Abstract
The transcription of tissue-specific genes is controlled by regulatory factors and cofactors and is suppressed in cardiac cells by the antineoplastic agent doxorubicin. Here we show that exposure of cultured cardiomyocytes to doxorubicin resulted in the rapid depletion of transcripts for MEF2C, dHAND, and NKX2.5, three pivotal regulators of cardiac gene expression. Delivery of exogenous p300, a coactivator of MEF2C and NKX2.5 in cardiomyocytes, restored cardiac transcription despite the presence of doxorubicin. Furthermore, p300 also restored the accumulation of transcripts for MEF2C itself. Importantly, cardiocytes exposed to doxorubicin displayed reduced levels of p300 proteins. This was not due to alterations in the level of p300 transcripts; rather, and surprisingly, doxorubicin promoted selective degradation of p300 mediated by the 26S-proteasome machinery. Doxorubicin had no effect on the general level of ubiquitinated proteins or on the levels of β-catenin, a protein known to be degraded by proteasome-mediated degradation. These results provide evidence for a new mechanism of transcriptional repression caused by doxorubicin in which the selective degradation of p300 results in reduced p300-dependent transcription, including production of MEF2C mRNA.
The regulation of tissue specification and determination involves a network of regulatory factors that modulate the activity of target genes. In cardiac muscle, transcription of cardiac genes is mediated by the interaction of specific transcription factors such as the MEF2 family of MADS-box proteins (10) and the homeodomain protein NKX2.5/Csx (27). The regulation of transcription is also controlled by cofactors that bind and support the function of these regulatory proteins. For example, the coactivator p300 (7) is recruited on promoter regions via direct interaction with a variety of nuclear factors (18), including MEF2 (9, 35). This binding is critical for supporting differentiation, cellular growth, and homeostasis of different cell types (1, 8). The proper expression of p300 appears to be necessary to prevent specific transcriptional defects. Disruption of the p300 gene leads to inappropriate heart development, which may explain its embryonic lethality (43), and mutation of the p300-related protein CBP in Rubinstein-Taybi syndrome is associated with developmental defects and mental retardation (32).
The transcription of cardiac genes is suppressed rapidly and selectively by the anticancer agent doxorubicin (17). In skeletal muscle cells, such disruption of gene expression is paralleled by the rapid fall in the expression of myogenic regulatory factor genes (24) as well as by the rapid induction of the negative regulator Id (22, 24). Because p300 has been implicated in regulating transcription engendered by MEF2C, we sought to investigate the possibility that transcriptional repression following doxorubicin exposure may result from a deregulation of the activity of cardiac transcription factors and of the coactivator p300.
We show here that exposure of cardiac cells to doxorubicin indeed leads to the rapid depletion of mRNAs for the regulatory factors MEF2C, dHAND, and NKX2.5. Like MEF2C, NKX2.5 is also regulated by p300. p300 is required for full MEF2C activity, and delivery of p300 counteracted the transcriptional repression induced by doxorubicin. Interestingly, this protective effect of p300 is due not only to its coactivation of MEF2C but also to transcriptional induction of the MEF2C gene itself. Importantly, exposure of cardiac cells to doxorubicin causes a depletion of p300 protein. Using two specific proteasome inhibitors, MG-132 and PSI, we demonstrate that the reduced levels of p300 protein in cells treated with doxorubicin result from a selective increase in proteasome-mediated degradation of the protein. These results imply a new mechanism of the disregulation of transcription mediated by doxorubicin whereby selective degradation of the p300 coactivator impairs the rate of transcription of p300-dependent genes.
MATERIALS AND METHODS
Plasmids.
The plasmids 2xA/Temb-CAT, CMV-p300, pCDNA3MEF2C, and E1A were described previously (35). The plasmids E1Aδ2-36 and E1AR2G were also described previously (34). The reporter gene 3xMEF2-luciferase was generously provided by Mona Nemer (Institut de Recherches Cliniques de Montréal, Montreal, Canada). The cytomegalovirus-green fluorescent protein (CMV-GFP) construct pEGFP-N1 was from Clontech. The construct G5E1b-luciferase has been described previously (16). The constructs Gal-MEF2C(1–465) and Gal-MEF2C(247–327) were gifts from Eric Olson (University of Texas Southwestern Medical Center, Dallas, Tex.).
Cell culture, DNA transfection, and doxorubicin treatment.
Neonatal rat cardiac myocytes from Sprague-Dawley rats (2 to 3 days old) were prepared as previously described (3). Briefly, cells were obtained by trypsinization of the hearts. After gentle mechanical disruption, the cells were washed, replated to reduce nonmyocardial cell contamination, and plated at a density of 4 × 106 cells per six-well dish. The cells were grown at 37°C with 5% CO2 in modified Eagle's medium containing 5% calf serum, 2 mM glutamine, 1% penicillin-streptomycin, and 1% 5-bromodeoxyuridine. Transfections were carried out by calcium phosphate precipitation. The cells were then maintained in the absence or presence of 1 μM doxorubicin for 72 h. Chloramphenicol acetyltransferase (CAT) activity was quantitated on an AMBIS Dual Radioanalytic Imaging System.
Luciferase activity was measured with an LKB luminomiter. The mouse myogenic C2C12 cell line was grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% fetal bovine serum. Doxorubicin treatment was performed as previously described (23). 293 cells were grown in DMEM supplemented with 10% fetal bovine serum. When the cells reached 60% confluence, doxorubicin treatment was started and maintained for 48 h. All transfections were performed in triplicate with at least two independent cardiocyte preparations, using two independent DNA plasmid preparations. Results were normalized to the total amount of protein.
Measurement of the rate of contraction of cardiac cells.
Contraction frequency of neonatal cardiocytes was measured by microscopic examination of three fields chosen randomly in the culture. The measurement was repeated on three independent cardiocyte preparations. The averages and mean values of all measurements were calculated.
RNA isolation and Northern blotting.
Total RNA was isolated from control and doxorubicin-treated cardiomyocytes or C2C12 cells as previously described (19). RNA concentration was measured by spectrophotometry, and the integrity of the RNA was ensured by analysis on a 1.2% formaldehyde–agarose gel. After RNA transfer, the membranes were hybridized at 55°C with NKX2.5 cDNA radiolabeled with [α-32P]dCTP, washed in 1% sodium dodecyl sulfate (SDS)–50 mM NaCl–1 mM EDTA three times at 55°C, and exposed to Amersham Hyperfilm film at −80°C.
RT-PCR.
The reverse transcriptase-PCR (RT-PCR) assay was carried out using the RETROscript and QuantumRNA kits (Ambion). Briefly, 2 μg of total RNA from neonatal rat cardiac myocytes and from C2C12 untreated or treated with 1 μM doxorubicin was used for first-strand cDNA synthesis using 100 U of Moloney murine leukemia virus reverse transcriptase. Random primers were used in a 20-μl reaction volume in the presence of 10 μM deoxynucleoside triphosphates (dNTPs) and of reverse transcription buffer (10 mM Tris-Cl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2). Then 5 μl of the reverse transcription reaction was used for PCR amplification in a volume of 50 μl containing gene-specific primers (5 μM), 10 mM Tris (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 2 μl of 5 mM dNTPs, and 1.25 μl of thermophilic polymerase (Gibco-BRL). PCR amplifications were carried out using gene-specific primers under conditions of linear range. To confirm that genomic DNA did not contaminate the samples, PCRs were also performed without reverse transcription. To compensate for differences in RNA quality and random tube-to-tube variation, rRNA primers were used as an internal control. The factors that we wanted to amplify were less abundant than the rRNA. To amplify them in the same linear range as the internal control, 18S primers were mixed with competimers. All reactions were run in duplicates on a 2% agarose gel. The images were captured electronically and quantitated using an AlphaImager (Alpha Innotech Corporation). Radioactive PCR was performed by adding traces of [32P]dCTP to the cocktail. PCR products were separated on a 5% acrylamide gel and quantitated with a STORM scanner (Molecular Dynamics). Results were normalized to 18S RNA expression.
