Abstract
Background
Molecular‐based tests used to identify symptomatic or asymptomatic patients infected by SARS‐CoV‐2 are characterized by high specificity but scarce sensitivity, generating false‐negative results. We aimed to estimate, through a systematic review of the literature, the rate of RT‐PCR false negatives at initial testing for COVID‐19.
Methods
We systematically searched Pubmed, Embase and CENTRAL as well as a list of reference literature. We included observational studies that collected samples from respiratory tract to detect SARS‐CoV‐2 RNA using RT‐PCR, reporting the number of false‐negative subjects and the number of final patients with a COVID‐19 diagnosis. Reported rates of false negatives were pooled in a meta‐analysis as appropriate. We assessed the risk of bias of included studies and graded the quality of evidence according to the GRADE method. All information in this article is current up to February 2021.
Results
We included 32 studies, enrolling more than 18,000 patients infected by SARS‐CoV‐2. The overall false‐negative rate was 0.12 (95%CI from 0.10 to 0.14) with very low certainty of evidence. The impact of misdiagnoses was estimated according to disease prevalence; a range between 2 and 58/1,000 subjects could be misdiagnosed with a disease prevalence of 10%, increasing to 290/1,000 misdiagnosed subjects with a disease prevalence of 50%.
Conclusions
This systematic review showed that up to 58% of COVID‐19 patients may have initial false‐negative RT‐PCR results, suggesting the need to implement a correct diagnostic strategy to correctly identify suspected cases, thereby reducing false‐negative results and decreasing the disease burden among the population.
Keywords: evidence, false negative, RT‐PCR, SARS‐CoV‐2
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a new human coronavirus causing the Coronavirus Disease 19 (COVID‐19) pandemic. Timely and accurate SARS‐CoV‐2 infection diagnosis is crucial for patient and population management, in order to contribute to outbreak prevention, guaranteeing diagnostic accuracy, public health surveillance, tracing, prevention and control measures. 1
Real‐time reverse transcriptase polymerase chain reaction (RT‐PCR) on clinical specimens is considered the first‐line test for the diagnosis of SARS‐CoV‐2 infection, and the results are used to rule out disease. 2 Target genes for most tests include the nucleocapsid (N), spike (S) and envelope (E) proteins or the RNA‐dependent RNA polymerase gene (RdRp), with different analytical sensitivities. 3 Although RT‐PCR has high analytical specificity, resulting in minimal false‐positive rates, its diagnostic sensitivity remains suboptimal. 4 Diagnostic efficiency of RT‐PCR depends not only on analytic performances of the adopted PCR assays, but also on other factors, including viral load, type of sample, stage of infection and time from symptom onset, skill of the healthcare professionals performing the sample collection, and mutations in the viral genome. 5 , 6
Molecular tests are used to identify symptomatic or asymptomatic patients infected by SARS‐CoV‐2, and fundamental criterion for this performance is the high clinical sensitivity to avoid false‐negative results.
However, many studies have shown that false‐negative results can be generated with RT‐PCR, putting the correct identification of infected patients at risk, subsequently leaving a significant repercussion on the entire community. 7 , 8 , 9 , 10
False‐negative results can have a serious impact on pandemic control, public health policies and contact‐tracing programmes, because a proportion of cases are categorized as uninfected and can unintentionally transmit the disease. So, we aimed to estimate the rate of false negatives for the detection of SARS‐CoV‐2 RNA performed for COVID‐19 diagnosis with RT‐PCR through a systematic review of the literature.
2. METHODS
2.1. Registered protocol and reporting guidelines
The protocol of this systematic review was registered in the International Prospective Register of Systematic Reviews database (PROSPERO identifier: CRD42021236950). The report of this review followed the Preferred Reporting Items for Systematic Reviews and Meta‐Analyses of Diagnostic Test Accuracy Studies (PRISMA‐DTA). 11 Reporting of the study conforms to broad EQUATOR guidelines. 12
2.2. Criteria for considering studies for this review
This systematic review included observational studies (including diagnostic test accuracy studies) reporting the number of subjects with suspected or confirmed SARS CoV‐2 infection for whom the detection of viral RNA with RT‐PCR was performed. We considered studies enrolling patients receiving an additional RT‐PCR test as confirmation of viral infection after an initial negative result. Eligibility was not restricted by language, patient age or study setting. We included all types of RT‐PCR kits and evaluated target genes.
We excluded studies that (1) did not report the number of subjects who received RT‐PCR for further confirmation of SARS‐CoV‐2 infection following initial negative results, (2) aimed to validate a methodology, (3) evaluated sample specimens, (4) included case series and case reports, (5) included abstracts only, (6) were editorials and (7) included animal models.
2.3. Search strategy
In order to identify all primary studies, we searched the following electronic databases: Pubmed, Embase and Cochrane Central Register of Controlled Trials (CENTRAL). The search strategy was developed for PubMed (Table S1) and adapted for all databases; the adopted search strategy included the keywords “sars cov 2", "2019 ncov", "Real‐Time Polymerase Chain Reaction", "COVID‐19 Nucleic Acid Testing", "Reverse Transcriptase Polymerase Chain Reaction", “RT‐PCR”. Reference lists of potentially eligible studies were also screened. We limited the search to studies published in 2020–2021. The literature search was conducted by one investigator in February 2021.
2.4. Study selection and data collection
One author screened titles and abstracts retrieved from the database searches and selected the studies for inclusion according to eligibility criteria. A second author checked the selection. Disagreements were resolved by consensus. From each included study, one author extracted the data necessary, and a second author validated the data. The following information was recorded: (1) study design type (i.e., cross‐sectional, cohort); (2) study characteristics (authors, year, country); (3) characteristics of trial participants (i.e., sample size, age, gender, number of patients with COVID 19 diagnosis); (4) characteristics of RT‐PCR test type, the cycle threshold value for positivity, target gene); (5) investigated outcomes (false‐negative subjects). Disagreement between reviewers was resolved by consensus.
2.5. Quality assessment in individual studies
Two researchers independently assessed the methodological quality of the included studies. Diagnostic studies were evaluated with the Quality Assessment of Diagnostic Accuracy Studies (QUADAS‐2) tool. 13 Four domains were considered (patient selection, index test, reference standard and flow and timing), each rated in terms of their risk of bias and applicability to the research question. Risk of bias and applicability were judged as ‘low’, ‘high’ or ‘unclear’. Any disagreements were resolved through discussion.
Cohort studies were assessed with an adapted National Institute of Health (NIH) Quality assessment tool for the NIH for Observational cohort and cross‐sectional studies. 14 The ad hoc checklist included 12 questions. Possible answers included ‘yes’, ‘no’ ‘unclear’. Each study was rated for overall quality as either good (≥7 ‘yes’), fair (≥4 ‘unclear’) or poor (≥3 ‘no/unclear’).
