Abstract
Severe COVID‐19 is often accompanied by coagulopathies such as thrombocytopenia and abnormal clotting. Rarely, such complications follow SARS‐CoV‐2 vaccination. The cause of these coagulopathies is unknown. It is hypothesized that coagulopathies accompanying SARS‐CoV‐2 infections and vaccinations result from bacterial co‐infections that synergize with virus‐induced autoimmunity due to antigenic mimicry of blood proteins by both bacterial and viral antigens. Coagulopathies occur mainly in severe COVID‐19 characterized by bacterial co‐infections with Streptococci, Staphylococci, Klebsiella, Escherichia coli, and Acinetobacter baumannii. These bacteria express unusually large numbers of antigens mimicking human blood antigens, as do both SARS‐CoV‐2 and adenoviruses. Bacteria mimic cardiolipin, prothrombin, albumin, and platelet factor 4 (PF4). SARS‐CoV‐2 mimics complement factors, Rh antigens, platelet phosphodiesterases, Factors IX and X, von Willebrand Factor (VWF), and VWF protease ADAMTS13. Adenoviruses mimic prothrombin and platelet factor 4. Bacterial prophylaxis, avoidance of vaccinating bacterially infected individuals, and antigen deletion for vaccines may reduce coagulopathy risk. Also see the video abstract here: https://youtu.be/zWDOsghrPg8
Keywords: cardiolipin, coagulation factors, phosphodiesterase, platelet, streptococci, thrombocytopenia, thrombosis
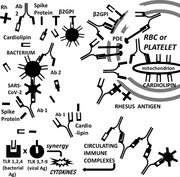
SARS‐CoV‐2 and bacteria synergize to induce COVID‐19 autoimmune coagulopathies. VIral proteins mimic blood proteins including Rh antigens, phosphodiesterases (PDE), and cardiolipin‐binding proteins (β2GPI) while bacteria express cardiolipin, complement‐, and coagulation‐factor‐like proteins. These antigens induce complementary antibodies (Ab), circulating immune complexes, cytokine over‐production via Toll‐like receptor (TLR) hyperactivation and autoimmune disease.
INTRODUCTION
SARS‐CoV‐2 is a new coronavirus causing a pandemic accompanied by significant risk of mortality. While SARS‐CoV‐2 infections generally cause minor symptoms such as fever, head‐, and muscle aches, dyspnea and temporary loss of smell and/or taste. severe cases of COVID‐19 can present with coagulopathies including viscous blood and microclotting, that can lead to impaired circulation, stroke or heart attack, respiratory complications, and other types of organ failure.[ 1 , 2 , 3 ] While mild cases of COVID‐19 have no increased risk of coagulopathies,[ 4 ] coagulopathies are estimated to affect about 10% of hospitalized patients,[ 5 ] 25% of critically ill COVID‐19 patients and up to 48% of those who end up in intensive care;[ 3 , 6 , 7 , 8 ] to affect the elderly more often than the young;[ 9 ] and to occur about ten times more frequently than among hospitalized influenza patients.[ 5 , 9 , 10 ] Such thrombotic complications are also seen rarely in people vaccinated against SARS‐CoV‐2.[ 5 , 11 , 12 , 13 ]
The cause of increased blood viscosity, microclotting, and thrombosis in COVID‐19 is unknown. Possible contributors include genetics,[ 14 ] defects in the renin‐angiotensin system,[ 15 ] defective platelet gene expression,[ 16 ] endotheliitis,[ 17 ] or cytokine storm accompanied by inappropriate complement activation.[ 6 ] Vaccine‐associated thrombosis may be linked to ethylenediaminetetraacetic acid (EDTA) preservative in the AstraZenaca formulation.[ 18 ] However, increasing evidence strongly supports an autoimmune pathogenesis for COVID‐19‐associated coagulopathies.[ 19 , 20 ]
Autoantibodies directed at phospholipids and phospholipid‐binding proteins have been identified in the majority of COVID‐19 patients affected by coagulopathies but rarely in mild cases[ 21 ]; these include lupus anticoagulant, antibodies against cardiolipin (CL), and antibodies against the cardiolipin‐binding proteins phosphatidylserine/prothrombin (Factor 2) and beta‐2 glycoprotein I (β2GPI).[ 7 , 21 , 22 , 23 , 24 ] However, these antibodies are also found transiently in many COVID‐19 patients who do not develop coagulopathies, calling their causal relationship into question,[ 25 ] a point of great significance to be discussed below. Autoantibodies against platelet factor 4 (PF4), a platelet activating factor that binds heparin (as well as bacterial antigens), have also been documented in thrombotic COVID‐19 patients[ 26 , 27 ] as well as in vaccinees who have developed thrombotic thrombocytopenia[ 28 , 29 ] but, again, are found in many patients who do have not developed clinically‐evident coagulopathies.[ 28 , 30 ] Severity of COVID‐19 also correlates with significant alterations in the function and expression of a range of other clotting factors including significant increases in von Willebrand factor (VWF), Factors IX, X, and Xa, and significant decreases in ADAMTS13 (VWF‐cleaving protease or VWFCP)[ 31 , 32 , 33 ] and autoantibodies against these proteins are found in some SARS‐CoV‐2 infected patients.[ 24 ] This range of autoantigen targets is a key point that any explanation of COVID‐19 coagulopathies must address.
Hypothesis: COVID‐19 coagulopathies are due to autoimmunity induced by virus‐bacteria synergy
I propose that the autoimmune blood‐related coagulopathies associated with SARS‐CoV‐2 infection are a result of two, synergistic phenomena: the first is molecular mimicry between SARS‐CoV‐2 proteins and human blood proteins; the second is immunological hyperactivation due to SARS‐CoV‐2 synergy with specific viral and/or bacterial co‐infections. Both are necessary but neither is sufficient to induce autoimmune coagulopathies. SARS‐CoV‐2 mimicry of host blood proteins sets the stage for the potential production of antibodies capable of cross‐reacting with host proteins. Such autoantibodies need not, however, progress to autoimmune disease. The fact that only severely ill COVID‐19 patients develop coagulopathies[ 6 , 7 , 8 ] demonstrates that such molecular mimicry is not, in and of itself, sufficient to trigger autoimmune disease, which requires innate immune system hyper‐activation. Co‐infections may be necessary to create this hyperactivation and are common in severe COVID‐19, most often involving bacteria, less frequently, viruses such as influenza and adenoviruses, or fungi. The bacteria that are most often found in severe COVID‐19 cases include Streptococci, Staphylococcus aureus or hemolyticus, Hemophilus influenzae or parainfluenzae, Klebsiella pneumoniae, Mycoplasma pneumoniae, Escherichia coli, Acinetobacter baumanii, and Pseudomonas aeruginosa (e.g.,[ 2 , 34 , 35 , 36 , 37 , 38 ]).
Additional hypothesis: Viral and bacterial mimicry of coagulation and complement proteins may directly alter function
Autoimmunity may not be the only factor at work in COVID‐19‐associated coagulopathies: viral and/or bacterial mimics may directly modify blood coagulation or platelet activation. For example, SARS‐CoV‐2 spike protein is structurally similar enough to some blood coagulation factors that Factors Xa and FIIa are able to cleave it, stimulating enhanced viral entry into susceptible cells.[ 39 ] The implications of this similarity for direct intervention by SARS‐CoV‐2 proteins or COVID‐19‐associated bacterial antigens by Factors Xa and FIIa have not yet been investigated and may extend to other blood protein mimics.
The purpose of this paper is to explore the range of similarities between human blood protein antigens and SARS‐CoV‐2 compared with other respiratory viruses such as influenza and adenoviruses, as well as the range of similarities between human blood proteins and bacteria highly associated with severe COVID‐19 such as Streptococci, Staphylococci, E. coli, Pseudomonas, and Acinetobacter baumannii.
METHODS
Similarity search procedures
Two types of similarity searches were carried out to identify likely molecular mimics shared by SARS‐CoV‐2 proteins (accessed on 2 May 2021 from https://viralzone.expasy.org/8996) and human blood proteins. The first type of search utilized BLASTP (version 2.2.31+) on the www.expasy.org server. BLOSUM80 was used to identify the type of short, continuous sequences approximately 10–15 amino acids in length that are presented by Human Leukocyte Antigens (HLA) to T and B cells.[ 40 , 41 ] The E value was set to 1000; filter low complexity regions on; no gaps; 3000 best scoring and best alignments to show. Only matches that had a Waterman‐Eggert score of at least 50, an E value of less than 1.0 and which contained a sequence of 10 amino acids in which at least six were identical, were counted as sufficiently similar to induce possible cross‐reactive immunity; this criterion is based on substantial research demonstrating that sequences exhibiting this degree of similarity have a high probability of being cross‐reactive under experimental conditions.[ 42 , 43 , 44 , 45 ]
The second search method employed LALIGN (www.expasy.org) to do a deeper dive into the SARS‐CoV‐2 protein similarities identified by the BLAST searches. The search algorithm was set to BLOSUM80; gap penalty at ‐10.0; E value, 10; 20 best matches displayed. The control viruses were poliovirus type 1, coxsackievirus B3, hepatitis A virus, rhinovirus 2, adenovirus 5, and influenza virus H1N1 (Wilson). UniProt accession numbers for the viruses and for the human blood proteins, as well as a list of the blood proteins, are provided in the TABLE captions. As with the BLAST searches, and for the same reasons, the LALIGN results were further culled for sequences with E < 1, Waterman‐Eggert score >45 and sequence similarity having a region containing at least six out of ten identities. The number of matches simultaneously satisfying the E value, Waterman‐Eggert, and 6‐of‐10 criteria were tabulated (Table 1) and representative matches provided (Figure 1).
TABLE 1.
