Abstract
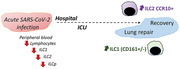
Accelerate lung repair in SARS‐CoV‐2 pneumonia is essential for pandemic handling. Innate lymphoid cells (ILCs) are likely players, given their role in mucosal protection and tissue homeostasis. We studied ILC subpopulations at two time points in a cohort of patients admitted in the hospital due to SARS‐CoV‐2 infection. COVID‐19 patients with moderate/severe respiratory failure featured profound depletion of circulating ILCs at hospital admission, in agreement with overall lymphocyte depletion. However, ILCs recovered in direct correlation with lung function improvement as measured by oxygenation index and in negative association with inflammatory and lung/endothelial damage markers like RAGE. While both ILC1 and ILC2 expanded, ILC2 showed the most striking phenotype changes, with CCR10 upregulation in strong correlation with these parameters. Overall, CCR10+ ILC2 emerge as relevant contributors to SARS‐CoV‐2 pneumonia recovery.
Keywords: COVID‐19, SARS‐CoV2, Lung recovery, Innate lymphoid cells, CCR10
Moderate to severe COVID‐19 is associated with an overall decrease in all innate lymphoid cells’ subsets in parallel with the well‐known lymphopenia. We report, in association with the oxygenation improvement, a significant expansion of ILC1, irrespectively of CD161 expression, and of CCR10 expressing ILC2, suggesting their involvement in lung regeneration.
Introduction
Lung immunopathology is the bottleneck of the SARS‐CoV‐2 infection, leading to prolonged hospital and intensive care unit (ICU) stay [1, 2]. In a quarter of patients infected by SARS‐CoV‐2, there is major lung pathology involving both the epithelia and endothelium [3], which has been associated with high levels of inflammatory cytokines [4]. It is of utmost importance to better understand not only the immunological mechanisms that limit both viral and immune‐mediated damage, but also those involved in lung repair.
Innate lymphoid cells (ILCs) are emerging as key populations in mucosal homeostasis and repair [5, 6]. Regarding the lung, ILC type 2 (ILC2) have been implicated in asthma [7], chronic obstructive pulmonary disease [8], and respiratory infections [9], and their levels in the blood [10] or related cytokines were reported as biomarkers in these diseases [11]. Moreover, increasing evidence suggest that ILC3 through IL‐22 [12] as well as ILC1 via interferon‐γ (IFN‐γ) may also play a role [11].
Therefore, it is plausible that ILCs contribute both to COVID‐19 pathology and to patient recovery. In agreement, loss of circulating ILCs has been associated with hospitalization requirement [13], and the few studies available support changes in ILC phenotype according to COVID‐19 severity [14, 15].
To specifically address the putative role of ILC in COVID‐19 recovery, we designed a longitudinal study involving individuals with SARS‐CoV‐2 pneumonia requiring hospitalization and respiratory support. We found that the recovery phase is linked to an expansion of ILC2 expressing high levels of the chemokine receptor CCR10, in correlation with serum markers implicated in lung injury/repair.
Results and discussion
ILCs recover in association with lung improvement during COVID‐19 recovery
We included 20 SARS‐CoV‐2‐infected individuals with moderate/severe pneumonia all having hypoxemia between April and October 2020, 11 of which were admitted to the ICU due to severe respiratory failure (Table 1).
Table 1.
