Abstract
Mutations in the human LMNA gene cause a collection of diseases called laminopathies, which includes muscular dystrophy and dilated cardiomyopathy. The LMNA gene encodes lamins, filamentous proteins that form a meshwork on the inner side of the nuclear envelope. How mutant lamins cause muscle disease is not well understood, and treatment options are currently limited. To understand the pathological functions of mutant lamins so that therapies can be developed, we generated new Drosophila models and human iPS cell-derived cardiomyocytes. In the Drosophila models, muscle-specific expression of the mutant lamins caused nuclear envelope defects, cytoplasmic protein aggregation, activation of the Nrf2/Keap1 redox pathway, and reductive stress. These defects reduced larval motility and caused death at the pupal stage. Patient-derived cardiomyocytes expressing mutant lamins showed nuclear envelope deformations. The Drosophila models allowed for genetic and pharmacological manipulations at the organismal level. Genetic interventions to increase autophagy, decrease Nrf2/Keap1 signaling, or lower reducing equivalents partially suppressed the lethality caused by mutant lamins. Moreover, treatment of flies with pamoic acid, a compound that inhibits the NADPH-producing malic enzyme, partially suppressed lethality. Taken together, these studies have identified multiple new factors as potential therapeutic targets for LMNA-associated muscular dystrophy.
Keywords: Drosophila, Lamins, Muscular dystrophy, Nuclear envelope, Reductive stress
Highlights
-
•
Drosophila models of LMNA muscular dystrophy exhibited redox imbalance.
-
•
Amino acid substitutions in all three domains of lamins caused reductive stress.
-
•
iPS cell-derived cardiomyocytes showed an altered response to NAC treatment.
-
•
Genetic manipulation of autophagy regulators suppressed lethality caused by mutant lamins.
-
•
Pamoic acid, a malic enzyme inhibitor, suppressed lethality caused by mutant lamins.
1. Introduction
Maintaining redox homeostasis is essential for many cellular functions, including gene expression, immune response, and central carbon metabolism [1]. Muscle cells are particularly sensitive to imbalances in redox homeostasis due to the large quantities of reactive oxygen species (ROS) produced during muscle contractions [2]. Reactive oxygen species can damage nucleic acids, lipids, and proteins, resulting in cellular dysfunctions [[3], [4], [5]]. Antioxidant systems are necessary to combat reactive oxygen species and maintain homeostasis [6]. Thus, it is not surprising that skeletal muscle and cardiac diseases are often associated with loss of redox homeostasis.
Skeletal muscles of individuals with Duchenne Muscular Dystrophy (DMD) exhibit increased levels of ROS that are implicated in tissue damage [7]. Treatment of DMD individuals with antioxidants partially reduced inflammation, skeletal muscle weakness, and fibrosis [8]. Additionally, mouse models of Emery-Dreifuss muscular dystrophy (EDMD) exhibit increased levels of oxidative stress in cardiac tissue [9]. Treatment with the antioxidant N-acetyl cysteine improved cardiac structure and function [9]. These findings demonstrate the importance of redox homeostasis for overall muscle health and suggest potential benefits of antioxidant therapies.
How redox status is altered by mutations that cause muscular dystrophy is not well understood. Moreover, these mutations affect proteins that reside in multiple cellular compartments. DMD is caused by mutations in the gene encoding dystrophin, a cytoplasmic protein that is part of a cell membrane-spanning complex that connects the cytoskeleton to the extracellular matrix [10]. EDMD is caused by mutations in the LMNA gene that encodes A-type lamins [11]. Lamins are intermediate filaments that form a meshwork lining the inside of the nuclear envelope. They interact with components of the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex that spans the double membrane surrounding the nucleus and connects the nuclear lamina to the cytoskeleton [12]. Thus, proteins involved in communication across different cellular membranes are important for muscle cell redox homeostasis.
To investigate the role of lamins in muscle redox homeostasis, we developed Drosophila models of LMNA-associated muscular dystrophy [13]. The Drosophila melanogaster genome encodes an orthologue of LMNA, designated Lamin C (LamC). Mutations in LMNA that cause disease affect amino acid residues conserved between human lamin A/C and Drosophila LamC. Lamins possess a conserved protein structure consisting of an N-terminal globular domain, a central coiled coil domain, and an Ig-like fold domain at the carboxy terminus [14]. Lamins dimerize, form head-to-tail filaments, and interact laterally to form higher order structures [[15], [16], [17]]. The majority of laminopathies are caused by single amino acid substitutions found throughout the lamin protein. It is unclear if these amino acid substitutions share pathological mechanisms, as lamins play a multitude of roles in the nucleus, and partner proteins interact with specific domains [15,18,19].
Our prior studies focused on the Ig-like fold domain of A-type lamins [20,21]. When disease-causing mutations affecting the Ig-like fold were modeled into LamC, the flies developed muscular defects [20]. Expression of mutant lamins specifically in larval body wall muscles, which share developmental and physiological properties with human skeletal muscles, reduced larval motility and led to premature death at the pupal stage [22]. At the cellular level, larval body wall muscles showed abnormal nuclear morphology, cytoplasmic aggregation of nuclear envelope proteins, and activation of the nuclear factor erythroid 2-related factor 2 (Nrf2)/kelch-like ECH-associated protein 1 (Keap1) redox pathway [23]. Surprisingly, activation of the Nrf2/Keap1 pathway was not caused by oxidative stress, as it is in many disease states [24]. Instead, it appeared to be caused by cytoplasmic protein aggregation, and resulted in reductive stress, a condition associated with elevated levels of reducing equivalents [25,26]. Cytoplasmic aggregation of nuclear envelope proteins and activation of the Nrf2/Keap1 pathway were also discovered in human muscle biopsy tissue from individuals with LMNA-associated muscular dystrophy, demonstrating human disease relevance of the Drosophila models [23].
Here, we expand on these findings by determining the consequences of disease-causing amino acid substitutions in additional domains of lamin. We found that mutations affecting each of the three domains cause reductive stress and premature lethality. Manipulating expression of genes involved in autophagy and redox biology partially suppressed lethality caused by mutant lamins. Supporting these analyses, treatment with a compound that targets malic enzyme, which generates reducing equivalents, partially suppressed lethality. Taken together, these findings suggest that elimination of cytoplasmic aggregates and lowering levels of reducing equivalents in skeletal muscles are potential therapeutic avenues for LMNA muscular dystrophy.
2. Materials and methods
2.1. Drosophila cultures
Stocks were cultured using standard cornmeal/sucrose media and grown at 25 °C [27]. Transgenic stocks were generated as previously described [20,23]. Wild-type and mutant transgenes were expressed using the Gal4/UAS system [28]. The C57 body wall muscle-specific Gal4 driver was used to express the LamC transgenes [[28], [29], [30]]. Stocks possessing either RNAi or over-expressed transgenes were obtained from Bloomington Stock Center (https://bdsc.indiana.edu/) or the Vienna Drosophila Resource Center (https://stockcenter.vdrc.at/control/main) and are listed in Table S1.
2.2. Western analysis
LamC protein levels were determined by performing western analysis as previously described [27]. Three independent biological samples of protein extracted from larval body wall muscles of two larvae were analyzed. LamC was detected with anti-LamC antibody (LC28.36; DHSB, Iowa City) and secondary antibody (A21202; Invitrogen, Waltham). Total protein was detected using LI-COR Revert (926–11010; LI-COR Bioscience, Lincoln) and used for normalization of protein loading. Quantification was performed using an Odyssey LI-COR imaging system (LI-COR, Lincoln).
