Abstract
Coronavirus disease 2019 (Covid‐19) is an emerging novel respiratory infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) that rapidly spread worldwide. In addition to lung injury, Covid‐19 patients may develop extrapulmonary symptoms, including cardiac, liver, kidney, digestive tract, and neurological injuries. Angiotensin converting enzyme 2 is the major receptor for the entry of SARS‐CoV‐2 into host cells. The specific mechanisms that lead to cell death in different tissues during infection by SARS‐CoV‐2 remains unknown. Based on data of the previous human coronavirus SARS‐CoV together with information about SARS‐CoV‐2, this review provides a summary of the mechanisms involved in cell death, including apoptosis, autophagy, and necrosis, provoked by severe acute respiratory syndrome coronavirus.
Keywords: cell death, pathways, Sars‐CoV‐2
Abbreviation
- ACE2
Angiotensin converting enzyme 2
- AIF
Apoptosis inducing factor
- AMBRA1
Activating molecule in BECLIN‐regulated autophagy 1
- ARDS
Acute respiratory distress syndrome
- ASK
Apoptosis signal‐regulating kinase
- ATG
Activing transcription factor
- βCov
Betacoronavirus
- BcL‐2
B‐cell lymphoma
- Bcl‐XL
B‐cell lymphoma extra‐large
- CCL2
C‐C motif chemokine ligand 2
- CD
Cluster of differentiation
- CHOP
C/EB homologous protein
- COPD
Chronic obstructive pulmonary disease
- Covid‐19
Coronavirus disease 2019
- CoVs
Coronaviruses
- CQ
Chloroquine
- CTLA4
Cytotoxic T‐lymphocyte associated protein 4
- CypD
Cyclophilin D
- Cyt c
Cytochrome c
- DAMPS
Damage‐associated molecular patterns
- DFCP1
Double FYVE‐containing protein 1
- DNA
Deoxyribonucleic acid
- E
Envelope protein
- EFGR
Epidermal Growth factor receptor
- ER
Endoplasmatic reticulum
- FADD
Fas associated via death domain
- FASL
Fas Ligand
- FKHRL1
Forkhead transcription factor
- GAS6
Growth arrest specific 6
- GRP78
Glucose‐regulated 78
- GRP94
Glucose‐regulated 94
- GSDM
gasdermin D‐regulated
- HcoV
Human coronavirus
- HCQ
Hydroxychloroquine
- IL
Interleukin
- INF
interferon
- IRE‐1
inositol‐requiring enzyme 1
- IRF
Interferon regulatory factor
- ISG
Interferon‐stimulated genes
- JNK
c‐Jun N‐terminal Kinase
- LDH
Lactate dehydrogenase
- MAPK
Mitogen activated Protein Kinase
- MERS‐CoV
Middle East respiratory syndrome coronavirus
- MERTK
MER proto‐oncogene, tyrosine Kinase
- MHV
Mouse hepatitis virus
- MLKL
Mixed‐lineage Kinase domain‐like pseudo Kinase
- MPT
Mitochondrial permeability transition
- MPTP
Mitochondrial permeability transition pore
- NFκB
Nuclear factor kappa B
- NSP
non‐structural protein
- ORF
Open reading frame
- PDK1
3 3‐phosphoinositide‐dependent protein kinase‐1
- PERK
Protein Kinase RNA‐like endoplasmatic reticulum Kinase
- PKB
Protein Kinase B/Akt
- PtdIns3KC3
Phofatidilinositol 3‐Kinase class 3
- RIPK
Receptor‐interacting protein Kinase
- RNA
Ribonucleic acid
- S
Spike protein
- SARS‐CoV‐2
Severe‐acute respiratory syndrome coronavirus 2
- SIRT4
Sirtuin 4
- TFEB
Transcription factor EB
- TGF‐β
Transforming growth factor beta
- TLR4
Toll‐like receptor 4
- TNF
Tumor necrosis factor
- TYRO3
Tyrosine‐protein Kinase 3
- UBC
Ubiquitin C
- WHO
World Health Organization
1. INTRODUCTION
Covid‐19 (Coronavirus disease 2019) is a new emerging respiratory infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). 1 The virus was discovered in late December 2019 in Wuhan, Hubei Province, China, and quickly spread around the world. 2 , 3 On 30 January 2020, WHO (World Health Organization) declared this outbreak a Public Health Emergency of International Concern. 4 As of 16 August 2021, the disease has affected more than 206, 693, 357 million people, being responsible for 4,352, 488 deaths. 5
Coronaviruses (CoVs) are enveloped, positive‐sense, single‐stranded RNA viruses. They are genetically classified into four major genera: Alphacoronavirus, Betacoronavirus (βCoV), Gammacoronavirus, and Deltacoronavirus. Coronaviruses possess the largest RNA genomes (27–32 kb) among the RNA viruses, the 5′ and 3′ ends of coronaviruses genomes contain short untranslated regions. For the coding regions, CoV viral genome encodes four major structural proteins: the spike (S) protein, the envelope (E) protein, the membrane (M) protein, and the nucleocapsid (N) protein. Sars‐CoV‐2 is classified in the genus βCoV, like SARS‐CoV and Middle East respiratory syndrome coronavirus (MERS‐CoV), with evidence indicating that the novel CoV is of bat‐origin. In addition, next‐generation sequencing shows that SARS‐CoV‐2 shares 79% sequence identity to SARS‐CoV and 50% to MERS‐CoV. 6 , 7 , 8 , 9 , 10 , 11 , 12
The clinical manifestations of Covid‐19include fever, cough, myalgia or fatigue, dyspnea and radiographic evidence of pneumonia. 13 , 14 The diffuse alveolar damage and acute respiratory failure are the main features of Covid‐19. 13 , 15 Studies suggest that previous comorbidities such as hypertension, diabetes, chronic obstructive pulmonary disease (COPD) and other cardiovascular diseases are associated with severe cases of the disease. 16 , 17 , 18 , 19 The fatal cases present complications as acute respiratory distress syndrome (ARDS), acute cardiac injury, acute kidney injury, and shock, eventually followed by multiple organ failure. 16 , 20 , 21
The cell death mechanisms associated with cell injury caused by the SARS‐CoV‐2 are not well understood nor established. Previous studies have shown that different apoptosis mechanisms are activated in coronavirus infection 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 . Previous finds from SARS CoV and MERS studies can be used as reference to investigate molecular cell death mechanisms involved in SARS‐CoV‐2 infection. Therefore, two reviewers conducted independent literature research at Medline via Pubmed up to March 30 using the following strategy: “Severe Acute Respiratory Syndrome Virus” OR “SARS‐Related Coronavirus” OR “SARS‐CoV” OR “SARS Coronavirus” OR “Coronavirus, SARS” OR “Severe acute respiratory syndrome‐related coronavirus” OR “Severe acute respiratory syndrome related coronavirus” OR “SARS‐Associated Coronavirus” OR “Coronavirus, SARS‐Associated” OR “SARS Associated Coronavirus”) AND (“cell death” OR “apoptosis” OR “Autophagic cell death” OR “immunogenic cell death” OR “regulated cell death”. A total of 370 studies were identified and then filtered by title and Abstract. After reviewing the remaining articles, 33 studies were included in this review. The two authors (Maríllya Morais da Silva and Priscilla Stela Santana de Oliveir) resolved disagreements by consensus and a third author (Michelly Cristiny Pereira) did the final revision.
2. CORONAVIRUS AND APOPTOSIS
Apoptosis is a process of programmed cell death, morphologically characterized by cellular shrinkage, condensation and margination of chromatin, a dense cytoplasm with tightly packed organelles and the formation of apoptotic bodies. Apoptosis is a natural phenomenon that occurs during growth, ageing, and cellular homeostasis. It may also happen as a result of infections or cell injury. 35 , 36 , 37 , 38
The mechanisms responsible for apoptosis can be grouped into two main pathways: the extrinsic pathway of apoptosis and the intrinsic pathway. In the extrinsic pathway, transmembrane receptor‐mediated interactions initiate the process of apoptosis, the sequence of events that integrates this pathway can be demonstrated by Fas Ligand (FasL)/Fas models. The binding of Fas ligand to Fas receptor results in the binding of the adapter protein FADD. FADD then associates with procaspase‐8 via dimerization of the death effector domain, when caspase‐8 is activated, the execution phase of apoptosis is triggered. While in the intrinsic way, stimuli cause changes in the inner mitochondrial membrane that results in an opening of the mitochondrial permeability transition (MPT) pore, loss of the mitochondrial transmembrane potential and release of two main groups of normally sequestered pro‐apoptotic proteins. The first group includes Cytochrome c (CytC), a molecule that binds and activates Apaf‐1 and procaspase‐9, forming the apoptosome, which later results in caspase 9 activation. Both paths converge to an execution path, where Caspase‐3 is activated by any of the initiator caspases (caspase‐8, caspase‐9, or caspase‐10), activating caspase 3 results in events like endonuclease activation, degradation of chromosomal DNA, protease activation and degradation of nuclear and cytoskeletal proteins, culminating in the cytomorphological changes characteristic of apoptosis. 35 , 39 , 40 , 41 , 42
2.1. Apoptosis induction by SARS‐CoV open reading frames
Coronaviruses can cause apoptosis via a variety of signaling pathways. SARS‐CoV and SARS‐CoV‐2 have two long open reading frames, orf1a and orf1ab, encoding polyproteins 1a and 1ab. The polyproteins are cleaved by viral proteases into 16 non‐structural proteins, Nsp1–Nsp16. 43 Genome contains eight accessory genes whose open reading frames are interspersed among the structural genes; two between S and E genes (ORFs 3a and 3b), five are located between the M and N genes (6, 7a, 7b, 8a, 8b) and one within the N gene (9b) (Figure 1). 44 . Schaecher, Scott R., and Andrew Pekosz has summarized the functions and the subcellular localization of the SARS‐CoV accessory proteins 45 . ORF3a is localized mostly in Golgi and plasma membrane, data propose that it has a role in virus release and lifecycle of SARS‐CoV. 46 , 47 , 48 On the other hand, ORF3b is localized in nucleolus and mitochondria, and induces cell death and prevents interferon‐β (IFN‐β) production. 49 , 50 , 51 , 52
FIGURE 1.