Summary table of LALIGN search results comparing human blood proteins with SARS‐COV‐2 proteins (https://viralzone.expasy.org/8996), poliovirus type 1 P03300; coxsackievirus B3 P03313; hepatitis A virus P06441; Rhinovirus 2 P04936; Adenovirus C5 complete genome https://www.ncbi.nlm.nih.gov/nuccore/AC_000008.1; and Influenza HI1N1 (Wilson): HIN1 Neuraminidase P03470: H1N1 Matrix Protein P05777; H1N1 Hemagglutinin P03454;H1N1 PBP2 P03427;H1N1 HDRP P03430; H1N1 Non‐Struct. Q82506;H1N1 PAP P15659’H1N1 Nucleoprotein P15682
| VIRUS | TOT | CL | SerAlb | C1q | C3 | C4 | C5 | PDE2–5 | RhA‐D | F2 | VWF | F IX | F10 | ADTS13 | Β2GP | CD55 | PF4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Poliovirus type 1 polyprotein (13 proteins) | 22 | 0 | 0 | 0 | 2 | 3 | 0 | 2 | 0 | 0 | 7 | 0 | 0 | 2 | 2 | 4 | 0 |
| Coxsackievirus B3 polyprotein (13 proteins) | 24 | 0 | 0 | 3 | 1 | 6 | 2 | 3 | 0 | 3 | 2 | 0 | 1 | 1 | 0 | 2 | 0 |
| Hepatitis A virus polyprotein (14 proteins) | 23 | 0 | 3 | 2 | 2 | 0 | 0 | 2 | 2 | 2 | 3 | 2 | 0 | 1 | 1 | 2 | 1 |
| Rhinovirus C3 poly‐protein (17 proteins) | 34 | 0 | 3 | 1 | 2 | 3 | 2 | 4 | 5 | 0 | 5 | 0 | 0 | 2 | 2 | 4 | 1 |
| Adenovirus 5 (36 proteins) | >69 | 0 | 2 | 1 | 2 | 2 | 7 | 13* | 6 | >20* | 3 | 2 | 2 | 2 | 0 | 0 | 7* |
| Influenza H1N1 (Wilson) (9 proteins) | 66 | 0 | 1 | 3 | 10* | 5 | 1 | 6 | 3 | 7 | 6 | 5 | 6 | 9* | 3 | 0 | 1 |
| AVERAGE # of MATCHES (17 proteins) | 36.2 | 0 | 1.5 | 1.8 | 1.8 | 3.2 | 2.0 | 3.4 | 2.7 | 2.4 | 4.3 | 1.5 | 1.5 | 1.6 | 2.0 | 0.9 | 0.8 |
| COVID‐19 (13 proteins) Totals | 169 | 0 | 6 | 2 | 11 | 8 | 6 | 42 | 32 | 9 | 15 | 7 | 7 | 12 | 3 | 6 | 2 |
| COVID‐19 Replicase 1a P0DTC1 | 57 | 0 | 2 | 1 | 1 | 3 | 5 | 16 | 15 | 5 | 1 | 1 | 0 | 2 | 1 | 2 | 2 |
| COVID‐19 Spike Protein P0DTC2 | 30 | 0 | 0 | 0 | 3 | 1 | 0 | 9 | 6 | 3 | 3 | 2 | 1 | 2 | 0 | 0 | 0 |
| COVID‐19 Protein 3a P0DTC3 | 10 | 0 | 2 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 2 | 2 | 0 | 0 | 0 |
| COVID‐19 Small Envelope P0DTC4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| COVID‐19 Membrane protein P0DTC5 | 13 | 0 | 1 | 0 | 2 | 1 | 1 | 1 | 3 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| COVID‐19 Non‐Struct P0DTC6 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| COVID‐19 Protein 7a P0DTC7 | 6 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| COVID‐19 Protein 8 P0DTC8 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 |
| COVID‐19 Nucleoprotein P0DTC9 | 13 | 0 | 0 | 1 | 1 | 1 | 0 | 3 | 0 | 0 | 3 | 1 | 0 | 2 | 0 | 1 | 0 |
| COVID‐19 Replicase 1ab P0DTD1 & | 17 | 0 | 0 | 0 | 1 | 2 | 0 | 3 | 4 | 0 | 2 | 1 | 2 | 0 | 1 | 1 | 0 |
| COVID‐19 Protein 9b P0DTD2 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| COVID‐19 NS14 P0DTD3 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| COVID‐19 Protein 7b P0DTD8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
These viral proteins were compared with the following human proteins: Ser Alb = serum albumin P02768; # PDE = Platelet phosphodiesterase: PDE2 O95551; PDE2a O00408; PDE3a Q92484; PDE3b Q13370; PDE5a O76074; ^ Rh = Rhesus blood types: Rhesus A Q02094; Rhesus B Q9H310; Rhesus C Q9UBD6; Rhesus CE P18577; Rhesus D Q02161; & C = Complement: C1q P02745; C3 P01024; C4 P0C0L4; C5 P01031;; F2 = prothrombin (Factor 2) P00734; VWF = von Willebrand Factor P04275; F IX = Factor IX P00740.; Factor X P00742; ADAMTS13 (von Willebrand factor‐cleaving protease or VWFCP) Q76LX8; β2GP (beta‐2 glycoprotein I) P02749; CD55/DAF P08174; PF4 (platelet factor 4) P02776; CL = cardiolipin. & = P0DTD1 (Replicase 1ab) overlaps P0DTC1 (Replicase 1a), so only additional (1b) protein matches are recorded.# = number; > = greater than; * = not counted in averages because significantly out of range of other data points. Out‐of‐range data were defined as being values at least three times the value of the averages of the other values for that protein. Underlined, numbers are at least five times the average for those proteins.
FIGURE 1.

Selected similarities between SARS‐CoV‐2 proteins and human blood proteins. WE = Waterman‐Eggert; lines represent identical amino acids in the compared sequences while colons represent amino acid similarities. Blood protein abbreviations can be found in the caption to Table 1
The same BLAST protocol described above was employed to analyze possible bacterial similarities to human blood proteins. Each blood protein listed above was used as a search string against the entire UniProtKB bacterial database. The results were then screened for the presence of bacteria associated with COVID‐19 (see Introduction): Acinetobacter baumanii, E. coli, H. influenzae and parainfluenzae, Klebsiella, M. pneumoniae, Mycobacteria (tuberculosis as well as atypical forms), P.aeruginosa, S. aureus, and pathogenic or commensal streptococci. The results were, as above, screened for significance using the criterion of six identities in a sequence of 10 amino acids.
Cardiolipin could not be searched using either BLAST or LALIGN since it is not a protein but its presence in each bacterium was determined from existing experimental literature.[ 46 , 47 ]
Statistics
Statistics were applied to the tabulated LALIGN results using a paired T‐test to explore pairwise comparisons between each class of virus‐human protein combination and every other (https://www.graphpad.com/quickcalcs/ttest2/). Since all possible (28) permutations of the results were explored, a Bonferroni correction was applied to the resulting p values (https://www.easycalculation.com/statistics/bonferroni‐correction‐calculator.php). To satisfy p = 0.05 after a Bonferroni correction, the uncorrected p value had to be <0.0024 (T > 3.75) and to satisfy p = 0.01, the uncorrected value had to be <0.0005.
RESULTS
SARS‐CoV‐2 mimicry of human blood proteins
Table 1 displays the LALIGN results comparing each viral protein with the human blood‐related proteins. One hundred and sixty‐nine matches that satisfied the criteria laid out in the Methods (briefly, a WE score over 50, E less than 1.0, and at least six amino acid identities in a sequence of 10) were found between SARS‐CoV‐2 proteins and the human blood and serum proteins. The SARS‐CoV‐2 total compared with an average of 26 matches for poliovirus type 1, coxsackievirus B3, hepatitis A virus, and rhinovirus C – a six point five‐fold difference – and an average of about 66 matches for the adenovirus 5 and Influenza virus H1N1 (Wilson) pair – a three‐fold difference. In short, SARS‐CoV‐2 incorporates many times the number of human blood mimics than any other respiratory virus. Two SARS‐CoV‐2 proteins accounted for the majority of these matches: the replicase 1a (P0DTC1) and spike protein (P0DTC2); these matches occurred more than six times as frequently as in any of the control viruses. The spike protein (P0DTC2) displays as many similarities to human blood proteins as does the entire proteome of the average virus while replicase 1a exhibits as many similarities as the entire proteome of adenovirus 5 and influenza virus H1N1.
The statistical significance of differences in incidence of human blood protein in Table 1 was evaluated using a paired T‐test with a Bonferroni correction (Table 2). SARS‐CoV‐2 exhibits antigenic mimicry with human blood proteins at a rate statistically significantly greater than the control virus average and from each of the viruses individually except adenovirus 5. Of the control comparisons, only polio, adenovirus, and influenza virus differed significantly from the virus average and also differed significantly from each other. The remaining control comparisons were statistically non‐significant compared with each other and with the virus average.
TABLE 2.
Paired T‐test statistics for Table 1 data
| Paired T‐test | Poliovirus type 1 | Coxsackie B3 | Hepatitis A | Rhinovirus C | Influenza H1N1 | Adeno‐virus 5 | Virus Average |
|---|---|---|---|---|---|---|---|
| Coxsackie B3 | t = 1.6653, P = 0.12 | ||||||
| Hepatitis A | t = 0.2215, P = 0.83 | t = 0.9787, P = 0.35 | |||||
| Rhinovirus C | t = 2.9245, P = 0.01 | t = 0.3801, P = 0.71 | t = 2.0903, P = 0.06 | ||||
| Influenza H1N1 | t = 2.6687, P = 0.02 | t = 1.3756, P = 0.19 | t = 2.2191, P = 0.05 | t = 1.1825, P = 0.26 | |||
| Adenovirus 5 | t = 2.0368, P = 0.050 | t = 1.9858, P = 0.056 | t = 2.0872, P = 0.045 | t = 1.5385, P = 0.13 | t = 0.1201, P = 0.91 | ||
| Virus Average | t = 3.8000, P = 0.002 | t = 0.0957, P = 0.93 | t = 1.7165, P = 0.11 | t = 0.8062, P = 0.43 | t = 1.8953, P = 0.08 | t = 1.6388, P = 0.11 | |
| SARS‐CoV‐2 | t = 8.9314, P < 0.0001 | t = 6.7729, P < 0.0001 | t = 7.7645, P < 0.0001 | t = 7.2111, P < 0.0001 | t = 6.0688, P < 0.0001 | t = 2.4551, P = 0.027 | t = 7.8312, P < 0.0001 |
To satisfy p = 0.05 after a Bonferroni correction for the 28 pairwise comparisons made in this Table, the p value must be <0.002; to satisfy p = 0.01, the corrected value must be <0.0005. Of the control comparisons, only polio as compared with the virus average is statistically significant after correction. All SARS‐CoV‐2 comparisons with other viruses and the virus average are highly statistically significant by satisfying a Bonferroni‐corrected P value of <0.002 (T > 3.75). Significant results are highlighted in bold.
Examples of the SARS‐CoV‐2 protein‐human protein matches are provided in Figure 1, which includes additional statistical measures (Waterman‐Eggert or WE scores as well as E values). WE scores above 50 and E values below 1.0 are generally considered to be statistically significant when the E value for the search has been set at 1000, as it was here. The largest group of similarities (Table 1) involves similarities between the SARS‐CoV‐2 Replicase 1a or spike protein and platelet phosphodiesterases or Rh blood group proteins. Additional matches above the statistical virus average occur between these two SARS‐CoV‐2 proteins and complement C3, C4, and C5 and prothrombin. The overall SARS‐CoV‐2 proteome exhibits significantly increased (three‐fold or more) similarities to serum albumin, clotting factors, platelet phosphodiesterases, Rh blood group proteins, prothrombin, VWF, Factor IX, Factor Xa, ADAMTS13, and CD55/DAF.
Notably, adenovirus 5, which is used as a vector for the SARS‐CoV‐2 spike protein in the AstraZeneca COVID‐19 vaccine, also has an unusually large number of similarities to some human blood proteins, including complement factor C5, various phosphodiesterases, prothrombin, and especially platelet factor 4 (Table 1). More than 20 sequences (the limit that LALIGN can identify) within the adenovirus proteome match prothrombin (Figure 2).
FIGURE 2.

Selected similarities between adenovirus 5 (“ADENO”) and human blood proteins. WE = Waterman‐Eggert; lines represent identical amino acids in the compared sequences while colons represent amino acid similarities. Blood protein abbreviations can be found in the caption to Table 1
Influenza A virus H1N1 exhibits an unusually large number of similarities to complement factor C3 and to ADAMTS13 (Table 1).
Overall, both influenza and adenoviruses have significantly more similarities with human blood proteins than do rhinoviruses, coxsackieviruses, polioviruses, or hepatitis A viruses or the overall virus average. Both viruses, however, display significantly fewer similarities than does SARS‐CoV‐2.
COVID‐19‐associated bacteria mimicry of human blood proteins
While pairwise similarity searches between the viruses and human blood proteins were performed using LALIGN, bacteria have thousands of proteins making such pairwise impossibly laborious, so BLAST was used instead. Thus, the bacteria results are not directly comparable to the virus results. Rather than investigating how many distinct bacterial proteins matched human blood proteins, Table 3 displays instead only whether any particular bacterial species displayed at least one significant similarity that satisfied the stringent criteria employed (a Waterman‐Eggert score above 50 and at least six amino acid identities in a 10 amino acid sequence).
TABLE 3.