2.3. Real-time quantitative PCR (RT-qPCR)
Body wall muscles from two third-instar larvae per sample were dissected, placed in a microcentrifuge tube and flash frozen with liquid nitrogen. RNA was extracted using RNeasy Micro Kit (ID: 74,004; Qiagen, Germantown). Briefly, muscles were thawed, immediately lysed with a mortar and pestle, and homogenized by pipetting. Lysates were centrifuged for 3 min, and supernatants were transferred to fresh microfuge tubes. The remainder of the RNA isolation was performed according to Qiagen instructions. RNA purity and concentration were measured using a NanoDrop 200 (Thermofisher, Waltham) at wavelengths 260 nm and 280 nm.
cDNA was produced using a High-Capacity cDNA Reverse Transcription Kit (f#4368814; Applied Biosystems, Foster City) according to the manufacturer's instructions. Starting with 600 ng of RNA in a final volume of 50 μl, cDNA was generated using a PTC-200 Peltier Thermo Cycler (MJ Research, Waltham) with the following settings: 25 °C for 10 min, 37 °C for 2 h, and 95 °C for 10 min. Finally, 150 μl of DEPC H2O was added to bring the final volume to 200 μl.
RT-qPCR was performed using iQ SYBR Green Supermix (#1708880; BioRad, Hercules) with primers generated by Integrated DNA Technologies (Coralville, IA). Each reaction contained a final concentration of 400 nM of each primer, 9 μL of cDNA, and 10 μL of the SYBR Green Supermix, for a total volume of 20 μL. PCR was performed by the Iowa Institute of Human Genetics on a QuantStudio-7 Flex qPCR (Life Technologies, Waltham). Relative mRNA levels were calculated using the Delta Delta Ct method, with a Ct threshold of 1.0. Expression levels of GAPDH and ribosomal protein L32 were used for normalization. Unpaired Student-T tests were performed on a minimum of three biological replicates.
2.4. Larval motility assays
Adults possessing UAS driven LamC transgenes and the host stock (w1118) were crossed to the C57 Gal4 driver stock. Resulting third Instar larvae were removed from the culture vials at 25 °C and placed on a Petri dish containing 1.8% agarose and allowed to equilibrate to room temperature for 10 min. Five larvae were transferred to a second Petri dish containing 1.8% agarose that had been marked with concentric circles half an inch apart. Larval motility was recorded for 2 min using a cell phone and videos were analyzed using wrMTack, a plug-in for ImageJ [20]. A calibration line was drawn from the first ring to the second ring using the “line tool” to produce the calibration length value. The path crawled by each larva was traced by generating a Z-project using the max intensity function. Distances traveled were measured by tracing larval paths with the segmented line drawing tool. These distances were then divided by the calibration length value to produce distance in millimeters. Velocities were calculated by dividing distance traveled by recording time (120 s). Values were plotted using GraphPad Prism (GraphPad Prism version 8.0.0 for Mac, GraphPad Software, San Diego, California USA) and velocities were compared to those of wild type using ANOVA analysis.
2.5. Adult viability
Adults possessing LamC transgenes and the host stock (w1118) were crossed to adults of the C57 stock in eight independent vials. Newly eclosed adult progeny were collected and counted daily. After ten days, the number of dead pupae in each vial were counted. Percent viability was calculated by dividing the number of adults by the number of dead pupae plus live adults, multiplied by 100. Fisher's Exact test was used for comparison of each genotype to the wild-type LamC control.
2.6. Immunohistochemistry
Third Instar larvae with muscle-specific expression of either wild type or mutant LamC were dissected and fixed with 4% formaldehyde for 20 min as previously described [20]. Larval body wall muscles were stained with anti-LamC antibodies [1:400, Developmental Studies Hybridoma Bank (DSHB), U of Iowa] and detected with either Alexa-Flour 488 goat anti-mouse (1:400, Invitrogen) or Rhodamine Red-X anti-mouse (1:400, Thermo Fisher). Larval body wall muscles were stained with Drosophila anti-Ref(2)P antibodies (1:3,000, a kind gift of G. Juhász, Eötvös Loránd University, Budapest) and detected with Alexa Fluor 488 goat anti-rabbit (1:500; Invitrogen). F-actin was detected with Texas Red Phalloidin (1:400, Invitrogen). Slides were mounted with Vectashield mounting medium containing DAPI (Vector Laboratories) and imaged using a Leica DMLB fluorescent microscope. Images were captured with a Leica CCD camera.
LamC staining intensity was measured using tools provided in ImageJ. Briefly, the nuclei were first traced and designated as Regions of Interest (ROI) in the DAPI channel. These ROI were then superimposed on the LamC channel and the fluorescence measured. Cytoplasmic intensity was calculated by measuring all the fluorescence within a muscle fiber and subtracting the nuclear intensity previously measured.
iPS cell-derived cardiomyocytes were fixed with 4% paraformaldehyde (Sigma) on coverslips, permeabilized with 0.1% Triton X-100 (Sigma) and blocked with 3% bovine serum albumin (Ultrapure BSA, Cell Signaling Technology). Primary antibodies included: SSEA-4 (1:40, Stem Cell Technologies), OCT-4 (1:400, Cell Signaling Technology), goat anti-lamin A/C polyclonal antibody (1:50, SantaCruz), rabbit anti-Nrf2 polyclonal antibody (1:100, SantaCruz), rabbit anti-SQSM1/p62 monoclonal antibody (1:100, Cell Signaling) and mouse anti-cTnT monoclonal antibody (1:100, NeoMarkers). Secondary antibodies were Alexa Fluor 555 anti-goat IgG, Alexa Fluor 488 donkey anti-rabbit IgG and Alexa Fluor 555 donkey anti-mouse IgG (all used at 1:500, Life Technologies). Slides were mounted with Ultracruz Hard Set mounting medium plus DAPI (Santa Cruz Biotechnology).
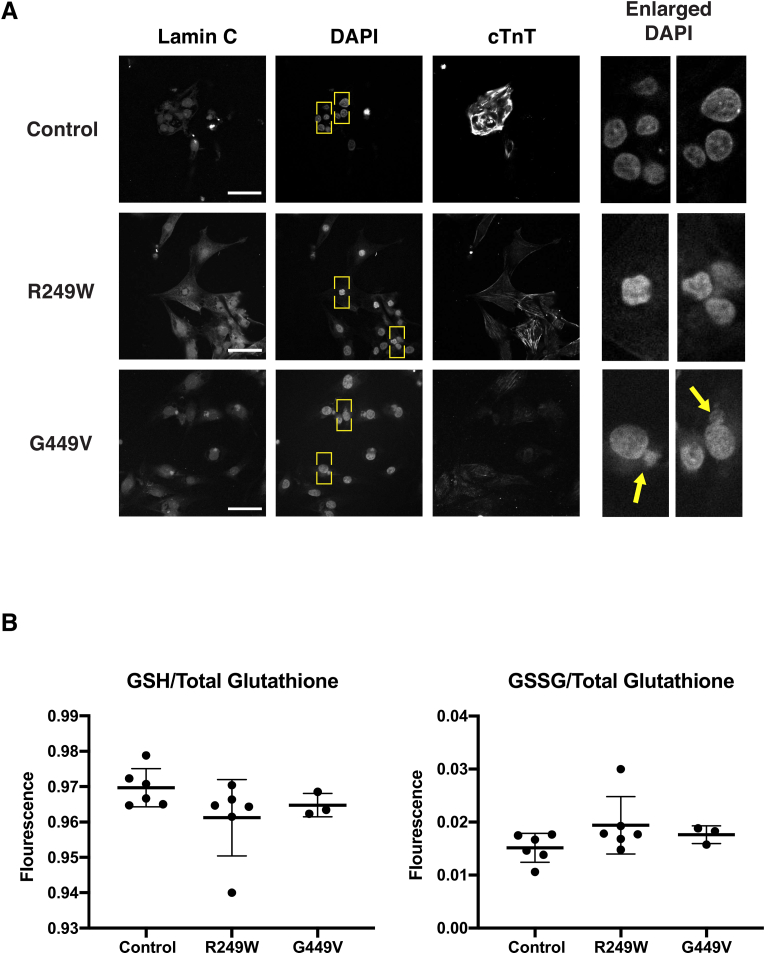
Unlabeled images of iPS cell derived cardiomyocytes were presented in a random order to two individuals who had never seen these images before. These individuals were then asked to count the total number of nuclei in each image, and the number of deformed nuclei. These counts were then averaged and the percent of nuclei containing the deformities was calculated. Please see Fig. 4 for an example of a nuclear deformity.