Epidemiological and clinical characterization of Covid‐19 patients
| Patient ID | Age | Male | Arterial hypertension | Diabetes type 2 | Obesity | Lung emphysema | Maximum WHO disease severity score | Days of symptoms to admission | Days of symptoms to recovery | Days of hospital to admission time point | Days of hospital to recovery time point | ICU | Respiratory Support | Remdesivir | Dexamethasone | Steroids | Tocilizumab | Lopinavir/ritonavir |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 64 | Y | Y | N | Y | Y | 8 | 13 | 46 | 6 | 39 | Y | MV | N | N | N | N | Y |
| 2 | 64 | Y | Y | N | N | N | 5 | 3 | 11 | 2 | 10 | N | OM | N | N | N | N | Y |
| 3 | 65 | N | N | Y | Y | N | 5 | 7 | NA | 1 | NA a | N | OM | N | N | N | N | Y |
| 4 | 58 | Y | Y | Y | N | N | 9 | 10 | NA | 6 | NA b | Y | MV | N | N | N | Y | N |
| 5 | 40 | N | N | N | N | N | 5 | 16 | NA | 2 | NA a | N | OM | N | N | N | N | Y |
| 6 | 37 | Y | N | N | N | N | 5 | 11 | NA | 3 | NA a | N | OM | N | N | N | N | Y |
| 7 | 50 | Y | N | N | N | N | 5 | 10 | 17 | 3 | 10 | N | OM | N | N | N | N | Y |
| 8 | 39 | Y | Y | Y | N | N | 5 | 9 | NA | 3 | NA a | N | OM | N | N | N | N | N |
| 9 | 24 | Y | N | N | N | N | 5 | 11 | 17 | 2 | 9 | N | OM | N | N | Y | N | Y |
| 10 | 57 | N | Y | N | Y | Y | 8 | 6 | 18 | 3 | 15 | Y | MV | N | N | Y | N | N |
| 11 | 79 | N | Y | Y | N | N | 8 | 14 | NA | 7 | NA b | Y | MV | N | N | Y | N | N |
| 12 | 44 | Y | N | N | N | N | 6 | 15 | 22 | 5 | 12 | Y | HFNO | N | N | Y | N | Y |
| 13 | 72 | Y | N | N | Y | N | 6 | 13 | 20 | 3 | 11 | Y | HFNO | Y | Y | N | N | N |
| 14 | 47 | Y | Y | Y | Y | N | 6 | 11 | 21 | 5 | 15 | Y | HFNO | Y | Y | N | N | N |
| 15 | 35 | Y | N | N | N | N | 6 | 8 | 16 | 1 | 9 | Y | HFNO | Y | Y | N | N | N |
| 16 | 67 | Y | Y | N | N | N | 5 | 5 | 12 | 1 | 8 | N | OM | Y | Y | N | N | N |
| 17 | 68 | Y | Y | Y | N | N | 5 | 5 | 11 | 2 | 8 | N | OM | Y | N | N | N | N |
| 18 | 38 | Y | N | N | Y | N | 7 | 14 | 31 | 2 | 18 | Y | MV | N | N | Y | Y | N |
| 19 | 54 | Y | N | N | N | N | 8 | 9 | 20 | 2 | 13 | Y | MV | Y | Y | N | N | N |
| 20 | 64 | Y | N | N | N | N | 6 | 12 | 20 | 2 | 10 | Y | HFNO | N | N | Y | N | N |
| % in cohort | NA | 80 | 40 | 30 | 30 | 10 | NA | NA | NA | NA | NA | 55 | NA | 30 | 25 | 30 | 10 | 40 |
| Cohort Median [IQ] | 56 [40‐65] | NA | NA | NA | NA | NA | 6 [5–8] | 11 [7–13] | 19 [15–21] | 3 [2–5] | 11 [9–15] | NA | NA | NA | NA | NA | NA | NA |
N: no; NA: not applicable; MV: mechanical ventilation; HFNO: high flux nasal O2; OM: oxygen mask; Y: yes.
No recovery time point sample collection.
Patient died.
We identified ILC as lymphocytes lacking lineage specification markers while expressing CD127 and annotated as ILC precursors (ILCp) cKit+CRTH2neg, ILC2 CRTH2+, and ILC1– cKitnegCRTH2neg (Fig. 1A). Both ILC1 and ILC2 can be further divided according to CD161 expression, which is not observed in ILCp (Fig. 1A). This allowed us to include in our analysis a CD161 negative population previously ignored in other COVID‐19 studies [7, 8].
Figure 1.
