (P = .003, .004 and .010, respectively).
FIGURE 3.
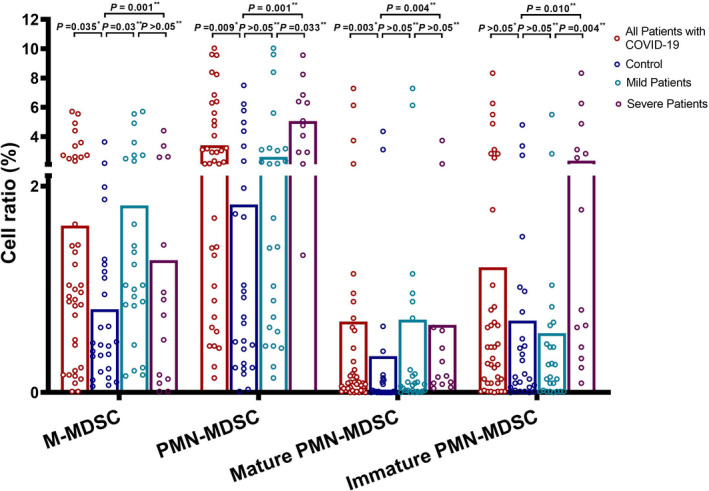
Comparison of M‐MDSC, PMN‐MDSC and PMN‐MDSC subsets between the study groups. The data were assessed statistically using the *Mann‐Whitney U test and the **Kruskal‐Wallis test. M‐MDSCs, monocytic myeloid‐derived suppressor cells; MDSCs, Myeloid‐derived suppressor cells; PMN‐MDSCs, polymorphonuclear myeloid‐derived suppressor cells
In the correlation analysis, MDSCs and PMN‐MDSCs were correlated with CRP, ferritin and LDH levels in the patients with COVID‐19. There was no significant correlation between mature PMN‐MDSCs, M‐MDSCs and the levels of CRP, LDH and ferritin, but immature PMN‐MDSCs were significantly correlated with these acute‐phase reactants (Figure 4, Table S2).
FIGURE 4.
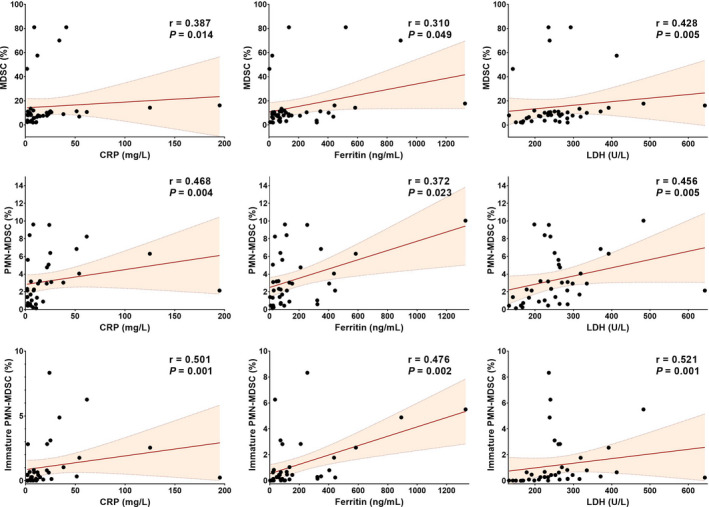
Correlation plots between MDSCs and the levels of CRP, ferritin and LDH. Total MDSCs, PMN‐MDSCs and immature PMN‐MDSCs were significantly positively correlated with CRP, ferritin and LDH levels. The Spearman correlation test was used for data. CRP, C‐reactive protein; LDH, lactate dehydrogenase; MDSCs, Myeloid‐derived suppressor cells; PMN‐MDSCs, polymorphonuclear myeloid‐derived suppressor cells.