Selected similarities between COVID‐19‐associated bacterial proteins and human blood proteins and summarized SARS‐CoV‐2 and adenovirus data from Table 1
| Bacterial species | TOTCAT | CL | SerAlb | C1q | C3 | C4 | C5 | PDE2–5 | RhA‐D | F2 | VWF | F IX | FX | ADAMTS13 | β2GP | CD55 | PF4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acinetobacter | 7 | x | X | x | x | x | x | X | |||||||||
| Chlamydia | 1 | x | |||||||||||||||
| Clostridium | 3 | x | x | x | |||||||||||||
| Escherichia coli | 7 | x | X | x | x | x | x | x | x | ||||||||
| Hemophilus | 2 | x | x | ||||||||||||||
| Klebsiella | 4 | x | X | x | x | ||||||||||||
| Mycobacterium | 3 | x | X | x | |||||||||||||
| Mycoplasma | 1 | x | |||||||||||||||
| Pseudomonas | 3 | x | X | x | |||||||||||||
| Staphylococci | 4 | x | X | X | x | x | |||||||||||
| Streptococci | 8 | x | X | x | x | X | x | x | |||||||||
| SARS‐CoV‐2 | 11 | X | x | x | x | x | x | x | x | x | X | x | |||||
| Spike Protein | 5 | x | x | x | x | x | |||||||||||
| Adenovirus 5 | 3 | x | x | x |
The following human proteins were compared to the entire UniProtKB bacterial protein database using BLAST 2.1: Ser Alb = serum albumin P02768; # PDE = phosphodiesterase: PDE2 O95551; PDE2a O00408; PDE3a Q92484; PDE3b Q13370; PDE5a O76074; Rh = Rhesus blood types: Rhesus A Q02094; Rhesus B Q9H310; Rhesus C Q9UBD6; Rhesus CE P18577; Rhesus D Q02161; C = Complement: C1q P02745; C3 P01024; C4 P0C0L4; C5 P01031; F2 = prothrombin (Factor 2) P00734; VWF = von Willebrand Factor P04275; F IX = Factor IX P00740.; Factor X P00742; ADAMTS13 (von Willebrand factor‐cleaving protease or VWFCP) Q76LX8; β2GP = beta‐2 glycoprotein I P02749; CD55/DAF (decay accelerating factor) P08174; PF4 (platelet factor 4) P02776; CL = cardiolipin.
As can be seen from Table 3, only some of the bacteria identified as co‐infections of SARS‐CoV‐2 display significant similarities with human blood proteins. Streptococci, E. coli, and A. baumannii each exhibited similarities to six or seven classes of blood proteins, including serum albumin, VWF, prothrombin, beta‐2‐glycoprotein I, CD55/DAF, and platelet factor 4. Some of these similarities are illustrated in Figure 3. Figure 3 also displays two sets of extraordinary similarities, one between A. baumannii and complement factor 4 (C4), and the other between both S. pneumoniae and E. coli with complement factor C1q, which repeats dozens of times within the protein.
FIGURE 3.

Selected BLAST similarities between COVID‐19‐associated bacterial proteins and human blood proteins. WE is Waterman‐Eggert score; + represents amino acid similarity. Protein abbreviations can be found in the caption to Table 1
Staphylococci (pyogenes and aureus species) and Klebsiella (pneumoniae and m ichiganiensis) each displayed similarities with three classes of human blood proteins (Table 3 and Figure 3), while P. aeruginosa, Mycobacterium (tuberculosis and avium), and Clostridium (perfringens, clostridioforme, and difficile) displayed similarities to only two classes; Haemophilus influenzae exhibited only one, and Chlamydia pneumoniae and Mycoplasma species, none. Some bacteria associated with COVID‐19 are therefore significantly more likely to mimic human blood proteins than others and therefore to participate as possible inducers of autoimmune coagulopathies in the presence of SARS‐CoV‐2.
Note that all the bacteria listed in Table 3 are known to incorporate cardiolipin in their cell membranes[ 46 , 47 ] and therefore to have an antigenic mimic to human cardiolipin. No virus is known to do so (Table 1). Since anti‐cardiolipin antibodies are found in COVID‐19 coagulopathy patients, but rarely in mild and asymptomatic patients (see Introduction), the presence of anti‐cardiolipin antibodies is strong evidence for bacterial coinfections in COVID‐19 coagulopathies.
DISCUSSION
SARS‐CoV‐2 proteins contain extraordinarily large numbers of blood protein mimics
SARS‐CoV‐2 proteins contain an extraordinarily large number of antigens that mimic human blood proteins compared with any of the other viruses (mostly respiratory) that were examined. While mimicry between proteins does not translate directly into increased risk of autoimmune disease (an important fact that will be discussed further below), the unusual number of such similarities certainly increases the probability of inducing cross‐reactive antibodies – in this case, up to six times the probability of such cross‐reactivity occurring following other virus infections. This result correlates well with the fact that hospitalized SARS‐CoV‐2‐infected patients are about 10 times more likely to develop coagulopathies than are hospitalized influenza patients.[ 5 , 10 , 11 , 12 ]
The most common SARS‐CoV‐2 similarities with blood proteins involved the virus spike protein and replicase 1a protein and these displayed numerous similarities to platelet phosphodiesterases and Rhesus blood antigens as well as prothrombin and VWF (Tables 1 and 2). The nucleoprotein and membrane protein also displayed an unusually large number of blood proteins similarities.
The unexpectedly large number of antigenic similarities between SARS‐CoV‐2 proteins and blood proteins discovered in this study provides two possible mechanisms by which SARS‐CoV‐2 may cause blood coagulation. One is to induce antibodies against SARS‐CoV‐2 proteins that cross‐react with red blood cells (RBC), platelet proteins and serum albumin resulting in thickening of blood, microclotting, and/or thrombosis. The other is by directly participating in blood coagulation pathways, either as agonists or antagonists. Antibodies against SARS‐CoV‐2 proteins, particularly the replicase 1ab (P0DTC1 and P0DTD1), spike protein (P0DTC2), membrane protein (P0DTC5), and nucleoprotein (P0DTC9), have a significant probability of being cross‐reactive with complement factors C3, C4, and C5 (but not C1q), all Rhesus antigens and platelet phosphodiesterases 2, 3, and 5 (Table 1 and Figure 1). It is also possible that the mimicry between SARS‐CoV‐2 proteins and complement factors, VWF and Factor IX might result in direct viral stimulation of inappropriate blood clotting or, alternatively, to idiopathic thrombocytopenia resulting from SARS‐CoV‐2 protein interference in the blood clotting pathway. Each of these possibilities will be dissected further below.
The observed antigenic similarities make sense as triggers of SARS‐CoV‐2‐induced blood clotting in terms of the observed pathology. Phosphodiesterases would make targets of both RBC and platelets. While phosphodiesterases 2, 3, and 5 are generally characterized as being intracellular proteins within platelets (e.g.,[ 48 ]) and erythrocytes (e.g.,[ 49 ]), phosphodiesterases have been found to be incorporated into lipid rafts and cellular membranes and may therefore present antibody‐accessible epitopes not only on RBC and platelets but also smooth and cardiac muscle (e.g.,[ 50 ]).
A role for Rh factors as a determinant of COVID‐19 coagulopathies is also strongly suggested by the large number of SARS‐CoV‐2 similarities revealed by this study. Rh proteins are involved in ammonia transport and regulation in both erythrocytes and in the kidneys of mammals[ 51 , 52 ] and distant evolutionary relatives of these transporters are present in the cell membranes of all bacteria.[ 53 ] In several large studies, Rh‐negative individuals (particularly type O‐negative) have substantially lower risk of severe COVID‐19 than do Rh‐positive individuals, after controlling for other risk factors[ 54 , 55 , 56 , 57 ] though three studies found no significant difference in COVID‐19 susceptibility associated with Rh status.[ 58 , 59 , 60 ] SARS‐CoV‐2 mimicry of Rh antigens and their glycosylations may camouflage SARS‐CoV‐2 from immune surveillance since glycans like those that N‐glycosylate position 37 in Rh proteins function as T‐cell checkpoints.[ 61 ] Should tolerance to Rh antigens be broken, however, the resulting immunity induced by SARS‐CoV‐2 would be likely to cross‐react with erythrocytes, inducing an autoimmune response. The bacteria listed in Table 3 may stimulate tolerance abrogation because all express a V8 proteinase[ 62 ] that directly cleaves Rh proteins at position 34, just to the N‐terminal side of the N‐glycosylation position.[ 63 ] This cleavage is likely to result in the production of unusual proteolytic fragments with higher‐than‐normal auto‐antigenicity. Because Rh proteins are involved in ammonia transport and regulation, autoimmunity directed at Rh proteins should adversely affect not only erythrocytes but also kidney function,[ 51 , 52 ] the latter problem occurring in up to 25% of severely ill COVID‐19 patients concurrent with coagulopathies.[ 64 , 65 ]
Probable roles of bacteria in triggering COVID‐19 coagulopathies
Similarities between SARS‐CoV‐2 proteins and human blood proteins do not account for some of the key autoantibodies observed in COVID‐19 patients with coagulopathies including anti‐phospholipid antibodies observed in most coagulopathy patients.[ 7 , 21 , 22 , 23 , 24 ] Cardiolipin is a diphosphatidylglycerol lipid found in human mitochondrial membranes, including those of RBC, but as such it is a “hidden antigen” unlikely to trigger autoimmunity directly. Cardiolipin is not present in any known virus but is almost ubiquitous in the cell membranes of all the bacteria associated with severe COVID‐19.[ 46 , 47 ] Additionally, cardiolipin antibodies have been found to activate the formation of NETs,[ 7 , 23 ] neutrophil extracellular traps, which are often considered to be diagnostic for the presence of bacterial infection and which were highly associated with onset of thrombocytopenia even before COVID‐19.[ 66 , 67 ] NETs — webs of chromatin, microbicidal proteins, and oxidant enzymes — are released in response to bacterial infections – stimulating cytokine release.[ 68 ] Cytokine over‐production, in turn, is correlated with the presence of bacterial coinfections of SARS‐CoV‐2 and the severity of COVID‐19 (reviewed in Ref. [69]). Other evidence of bacterial co‐infections in severe COVID‐19 are the presence of elevated ferritin,[ 70 , 71 ] C‐reactive protein,[ 71 , 72 , 73 , 74 ] procalcitonin levels,[ 71 , 74 ] as well as eosinopenia and lymphopenia[ 74 , 75 ] and cytokine overproduction syndrome (reviewed in[ 69 ]), all of which are independently diagnostic for bacterial infections and differentiate severe cases from mild and asymptomatic ones. The preceding considerations along with direct evidence of bacterial infections in non‐COVID thrombocytopenias[ 76 , 77 ] and some COVID‐19 coagulopathy cases[ 78 , 79 , 80 , 81 ] have led Di Micco et al.[ 82 ] to conclude that, “Patients with COVID‐19, because of its tendency to induce leucopenia and overlapping of bacterial infection, may experience sudden disseminated intravascular coagulation (DIC).”
Why have these results not been reported before? Relationship of results to previous studies
The results of the current study differ from some previous ones. Dotan et al.[ 19 ] searched previously for SARS‐CoV‐2 similarities to human proteins but identified none of those listed in this paper. This difference is due to their using a different search algorithm that limited results to sequential heptapeptide identities thereby missing the significant mimics reported here. On the other hand, Greinacher et al.[ 18 ] have previously reported similarities between platelet factor 4/heparin and the SARS‐CoV‐2 spike protein but found experimentally that these spike protein regions were not recognized by PF4 antibodies nor did antibodies against the spike protein recognize PF4 peptides. None of the Greinacher et al., similarities are among those reported here, however. Once again, methodology may matter.
Finally, it is important to emphasize that no one has previously explored COVID‐19‐associated bacteria for human‐blood protein similarities, which may provide the most important clues as to the origins of coagulopathies in COVID‐19.