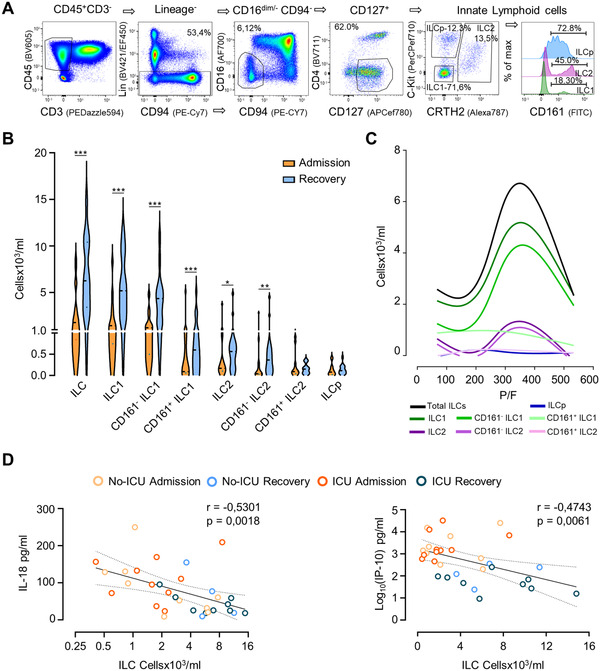
Circulating ILCs counts in COVID‐19 patients. (A) Illustrative whole blood ILC flow cytometry analysis in a representative healthy control showing the sequential manual gating in lymphocytes, CD45+, cells negative for CD3, other lineage markers (CD19, CD14, CD11c, and CD123), CD94, CD16, and CD4, followed by gating on CD127+ cells to define total ILCs; subsequently cKit and CRTH2 were used to define ILCp, ILC2 and ILC1; the histogram illustrates the distinct CD161 expression in these subsets. (B) Comparison of the counts of the main ILC populations at hospital/ICU admission and at recovery; data refer to 20 different patients at admission, with 14 patients also contributing with recovery time points; bars in violin plots refer to median and interquartile range; Wilcoxon matched‐pairs signed ranked test: ***p < 0.001; **p < 0,01; *p < 0.05. (C) Variation of the counts of ILC subsets according to the P/F (Smoothing spline curve fit using data shown in Supporting Information Fig. 2) in the same patients. (D) Correlation of the total ILC counts with IL‐18 and IP‐10 serum levels using Spearman correlation analysis; curve shown with 95% confidence interval (n = 19, with 13 patients contributing with the two time points). The healthy control values for all parameters are listed in Supporting Information Table S2. Samples from patients and controls were processed immediately after blood collection.
We found that total ILC counts were significantly reduced at admission in comparison with age and gender matched noninfected individuals (1955 [1010–3908] cells/mL in the 20 patients, as compared to 7700 [5140–14640] cells/mL controls, p = 0.0003), as previously reported [14, 15, 16]. The stratification of the patients according to ICU requirement showed no significant differences between total ILC counts in the two groups (2120 [860–6050] cells/mL in the non‐ICU group vs. 1670 [540–2300] in the ICU group, p = 0.44). Interestingly, we also found no association between ILC counts and plasma viral load, despite the significantly higher viremia in the ICU group (100 [32–568] RNAcp/mL vs. 11 [11–61] RNAcp/mL in non‐ICU, p = 0.014).
ILC counts according to time since beginning of symptoms were evaluated and compared with the cellular dynamics of the main lymphocyte subsets listed in Supporting Information Table S1 (Fig. 1B and Supporting Information Fig. S1). Total ILCs increased significantly at recovery, which was mostly attributed to ILC1, irrespectively of CD161 expression, and to ILC2, particularly those lacking CD161 (Fig. 1B and Supporting Information Fig. S1). Remarkably, peripheral blood ILCp counts did not change significantly throughout the follow‐up (Fig. 1B and Supporting Information Fig. S1) and were still significantly reduced at discharge when compared to noninfected (p = 0.014; Supporting Information Table S2). Since we only performed peripheral blood analysis, we cannot exclude rapid tissue migration to explain the lack of change in ILCp counts.
We next investigated whether the ILC increase was associated with lung recovery, using the P/F ratio (partial pressure of arterial oxygen/fraction of inspired oxygen) as a measurement of respiratory insufficiency. To evaluate the relationship between P/F and ILCs, we performed mathematical modeling using spline curves since the data were not linearly correlated [ 17 ]. Interestingly, ILC curves peaked around 300 (Fig. 1C and Supporting Information Fig. S2), which is considered the threshold for respiratory failure [18], supporting a close association between circulating ILC counts and lung function.
Moreover, significantly negative correlations were found with serum markers linked with COVID‐19 severity (Supporting Information Fig. S3), particularly illustrated for the consensual prognostic markers IP‐10 and IL‐18 [16, 19] (Fig. 1D).
ILCs upregulate CCR10 at recovery
Detailed ILC phenotype analysis using an unsupervised dimensionality reduction analysis (Fig. 2A) revealed a higher expression of the activation marker CD69 at admission, confirmed by manual gating in all subsets (Supporting Information Fig. S4A). Remarkably, although we observed a major increase in CD69 expression in ILCp at admission, in agreement with previous reports [14], the fact that we found no changes in ILCp counts during disease course suggests that ILCp expansion is not required for severe COVID‐19 recovery, though we cannot exclude a preferential recruitment of this population to the tissues. Conversely, an increase in CCR10 expression was apparent at recovery (Fig. 2A), particularly in ILC2. These findings were confirmed by our manual analysis also revealing a significant increase in the CCR10 MFI in the CD161negILC2 (Fig. 2B), which was inversely correlated with the decline of RAGE, a marker of endothelium lung damage [20] and positively correlated with EGF, a cytokine that promotes epithelial proliferation [21] (Fig. 2C).