2.6. Overall certainty of evidence
Two authors independently assessed the certainty of evidence for the primary outcomes using the Grading of Recommendation, Assessment, Development and Evaluation (GRADE) framework methodology. 15 , 16
Cross‐sectional and prospective studies were initially considered at high quality but were downgraded according to: risk of bias, directness of evidence, consistency or imprecision. Directness refers to the link between test of interest and disease or populations evaluated. Consistency concerns the degree of homogeneity (direction and magnitude) of results across the different studies. Imprecision describes the grade of uncertainty across the effects estimate, in other words, the width of confidence intervals for diagnostic accuracy measurement. The quality of evidence for the main outcome of interest was rated as high, moderate, low or very low, depending on evaluated domains. 17
2.7. Statistical analysis
Data were analysed with Stata V.15.1 (StataCorp). We presented data from eligible studies in evidence tables which were summarized using descriptive statistics. The percentage of false negatives was calculated using the Metaprop_one, a command to perform meta‐analysis of proportions in Stata. The false‐negative rate was calculated together with the corresponding 95% confidence interval (CI) and a forest plot was generated to show the individual and pooled false‐negative rate with 95% CI. Heterogeneity between primary studies was assessed using the Cochran's Q test and quantified with the I 2 statistic: I 2<25% reveals low heterogeneity, ≥25% I 2 <75% indicates moderate heterogeneity, I 2≥75% expresses substantial heterogeneity. We performed subgroup analysis according to study design (accuracy or cohort studies), age of participants (adults or children), time interval between initial negative to positive RT‐PCR (>3 days and ≤3 days) and type of specimen (nasopharyngeal, oropharyngeal or others). p‐value <.05 was considered statistically significant.
3. RESULTS
3.1. Studies identification and selection
The literature search, after the exclusion of duplicates and irrelevant records, identified 5,611 references. Of these, 5,530 were excluded because they did not meet the inclusion criteria. There were 81 studies considered eligible for inclusion and details were obtained from full texts. From full‐text analysis, further 49 texts were excluded, leaving a total of 32 studies 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 included in this systematic review (Figure 1).
FIGURE 1.
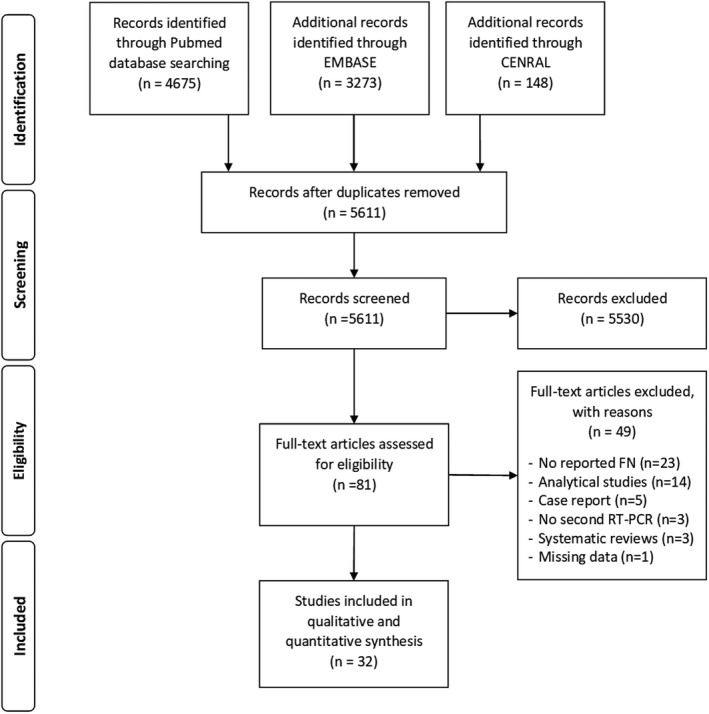
PRISMA flow diagram of the study selection process for this systematic review
3.2. Characteristics of included studies
We included 32 studies (enrolling 146,454 participants), of whom 18,565 (12.7%) were COVID‐19 confirmed cases. The number of participants ranged from 18 to 95,919. We included 15 cohort studies, 19 , 27 , 31 , 32 , 33 , 34 , 36 , 38 , 39 , 40 , 41 , 42 , 44 , 45 , 46 , 47 , 48 and 17 diagnostic accuracy studies, 18 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 28 , 29 , 30 , 35 , 37 , 43 , 49 details are reported in Table 1. Participants’ ages were reported in different ways: average mean ages ranged from 35 to 59 years, median ages ranged from 3 to 98 years. Overall, included studies reported that most patients were men (n = 9,821; 52%). The majority of studies were conducted in China (n = 19; 59%).
TABLE 1.
Characteristics of studies included in the systematic review
| Study | Country | Recruiting time | N° patients included | Men (%)/Female (%) | Age, years mean (SD) or median (range) | Type of specimen | Type of RT‐PCR (Producer) | Target gene | CT value | Day from symptom onset; mean (SD) or median(range) | Time interval between initial negative to positive PCR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AI 2020 | China | January 6‐ February 6, 2020 | 1014 | 467(46%)/547 (54%) | 51(15) | Throat swab | TaqMan One‐StepRT‐PCR Kits (HuiruiBiotechnology Co., Ltd, Shanghai) | Nr | Nr | Nr | mean 5.1±1.5 day (median 4 days, range 4–8) |
| Albert 2020 | Spain | Until April 14, 2020 | 202 | 115 (57%)/87 (43%) | 65 (3–98) | Nasopharyngeal or oropharyngeal swabs, upper RT samples | LightCycle 480 real time PCR system version II (Roche diagnostics, Pleasanton, USA) | GUSB | 31.2 | 5 (1–14) | 24–72 hours |
| Baron 2020 | Liechtenstein | first wave of the COVID−19 pandemic, until April 23, 2020 | 151 | 66 (44%)/78 (56%) | 39 (range 3–84), | Nasopharyngeal swabs | COBAS 6800 (Roche diagnostics), BD max (Becton Dickinson), CepheidGenexpert (Axon Lav) | Nr | Nr | Nr | 5, 10, 13 and 31 days |
| Besutti 2020 | Italy | February 27‐ March 24, 2020 | 696 | 408 (59%)/288 (41%) | 59 (15.8) | Nasopharyngeal or oropharyngeal wabs | GeneFinderTM COVID−19 PLUS Real Amp Kit | Nr | Nr | Nr | within 15 days |
| Chen D 2021 | China | January 19 ‐ February 20, 2020 | 21 | 9 (43%)/12 (57%) | 49.7 (15.7) | Nasopharyngeal or oropharyngeal swabs | Nr | Nr | Nr | Nr | 2 days |
| Chen HJ 2020 | China | January 21 ‐ February 4, 2020 | 34 | 21 (62%)/13 (38%) | 54.5 (11.8) | Nr | Nr | Nr | Nr | 6.3 (5.6) | Nr |