Fig. 4.
LMNA patient-based cell models show nuclear deformities but no changes in glutathione levels. (A) iPS cell derived cardiomyocytes from patients were stained with DAPI and antibodies to lamin A/C and cardiac Troponin T (cTnT). The LMNA mutation producing lamin A/C G449V increased nuclear deformities (arrows) relative to controls expressing wild type lamin A/C. Yellow boxes indicate enlarged regions (right). Scale Bar: 30 μm. (B) Reduced and oxidized glutathione levels were measured in iPS cell derived cardiomyocytes. GSH and GSSG levels were measured in three to six independent biological samples. GSH was normalized to total glutathione and plotted as fluorescence intensity. Likewise, GSSG was normalized to total glutathione and plotted as fluorescence intensity. Unpaired Student's t-tests were then performed revealing no difference between patient-based cells and controls. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.7. GSH/GSSG quantification
GSSG and GSH measurements were performed on Drosophila larval body wall muscles as described (Dialynas, 2014) with assistance from the University of Iowa Free Radical and Radiation Biology Core. Briefly, muscle fillets were dissected from sets of 15 Drosophila larvae. Total glutathione was determined by oxidizing all of the glutathione in the sample with the addition of glutathione reductase. GSSG measurements were performed by adding a 1:1 mixture of 2-vinylpyridine and ethanol to the samples and incubating for 2 h. Then, total glutathione was determined by spectrophotometric measurements of the production of 5-thio-2-nitrobenzoic acid (TNB), a byproduct of the reaction between GSH and Ellman's reagent. The rate of TNB production is directly proportional to total glutathione. Concentrations were obtained by comparing enzymatic rates to those in standard curves obtained using control samples. GSH concentration was calculated by subtracting measured GSSG from measured total glutathione. GSH and GSSG amounts were normalized to the protein content of the insoluble pellet using a BCA Protein Assay Kit (Thermo Scientific, Waltham). Unpaired Student's t-tests were performed on a minimum of three biological replicates.
GSSG and GSH measurements were performed on iPSC cardiomyocytes using the GSH/GSSG-Glo Assay (V6611; Promega, Madison). Values were normalized to cell counts. Statistical analysis was performed as described above.
2.8. Compound feeding
Drosophila medium was prepared by mixing 1 g of instant Drosophila medium (Formula 4-24® Instant Drosophila, Carolina Biological) with 4.5 mL of water and allowing it to absorb for 1 h. Two females from the C57 Gal4 driver stock were placed in each vial with two males homozygous for a LamC transgene of each genotype. Once first instar larvae were observed (approximately four days after crosses were established) 1 mL of either 5 mM pamoic acid in water, or vehicle only (water), was added to each vial, reserving vials as untreated controls. Percent adult viability was calculated, and statistical analysis was performed as described above for adult viability measurements.
2.9. Statistics
Experimental results are reported for a minimum of three independent biological samples. Continuous numeric datasets were analyzed with either a one-way analysis of variance (ANOVA) or Student's t-test depending on whether multiple groups or only two groups were being compared, respectively. Prism (GraphPad) was used for all statistical analyses. In graphs, error bars represent mean ± standard deviation (SD).
3. Results
3.1. Mutant lamins reduced Drosophila larval motility and adult viability
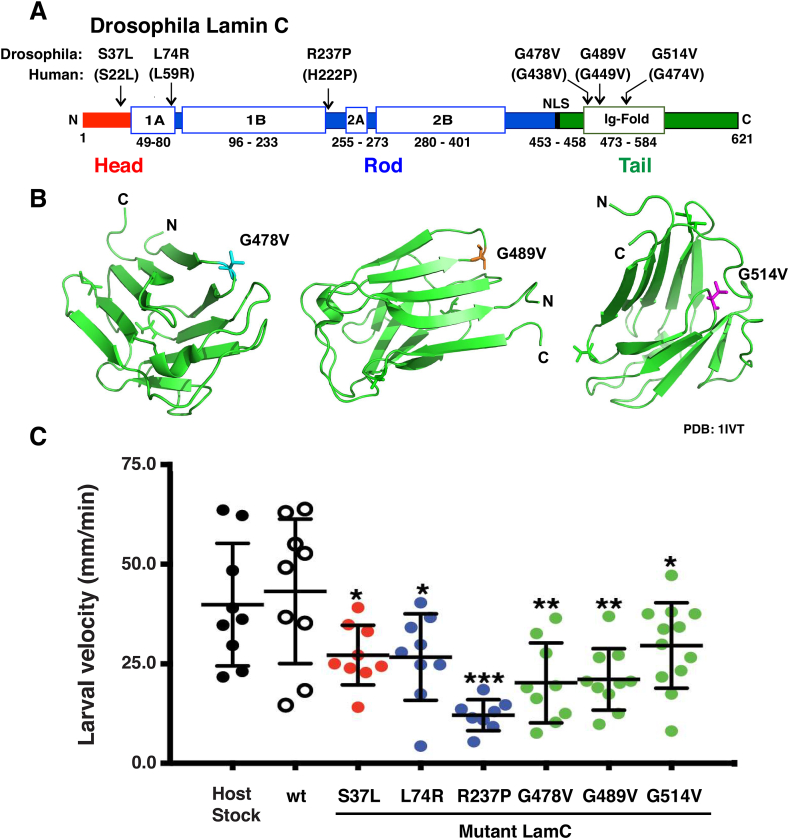
Our prior studies focused on mutations in LMNA that cause single amino acid substitutions in the lamin A/C Ig-like fold domain [21,23,27]. Here, we determined the generality of those findings by extending our studies to include amino acid substitutions in the head and rod domain. To this end, we modeled lamin A/C S22L, L59R and H222P that affect the head and rod domains in Drosophila LamC (Fig. 1A). These mutations were identified in individuals with cardiomyopathy, progeria features, and EDMD as shown in Table 1 [11,20,[31], [32], [33]].
Fig. 1.
Muscle-specific expression of mutant LamC reduced larval motility. (A) LamC protein domains are diagrammed, and amino acid substitutions used in this study are labeled. (B) Ribbon diagrams of the Ig-fold domain of human lamin A/C (PBD: 1IVT) displaying the positions of the amino acid substitutions used in this study. (C) Larval crawling velocity for each genotype tested is plotted. Larval velocity was measured using 2-min video clips and processed with ImageJ. One-way ANOVA analysis was used to determine statistical significance among the average values with the wild-type (wt) as the control.
Table 1.
Clinical information on individuals with LMNA mutations used in this study.
| Nucleotide Change | Domain Affected | Age of Diagnosis | Clinical Diagnosis | MSK | Cardiac | Contractures | Ref. |
|---|---|---|---|---|---|---|---|
| 65C > T | Head | 45 (NR) | Familial DCM | x | 30 | ||
| 176T > C | Rod | 16 (F) | Progeroid Features | x | x | 31 | |
| 176T > G | Rod | 10 (F) | “Malouf syndrome" like Laminopathy | x | x | 32 | |
| 665A > C | Rod | Birth (M) | EDMD 3 | x | x | x | 11 |
| 665A > C | Rod | Childhood (F) | EDMD 3 | x | x | ||
| 665A > C | Rod | 5–6 years old (M) | EDMD 3 | x | x | x | |
| 1346G > T | Tail | 3 (NR) | CMD | x | x | 19 |
M: Male; F: Female; NR: Not Reported; MSK: Musculoskeltal; x, indicates presence of disease phenotype.
Previously, we showed that lamin A/C G449V produced severe muscle disease phenotypes in humans and when modeled in Drosophila LamC. Therefore, this amino acid substitution was used in these studies as a positive control. Consistent with this finding, structural studies have shown that Lamin A/C G449V causes perturbations in the 3D structure of the Ig-fold [23]. We noted that Gly residues were present in other loops between beta-strands in the Ig-fold (Fig. 1B). With this in mind, we generated new Drosophila models containing G438V and G474V (Fig. 1A). LamC was expressed in transgenic host stocks at 0.8–2.2 times the levels of endogenous expression.