Figure 2.
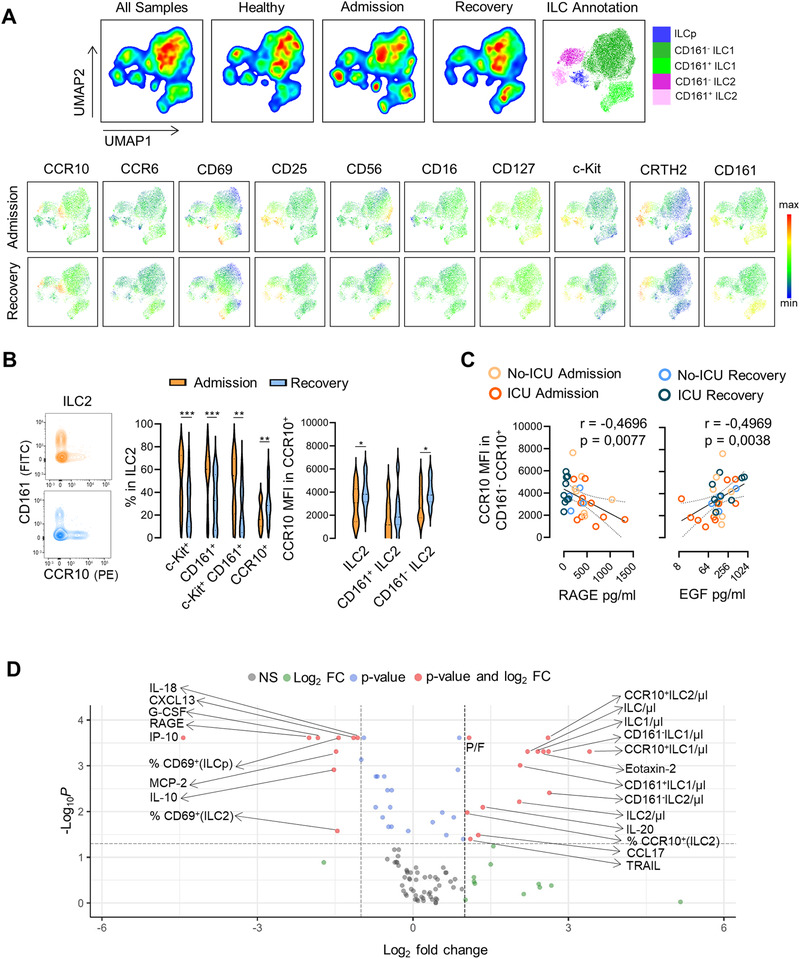
Expansion of CCR10 expressing ILC2 along with the recovery of COVID‐19. (A) Dimensionality reduction analysis of total ILCs was performed using unsupervised dimensionality reduction analysis (UMAP) algorithm; bi‐dimensional plots of all samples including eight healthy controls and 13 patients with both admission and recovery time points are shown on the top with the annotation of the main ILC populations represented; levels of expression of ILC markers on the grouped patients at admission and recovery are shown in the bottom. (B) Illustrative contour plots of the manual flow cytometry analysis of CD161 and CCR10 within ILC2 at admission and recovery in a representative patient; graphs show the frequency of c‐Kit+, CD161+, c‐Kit+CD161+, and CCR10+ cells within ILC2, as well as the MFI of CCR10 within total CCR10+ILC2, CCR10+CD161+ ILC2, and CCR10+CD161neg ILC2 subsets (n = 14); bars in violin plots refer to median and interquartile range; Wilcoxon matched pairs signed ranked test was used; ***p < 0,001; **p < 0,01; *p < 0,05. (C) Graphs show the correlation of the MFI of CCR10 within CCR10+CD161neg ILC2 with the serum levels of RAGE and EGF using Spearman correlation analysis (n = 18 at admission time point and 13 patients also contributing with recovery time point), curve is shown with the 95% confidence interval. (D) Volcano plot representing a fold change > |2| between admission and recovery of 108 parameters analyzed on the patient group (n = 13); frequencies refer to the gate mentioned in brackets. Patient and healthy control values for all parameters are listed in Supporting Information Table S2. Samples from patients and controls were processed immediately after blood collection.
Of note, the frequency of CCR10+ cells within ILC2 was positively associated with P/F, and CCR10+ILC2 counts correlates with serum levels of CCL17 (Supporting Information Fig. S3). Additionally, we found significant negative correlations between ILC2 or CCR10+ILC2 counts and serum levels of IL‐18, IP‐10, M‐CSF, G‐CSF, TGF‐α, FGF‐2, CXCL13, IL‐15, IL‐17, and IL‐17F (Supporting Information Fig. S3), factors known to contribute to lung inflammation and/or impact in ILC biology.
