The most severe forms of coronavirus disease 2019 (COVID‐19) are often associated with the presence of syncytia in the lungs resulting from cell–cell fusion mediated by the SARS‐CoV‐2 spike protein. In this issue, Rajah and colleagues show that the SARS‐CoV‐2 alpha, beta, and delta variants promote enhanced syncytia formation as compared to the original strain.
Subject Categories: Microbiology, Virology & Host Pathogen Interaction
New work finds the syncytia formation in lungs that is associated with severe forms of coronavirus disease to be enhance upon infection with recently emerged alpha, beta and delta viral variants.
Since severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) was first reported in late 2019 in Wuhan, China, the original strain has been supplanted by numerous variants derived from various mutations. Several mutations have typically occurred in the spike (S) protein that decorates the viral particles. The S protein consists of two subunits, S1 and S2, and is proteolytically activated by host proteases in a succession of cleavages involving furin in infected producer cells and TMPRSS2 or cathepsins in target cells (Koch et al, 2021). The S1 subunit comprises the receptor‐binding domain (RBD) and is responsible for SARS‐CoV‐2 attachment to host cells, notably through interactions with the receptor angiotensin‐converting enzyme 2 (ACE2). The S2 subunit contains the fusion peptide that ensures viral penetration by membrane fusion, a prerequisite for the release of the viral genome into the cytosol and initiation of viral replication.
The severity of COVID‐19 correlates closely with lung damage, and syncytia are often observed in the lungs of patients who have developed fatal pneumonia (Braga et al, 2021). Although syncytia have been reported to form in cell monolayers in vitro upon SARS‐CoV‐2 infection, their contribution to lung injury and disease development in patients infected with any emerging SARS‐CoV‐2 variants remains largely uncharacterized (Buchrieser et al, 2020). Rajah et al (2021a) examined the syncytia‐forming potential of the SARS‐CoV‐2 alpha, beta, and delta strains. The authors also assessed the contribution of the individual spike‐associated mutations to the fusogenicity of the alpha and beta variants, their binding to ACE2, and their evasion from the humoral immune response.
Rajah and colleagues first followed productive infection by the alpha and beta variants in cell lines and primary human airway epithelial cells and found that replication did not differ from that of a D614G strain, one of the earliest variants isolated with increased transmission. The authors then measured syncytia formation upon infection using split green fluorescent protein (GFP) technology combined with image‐based analysis (Foglieni et al, 2017). The method involves splitting GFP into two nonfluorescent fragments, each expressed in a distinct cell population; in this system, GFP does not fluoresce until infected cells fuse and the two fragments assemble. Syncytia sizes were normalized against the number of nuclei to allow a quantitative characterization of the differences in fusogenicity. With this elegant approach, the authors demonstrated that the alpha and beta variants produce increasingly larger syncytia in infected cells than both the original Wuhan strain and the D614G variant.
These results suggested a higher fusogenicity and syncytia‐forming potential of the spike proteins of emerging variants. To pursue this possibility, the authors overexpressed the spike protein from plasmids in their split GFP system. In this model, the spikes of the alpha and delta variants were the fastest to produce syncytia, followed by that of the beta strain. Based on these experiments, spike mutations appear to confer an advantage to SARS‐CoV‐2 not only for cell‐free infection and spread in cell monolayers but also for syncytia formation. During natural infection, syncytia may contribute to virus persistence, viral spread, and immune evasion (Rajah et al, 2021b).
Next, Rajah and colleagues evaluated the contribution of all individual mutations associated with the spike protein to the ability of alpha and beta variants to bind to cellular receptors, escape neutralizing antibodies (nAbs), and induce cell–cell fusion. To this end, the authors genetically engineered and then overexpressed the spike proteins of the D614G strain with each mutation of alpha and beta variants on the cell surface. The soluble ACE2 ectodomain showed the highest affinity for spike proteins with an RBD bearing the N501Y mutation. Of the two variants carrying N501Y, only ACE2 binding to the alpha spike was significantly enhanced, whereas that to the beta spike was increased to a much lesser extent. In addition, although it had no mutations at position 501, the delta variant showed increased binding to ACE2.
Similar results were obtained when the spike mutants were assessed for either antibody escape or cell–cell fusion. For instance, the K417N mutation prevented detection of the beta variant spike by mAb48, but not by mAb98, while both antibodies targeted the RBD, and more mutations had a negative rather than a positive effect on syncytia formation. No clear correlation could be established between syncytia formation and the impact of spike mutations on receptor binding or antibody escape. Collectively, these results provide important evidence that combinations of mutations, not single mutations, underlie enhanced receptor binding, nAb evasion, and fusogenicity of the emerging SARS‐CoV‐2 variants. This phenomenon presumably occurs through conformational structural changes in the spike, the details of which remain to be explored. Although the authors did not test nAbs for their ability to prevent syncytia formation, their work suggests an evolutionary trade‐off of the spike protein between receptor binding, immune escape, and fusogenicity.