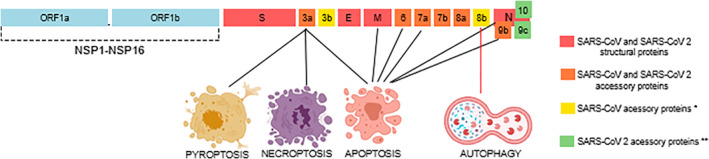
Illustration of the genome of severe acute respiratory syndrome coronavirus (SARS‐CoV) and SARS‐CoV‐2 showing the location of structural proteins, open reading frames (orf1a and orf1ab), accessory proteins which them are known to trigger different forms of cell death. *ORF3b and ORF8b is only present in SARS‐CoV. **ORF10 is only present in SARS‐CoV‐2
The ORF6 protein is subcellular, found in ER and Golgi, and packaged into virus particles and incorporated into virus‐like particles. 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 Other studies suggest that ORF6 is also related to virus assembly or replication processes and antagonizes IFN‐β production. 55 , 56 Overexpression of ORF6 of SARS‐CoV induced apoptosis via caspase‐3–mediated, dependent pathways, and caspase‐3 inhibitor and c‐Jun N‐terminal kinase (JNK) inhibitor blocked ORF‐6 induced apoptosis. The protein level of ER chaperon protein, GRP94, was up‐regulated which suggests that overexpression of ORF‐6 could induce apoptosis via the ER‐stress pathway too. 57 , 58
Gene 7 encodes two accessory proteins, ORF7a and ORF7b. ORF7a is subcellular localized in the Golgi region of transfected and infected cells and its specific function remains unknown. 59 ORF7a may play a role in different processes such as apoptosis induction, activation of signaling pathways and others. 21 , 60 ORF7b is also localized in the Golgi region using cDNA transfected and SARS‐CoV infected cells. 59 , 61 ORF7b is associated with intracellular virus particles and with apoptosis. 54 , 55 Furthermore, Tan et al. suggests SARS‐CoV 7a protein triggers apoptosis by interfering directly with the function of Bcl‐XL, a prosurvival member of the Bcl‐2 family. The results showed that overexpression of Bcl‐XL, blocks 7a‐induced apoptosis, implying that the mechanism behind apoptosis induced by the 7a protein is at the level of or upstream from the Bcl‐2 family. 62
Gene 8 also contains two ORFs. ORF8a is found in the cytoplasm and mitochondria and is reported to have proapoptotic effects. 63 , 64 ORF8b is localized in cytoplasm and its function is still a mystery. A study has suggested a potential role for ORF8b in modulating degradation or stability of the SARS‐CoV E protein. 65 Lastly, ORF9b is localized to the ER region of transfected cells and may interact with viral proteins nsp8, nsp14, and ORF7b. 66 In addition, it was proposed that 9b protein is associated with nucleocytoplasmic export linked apoptosis. The study showed that the 9b protein, known to be localized in the extra‐nuclear region, was also present in the nucleus, and blocking 9b nuclear expor led to caspase‐3 dependent apoptosis of the host cells. 67
Another open reading frame of SARS‐CoV‐2 was annotated, the ORF9c. 68 Dominguez et al. has proposed that ORF9c is a membrane‐associated protein that suppresses antiviral responses in cells. ORF9c enables cells to escape from immune surveillance through reduced HLA abundance and antigen presentation, also slowing cell replication. 69 SARS‐CoV‐2 has another accessory protein, ORF10 is located within the N gene, evidence shows that ORF10 might bind to CULZYG11Bcomplexand hijack its ubiquitination of restriction factors. 43
Some of these accessories proteins trigger apoptosis by different mechanisms. Tan et al. propose that 7a protein of SARS‐CoV can induce apoptosis when overexpressed, as evidenced by an increase in caspase‐3 protease activity, a hallmark of apoptosis. 23 Kopecky‐Bromberg et al. have described that the 7a protein has the ability to inhibit cellular translation and activate p38 Mitogen Activated Protein Kinase (MAPK); this may be the mechanism that leads to apoptosis induction. 60
The 3a protein participates in several signaling pathways that lead to apoptosis. Padhan et al. described that SARS‐CoV 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. 22 Activation of p38 kinase, leads to upregulation of p53, which can increase nuclear transcription of the Bax gene, promoting cytoplasmic oligomerization of the Bax protein. Bax multimers insert in the mitochondrial membrane, provoke loss of potential and release of CytC, which promotes formation of the apoptosome and activation of caspase‐9. 70
Other in vitro studies confirmed the role of orf‐3a in inducing apoptosis. Law et al. examined the expression levels of Bcl‐2 family proteins and caspase‐8, which mediate the intrinsic and extrinsic pathway of apoptosis, respectively. Cleavage of pro‐caspase‐8 was increased in 3a‐transfected Vero E6 cells, indicating that the protein activates apoptosis by its extrinsic pathway. 71 In a more recent study, Ren et al. evaluated the effects of SARS‐CoV‐2 ORF‐3a protein in three cell strains: HEK293T cell line, a human embryonic kidney cell line; HepG2, a human liver cancer cell line; and VeroE6, the Vero lineage derived from kidney epithelial cells extracted from an African green monkey. Results imply that after transfection, SARS‐CoV‐2 ORF‐3a protein can induce apoptosis by activating caspase‐8, which when activated cleaves BH3‐interacting domain death agonist (Bid) to tBid, which in turn induces the release of CytC, resulting in the formation of apoptosome and cleavage of caspase‐9. 72
Furthermore, 3a protein can form ion channels on membranes. Chan et al. reported that 3a induced apoptosis is related to the disturbance of intracellular ion homeostasis, implying that perturbation of intracellular ion flux could be one of the SARS‐CoV pathogenic mechanisms. 31 The 3a protein also causes ER stress by activation of the protein kinase RNA‐like endoplasmic reticulum kinase (PERK) pathway, which enhances protein folding in the ER as described by Minakshi et al. When it activates only the PERK, but not the Inositol‐Requiring Enzyme 1 and Activating Transcription Factor 6 pathways during the unfolded protein response (UPR), the 3a protein protects itself and other viral proteins from ER‐associated protein degradation. Prolonged PERK activation would lead to apoptosis. The activation of downstream effectors such as activating transcription factor 4 and C/EBP homologous protein may be yet another mechanism through which the 3a protein can be apoptotic. 28
ORF3a is involved with the formation of intracellular vesicles, but the mechanism of membrane rearrangement and vesicle formation remains poorly understood. Freundt et al. reported that ORF 3a is necessary for fragmentation of the Golgi apparatus and suggested that this could be explained by the 3a protein ability to disturb Golgi regulator protein, Arf1, function. 73 They also showed that the 3a protein accumulates and localizes to vesicles containing markers for late endosomes and its overpression is sufficient to determine cell death.
2.1.1. Apoptosis induction by SARS‐CoV structural proteins
Coronaviruses have four major structural proteins. The membrane (M) protein and the nucleocapsid (N) protein can trigger apoptosis by several mechanisms. The SARS‐CoV N protein has as primary function the packaging of the genomic RNA in a protective covering. 74 Surjit et al. discovered that the N protein also has the pro‐apoptotic ability to up‐regulate JNK and p38 MAPK activities. Expression of the protein also down‐regulates the levels of phospho (p)‐Akt and Bcl‐2, and activates caspases 3 and 7, which leads to apoptosis. 29
Some of the SARS‐CoV M protein roles include mediating nucleocapsid incorporation into the newly formed virions, regulating viral replication, and packing genomic RNA into viral particles. 37 , 38 . M protein also has a role in induction of apoptosis in SARS‐CoV infected cells. Chan et al. observed that M protein over‐expression induced apoptotic cell death with nuclear condensation and release of mitochondrial CytC. 38 Beyond that, over‐expression of the M protein also caused down‐regulation of Akt phosphorylation, which would further reduce the cell survival signal, consequently, leading to induction of apoptosis.