Although there was no correlation with CCL28 or CCL27 (Supporting Information Fig. S3), both CCR10 ligands, serum CCL28 levels were persistently elevated as compared to healthy controls (admission, p = 0.009; and recovery p = 0.003, Supporting Information Table S2). Moreover, the levels of cytokines known to be involved in ILC2 biology, like IL‐33, were also significantly elevated in the patients both at admission and recovery (admission, p = 0.0086 and recovery, p = 0.0131, Supporting Information Table S2). CCL28 is thought to be mainly produced by lung and gut epithelia [22], and possibly contributing to attract CCR10+ILC2. CCL27, whose serum levels were not elevated in our cohort, has been mostly implicated in the homing to CCR10+ cell to the skin [22], where they have been increasingly involved in repair, for instance after radiation injury, and in psoriasis [23]. On the other hand, also in psoriasis, cKit+ILC2 have been associated with a shift toward IL‐17 production in an ILC3‐like behavior with pro‐inflammatory function [24]. In this regard, it is worth emphasizing that the CCR10 + ILC2 that we identified featured low levels of both cKit (Fig. 2B) and CCR6, the latter being more expressed in ILCp (Supporting Information Fig. S4B). Of note, we confirm that CCR10 + ILC2 express CCR4 and GATA‐3, though at lower levels than CCR10negILC2 (Supporting Information Fig. S5A). Moreover, there was no enrichment of these markers in both CD161+/– ILC1 subsets, which express comparable increased levels of Tbet, CXCR3, and Eomes (Supporting Information Fig. S5B).
Regarding ILC1, we found that the relative proportion of CCR10+ did not change, although the counts of this subset significantly increase in parallel with the ILC1 expansion (Supporting Information Fig. S4C). In fact, and despite the ILC1‐marked increase, we did not find major changes in their phenotype, in agreement with a previous report in severe COVID‐19 [14].
ILC expansion and CCR10 upregulation are linked to COVID‐19 recovery
In order to further shed light on factors associated with disease recovery, we plotted the changes between admission and discharge of ILC counts and phenotype in conjunction with the serum panel and clinical parameters (listed in Supporting Information Tables S1 and S2) in a volcano plot (Fig. ). Recovery was associated with higher counts of total ILCs, ILC1, ILC2, and CCR10+ subsets, as well as the frequency of CCR10+ cells within ILC2, in parallel with the expected increase in P/F and a striking decrease in levels of RAGE, IP‐10, IL‐18, and IL‐10, and decline in CD69‐expressing ILC2 and ILCp.
These data suggest a role for CCR10 in COVID‐19. A principal component analysis of the parameters that had significant fold changes further supported this concept, since ILC subsets expressing CCR10 are among the parameters separating the admission and recovery time points irrespectively of ICU requirement (Supporting Information Fig. S6).
Concluding remarks
We showed here that the recovery of SARS‐CoV‐2 pneumonia was linked with CCR10+ILC2 expansion, which negatively correlated with inflammation and lung injury markers. The mechanisms underlying lung restoration have not been fully deciphered, but ILCs have been emerging as key players after acute insults [9]. In fact, there was a timely association of ILC expansion and the resolution of oxygenation impairment, supporting an important role of ILCs in lung regeneration.
The expansion of ILC2, in contrast to ILC1, was associated with significant upregulation of CCR10. ILC2 have been pointed as a double‐edge sword in respiratory disease, contributing both to lung homeostasis and to pathogenicity in chronic and acute settings. In animal models submitted to pneumectomy, IL‐33 activates ILC2 that promote alveologenesis and lung regeneration through IL‐13 [25]. Notwithstanding, ILC2‐dependent mechanisms of lung regeneration if sustained for a long period have been associated with evolution toward lung fibrosis, a process that has also been linked to IL‐13 production. Interestingly, CCR10 expressing ILC2 was linked in asthma with decreased bronchoreactivity through a shift from IL‐13 production to T‐bet upregulation and IFN‐y production [10], which may be in line with our observation of relatively lower GATA‐3 levels. Of note, ILC2 have been shown to upregulate NKG2D, a cytotoxic marker, in SARS‐CoV2 infection, possibly mediated by IL‐18 [7]. In this cross‐sectional study, the presence of this phenotype was significantly higher in hospitalized patients that did not required mechanical ventilation [7]. It is, therefore, likely that this population overlaps with the CCR10+ILC2 cells identified in our study. Functional studies including lung samples would be very important to uncover the mechanisms underlying ILC2 plasticity and its relative contribution for both viral control and alveolar repair, and our study suggests CCR10 to be an important marker in order to understand these processes.