To determine if these newly generated LamC transgenes altered muscle function, they were expressed in larval body wall muscles using the Gal4/UAS system with the C57 Gal4 driver stock [34]. In this stock, the Gal4 transcription factor is expressed in larval body wall muscles during late embryonic larval stages [34]. LamC levels in these transgenic larvae were measured by western analysis (Fig. S1). These data showed that mutant LamC levels were similar or less than two-fold greater than wild-type.
To determine effects of the mutant transgenes on muscle function, third instar larvae expressing either wild-type or mutant LamC were placed in the center of an agar plate, and their movement was recorded for 2 min. The resulting videos were used to determine larval velocities. Larvae expressing mutant LamC showed decreased motility relative to control larvae expressing wild-type LamC (Fig. 1C). Reduced motility is consistent with impaired muscle function.
A second indication of the loss of muscle function came from adult survival studies. Larval body wall muscle functions in metamorphosis during the pupal stage prior to undergoing developmental histolysis [35]. As a result of poor muscle function, pupal death is observed. Flies possessing either wild type or mutant LamC transgenes were crossed to flies expressing Gal4 in larval body wall muscles. The total number of living adult progeny that resulted was divided by the total number of progeny (live adults plus dead pupae) and used to calculate the percent adult viability. Expression of LamC R237P and G514V resulted in no surviving adults. In contrast, expression of LamC G478V, G489V, L74R and S37L caused semi-lethality, with survival ranging from 0.3 to 39.6% (Table 2). The adult “escapers” that survived exhibited no overt abnormal phenotypes.
Table 2.
Viability of adults expressing mutant lamins in larval body wall muscle.
| LamC | # Live Adults | # Dead Pupae | % Adult Viability | n | p-Value |
|---|---|---|---|---|---|
| wt | 709 | 7 | 99.0 | 716 | |
| S37L | 282 | 431 | 39.6 | 713 | <0.00001 |
| L74R | 253 | 466 | 35.2 | 719 | <0.00001 |
| R237P | 0 | 1162 | 0.0 | 1162 | NA |
| G478V | 1 | 384 | 0.3 | 385 | <0.00001 |
| G489V | 20 | 1427 | 1.4 | 1447 | <0.00001 |
| G514V | 0 | 17 | 0.0 | 17 | NA |
3.2. Mutant lamins mislocalized and caused nuclear shape changes
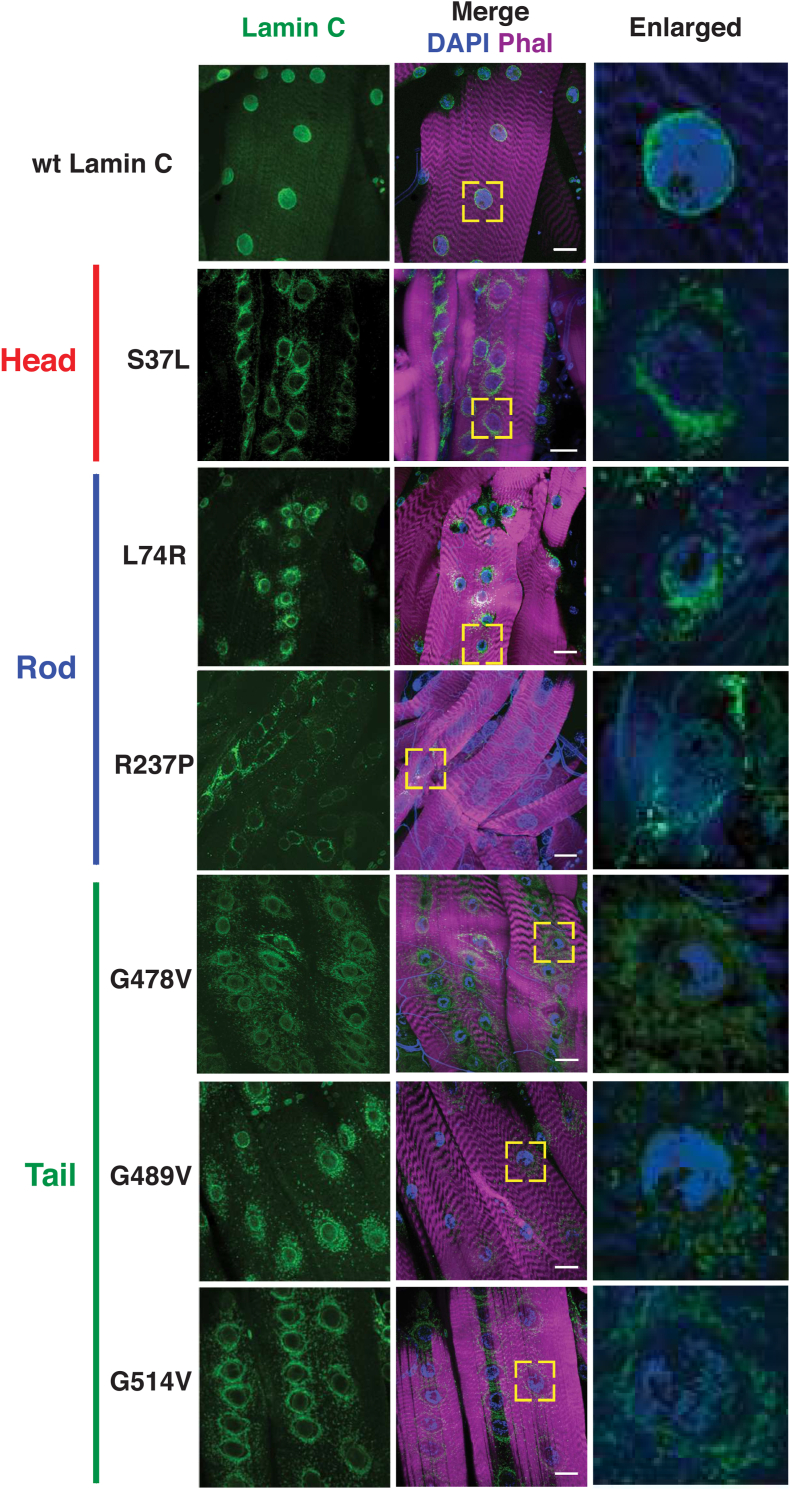
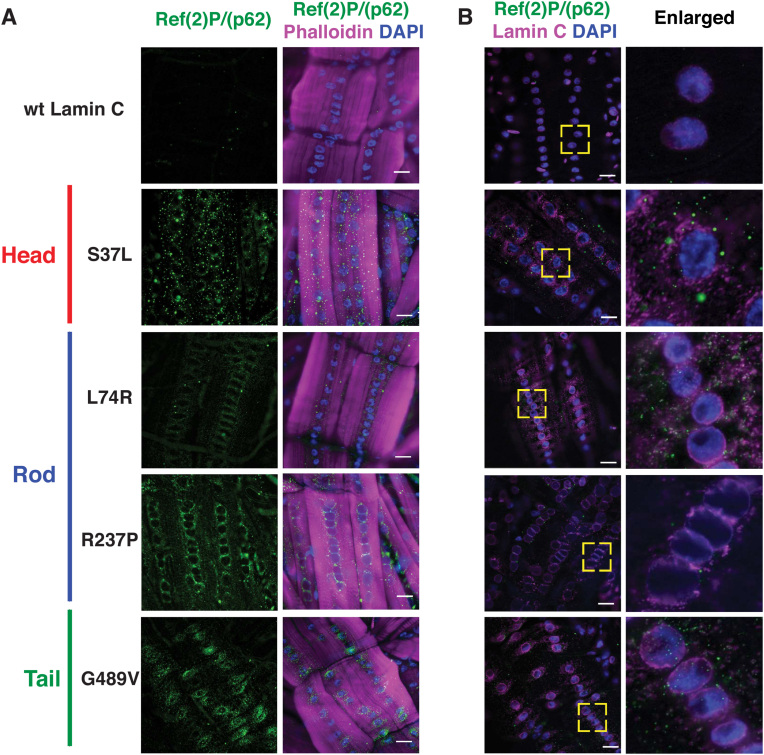
The motility and survival assays were consistent with loss of muscle function; therefore, we examined larval body wall muscles at the cellular level. Third instar larvae were dissected and stained with phalloidin (filamentous actin), DAPI (DNA) and antibodies to nuclear envelope proteins. The muscles of larvae expressing wild-type LamC showed fairly evenly spaced nuclei along the length of the fiber and nuclear envelope localization of LamC as anticipated (Fig. 2). By contrast, muscles expressing mutant LamC showed a range of defects depending on the particular mutation. Relative to wildtype, all mutants displayed increased staining with anti-LamC antibodies in the cytoplasm (Table S2). Muscles expressing LamC S37L showed increased perinuclear lamin localization. Similarly, LamC L74R showed perinuclear localization and LamC aggregation throughout the cytoplasm. Muscles expressing LamC R237P showed enriched localization between nuclei, with little staining at the nuclear envelope. Expression of LamC G478V, G489V, and G514V, which all map to the Ig-like fold domain, showed enriched rings of lamin localization around the nuclei. Taken together, each of the mutant lamins displayed abnormal localization.