Cell sorting.
Neonatal rat cardiocytes were cotransfected with the CMV-GFP construct and a plasmid encoding full-length p300 or CMV backbone alone. After 24 h, doxorubicin treatment was started and maintained for an additional 48 h. Cardiac cells expressing GFP were sorted by fluorescence-activated cell sorting (FACS) analysis, and total RNA was extracted to quantitate MEF2C mRNA by RT-PCR technology as previously described.
Western blot analysis.
Neonatal rat cardiomyocytes and C2C12 cells were maintained in doxorubicin-free medium or treated with 1 μM doxorubicin for 48 h. Nuclear extracts were prepared as previously described (13). Briefly, the cells were washed three times in ice-cold phosphate-buffered saline and then scraped into lysis buffer (20 mM HEPES [pH 7.6], 20% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 10 mM NaCl) supplemented with freshly prepared protease and phosphate inhibitors (1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], leupeptin and pepstatin [10 μg/ml], and aprotinin [100 μg/ml]). Cells were lysed in a Dounce homogenizer using 10 strokes with an A pestle on ice. Samples were centrifuged for 10 min at 2,000 rpm, and the supernatant fraction was discarded. The pellet was resuspended in cold nuclear extract buffer (20 mM HEPES [pH 7.6], 20% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 500 mM NaCl) supplemented with freshly prepared protease inhibitors (1 mM DTT, 1 mM PMSF, leupeptin and pepstatin [10 μg/ml each], and aprotinin [100 μg/ml]) and incubated on ice for 1 h. Cellular debris was removed by centrifugation at 10,000 rpm for 10 min at 4°C, and the supernatant containing nuclear proteins was assayed for protein (Bradford assay). Equal amounts of nuclear proteins were electrophoresed on a 4 to 20% gradient gel and transferred to nitrocellulose membrane (Hybond ECL; Amersham) at 50 V for 12 h at 4°C. The membranes were blocked for 30 min at room temperature in TBS (10 mM Tris-HCl [pH 8], 150 mM NaCl)–5% nonfat dry milk–0.05% Tween 20 and incubated with a primary antibody directed against p300 (anti-human p300; power clonal [UBI] or N-15X [Santa Cruz], dilution 1:1,000,), β-catenin (Santa Cruz; dilution 1:500), or ubiquitin (Santa Cruz; dilution 1:200) overnight at 4°C. Incubation with a secondary antibody was carried out for 1 h at room temperature. After washing the membrane three times with TBS–0.05% Tween 20, antigen-antibody reaction was visualized with chemiluminescent reagent.
Immunoprecipitation.
Nuclear extracts (100 μg) were incubated with 4 μg of anti-p300 antibody N-15X or power clonal for 2 h at 4°C in a buffer containing 20 mM NaH2PO4 (pH 7.8), 160 mM NaCl, 0.1% NP-40, 5 mM EDTA, and 1 mM DTT, supplemented with freshly made protease and phosphatase inhibitors (1 mM PMSF plus aprotinin, leupeptin, and pepstatin at 10 μg/ml each). The extracts were then preadsorbed on protein A/G PLUS-agarose (Santa Cruz) for 2 h at 4°C with rocking and washed three times. The agarose resin was recovered by centrifugation and resuspended in 20 μl of SDS-loading dye. The samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on a 5% polyacrylamide gel run overnight at 4°C. After transfer at 70 V for 6 h at 4°C, membranes were blocked in 5% nonfat milk in TBS–0.05% Tween 20 and incubated overnight at 4°C with anti-p300 antibody (dilution 1:1,000). Immunocomplexes were detected by chemiluminescence.
Gel mobility shift assay.
Nuclear extracts were incubated with an anti-p300 antibody (2) or with preimmune serum. An end-labeled DNA probe corresponding to an MEF2 binding site derived from the muscle creatine kinase enhancer was incubated with the extract for 15 min at room temperature. The DNA-protein complexes were fractionated on a 5% polyacrylamide gel. The gels were dried and processed for autoradiography. To ascertain that the MEF2C-DNA binding complex was specific, cardiac nuclear extracts were incubated with a polyclonal anti-MEF2C antibody (Santa Cruz) or with an anti-T antigen antibody used as a control.
Cycloheximide treatment.
Three days after isolation, neonatal cardiomyocytes were treated with cycloheximide (10 μM) for 5 h to inhibit de novo protein synthesis. Half of the cells were then maintained in the presence of cycloheximide (control cells) for various times, and the other half were treated with cycloheximide supplemented with doxorubicin (1 μM). Nuclear extracts were prepared at the end of the treatment, and 30 μg of nuclear protein was separated by SDS-PAGE on a 4 to 20% gradient gel. After protein transfer overnight at 4°C at 50 V, the membranes were blocked for 1 h at room temperature in 5% nonfat dry milk in TBS–0.05% Tween 20 and incubated with an anti-p300 antibody (N-15X) overnight at 4°C. Incubation with a secondary antibody was carried out for 1 h at room temperature. After washing the membrane three times with TBS–0.05% Tween 20, antigen-antibody reaction was visualized by chemiluminescence.
Duplicates of the nuclear extracts were separated on a 4 to 20% gradient gel and stained with Coomassie blue to visualize total nuclear proteins.
Proteasome inhibitors.
The proteasome inhibitors MG-132 and PSI were purchased from Peptides International Inc. Neonatal cardiocytes were prepared as described earlier and treated with the proteasome inhibitors for the indicated times at concentrations of 30 μM. Control cells were treated with the vehicle solvent dimethyl sulfoxide (DMSO).
RESULTS
Effects of doxorubicin on cardiocytes in culture.
Since we postulated that the early changes in gene expression induced by doxorubicin precede wide-scale cytotoxic events, we first selected a duration of doxorubicin exposure that would lead to changes in gene expression before overt signs of cell damage. Neonatal rat cardiocytes were cultured for various times in the presence of 1 μM doxorubicin, a minimal concentration, to inhibit transcription. The onset of doxorubicin toxicity was evaluated by measuring by light microscopy both the number of cells in the culture and their intrinsic rate of contraction. The cell density and the rate of beating were indistinguishable between control cells and cells treated with doxorubicin for 24 h (Table 1). After 48 h of drug administration, while cell density and morphology remained unchanged, the contraction rate of the treated cells dropped significantly. After 72 h of culture, the majority of the cells treated with doxorubicin lost their ability to contract and exhibited visible signs of cell damage. Therefore, we limited our observations on gene expression to the first 48 h of doxorubicin treatment.
TABLE 1.
Rate of contraction and cell density of cardiocytes exposed to doxorubicina
| Exposure (h) | Beats/min
|
Cell density (%)
|
||
|---|---|---|---|---|
| Control | Treated | Control | Treated | |
| 24 | 166 ± 15 | 146 ± 21 | 100 | 100 |
| 48 | 221 ± 25 | 24 ± 12* | 100 | 100 |
| 72 | 256 ± 37 | ∼0* | 100 | 50* |
Cardiac cells were untreated (control) exposed to 1 μM doxorubicin (treated) for the indicated times. Doxorubicin toxicity was evaluated by counting the number of cells present in the culture and by measuring their intrinsic rate of contraction by microscopic examination of three different fields chosen randomly in the culture. The data represent the mean and standard error of three measurements from three separate cell preparations. ∗, statistically significant difference (P < 0.001, Student t test) between control and doxorubicin-treated cardiocytes.
Transcription of MEF2C, dHAND, and NKX2.5 is inhibited by doxorubicin in cardiac cells.