Interpreting the results in terms of different autoimmune disease theories
The major question these results leave unresolved is why the abundance of autoantigen‐inducing targets presented by SARS‐CoV‐2 proteins and its associated bacterial co‐infections fail to result in autoimmune complications such as microclotting or thrombocytopenia in all, or at least the majority of, individuals infected with these microbes. The obvious conclusion, which has been reached previously in studies of other autoimmune diseases, is that molecular mimicry may be necessary to induce auto‐reactive B and T cells but is clearly not sufficient to induce autoimmune disease.[ 22 , 83 , 84 , 85 , 86 ] This point is essential for understanding how many hospitalized COVID‐19 patients are found transiently to express anti‐phospholipid antibodies (aPL), anti‐PL4, b2GPI, and other blood cell antibodies[ 7 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 87 , 88 ] but only a fraction of these develop coagulopathies[ 25 , 30 , 87 , 88 ] and also why SARS‐CoV‐2 vaccines (to be discussed in “Implications of bacterial and viral coinfections for understanding vaccine‐induced coagulopathies”) often induce autoantibodies but rarely autoimmune disease. Mimicry may frequently induce autoantibody production but rarely leads to overt autoimmune disease or, alternatively, mimics may be perceived by the immune system as “self” antigens resulting in T cell tolerance. The difficulty with this explanation is that it leaves open why such tolerance should occur in the majority of people but not in the minority that develop coagulopathies.
Two other theories of autoimmune disease may explain how autoimmunity develops into autoimmune disease in this minority of cases. One is the bystander activation theory which proposes a non‐specific secondary infection or adjuvant causes hyper‐activation the innate immune system, preventing the development of tolerance in T‐cells, thereby setting the stage for autoimmune disease when molecular mimicry is present.[ 84 , 89 , 90 ] Certainly, one result of bacterial co‐infections of SARS‐CoV‐2 is dramatically up‐regulated Toll‐like receptor (TLR) activation. Briefly, bacterial antigens primarily activate TLRs 1, 2, and 4 while viral antigens primarily activate TLRs 3, 7, 8, and/or 9 and the many of these viral and bacterial pathways are synergistic, producing cytokine overproduction (reviewed in[ 69 , 91 ]).
The complementary antigen theory also suggests that SARS‐CoV‐2 requires a co‐infection to hyper‐stimulate innate immunity and break tolerance to molecular mimics[ 69 , 91 ] but differs from the bystander theory in presupposing that the co‐infection must also express antigens that mimic the target tissue and are complementary to the viral ones.[ 76 , 85 , 86 , 91 , 92 , 93 , 94 ] Because the viral and bacterial antigenic triggers are molecularly complementary, the resulting immune responses will also be molecularly complementary resulting effectively in idiotype‐anti‐idiotype antibody pairs that will form circulating immune complexes (CIC) that stimulate cytokine production. Platelet‐activating CIC are found in all severely ill COVID‐19 patients but not mild cases.[ 95 , 96 , 97 ] Notably, neither molecular mimicry theory nor bystander theory predicts the formation of CIC. Complementary antibodies will, in turn, target molecularly complementary host antigens of which many examples exist in Tables 1 and 2. For example, both beta‐2‐glycoprotein I (β2GPI) and phosphatidylserine/prothrombin (Factor 2) bind to cardiolipin; cardiolipin can be provided by any of the bacteria listed in Table 3, while SARS‐CoV‐2 can provide antigenic mimics to both β2GPI and Factor 2 (Table 1 and Figure 1). Thus, SARS‐CoV‐2 could synergize with any of the bacteria to trigger autoimmunity directed at these antigenic pairs. Another example of complementary antigens consists of PF4 binding to VWF producing an antigenic complex that induces thrombus formation[ 98 ]; Streptococcus, Staphylococcus, Klebsiella, and E. coli all express PF4 mimics, while SARS‐CoV‐2 is a source of VWF mimics (Table 3), potentially producing an antigenic complex mimicking the PF4‐heparin complex responsible for heparin‐induced thrombocytopenia (HIT).[ 99 , 100 ] A very important implication of the complementary antigen theory is that SARS‐CoV‐2 is not sufficient to induce coagulopathies and there are likely to be multiple molecular targets involved in coagulopathy pathogenesis.
Implications of bacterial and viral coinfections for understanding vaccine‐induced coagulopathies
Vaccine‐related coagulopathies may also be explained by molecular mimicry combined with either bystander activation or complementary antigens. The mRNA‐based vaccines (Pfizer‐Biontech and Moderna) and the AstraZeneca and Johnson‐and‐Johnson adenovirus‐based vaccines all employ the SARS‐CoV‐2 spike protein as their main antigen. The spike protein (Table 1) is particularly rich in PDE and Rh mimics and also expresses mimics of VWF and prothrombin and other blood proteins. Adenovirus 5, which is used in the AstraZeneca vaccine, is particularly rich in PDE, prothrombin, and PF4 mimics. Additionally, adenoviruses bind to coxsackie and adenovirus receptors (CXAR) on both red blood cells and platelets,[ 101 ] cause the release of VWF from endothelial cells,[ 102 ] and the virus can bind directly to Factors IX and X.[ 102 , 103 ] Thus, intravenous delivery of adenovirus vectors[ 101 , 104 , 105 ] and adenovirus pneumonia[ 106 ] are both highly associated with thrombocytopenia. While intramuscular injection of replication‐impaired adenovirus vectors is presumably much safer than intravenous delivery or actively replicating virus, direct adenoviral interactions with coagulation‐related proteins and cells may be important in understanding vaccine‐associated coagulopathy risks.
Fortunately, the development of thrombotic thrombocytopenia and other coagulopathies following COVID‐19 vaccination is rare despite a significant rate of autoantibody production against blood proteins. Transient but significant titers of anti‐PF4 antibodies were found in 5.6% of BNT162β (the Pfizer‐Biontech mRNA vaccine) and around 8.0% of ChAdOx1 nCoV‐19 (the AstraZeneca adenovirus 5‐based vaccine) in several studies[ 107 , 108 , 109 ] and low‐titer antibodies in 67% of vaccinees in another AstraZeneca vaccine study.[ 110 ] No one in these studies developed clinically overt coagulopathies, again emphasizing the point that molecular mimicry may induce autoantibodies without inducing autoimmune disease and making antibody positivity studies of limited value in predicting complications.[ 107 , 108 , 109 , 110 , 111 ] However, these transient anti‐PF4 antibodies could result in sub‐clinical interference with blood clotting by blocking PF4's antagonism of heparin, thereby resulting in some SARS‐CoV‐2 vaccinees experiencing longer blood‐clotting times, developing blood blisters or bruises more easily, and causing some women to experience unusually heavy and early menstrual periods.[ 112 ] Clinically evident coagulopathy following vaccination does involve PF4 antibodies producing a heparin‐induced thrombocytopenia (HIT)‐like syndrome characterized by activation of platelet aggregation but in the absence of previous exposure to heparin.[ 18 , 20 , 112 , 113 , 114 , 115 ] Current evidence suggests that mRNA‐based vaccines provoke autoimmune reactions less often than adenovirus‐vectored vaccines[ 116 ] as predictable from the many adenovirus‐PF4 mimics listed Table 1 and Figure 2. The rarity of these complications, however, argues once again that molecular mimicry is not sufficient to induce vaccine‐associated coagulopathies which may require concomitant bacterial co‐infections or other causes of hyperactivation of innate immunity in vaccinees.
Direct interference in coagulopathy pathways by SARS‐CoV‐2 blood protein mimics
Molecular mimics may also interfere directly with coagulation pathways. For example, C1q binds to C4 and C3b binds to C5 in the complement pathway; prothrombin binds to Factor Xa; VWF binds to Factors VIII, and Factor VIII interacts with IX and X. Various combinations of viruses and bacteria might directly interfere in all of these pathways (Tables 1 and 2).
Additionally, all group A streptococci, E. coli, M. pneumonia, and S. aureus express a plasminogen (plasmin) receptor (glyceraldehyde‐3‐phosphate dehydrogenase) on their cell membranes that binds up serum plasminogen, blocking plasminogen's conversion to plasmin and its fibrinolytic activity.[ 117 , 118 ] Thus, these bacteria may participate both in the induction of coagulopathies and by actively preventing fibrinolysis. Notably, plasminogen levels, and consequently plasmin levels, increase significantly in COVID‐19 patients experiencing thrombosis, resulting in facilitated production of the fibrin‐breakdown product, D‐Dimer,[ 119 ] which characterizes such cases.
Proposed experimental and clinical tests of the hypotheses
In sum, it is proposed that coagulopathies following SARS‐CoV‐2 infection or vaccination are due to viral proteins expressing antigenic mimics of human blood proteins that require either a generalized bystander activation or a specific complementary activation by a bacterial (or possibly viral) coinfection to break self‐tolerance and induce autoimmune disease. Some SARS‐CoV‐2 proteins may also interfere directly with coagulation processes through such mimics. The hypotheses proposed here are experimentally testable in many complementary ways.
Animals susceptible to SARS‐CoV‐2, such as the Syrian Gold Hamster, might be infected with both the virus and a group A streptococcus, Staphylococcus, A. baumannii, or other bacteria and viruses listed here as potential blood protein mimics, with the prediction that the combinations, but not the individual agents, will induce coagulopathies. The potential for bacterial or viral infections to stimulate coagulopathies when present at the time of SARS‐CoV‐2 vaccination can be tested similarly. Direct interference with coagulation by SARS‐CoV‐2 mimics of blood proteins can be tested by introducing the appropriate SARS‐CoV‐2 proteins, or the peptide mimics illustrated in Figures 1, 2, 3, intravenously into an appropriate animal model or adding them to freshly drawn human blood, where their presence should alter coagulation parameters. The coagulant effects of SARS‐CoV‐2 antibodies might be tested similarly, both alone and in combination with bacterial, adenovirus or influenza antibodies that cross‐react with blood protein mimics. Once again, it is predicted that SARS‐CoV‐2 antibodies combined with bacterial or viral antibodies from the mimics identified here will result in the induction of coagulopathies. The detailed mechanisms might be tested by using such microbial antibodies to determine whether they bind to the particular human blood protein mimics identified here. Conversely, anti‐PF4, anti‐cardiolipin, and other autoantibodies isolated from human COVID‐19 coagulopathy patients, or raised against purified proteins in rodents, may be tested for binding to the SARS‐CoV‐2 proteins using enzyme‐linked immunoadsorption assays, Western blots or similar immunological techniques. A final implication of the results reported here is that some SARS‐CoV‐2 antigens are complementary to some bacterial antigens so that polyclonal antibodies against SARS‐CoV‐2 may precipitate (by acting like anti‐idiotypes) the polyclonal antibodies against COVID‐19‐associated bacteria; such complementarity has previously been demonstrated for influenza A virus and Hib bacterial antigens as well as their antibodies.[ 43 ]
CONCLUSIONS: TESTING AND PREVENTING COVID‐19 AUTOIMMUNITY AND MAKING VACCINES SAFER
If the hypotheses proposed here are valid, then several important diagnostic and therapeutic conclusions follow. One is that hospitalized patients need to be screened for potential bacterial or viral coinfections that might trigger coagulopathies. Sites of coinfection such as gastrointestinal, bladder, kidney, and gums should be considered in addition to blood stream and respiratory system.[ 120 , 121 , 122 , 123 ] Because SARS‐CoV‐2 might synergize with a variety of co‐infections, there will be no single cause for coagulopathies and therefore no single treatment that is optimal for all patients; the particular spectrum of autoantibodies needs to be determined. However, steroids along with either intravenous immunoglobulins[ 124 , 125 ] or plasmapheresis[ 126 , 127 ] are each effective treatments for COVID‐19‐associated coagulopathies (additional evidence that these are autoimmune in origin). Vaccination against SARS‐CoV‐2 as well as against recognized coinfections such as Streptococci, Haemophilus, and influenza virus should decrease risk of coagulopathies by preventing bystander or complementary co‐infections, just as they decrease risk of severe COVID‐19 in general.[ 128 , 129 , 130 , 131 , 132 , 133 , 134 , 135 , 136 ] Additionally, the results reported here may provide insight into the causes of menstruation alterations reported by some women following SARS‐CoV‐2 vaccination.[ 112 ] Finally, the present work has implications for future SARS‐CoV‐2 vaccine design, implying that whole virus SARS‐CoV‐2 vaccines may present an extraordinary risk of inducing coagulopathies compared with the mRNA, peptide, or subunit vaccines because of the very large number of blood protein mimics present; however removing molecular mimicry regions from SARS‐CoV‐2 mRNAs, proteins and their virus vectors may significantly improve vaccine safety.