Zhang et al. transfected COS‐1, Huh‐7 and HepG2 cells with pCDNA3.1(‐)/his‐myc vector containing the SARS coronavirus N, M and S genes. 75 The COS‐1 lineage, a fibroblast‐like cell line derived from monkey kidney tissue, when transfected SARS coronavirus N protein increased intracellular reactive oxygen species and CytC release, with caspase‐3, 9 activation and PARP cleavage, leading to apoptosis. While SARS coronavirus M, S protein cannot induce apoptosis in COS‐1, HepG2 and Huh‐7, SARS coronavirus N protein also did not induce apoptosis in HepG2 and Huh‐7. These findings imply that coronavirus proteins behave uniquely in different cell types.
A more recent study proposes that the M‐protein compromises the protein kinase B/Akt (PKB) cell survival‐signaling pathway through interfering with its upstream activator 3‐phosphoinositide‐dependent protein kinase‐1(PDK1). Tsoi et al. described that the M‐protein can interact with PDK1, PDK1 mediates PKB/Akt phosphorylation. 27 PKD1 interaction with the M‐protein reduces PKB/Akt phosphorylation. The forkhead transcription factor (FKHRL1) regulates FasL expression and its phosphorylation is regulated by PKB/Akt activity. When phosphorylated, FKHRL1 is hold in cytoplasm, where it cannot mediate FasL expression. With the activity of PKB/Akt compromised, FKHRL1 translocates to the nucleus and induces FasL expression. Increased FasL leads to the activation of caspase 8, an apoptotic mediator. Furthermore, PKB/Akt also regulates ASK (apoptosis signal‐regulating kinase), down‐regulated PKB/Akt activity results in reduction in the level of phosphorylated ASK, which leads to caspase 9 activation.
The mechanisms that leads to apoptosis triggered by structural and accessory proteins of SARS‐CoV is summarized in Figure 2.
FIGURE 2.
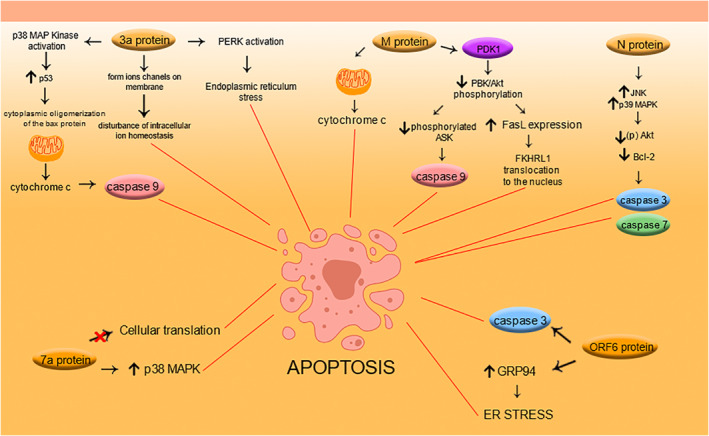
Summary of the mechanisms that lead to apoptosis triggered by the structural and accessory proteins of severe acute respiratory syndrome coronavirus (SARS‐CoV)
2.1.2. Other coronavirus mechanisms that lead to apoptosis
Besides the structural and accessory proteins, coronavirus can induce apoptosis by other mechanisms. Chu et al. suggested that MERS‐CoV infected T cells from the peripheral blood and from human lymphoid organs, including the spleen and the tonsil induced apoptosis in T cells by the activation of caspases 3, 8 and 9, triggering activation of the extrinsic and intrinsic apoptosis pathways. 76 Bellesi et al. observed increased CD95 (Fas) expression in peripheral blood T lymphocytes in COVID‐19 patients. It suggests that CD95 induced apoptosis could be a possible mechanism for COVID‐19‐induced lymphopenia 77 . CD95 is a cell surface protein that can mediate apoptosis bound to its ligand, CD95L. CD95L is mainly expressed in activated T lymphocytes and natural killer cell. 78
Yeung et al. revealed that MERS‐CoV induces apoptosis in kidney and lung by upregulating the mothers against decapentaplegic homolog 7—a homolog of the drosophila gene (Smad7) and the fibroblast growth factor 2 (FGF2) proteins. 79 Other studies have also linked smad7 with apoptosis in renal disease pathogenesis by inducing apoptosis in mesangial cells and podocytes. 80 , 81 Smad7 also induced apoptosis in hepatocarcinogenesis in vitro through the attenuation of NFκB and TGFβ signaling, 98 In its turn, FGF2 induced podocyte injury and glomerulosclerosis in rat model. 82
As has been shown, depending on the cell, the tissue in its microenvironmental context, the SARS family virus can induce different apoptosis mechanisms. Favreau et al. suggested that some HCoV strains can invade the central nervous system, where they induce neuronal programmed cell death cyclophilin D (CypD) dependent and potentially caspase dispensable. 83 CypD is responsible for modulation of mitochondrial permeability transition pore (mPTP) in various types of cell death. 84 When it opens, mPTP allows the release of proapoptotic factors such as CytC and apoptosis‐inducing factor (AIF). 85 AIF translocates to the nucleus and promotes high‐molecular‐weight DNA fragmentation and chromatin condensation. 86 However, it is still poorly understood which cellular proteins interacts with CypD to promote mPTP formation.
3. CORONAVIRUSES AND AUTOPHAGY
Autophagy is a cytoprotective tool that enables cellular decomposition and recycling. This process is essential for normal cellular development and growth, having a role in regulating the balance between protein synthesis and degradation, aging, cancer, neurodegenerative disorders, and lysosomal disorders. It can be initiated by several stimuli sux as nutrient deprivation, oxidative stress, hypoxia, protein aggregates and toxic molecules to mitigate stress. 36 , 87 , 88 , 89 Autophagosomes are double membrane vesicles that engulf part of the cytoplasm that contains long‐lived proteins, pathogens, and damaged organelles. To complete the self‐digesting mechanism, the autophagosome fuses with endosomal and/or lysosomal vesicles in a maturation process, resulting in autolysosomes. 90 , 91 , 92
Previous studies using mouse hepatitis virus as a prototype, showed that (βCoV) induce autophagy related 5 (ATG5) ‐dependent autophagy and ATG5‐dependent autophagosome formation via nps6. 93 NSP6 is also present in other CoVs, including SARS‐CoV‐2, but intriguingly, other researchers reported that this protein limits further autophagosomal expansion, compromising autophagic delivery of viral components to lysosomes. 94 Autophagy‐related proteins are responsible for autophagy regulation and ATG5 has important roles in canonical and non‐canonical autophagy. It is indispensable for autophagic vesicle formation, during the canonical autophagy, ATG5 binds with ATG12 and ATG16 forming a complex that contributes to autophagosome maturation. 95 The canonical formation of autophagosomes consists of four steps: 1. initiation; 2. nucleation; 3. elongation and closure; 4. recycling. Each of them involves the activity of ATG proteins, as well as non‐ATG proteins such as phosphatidylinositol 3‐kinase class III (PtdIns3KC3), p150, activating molecule in BECLIN1‐regulated autophagy 1 (AMBRA1) and double FYVE‐containing protein 1 (DFCP1). While the non‐canonical autophagy can bypass some of these steps, the formation of the double‐membraned autophagosome does not require the hierarchical intervention of all of the ATG proteins; the alternative mechanisms are currently under debate. 96
Moreover, the viral membrane‐anchored papain‐like protease/PLproTM polyprotein produced by MERS‐CoV and SARS‐CoV induces the formation of autophagosomes, but inhibits their maturation, blocking the autophagosome‐lysosome fusion and autophagic flux. Thus, these data suggest an antiviral role for autophagy. 97 , 98 SARS‐CoV papain‐like protease (PLpro) and SARS‐CoV‐2 PLpro share 82.9% sequence identity. 99 PLpro functions also include the processing of the large polyproteins, pp1a and pp1ab produced by (ORF1a and ORF1ab), processing is essential for the release of 16 non‐structural proteins (nsps1–16) of HCoVs. The replicase complex formation essential for viral genome replication is dependent on nsps. 100 , 101
Shi et al. suggested that SARS‐Coronavirus Open Reading Frame‐8b (ORF8b) triggers intracellular stress pathways 32 . They reported that ORF8b forms intracellular aggregates dependent on a valine at residue 77, those intracellular protein aggregates and misfolded proteins leads to ER stress and induction of the UPR, which contributes to the induction of lysosomal stress, autophagy, and eventual cell death. Furthermore, under ER stress, glucose‐regulated protein 78 (GRP78) is overexpressed, which may result in its translocation to the cell surface. Ibrahim et al. proposed that the Spike protein has the capacity to invade the cell through molecules other than ACE2 102 . The S‐protein of SARS‐Cov‐2 can bind to GRP78 on the cell surface, thus mediating virus entry into the cell. The mechanisms associated with autophagy triggered by coronaviruses is summarized in Figure 3.
FIGURE 3.