4. DISCUSSION
In this study, peripheral MDSC subsets were evaluated in patients with COVID‐19, and the relationship between MDSC subsets and the severity of the disease was investigated for the first time. Increased total MDSCs, PMN‐MDSCs, mature PMN‐MDSCs and M‐MDSCs were found in all patients with COVID‐19 and the severe subgroup compared to the control group. Patients in the severe subgroup had increased PMN‐MDSCs and immature PMN‐MDSCs compared to patients in the mild subgroup. Also, total MDSCs, PMN‐MDSCs and immature MDSCs were significantly correlated with CRP, LDH and ferritin levels, which are associated with the severity of COVID‐19 disease.
The role of MDSCs and their subsets in disease severity has been demonstrated by some functional studies on sepsis and COVID‐19. 17 , 22 , 30 , 31 Mathias et al reported that MDSCs are increased in patients with severe sepsis and septic shock, and these cells are responsible for chronic immune suppression through upregulated Arg‐1 expression. 30 Takano et al reported that PMN‐MDSCs, but not other MDSC subsets, transiently expanded in patients with severe COVID‐19 in their study. 31 Agrati et al studied 18 patients with COVID‐19 and reported an increase in PMN‐MDSCs in the patients with COVID‐19 compared to the control group and a higher frequency among the patients with severe COVID‐19 than those with mild COVID‐19. 22 In a study by Sacchi et al, PMN‐MDSCs increased in patients with COVID‐19 and were higher among the patients in the ICU. 17 The researchers also found that PMN‐MDSCs inhibited IFN‐γ production of T cells after SARS‐CoV‐2 peptide stimulation. 17 In this study, we found increased PMN‐MDSCs in patients with COVID‐19, especially in the severe subgroup. Thus, we infer that PMN‐MDSCs are involved in the pathogenesis of COVID‐19 and responsible for severe COVID‐19.
We classify PMN‐MDSCs as mature and immature, according to CD16 expressions, for the first time in patients with COVID‐19. Mature PMN‐MDSCs were increased in all patients, especially in the severe subgroup, compared to the controls. In immature PMN‐MDSCs, there was no significant difference between all patients with COVID‐19 and the controls, but they were elevated in patients in the severe subgroup compared to those in the mild subgroup. Lang et al reported that mature PMN‐MDSCs suppress T cell proliferation and cytokine expression by expressing Arg‐1 at a higher level and activity than other subsets in cancer patients. 28 In a study involving patients with metastatic melanoma, Gondois‐Rey et al reported that mature PMN‐MDSCs suppressed the proliferation of T cells, while immature PMN‐MDSCs showed a cytotoxic effect on T cells and suppressed them. 11 Our previous study showed that SARS‐CoV‐2 infection induced T cell apoptosis and caused lymphopenia. 32 Also, T cell apoptosis was increased in patients with severe COVID‐19. This study suggested that SARS‐CoV‐2 might suppress T cell proliferation and responses through mature PMN‐MDSCs and is responsible for lymphopenia by the cytotoxic effects of immature PMN‐MDSCs on T cells in patients with severe COVID‐19. The positive correlation between immature PMN‐MDSCs and acute‐phase reactants, such as CRP, LDH and ferritin, confirmed the role of these cells in COVID‐19 severity. Functional studies are needed to elucidate the mechanism of immature PMN‐MDSCs and lymphopenia in COVID‐19.
M‐MDSCs are known to specifically suppress virus‐specific CD8+ T cell activation and lead to delayed viral clearance. 33 Also, they suppress antigen presentation to antigen‐specific CD4+ T cells by inhibiting CD40 and HLA‐DR expression on stimulated B cells. 34 An increase in M‐MDSCs has been reported in patients with hepatitis B virus, hepatitis C virus and human immunodeficiency virus. 21 , 35 , 36 The expansion of M‐MDSCs has been observed in chronic viral diseases, indicating that MDSCs contribute to the establishment of chronic infection. 13 Several studies have reported that M‐MDSCs are increased in patients with COVID‐19, especially in patients with severe COVID‐19. 23 , 37 , 38 In a functional study, Falck‐Jones et al reported that M‐MDSCs suppressed the proliferation of T cells and IFN‐γ production through the Arg‐1‐dependent mechanism in COVID‐19. 37 In our study, M‐MDSCs were increased in patients with COVID‐19. There was no significant difference between the mild and severe subgroups, but M‐MDSCs were found to be increased in patients with mild COVID‐19 compared to the controls. In this context, we suggest that increased M‐MDSCs play a role in the pathogenesis of COVID‐19 but are not related to the severity of the disease. The reason for the inconsistency regarding M‐MDSCs compared to other studies may be that among the patients with COVID‐19 included in our study, the number of patients in need of intensive care and/or fatal patients was lower than in other studies.