This study provides significant insights into the fusogenic potential of the alpha, beta, and delta variants of SARS‐CoV‐2, and as is often the case in science, has raised new questions: How the variants elicit a higher rate of transmissibility globally remains to be addressed. TMPRSS2 expression was found to abolish the apparent enhanced fusogenicity of the variants compared to the original Wuhan isolate. Beyond the question about the role of TMPRSS2 in producer and target cells in activating variant spikes on the cell surface, future work should address whether syncytia formation upon infection with the emerging SARS‐CoV‐2 variants contributes to disease severity or is a consequence (Fig 1).
Figure 1. Emerging SARS‐CoV‐2 variants induce enhanced syncytia formation.
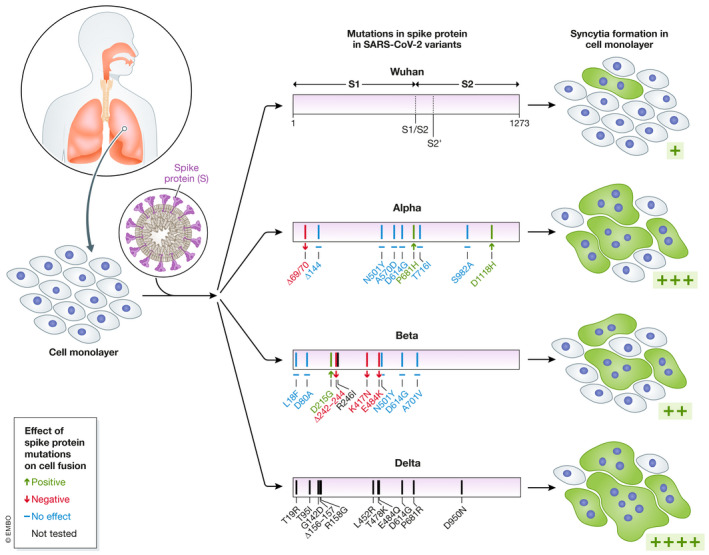
Lung injury correlates closely with COVID‐19 severity. The original SARS‐CoV‐2 strain isolated in Wuhan in 2019 has been supplanted by many variants, most often with numerous mutations in the viral envelope glycoprotein spike. In infected cells, a fraction of the spike protein is expressed on the surface and can induce fusion between neighboring cells, leading to syncytia formation. In this study, Rajah and colleagues showed that spike proteins of emerging SARS‐CoV‐2 variants produce more and larger syncytia (shown in green) than the original Wuhan strain upon infection of cell monolayers. Spike mutations with a positive, negative, or no effect appear in green, red, and blue, respectively. Mutations in black were not tested in this study.
The EMBO Journal (2021) 40: e110041.
See also: MM Rajah et al (December 2021)
References
- Braga L, Ali H, Secco I, Chiavacci E, Neves G, Goldhill D, Penn R, Jimenez‐Guardeno JM, Ortega‐Prieto AM, Bussani R et al (2021) Drugs that inhibit TMEM16 proteins block SARS‐CoV‐2 spike‐induced syncytia. Nature 594: 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C, Porrot F, Guivel‐Benhassine F, Van der Werf S et al (2020) Syncytia formation by SARS‐CoV‐2‐infected cells. EMBO J 39: e106267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foglieni C, Papin S, Salvade A, Afroz T, Pinton S, Pedrioli G, Ulrich G, Polymenidou M, Paganetti P (2017) Split GFP technologies to structurally characterize and quantify functional biomolecular interactions of FTD‐related proteins. Sci Rep 7: 14013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J, Uckeley ZM, Doldan P, Stanifer M, Boulant S, Lozach PY (2021) TMPRSS2 expression dictates the entry route used by SARS‐CoV‐2 to infect host cells. EMBO J 40: e107821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajah MM, Bernier A, Buchrieser J, Schwartz O (2021a) The mechanism and consequences of SARS‐CoV‐2 spike‐mediated fusion and syncytia formation. J Mol Biol 10.1016/j.jmb.2021.167280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajah MM, Hubert M, Bishop E, Saunders N, Robinot R, Grzelak L, Planas D, Dufloo J, Gellenoncourt S, Bongers A et al (2021b) SARS‐CoV‐2 alpha, beta and delta variants display enhanced spike‐mediated syncytia formation. EMBO J 40: e108944 [DOI] [PMC free article] [PubMed] [Google Scholar]