Fig. 2.
Amino acid substitutions in all three LamC protein domains disrupt localization of LamC in muscle. Immunohistochemistry was performed on third instar larval body wall muscles using antibodies to LamC (green), Phalloidin (magenta), and DAPI (blue). The larvae expressed either wild-type LamC or mutant LamC driven by the C57 Gal4 driver. Abnormal nuclear migration and perinuclear localization were observed with all mutant lamins, but not with wild-type. The yellow boxes indicate the magnified regions shown at the right. Scale bar, 30 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Mutations impacting each of the three lamin protein domains caused reductive stress
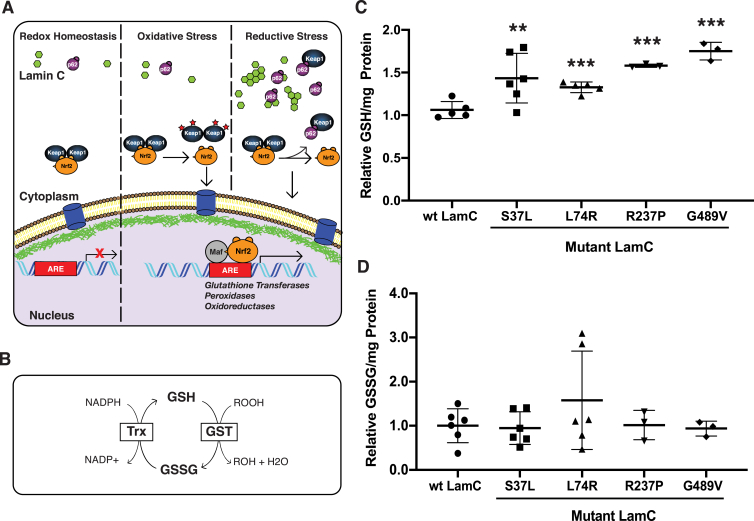
In our prior studies, mutations that impacted the LamC Ig-like fold [23] activated genes regulated by Cap-n-CollarC (CncC), a transcription factor that is the orthologue of human Nrf2 [36,37]. CncC/Nrf2, binds antioxidant response elements (AREs) in the promoter regions of redox modulating genes (Fig. 3A) [36]. Under redox homeostasis, CncC/Nrf2 is anchored in the cytoplasm by Keap1 [38]. Upon redox imbalance CncC/Nrf2 translocates to the nucleus where it activates redox regulating genes in combination with its partner small Maf [38,39]. Translocation of CncC/Nrf2 into the nucleus occurs under conditions of redox imbalance (Fig. 3A) [38].
Fig. 3.
Mutant LamC alters redox homeostasis. (A) A model illustrating the Nrf2/Keap1 pathway under redox homeostasis, oxidative stress, and reductive stress. Under redox homeostasis, the pathway is inactive, with Nrf2 (orange) being rapidly degraded through the ubiquination pathway. Under oxidative stress, cysteine residues in Keap1 are oxidized, preventing Nrf2 binding. This permits Nrf2 to translocate into the nucleus, form a heterodimer with small Maf (grey) at antioxidant responsive elements (AREs), and activate Nrf2 target genes. Under reductive stress, cytoplasmic aggregates (green) cause increased levels of the autophagy chaperone p62 (purple). p62 competes with Nrf2 for Keap1 binding, leading to free Nrf2 in the cytoplasm, which translocates into the nucleus, forms a heterodimer with small Maf (grey) at antioxidant responsive elements (AREs), and activates antioxidant genes. (B) A diagram of the pathway for production of reduced glutathione (GSH) and oxidized glutathione (GSSG) in Drosophila is shown. (C) The relative amount of GSH per milligram of total protein in third instar larval muscle expressing either wild-type or mutant LamC is shown. Each point represents an independent biological replicate. (D) The relative amount of GSSG per milligram of total protein in third instar larval muscle expressing either wild-type or mutant LamC is shown. Concentrations were determined via a GSH→GSSG recycling assay using three to five independent biological samples from separate genetic crosses. An ANOVA analysis was performed to test for significance: **p < 0.01; ***, p < 0.001. LamC L74R showed the most variability among the independent biological replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To determine the redox status of muscles expressing mutant LamC, we measured the redox state of glutathione. Glutathione is a major antioxidant within cells and is used by the cell to prevent fluctuations in redox status [40]. Relative amounts of reduced glutathione (GSH) and glutathione disulfide (GSSG) are indicators of the redox status of cells [41]. Oxidation of GSH to GSSG occurs via glutathione S-transferases (GST), with an oxygen radical reduced to a less reactive product in the process. In humans, the reaction from GSSG to GSH is catalyzed by glutathione reductase [40]. In Drosophila, however, this reaction is carried out by thioredoxin [[42], [43], [44]]. In both cases, reduction consumes NADPH (Fig. 3B).
To quantitate GSH and GSSG, enzymatic assays were carried out on dissected third instar larval body wall muscles expressing either wild-type LamC or mutant LamC. GSH and GSSG were measured according to published protocols [26]. Measurements were made on three to five independent biological samples per genotype. All four LamC mutants tested showed elevated levels of GSH relative to the control (Fig. 3C). By contrast, no significant differences in GSSG levels relative to the control were observed among the muscles expressing mutant LamC (Fig. 3D). Collectively, these data demonstrate that mutations impacting any of the three LamC domains can alter redox status. Furthermore, the elevated GSH levels are indicative of reductive stress [[45], [46], [47]].
3.4. Patient-derived cardiomyocytes show nuclear deformities and abnormal response to NAC
Given the cellular defects caused by mutant lamins in Drosophila, we asked whether similar defects were observed in human cells possessing LMNA mutations. To accomplish this, dermal fibroblasts were obtained from two individuals with muscular dystrophy-causing point mutations in LMNA and from two healthy controls. The LMNA point mutations caused amino acid substitutions in the rod and Ig-like fold domain, R249W and G449V, respectively. Fibroblasts were dedifferentiated into induced pluripotent (iPS) cells using standard procedures [[48], [49], [50]]. These iPS cells expressed the appropriate stem cell markers (Fig. S2) and showed the expected karyotype (Fig. S3) [51]. The iPS cells were differentiated into cardiomyocytes using standard procedures and stained with antibodies to Troponin T used as a marker of cardiomyocyte differentiation (Fig. S2) [48]. The iPS cell derived cardiomyocytes possessing LMNA mutations exhibited increased nuclear lobulations relative to controls (Fig. 4A and Table S3). Similar nuclear lobulations have been observed in human cells from individuals with Hutchinson-Gilford progeria, an early onset aging syndrome caused by mutations in LMNA [[52], [53], [54], [55]].