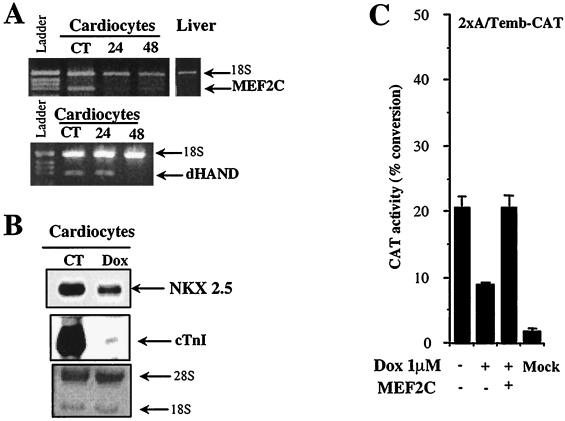
To understand the mechanisms of the transcriptional repression induced by doxorubicin, we examined three transcriptional activators known to be critical for cardiac transcription: MEF2C, NKX2.5, and dHAND. We measured the relative levels of their mRNA transcripts at various times after exposure to doxorubicin. Members of the myocyte enhancer factor 2 (MEF2) family of transcription factors bind a conserved A/T-rich DNA sequence found in the control region of numerous muscle-specific genes and are expressed in precursors of the cardiac lineage as well as in differentiated cardiomyocytes (10). Among the four members of this multigene family, MEF2C appears to be essential for cardiac myogenesis and for the development of the right ventricle (26). Doxorubicin inhibits MEF2C mRNA more than 50% in cardiac cells treated with the drug for 24 h (Fig. 1A). When the MEF2C gene is inactivated in mouse embryos, heart looping does not occur, the future right ventricle does not form, and mutant embryos lack expression of the basic helix-loop-helix (bHLH) protein dHAND. This suggests that MEF2C is an upstream regulator of dHAND expression in the future right ventricle (26). Therefore, we determined whether doxorubicin affects the expression of dHAND and found that its expression was reduced by as much as 85% after 48 h of drug administration (Fig. 1A). The importance in the heart of a third activator, NKX2.5, has been demonstrated by targeted disruption of its gene, which causes severe morphologic defect of the heart and is lethal to the embryo (28). NKX2.5 transcripts were reduced by 54% after 48 h of doxorubicin exposure (Fig. 1B).
FIG. 1.
Inhibition of expression of cardiac transcription factors in neonatal cardiomyocytes treated with doxorubicin. (A) Doxorubicin inhibits MEF2C mRNA in cardiocytes. Quantitative RT-PCR analysis of MEF2C and dHAND in control (CT) and doxorubicin-treated cardiocytes. Total RNA (2 μg) from either control neonatal rat cardiocytes or cardiocytes exposed to 1 μM doxorubicin for 24 or 48 h was reverse transcribed and PCR amplified with gene-specific primers for the indicated transcripts. PCR products were separated on a 2% agarose gel and quantitated with AlphaImager software. In all PCRs, the level of expression of the gene of interest was normalized against 18S cDNA. Each experiment was repeated at least twice with RNA template from two independent cardiocyte preparations. (B) Doxorubicin downregulates NKX2.5 transcription in cardiocytes. Northern blot analysis of NKX2.5 transcription in control (CT) cardiocytes and cardiocytes exposed to doxorubicin (Dox) for 48 h. Cardiac troponin I (cTnI) was measured in parallel to ensure that doxorubicin treatment was effective. The methylene blue staining of the blot shows 28S and 18S RNA and indicates that an equal amount of RNA was loaded in each lane. (C) Doxorubicin inhibits MEF2C transcriptional activity in cardiocytes. Neonatal rat cardiocytes were transfected with 2 μg of 2xA/Temb-CAT plasmid DNA, which contains two copies of the MEF2 binding site from the embryonic myosin heavy-chain enhancer placed upstream of a CAT reporter gene, and the cells were then were exposed to 1 μM doxorubicin (Dox). CAT activity was measured after 48 h of drug exposure as described in the text and corrected for protein content. The data represent the mean ± standard deviation of three independent experiments carried out in triplicate.
The functional consequences of decreased expression of MEF2C caused by doxorubicin were investigated using an MEF2-dependent reporter construct. Neonatal cardiocytes were transiently transfected with a construct bearing two copies of an MEF2-binding site (2xA/Temb-CAT) (35). Doxorubicin suppressed MEF2C-dependent transcription by 54% (Fig. 1C). To distinguish between a direct and indirect effect of doxorubicin on MEF2C activity, we tested the ability of forced expression of MEF2C to prevent the doxorubicin inhibition. Overexpression of MEF2C in cardiocytes fully rescued the ability of doxorubicin to suppress expression from the 2xA/Temb promoter construct (Fig. 1C). These results are consistent with the notion that doxorubicin causes a selective reduction of MEF2C-dependent transcripts.
p300 coactivates MEF2C and NKX2.5 in cardiomyocytes.
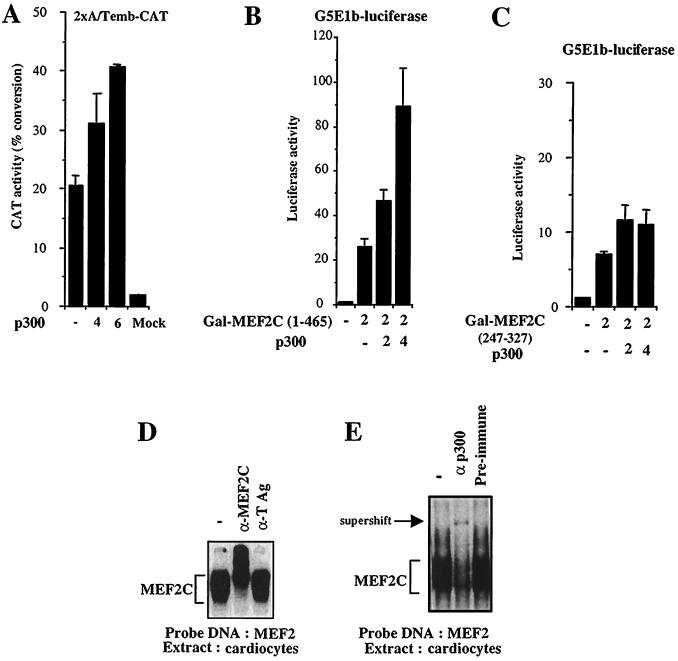
In muscle cells, transcription activated by MEF2C is dependent on its interaction with p300 (9, 33, 35). p300 is expressed in the myocardium (42). Thus, we explored the possibility that the activity of MEF2C in the heart is also dependent on p300. Indeed, forced expression of p300 augmented transcription from the MEF2C-dependent 2xA/Temb promoter in a dose-dependent manner (Fig. 2A). To begin to determine whether this transactivation is mediated by a direct interaction of the p300 and MEF2C proteins, we cotransfected cardiomyocytes with plasmid DNA encoding p300 plus a construct in which the DNA-binding domain of GAL4 (amino acids 1 to 147) is fused to full-length MEF2C. We measured transcriptional activity on the GAL4-dependent reporter construct G5E1b-luciferase. Overexpression of p300 caused a vigorous dose-dependent increase in MEF2C transcriptional activity (Fig. 2B). In comparison, p300 had only a minimal effect on the transcriptional activity of a deletion construct lacking the MADS domain of MEF2C (Fig. 2C). These data are consistent with our previous results that indicate that the MADS domain of MEF2C serves as an interface for interaction with p300 (35).
FIG. 2.