CONFLICT OF INTEREST
None.
ACKNOWLEDGMENTS
None.
Root‐Bernstein, R. (2021). COVID‐19 Coagulopathies: Human Blood Proteins Mimic SARS‐CoV‐2 Virus, Vaccine Proteins and Bacterial Co‐Infections Inducing Autoimmunity. BioEssays, 43, e2100158. 10.1002/bies.202100158
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Zhou, F. , Yu, T. , Du, R. , Fan, G. , Liu, Y. , Liu, Z. , Xiang, J. , Wang, Y. , Song, B. , Gu, X. , Guan, L. , Wei, Y. , Li, H. , Wu, X. , Xu, J. , Tu, S. , Zhang, Y. , Chen, H. , & Cao, B. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan; China: A retrospective cohort study. Lancet, 395, 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang, J.‐J. , Dong, X. , Cao, Y.‐Y. , Yuan, Y.‐D. , Yang, Y.‐B. , Yan, Y.‐Q. , Akdis, C. A. , & Gao, Y. D. (2020). Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy, 75, 1730–1741. [DOI] [PubMed] [Google Scholar]
- 3. Connors, J. M. , & Levy, J. H. (2020). COVID‐19 and its implications for thrombosis and anticoagulation. Blood, 135(23), 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lund, L. C. , Hallas, J. , Nielsen, H. , Koch, A. , Mogensen, S. H. , & Brun, N. C. , Christiansen, C. F. , Thomsen, R. W. , Pottegård, A. (2021).Post‐acute effects of SARS‐CoV‐2 infection in individuals not requiring hospital admission: A Danish population‐based cohort study. The Lancet Infectious Diseases, 21(10), 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taquet, M. , Husain, M. , Geddes, J. R. , Luciano, S. , & Harrison, P. J. (2021).Cerebral venous thrombosis and portal vein thrombosis: A retrospective cohort study of 537,913 COVID‐19 cases. EClinicalMedicine, 39, 101061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abou‐Ismail, M. Y. , Diamond, A. , Kapoor, S. , Arafah, Y. , & Nayak, L. (2020).The hypercoagulable state in COVID‐19: Incidence, pathophysiology, and management. Thrombosis Research, 194, 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taha, M. , & Samavati, L. (2021).Antiphospholipid antibodies in COVID‐19: A meta‐analysis and systematic review. RMD Open, 7(2), e001580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Najim, M. , Rahhal, A. , Khir, F. , Aljundi, A. H. , Abu Yousef, S. , Ibrahim, F. , Amer, A. , Mohamed, A. S. , Saleh, S. , Alfaridi, D. , Mahfouz, A. , Alyafei, S. , Howady, F. , Khatib, M. , & Alemadi, S. A. (2021).Prevalence and clinical significance of antiphospholipid antibodies in patients with coronavirus disease 2019 admitted to intensive care units: A prospective observational study. Rheumatology International, 41(7), 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bhattacharjee, S. , & Banerjee, M. (2020). Immune thrombocytopenia secondary to COVID‐19: A systematic review. SN Comprehensive Clinical Medicine, (19), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burkhard‐Koren, N. M. , Haberecker, M. , Maccio, U. , Ruschitzka, F. , Schuepbach, R. A. , Zinkernagel, A. S. , Hardmeier, T. , Varga, Z. , & Moch, H. (2021). Higher prevalence of pulmonary macrothrombi in SARS‐CoV‐2 than in influenza A: Autopsy results from ‘Spanish flu’ 1918/1919 in Switzerland to Coronavirus disease 2019. Journal of Pathology: Clinical Research, 7(2), 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nicolai, L. , Leunig, A. , Brambs, S. , Kaiser, R. , Joppich, M. , Hoffknecht, M. L. , Gold, C. , Engel, A. , Polewka, V. , Muenchhoff, M. , Hellmuth, J. C. , Ruhle, A. , Ledderose, S. , Weinberger, T. , Schulz, H. , Scherer, C. , Rudelius, M. , Zoller, M. , Keppler, O. T. , … Stark, K. (2021).Vascular neutrophilic inflammation and immunothrombosis distinguish severe COVID‐19 from influenza pneumonia. Journal of Thrombosis and Haemostasis, 19(2), 574–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mahase, E. (2021). Covid‐19: AstraZeneca vaccine is not linked to increased risk of blood clots, finds European Medicine Agency. British Medical Journal (Clinical Research Edition), 372, n774. [DOI] [PubMed] [Google Scholar]
- 13. The Lancet Haematology . (2021). COVID‐19 vaccines: Building and maintaining confidence. The Lancet Haematology, 8(5), e305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gemmati, D. , Bramanti, B. , Serino, M. L. , Secchiero, P. , Zauli, G. , & Tisato, V. (2020).COVID‐19 and individual genetic susceptibility/receptivity: Role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double x‐chromosome in females be protective against SARS‐CoV‐2 compared to the single X‐chromosome in males? International Journal of Molecular Sciences, 21(10), 3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henry, B. M. , Vikse, J. , Benoit, S. , Favaloro, E. J. , & Lippi, G. (2020).Hyperinflammation and derangement of renin‐angiotensin‐aldosterone system in COVID‐19: A novel hypothesis for clinically suspected hypercoagulopathy and microvascular immunothrombosis. Clinica Chimica Acta, 507, 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manne, B. K. , Denorme, F. , Middleton, E. A. , Portier, I. , Rowley, J. W. , Stubben, C. , Petrey, A. C. , Tolley, N. D. , Guo, L. , Cody, M. , Weyrich, A. S. , Yost, C. C. , Rondina, M. T. , & Campbell, R. A. (2020).Platelet gene expression and function in patients with COVID‐19. Blood, 136(11), 1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamel, M. H. , Yin, W. , Zavaro, C. , Francis, J. M. , & Chitalia, V. C. (2020).Hyperthrombotic milieu in COVID‐19 patients. Cells, 9(11), 2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greinacher, A. , Selleng, K. , Mayerle, J. , Palankar, R. , Wesche, J. , Reiche, S. , Aebischer, A. , Warkentin, T. E. , Muenchhoff, M. , Hellmuth, J. C. , Keppler, O. , Duerschmied, D. , Lother, A. , Rieg, S. , Gawaz, M. , Mueller, K. A. L. , Scheer, C. , Napp, M. , Hahnenkamp, K. , … Thiele, T. (2021).Anti‐platelet factor 4 antibodies causing VITT do not cross‐react with SARS‐CoV‐2 spike protein. Blood, 138(14), 1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dotan, A. , Muller, S. , Kanduc, D. , David, P. , Halpert, G. , & Shoenfeld, Y. (2021).The SARS‐CoV‐2 as an instrumental trigger of autoimmunity. Autoimmunity Reviews, 20(4), 102792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dotan, A. , & Shoenfeld, Y. (2021).Perspectives on vaccine induced thrombotic thrombocytopenia. Journal of Autoimmunity, 121, 102663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiao, M. , Zhang, Y. , Zhang, S. , Qin, X. , Xia, P. , Cao, W. , Jiang, W. , Chen, H. , Ding, X. , Zhao, H. , Zhang, H. , Wang, C. , Zhao, J. , Sun, X. , Tian, R. , Wu, W. , Wu, D. , Ma, J. , Chen, Y. , … Zhang, S. (2020). Antiphospholipid antibodies in critically ill patients with COVID‐19. Arthritis & Rheumatology, 72(12), 1998–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Borghi, M. O. , Beltagy, A. , Garrafa, E. , Curreli, D. , Cecchini, G. , Bodio, C. , Grossi, C. , Blengino, S. , Tincani, A. , Franceschini, F. , Andreoli, L. , Lazzaroni, M. G. , Piantoni, S. , Masneri, S. , Crisafulli, F. , Brugnoni, D. , Muiesan, M. L. , Salvetti, M. , Parati, G. , … Meroni, P. L. (2020).Anti‐phospholipid antibodies in COVID‐19 are different from those detectable in the anti‐phospholipid syndrome. Frontiers in Immunology, 11, 584241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zuo, Y. , Estes, S. K. , Ali, R. A. , Gandhi, A. A. , Yalavarthi, S. , Shi, H. , Sule, G. , Gockman, K. , Madison, J. A. , Zuo, M. , Yadav, V. , Wang, J. , Woodard, W. , Lezak, S. P. , Lugogo, N. L. , Smith, S. A. , Morrissey, J. H. , Kanthi, Y. , & Knight, J. S. (2020). Prothrombotic autoantibodies in serum from patients hospitalized with COVID‐19. Science Translational Medicine., 12(570), eabd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grobler, C. , Maphumulo, S. C. , Grobbelaar, L. M. , Bredenkamp, J. C. , Laubscher, G. J. , Lourens, P. J. , Steenkamp, J. , Kell, D. B. , & Pretorius, E. (2020).Covid‐19: The Rollercoaster of Fibrin(Ogen), D‐Dimer, Von Willebrand Factor, P‐Selectin and their interactions with endothelial cells, platelets and erythrocytes. International Journal of Molecular Sciences, 21(14), E5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Favaloro, E. J. , Henry, B. M. , & Lippi, G. (2021).Is lupus anticoagulant a significant feature of COVID‐19? A critical appraisal of the literature. Seminars in Thrombosis and Hemostasis, 10.1055/s-0041-1729856 [DOI] [PubMed] [Google Scholar]
- 26. Dragonetti, D. , Guarini, G. , & Pizzuti, M. (2020).Detection of anti‐heparin‐PF4 complex antibodies in COVID‐19 patients on heparin therapy. Blood Transfus, 18(4), 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brodard, J. , Kremer Hovinga, J. A. , Fontana, P. , Studt, J. D. , Gruel, Y. , & Greinacher, A. (2021).COVID‐19 patients often show high‐titer non‐platelet‐activating anti‐PF4/heparin IgG antibodies. Journal of Thrombosis and Haemostasis, 19(5), 1294–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Greinacher, A. , Thiele, T. , Warkentin, T. E. , Weisser, K. , Kyrle, P. A. , & Eichinger, S. (2021).Thrombotic thrombocytopenia after ChAdOx1 nCov‐19 vaccination. New England Journal of Medicine, 384(22), 2092–2101. 10.1056/NEJMoa2104840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scully, M. , Singh, D. , Lown, R. , Poles, A. , Solomon, T. , Levi, M. , Goldblatt, D. , Kotoucek, P. , Thomas, W. , & Lester, W. (2021).Pathologic antibodies to platelet factor 4 after ChAdOx1 nCoV‐19 vaccination. New England Journal of Medicine, 384(23), 2202–2211. 10.1056/NEJMoa2105385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Favaloro, E. J. , Henry, B. M. , & Lippi, G. (2021).The complicated relationships of heparin‐induced thrombocytopenia and platelet factor 4 antibodies with COVID‐19. International Journal of Laboratory Hematology, 43(4), 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frydman, G. H. , Streiff, M. B. , Connors, J. M. , & Piazza, G. (2020).The potential role of coagulation factor Xa in the pathophysiology of COVID‐19: A role for anticoagulants as multimodal therapeutic agents. TH Open, 4(4), e288–e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mancini, I. , Baronciani, L. , Artoni, A. , Colpani, P. , Biganzoli, M. , Cozzi, G. , Novembrino, C. , Boscolo Anzoletti, M. , De Zan, V. , Pagliari, M. T. , Gualtierotti, R. , Aliberti, S. , Panigada, M. , Grasselli, G. , Blasi, F. , & Peyvandi, F. (2021). The ADAMTS13‐von Willebrand factor axis in COVID‐19 patients. Journal of Thrombosis and Haemostasis, 19(2), 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mir, T. H. (2021).Thrombotic microangiopathy (aHUS/iTTP) reported so far in Covid‐19 patients: The virus alone or an omnium gatherum of mechanisms and etiologies? Critical Reviews in Oncology/Hematology, 162, 103347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fontana, C. , Favaro, M. , Minelli, S. , Bossa, M. C. , & Altieri, A. (2021).Co‐infections observed in SARS‐CoV‐2 positive patients using a rapid diagnostic test. Scientific Reports, 11(1), 16355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rawson, T. M. , Moore, L. S. P. , Zhu, N. , Ranganathan, N. , Skolimowska, K. , Gilchrist, M. , Satta, G. , Cooke, G. , & Holmes, A. (2020). Bacterial and fungal coinfection in individuals with coronavirus: A rapid review to support COVID‐19 antimicrobial prescribing. Clinical Infectious Diseases, 71(9), 2459–2468. 