To determine the redox status of the iPS cell derived cardiomyocytes, GSH and GSSG levels were measured using the GSH/GSSG-Glo Assay (V6611; Promega, Madison) according to manufacturer's instructions. The LMNA patient derived cardiomyocytes showed no difference in GSH and GSSG levels relative to controls (Fig. 4B). However, when treated with N-Acetyl Cysteine (NAC), they exhibited an abnormal response. While NAC treatment led to increased levels of total glutathione in control cardiomyocytes (Fig. S4), mutant LMNA patient-derived cardiomyocytes paradoxically displayed decreased levels of total glutathione. Taken together, these findings suggest that the cultured cardiomyocytes possess an altered ability to maintain redox homeostasis upon addition of a neutralizing agent.
3.5. Mutant lamins increased cytoplasmic levels of p62/Ref(2)P
Reductive stress has been observed in many different disease conditions in a variety of organisms and cell types, all of which are associated with protein aggregation [26,46,56,57]. Organisms have evolved many pathways to degrade protein aggregates [58]. The exact mechanisms of A-type lamin degradation are not well understood; however, B-type lamins are degraded by autophagy [59]. The autophagic process is facilitated by association of unfolded and/or aggregated proteins with the autophagy chaperone p62 (also known as SQSTM1) [[60], [61], [62]]. The Drosophila orthologue of p62 is refractory to Sigma P [Ref(2)P] [63,64]. Given the cytoplasmic aggregation of mutant LamC in larval body wall muscles, dissected and fixed muscles were stained with antibodies to Ref(2)P/p62. Muscles expressing wild-type LamC showed little to no staining for Ref(2)P/p62. This was anticipated, given that under normal circumstances p62 is rapidly turned over through ubiquitination [65]. By contrast, muscles expressing mutant LamC showed increased cytoplasmic staining for Ref(2)P/p62 (Fig. 5A). Given that Ref(2)P/p62 is a target gene for CncC/Nrf2 [66,67], we wondered if the increased protein levels of Ref(2)P/p62 resulted from increased transcription. RT-qPCR analysis revealed that transcription of Ref(2)P/p62 was increased, consistent with increased levels of nuclear Cnc/Nrf2 (Fig. S5).
Fig. 5.
Mutant LamC caused cytoplasmic accumulation of Ref(2)P/p62. (A) Immunohistochemistry of larval body wall muscles stained with antibodies to Ref(2)P/p62 (green), Phalloidin (magenta) and DAPI (blue). All LamC mutants caused increased staining with antibodies to Ref(2)P/p62 relative to the wild-type control. (B) Immunohistochemistry of larval body wall muscles stained with antibodies to Ref(2)P/p62 (green), Lamin C (Magenta), and DAPI (blue). Only a subset of the Ref(2)P/p62 foci overlap with staining for LamC as seen in the enlarged images. The yellow boxes indicate the region magnified (right). Scale bar; 30 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We noted that perinuclear localization of Ref(2)P/p62 was reminiscent of the perinuclear localization of mutant LamC, suggestive of co-localization. To test for this, larval body wall muscles were co-stained with antibodies for LamC and Ref(2)P/p62. The two antibodies showed partial overlap (Fig. 5B), suggesting the possibility that additional proteins might be accumulating in the cytoplasm. This idea is supported by the fact that mutant lamins cause cytoplasmic localization of nuclear pores and other nuclear envelope proteins [20,54].
3.6. Genetic and pharmacological treatments suppressed lethality caused by mutant LamC
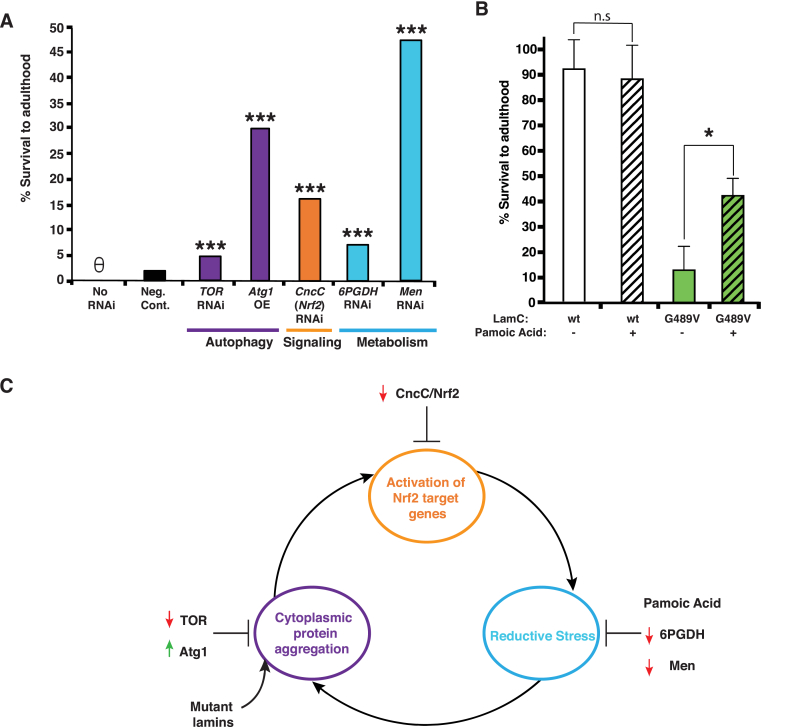
The current hypothesis for the underlying mechanism of reductive stress is that it is triggered by cytoplasmic protein aggregation (Fig. 3A) [25,57]. As an in vivo test of this hypothesis, an RNAi transgene against target of rapamycin (TOR) was expressed in larval body wall muscles with mutant LamC. It is well documented that decreasing TOR activity increases autophagy [[68], [69], [70], [71]]. In a wild-type genetic background, expression of LamC G489V reduced adult viability to 1.4%. Therefore, an increased percentage of adult survivors would indicate genetic suppression of lethality caused by mutant LamC. Expression of the RNAi against TOR partially suppressed lethality, increasing adult viability to 4.8% (Fig. 6A). As a second means of increasing autophagy, a transgene encoding AuTophaGy related (Atg1), a serine/threonine kinase essential for construction of the autophagosome, was over-expressed in muscles expressing LamC G449V (Fig. 6A) [72]. Over-expression of Atg1 partially suppressed lethality, increasing adult viability to 30% (Fig. 6A). Thus, two independent genetic manipulations that increased autophagy partially suppressed lethality.
Fig. 6.
Genetic and pharmacological suppression of muscle defects caused by mutant Lamin C. (A) Graph of the percentage of live adults produced from crosses between flies expressing mutant LamC (G489V) via the C57 larval muscle-specific Gal4 driver and a stock possessing either a Gal4-activated RNAi or an over-expressed transgene as indicated. Expression of mutant LamC (no RNAi) caused death at the pupal stage with no surviving adults. Expression of a non-specific luciferase RNAi (neg. control) allowed survival of only a few adults. By contrast, partial suppression of lethality was observed for the RNAi and over-expression transgenes. Approximately 200–300 total progeny were scored per genotype. Unpaired Student's t-tests were performed for statistical analysis. θ = no survivors. (B) The percent of viable adults after treatment with either 5 mM pamoic acid or vehicle (dH20) only was plotted. Viability was calculated as described for (A). Pamoic acid treatment had no effect on the viability of flies expressing wild-type LamC. By contrast, treatment increased the survival of flies expressing mutant LamC. Unpaired Student's t-tests were performed for statistical analysis. (C) A model illustrating a self-perpetuating cycle that is initiated by cytoplasmic aggregation of nuclear envelope proteins, leads to the activation of the Nrf2/Keap1 pathway, resulting in reductive stress, which promotes cytoplasmic aggregation. The reductive environment promotes additional protein aggregation, causing the cycle to repeat. Red arrows indicate suppression via RNAi, and green arrows indicate overexpression. *, p < 0.05; ***, p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The Drosophila models of LMNA muscular dystrophy exhibited properties consistent with reductive stress (Fig. 3), therefore, we tested redox regulators for suppression of lethality. To shift the redox balance towards an oxidative state, RNAi transgenes against 6-phosphogluconate dehydrogenase (6PGDH) and malic enzyme (MEN) were independently expressed in muscle with LamC G489V (Fig. 6A). Both enzymes produce NADPH; therefore, knock-down is predicted to lower the reducing potential in the muscles. Expression of MEN and 6PGDH RNAi transgenes partially suppressed adult lethality, yielding 44% and 7% adult viability, respectively. Thus, lowering the reducing potential in muscle might be a novel therapeutic approach to LMNA muscular dystrophies.