(A) p300 coactivates MEF2C in cardiocytes. Rat neonatal cardiocytes were cotransfected with 2 μg of the 2xA/Temb-CAT construct and with increasing amounts of a plasmid encoding either full-length p300 or just the CMV backbone. In all transfections, the total amount of DNA was kept constant. CAT activity measurements represent the means of three independent experiments carried out in triplicate and normalized to protein content. (B and C) Stimulation of MEF2C transcriptional activity by p300 is dependent on the MADS domain of MEF2C. Neonatal cardiocytes were cotransfected with a GAL4 construct fused to full-length MEF2C [Gal-MEF2C(1-465)] or with a construct lacking the MADS domain [Gal-MEF2C(247-327)] but retaining transcriptional activity and with increasing concentration of CMV-p300. Transcriptional activity was measured 48 h later on a GAL4-dependent reporter construct, G5E1b-luciferase. (D and E) MEF2C binds p300 in cardiocytes. A binding assay was performed with a DNA fragment containing an MEF2 binding site derived from the muscle creatine kinase promoter and with nuclear extracts prepared from cardiac cells. A major complex corresponding to the binding of MEF2C protein to the DNA template was observed (D). The binding reaction was specific to MEF2C, since an antibody specifically recognizing MEF2C resulted in a further reduction in the mobility of the complex (supershift). An antibody directed against large T antigen (TAg) had no effect. The binding of p300 with MEF2C was detected in cardiac cells, since the complex was supershifted by an anti-p300 antibody (E).
To conclusively address whether or not MEF2C and p300 form a complex in cardiomyocytes, we performed a series of electromobility shift assays using an oligonucleotide containing a natural MEF2 binding site derived from the muscle creatine kinase promoter. Nuclear extracts from cardiomyocytes gave rise to a major complex (Fig. 2D, leftmost lane). This complex is specific for MEF2C, since an anti-MEF2C antibody resulted in a supershift, whereas a control antibody had no effect (Fig. 2D, middle and rightmost lanes). p300 is present in the MEF2C complex in cardiac nuclear extracts, since the complex was supershifted by an anti-p300 antibody but not by preimmune serum (Fig. 2E). The results of these experiments established that MEF2C and p300 are associated in a DNA-binding complex in nuclear extracts derived from cardiocytes.
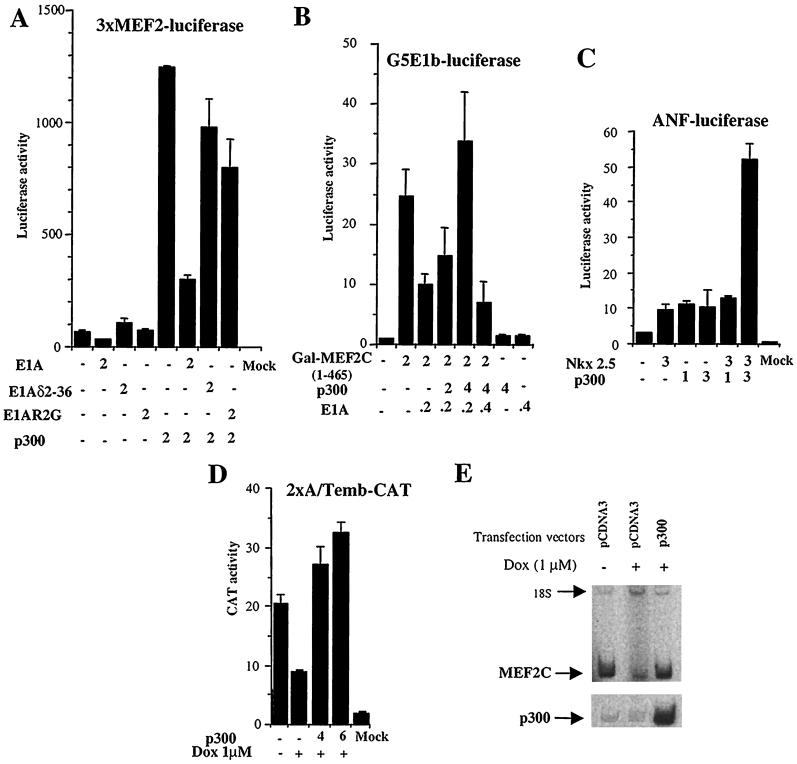
While these experiments show that p300 coactivates and is associated with MEF2C in cardiocytes, they do not clarify whether p300 is a necessary cofactor for MEF2-dependent transcription. Therefore, we decided to inhibit endogenous p300 function by expressing the oncoprotein E1A. Neonatal cardiomyocytes were transiently transfected with a plasmid expressing wild-type E1A and a 3xMEF2-luciferase reporter gene. The presence of E1A resulted in an inhibition of MEF2C activity that could be counteracted by forced expression of p300 (Fig. 3A). E1A mutants lacking p300 binding activity, E1Aδ2-36 and E1AR2G, had almost no effect on MEF2C transcriptional activity, consistent with our previous report (34). Taken together, these results indicate that p300 is a required cofactor for full MEF2C activity in cardiomyocytes. To verify that MEF2C transactivation is mediated by direct binding of p300 and MEF2C, we investigated the effect of E1A in a GAL4 assay (Fig. 3B). MEF2C-activated transcription was repressed by E1A. When increasing amounts of p300 were cotransfected in the cells, MEF2C transcriptional activity was restored.
FIG. 3.
(A and B) Disruption of p300 function inhibits MEF2C activity. Neonatal cardiocytes were cotransfected with 2 μg of 3xMEF2-luciferase construct or GAL-MEF2C(1-465) and with the indicated amount of expression vectors. After 48 h, transcriptional activity was measured in the transfected cells. Each value represents the mean ± standard deviation (SD) of three independent experiments carried out in triplicate and normalized to protein content. (C) p300 enhances NKX2.5-dependent transcription. Neonatal cardiocytes were transiently transfected with plasmids encoding either or both full-length NKX2.5 or p300 plus the ANF promoter. Luciferase activity was measured 24 h later and corrected for protein content. The data represent the mean ± SD of three independent experiments carried out in triplicate. (D) Overexpression of p300 rescues MEF2C inhibition by doxorubicin. Control cardiocytes or cardiocytes exposed to 1 μM doxorubicin (Dox) were cotransfected with 2 μg of 2xA/Temb-CAT and with increasing concentrations of CMV-p300 or CMV backbone. CAT activity was measured 48 h after transfection. The total amount of DNA was kept constant in all transfections. The data represent the mean ± SD of three independent experiments carried out in triplicate. (E) Forced expression of p300 in doxorubicin-treated cardiocytes upregulates endogenous MEF2C. Neonatal cardiocytes untreated or treated with 1 μM doxorubicin (Dox) were cotransfected with a plasmid expressing GFP and CMV-p300 or CMV alone. At 72 h after transfection, GFP-positive cells were sorted by FACS. Total RNA was extracted from the transfected cells, and the relative amounts of MEF2C and p300 mRNAs were determined by RT-PCR. 18S RNA mixed with competimers was used as an internal control. This experiment was done twice with two independent cardiac cell preparations.
Members of the NK class of homeodomain proteins are key regulators in the establishment and maintenance of the cardiac phenotype (28). NKX2.5 binds to the NKE element found in the promoters for the atrial natriuretic (ANF) promoter (6) and cardiac α-actin (37) and cooperates with cardiac transcription factors to optimally drive transcription (5). To determine whether p300 can serve as a coactivator for NKX2.5, we tested the effect of overexpression of p300 on transcription driven from the ANF promoter. Forced expression of NKX2.5 only weakly stimulated the ANF promoter following transient transfection of cardiomyocytes (Fig. 3C). Overexpression of p300 alone increased ANF promoter activity similarly. Such effects of p300 on promoter activity have been observed previously with the p21 promoter, suggesting that p300 coactivates additional transcription factors. However, when NKX2.5 and p300 were coexpressed, NKX2.5-dependent transcription increased as much as fivefold, suggesting cooperation between the two factors (Fig. 3C). Similar results were obtained in the mouse embryonic cell line 10T1/2 (data not shown). Further studies will be needed to determine whether this transactivation is mediated by direct interaction between p300 and NKX2.5.
Forced expression of p300 overrides MEF2C inhibition by doxorubicin.