10.1093/cid/ciaa530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Foschi, C. , Zignoli, A. , Gaibani, P. , Vocale, C. , Rossini, G. , Lafratta, S. , Liberatore, A. , Turello, G. , Lazzarotto, T. , & Ambretti, S. (2021). Respiratory bacterial co‐infections in intensive care unit‐hospitalized COVID‐19 patients: Conventional culture vs BioFire FilmArray pneumonia Plus panel. Journal of Microbiological Methods, 186, 106259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elabbadi, A. , Turpin, M. , Gerotziafas, G. T. , Teulier, M. , Voiriot, G. , & Fartoukh, M. (2021).Bacterial coinfection in critically ill COVID‐19 patients with severe pneumonia. Infection, Jan 3, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu, G. S. , Li, H. , Zhao, S. C. , Lu, R. J. , Niu, P. H. , & Tan, W. J. (2019).Viral and bacterial etiology of acute febrile respiratory syndrome among patients in Qinghai; China. Biomedical and Environmental Sciences, 32, 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kastenhuber, E. R. , Jaimes, J. A. , Johnson, J. L. , Mercadante, M. , Muecksch, F. , Weisblum, Y. , Bram, Y. , Schwartz, R. E. , Whittaker, G. R. , & Cantley, L. C. (2021).Coagulation factors directly cleave SARS‐CoV‐2 spike and enhance viral entry. bioRxiv, 1:2021.03.31.437960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Powell, P. D. , DeMartini, J. C. , Azari, P. , Stargell, L. A. , Cordain, L. , & Tucker, A. (2000).Evolutionary stable strategy: A test for theories of retroviral pathology which are based upon the concept of molecular mimicry. Journal of Theoretical Biology, 202(3), 213–229. [DOI] [PubMed] [Google Scholar]
- 41. Rudensky, A. , Preston‐Hurlburt, P. , Hong, S. C. , Barlow, A. , & Janeway, C. A., Jr. (1991).Sequence analysis of peptides bound to MHC class II molecules. Nature, 353(6345), 622–627. [DOI] [PubMed] [Google Scholar]
- 42. Cunningham, M. W. , McCormack, J. M. , Fenderson, P. G. , & Ho, M. K. (1989).Human and murine antibodies cross‐reactive with streptococcal M protein and myosin recognize the sequence GLN‐LYS‐SER‐LYS‐GLN in M protein. Journal of Immunology, 143(8), 2677–2683. [PubMed] [Google Scholar]
- 43. Root‐Bernstein, R. S. , Podufaly, A. , & Aimone, F. (2013).Antigenic complementarity between influenza A virus and Haemophilus influenzae may drive lethal co‐infection such as that seen in 1918–19. J Virol Antiviral Research, 2:1 10.4172/2324-8955.1000104 [DOI] [Google Scholar]
- 44. Root‐Bernstein, R. (2017).Human immunodeficiency virus proteins mimic human T cell receptors inducing cross‐reactive antibodies. International Journal of Molecular Sciences, 18(10), 2091. 10.3390/ijms18102091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Root‐Bernstein, R. S. (2014).Rethinking molecular mimicry in rheumatic heart disease and autoimmune myocarditis: Laminin, collagen IV, CAR, and B1AR as initial targets of disease. Frontiers in Pediatric Rheumatology, 2:85. 10.3389/fped.2014.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sohlenkamp, C. , & Geiger, O. (2016).Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiology Reviews, 40(1), 133–159. [DOI] [PubMed] [Google Scholar]
- 47. López‐Lara, I. M. , & Geiger, O. (2017).Bacterial lipid diversity. Biochimica et Biophysica Acta ‐ Molecular and Cell Biology of Lipids, 1862(11), 1287–1299. [DOI] [PubMed] [Google Scholar]
- 48. Rondina, M. T. , & Weyrich, A. S. (2012).Targeting phosphodiesterases in anti‐platelet therapy. Handbook of Experimental Pharmacology, 210, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hanson, M. S. , Stephenson, A. H. , Bowles, E. A. , Sridharan, M. , Adderley, S. , & Sprague, R. S. (2008).Phosphodiesterase 3 is present in rabbit and human erythrocytes and its inhibition potentiates iloprost‐induced increases in cAMP. American Journal of Physiology. Heart and Circulatory Physiology, 295(2), H786–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ivorra, M. D. , Bec, A. Le , & Lugnier, C. (1992).Characterization of membrane‐bound cyclic nucleotide phosphodiesterases from bovine aortic smooth muscle. Journal of Cardiovascular Pharmacology, 19(4), 532–540. [DOI] [PubMed] [Google Scholar]
- 51. Brown, A. C. , Hallouane, D. , Mawby, W. J. , Karet, F. E. , Saleem, M. A. , Howie, A. J. , & Toye, A. M. (2009).RhCG is the major putative ammonia transporter expressed in the human kidney, and RhBG is not expressed at detectable levels. American Journal of Physiology. Renal Physiology, 296(6), F1279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gruswitz, F. , Chaudhary, S. , Ho, J. D. , Schlessinger, A. , Pezeshki, B. , Ho, C. M. , Sali, A. , Westhoff, C. M. , & Stroud, R. M. (2010).Function of human Rh based on structure of RhCG at 2.1 A. Proceedings of the National Academy of Sciences of the United States of America, 107(21), 9638–9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Blakey, D. , Leech, A. , Thomas, G. H. , Coutts, G. , Findlay, K. , & Merrick, M. (2002).Purification of the Escherichia coli ammonium transporter AmtB reveals a trimeric stoichiometry. Biochemical Journal, 364(Pt 2), 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Niles, J. K. , Karnes, H. E. , Dlott, J. S. , & Kaufman, H. W. (2021).Association of ABO/Rh with SARS‐CoV‐2 positivity: The role of race and ethnicity in a female cohort. American Journal of Hematology, 96(1), E23–E26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yaylacı, S. , Dheir, H. , İşsever, K. , Genc, A. B. , Şenocak, D. , Kocayigit, H. , Guclu, E. , Suner, K. , Ekerbicer, H. , & Koroglu, M. (2020).The effect of ABO and Rh blood group antigens on admission to intensive care unit and mortality in patients with COVID‐19 infection. Revista Da Associacao Medica Brasileira, 66(Suppl 2), 86–90. [DOI] [PubMed] [Google Scholar]
- 56. Latz, C. A. , DeCarlo, C. , Boitano, L. , Png, C. Y. M. , Patell, R. , Conrad, M. F. , Eagleton, M. , & Dua, A. (2020).Blood type and outcomes in patients with COVID‐19. Annals of Hematology, 99(9), 2113–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ray, J. G. , Schull, M. J. , Vermeulen, M. J. , & Park, A. L. (2021).Association between ABO and Rh blood troups and SARS‐CoV‐2 infection or severe COVID‐19 illness: A population‐based cohort study. Annals of Internal Medicine, 174(3), 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Abdollahi, A. , Mahmoudi‐Aliabadi, M. , Mehrtash, V. , Jafarzadeh, B. , & Salehi, M. (2020).The novel coronavirus SARS‐CoV‐2 vulnerability association with ABO/Rh blood types. Iranian Journal of Pathology, 15(3), 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. El‐Shitany, N. A. , El‐Hamamsy, M. , Alahmadi, A. A. , Eid, B. G. , Neamatallah, T. , Almukadi, H. S. , Arab, R. A. , Faddladdeen, K. A. , Al‐Sulami, K. A. , Bahshwan, S. M. , Ali, S. S. , Harakeh, S. , & Badr‐Eldin, S. M. (2021).The impact of ABO blood Grouping on COVID‐19 vulnerability and seriousness: A retrospective cross‐sectional controlled study among the Arab community. International Journal of Environmental Research and Public Health, 18(1), 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Coluk, Y. , Hizli, O. , Gunaydın, S. , Yildirim, G. , Baysal, E. , & Ozgen Hergul, G. (2021).Association of blood subgroups with PCR test positivity and lung involvement in patients with COVID‐19. Cureus, 13(3), e14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pereira, M. S. , Alves, I. , Vicente, M. , Campar, A. , Silva, M. C. , Padrão, N. A. , Pinto, V. , Fernandes Dias, Â. A. M. , & Pinho, S. S. (2018).Glycans as key checkpoints of T cell activity and function. Frontiers in Immunology, 9, 2754. 10.3389/fimmu.2018.02754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Prasad, L. , Leduc, Y. , Hayakawa, K. , & Delbaere, L. T. (2004).The structure of a universally employed enzyme: V8 protease from Staphylococcus aureus . Acta Crystallographica. Section D: Biological Crystallography, 60(Pt 2), 256–259. [DOI] [PubMed] [Google Scholar]
- 63. Eyers, S. A. , Ridgwell, K. , Mawby, W. J. , & Tanner, M. J. (1994).Topology and organization of human Rh (rhesus) blood group‐related polypeptides. Journal of Biological Chemistry, 269(9), 6417–6423. [PubMed] [Google Scholar]
- 64. Gabarre, P. , Dumas, G. , Dupont, T. , Darmon, M. , Azoulay, E. , & Zafrani, L. (2020).Acute kidney injury in critically ill patients with COVID‐19. Intensive Care Medicine, 46(7), 1339–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Molina Barragan, A. M. , Pardo, E. , Galichon, P. , Hantala, N. , Gianinazzi, A. C. , Darrivere, L. , Tsai, E. S. , Garnier, M. , Bonnet, F. , Fieux, F. , & Verdonk, F. (2021).SARS‐CoV‐2 renal impairment in critical care: An observational study of 42 Cases (Kidney COVID). Journal of Clinical Medicine, 10(8), 1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kimball, A. S. , Obi, A. T. , Diaz, J. A. , & Henke, P. K. (2016).The emerging role of NETS in venous thrombosis and immunothrombosis. Frontiers in Immunology, 7, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li, R. H. L. , & Tablin, F. (2018).A comparative review of neutrophil extracellular traps in sepsis. Frontiers in Veterinary Science, 5, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brinkmann, V. , Reichard, U. , Goosmann, C. , Fauler, B. , Uhlemann, Y. , Weiss, D. S. , Weinrauch, Y. , & Zychlinsky, A. (2004).Neutrophil extracellular traps kill bacteria. Science, 303(5663), 1532–1535. [DOI] [PubMed] [Google Scholar]