| ChenZH 2020 | China | January 24‐ February 6, 2020 | 33 | 18 (54%)/15 (46%) | 46.9 (11). | Nr | Nr | Nr | Nr | 3.12 | median 2 days |
| Çinkooğlu 2020 | Turkey | March 15‐ April 15, 2020 | 185 | 87 (47%)/ 98 (53%) | 48.7 (18–95) | Nr | Nr | Nr | Nr | Nr | mean 1.7±0.8 d |
| Fang 2020 | China | January 19‐February 4, 2020 | 51 | 29 (57%)/22 (43%) | 45 (39–55) | Throats wabs | Shanghai ZJ Bio‐Tech Co, Ltd, China | Nr | Nr | 3±3 | range 1–7 days |
| Ghazi 2020 | USA | October 3 2020‐ January 9, 2021 | 2727 | 1369 (50.3%)/1358 (49.7%) | Nr | Nasopharyngeal swab | Nr | Nr | Nr | Nr | Nr |
| Gietema 2020 | Netherlands | March 13 ‐ March 24, 2020 | 193 | 113 (59%)/80 (41) | 66 years (55–76) | Nasopharyngeal and/or oropharyngeal swabs | Quantstudio 5 (AppliedBiosystems, US) | RdRp‐gene and E‐gene | Nr | Nr | Nr |
| He JL 2020 | China | January ‐ March, 2020 | 82 | 49 (60%)/33 (40%) | 52 (8–74) 37(1–76) | Nasopharyngeal or oropharyngeal swabs, end tracheal aspirate, or bronchoalveolar lavage | BGI Genomics (Shenzhen, China) | Nr | Nr | Nr | within 14 days |
| Holborow 2020 | UK | Nr | 127 | Nr | Nr | Throat swab | Nr | E, RdRp, N1/N2 | Nr | Nr | Nr |
| Kanji 2020 | Canada | January 21 ‐April 18, 2020 | 95919 | Nr | Nr | Nasopharyngeal or oropharyngeal deep nasal turbinate swabs, endotracheal aspirates, bronchoalveolar lavages | Centers for Disease Control (Atlanta, USA) SARS‐CoV−2 assay, RNAse P rtRT‐PCR kit (Integrated DNA Technologies, Coralville, USA) | E, RdRp, N1/N2 | >35 | Nr | 6.1 days? |
| Lan FY 2020 | USA | March 9 ‐ April 15, 2020 | 592 | 125 (21%)467 (79%) | 43.6 (12.9) | Nasopharyngeal swabs | MADPH, CDC 2019‐NovelRT‐PCR; commercial laboratory, Roche Cobas SARS‐CoV−2; and hospital partner, AbbottReal Time SARS‐CoV−2 | Nr | Nr | Nr | Nr |
| Lee 2020 | Singapore | February 2020 | Nr | Nr | Nr | Nasopharyngeal swabs, sputum, stool if diarrhoea if present | LightCycler 2.0 instrument (Roche); NucliSensEasyMAG (Biomerieux); A*STAR Fortitude Kit (Accelerate Technologies, Singapore) |
N ORF1ab |
5.5 (2–22) | Nr | |
| Li 2020 | China | February 2 – February 17, 2020 | 610 | 340 (55.8%)/270 (44.2%) | 52.7 (20–88) | Pharyngeal swabs | Nr | Nr | Nr | Nr | 1‐2days |
| Long C 2020 | China | January 20 ‐ February 8, 2020 | 87 | 20 (56%)/16 (44%) | 44.8 (18.2) | Nr | Nr | Nr | Nr | Nr | Nr |
| Long D 2021 | USA | March 2 ‐April 7, 2020 | 20912 | 3287 (42%)/ 4520 (58%) | 46.6 (17.7) | Nasopharyngeal swabs | Laboratory‐developed 2‐target/2‐control assay modified from the CDC; | N1, N2 | Nr | Nr | 4±2 days? |
| 4920 (46.4%)/ 5682 (53.6%) | 46.6 (21.1) | Panther Fusion SARSCoV−2 assay (Hologic, Marlborough, MA) | ORF1ab | ||||||||
| Roche RT‐PCR (Basel, Switzerland) | E | ||||||||||
| DiaSorin (Saluggia, Italy,) |
ORF1ab S |
||||||||||
| SHC Emergency Use Authorization laboratory‐developed test | E | ||||||||||
| Lu 2020 | China | January ‐ February 2020 | 18 | 11 (61%)/7 (39%) | 35.94 (16.32) | Throat swabs | Sansure Biotech Inc (Hunan, China; Lot No. 2 020 007) |
ORF1ab N |
>40 | Nr | Nr |
| Shanghai BioGerm Medical Biotechnology Co., Ltd. (Lot No. 20200304A). |
ORF1ab N |
>38 | |||||||||
| Ma 2020 | China | January 21 ‐ February 14, 2020 | 50 | 28 (56%)/22 (44%) | 2.5 (0.9–7.0) | Respiratory secretion | Nr | Nr | Nr | Nr | Nr |
| Richardson 2020 | USA | March 1, ‐ April 4, 2020 | 5700 | 3437 (60%) 2263 (40%) | 63 (52–75) | Nasopharyngeal swabs | Nr | Nr | Nr | Nr | Nr |
| Shen 2020 | China | January 22 ‐ February 18, 2020 | 5630 | 2631 (47%)2999 (53%) | 51 (36–63) | Throat swabs | SARS‐CoV−2 nucleic acid detection kit (Shanghai Huirui Biotechnology Co. Ltd) |
N ORF1ab |
≥35 | Nr | Nr |
| Valent 2020 | Italy | March 1 ‐ April 12, 2020 | 10482 | Nr | Nr | Throat swabs | Nr | Nr | Nr | Nr | Nr |
| Wang 2020 | China | February 9 ‐ March 28, 2020 | 37 | 17 (46%)/20 (54%) | 62 | Upper respiratory tract sampling | Nr | Nr | Nr | 25 (14–37) | Nr |
| Wen 2020 | China | January 21 ‐ February 14, 2020 | 103 | 48 (47%)/55 (53%) | 46 (15) | Throat swabs, sputum or alveolar lavage fluids | Nr | Nr | Nr | Nr | 1–3 days |
| Wong 2020 | China | January 1 2020 ‐March 5, 2020 | 64 | 26 (41%)/38 (59%) | 56 (16–96) | Nasopharyngeal and throat swabs | QuantiNova Probe RT‐PCR Kit (QIAGEN, Hilden, Germany) | RdRp/helicase (Hel) | Nr | Nr | Nr |
| Wu 2020 | China | January 22 ‐ February 14, 2020 | 80 | 39 (49%)/41 (51%) | 46.1 (30.7–61.5) | Nose and/orthroat swab | Bio‐germ, Shanghai |
N ORF1ab |
Nr | Nr | Nr |
| Xiao 2020 | China | January 21 ‐ February 12, 2020 | 70 | 31 (44%)/39 (56%) | 57 (44–65) | Throat swab | Shanghai Huirui Biotechnology Co., Ltd | Nr | Nr | 22 (19–32) | Nr |
| Zhang H 2020 | China | January 22 ‐ February 28, 2020 | 194 | 108 (56%)/86 (44%) | 48.3 (33–56) | Nr | The Beijing Genomics Institute (BGI, Beijing, China) | Nr | Nr | Nr | Nr |
| Zhang JJ 2020 | China | December 29, 2019 ‐February 16, 2020 | 290 | 155 (53%)/135 (47%) | 57 (22–88) | Pharyngeal swabs | Shanghaibio‐germ Medical Technology Co Ltd). |
N ORF1ab |
Nr | Nr | Nr |
| Zhou 2020 | China | January 16 ‐February 12, 2020 | 100 | 54 (54%)/46 (46%) | 52.3 (13.1) | Pharyngeal swabs | Nr | Nr | Nr | 14 | Nr |
Abbreviations Ct, cycle threshold; Nr, not reported; SD, standard deviations.