Since p300 is required for full MEF2C activity, we asked whether p300 would also rescue the inhibitory effect of doxorubicin on MEF2C-dependent transcription. Using the MEF2C-dependent 2xA/Temb promoter, we tested the ability of p300 to effect transcription in cardiomyocytes treated with doxorubicin. Strikingly, forced expression of p300 was able to overcome fully the transcriptional inhibition by doxorubicin (Fig. 3D). This suggests that the inhibition of MEF2C activity by doxorubicin is due at least in large part to a loss of p300 activity.
The experiments presented earlier demonstrate that p300 acts in part through protein-protein interactions with MEF2C on promoters, but the protective effect of p300 in the presence of doxorubicin could also be due to increased levels of MEF2C itself. To test whether p300 possibly effects transcription of the MEF2C gene, we measured the relative levels of MEF2C mRNA in cardiac cells treated with doxorubicin in the presence and absence of a p300 expression vector. Neonatal cardiomyocytes were transfected with p300 or control plasmids and then treated with doxorubicin. All cells were cotransfected with a GFP plasmid to help identify transfected cells. At 48 h, GFP-positive cardiocytes were sorted by FACS and analyzed for expression of endogenous MEF2C and p300 using RT-PCR (Fig. 3E). Doxorubicin treatment resulted in a marked decrease in endogenous MEF2C mRNA, as expected (see also Fig. 1A). Exogenous p300 was expressed manyfold over endogenous p300. The overexpression of p300 in doxorubicin-treated cardiocytes induced endogenous MEF2C transcripts by at least sixfold compared to cells transfected with an empty plasmid (Fig. 3E). We conclude, therefore, that doxorubicin exerts its effect not only by targeting the transcription of the genes for factors such as MEF2C but also by interfering with p300 function. We also infer from these results that p300 is a cofactor not only for transcription driven by MEF2C but also for cardiac transcription of the MEF2C gene itself.
Doxorubicin reduces p300 protein but not p300 mRNA.
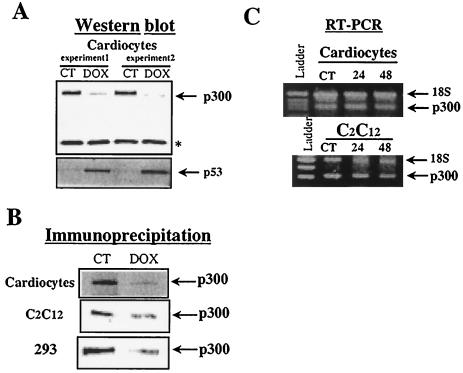
The involvement of p300 in MEF2C activity and the ability of p300 to rescue doxorubicin inhibition of MEF2C-dependent transcription strongly suggest that p300 is a target for doxorubicin. Indeed, when the amount of p300 protein was estimated by Western blot analysis, its levels were significantly reduced in doxorubicin-treated cardiac cells (Fig. 4A). To establish that these biological effects of doxorubicin on cardiomyocytes were appropriate, we confirmed the levels of the tumor suppressor p53, which are known to rise in response to doxorubicin (25). Furthermore, immunoprecipitation of nuclear extracts prepared from cardiocytes, C2C12 cells, and 293 cells confirmed that p300 protein levels are reduced in doxorubicin-treated cells (Fig. 4B). We expected to find that doxorubicin reduced the expression of p300 when we measured the relative abundance of p300 mRNA in control and doxorubicin-treated cells. Remarkably, p300 transcript levels were identical in control cells and in cells exposed to the drug (Fig. 4C). This surprising result again underscores the selective effect of doxorubicin on transcription. The failure of doxorubicin-treated cardiocytes to express normal levels of p300 protein despite high levels of p300 mRNA raised the intriguing possibility that doxorubicin modifies the translation of specific mRNAs or induces their selective degradation.
FIG. 4.
p300 protein levels but not mRNA levels are decreased by doxorubicin. (A) Nuclear extracts (100 μg) from neonatal cardiomyocytes cultured in the absence (control, CT) or presence of 1 μM doxorubicin (Dox) for 48 h were electrophoresed on a 4 to 20% gradient gel. After transfer of the proteins, the membranes were incubated with a primary antibody directed against p300 or p53 overnight at 4°C. After incubation with a secondary antibody, visualization of antigen-antibody reaction was performed with a chemiluminescent reagent. ∗, unknown protein not sensitive to doxorubicin. (B) Nuclear extracts (100 μg) from control and doxorubicin-treated cardiocytes, C2C12 cells, and 293 cells were incubated with an anti-p300 antibody (power clonal [UBI] or N-15X [Santa Cruz]). The extracts were then preadsorbed on protein A/G PLUS-agarose and washed. After centrifugation, the supernatant was analyzed by SDS-PAGE on a 5% polyacrylamide gel. After transfer of the proteins, membranes were blocked and incubated with anti-p300 antibody. Immunocomplexes were detected by chemiluminescence. (C) Relative quantitative RT-PCR of p300 in control and doxorubicin-treated cardiocytes and C2C12 cells. Total RNA (2 μg) was reverse transcribed and PCR amplified with specific primers for p300. PCR products were separated on a 2% agarose gel and quantitated with AlphaImager software. In all PCRs, the level of expression of p300 was normalized against 18S cDNA. Each experiment was repeated at least twice with RNA template from two independent cardiocyte preparations.
p300 is degraded in cardiocytes treated with doxorubicin.
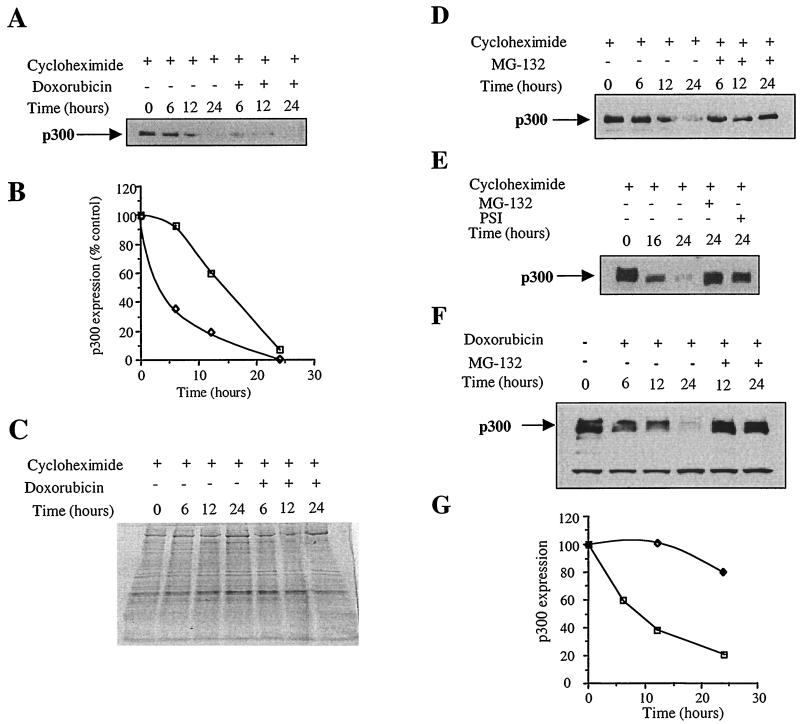
To determine whether the stability of p300 protein is affected following doxorubicin exposure, de novo protein synthesis was inhibited with cycloheximide and the rate of p300 degradation was followed over time in control and doxorubicin-treated cardiocytes. Doxorubicin exposure markedly accelerated proteolysis of p300 (Fig. 5A). In this assay, the relative half-life of p300 was reduced from about 15 h to less than 5 h (Fig. 5B). Indeed, 65% of p300 was degraded in the cells treated with doxorubicin for 6 h, whereas 92% of p300 remained in control cells. After 12 h of doxorubicin exposure, p300 levels were only 20% of the initial value, while 60% persisted in control cells.
FIG. 5.