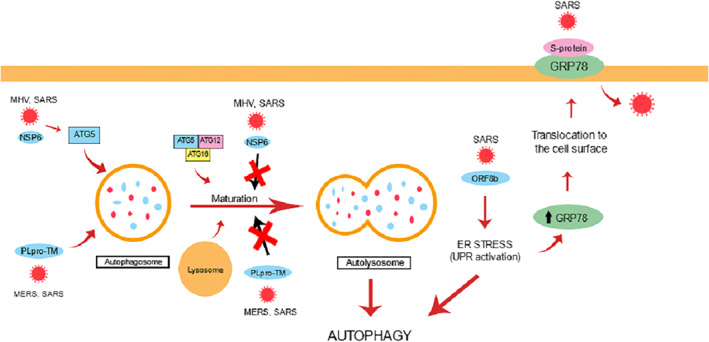
Mechanisms associated with autophagy triggered by coronaviruses
Several potential drug candidates against COVID‐19as chloroquine (CQ) and hydroxychloroquine (HCQ) are autophagy modulators. 97 , 98 , 102 , 103 , 104 , 105 But interestingly those drugs seem to inhibit autophagy flux, similarly to the virus effects. Shojaei et al. suggest in a recent study that the effect against the disease of these drugs is “possibly due to the over‐accumulation of autophagosomes that can potentially induce apoptotic cell death of virally infected cells and disrupt the virus replication cycle” 98 . In addition to the anti‐inflammatory effects by blocking the signaling of endosomal toll‐like receptors, increasing the endolysosomal pH would also make it difficult to fuse the virus membrane with the endosomal membrane, preventing the virus from entering the cell. Unfortunately, clinical studies have shown that there is no improvement when these antimalarial drugs are given to hospitalized patients or in post‐exposure prophylaxis 106 .
The role of autophagy in coronavirus infection remains controversial. More work is needed to clarify its function in SARS‐CoV‐2 cell injury.
4. CORONAVIRUS AND PROGRAMMED NECROTIC CELL DEATH
4.1. Pyroptposis
Pyroptosis is a form of necrotic cell death that has been identified as caspase 1‐dependent necrosis for a long time. Caspase 1 activation induces the maturation and secretion of proinflammatory cytokines, and consequently cell death occurs as result of gasdermin D–regulated (GSDMD‐regulated) membranous pore formation in the plasma membrane. This process eventually leads to cell swelling and osmotic lysis with release of inflammatory intracellular contents. Caspase 1 is activated by several inflammasome, multiprotein complexes that assemble in response to host or infectious agent derived danger signals. 107 , 108 , 109
SARS‐CoV Viroporin 3a activates the Nod‐like receptor family, NOD‐, LRR‐ and pyrin domain‐containing protein 3(NLRP3) inflammasome by its ion channel (IC) activity. Based on data of previous study, it is known that 3a protein of SARS‐CoV acts as a K+ channel and K+ efflux is a well‐known activator of the NLRP3 inflammasome. Chen et al. suggested that SARS‐CoV 3a protein disrupts intracellular ionic concentrations and causes mitochondrial damage, leading to NLRP3 inflammasome activation, which may result in pyroptosis (Figure 4a). 26 SARS 3a also has the ability to activate caspase‐1 directly by its monomers or via K+ efflux by its oligomers, which results in the activation of NLRP3 inflammasome, contributing to pyroptotic cell death. 24
FIGURE 4.
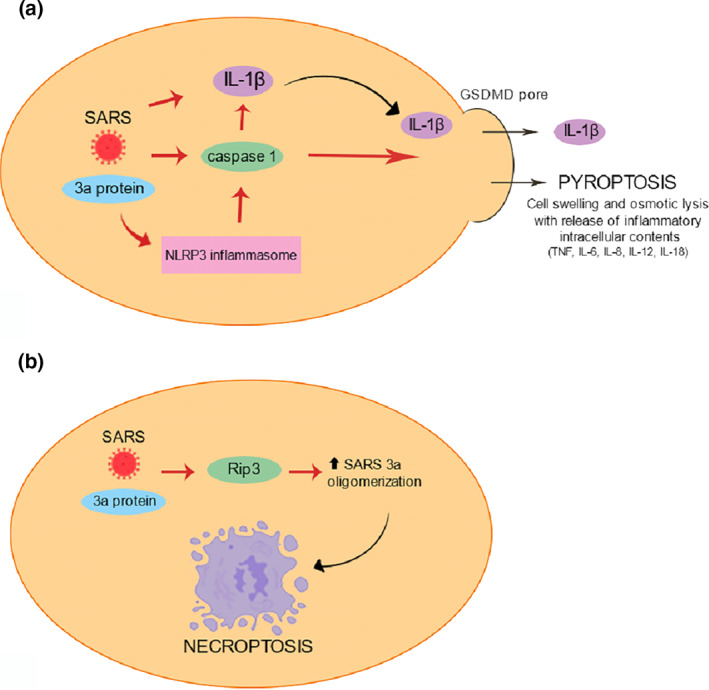
(a)–Pyroptosis triggered by the 3a protein of SARS‐CoV; (b)—3a protein of SARS‐CoV leads to necroptotic cell death by the ability of the SARS 3a protein to oligomerize and insert into membranes
Nieto‐Torres et al. showed that SARS‐CoV envelope (E) gene encodes a small multifunctional protein that possesses IC activity, an important function in virus‐host interaction. 110 The results suggest that E protein IC activity induced edema accumulation and triggers inflammasome resulting in the production of IL‐1β, TNF and IL‐6. Jiang et al. described that MERS‐CoV infection also induced pyroptosis. 111 When infected with MERS‐CoV, human DPP4 transgenic mouse model overexpressed caspase‐1 in the spleen and showed high IL‐1β levels in serum. Inhibition of complement receptor C5aR1 reduced pyroptosis, suggesting that MERS‐CoV infection induces overactivation of complement, which may contribute to pyroptosis and inflammation.
A clinical and pathological investigation of patients with severe COVID‐19 performed by Li et al. showed that levels of TNFα and other factors were dramatically upregulated in blood, as well as severe inflammatory infiltrates in lung tissues of those patients. 25 The release of such molecules indicates that pyroptosis is the most frequent pattern of pulmonary injury. More recently, Rodrigues S et al. suggested that NLRP3 inflammasome is activated in response to SARS‐CoV‐2 infection and it is active during the disease. 112 Besides finding active NLRP3 inflammasome in PBMCs and tissues of post‐mortem patients upon autopsy, the inflammasome‐derived products such as Casp1p20 and IL‐18 in the sera, correlated with the markers of COVID‐19 severity, including IL‐6 and lactate dehydrogenase. Ferreirra et al. also showed that SARS‐CoV‐2 engages inflammasome and pyroptosis in human primary monocytesm. 113 According to their findings, SARS‐CoV‐2 induced pro‐caspase‐1 cleavage, which is known to trigger IL‐1β production and GSDMD cleavage, leading to pyroptosis.
4.2. Necroptosis
Necroptosis is a caspase‐independent form of programmed necrotic cell death, and its pathway is triggered by several stimuli, including death receptor ligands. Using standard histological techniques, morphologically there are no differences between cells that are undergoing necrosis and necroptosis. While apoptosis and other forms of programmed cell death have caspases as key role, necroptosis is regulated by receptor‐interacting protein kinases (RIPK). The cell death mechanism is activated by RIPK1 and requires RIPK3‐dependent phosphorylation of mixed‐lineage kinase domain–like pseudokinase (MLKL), resulting in MLKL oligomerization. It induces plasma membrane rupture and consequently cell death with release of damage‐associated molecular patterns. 114 , 115 , 116
Yue et al. described that SARS‐Coronavirus open Reading Frame‐3a (SARS 3a) drives multimodal necrotic cell death. The study showed that SARS 3a protein interacts with receptor interacting protein 3 (Rip3), which augments the oligomerization of SARS 3a protein. This mechanism probably leads to necroptotic cell death by the ability of the SARS 3a protein to oligomerize and insert into membranes as phosphorylated MLKL (Figure 4b). Furthermore, SARS 3a causes lysosome damage and dysfunction by inserting into lysosomal membranes, which leads to activation of transcription factor EB, increasing the transcription of autophagy‐ and lysosome‐related genes. 24
The studies that focus on the cell death caused by Coronaviruses used in this review is summarized in Table 1.
TABLE 1.