According to the study inclusion criteria, the presence of infection in all studies was confirmed after detection of SARS‐CoV‐2 RNA using RT‐PCR assay, repeated following an initial negative result. The specimens collected for the RT‐PCR assessment were heterogeneous among included studies; most studies reported nasopharyngeal swabs (n = 13) and oropharyngeal swabs (n = 7). The SARS‐CoV‐2 RNA was detected using different kits (n = 18 studies), and 11 studies reported the specific target gene, which was most frequently the ORF1ab gene (n = 8 studies) (Table 1).
3.3. Methodological quality of included studies
We evaluated the 15 cohort studies with the QUADAS‐2 tool. Eight studies were retrospective, five studies enrolled consecutive patients and in 10 studies the enrolment was unclear. In 6 studies, the authors were blinded to results of the index test and the defined threshold value. Because the interpretation of reference standards was objective, we evaluated this domain as low risk of bias for all studies. In 6 studies, the time interval between index test and reference standard was appropriate, but in all studies except one, all patients received the same reference standard (Figure 2, Table S2).
FIGURE 2.
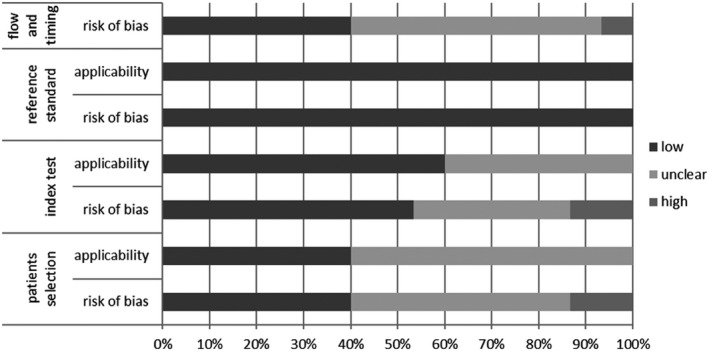
Summary of risk of bias assessment with the QUADAS‐2 tool. The x‐axis represents the percentage of studies graded to a specific risk of bias: low, moderate or high risk of bias. The y‐axis represents the 4 domains that were graded: patient selection, index test, reference standard, flow and timing
The 17 diagnostic accuracy studies were evaluated with the NIH tool. The overall methodological quality of included studies was classified as good in 12, fair in three and poor in two studies. All studies except three clearly defined the research question, the study population and the intervention. Twelve (70%) studies reported that the subjects were recruited from similar populations, 6 (35%) clearly described the inclusion and exclusion criteria, patients were enrolled consecutively in only 5 studies, and only 1 study provided a sample size justification. All studies clearly defined the outcome measures and described results adequately. The blinding of the outcome assessor was unclear in all studies, and 10 studies (59%) clearly described the statistical methods and considered an adequate follow‐up (Figure 3, Table S3).
FIGURE 3.
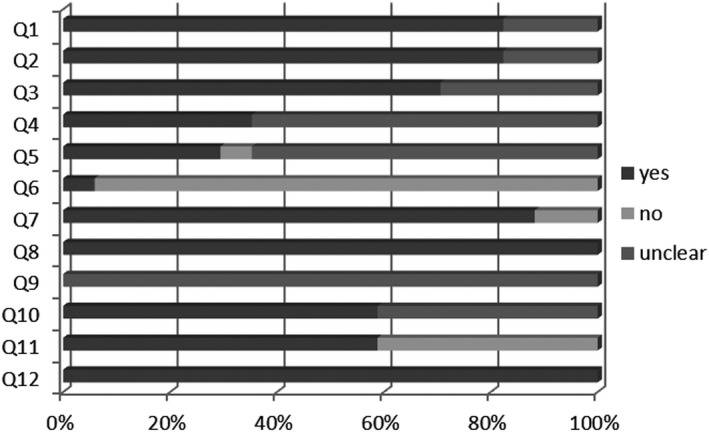
Summary of risk of bias assessment with the NIH tool. The x‐axis represents the percentage of answers: yes, no, unclear. The y‐axis reported the 12 questions considered in the evaluation. Q1: Was the research question or objective in this paper clearly stated?; Q2: Was the study population clearly specified and defined?; Q3: Were all the subjects selected or recruited from the same or similar populations (including the same time period)?; Q4: Were inclusion and exclusion criteria for being in the study prespecified and applied uniformly to all participants?; Q5: Were the cases consecutive?; Q6: Was a sample size justification, power description, or variance and effect estimates provided?; Q7: Was the intervention clearly described?; Q8: Were the outcome measures (dependent variables) clearly defined, valid, reliable, and implemented consistently across all study participants?; Q9: Were the people assessing the outcomes blinded to the participants’ exposures/interventions?; Q10: Was the length of follow‐up adequate?; Q11: Were the statistical methods well described?; Q12: Were the results well described?
3.4. False‐negative rate of SARS‐CoV‐2 RT‐PCR testing
In quantitative analyses, we included all 32 studies reporting data from 18,565 patients affected by SARS‐CoV‐2. False‐negative rates ranged from 2% 37 to 58%. 40 The summary estimate of the overall false‐negative rate was 12% (95% CI 0.10–0.14; p < .001). We observed substantial heterogeneity among included studies (I 2 = 96%) (Table 2; Figure 4).
TABLE 2.
Meta‐analyses of false‐negative rate of SARS‐CoV‐2 RT‐PCR testing
| N studies | False‐negative rate | 95%CI | I 2 | p | |
|---|---|---|---|---|---|
| False‐negative rate | |||||
| Overall | 32 | 0.12 | 0.10–0.14 | 96.2% | <.0001 |
| Study design | |||||
| Accuracy | 15 | 0.14 | 0.10–0.18 | 92.7% | <.0001 |
| Cohort | 17 | 0.12 | 0.10–0.14 | 97.4% | <.0001 |
| Age | |||||
| Adult | 28 | 0.12 | 0.10–0.14 | 96.4% | <.0001 |
| Child | 1 | 0.10 | 0.04–0.21 | Na | Na |
| Adult +child | 3 | 0.09 | 0.04–0.14 | Na | Na |
| Time interval between initial negative to positive PCR | |||||
| >3 days | 6 | 0.02 | 0.01–0.03 | 75.9% | <.0001 |
| <=3 days | 6 | 0.28 | 0.14–0.43 | 96.4% | <.0001 |
| Not reported | 20 | 0.14 | 0.12–0.17 | 96.05% | <.0001 |
| Type of specimen | |||||
| Throat swab | 7 | 0.13 | 0.08–0.18 | 95% | <.0001 |
| Nasopharyngeal | 5 | 0.03 | 0.01–0.04 | 90.8% | <.0001 |
| Pharyngeal | 3 | 0.16 | 0.02–0.3 | Na | Na |
| Mix (nasopharyngeal or oropharyngeal or others) | 12 | 0.17 | 0.13–0.21 | 95.6% | <.0001 |
| Not reported | 5 | 0.26 | 0.07–0.45 | 96% | <.0001 |
| Country | |||||
| China | 19 | 0.24 | 0.18–0.29 | 96% | <.0001 |
| Others | 13 | 0.04 | 0.03–0.06 | 90.9% | <.0001 |
Abbreviations: FN, false negative; NA, not availableNR, not reported.