Doxorubicin increases p300 degradation and inhibitors of the proteasome stabilize p300 protein and prevent p300 degradation by doxorubicin. (A) Neonatal cardiomyocytes were treated with 10 μM cycloheximide for 5 h to inhibit general protein synthesis. Half of the cells were exposed to doxorubicin. Cycloheximide was always present in the medium. Nuclear proteins (30 μg) were separated by SDS-PAGE on a 4 to 20% gradient gel. After transfer of the proteins, the levels of p300 protein were determined by Western blot analysis using an anti-p300 antibody. (B) Relative amounts of p300 protein were determined using an AlphaImager. □, p300 protein level in cardiocytes treated with cycloheximide only; ◊, p300 protein level in cardiocytes treated with cycloheximide plus doxorubicin. (C) Coomassie staining of the nuclear proteins separated by SDS-PAGE. (D) Neonatal cardiomyocytes exposed to 10 μM cycloheximide were treated with DMSO or with 30 μM MG-132 proteasome inhibitor for the indicated times. Nuclear extracts were prepared and resolved on a 4 to 20% gradient gel. Following transfer of the protein to nitrocellulose membranes, p300 levels were determined by Western blot using an anti-p300 antibody. (E) Two different inhibitors of the proteasome stabilize p300 protein. Cardiomyocytes treated with cycloheximide were coincubated with 30 μM MG-132 or 30 μM PSI. Nuclear extracts were prepared before the addition of inhibitors of the proteasome (time zero) or after 16 or 24 h of incubation. Proteins were analyzed on a 4 to 20% gradient gel, and p300 expression was detected by Western blot using an anti-p300 antibody. These experiments were repeated three times with three independent cardiocyte preparations. (F) Neonatal cardiocytes were exposed to 1 μM doxorubicin. Half the cells were also exposed to MG-132. Nuclear extracts were prepared after various times of incubation and resolved on a 4 to 20% gradient gel. Following transfer of the protein on nitrocellulose membranes, p300 levels were determined by Western blot using an anti-p300 antibody. (G) Quantitative analysis of experiment C. □, p300 protein level in cardiocytes treated with doxorubicin only; ◊, p300 protein level in cardiocytes treated with doxorubicin plus MG-132.
Nuclear extracts prepared from cycloheximide-treated cardiocytes and from cells treated with cycloheximide and doxorubicin were compared by SDS-PAGE and stained by Coomassie blue. Interestingly, the large majority of cellular proteins were not affected by doxorubicin treatment (Fig. 5C). This observation is consistent with the notion that doxorubicin does not induce a general proteolysis but might affect the degradation of specific proteins. Induction and stabilization of p53 proteins were used as indicators of effective doxorubicin treatment (25). This was confirmed by evaluating the degradation of p53 in cardiocytes treated with the drug. Following exposure of the cells to doxorubicin, p53 protein levels rose dramatically from very low levels, as expected. When de novo protein synthesis was inhibited with cycloheximide, p53 degradation was not altered by doxorubicin (data not shown). This observation underscores the view that doxorubicin does not induce a general proteolysis but only enhances the degradation of specific nuclear proteins, of which p300 is the first example.
Inhibitors of the proteasome stabilize p300 protein.
Mammalian cells contain multiple pathways for intracellular protein breakdown (4). If doxorubicin accelerates the rate of degradation of p300 protein, which proteolytic system is involved? The nonlysosomal ubiquitin-proteasome system is known to be active in the heart as a major pathway for selective proteolysis in the cytoplasm and the nucleus (15). Following conjugation by multiple chains of ubiquitin, these proteins are degraded by the proteasome. Furthermore, p300 is posttranscriptionally regulated by ubiquitination (1). Since ubiquitinated proteins are often degraded by the proteasome, we investigated whether the ubiquitin-proteasome pathway is involved in p300 degradation both under basal metabolic conditions and after doxorubicin treatment.
We compared the rate of degradation of p300 over time in cardiocytes treated with cycloheximide and in cardiocytes cotreated with cycloheximide and specific inhibitors of the proteasome. Equal amounts of nuclear protein extracts were separated by SDS-PAGE, and p300 levels were determined by Western blot. p300 expression decreased with time in cardiocytes treated with cycloheximide, as expected (Fig. 5D). However, coincubation of the cells with the proteasome inhibitor MG-132 resulted in stabilization of p300 protein. A second inhibitor of the proteasome, PSI, also prevented p300 degradation (Fig. 5E). MG-132 may be slightly more efficient in blocking p300 degradation than PSI. p300 protein levels remained at 82% after 24 h with MG-132 but fell to 62% in the presence of PSI. The Coomassie stains of nuclear proteins separated by SDS-PAGE revealed that the majority were not affected by addition of inhibitors of the proteasome (data not shown). This observation is consistent with the hypothesis that ubiquitin-proteasome proteolysis is responsible for the highly selective degradation of intracellular proteins.
Proteasome inhibitor prevents p300 degradation by doxorubicin.
To determine whether the accelerated loss of p300 protein following doxorubicin exposure is caused by proteasome-mediated proteolysis, the ability of proteasome inhibitor to prevent p300 degradation in response to doxorubicin was examined. Cardiomyocytes were maintained in the presence of either doxorubicin alone or doxorubicin and 30 μM MG-132 for various times. Nuclear extracts obtained from the cultured cardiomyocytes were analyzed by SDS-PAGE, and the expression of p300 was measured by Western blot. p300 expression decreased as early as 6 h with doxorubicin treatment and fell to less than 40% by 12 h and to only 20% at 24 h (Fig. 5F and G). Addition of the proteasome inhibitor dramatically prevented p300 degradation by the anticancer agent; its level was unchanged at 12 h and remained above 80% at 24 h. Most nuclear proteins were not affected by either doxorubicin or proteasome inhibitor treatment (data not shown), again suggesting the specificity of both agents.
Proteasome-mediated degradation induced by doxorubicin is a selective process.
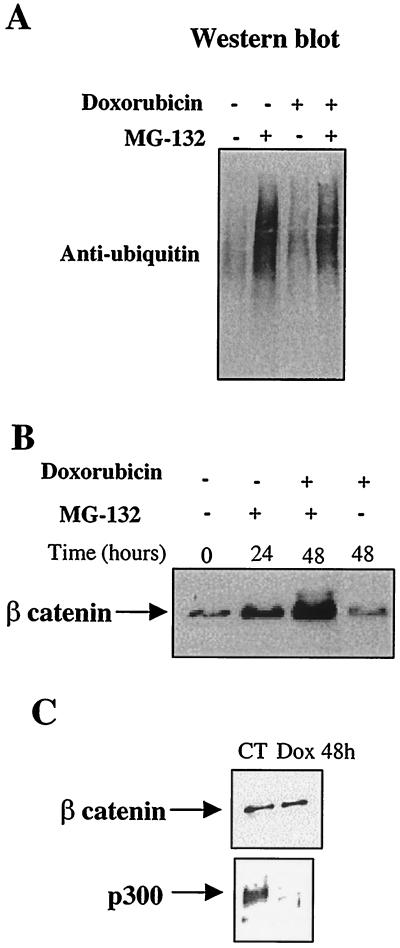
Next, we investigated whether the proteasome-mediated degradation of p300 engendered by doxorubicin is a selective process or whether it results from a general increase in the ubiquitin-proteasome pathway. To this end, we inhibited proteasomes by treating cardiocytes with MG-132 and compared levels of expression of total ubiquitinated proteins in control and doxorubicin-treated cells. Identical levels of ubiquitinated proteins were found (Fig. 6A). To establish that the degradation of p300 by the proteasome following doxorubicin exposure is selective, we investigated the effect of doxorubicin on a protein known to be regulated by this proteolytic system, β-catenin (31). As expected, the treatment of cardiac cells with MG-132 resulted in the accumulation of β-catenin (Fig. 6B). However, β-catenin protein levels in cardiocytes treated with doxorubicin for 48 h were similar to those in control cells (Fig. 6C), emphasizing that the proteasome-mediated degradation induced by doxorubicin is a selective event.