In summary, our data support that the recovery of severe SARS‐CoV‐2 is linked to expansion of CCR10 expressing ILC2, reinforcing the relevance of targeting this subset to accelerate lung recovery in COVID‐19, as well as in other diseases.
Materials and methods
Patients
We performed a prospective longitudinal study including 20 SARS‐CoV‐2‐infected individuals (confirmed by RT‐PCR of nasopharyngeal swabs), with pneumonia admitted to Centro Hospitalar Universitário Lisboa Norte (CHULN, Portugal), between April and October 2020. Of note, eight of the patients were included between April and June before standard use of corticosteroid therapy. The clinical and epidemiological data are depicted in Table 1. The study was approved by the Ethical Board of CHULN/Centro Académico de Medicina de Lisboa. Oral informed consent was provided by all participants.
Study design
Twenty patients were assessed at hospital admission and 14 at discharge from the ICU or ward (non‐ICU). Healthy controls, recruited from health professionals, were studied in parallel as a reference (n = 9, with a mean age of 58 (IQ range: 48–61); six male and three female). Whole blood was processed immediately after collection for flow cytometry and for plasma and serum storage.
Flow cytometry analysis
Whole blood staining (15M leukocytes/tube) was performed after FcR blocking, with the panel listed in Supporting Information Table S3, acquired on a LSRFortessa X‐20 flow cytometer (BD Biosciences, CA), and analyzed using Flowjo software (version 10.7, Tree Star, Ashland, OR), applying both sequential manual gating (Fig. 1A) and unsupervised algorithms (unsupervised dimensionality reduction analysis, version 3.1). ILCs were identified as CD45+ CD127+ cells in the lymphogate, after exclusion of the cells expressing CD3,CD14, CD19, CD123, CD11c, CD4, CD94, or high levels of CD16.
Plasma viral load quantification
Plasma was used to quantify for SARS‐CoV‐2 viral load upon total RNA extraction (560 μL plasma, QIAamp Viral‐RNA MiniKit, QIAGEN) by droplet digital PCR (ddPCR, SARS‐CoV‐2 ddPCR Test Kit; Bio‐Rad, Hercules, CA, USA) on QX200 ddPCR System (Bio‐Rad), following manufacturer's instructions. Duplicates of 20 μL ddPCR reaction using 5 μL RNA were analyzed on QuantaSoft Analysis Pro (1.0.596). Plasma samples with N1 or N2 regions, or both regions detected, were considered positive, and SARS‐CoV‐2 RNA concentrations (cp/mL) were calculated considering the extracted volume of plasma.
Serum analysis
Serum levels of 71 soluble analytes were determined using a Multiplexing LASER Bead Assay (Human Cytokine Array/Chemokine Array 71‐Plex Panel (HD71); Eve Technologies, Canada) and serum levels of CCL28, RAGE, SP‐D, and IL‐22BP were determined by Sandwich ELISA kits (RayBiotech, GA) using duplicates. Serum IgA, IgM, and IgG anti‐SARS‐CoV‐2 titers were assessed by ELISA as previously described [26].
Statistical analysis
Data were analyzed with R version 4.0.2., using the packages heatmaply, EnhancedVolcano, corrplot, ggplot2 for data visualization and GraphPad Prism version 8 (GraphPad Software, San Diego, CA, USA) using Wilcoxon matched‐pairs signed ranked test, Mann–Whitney U test, Spearman correlation, and Smoothing spline [17]. Results are shown as median and interquartile ranges. All p‐values below 0.05 were considered statistically significant.
Conflict of interest
The authors report no commercial or financial conflict of interest.
Author contributions
ACT, AGS, PR, ARMA, AES, and SMF designed the study; AMCG, GBF, JL, ACT, AGS, PR, DFS, CMC, and ARMA performed experiments; AMCG, GBF, MDS, CMC, and ARMA were involved with data analysis; RCR, MAS, CM, and SMF were associated with patient recruitment, patient follow‐up, and clinical data registration; ACT, ARMA, AES, and SMF supervised the study; ACT and SMF acquired funding for the study; AMCG, MDS, AES, and SMF wrote the original draft; all authors were involved in the review and editing of the final manuscript.
Ethical statement
The study was approved by the Ethical Board of CHULN/Centro Académico de Medicina de Lisboa.