FIGURE 4.
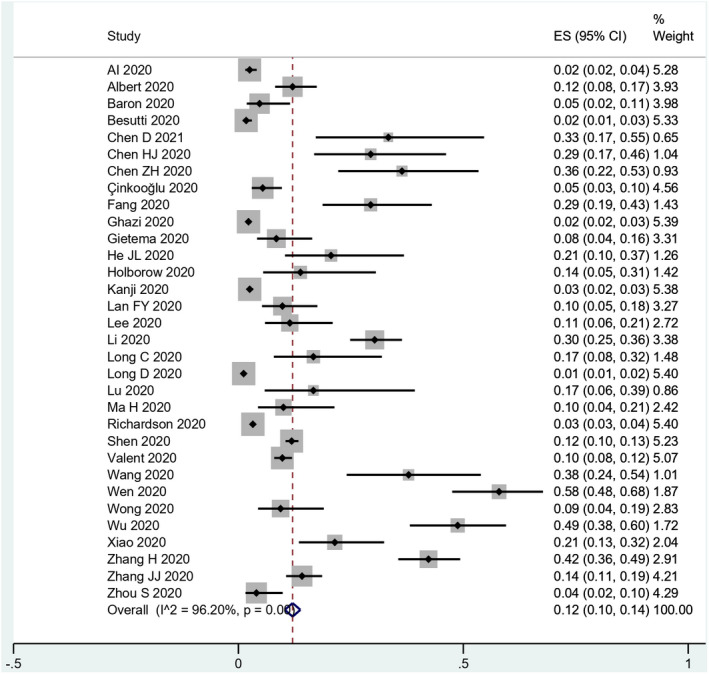
Forest plots of the false‐negative rate of RT‐PCR for SARS‐CoV‐2 infection
After subgroup analyses, we observed that the rate of false negatives was similar in diagnostic and cohort studies (14% and 12% respectively) with high heterogeneity among the included studies (Table 2, Figure S1). According to participant ages, the false‐negative rates varied: 12% in studies enrolling adults only (n = 28), 9% in studies including both adults and children (n = 3), and 10% in a single study enrolling children only (n = 1). High heterogeneity was found among included studies (Table 2, Figure S2).
The false‐negative rate was lower among studies who performed the second RT‐PCR >3 days (2%) from the first RT‐PCR, compared to studies performing test <3 days (28%), with high heterogeneity among included studies (76% vs. 96%) (Table 2, Figure S3). In the sensitivity analysis, excluding studies of He et al. 29 and Çinkooğluet al., 25 heterogeneity remained high (70% and 95%).
According to the specimen type used for RT‐PCR assessment, false‐negative rates were different. Most studies reported a mix of specimen types (n = 12) and the cumulative false‐negative rate was 17%. False‐negative rates for nasopharyngeal specimens (n = 5) were 3%, and for oropharyngeal swab (n = 3) was 16% (Table 2, Figure S4). However, 8 (25%) out of 32 included studies reporting the false‐negative rate varied according to the number of days from symptom onset, decreased from 37% (day 3) 24 to 4% after 14 days. 49
The analysis by country showed a higher false‐negative rate in studies conducted in China (24%) compared to other countries (4%), and the heterogeneity among included studies remains high (96% vs. 90%) (Table 2, Figure S5) probably due to the high prevalence of disease in this region at the beginning of 2020. 50
3.5. The evidence profile
Using the GRADE approach, we assessed the overall quality of evidence using the range of sensitivity to estimate the true value of false‐negative rates. The quality of the evidence was downgraded for the risk of bias, indirectness and inconsistency and was judged overall to be very low (Table 3). We subtracted 1 point for risk of bias for retrospective study design, 1 point for inconsistency as our meta‐analysis showed substantial heterogeneity, and an additional 1 point for indirectness as the detection of false‐negative results was not the primary aim of many of the included studies.
TABLE 3.
GRADE assessment
| Range of sensitivities: 0.42 to 0.98 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Test result | Study design | Factors that may decrease certainty of evidence | Number of results per 1.000 patients tested | Number of infections (studies) | Certainty of the Evidence (GRADE) | ||||||
| Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | Prevalence | ||||||
| 10% | 30% | 50% | |||||||||
| False negatives | Diagnostic accuracy and cohort | Serious a | Serious b | Serious c | Not serious | none | 2 to 58 | 6 to 174 | 10 to 290 | 18,565 (32) | ⨁◯◯◯ VERY LOW |
We included 17 cohort studies and 15 diagnostic accuracy studies.
High heterogeneity among included studies.
Unclear or high concerns about applicability of selected populations enrolled in studies.
To assess the impact of false‐negative outcomes, we calculated a range of patients with misdiagnosis, based on disease prevalence among the population studies. We considered three different values of disease prevalence (10%, 30% and 50%) to describe the range of false‐negative rates/1,000 tested subjects. Subject misdiagnosed according to prevalence were calculated to range from 2 to 58 for 10% prevalence, 6–164 for 30% prevalence and 10–290 for 50% prevalence (Table 3).
4. DISCUSSION
This systematic review and meta‐analysis, including data from 32 studies and more than 18,000 patients with SARS‐Cov‐2 infection, showed that the false‐negative rate for RT‐PCR detection of viral RNA for the COVID‐19 diagnosis was 12% (range 2%–58%), with a very low certainty of evidence. Many of the included studies were affected by several sources of potential bias, especially related to patient selection, description of test characteristics and data analysis.
Usually, the diagnosis of COVID‐19 is based on clinical and laboratory tests results, including chest X‐ray and chest tomography revealing images suggestive of disease, and the research of antibodies against viral proteins or the viral RNA in respiratory samples using RT‐PCR. This molecular method is considered the gold standard for COVID‐19 diagnosis, and includes the RNA extraction from samples, followed by a reverse transcription step to convert RNA into complementary DNA (cDNA) that will undergo quantitative amplification using fluorescent probes that will recognize and hybridize to segments of the amplification products. This assay has the capacity to detect the viral nucleic acids in different sample types, making it the best diagnostic test available for adequate detection of infection. This test has high specificity, but the sensitivity is imperfect, 4 making questionable the accuracy of RT‐PCR and the production of false‐negative results that impact patients’ management. The time window in which it is more likely to observe false‐negative results is not clear.