In viral infections including SARS‐CoV‐2, TNF‐α causes elevated serum ferritin and LDH levels, as well as thrombocytopenia, lymphopenia and haemolysis. MDSCs themselves contribute to inflammation and coagulation by producing IL‐1β, TNF‐α, IL‐6 and IL‐8. 39 Takano et al reported that PMN‐MDSCs are positively correlated with IL‐8 levels in patients with COVID‐19 and that IL‐8 plays a role in the recruitment of PMN‐MDSCs to peripheral blood following the onset of severe COVID‐19. 31 Increased IL‐1β, TNF‐α, IL‐6 and IL‐8 levels are associated with increased CRP, D‐dimer and ferritin, causing severe inflammation and coagulation in COVID‐19. It has been reported that the presence of lymphopenia and high serum levels of CRP, D‐dimers, ferritin and IL‐6 can be used in the prediction of severe COVID‐19 and fatality.
CRP and LDH are involved in the development and suppressive functions of MDSCs. Jimenez et al, in their in vitro study with mice, reported that CRP increased the generation of MDSCs from bone marrow progenitors and enhanced the cells’ production of intracellular ROS and, thus, their suppressive function. 18 High serum LDH levels indicate tumour glycolytic activity and a more immunosuppressive, MDSC‐enriched tumour microenvironment in cancer studies. 19 In our study, the positive correlation between MDSCs and the levels of CRP, ferritin and LDH reveals that MDSCs contribute to inflammation and coagulation caused by SARS‐CoV‐2 infection. Also, PMN‐MDSCs, particularly immature PMN‐MDSC subsets, were increased in patients with severe COVID‐19 compared to patients with mild COVID‐19. Consistent with these results, the positive correlation of immature PMN‐MDSCs with the levels of CRP, LDH and ferritin might be an indication that these cells have a dominant role in COVID‐19 severity.
The limitation of this study was the lack of a functional analysis. Further studies and a standard approach are needed to identify which MDSC subsets, as well as acute‐phase reactants, are essential for early diagnosis, prediction and follow‐up.
The findings of this study demonstrate that MDSCs, including PMN‐MDSCs and M‐MDSCs, play a role in the pathogenesis of COVID‐19 and that PMN‐MDSCs are associated with the severity of the disease. PMN‐MDSCs, especially immature PMN‐MDSCs, were positively correlated with acute‐phase reactants as a predictor of COVID‐19 severity. Thus, we suggest that MDSC subsets constitute an important immunological parameter contributing to the severity of SARS‐CoV‐2 infection. Further clinical studies are needed to determine their clinical relevance.
CONFLICT OF INTEREST
The authors declare they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
HA, YK and HV conceived and designed the project, as well as contributed to writing the manuscript. SS, BT, HC, MHG and FK provided the diagnosis, treatment and follow‐up of the COVID‐19 patients. HV contributed to the biochemical testing of blood samples and to obtaining the control group. UA contributed to the diagnosis of COVID‐19 by polymerase chain reaction testing. AE performed the flow cytometry‐based experiment, analysed the results and wrote the manuscript. All authors revised the manuscript and approved the final version of the manuscript.
ETHICAL APPROVAL
This study was approved by the Ethical Committee of Selcuk University Medical Faculty (2020/235).