Reductive stress is associated with activation of CncC/Nrf2 target genes, such as Gpx2 and G6PD [73]. To determine if this altered gene expression is a contributing factor to lethality, an RNAi transgene against CncC/Nrf2 was expressed in muscles with LamC G489V. The result was a partial suppression of lethality, giving rise to 36% adult viability. This finding suggests the Nrf2/Keap1 pathway as a potential therapeutic target for LMNA muscular dystrophies. Efficiency of RNAi and overexpression was evaluated by RT-qPCR (Fig. 6S). All genetic interventions achieved significant suppression/overexpression except for CncC/Nrf2 which showed a trend towards statistical significance with a p-value of 0.06.
Given that the RNAi transgene against Men was the most effective in suppressing lethality caused by mutant LamC, we took a complementary pharmacological approach. Pamoic acid is an allosteric inhibitor of malic enzyme. The compound binds at the Men dimer interface, adjacent to the fumarate binding site, and inhibits enzymatic activity [74]. To test for pamoic acid effects, third instar larvae expressing either wild-type or LamC G489V were fed standard food containing either 5 mM pamoic acid or vehicle (dH2O) only throughout the larval stages. Pamoic acid had no overt deleterious or beneficial effects on larvae expressing wild-type LamC compared to untreated controls. By contrast, larvae expressing LamC G489V showed 42% adult viability compared to controls exhibiting 13% adult viability (Fig. 6B). Thus, two independent methods of reducing Men activity suppressed lethality caused by mutant LamC, suggesting Men inhibition as a potential therapy for LMNA muscular dystrophies.
4. Discussion
A-type lamins have been linked to many cellular functions, including supporting nuclear structure, serving as a scaffold for interacting proteins, and organizing the genome [[13], [14], [15]]; however, how mutant lamins cause muscle disease remains poorly understood. Here we show using Drosophila models that amino acid substitutions in each of the three lamin domains (head, rod and tail) cause unique patterns of lamin mislocalization and distinct nuclear defects; however, they share a common disruption of redox homeostasis (Fig. 2, Fig. 3). The maintenance of redox homeostasis is critical for muscle cells as ROS is produced during muscle contractions [2]. Given the damaging effects of ROS on cellular components and functions, redox homeostasis is tightly regulated in healthy muscles [75].
The Nrf2/Keap1 pathway is the main cellular defense mechanism against oxidative stress. During conditions of oxidative stress, Nrf2 deploys the action of ∼500 gene products to combat reactive oxygen species [1,76]. These gene products protect against neurodegeneration, improve cardiovascular health, reduce inflammation, and slow aging [76] Under pathological conditions, constitutive activation of Nrf2 target genes causes reductive stress [25]. This often-over-looked redox state is characterized by increased levels of reducing equivalents such as GSH and NADPH [[77], [78], [79]]. Reductive stress has been observed in an increasing number of instances including Drosophila models of cardiac skeletal muscle and retinal degeneration disease, mouse models of cardiac disease, and in the serum of humans predisposed to Alzheimer's Disease [23,26,46,80].
The mechanisms by which reductive stress cause pathogenesis are not well understood although non-mutually exclusive hypotheses have been proposed. These include the hypothesis that high levels of reducing equivalents, associated with increased levels of total glutathione by NAC treatment, enters the mitochondria, altering electron transport chain function, which leads paradoxically to net mitochondrial superoxide generation and cytotoxicity [47]. Alternatively, during periods of limited GSSG availability, reducing equivalents such as NADPH/FADH2 initiate the shift of electrons to molecular oxygen as an alternative electron acceptor to produce superoxide and hydrogen peroxide [47,81]. Consistent with these hypotheses, we showed elevated reducing equivalent levels in control cardiomyocytes by NAC treatment and decreased total glutathione by mutant lamins, respectively (Fig. S4). Another hypothesis posits that excessive glutathionylation of target proteins contributes to mitochondrial oxidation [82]. Accordingly, we showed that muscle-specific expression of RNAi transgenes that knock-down levels of NADPH-producing enzymes suppressed lethality caused by mutant lamins (Fig. 6). In support of this finding, treatment with pamoic acid (also known as embonic acid), an allosteric inhibitor of malic enzyme inhibitor, partially suppressed lethality caused by mutant lamins (Fig. 6). Pamoic acid is currently used as an additive to psychoactive compounds, anthelmintic medications, and peptide hormones to increase their duration-of-action [74,83]. Taken together, our findings warrant further investigations into pamoic acid and other malic enzyme inhibitors as potential therapies for LMNA-associated muscle disease.
Reductive stress seen in disease conditions is associated with cytoplasmic protein aggregation. For example, mouse models revealed that mutant forms of the chaperone CryAB aggregate in the cytoplasm of cardiomyocytes, leading to reductive stress and dilated cardiomyopathy [26,84,85]. Likewise, in Drosophila models, mutant forms of lamin cause cytoplasmic aggregation of envelope proteins in muscle, leading to a plethora of muscle defects and reduced muscle function [20,86]. In these cases, protein aggregation is associated with increased cytoplasmic levels of the autophagy chaperone Ref(2)P/p62 [23], a factor used to quantitatively assess aggregate clearance in tissues (Fig. 5) [87]. Increased Ref(2)P/p62 is an indicator of reduced rates of autophagy, a process that maintains the balance between newly synthesized proteins and the elimination of improperly folded and damaged proteins [87]. Our finding that over-expression of Atg1 partially suppressed lethality caused by mutant lamins suggests that increasing the rate of autophagy offers benefits for LMNA-associated muscle disease (Fig. 6). These results are consistent with those observed upon treatment of laminopathy mouse models with rapamycin, a compound that inhibits mTOR activity [88]. In this case, increasing autophagy partially suppressed muscle defects caused by loss of lamin A/C [88,89]. These findings emphasize the importance of protein aggregation in the pathogenesis of muscle disease.
Protein aggregation has been observed for many diseases including liver injury and neurological disorders such as Parkinson's and Alzheimer's [78,90,91]. Reasons for cytoplasmic aggregation are often unclear. It is postulated that when machinery responsible for proteostasis is compromised, misfolded and damaged proteins accumulate in the cytoplasm. Interestingly, cytoplasmic, but not nuclear, protein aggregation disrupts nucleocytoplasmic transport of proteins and RNA [92], suggesting the existence of a cytoplasmic sensor that regulates nuclear pore function.
There are interconnected relationships among cytoplasmic protein aggregation, Ref2(P)/p62 accumulation, CncC(Nrf2)/Keap1 pathway activation, autophagy, and reductive stress. For example, CncC/Nrf2 binds to ARE elements in the promoter region of the Ref(2)P/p62 gene, resulting in production of additional p62 that competes with CncC/Nrf2 for association with Keap1 [66,67,93]. In addition, mTOR -dependent phosphorylation of p62 increases the binding affinity of p62 for Keap1 [94]. Both of these events generate a positive feedback loop reinforcing the expression of CncC/Nrf2 target genes and promoting reductive stress. Based on the relatively small increase in expression relative to the large increase seen by immunohistochemistry, the increase in Ref(2)P/p62 is likely a result of aggregation more than increased transcription. (Fig. 5 and S5). The reducing environment associated with reductive stress can cause aberrant protein folding and aggregation. Relevant to our study, single amino acid substitutions in the lamin A/C Ig-fold domain caused abnormal tetramerization under reducing conditions [95]. Collectively, multiple positive feedback loops provide mechanisms that make it challenging to re-establish redox homeostasis (Fig. 6C).