Our meta‐analysis showed high heterogeneity among included studies, which was not explained by some studies’ characteristics, limiting the interpretation of summary estimates of the proportion of the false‐negative results. We investigated plausible sources of heterogeneity, such as the type of study design, the specimen used, the age, the time to onset of symptoms, the country and the time between initial negative to positive RT‐PCR, but not all included studies reported these details. We were able to find a small reduction of this variability in subgroups; studies collecting nasopharyngeal swabs, not Chinese studies and studies with the second RT‐PCR performed >3 days from the initial tests, with false‐negative rates of 3%, 4% and 2% respectively. However, we observed that the range of false‐negative rates decreased as times increased from disease onset.
Wikramaratna et al. 51 using a statistical model, previously determined that the probability of obtaining a false‐negative result in infected patients is affected by time since symptom onset and varying swab type sensitivities (nasopharyngeal swabs are more sensitive than oropharyngeal). The authors reported that the probability of incorrectly identifying an uninfected individual due to a false‐negative test was considerably reduced if negative tests were repeated 24 h later. 51
In clinical practice, the accuracy of molecular tests is influenced by the stage of the disease 52 and the type and quality of sampling. 53 Viral RNA becomes detectable in the nasopharyngeal already from the first day of symptoms onset, reaches the peak within the first week from symptoms onset, and then decline. 52 However, the sensitivity estimated was 93% for broncho‐alveolar lavage, 72% for sputum, 63% for nasal and 32% for throat swabs. 54
RT‐PCR are often used to ‘rule out’ infection, especially among high‐risk participants, such as exposed inpatients and health care workers: In these cases, a negative RT‐PCR result is often interpreted as the absence of disease. 55 Using the GRADE approach, we evaluated the impact of the rate of false negative on 1,000 subjects tested, considering three different values of prevalence (10%, 30% and 50%). Even if the sensitivity is as high, the risk of false‐negative test results will be substantial, as testing becomes more widespread and the prevalence of COVID‐19 infection rises. Otherwise, if the sensitivity of the test was poor, the number of false‐negative results will be very high, even with low disease prevalence.
Studies included in this systematic review observed a wide range of false‐negative RT‐PCR results for SARS‐CoV‐2 infection according to a previous systematic review reporting that up to 36% of patients with COVID‐19 may have an initial false‐negative result. 56 Recent findings showed that these false‐negative results may be determined by several factors, such as the type of specimen type, 57 temporal variation in viral shedding, 58 or diagnostic primer/probe mismatches with infecting virus sequence. 59 These false‐negative rates have several implications on correct diagnosis, for public safety, health care worker safety, subsequent community transmission, and on health and economic policies to contain the SARS‐Cov‐2 pandemic. The magnitude of this concern is difficult to determine due to scarce information about test performance. Possible causes for false‐negative tests could include (1) pre‐analytical errors, which occur during the sampling procedure (i.e., skills of healthcare worker, patients cooperation) or during the sample transport (sample degradation), (2) laboratory errors due to scarce analytical sensibility, or presence of RT‐PCR inhibitors, (3) imperfect timing of sampling during the course of the disease (if samples are taken in the early infection phase false‐negative results are increased; variability due to the disease severity) due to the variability in viral shedding (viral nucleic acid in the tissue usually declines after the peak of symptoms).
In light of the data produced from this meta‐analysis, the correct interpretation of RT‐PCR results requires (1) clear definitions of pre‐analytical risk levels prior to molecular testing; (2) cautious evaluation of negative results for subjects at high risk of SARS‐CoV‐2 infection; (3) second sample testing for subjects with high suspicion of SARS‐Cov‐2 infection following negative results; (4) the development of tests or combination of tests (detection of antibodies and viral genome) with high sensitivity to minimize the risk of false‐negative results.
In order to be a reliable test for SARS‐CoV‐2, it is necessary that assays identifying viral genome should be accurate. RT‐PCR sensitivity is estimated to range between 70% and 98%, while the specificity is approximately 95%, 60 , 61 and for the diagnosis of other coronavirus infections, the overall summary of sensitivity and specificity was 89% and 99% respectively. 62 Likewise, the sensitivity and specificity of RT‐PCR is not 100% for the determination of other pathogens, the sensitivity ranges from 65% for Cytomegalovirus 63 to 97% for Legionella 64 and 96% for Mycobacterium tuberculosis, 65 suggesting a similar false‐negative rate observed for SARS‐CoV‐2 detection. The specificity ranges from 92% for Mycobacterium tuberculosis 65 to 94% for Cytomegalovirus 63 and 98% for Legionella, 64 minimizing the impact of false‐positive results.
This systematic review has a number of limitations that call for caution in result interpretation. Our meta‐analyses were afflicted by substantial heterogeneity among included studies which were difficult to explain by studies’ characteristics. Furthermore, due to very‐low quality of evidence, the results should be interpreted with additional caution. Included studies were affected by several sources of potential bias, limiting their applicability. In addition, most included studies did not aim to evaluate the number of false‐negative results of RT‐PCT for COVID‐19 diagnosis. Finally, included studies used various methods for SARS‐CoV‐2 testing, reducing the standardization for the molecular diagnosis of COVID‐19. However, we assumed the second RT‐PCR as a gold standard, and this could underestimate the rate of false‐negative. RT‐PCR is a test with high specificity, resulting in a small false‐positive rate. 66 , 67 Cohen et al. 68 showed evidence indicating that COVID‐19 RT‐PCR tests have a low but significant false‐positive rate (between 0.2% and 0.9%) calculated with real‐world data. These rates may seem low, but when the rate of infection is low, even a small false‐positive rate can greatly diminish the reliability of positive test results. Although false‐negative results having a substantial impact on patients’ management and influencing the propagation of the virus, the consequence of false‐positive results could be significant. In fact, a false‐positive test result can lead not only an unnecessary quarantine and contact tracing, but also an incorrect diagnosis, delaying or depriving patients of appropriate treatment. 68
A last challenge is related to time between first and second RT‐PCR. Our analysis showed that the false‐negative rate was lower among studies performing the second RT‐PCR >3 days from the first RT‐PCR, compared to studies performing test <3 days. This result is based on data reported by only 12 studies (37%) while the most of included studies not reported information about additional RT‐PCR within the appropriate days to the first result, and not all patients enrolled in included studies received a second molecular testing.
In conclusion, up to 58% of COVID‐19 patients may have an initial false‐negative RT‐PCR result, suggesting the need to implement a correct diagnostic strategy to correctly identify cases at high risk, reduce false‐negative results and decrease the disease burden among the population.
CONFLICT OF INTEREST
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
AUTHOR CONTRIBUTIONS
All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission. VP conceived and designed the systematic review. VP and AN wrote the protocol. VP designed and implemented the search strategy. VP and AN extracted and analysed the data. VP, AN and TT wrote the paper. All authors were involved in the critical revision of the intellectual content of the manuscript.
Supporting information
Supplementary Material
ACKNOWLEDGEMENT
The authors thank Johanna Chester for her critical revision and editing assistance.