Supporting information
Supplementary Material
Emsen A, Sumer S, Tulek B, et al. Correlation of myeloid‐derived suppressor cells with C‐reactive protein, ferritin and lactate dehydrogenase levels in patients with severe COVID‐19. Scand J Immunol. 2022;95:51–60. 10.1111/sji.13108
Funding information
This study was supported by the Scientific Research Projects Committee of Selcuk University (18611484)
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Azkur AK, Akdis M, Azkur D, et al. Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy. 2020;75:1564‐1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. WHO Coronavirus (COVID‐19) . Dashboard. 2021. [cited 2021 09.13.2021]; Available from: https://covid19.who.int/
- 3. Republic of Turkey, Ministry of Health . COVID‐19 Weekly Situation Report. 2020 21.02.2021]; Available from: https://covid19.saglik.gov.tr/Eklenti/39230/0/covid‐19‐weekly‐situation‐report‐‐‐43pdf.pdf?_tag1=D3D202441F1F5165A33D16981E6544EF7FC0A32F
- 4. Gao YD, Ding M, Dong X, et al. Risk factors for severe and critically ill COVID‐19 patients: a review. Allergy. 2021;76(2):428‐455. 10.1111/all.14657 [DOI] [PubMed] [Google Scholar]
- 5. Wolff D, Nee S, Hickey NS, Marschollek M. Risk factors for Covid‐19 severity and fatality: a structured literature review. Infection. 2021;49:15‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang P, Sha J, Meng M, et al. Risk factors for severe COVID‐19 in middle‐aged patients without comorbidities: a multicentre retrospective study. J Transl Med. 2020;18(1):461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Terpos E, Ntanasis‐Stathopoulos I, Elalamy I, et al. Hematological findings and complications of COVID‐19. Am J Hematol. 2020;95:834‐847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Samprathi M, Jayashree M. Biomarkers in COVID‐19: an up‐to‐date review. Front Pediatr. 2020;8:607647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Veglia F, Perego M, Gabrilovich D. Myeloid‐derived suppressor cells coming of age. Nat Immunol. 2018;19:108‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bronte V, Brandau S, Chen S‐H, et al. Recommendations for myeloid‐derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gondois‐Rey F, Paul M, Alcaraz F, et al. Identification of an immature subset of PMN‐MDSC correlated to response to checkpoint inhibitor therapy in patients with metastatic melanoma. Cancers. 2021;13:1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goh C, Narayanan S, Hahn YS. Myeloid‐derived suppressor cells: the dark knight or the joker in viral infections? Immunol Rev. 2013;255(1):210‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Connor MA, Rastad JL, Green WR. The role of myeloid‐derived suppressor cells in viral infection. Viral İmmunol. 2017;30(2):82‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kusmartsev S, Gabrilovich DI. Inhibition of myeloid cell differentiation in cancer: the role of reactive oxygen species. J Leukoc Biol. 2003;74(2):186‐196. [DOI] [PubMed] [Google Scholar]
- 15. Kusmartsev S, Gabrilovich DI. STAT1 signaling regulates tumor‐associated macrophage‐mediated T cell deletion. J Immunol. 2005;174(8):4880‐4891. [DOI] [PubMed] [Google Scholar]
- 16. Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid‐derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37(3):208‐220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sacchi A, Grassi G, Bordoni V, et al. Early expansion of myeloid‐derived suppressor cells inhibits SARS‐CoV‐2 specific T‐cell response and may predict fatal COVID‐19 outcome. Cell Death Dis. 2020;11(10):921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jimenez RV, Kuznetsova V, Connelly AN, Hel Z, Szalai AJ. C‐Reactive protein promotes the expansion of myeloid derived cells with suppressor functions. Front Immunol. 2019;10:2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cona MS, Lecchi M, Cresta S, et al. Combination of baseline LDH, performance status and age as integrated algorithm to identify solid tumor patients with higher probability of response to anti PD‐1 and PD‐L1 monoclonal antibodies. Cancers. 2019;11:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vollbrecht T, Roider J, Stirner R, et al. Chronic progressive HIV‐1 infection is associated with elevated levels of myeloid‐derived suppressor cells. Retrovirology. 2012;9:P283. [DOI] [PubMed] [Google Scholar]