Cell death and CoVs
| Autor | Ano | CoV | CellLine | Tissue | Cell death | Reference |
|---|---|---|---|---|---|---|
| Surjit, Milan et al. | 2004 | SARS‐CoV | COS‐1, HuH7 | Kidney | Apoptosis | 29 |
| Tan, Yee‐Joo et al. | 2004 | SARS‐CoV | HEK293T, HeLa, A549, HepG2, Vero E6, COS‐7 | Kidney, cervix, lung, liver, | Apoptosis | 23 |
| Ren, Lili et al. | 2005 | SARS‐CoV | Vero | Kidney | Apoptosis | 70 |
| Law, Patrick T W et al. | 2005 | SARS‐CoV | Vero E6 | Kidney | Apoptosis | 71 |
| Kopecky‐Bromberg, Sarah A et al. | 2006 | SARS‐CoV | HEK293T, A549, and HeLa | Human embryonic kidney, lungs, cervix | Apoptosis | 60 |
| Khan, Sehaam et al. | 2006 | SARS‐CoV | Vero E6 | Kidney | Apoptosis and neccrosis | 52 |
| Chan, Chak‐Ming et al. | 2007 | SARS‐CoV | HEK293T, TransgenicDrosophila | Kidney | Apoptosis | 24 |
| Chen, Chia‐Yen et al. | 2007 | SARS‐CoV | VeroE6, HEK 293T, and HuH‐7 | Kidney human embryonic kidney | Apoptosis | 52 |
| Zhang, Lu et al. | 2007 | SARS‐CoV | COS‐1 | Kidney | Apoptosis | 75 |
| Tan, Ying‐Xim et al. | 2007 | SARS‐CoV | VeroE6, HEK293T | Kidney | Apoptosis | 62 |
| Chan, E et al. | 2008 | SARS‐CoV | Vero E6 e 3a TransgenicDrosophilamodel | Kidney | Apoptosis | 31 |
| Padhan, Kartika et al. | 2008 | SARS‐CoV | Huh7 | Kidney | Apoptosis | 22 |
| Ye, Zhongde et al. | 2008 | SARS‐CoV | Vero E6, COS‐7 cells and HEK293T cells | Kidney and human embryonic kidney | Apoptosis | 58 |
| Minakshi, Rinki et al. | 2009 | SARS‐CoV | COS‐1, Vero, Huh7 | Kidney | Apoptosis | 28 |
| Freundt, Eric C et al. | 2009 | SARS‐CoV | Vero cells | Kidney | Apoptosis | 73 |
| Ye et al. | 2010 | SARS‐CoV | Vero E6 and COS‐7 cells | Kidney | Apoptosis | 57 |
| Sharma, Kulbhushan et al. | 2011 | SARS‐CoV | Vero cells | Kidney | Apoptosis | 67 |
| Tsoi, Ho et al. | 2014 | SARS‐CoV | HEK293FT | Kidney | Apoptosis | 27 |
| Nieto‐Torres, Jose L et al. | 2014 | SARS‐CoV | Vero E6, BHK‐21 cells | Kidney | Pyroptosis | 110 |
| Chu, Hin et al. | 2016 | MERS‐CoV | Peripheral blood mononuclear cells, Tcells and tonsil and spleen‐dissociated cells | Blood | Apoptosis | 76 |
| Yeung, Man‐lung et al. | 2016 | MERS‐CoV | Calu‐3 and normal human mesangial cells | Lungs, kidney | Apoptosis | 79 |
| Yue, Yuan et al. | 2018 | SARS‐CoV | HeLa, HEK 293T, A549, Thp‐1 | Cervix, kidney, lung, peripheral blood | Necrosis | 24 |
| Chen, I‐Yin et al. | 2019 | SARS‐CoV | BMMs, HEK293FT, HeLa, HT‐1080 | Kidney, Cervix, connective tissue | Pyroptosis | 26 |
| Shi, Chong‐Shan et al. | 2019 | SARS‐ CoV | THP‐1, HEK293, A549, HeLa | Peripheral blood, kidney, lung, cervix | Autophagy | 32 |
| Jiang, Yuting et al. | 2019 | MERS‐CoV | Human monocytic cells (THP‐1) | Blood | Pyroptosis | 111 |
| Carmona‐Gutierrez, Didac et al. | 2020 | MERS‐CoV,SARS‐CoV | ‐ | ‐ | Autophagy | 54 |
| Li, Shaohua et al. | 2020 | SARS‐CoV‐2 | ‐ | ‐ | Pyroptosis | 25 |
| Li, Shufen et al. | 2020 | SARS‐CoV‐2 | Calu‐3, Vero, Vero E6, THP‐1 | Lung | ApoptosisNecroptosis | 91 |
| Shojaei, Shahla et al. | 2020 | SARS‐CoV‐2 | ‐ | ‐ | Autophagy | 53 |
| Bellesi, Silvia et al. | 2020 | SARS‐CoV‐2 | Peripheral blood T Lympocytes | Blood | Apoptosis | 77 |
| Ren, Yujie et al. | 2020 | SARS‐CoV‐2 | VeroE6, HEK293T and HepG2 | Kidney, lung | Apoptosis | 72 |
| Rodrigues, Tamara S et al. | 2021 | SARS‐CoV‐2 | Peripheral blood mononuclear cells | Blood | Pyroptosis | 112 |
| Ferreira, André C et al. | 2021 | SARS‐CoV‐2 | Vero E6, human primary monocytes | Kidney, blood | Pyroptosis | 113 |
Abbreviations: CoVs, coronaviruses; MERS‐CoV, Middle East respiratory syndrome coronavirus; SARS‐CoV, severe acute respiratory syndrome coronavirus.
5. IN SILICO ANALYSIS TO IDENTIFY CELL DEATH PATHWAYS INVOLVED IN SARS‐CoV INJURY
To identify the key cell death pathways related with SARS‐CoV infection, we used text mining, using pubmed. mineR 117 , 118 from PubMed abstracts. We performed the search using the same terms cited above and found 105 articles. Gene ontology term enrichment analysis of 19 genes and biological process were performed, and results were plotted in Figure 5.
FIGURE 5.
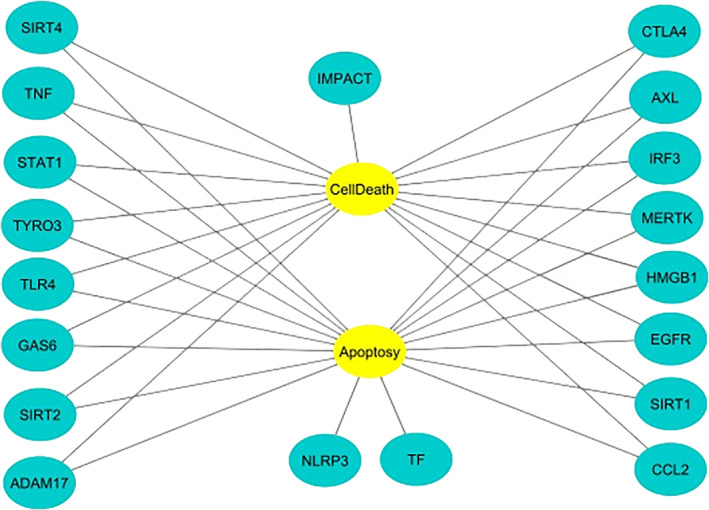
Biomolecular interaction network with genes found by text‐mining analysis
Some of these genes such as SIRT4, TYRO3, GAS6, AXL, MERTK, EGFR and CCL2 are negative regulators of apoptosis, while the TLR4, SIRT2, CTLA4 and HMGB1 genes are positive regulators. In contrast, the TNF and SIRT1 genes are concomitantly positive and negative regulators of apoptosis. In addition, the TNF, TLR4 and IRF3 genes appear to be involved in the regulation of necrosis and apoptosis. The specific functions of each gene in the figure above are described in Table S2 of the Supplementary Material.
5.1. Enrichment analysis for SARS‐Human proteins interactions
To better understand what interactions the SARS proteins could potentially have with the human host and its death‐processes associated proteins, we performed another in silico analysis. We used interaction data from the p‐HIPSTer tool, which is an algorithm for predicting interactions between viral and human proteins based on similar interactions of reference viral proteins and the human protein. Viral protein data from p‐HIPSTer corresponds to SARS‐CoV, not yet to SARS‐CoV 2, and because of this our results should have preliminary and exploratory purposes only.
For each viral protein a list of targets was predicted by P‐HIPSTer as possible interactors. With these list of targets, an enrichment analysis for Gene Ontology Biological Processes was performed using g:Profiler and a heat map was made to show the negative log10 of adjusted p‐value for enrichment of death‐related biological process in these interactors lists.
According to p‐HIPSTER results, the ORF7b had only one interaction (PCNA protein) and no interactors resulted from M protein prediction on p‐HIPSTer. For both of these two, no enrichment analysis could be performed on g:Profiler due to not enough predicted interacting proteins.
As presented in Figure 6, many SARS‐CoV proteins (N, NSP1, NSP10, NSP4, NSP5, NSP7, ORF7a and ORF9b) show interactions with human proteins strongly associated with death biological processes. Some others (NSP12, NSP13, NSP9, NSP8 and Spike) were just slightly associated to death biological processes. However, many other SARS proteins are not associated with any death biological processes (E, NSP14, NSP15 and NSP16). Although NSP14 shows no death‐related biological processes enriched among its predicted interactors, it was due to only few human targets predicted as interactors (ISG15 and UBC).
FIGURE 6.