Pecoraro V, Negro A, Pirotti T, Trenti T. Estimate false‐negative RT‐PCR rates for SARS‐CoV‐2. A systematic review and meta‐analysis. Eur J Clin Invest.2022;52:e13706. doi: 10.1111/eci.13706
Funding information
The authors received no financial support for the research, authorship, and/or publication of this article
REFERENCES
- 1. World Economic Forum ‐ Global Agenda. https://www.weforum.org/agenda/2020/03/testing‐tracing‐backbonewho‐coronavirus‐wednesdays‐briefing. Last accessed February 2021
- 2.Novel coronavirus (SARS‐CoV‐2) Discharge criteria for confirmed COVID‐19 cases – When is it safe to discharge COVID‐19 cases from the hospital or end home isolation?TECHNICAL REPORT available on https://www.ecdc.europa.eu/en/publications‐data/novel‐coronavirus‐sars‐cov‐2‐discharge‐criteria‐confirmed‐covid‐19‐cases. Last access February 2021
- 3. Udugama B, Kadhiresan P, Kozlowski HN, et al. Diagnosing COVID‐19: the disease and tools for detection. ACS Nano. 2020;14(4):3822‐3835. [DOI] [PubMed] [Google Scholar]
- 4. Dinnes J, Deeks JJ, Adriano A, et al. Rapid, point‐of‐care antigen and molecular‐based tests for diagnosis of SARS‐CoV‐2 infection. Cochrane Database Syst Rev. 2020;8:CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zou LR, Ruan F, Huang MX, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Winichakoon P, Chaiwarith R, Liwsrisakun C, et al. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID‐19. J Clin Microbiol. 2020;58:e00297–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang Y, Yang M, Yuan J, et al. Laboratory Diagnosis and Monitoring the Viral Shedding of SARS‐CoV‐2 Infection. Innovation (N Y). 2020;1(3):100061. 10.1016/j.xinn.2020.100061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581:465‐469. [DOI] [PubMed] [Google Scholar]
- 9. Zou L, Ruan F, Huang M, et al. SARSCoV‐ 2 Viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woloshin S, Patel N, Kesselheim AS. False negative tests for SARS‐CoV‐2 infection ‐ challenges and implications. N Engl J Med. 2020;383(6):e38. [DOI] [PubMed] [Google Scholar]
- 11. McInnes MDF, Moher D, Thombs BD, et al. Preferred reporting items for a systematic review and meta‐analysis of diagnostic test accuracy studies: the PRISMA‐DTA statement. JAMA. 2018;319:338‐396. [DOI] [PubMed] [Google Scholar]
- 12. Simera I, Moher D, Hoey J, Schulz KF, Altman DG. A catalogue of reporting guidelines for health research. Eur J Clin Invest. 2010;40(1):35‐53. [DOI] [PubMed] [Google Scholar]
- 13. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529‐536. [DOI] [PubMed] [Google Scholar]
- 14. NIH Study Quality Assessment Tools. Available on http://www.nhlbi.nih.gov/health‐pro/guidelines/in‐develop/cardiovascularrisk‐reduction/tools/cohort
- 15. Schunemann HJ, Mustafa RA, Brozek J, et al. GRADE guidelines: 21 part 1. Study design, risk of bias and indirectness in rating the certainty across a body of evidence for test accuracy. J Clin Epidemiol. 2020;122:129‐141. [DOI] [PubMed] [Google Scholar]
- 16. Schunemann HJ, Mustafa RA, Brozek J, et al. 21 part 2. Inconsistency, Imprecision, publication bias and other domains for rating the certainty of evidence for test accuracy and presenting it in evidence profiles and summary of findings tables. J Clin Epidemiol. 2020;122:142‐152. [DOI] [PubMed] [Google Scholar]
- 17. Schunemann H, Oxman A, Brozek J, et al. Grading quality of evidence and strength of recommendations for diagnostic test and strategies. BMJ. 2008;336:1106‐1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT‐PCR testing for coronavirus disease 2019 (COVID‐19) in China: a report of 1014 cases. Radiology. 2020;296:E32‐E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Albert E, Ferrer B, Torres I, et al. Amplification of human β‐glucuronidase gene for appraising the accuracy of negative SARS‐CoV‐2 RT‐PCR results in upper respiratory tract specimens. J Med Virol. 2021;93:48‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baron RC, Risch L, Weber M, et al. Frequency of serological non‐responders and false‐negative RT‐PCR results in SARS‐CoV‐2 testing: a population‐based study. Clin Chem Lab Med. 2020;58:2131‐2140. [DOI] [PubMed] [Google Scholar]
- 21. Besutti G, Giorgi Rossi P, Iotti V, et al. Accuracy of CT in a cohort of symptomatic patients with suspected COVID‐19 pneumonia during the outbreak peak in Italy. Eur Radiol. 2020;30:6818‐6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen D, Jiang X, Hong Y, et al. Can chest CT features distinguish patients with negative from those with positive initial RT‐PCR results for coronavirus disease (COVID‐19)? Am J Roentgenol. 2021;216:66‐70. [DOI] [PubMed] [Google Scholar]
- 23. Chen HJ, Qiu J, Wu B, et al. Early chest CT features of patients with 2019 novel coronavirus (COVID‐19) pneumonia: relationship to diagnosis and prognosis. EurRadiol. 2020;30:6178‐6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen ZH, Li YJ, Wang XJ, et al. Chest CT of COVID‐19 in patients with a negative first RT‐PCR test: comparison with patients with a positive first RT‐PCR test. Medicine (Baltimore). 2020;99(26):e20837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Çinkooğlu A, Hepdurgun C, Bayraktaroğlu S, et al. CT imaging features of COVID‐19 pneumonia: initial experience from Turkey. Diagn Interv Radiol. 2020;26:308‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fang Y, Zhang H, Xie J, et al. Sensitivity of chest CT for COVID‐19: comparison to RT‐PCR. Radiology. 2020;296:E115‐E117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ghazi L, Simonov M, Mansour S, et al. Predicting patients with false negative SARS‐CoV‐2 testing at hospital admission: a retrospective multi‐center study. medRxiv 2020 Dec 2:2020:2020.11.30.20241414. [DOI] [PMC free article] [PubMed]
- 28. Gietema HA, Zelis N, Nobel JM, et al. CT in relation to RT‐PCR in diagnosing COVID‐19 in The Netherlands: a prospective study. PLoS One. 2020;15:e0235844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He JL, Luo L, Luo ZD, et al. Diagnostic performance between CT and initial real‐time RT‐PCR for clinically suspected 2019 coronavirus disease (COVID‐19) patients outside Wuhan, China. Respir Med. 2020;168:105980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holborow A, Asad H, Porter L, et al. The clinical sensitivity of a single SARS‐CoV‐2 upper respiratory tract RT‐PCR test for diagnosing COVID‐19 using convalescent antibody as a comparator. Clin Med (Lond). 2020;20:e209‐e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanji JN, Zelyas N, MacDonald C, et al. False negative rate of COVID‐19 PCR testing: a discordant testing analysis. Virol J. 2021;18:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lan FY, Filler R, Mathew S, et al. COVID‐19 symptoms predictive of healthcare workers’ SARS‐CoV‐2 PCR results. PLoS One. 2020;15:e0235460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee TH, Junhao Lin R, Lin RTP, et al. Testing for SARS‐CoV‐2: can we stop at 2? Clin Infect Dis. 2020;71:2246‐2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Y, Yao L, Li J, et al. Stability issues of RT‐PCR testing of SARS‐CoV‐2 for hospitalized patients clinically diagnosed with COVID‐19. J Med Virol. 