FIG. 6.
Proteasome-mediated degradation of p300 is a specific process. Neonatal cardiocytes were maintained in culture in doxorubicin-free medium (CT), in medium containing 10 μM MG-132, in medium supplemented with 1 μM doxorubicin (Dox), or in medium containing both doxorubicin and MG-132 for 48 h. Equal amounts of nuclear extracts were electrophoresed on 4 to 20% gradient gels. After transfer of the proteins, ubiquitinated proteins were detected with an antiubiquitin antibody (A). β-Catenin levels were determined by Western blot using an anti-β-catenin antibody (B and C).
Although the ubiquitin-proteasome pathway is not generally enhanced after doxorubicin exposure, p300 degradation could still result from a selective increase in the ubiquitination of p300. We investigated this possibility but were unable to detect any ubiquitinated p300 in either control or doxorubicin-treated cells. Therefore, it remains unclear whether proteasome-mediated degradation of p300 following doxorubicin exposure requires increased ubiquitination (40).
DISCUSSION
p300 integrates intracellular signals that result from a wide variety of transduction pathways (reviewed in reference 39). The activity of this critical integrator is regulated by its interaction with other cellular factors, including transcription activators and regulators of the cell cycle (39) as well as the viral oncoprotein E1A (11, 41) and the embryonic differentiation inhibitor Twist (our unpublished data). In addition, p300 is itself the target of phosphorylation (1, 21) and acetylation (30), raising the possibility that its activity is regulated by posttranslational modification. The results of the studies reported here establish regulated protein degradation as a new and unexpected mode of control of p300. While uncovering fundamental mechanisms responsible for cardiac transcriptional repression by the antineoplastic agent doxorubicin, we found that doxorubicin induces the degradation of p300 mediated by the proteasome. p300 transcript levels were unaffected by doxorubicin, whereas the level of p300 protein was markedly reduced. Several considerations suggested that these observations were more likely to be the result of induction degradation of proteins than the repression of their translation. First, cardiocytes exposed to doxorubicin continue to express a number of housekeeping genes at normal or nearly normal levels. Thus the effect on p300 appears to be the result of a selective, not a general, process. Second, effects on translation are more likely to be general than specific, and doxorubicin seemed not to exert a general effect on protein degradation. Indeed, while doxorubicin does suppress overall protein synthesis in cardiocytes, it does so with a slow time course, consistent with the loss of mRNAs for the major contractile proteins (17). Thus, following doxorubicin, the decrease in p300 protein levels despite persistent levels of its transcripts is more likely due to an increase in degradation of p300 protein. Indeed, specific inhibitors of the proteasome dramatically stabilize p300 protein (Fig. 6). Clearly the large majority of other cellular proteins were not sensitive to the proteasome inhibitor. Such selectivity was manifested by the observation that doxorubicin had no effect on β-catenin, which is normally degraded by the ubiquitin-proteasome pathway. In addition, doxorubicin induces p53 protein expression. Because p300 is a critical component of p53 activity (2) and is required for p53 degradation (12), one might speculate that the induction of p53 protein by doxorubicin (or DNA-damaging agents) results from the loss of p300. Thus, normal p53 turnover might be restored by forced expression of p300 in cells treated with doxorubicin. Whether such strategies can protect against cell damage by doxorubicin remains to be determined.
p300, as well as several other transcription factors, is modified posttranslationally by phosphorylation and ubiquitination (1). After ubiquitination, a protein is generally marked for degradation by the 26S proteasome (4, 14), and p300 is likely similarly targeted. However, we were unable to determine whether ubiquitination is a prerequisite for proteasomal degradation of p300. There are examples of proteins whose turnover by the proteasome is ubiquitin independent, including ornithine decarboxylase (29) and p21 (38), and their numbers may increase (40). Thus, the possibility remains that p300 might be another example of a protein degraded by the proteasome independently of ubiquitin attachment.
The increased proteasome-mediated degradation of p300 is likely to be central to the mechanisms underlying inhibition of cardiac transcription by doxorubicin. The finding that MEF2C and NKX2.5 are coactivated by p300 and that the levels of mRNA for both factors are repressed by doxorubicin support this concept. In keeping with this view, our previous studies conducted in skeletal muscle cells demonstrated that doxorubicin inhibits the expression of the myogenic factor MyoD (24). MyoD activity is also regulated by p300. Therefore, MyoD, in addition to MEF2C and NKX2.5, is a third example of a transcription factor dependent on p300 and targeted by doxorubicin.
p300 appears to play at least two major roles in the mechanism of doxorubicin suppression of tissue-specific gene expression. First, loss of p300 protein deprives several tissue-specific factors of this necessary coactivator, diminishing the expression of tissue-specific target genes, such as contractile proteins. Second, and unexpectedly, p300 appears to be an activator for the transcription of the transcription factor mRNAs themselves. In the case of MEF2C, overexpression of p300 stimulates MEF2C mRNA accumulation and counteracts the doxorubicin suppression of MEF2C-dependent transcription. This suggests that p300, in concert with yet unidentified transcription factors, drives transcription of the MEF2C gene itself. MyoD requires p300 for its activity (33, 35, 44). Since MyoD autoactivates its own transcription, loss of p300 activity likely affects MyoD mRNA polymerization. The concept of gene dosage as critical for p300 function has been evoked before (42) and is consistent with the hypothesis that p300 is expressed in limiting amounts in the cell (20). Thus, the depletion of p300 following degradation by the proteasome would result in an impairment of MEF2C, NKX2.5, and eventually all p300-dependent transcription (Fig. 7). In keeping with this view, p300 and its related factor CREB-binding protein are required for the activity of many transcription factors (39). Therefore, we speculate that p300 might drive transcription of other transcriptional regulators and that the mechanism of regulation that we describe here for the MEF2C gene might be common for a variety of nuclear factors.
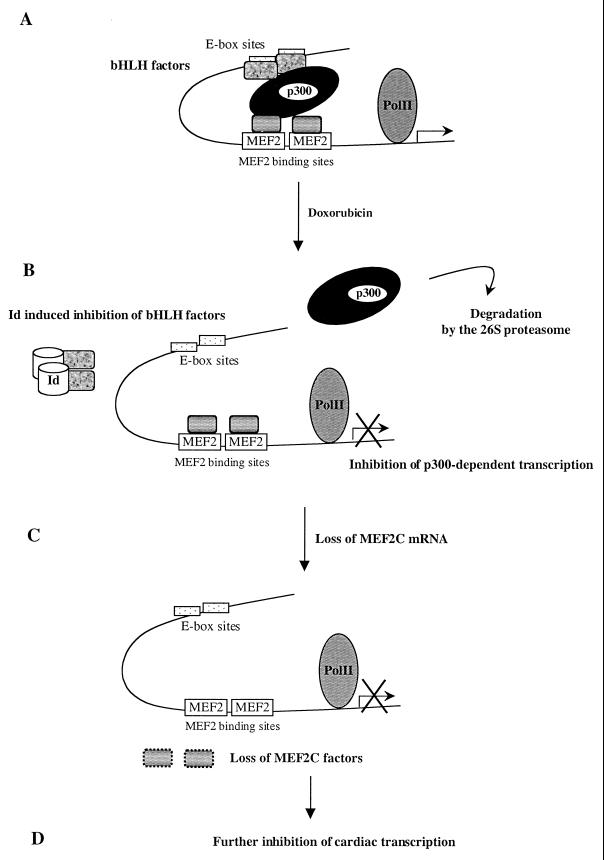
FIG. 7.