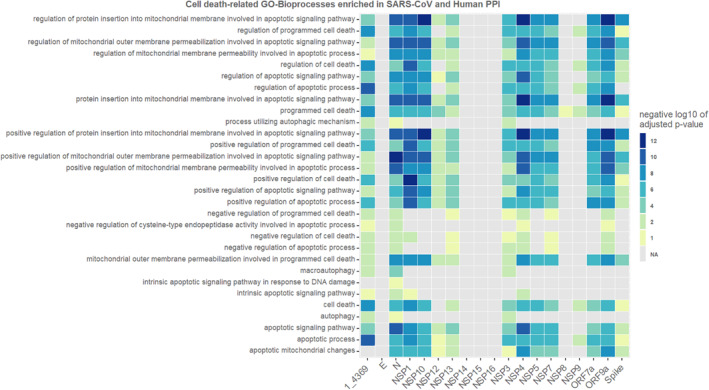
Heat map enrichment of death‐related biological process and genes of Sars‐CoV (negative log10 of adjusted p‐value)
All SARS proteins which had their targets enriched with apoptotic functions were more associated with its positive regulation than negative. Although not closely related to it, the nucleocapsid protein and NSP3 had a mild enrichment with human protein interactors functionally associated with autophagy biological processes. Also, according to our analysis necrosis‐related biological processes were enriched for none of the viral protein interactors lists.
Many of the proteins identified in the in silico analyses are not the same as those addressed in the first section of the study, indicating that this topic still needs to be explored further.
6. CONCLUSIONS
Coronaviruses can induce different forms of cell death, triggered by several signaling pathways. Understanding the molecular mechanisms of cell death that cause tissue and cell injuries by SARS‐CoV‐2 may contribute to finding potential therapeutic targets against the current pandemic. Functional studies with recombinant viruses with specific deletions or mutations, or with transfection vectors containing SARS proteins will be important to determine the relative contribution of these proteins to the cell death mechanism of SARS‐CoV‐2 injuries.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
Maríllya Morais da Silva, Vanessa Mylenna Florêncio De Carvalho, Priscilla Stela Santana de Oliveira ‐writing–original draft and editing. André Silva Lira de Lucena, Sergio de Sá Leitão Paiva Júnior ‐ in silico analysis and writing; Michelle Melgarejo da Rosa, Moacyr Jesus Barreto de Melo Rego, Maira Galdino da Rocha Pitta ‐ reviewing, and editing; Michelly Cristiny Pereira ‐ conceptualization, preparation, writing, reviewing, and editing the final manuscript.
Supporting information
Supplementary Material
ACKNOWLEDGEMENT
No funding was obtained for this study. Michelly C Pereira, Moacyr J. Melo Rego, Maira G. Pitta reports grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico, Fundação de Amparo a Ciência e Tecnologia de PE, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior outside the submitted work.
Morais da Silva M, Lira de Lucena AS, Paiva Júnior SdSL, et al. Cell death mechanisms involved in cell injury caused by SARS‐CoV‐2. Rev Med Virol. 2022; 32(3):e2292. 10.1002/rmv.2292
DATA AVAILABILITY STATEMENT
All data used are available in this review.
References
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol. 2020;92(4):401‐402. doi: 10.1002/jmv.25678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodríguez‐Morales AJ, MacGregor K, Kanagarajah S, Patel D, Schlagenhauf P. Going global ‐ travel and the 2019 novel coronavirus. Trav Med Infect Dis. 2020;33:101578. doi: 10.1016/j.tmaid.2020.101578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rolling updates on coronavirus disease (COVID‐19) June, 2020. https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019/events‐as‐theyhappen
- 5. Weekly operational update on COVID‐19 . World Health Organization (WHO). https://www.who.int/publications/m/item/weekly‐operational‐update‐on‐covid‐19‐‐‐16‐august‐2021
- 6. Chan JF, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human‐pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan [published correction appears in Emerg Microbes Infect. 2020 Dec;9(1):540]. Emerg Microb Infect. Published 2020. Jan 28;9(1):221‐236. doi: 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019‐nCoV) originating in China. Cell Host Microbe. 2020;27(3):325‐328. doi: 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ren LL, Wang YM, Wu ZQ, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020;133:1015‐1024. doi: 10.1097/CM9.0000000000000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang H, Li X, Li T, et al. The genetic sequence, origin, and diagnosis of SARS‐CoV‐2. Eur J Clin Microbiol Infect Dis. 2020;39(9):1629‐1635. doi: 10.1007/s10096-020-03899-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hasöksüz M, Kiliç S, Saraç F. Coronaviruses and SARS‐COV‐2. Turk J Med Sci. 2020;50(SI‐1):549‐556. doi: 10.3906/sag-2004-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID‐19: their roles in pathogenesis. J Microbiol Immunol Infect. 2021;54(2):159‐163. doi: 10.1016/j.jmii.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. doi: 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China [published correction appears in Lancet 2020 Jan 30]. Lancet. 2020;395(10223):497‐506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhai P, Ding Y, Wu X, Long J, Zhong Y, Li Y. The epidemiology, diagnosis and treatment of COVID‐19. Int J Antimicrob Agents. 2020;55(5):105955. doi: 10.1016/j.ijantimicag.2020.105955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheng Y, Luo R, Wang K, et al. Kidney disease is associated with in‐hospital death of patients with COVID‐19. Kidney Int. 2020;97(5):829‐838. doi: 10.1016/j.kint.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Du Y, Tu L, Zhu P, et al. Clinical features of 85 fatal cases of COVID‐19 from Wuhan. A retrospective observational study. Am J Respir Crit Care Med. 2020;201(11):1372‐1379. doi: 10.1164/rccm.202003-0543OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guan WJ, Liang WH, Zhao Y, et al. Comorbidity and its impact on 1590 patients with COVID‐19 in China: a nationwide analysis. Eur Respir J. 2020;55(5):2000547. doi: 10.1183/13993003.00547-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. J Am Med Assoc. 2020;323(11):1061‐1069. doi: 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang B, Li R, Lu Z, Huang Y. Does comorbidity increase the risk of patients with COVID‐19: evidence from meta‐analysis. Aging (Albany NY). 2020;12(7):6049‐6057. doi: 10.18632/aging.103000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rodriguez‐Morales AJ, Cardona‐Ospina JA, Gutiérrez‐Ocampo E, et al. Clinical, laboratory and imaging features of COVID‐19: a systematic review and meta‐analysis. Trav Med Infect Dis. 2020;34:101623. doi: 10.1016/j.tmaid.2020.101623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study [published correction appears in Lancet 2020 Mar 28;395(10229):1038] [published correction appears in Lancet. 2020 Mar 28;395(10229):1038]. Lancet. 2020;395(10229):1054‐1062. doi: 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Padhan K, Minakshi R, Towheed MAB, Jameel S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J Gen Virol. 2008;89(Pt 8):1960‐1969. doi: 10.1099/vir.0.83665-0 [DOI] [PubMed] [Google Scholar]
- 23. Tan YJ, Fielding BC, Goh PY, et al. Over expression of 7a, a protein specifically encoded by these severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase‐dependent pathway. J Virol. 2004;78(24):14043‐14047. doi: 10.1128/JVI.78.24.14043-14047.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yue Y, Nabar NR, Shi CS, et al. SARS‐Coronavirus Open Reading Frame‐3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9(9):904. doi: 10.1038/s41419-018-0917-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li S, Jiang L, Li X, et al. Clinical and pathological investigation of patients with severe COVID‐19. JCI Insight. 2020;5(12):e138070. doi: 10.1172/jci.insight.138070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe acute respiratory syndrome coronavirus Viroporin 3a activates the NLRP3 inflammasome. Front Microbiol. 2019;10(50). doi: 10.3389/fmicb.2019.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsoi H, Li L, Chen ZS, Lau KF, Tsui SK, Chan HY. The SARS‐coronavirus membrane protein induces apoptosis via interfering with PDK1‐PKB/Akt signalling. Biochem J. 2014;464(3):439‐447. doi: 10.1042/BJ20131461 [DOI] [PubMed] [Google Scholar]
- 28. Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand‐independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4(12):e8342. doi: 10.1371/journal.pone.0008342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Surjit M, Liu B, Jameel S, Chow VT, Lal SK. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS‐1 cells in the absence of growth factors. Biochem J. 2004;383(Pt 1):13‐18. doi: 10.1042/BJ20040984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chan CM, Ma CW, Chan WY, Chan HY. The SARS‐Coronavirus Membrane protein induces apoptosis through modulating the Akt survival pathway. Arch Biochem Biophys. 2007;459(2):197‐207. doi: 10.1016/j.abb.2007.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan E, Tsui S, Chan CM, et al. Molecular and genetic characterisation of the SARS coronavirus auxiliary protein X1 in Drosophila. Hong Kong Med J. 2008;14(Suppl 4):14‐16. [PubMed] [Google Scholar]
- 32. Shi CS, Nabar NR, Huang NN, Kehrl JH. SARS‐Coronavirus Open Reading Frame‐8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:101. doi: 10.1038/s41420-019-0181-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Norbury CJ, Hickson ID. Cellular responses to DNA damage. Annu Rev PharmacolToxicol. 2001;41:367‐401. doi: 10.1146/annurev.pharmtox.41.1.367 [DOI] [PubMed] [Google Scholar]