- 69. Root‐Bernstein, R. (2021). Innate receptor activation patterns involving TLR and NLR synergisms in COVID‐19, ALI/ARDS and sepsis cytokine storms: A review and model making novel predictions and therapeutic suggestions. International Journal of Molecular Sciences, 22(4), 2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vargas‐Vargas, M. , & Cortés‐Rojo, C. (2020).Ferritin levels and COVID‐19. Revista Panamericana de Salud Pública, 44, e72, 10.26633/rpsp.2020.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gonçalves, J. M. F. , Pérez, J. M. H. , Sorensen, M. A. , Pérez, A. L. W. , De La Rosa, E. M. R. , Castilla, J. L. T. , Pérez, D. D. , & Ramallo‐Fariña, Y. (2020).Biomarkers of acute respiratory distress syndrome in adults hospitalised for severe SARS‐CoV‐2 infection in Tenerife Island, Spain. BMC Res. Notes, 13, 1–7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Terpos, E. , Ntanasis‐Stathopoulos, I. , Elalamy, I. , Kastritis, E. , Sergentanis, T. N. , Politou, M. , Psaltopoulou, T. , Gerotziafas, G. , & Dimopoulos, M. A. (2020).Hematological findings and complications of COVID ‐19. American Journal of Hematology, 95, 834–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tan, C. , Huang, Y. , Shi, F. , Tan, K. , Ma, Q. , Chen, Y. , Jiang, X. , & Li, X. (2020).C‐reactive protein correlates with computed tomographic findings and predicts severe COVID‐19 early. Journal of Medical Virology, 92, 856–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhu, X. , Ge, Y. , Wu, T. , Zhao, K. , Chen, Y. , Wu, B. , Zhu, F. , Zhu, B. , & Cui, L. (2020).Co‐infection with respiratory pathogens among COVID‐2019 cases. Virus Research, 285, 198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lavoignet, C.‐E. , Network, A. T. C. , Le Borgne, P. , Chabrier, S. , Bidoire, J. , Slimani, H. , Chevrolet‐Lavoignet, J. , Lefebvre, F. , Jebri, R. , Sengler, L. , Bilbault, P. , & the CREMS network (2019).White blood cell count and eosinopenia as valuable tools for the diagnosis of bacterial infections in the ED. European Journal of Clinical Microbiology & Infectious Diseases, 38, 1523–1532. [DOI] [PubMed] [Google Scholar]
- 76. Root‐Bernstein, R. , & Couturier, J. (2006).Antigenic complementarity in the origins of autoimmunity: A general theory illustrated with a case study of idiopathic thrombocytopenia purpura. Clinical and Developmental Immunology, 13, 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Loof, T. G. , Deicke, C. , & Medina, E. (2014).The role of coagulation/fibrinolysis during Streptococcus pyogenes infection. Frontiers in Cellular and Infection Microbiology, 4, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Perrotta, F. , & Perrini, M. P. (2021).Successful treatment of Klebsiella pneumoniae NDM sepsis and intestinal decolonization with Ceftazidime/Avibactam plus Aztreonam combination in a patient with TTP complicated by SARSCoV‐2 nosocomial infection. Medicina (Kaunas, Lithuania), 57(5), 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rokkam, V. R. P. , Kutti Sridharan, G. , Vegunta, R. , Vegunta, R. , Boregowda, U. , & Mohan, B. P. (2021). Clostridium difficile and COVID‐19: Novel risk factors for acute portal vein thrombosis. Case Reports in Vascular Medicine. 2021, 8832638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Khanna, S. , & Kraft, C. S. (2021).The interplay of SARS‐CoV‐2 and Clostridioides difficile infection. Future Microbiol, 16, 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Llitjos, J. F. , Leclerc, M. , Chochois, C. , Monsallier, J. M. , Ramakers, M. , Auvray, M. , & Merouani, K. (2020).High incidence of venous thromboembolic events in anticoagulated severe COVID‐19 patients. Journal of Thrombosis and Haemostasis, 18(7), 1743–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Di Micco, P. , Imparato, M. , Lubrano, G. , Iannuzzo, D. , Fontanella, L. , Improta, L. , Poggiano, M. R. , Salzano, C. , Rodolico, A. , & Fontanella, A. (2021).Resolution of disseminated intravascular coagulation in a patient with COVID‐19 and associated sepsis‐induced neutropenia. Medicina (Kaunas, Lithuania), 57(2), 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rose, N. R. , & Mackay, I. R. (2000).Molecular mimicry: A critical look at exemplary instances in human diseases, Cellular and Molecular Life Sciences, 57, 542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fujinami, R. S. , von Herrath, M. G. , Christen, U. , & Whitton, J. L. (2006).Molecular mimicry, bystander activation, or viral persistence: Infections and autoimmune disease. Clinical Microbiology Reviews, 19, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Root‐Bernstein, R. , & Fairweather, D. (2014).Complexities in the relationship between infection and autoimmunity. Current Allergy and Asthma Reports, 14(1), 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Root‐Bernstein, R. , & Fairweather, D. (2015).Unresolved issues in theories of autoimmune disease using myocarditis as a framework. Journal of Theoretical Biology, 375, 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pascolini, S. , Vannini, A. , Deleonardi, G. , Ciordinik, M. , Sensoli, A. , Carletti, I. , Veronesi, L. , Ricci, C. , Pronesti, A. , Mazzanti, L. , Grondona, A. , Silvestri, T. , Zanuso, S. , Mazzolini, M. , Lalanne, C. , Quarneti, C. , Fusconi, M. , Giostra, F. , Granito, A. , … Muratori, P. (2021).COVID‐19 and immunological dysregulation: Can autoantibodies be useful? Clinical and Translational Science, 14(2), 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Foret, T. , Dufrost, V. , Salomon Du Mont, L. , Costa, P. , Lefevre, B. , Lacolley, P. , Regnault, V. , Zuily, S. , & Wahl, D. (2021).Systematic review of antiphospholipid antibodies in COVID‐19 patients: Culprits or bystanders? Current Rheumatology Reports, 23(8), 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. von Herrath, M. G. , Fujinami, R. S. , & Whitton, J. L. (2003).Microorganisms and autoimmunity: Making the barren field fertile? Nature Reviews Microbiology, 1, 151–157. [DOI] [PubMed] [Google Scholar]
- 90. McCoy, L. , Tsunoda, I. , & Fujinami, R. S. , (2006).Multiple sclerosis and virus induced immune responses: Autoimmunity can be primed by molecular mimicry and augmented by bystander activation, Autoimmunity, 39, 9–19. [DOI] [PubMed] [Google Scholar]
- 91. Root‐Bernstein, R. (2020).Synergistic activation of toll‐like receptors by complementary antigens as a facilitator of autoimmune disease and mediators of sex hormone‐associated susceptibility: Review, theory and novel predictions. International Journal of Molecular Sciences, 21(13), 4645; 10.3390/ijms21134645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Root‐Bernstein, R. (2007).Antigenic complementarity in the induction of autoimmunity: A general theory and review. Autoimmunity Reviews, 6(5), 272–277. [DOI] [PubMed] [Google Scholar]
- 93. Root‐Bernstein, R. (2015).How to make a non‐antigenic protein (auto) antigenic: Molecular complementarity alters antigen processing and activates adaptive‐innate immunity synergy. Anti‐Cancer Agents in Medicinal Chemistry, 15(10), 1242–1259. [DOI] [PubMed] [Google Scholar]
- 94. Westall, F. C. , & Root‐Bernstein, R. (1986).Cause and prevention of postinfectious and postvaccinal neuropathies in light of a new theory of autoimmunity. Lancet, 2(8501), 251–252. [DOI] [PubMed] [Google Scholar]
- 95. Nazy, I. , Jevtic, S. D. , Moore, J. C. , Huynh, A. , Smith, J. W. , Kelton, J. G. , & Arnold, D. M. (2021).Platelet‐activating immune complexes identified in critically ill COVID‐19 patients suspected of heparin‐induced thrombocytopenia. Journal of Thrombosis and Haemostasis, 19(5), 1342–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cristiano, A. , Fortunati, V. , Cherubini, F. , Bernardini, S. , & Nuccetelli, M. (2021).Anti‐phospholipids antibodies and immune complexes in COVID‐19 patients: A putative role in disease course for anti‐annexin‐V antibodies. Clinical Rheumatology, 19, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mazzitelli, I. , Bleichmar, L. , Ludueña, M. G. , Pisarevsky, A. , Labato, M. , Chiaradia, V. , & Finocchieto, P. et al. (2021).IgG immune complexes may contribute to neutrophil activation in the course of severe COVID‐19. Journal of Infectious Diseases, 2:jIab174. 10.1093/infdis/jiab174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Johnston, I. , Sarkar, A. , Hayes, V. , Koma, G. T. , Arepally, G. M. , Chen, J. , Chung, D. W. , López, J. A. , Cines, D. B. , Rauova, L. , & Poncz, M. (2020).Recognition of PF4‐VWF complexes by heparin‐induced thrombocytopenia antibodies contributes to thrombus propagation. Blood, 135(15), 1270–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Krauel, K. , Pötschke, C. , Weber, C. , Kessler, W. , Fürll, B. , Ittermann, T. , Maier, S. , Hammerschmidt, S. , Bröker, B. M. , & Greinacher, A. (2011).Platelet factor 4 binds to bacteria, [corrected] inducing antibodies cross‐reacting with the major antigen in heparin‐induced thrombocytopenia. Blood, 117(4), 1370–1378. [DOI] [PubMed] [Google Scholar]
- 100. Brandt, S. , Krauel, K. , Jaax, M. , Renné, T. , Helm, C. A. , Hammerschmidt, S. , Delcea, M. , & Greinacher, A. (2015).Polyphosphates form antigenic complexes with platelet factor 4 (PF4) and enhance PF4‐binding to bacteria. Thrombosis and Haemostasis, 114(6), 1189–1198. [DOI] [PubMed] [Google Scholar]
- 101. Othman, M. , Labelle, A. , Mazzetti, I. , Elbatarny, H. S. , & Lillicrap, D. (2007).Adenovirus‐induced thrombocytopenia: The role of von Willebrand factor and P‐selectin in mediating accelerated platelet clearance. Blood, 109(7), 2832–2839. [DOI] [PubMed] [Google Scholar]
- 102. Shirley, J. L. , de Jong, Y. P. , Terhorst, C. , & Herzog, R. W. (2020).Immune responses to viral gene therapy vectors. Molecular Therapy, 28(3), 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jonsson, M. I. , Lenman, A. E. , Frängsmyr, L. , Nyberg, C. , Abdullahi, M. , & Arnberg, N. (2009).Coagulation factors IX and X enhance binding and infection of adenovirus types 5 and 31 in human epithelial cells. Journal of Virology, 83(8), 3816–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hofherr, S. E. , Mok, H. , Gushiken, F. C. , Lopez, J. A. , & Barry, M. A. (2007).Polyethylene glycol modification of adenovirus reduces platelet activation, endothelial cell activation, and thrombocytopenia. Human Gene Therapy, 18(9), 837–848. [DOI] [PubMed] [Google Scholar]
- 105. Raddi, N. , Vigant, F. , Wagner‐Ballon, O. , Giraudier, S. , Custers, J. , Hemmi, S. , & Benihoud, K. (2016).Pseudotyping serotype 5 adenovirus with the fiber from other serotypes uncovers a key role of the fiber protein in adenovirus 5‐induced thrombocytopenia. Human Gene Therapy, 27(2), 193–201. [DOI] [PubMed] [Google Scholar]