Patient consent statement
Oral informed consent was required from all participants.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202149311
Abbreviations
- ICU
intensive care unit
- ILC
innate lymphoid cell
- ILCp
ILC precursor
- P/F
partial pressure of arterial oxygen/fraction of inspired oxygen
Supporting information
supporting information
Acknowledgements
First, we acknowledge all the participating patients and the health care professionals involved in patient care. We also acknowledge Marc Veldhoen for SARS‐CoV‐2 antibody quantification. We thank the team members of Flow Cytometry facility at Instituto de Medicina Molecular João Lobo Antunes (Lisboa, Portugal) for technical assistance. This work was funded by the following grants from Fundação para a Ciência e a Tecnologia (FCT), Portugal, through “APOIO ESPECIAL RESEARCH4COVID‐19,” project numbers 125 to SMF and 803 to ACT. AMCG and GBF received fellowships funded by FCT (DOCTORATES4COVID‐19, 2020.10202.BD) and JANSSEN‐ CILAG FARMACÊUTICA, respectively.
Data availability statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
- 1. Ferrando, C. , Suarez‐Sipmann, F. , Mellado‐Artigas, R. , Hernandez, M. , Gea, A. , Arruti, E. , Aldecoa, C. et al., Clinical features, ventilatory management, and outcome of ARDS caused by COVID‐19 are similar to other causes of ARDS. Intensive Care Med. 2020. 46: 2200–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Doidge, J. C. , Gould, D. W. , Ferrando‐Vivas, P. , Mouncey, P. R. , Thomas, K. , Shankar‐Hari, M. , Harrison, D. A. et al., Trends in intensive care for patients with COVID‐19 in England, Wales, and Northern Ireland. Am. J. Respir. Crit. Care Med. 2021. 203: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang, P. , Luo, R. , Zhang, M. , Wang, Y. , Song, T. , Tao, T. , Li, Z. et al., A cross‐talk between epithelium and endothelium mediates human alveolar‐capillary injury during SARS‐CoV‐2 infection. Cell Death Dis. 2020. 11: 1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gustine, J. N. and Jones, D. , Immunopathology of Hyperinflammation in COVID‐19. Am. J. Pathol. 2021. 191: 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fernandes, S. M. , Pires, A. R. , Ferreira, C. , Foxall, R. B. , Rino, J. , Santos, C. , Correia, L. et al., Enteric mucosa integrity in the presence of a preserved innate interleukin 22 compartment in HIV type 1‐treated individuals. J. Infect. Dis. 2014. 210: 630–640. [DOI] [PubMed] [Google Scholar]
- 6. Fernandes, S. M. , Pires, A. R. , Matoso, P. , Ferreira, C. , Nunes‐Cabaco, H. , Correia, L. , Valadas, E. et al., HIV‐2 infection is associated with preserved GALT homeostasis and epithelial integrity despite ongoing mucosal viral replication. Mucosal Immunol. 2018. 11: 236–248. [DOI] [PubMed] [Google Scholar]
- 7. Ealey, K. N. , Moro, K. and Koyasu, S. , Are ILC2s Jekyll and Hyde in airway inflammation? Immunol. Rev. 2017. 278: 207–218. [DOI] [PubMed] [Google Scholar]
- 8. Silver, J. S. , Kearley, J. , Copenhaver, A. M. , Sanden, C. , Mori, M. , Yu, L. , Pritchard, G. H. et al., Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat. Immunol. 2016. 17: 626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Monticelli, L. A. , Sonnenberg, G. F. , Abt, M. C. , Alenghat, T. , Ziegler, C. G. , Doering, T. A. , Angelosanto, J. M. et al., Innate lymphoid cells promote lung‐tissue homeostasis after infection with influenza virus. Nat. Immunol. 2011. 12: 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beuraud, C. , Lombardi, V. , Luce, S. , Horiot, S. , Naline, E. , Neukirch, C. , Airouche, S. et al., CCR10(+) ILC2s with ILC1‐like properties exhibit a protective function in severe allergic asthma. Allergy 2019. 74: 933–943. [DOI] [PubMed] [Google Scholar]