- 34. Van Cruchten S, Van Den Broeck W. Morphological and biochemical aspects of apoptosis, oncosis and necrosis. Anat Histol Embryol. 2002;31(4):214‐223. doi: 10.1046/j.1439-0264.2002.00398.x [DOI] [PubMed] [Google Scholar]
- 35. Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495‐516. doi: 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. D'Arcy MS. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int. 2019;43(6):582‐592. doi: 10.1002/cbin.11137 [DOI] [PubMed] [Google Scholar]
- 37. Hu Y, Wen J, Tang L, et al. The M protein of SARS‐CoV: basic structural and immunological properties. Dev Reprod Biol. 2003;1(2):118‐130. doi: 10.1016/s1672-0229(03)01016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siu YL, Teoh KT, Lo J, et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus‐like particles. J Virol. 2008;82(22):11318‐11330. doi: 10.1128/JVI.01052-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Horvitz HR. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Canc Res. 1999;59(7 Suppl):1701s‐1706s. [PubMed] [Google Scholar]
- 40. Wajant H. The Fas signaling pathway: more than a paradigm. Science. 2002;296(5573):1635‐1636. doi: 10.1126/science.1071553 [DOI] [PubMed] [Google Scholar]
- 41. Hill MM, Adrain C, Duriez PJ, Creagh EM, Martin SJ. Analysis of the composition, assembly kinetics and activity of native Apaf‐1 apoptosomes. EMBO J. 2004;23(10):2134‐2145. doi: 10.1038/sj.emboj.7600210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Slee EA, Adrain C, Martin SJ. Executioner caspase‐3, ‐6, and ‐7 perform distinct, non‐redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276(10):7320‐7326. doi: 10.1074/jbc.M008363200 [DOI] [PubMed] [Google Scholar]
- 43. Parlikar A, Kalia K, Sinha S, et al. Understanding genomic diversity, pan‐genome, and evolution of SARS‐CoV‐2. PeerJ. 2020;8:e9576. doi: 10.7717/peerj.9576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Narayanan K, Huang C, Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133(1):113‐121. doi: 10.1016/j.virusres.2007.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schaecher SR, Pekosz A. SARS coronavirus accessory gene expression and function. Mol Biol SARS‐Coronavirus. 2009;153:153‐166. doi: 10.1007/978-3-642-03683-5_10 [DOI] [Google Scholar]
- 46. Yuan X, Li J, Shan Y, et al. Subcellular localization and membrane association of SARS‐CoV 3a protein. Virus Res. 2005;109(2):191‐202. doi: 10.1016/j.virusres.2005.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu W, Zheng BJ, Xu K, et al. Severe acute respiratory syndrome‐associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc Natl Acad Sci U. S. A. 2006;103(33):12540‐12545. doi: 10.1073/pnas.0605402103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sharma K, Surjit M, Satija N, Liu B, Chow VT, Lal SK. The 3a accessory protein of SARS coronavirus specifically interacts with the 5'UTR of its genomic RNA, Using a unique 75 amino acid interaction domain. Biochemistry. 2007;46(22):6488‐6499. doi: 10.1021/bi062057p [DOI] [PubMed] [Google Scholar]
- 49. Yuan X, Yao Z, Shan Y, et al. Nucleolar localization of non‐structural protein 3b, a protein specifically encoded by the severe acute respiratory syndrome coronavirus. Virus Res. 2005;114(1‐2):70‐79. doi: 10.1016/j.virusres.2005.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yuan X, Shan Y, Yao Z, et al. Mitochondrial location of severe acute respiratory syndrome coronavirus 3b protein. Mol Cell. 2006;21(2):186‐191. [PubMed] [Google Scholar]
- 51. Yuan X, Shan Y, Zhao Z, Chen J, Cong Y. G0/G1 arrest and apoptosis induced by SARS‐CoV 3b protein in transfected cells. Virol J. 2005;2:66. doi: 10.1186/1743-422X-2-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khan S, Fielding BC, Tan TH, et al. Over‐expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 2006;122(1‐2):20‐27. doi: 10.1016/j.virusres.2006.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Geng H, Liu YM, Chan WS, et al. The putative protein 6 of the severe acute respiratory syndrome‐associated coronavirus: expression and functional characterization. FEBS Lett. 2005;579(30):6763‐6768. doi: 10.1016/j.febslet.2005.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang C, Peters CJ, Makino S. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion‐associated protein and is released from 6 protein‐expressing cells. J Virol. 2007;81(10):5423‐5426. doi: 10.1128/JVI.02307-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kumar P, Gunalan V, Liu B, et al. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007;366(2):293‐303. doi: 10.1016/j.virol.2007.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kopecky‐Bromberg SA, Martínez‐Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81(2):548‐557. doi: 10.1128/JVI.01782-06. Epub 2006 Nov 15. PMID: 17108024; PMCID: PMC1797484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ye ZD, Wong CK, Li P, Xie Y. The role of SARS‐CoV protein, ORF‐6, in the induction of host cell death. Hong Kong Med J. 2010;16(5 Suppl 4):22‐26. [PubMed] [Google Scholar]
- 58. Ye Z, Wong CK, Li P, Xie Y. A SARS‐CoV protein, ORF‐6, induces caspase‐3 mediated, ER stress and JNK‐dependent apoptosis. Biochim Biophys Acta. 2008;1780(12):1383‐1387. doi: 10.1016/j.bbagen.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pekosz A, Schaecher SR, Diamond MS, Fremont DH, Sims AC, Baric RS. Structure, expression, and intracellular localization of the SARS‐CoV accessory proteins 7a and 7b. Adv Exp Med Biol. 2006;581:115‐120. doi: 10.1007/978-0-387-33012-9_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kopecky‐Bromberg SA, Martinez‐Sobrido L, Palese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen‐activated protein kinase. J Virol. 2006;80(2):785‐793. doi: 10.1128/JVI.80.2.785-793.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schaecher SR, Mackenzie JM, Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS‐CoV) is expressed in virus‐infected cells and incorporated into SARS‐CoV particles. J Virol. 2007;81(2):718‐731. doi: 10.1128/JVI.01691-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tan YX, Tan TH, Lee MJ, et al. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl‐XL protein. J Virol. 2007;81(12):6346‐6355. doi: 10.1128/JVI.00090-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schaecher SR, Touchette E, Schriewer J, Buller RM, Pekosz A. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus‐induced apoptosis. J Virol. 2007;81(20):11054‐11068. doi: 10.1128/JVI.01266-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen CY, Ping YH, Lee HC, et al. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J Infect Dis. 2007;196(3):405‐415. doi: 10.1086/519166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Keng CT, Choi YW, Welkers MR, et al. The human severe acute respiratory syndrome coronavirus (SARS‐CoV) 8b protein is distinct from its counterpart in animal SARS‐CoV and down‐regulates the expression of the envelope protein in infected cells. Virology. 2006;354(1):132‐142. doi: 10.1016/j.virol.2006.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Von Brunn A, Teepe C, Simpson JC, et al. Analysis of intraviral protein‐protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2(5):e459. doi: 10.1371/journal.pone.0000459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sharma K, Åkerström S, Sharma AK, et al. SARS‐CoV 9b protein diffuses into nucleus, undergoes active Crm1 mediated nucleocytoplasmic export and triggers apoptosis when retained in the nucleus. PLoS One. 2011;6(5):e19436. doi: 10.1371/journal.pone.0019436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459‐468. doi: 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dominguez Andres A, Feng Y, Campos AR, et al. SARS‐CoV‐2 ORF9c is a membrane‐associated protein that suppresses antiviral responses in cells. Preprint. bioRxiv. Published Aug 2020;2020.08.18.256776. doi: 10.1101/2020.08.18.256776 Published [DOI] [Google Scholar]
- 70. Ren L, Yang R, Guo L, Qu J, Wang J, Hung T. Apoptosis induced by the SARS‐associated coronavirus in Vero cells is replication‐dependent and involves caspase. DNA Cell Biol. 2005;24(8):496‐502. doi: 10.1089/dna.2005.24.496 [DOI] [PubMed] [Google Scholar]
- 71. Law PTW, Wong CH, Au TCC, et al. The 3a protein of severe acute respiratory syndrome‐associated coronavirus induces apoptosis in Vero E6 cells. J Gen Virol. 2005;86(Pt 7):1921‐1930. doi: 10.1099/vir.0.80813-0 [DOI] [PubMed] [Google Scholar]