Hypothetical mechanism of tissue-specific transcriptional repression mediated by doxorubicin. A tissue-specific transcription factor complex (A) comprising tissue-specific and general transcription factors binds to cognate DNA enhancer sites, including bHLH factors and MEF2C. These transcription factors are capable of acting as a scaffold for the coactivator p300; the stoichiometry of the factors depicted is purely hypothetical. The curved line represents the chromosomal DNA, and the arrow marks the start of transcription. Doxorubicin exposure rapidly leads to degradation of p300 via the proteasome pathway (B). Doxorubicin also more slowly leads to induction of the bHLH inhibitor Id (22). As a result, p300-dependent transcription is diminished, including the transcription of the MEF2C gene itself (C), which in turn further inhibits MEF2C-dependent transcription (D). PolII, polymerase II.
The finding that doxorubicin inhibits MEF2C transcription by approximately half raises the possibility that the disruption of cardiomyocyte integrity by doxorubicin is mediated by moderate but concomitant decreased expression of several cardiac-relevant transcription factors rather than by the elimination of a single activator. Targeted disruption of the MEF2C gene has shown that the bHLH protein dHAND is downstream of MEF2C. Therefore, it is possible that the doxorubicin effect we observed on dHAND expression is a consequence of MEF2C depletion or inactivation. However, when MEF2C expression is abolished in mice, the cardiac homeobox gene NKX2.5 is expressed at normal levels. This suggests that the mechanisms contributing to the doxorubicin-induced decreased in NKX2.5 mRNA are independent of transcriptional repression of MEF2C. Nevertheless, the synergy exhibited by NKX2.5 and p300 on transcription of the ANF promoter emphasizes the pivotal position played by p300 as a target of doxorubicin-engendered transcriptional suppression.
Remarkably, inhibitors of the proteasome completely prevented p300 degradation by doxorubicin. A hypothesis that we are testing is that the transcription of MEF2C should be restored in doxorubicin-treated cells also exposed to proteasome inhibitors.
p300 is a histone acetylase (30), and many observations support the view of a relationship between acetylation and function of transcription factors (34, 35, 36). Accordingly, another implication of our findings is that doxorubicin likely diminishes acetyltransferase activity in cells, at least as a consequence of its depletion of p300 protein, if not by another mechanism.
What are the other implications of the altered rate of degradation of p300 mediated by the proteasome? This mechanism of regulation might play a role during normal cellular differentiation, proliferation, or homeostasis, since p300 is implicated in all these biological activities. Doxorubicin as an intercalating agent induces DNA damage. p53 protein is a major sensor of such cellular DNA damage. Association of p300 with MDM2 has been shown to participate in MDM2-regulated degradation of p53. Thus, proteasome-mediated degradation of p300 induced by doxorubicin might stabilize p53 levels. It will be of interest to determine whether other DNA-damaging agents such as UV light and γ-irradiation have similar effects on p300 stability. Finally, it would not be surprising to find that other transcriptional cofactors are subject to similar modes of regulation.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institutes of Health (to L.K.) and from the American Heart Association (to R.K.). C.P. was supported by a research fellowship from the American Heart Association-Greater Los Angeles.
We thank Henry Sucov for his suggestions and criticisms and members of the Kedes lab for helpful discussions.
REFERENCES
- 1.Avantaggiati M L, Carbone M, Graessman A, Nakatani Y, Howard B, Levine A S. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 1996;15:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- 2.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 3.Bishopric N H, Kedes L. Adrenergic regulation of the skeletal alpha-actin gene promoter during myocardial cell hypertrophy. Proc Natl Acad Sci USA. 1991;88:2132–2136. doi: 10.1073/pnas.88.6.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 5.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx2–5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durocher D, Chen C Y, Ardati A, Schwartz R J, Nemer M. The atrial natriuretic factor promoter is a downstream target for Nkx-2.5 in the myocardium. Mol Cell Biol. 1996;16:4648–4655. doi: 10.1128/mcb.16.9.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 8.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 10.Edmondson D G, Lyons G E, Martin J F, Olson E N. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 11.Egan C, Jelsma T N, Howe J A, Bayley S T, Ferguson B, Branton P E. Mapping of cellular protein-binding sites on the products of early-region 1A of human adenovirus type 5. Mol Cell Biol. 1988;8:3955–3959. doi: 10.1128/mcb.8.9.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 13.Gustafson T A, Kedes L. Identification of multiple proteins that interact with functional regions of the human cardiac alpha-actin promoter. Mol Cell Biol. 1989;9:3269–3283. doi: 10.1128/mcb.9.8.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 15.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 16.Huang J, Weintraub H, Kedes L. Intramolecular regulation of MyoD activation domain conformation and function. Mol Cell Biol. 1998;18:5478–5484. doi: 10.1128/mcb.18.9.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito H, Miller S C, Billingham M E, Akimoto H, Torti S V, Wade R, Gahlmann R, Lyons G, Kedes L, Torti F M. Doxorubicin selectively inhibits muscle gene expression in cardiac muscle cells in vivo and in vitro. Proc Natl Acad Sci USA. 1990;87:4275–4279. doi: 10.1073/pnas.87.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janknecht R, Hunter T. Transcription: a growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 19.Jeyaseelan R, Poizat C, Baker R K, Abdishoo S, Isterabadi L B, Lyons G E, Kedes L. A novel cardiac-restricted target for doxorubicin: CARP, a nuclear modulator of gene expression in cardiac progenitor cells and cardiomyocytes. J Biol Chem. 1997;272:22800–22808. doi: 10.1074/jbc.272.36.22800. [DOI] [PubMed] [Google Scholar]
- 20.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 21.Kitabayashi I, Eckner R, Arany Z, Chiu R, Gachelin G, Livingston D M, Yokoyama K K. Phosphorylation of the adenovirus E1A-associated 300 kDa protein in response to retinoic acid and E1A during the differentiation of F9 cells. EMBO J. 1995;14:3496–3509. doi: 10.1002/j.1460-2075.1995.tb07356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurabayashi M, Dutta S, Jeyaseelan R, Kedes L. Doxorubicin-induced Id2A gene transcription is targeted at an activating transcription factor/cyclic AMP response element motif through novel mechanisms involving protein kinases distinct from protein kinase C and protein kinase A. Mol Cell Biol. 1995;15:6386–6397. doi: 10.1128/mcb.15.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurabayashi M, Jeyaseelan R, Kedes L. Antineoplastic agent doxorubicin inhibits myogenic differentiation of C2 myoblasts. J Biol Chem. 1993;268:5524–5529. [PubMed] [Google Scholar]
- 24.Kurabayashi M, Jeyaseelan R, Kedes L. Doxorubicin represses the function of the myogenic helix-loop-helix transcription factor MyoD: involvement of Id gene induction. J Biol Chem. 1994;269:6031–6039. [PubMed] [Google Scholar]
- 25.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 26.Lin Q, Schwarz J, Bucana C, Olson E N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lints T J, Parsons L M, Hartley L, Lyons I, Harvey R P. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. . (Erratum, 119:969.) [DOI] [PubMed] [Google Scholar]
- 28.Lyons I, Parsons L M, Hartley L, Li R, Andrews J E, Robb L, Harvey R P. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 29.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 30.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 31.Orford K, Crockett C, Jensen J P, Weissman A M, Byers S W. Serine phosphorylation-regulated ubiquitination and degradation of β-catenin. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 32.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C, Masuno M, Tommerup N, van Ommen G J, Goodman R H, Peters D J, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 33.Puri P L, Avantaggiati M L, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puri P L, Sartorelli V, Yang X J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 35.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sartorelli V, Puri P L, Hamamori Y, Ogryzko V V, Chung G, Nakatani Y, Wang J Y, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 37.Sepulveda J L, Belaguli N, Nigam V, Chen C Y, Nemer M, Schwartz R J. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheaff R J, Singer J D, Swanger J, Smitherman M, Roberts J M, Clurman B E. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 39.Shikama N, Lyon J, La Thange N. The p300/CBP family:integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 40.Verma R, Deshaies R J. A proteasome howdunit: the case of the missing signal. Cell. 2000;101:341–344. doi: 10.1016/s0092-8674(00)80843-0. [DOI] [PubMed] [Google Scholar]
- 41.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 42.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 44.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]