- 11. Panda, S. K. and Colonna, M. Innate lymphoid cells in mucosal immunity. Front. Immunol. 2019. 10: 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Felton, J. M. , Duffin, R. , Robb, C. T. , Crittenden, S. , Anderton, S. M. , Howie, S. E. M. , Whyte, M. K. B. et al., Facilitation of IL‐22 production from innate lymphoid cells by prostaglandin E2 prevents experimental lung neutrophilic inflammation. Thorax 2018. 73: 1081–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Silverstein, N. J. , Wang, Y. , Manickas‐Hill, Z. , Carbone, C. C. , Dauphin, A. , Li, J. Z. , Boribong, B. P. et al., Innate lymphoid cells and disease tolerance in SARS‐CoV‐2 infection. medRxiv. 2021. [Google Scholar]
- 14. Garcia, M. , Kokkinou, E. , Garcia, A. C. , Parrot, T. , Medina, L. M. P. , Maleki, K. T. , Christ, W. et al., Innate lymphoid cell composition associates with COVID‐19 disease severity. Clin. Transl. Immunol. 2020. 9: e1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gomez‐Cadena, A. , Spehner, L. , Kroemer, M. , Khelil, M. B. , Bouiller, K. , Verdeil, G. , Trabanelli, S. et al., Severe COVID‐19 patients exhibit an ILC2 NKG2D(+) population in their impaired ILC compartment. Cell. Mol. Immunol. 2021. 18: 484–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flament, H. , Rouland, M. , Beaudoin, L. , Toubal, A. , Bertrand, L. , Lebourgeois, S. , Rousseau, C. et al., Outcome of SARS‐CoV‐2 infection is linked to MAIT cell activation and cytotoxicity. Nat. Immunol. 2021. 22: 322–335. [DOI] [PubMed] [Google Scholar]
- 17. Zhan, C. and Yeung, L. F. Parameter estimation in systems biology models using spline approximation. BMC Syst. Biol. 2011. 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Force, A. D. T. , Ranieri, V. M. , Rubenfeld, G. D. , Thompson, B. T. , Ferguson, N. D. , Caldwell, E. , Fan, E. et al., Acute respiratory distress syndrome: the Berlin definition. JAMA 2012. 307: 2526–2533. [DOI] [PubMed] [Google Scholar]
- 19. Laing, A. G. , Lorenc, A. , Del Molino Del Barrio, I. , Das, A. , Fish, M. , Monin, L. , Muñoz‐Ruiz, M. et al., A dynamic COVID‐19 immune signature includes associations with poor prognosis. Nat. Med. 2020. 26: 1623–1635. [DOI] [PubMed] [Google Scholar]
- 20. Oczypok, E. A. , Perkins, T. N. and Oury, T. D. All the “RAGE” in lung disease: the receptor for advanced glycation endproducts (RAGE) is a major mediator of pulmonary inflammatory responses. Paediatr. Respir. Rev. 2017. 23: 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Balnis, J. , Adam, A. P. , Chopra, A. , Chieng, H. C. , Drake, L. A. , Martino, N. , Ramos, R. B. et al., Unique inflammatory profile is associated with higher SARS‐CoV‐2 acute respiratory distress syndrome (ARDS) mortality. Am J Physiol Regul Integr Comp Physiol. 2021. 320: R250–R257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiong, N. , Fu, Y. , Hu, S. , Xia, M. and Yang, J. , CCR10 and its ligands in regulation of epithelial immunity and diseases. Protein Cell. 2012. 3: 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li, C. , Xu, M. , Coyne, J. , Wang, W. B. , Davila, M. L. , Wang, Y. and Xiong, N. , Psoriasis‐associated impairment of CCL27/CCR10‐derived regulation leads to IL‐17A/IL‐22‐producing skin T‐cell overactivation. J. Allergy Clin. Immunol. 2021. 147: 759–763. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bernink, J. H. , Ohne, Y. , Teunissen, M. B. M. , Wang, J. , Wu, J. , Krabbendam, L. , Guntermann, C. et al., c‐Kit‐positive ILC2s exhibit an ILC3‐like signature that may contribute to IL‐17‐mediated pathologies. Nat. Immunol. 2019. 20: 992–1003. [DOI] [PubMed] [Google Scholar]
- 25. Lechner, A. J. , Driver, I. H. , Lee, J. , Conroy, C. M. , Nagle, A. , Locksley, R. M. and Rock, J. R. , Recruited monocytes and type 2 immunity promote lung regeneration following pneumonectomy. Cell Stem Cell. 2017. 21: 120–134. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Figueiredo‐Campos, P. , Blankenhaus, B. , Mota, C. , Gomes, A. , Serrano, M. , Ariotti, S. , Costa, C. et al., Seroprevalence of anti‐SARS‐CoV‐2 antibodies in COVID‐19 patients and healthy volunteers up to 6 months post disease onset. Eur. J. Immunol. 2020. 50: 2025–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supporting information
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