- 72. Ren Y, Shu T, Wu D, et al. The ORF3a protein of SARS‐CoV‐2 induces apoptosis in cells. Cell Mol Immunol. 2020;17(8):881‐883. doi: 10.1038/s41423-020-0485-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Freundt E, Yu L, Goldsmith C, et al. The open reading frame 3a protein of severe acute respiratory syndrome‐associated coronavirus promotes membrane rearrangement and cell death. J Virol. 2009;84(2):1097‐1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Surjit M, Lal SK. The SARS‐CoV nucleocapsid protein: a protein with multifarious activities. Infect Genet Evol. 2008;8(4):397‐405. doi: 10.1016/j.meegid.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang L, Wei L, Jiang D, Wang J, Cong X, Fei R. SARS‐CoV nucleocapsid protein induced apoptosis of COS‐1 mediated by the mitochondrial pathway. Artif Cells Blood Substit Immobil Biotechnol. 2007;35(2):237‐253. doi: 10.1080/10731190601188422 [DOI] [PubMed] [Google Scholar]
- 76. Chu H, Zhou J, Wong BH, et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis. 2016;213(6):904‐914. doi: 10.1093/infdis/jiv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bellesi S, Metafuni E, Hohaus S, et al. Increased CD95 (Fas) and PD‐1 expression in peripheral blood T lymphocytes in COVID‐19 patients. Br J Haematol. 2020;191(2):207‐211. doi: 10.1111/bjh.17034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Krammer PH. CD95's deadly mission in the immune system. Nature. 2000;407(6805):789‐795. doi: 10.1038/35037728 [DOI] [PubMed] [Google Scholar]
- 79. Yeung ML, Yao Y, Jia L, et al. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat Microbiol. 2016;1(3):16004. doi: 10.1038/nmicrobiol.2016.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Schiffer M, Bitzer M, Roberts IS, et al. Apoptosis in podocytes induced by TGF‐beta and Smad7. J Clin Invest. 2001;108(6):807‐816. doi: 10.1172/JCI12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lan HY, Mu W, Tomita N, et al. Inhibition of renal fibrosis by gene transfer of inducible Smad7 using ultrasound‐microbubble system in rat UUO model. J Am Soc Nephrol. 2003;14(6):1535‐1548. doi: 10.1097/01.asn.0000067632.04658.b8 [DOI] [PubMed] [Google Scholar]
- 82. Floege J, Kriz W, Schulze M, et al. Basic fibroblast growth factor augments podocyte injury and induces glomerulosclerosis in rats with experimental membranous nephropathy. J Clin Invest. 1995;96(6):2809‐2819. doi: 10.1172/JCI118351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Favreau DJ, Meessen‐Pinard M, Desforges M, Talbot PJ. Human coronavirus‐induced neuronal programmed cell death is cyclophilin d dependent and potentially caspase dispensable. J Virol. 2012;86(1):81‐93. doi: 10.1128/JVI.06062-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Baines CP, Kaiser RA, Purcell NH, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434(7033):658‐662. doi: 10.1038/nature03434 [DOI] [PubMed] [Google Scholar]
- 85. Kumarswamy R, Chandna S. Putative partners in Bax mediated cytochrome‐c release: ANT, CypD, VDAC or none of them? Mitochondrion. 2009;9(1):1‐8. doi: 10.1016/j.mito.2008.10.003 [DOI] [PubMed] [Google Scholar]
- 86. Susin SA, Lorenzo HK, Zamzami N, et al. Molecular characterization of mitochondrial apoptosis‐inducing factor. Nature. 1999;397(6718):441‐446. doi: 10.1038/17135 [DOI] [PubMed] [Google Scholar]
- 87. Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self‐eating. Cell Death Differ. 2005;12(Suppl 2):1542‐1552. doi: 10.1038/sj.cdd.4401765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxidants Redox Signal. 2014;20(3):460‐473. doi: 10.1089/ars.2013.5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Saha S, Panigrahi DP, Patil S, Bhutia SK. Autophagy in health and disease: a comprehensive review. Biomed Pharmacother. 2018;104:485‐495. doi: 10.1016/j.biopha.2018.05.007 [DOI] [PubMed] [Google Scholar]
- 90. Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1509‐1518. doi: 10.1038/sj.cdd.4401751 [DOI] [PubMed] [Google Scholar]
- 91. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67‐93. doi: 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 2005;1(1):1‐10. doi: 10.4161/auto.1.1.1270 [DOI] [PubMed] [Google Scholar]
- 93. Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279(11):10136‐10141. doi: 10.1074/jbc.M306124200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10(8):1426‐1441. doi: 10.4161/auto.29309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ye X, Zhou XJ, Zhang H. Exploring the role of autophagy‐related gene 5 (ATG5) yields important insights into autophagy in autoimmune/autoinflammatory diseases. Front Immunol. 2018;9:2334. doi: 10.3389/fimmu.2018.02334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Codogno P, Mehrpour M, Proikas‐Cezanne T. Canonical and non‐canonical autophagy: variations on a common theme of self‐eating? Nat Rev Mol Cell Biol. 2011;13(1):7‐12. doi: 10.1038/nrm3249 [DOI] [PubMed] [Google Scholar]
- 97. Shojaei S, Suresh M, Klionsky DJ, Labouta HI, Ghavami S. Autophagy and SARS‐CoV‐2 infection: apossible smart targeting of the autophagy pathway. Virulence. 2020;11(1):805‐810. doi: 10.1080/21505594.2020.1780088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Carmona‐Gutierrez D, Bauer MA, Zimmermann A, et al. Digesting the crisis: autophagy and coronaviruses. Microb Cell. 2020;7(5):119‐128. doi: 10.15698/mic2020.05.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ibrahim TM, Ismail MI, Bauer MR, Bekhit AA, Boeckler FM. Supporting SARS‐CoV‐2 papain‐like protease drug discovery: in silico methods and benchmarking. Front Chem. 2020:8:592289. doi: 10.3389/fchem.2020.592289. PMID: 33251185; PMCID: PMC7674952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. The papain‐like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005;79(24):15189‐15198. doi: 10.1128/JVI.79.24.15189-15198.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1‐23. doi: 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA. COVID‐19 spike‐host cell receptor GRP78 binding site prediction. J Infect. 2020;80(5):554‐562. doi: 10.1016/j.jinf.2020.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bonam SR, Muller S, Bayry J, Klionsky DJ. Autophagy as an emerging target for COVID‐19: lessons from an old friend, chloroquine. Autophagy. 2020;16(12):2260‐2266. doi: 10.1080/15548627.2020.1779467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Gorshkov K, Chen CZ, Bostwick R, et al. The SARS‐CoV‐2 cytopathic effect is blocked with autophagy modulators. Preprint. bioRxiv. 2020:2020.05.16.091520. doi: 10.1101/2020.05.16.091520 [DOI] [Google Scholar]
- 105. Bello‐Perez M, Sola I, Novoa B, Klionsky DJ, Falco A. Canonical and noncanonical autophagy as potential targets for COVID‐19. Cells. 2020;9(7):1619. doi: 10.3390/cells9071619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bordallo B, Bellas M, Cortez AF, Vieira M, Pinheiro M. Severe COVID‐19: what have we learned with the immunopathogenesis? Adv Rheumatol. 2020;60(1):50. doi: 10.1186/s42358-020-00151-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7(2):99‐109. doi: 10.1038/nrmicro2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jorgensen I, Miao EA. Pyroptotic cell death defends against intracellular pathogens. Immunol Rev. 2015;265(1):130‐142. doi: 10.1111/imr.12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shi J, Gao W, Shao F. Pyroptosis: gasdermin‐mediated programmed necrotic cell death. Trends Biochem Sci. 2017;42(4):245‐254. doi: 10.1016/j.tibs.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 110. Nieto‐Torres JL, DeDiego ML, Verdiá‐Báguena C, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10(5):e1004077. doi: 10.1371/journal.ppat.1004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Jiang Y, Li J, Teng Y, et al. Complement receptor C5aR1 inhibition reduces pyroptosis in hDPP4‐transgenic mice infected with MERS‐CoV. Viruses. 2019;11(1):39. doi: 10.3390/v11010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Rodrigues TS, de Sá KSG, Ishimoto AY, et al. Inflammasomes are activated in response to SARS‐CoV‐2 infection and are associated with COVID‐19severity in patients. J Exp Med. 2021;218(3):e20201707. doi: 10.1084/jem.20201707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ferreira AC, Soares VC, de Azevedo‐Quintanilha IG, et al. SARS‐CoV‐2 engages inflammasome and pyroptosis in human primary monocytes. Cell Death Discov. 2021;7(1):43. doi: 10.1038/s41420-021-00428-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Linkermann A, Green DR. Necroptosis. N Engl J Med. 2014;370(5):455‐465. doi: 10.1056/NEJMra1310050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Choi ME, Price DR, Ryter SW, Choi AMK. Necroptosis: a crucial pathogenic mediator of human disease. JCI Insight. 2019;4(15):e128834. doi: 10.1172/jci.insight.128834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Khoury MK, Gupta K, Franco SR, Liu B. Necroptosis in the pathophysiology of disease. Am J Pathol. 2020;190(2):272‐285. doi: 10.1016/j.ajpath.2019.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Rani J, Shah AB, Ramachandran S. Pubmed.mineR: an R package with text‐mining algorithms to analyse PubMed abstracts. J Biosci. 2015;40(4):671‐682. doi: 10.1007/s12038-015-9552-2 [DOI] [PubMed] [Google Scholar]
- 118. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498‐2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
All data used are available in this review.