- 106. Kim, S. J. , Kim, K. , Park, S. B. , Hong, D. J. , & Jhun, B. W. (2015).Outcomes of early administration of cidofovir in non‐immunocompromised patients with severe adenovirus pneumonia. PLoS ONE, 10(4), e0122642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Platton, S. , Bartlett, A. , MacCallum, P. , Makris, M. , McDonald, V. , Singh, D. , Scully, M. , & Pavord, S. (2021).Evaluation of laboratory assays for anti‐platelet factor 4 antibodies after ChAdOx1 nCOV‐19 vaccination. Journal of Thrombosis and Haemostasis, 19(8), 2007. 10.1111/jth.15362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sørvoll, I. H. , Horvei, K. D. , Ernstsen, S. L. , Laegreid, I. J. , Lund, S. , Grønli, R. H. , Olsen, M. K. , Jacobsen, H. K. , Eriksson, A. , Halstensen, A. M. , Tjønnfjord, E. , Ghanima, W. , & Ahlen, M. T. (2021).An observational study to identify the prevalence of thrombocytopenia and anti‐PF4/polyanion antibodies in Norwegian health care workers after COVID‐19 vaccination. Journal of Thrombosis and Haemostasis, 19(7), 1813. 10.1111/jth.15352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Thiele, T. , Ulm, L. , Holtfreter, S. , Schönborn, L. , Kuhn, S. O. , Scheer, C. , Warkentin, T. E. , Bröker, B. , Becker, K. , Aurich, K. , Selleng, K. , Hübner, N. O. , & Greinacher, A. (2021). Frequency of positive anti‐PF4/polyanion antibody tests after COVID‐19 vaccination with ChAdOx1 nCoV‐19 and BNT162β. Blood, blood.2021012217.138(4), 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Terpos, E. , Politou, M. , Ntanasis‐Stathopoulos, I. , Karalis, V. , Merkouri, E. , Fotiou, D. , Gavriatopoulou, M. , Malandrakis, P. , Kastritis, E. , Trougakos, I. P. , & Dimopoulos, M. A. (2021).High prevalence of anti‐PF4 antibodies following ChAdOx1 nCov‐19 (AZD1222) vaccination even in the absence of thrombotic events. Vaccines (Basel), 9(7), 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Favaloro, E. J. (2021).Laboratory testing for suspected COVID‐19 vaccine‐induced (immune) thrombotic thrombocytopenia. International Journal of Laboratory Hematology, 43(4), 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hunter, P. R. (2021).Thrombosis after covid‐19 vaccination. British Medical Journal (Clinical Research Edition), 373, n958. 10.1136/bmj.n958 [DOI] [PubMed] [Google Scholar]
- 113. von Hundelshausen, P. , Lorenz, R. , Siess, W. , & Weber, C. (2021).Vaccine‐induced immune thrombotic thrombocytopenia (VITT): Targeting pathomechanisms with Bruton tyrosine kinase inhibitors. Thrombosis and Haemostasis, 10.1055/a-1481-3039 [DOI] [PubMed] [Google Scholar]
- 114. Thaler, J. , Ay, C. , Gleixner, K. V. , Hauswirth, A. W. , Cacioppo, F. , Jürgen, G. , Quehenberger, P. , Pabinger, I. , & Knöbl, P. (2021).Successful treatment of vaccine‐induced prothrombotic immune thrombocytopenia (VIPIT). Journal of Thrombosis and Haemostasis, 19(7), 1819. 10.1111/jth.15346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Althaus, K. , Möller, P. , Uzun, G. , Singh, A. , Beck, A. , Bettag, M. , Bösmüller, H. , Guthoff, M. , Dorn, F. , Petzold, G. C. , Henkes, H. , Heyne, N. , Jumaa, H. , Kreiser, K. , Limpach, C. , Luz, B. , Maschke, M. , Müller, J. A. , Münch, J. , … Sachs, U. (2021).Antibody‐mediated procoagulant platelets in SARS‐CoV‐2‐ vaccination associated immune thrombotic thrombocytopenia. Haematologica, 106(8), 2170–2179. 10.3324/haematol.2021.279000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Watad, A. , De Marco, G. , Mahajna, H. , Druyan, A. , Eltity, M. , Hijazi, N. , Haddad, A. , Elias, M. , Zisman, D. , Naffaa, M. E. , Brodavka, M. , Cohen, Y. , Abu‐Much, A. , Abu Elhija, M. , Bridgewood, C. , Langevitz, P. , McLorinan, J. , Bragazzi, N. L. , Marzo‐Ortega, H. … McGonagle, D. (2021).Immune‐mediated disease flares or new‐onset disease in 27 subjects following mRNA/DNA SARS‐CoV‐2 vaccination. Vaccines (Basel), 9(5), 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Winram, S. B. , & Lottenberg, R. (1996).The plasmin‐binding protein Plr of group A streptococci is identified as glyceraldehyde‐3‐phosphate dehydrogenase. Microbiology (Reading), 142 (Pt 8), 2311–2320. [DOI] [PubMed] [Google Scholar]
- 118. Kopeckova, M. , Pavkova, I. , & Stulik, J. (2020).Diverse localization and protein binding abilities of glyceraldehyde‐3‐phosphate dehydrogenase in pathogenic bacteria: The key to its multifunctionality? Frontiers in Cellular and Infection Microbiology, 10, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Medcalf, R. L. , Keragala, C. B. , & Myles, P. S. (2020).Fibrinolysis and COVID‐19: A plasmin paradox. Journal of Thrombosis and Haemostasis, 18(9), 2118–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Karaba, S. M. , Jones, G. , Helsel, T. , Smith, L. L. , Avery, R. , Dzintars, K. , Salinas, A. B. , Keller, S. C. , Townsend, J. L. , Klein, E. , Amoah, J. , Garibaldi, B. T. , Cosgrove, S. E. , & Fabre, V. (2020).Prevalence of co‐infection at the time of hospital admission in COVID‐19 patients, A multicenter study. Open Forum Infectious Diseases, 8(1), oFaa578. 10.1093/ofid/ofaa578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gudiol, C. , Durà‐Miralles, X. , Aguilar‐Company, J. , Hernández‐Jiménez, P. , Martínez‐Cutillas, M. , Fernandez‐Avilés, F. , Machado, M. , Vázquez, L. , Martín‐Dávila, P. , de Castro, N. , Abdala, E. , Sorli, L. , Andermann, T. M. , Márquez‐Gómez, I. , Morales, H. , Gabilán, F. , Ayaz, C. M. , Kayaaslan, B. , Aguilar‐Guisado, M. , … Carratalà, J. (2021).Co‐infections and superinfections complicating COVID‐19 in cancer patients: A multicentre, international study. Journal of Infection, 83(3), 306–313:S0163‐4453(21)00356‐X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sampson, V. , Kamona, N. , & Sampson, A. (2020).Could there be a link between oral hygiene and the severity of SARS‐CoV‐2 infections? British Dental Journal, 228(12), 971–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Soffritti, I. , D'Accolti, M. , Fabbri, C. , Passaro, A. , Manfredini, R. , Zuliani, G. , Libanore, M. , Franchi, M. , Contini, C. , & Caselli, E. (2021).Oral microbiome dysbiosis is associated with symptoms severity and local immune/inflammatory response in COVID‐19 patients: A cross‐sectional study. Frontiers in Microbiology, 12:687513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kewan, T. , Gunaratne, T. N. , Mushtaq, K. , Alayan, D. , Daw, H. , & Haddad, A. (2021).Outcomes and management of immune thrombocytopenia secondary to COVID‐19: Cleveland clinic experience. Transfusion, 61(7), 2014–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Alam, W. (2021).COVID‐19 vaccine‐induced immune thrombotic thrombocytopenia: A review of the potential mechanisms and proposed management. Science Progress, 104(2), 368504211025927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Balagholi, S. , Dabbaghi, R. , Eshghi, P. , Mousavi, S. A. , Heshmati, F. , & Mohammadi, S. (2020).Potential of therapeutic plasmapheresis in treatment of COVID‐19 patients: Immunopathogenesis and coagulopathy. Transfusion and Apheresis Science, 59(6), 102993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zachariah, U. , Nair, S. C. , Goel, A. , Balasubramanian, K. A. , Mackie, I. , Elias, E. , & Eapen, C. E. (2020).Targeting raised von Willebrand factor levels and macrophage activation in severe COVID‐19: Consider low volume plasma exchange and low dose steroid. Thrombosis Research, 192:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Thindwa, D. , Garcia Quesada, M. , Liu, Y. , Bennett, J. , Cohen, C. , Knoll, M. D. , von Gottberg, A. , Hayford, K. , & Flasche, S. (2020).Use of seasonal influenza and pneumococcal polysaccharide vaccines in older adults to reduce COVID‐19 mortality. Vaccine, 38(34), 5398–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Root‐Bernstein, R. (2020).Age and location in severity of COVID‐19 pathology: Do lactoferrin and pneumococcal vaccination explain low infant mortality and regional differences? Bioessays, 42(11), e2000076. [DOI] [PubMed] [Google Scholar]
- 130. Root‐Bernstein, R. (2021). Pneumococcal and influenza vaccination rates and pneumococcal invasive disease rates set geographical and ethnic population susceptibility to serious COVID‐19 cases and deaths. Vaccines (Basel), 9(5), 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Root‐Bernstein, R. (2020). Possible cross‐reactivity between SARS‐CoV‐2 proteins, CRM197 and proteins in pneumococcal vaccines may protect against symptomatic SARS‐CoV‐2 disease and death. Vaccines (Basel), 8(4), 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Jehi, L. , Ji, X. , Milinovich, A. , Erzurum, S. , Rubin, B. P. , Gordon, S. , Young, J. B. , & Kattan, M. W. (2020). Individualizing risk prediction for positive coronavirus disease 2019 testing: Results from 11,672 patients. Chest, 158(4), 1364–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Noale, M. , Trevisan, C. , Maggi, S. , Antonelli Incalzi, R. , Pedone, C. , Di Bari, M. , Adorni, F. , Jesuthasan, N. , Sojic, A. , Galli, M. , Giacomelli, A. , Molinaro, S. , Bianchi, F. , Mastroianni, C. , Prinelli, F ., & Group OBOTEW . (2020). The association between influenza and pneumococcal vaccinations and SARS‐Cov‐2 infection: Data from the EPICOVID19 web‐based survey. Vaccines (Basel), 8(3), 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pawlowski, C. , Puranik, A. , Bandi, H. , Venkatakrishnan, A. J. , Agarwal, V. , Kennedy, R. , O'Horo, J. C. , Gores, G. J. , Williams, A. W. , Halamka, J. , Badley, A. D. , & Soundararajan, V. (2021).Exploratory analysis of immunization records highlights decreased SARS‐CoV‐2 rates in individuals with recent non‐COVID‐19 vaccinations. Scientific Reports, 11(1), 4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Taghioff, S. M. , Slavin, B. R. , Holton, T. , & Singh, D. (2021).Examining the potential benefits of the influenza vaccine against SARS‐CoV‐2: A retrospective cohort analysis of 74,754 patients. PLoS ONE, 16(8), e0255541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Yang, M. J. , Rooks, B. J. , Le, T. T. , Santiago, I. O. 3rd , Diamond, J. , Dorsey, N. L. , & Mainous, A. G., 3rd. (2021). Influenza vaccination and hospitalizations among COVID‐19 infected adults. The Journal of the American Board of Family Medicine, 34(Suppl), S179–S182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