2020;92:903‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Long C, Xu H, Shen Q, et al. Diagnosis of the coronavirus disease (COVID‐19): rRT‐PCR or CT? Eur J Radiol. 2020;126:108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Long DR, Gombar S, Hogan CA, et al. Occurrence and timing of subsequent severe acute respiratory syndrome coronavirus 2 reverse‐transcription polymerase chain reaction positivity among initially negative patients. Clin Infect Dis. 2021;72:323‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu Y, Li L, Ren S, et al. Comparison of the diagnostic efficacy between two PCR test kits for SARS‐CoV‐2 nucleic acid detection. J Clin Lab Anal. 2020;34:e23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma H, Hu J, Tian J, et al. A single‐center, retrospective study of COVID‐19 features in children: a descriptive investigation. BMC Med. 2020;18:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Richardson S, Hirsch JS, Narasimhan M, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID‐19 in the New York City area. JAMA. 2020;323:2052‐2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen N, Zhu Y, Wang X, et al. Characteristics and diagnosis rate of 5630 subjects receiving SARS‐CoV‐2 nucleic acid tests from Wuhan, China . JCI Insight. 2020;510:e137662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Valent F, Doimo A, Mazzilis G, Pipan C. RT‐PCR tests for SARS‐CoV‐2 processed at a large Italian Hospital and false‐negative results among confirmed COVID‐19 cases. Infect Control Hosp Epidemiol. 2020;1:1‐2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang G, Yu N, Xiao W, et al. Consecutive false‐negative rRT‐PCR test results for SARS‐CoV‐2 in patients after clinical recovery from COVID‐19. J Med Virol. 2020;92:2887‐2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wen Z, Chi Y, Zhang L, et al. Coronavirus disease 2019: initial detection on chest CT in a retrospective multicenter study of 103 Chinese subjects. Radiol Cardiothorac Imaging. 2020;2:e200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wong HYF, Lam HYS, Fong AH, et al. Frequency and distribution of chest radiographic findings in patients positive for COVID‐19. Radiology. 2020;296:E72‐E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu J, Liu J, Zhao X, et al. Clinical Characteristics of Imported cases of coronavirus disease 2019 (COVID‐19) in Jiangsu Province: a multicenter descriptive study. Clin Infect Dis. 2020;71:706‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiao AT, Tong YX, Zhang S. False negative of RT‐PCR and prolonged nucleic acid conversion in COVID‐19: rather than recurrence. J Med Virol. 2020;92:1755‐1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang H, Shang W, Liu Q, et al. Clinical characteristics of 194 cases of COVID‐19 in Huanggang and Taian, China. Infection. 2020;48:687‐694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang JJ, Cao YY, Dong X, et al. Distinct characteristics of COVID‐19 patients with initial rRT‐PCR‐positive and rRT‐PCR‐negative results for SARS‐CoV‐2. Allergy. 2020;75:1809‐1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou S, Zhu T, Wang Y, Xia L. Imaging features and evolution on CT in 100 COVID‐19 pneumonia patients in Wuhan, China. Eur Radiol. 2020;30:5446‐5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and coronavirus disease‐2019 (COVID‐19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wikramaratna PS, Paton RS, Ghafari M, et al. Estimating the false‐negative test probability of SARS‐CoV‐2 by RT‐PCR. Euro Surveill. 2020;25:2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sethuraman N, Jeremiah SS, Ryo A. Interpreting diagnostic tests forSARS‐CoV‐2. JAMA. 2020;323:2249‐2251. [DOI] [PubMed] [Google Scholar]
- 53. Guo W, Zhou Q, Xu J. Negative results in nucleic acid test of COVID‐19 patients: assessment from the perspective of clinical laboratories. Ann Palliat Med. 2020;9:4246‐4251. [DOI] [PubMed] [Google Scholar]
- 54. Wang W, Xu Y, Gao R, et al. Detection of SARS‐CoV‐2 in different types of clinical specimens. JAMA. 2020;323(18):1843‐1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Usher‐Smith JA, Sharp SJ, Griffin SJ. The spectrum effect in tests for risk prediction, screening, and diagnosis. BMJ. 2016;353:i3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Arevalo‐Rodriguez I, Buitrago‐Garcia D, Simancas‐Racines D, et al. False‐negative results of initial RT‐PCR assays for COVID‐19: a systematic review. PLoS One. 2020;15(12):e0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vandenberg O, Martiny D, Rochas O, van Belkum A, Kozlakidis Z. Considerations for diagnostic COVID‐19 tests. Nat Rev Microbiol. 2021;19(3):171‐183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false‐negative rate of reverse transcriptase polymerase chain reaction‐based SARS‐CoV‐2 tests by time since exposure. Ann Intern Med. 2020;173(4):262‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang R, Hozumi Y, Yin C, Wei GW. Mutations on COVID‐19 diagnostic targets. Genomics. 2020;112(6):5204‐5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Goudouris ES. Laboratory diagnosis of COVID‐19. J Pediatr (Rio J). 2021;97(1):7‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Watson J, Whiting PF, Brush JE. Interpreting a covid‐19 test result. BMJ. 2020;369:m1808. [DOI] [PubMed] [Google Scholar]
- 62. Mustafa Hellou M, Górska A, Mazzaferri F, et al. Nucleic acid amplification tests on respiratory samples for the diagnosis of coronavirus infections: a systematic review and meta‐analysis. Clin Microbiol Infect. 2021;27(3):341‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Eguchi H, Horita N, Ushio R, et al. Diagnostic test accuracy of antigenaemia assay for PCR‐proven cytomegalovirus infection‐systematic review and meta‐analysis. Clin Microbiol Infect. 2017;23(12):907‐915. [DOI] [PubMed] [Google Scholar]
- 64. Avni T, Bieber A, Green H, Steinmetz T, Leibovici L, Paul M. Diagnostic accuracy of PCR alone and compared to urinary antigen testing for detection of legionella spp.: a systematic review. J Clin Microbiol. 2016;54(2):401‐411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wei Z, Zhang X, Wei C, et al. Diagnostic accuracy of in‐house real‐time PCR assay for Mycobacterium tuberculosis: a systematic review and meta‐analysis. BMC Infect Dis. 2019;19(1):701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Healy B, Khan A, Metezai H, Blyth I, Asad H. The impact of false positive COVID‐19 results in an area of low prevalence. Clin Med (Lond). 2021;21(1):e54‐e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Surkova E, Nikolayevskyy V, Drobniewski F. False‐positive COVID‐19 results: hidden problems and costs. Lancet Respir Med. 2020;8(12):1167‐1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cohen AN, Kessel B, Milgroom MG. Diagnosing SARS‐CoV‐2 infection: the danger of over‐reliance on positive test results medRxiv 2020.04.26.20080911 . 10.1101/2020.04.26.20080911 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