- 21. Cai W, Qin A, Guo P, et al. Clinical significance and functional studies of myeloid‐derived suppressor cells in chronic hepatitis C patients. J Clin Immunol. 2013;33(4):798‐808. [DOI] [PubMed] [Google Scholar]
- 22. Agrati C, Sacchi A, Bordoni V, et al. Expansion of myeloid‐derived suppressor cells in patients with severe coronavirus disease (COVID‐19). Cell Death Dis. 2020;27(11):3196‐3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kvedaraite E, Hertwig L, Sinha I, et al. Major alterations in the mononuclear phagocyte landscape associated with COVID‐19 severity. Proc Natl Acad Sci USA. 2021;118(6):e2018587118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Republic of Turkey, Ministry of Health . COVID‐19 guideline, adult patient management. 2020 [cited 2021 May 31]; Available from: https://covid19.saglik.gov.tr/Eklenti/40719/0/covid‐19rehberieriskinhastayonetimivetedavipdf.pdf
- 25. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323(13):1239‐1242. [DOI] [PubMed] [Google Scholar]
- 26. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis‐3). JAMA. 2016;315(8):801‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis: for the third international consensus definitions for sepsis and septic shock (Sepsis‐3). JAMA. 2016;315(8):762‐774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lang S, Bruderek K, Kaspar C, et al. Clinical relevance and suppressive capacity of human myeloid‐derived suppressor cell subsets. Clin Cancer Res. 2018;24(19):4834‐4844. [DOI] [PubMed] [Google Scholar]
- 29. Gustafson MP, Lin Y, Maas ML, et al. A method for identification and analysis of non‐overlapping myeloid immunophenotypes in humans. PLoS One. 2015;10(3):e0121546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mathias B, Delmas AL, Ozrazgat‐Baslanti T, et al. Human myeloid‐derived suppressor cells are associated with chronic immune suppression after severe sepsis/septic shock. Ann Surg. 2017;265(4):827‐834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takano T, Matsumura T, Adachi Y, et al. Myeloid cell dynamics correlating with clinical outcomes of severe COVID‐19 in Japan. Int Immunol. 2021;33(4):241‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cizmecioglu A, Akay Cizmecioglu H, Goktepe MH, et al. Apoptosis‐induced T‐cell lymphopenia is related to COVID‐19 severity. J Med Virol. 2021;93(5):2867‐2874. [DOI] [PubMed] [Google Scholar]
- 33. Fortin C, Yang Y, Huang X. Monocytic myeloid‐derived suppressor cells regulate T‐cell responses against vaccinia virus. Eur J Immunol. 2017;47(6):1022‐1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rastad JL, Green WR. LP‐BM5 retrovirus‐expanded monocytic myeloid‐derived suppressor cells alter B cell phenotype and function. ImmunoHorizons. 2018;2(3):87‐106. [DOI] [PubMed] [Google Scholar]
- 35. Garg A, Spector SA. HIV type 1 gp120‐induced expansion of myeloid derived suppressor cells is dependent on interleukin 6 and suppresses immunity. J Infect Dis. 2014;209(3):441‐451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fang Z, Li J, Yu X, et al. Polarization of monocytic myeloid‐derived suppressor cells by hepatitis B surface antigen is mediated via ERK/IL‐6/STAT3 signaling feedback and restrains the activation of T cells in chronic hepatitis B virus infection. J Immunol. 2015;195(10):4873‐4883. [DOI] [PubMed] [Google Scholar]
- 37. Falck‐Jones S, Vangeti S, Yu M, et al. Functional monocytic myeloid‐derived suppressor cells increase in blood but not airways and predict COVID‐19 severity. J Clin Invest. 2021;131(6):e144734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Silvin A, Chapuis N, Dunsmore G, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID‐19. Cell. 2020;182(6):1401‐18.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Signorini C, Pignatti P, Coccini T. How do inflammatory mediators, immune response and air pollution contribute to COVID‐19 disease severity? A lesson to learn. Life. 2021;11(3):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