Disruption of redox homeostasis has been implicated in several types of muscular dystrophy other than those caused by mutations in LMNA. Duchenne muscular dystrophy is associated with increased levels of oxidized glutathione and oxidative damage in muscles that correlates with increased severity of muscle defects and disease progression [96]. SEPN1‐related myopathy, caused by mutations in the gene encoding Selenoprotein N1, is characterized by high levels of basal ROS and increased susceptibility to H2O2 exposure [97]. Facioscapulohumeral muscular dystrophy myoblasts show decreased expression of antioxidant genes and increased lethality when exposed to H2O2 [98]. Interestingly, treatment with the ROS scavenger N-acetyl cysteine or antioxidants such as vitamin A, has improved muscle function in these types of muscular dystrophy [[96], [97], [98]]. Notwithstanding, we posit that pro-reducing redox states among heterogeneous clinical cohorts and/or the pro-reducing effects of antioxidant agents will, either alone or in combination, promote life-threatening reductive stress.
Mutant lamins cause redox imbalance in diseases other than muscle laminopathies including Hutchinson-Gilford Progeria and Dunnigan-type familial partial lipodystrophy (FPLD). In the early onset aging syndrome Hutchinson-Gilford progeria, an accumulation of farnesylated pre-lamin A occurs at the nuclear envelope, which correlates with increased activity of manganese superoxide dismutase activity and ROS production [99]. FPLD is characterized by decreased expression of cytochrome oxidase IV, the final electron acceptor of the electron transport chain, and over-production of ROS. These two altered properties were reversed by inhibition of farnesylation, which decreased the accumulation of pre-lamin A at the nuclear envelope [100]. Collectively, these findings show that disruption of redox homeostasis appears to be a common theme in laminopathies.
The mechanisms by which lamins regulate redox imbalance are not well understood. Mutation of three cysteine residues (Cys 522, 588 and 591) in A-type lamins to alanine residues increased susceptibility to mild oxidative stress and induced gene expression similar to that seen in chronic ROS exposure, suggesting lamins “buffer” the effects of ROS [101]. The buffering effect might be related to a 300 nm “nuclear shield” that surrounds the nucleus and is thought to provide protection from oxidative damage [102]. The nuclear shield consists of a high concentration of anti-ROS enzymes such as catalase, glutathione peroxidase, and GSTs [102]. In addition, Nesprins, components of the LINC complex, which make physical connections between the nucleus and cytoskeleton, are important for nuclear shield formation [103]. Given that mutant lamins cause the mislocalization of Nesprins, they might destabilize the nuclear shield and increase sensitivity to redox damage [104,105].
Powerful experimental tools have been developed in recent years to determine the functions of lamins, including their role in redox homeostasis. A major breakthrough has been the ability to generate induced pluripotent stem (iPS) cells from LMNA patient fibroblasts, which are then differentiated into relevant cell types [48]. This ability allows studies to be performed in the genetic background of the patient. This is important because pathological features of LMNA-associated diseases are modified by genetic background [106]. In addition, such cells are needed for testing of potential therapeutic compounds, which often fail due to unique features of an individual's metabolism. As a case in point, the use of patient-derived cardiomyocytes showed that inhibition of platelet derived growth factor (PDGF) eliminated arrythmias in pre-clinical models [107].
Here, we generated two unique patient-based LMNA iPS cell lines (Fig. 4). When differentiated into cardiomyocytes, these cells show nuclear lobulations, which are characteristic of other LMNA cell culture models, patient fibroblasts, and human muscle biopsy tissue [27,55,108,109]. We show that the LMNA patient-derived cardiomyocytes exhibited an abnormal response to NAC treatment compared to that of controls (Fig. S4), suggesting altered regulation across the oxido-reductive spectrum. Thus, the LMNA cell lines recapitulate features observed in humans and Drosophila models, collectively providing multiple experimental systems to test potential therapeutic compounds in the future. Further, targeting pathways linked to NRF2-dependent regulation and redox homeostasis remains an attractive hypothesis for individualized therapy in humans.
Author contributions
G.S.C., J. R-M., S.L., Q.D. participated in experimental design and execution and manuscript preparation. A.C.G., M.R.K., M.F.V. and N.U. participated in experimental execution. I.J.B. was involved in project oversight and manuscript preparation. L.L.W. performed project oversight, participated in experimental design and execution, and manuscript preparation.
Funding sources
Funding was provided by the Muscular Dystrophy Association (Award ID 477283) to L.L.W. and I.J.B. and NIH NIAMS (grant number AR075193) to L.L.W. The University of Iowa Carver College of Medicine Foster Undergraduate Talent Future – Uniting Research and Education in Biomedicine provided support for G.S.C. and M.F.V.
Declaration of competing interests
None
Acknowledgements
We thank D. Thiemann and the University of Iowa Free Radical and Radiation Biology Program Research Core for technical assistance and the Bloomington and Vienna Stock Centers for fly stocks.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102196.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies K.J., Quintanilha A.T., Brooks G.A., Packer L. Free radicals and tissue damage produced by exercise. Biochem. Biophys. Res. Commun. 1982;107:1198–1205. doi: 10.1016/s0006-291x(82)80124-1. [DOI] [PubMed] [Google Scholar]
- 3.Di Meo S., Reed T.T., Venditti P., Victor V.M. Role of ROS and RNS sources in physiological and pathological conditions. Oxid Med Cell Longev. 2016:1245049. doi: 10.1155/2016/1245049. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nissanka N., Moraes C.T. Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease. FEBS Lett. 2018;592:728–742. doi: 10.1002/1873-3468.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granger D.N., Kvietys P.R. Reperfusion injury and reactive oxygen species: the evolution of a concept. Redox Biol. 2015;6:524–551. doi: 10.1016/j.redox.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ursini F., Maiorino M., Forman H.J. Redox homeostasis: the Golden Mean of healthy living. Redox Biol. 2016;8:205–215. doi: 10.1016/j.redox.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kourakis S., Timpani C.A., de Haan J.B., Gueven N., Fischer D., Rybalka E. Targeting Nrf2 for the treatment of Duchenne muscular dystrophy. Redox Biol. 2021;38:101803. doi: 10.1016/j.redox.2020.101803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitehead N.P., Kim M.J., Bible K.L., Adams M.E., Froehner S.C. A new therapeutic effect of simvastatin revealed by functional improvement in muscular dystrophy. Proc. Natl. Acad. Sci. U. S. A. 2015;112:12864–12869. doi: 10.1073/pnas.1509536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez B.M., Khouzami L., Decostre V., Varnous S., Pekovic-Vaughan V., Hutchison C.J., Pecker F., Bonne G., Muchir A. N-acetyl cysteine alleviates oxidative stress and protects mice from dilated cardiomyopathy caused by mutations in nuclear A-type lamins gene. Hum. Mol. Genet. 2018;27:3353–3360. doi: 10.1093/hmg/ddy243. [DOI] [PubMed] [Google Scholar]
- 10.Gao Q.Q., McNally E.M. The dystrophin complex: structure, function, and implications for therapy. Comp. Physiol. 2015;5:1223–1239. doi: 10.1002/cphy.c140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonne G., Mercuri E., Muchir A., Urtizberea A., Becane H.M., Recan D., Merlini L., Wehnert M., Boor R., Reuner U., Vorgerd M., Wicklein E.M., Eymard B., Duboc D., Penisson-Besnier I., Cuisset J.M., Ferrer X., Desguerre I., Lacombe D., Bushby K., Pollitt C., Toniolo D., Fardeau M., Schwartz K., Muntoni F. Clinical and molecular genetic spectrum of autosomal dominant Emery-Dreifuss muscular dystrophy due to mutations of the lamin A/C gene. Ann. Neurol. 2000;48:170–180. [PubMed] [Google Scholar]
- 12.Lu J.T., Muchir A., Nagy P.L., Worman H.J. LMNA cardiomyopathy: cell biology and genetics meet clinical medicine. Dis Model Mech. 2011;4:562–568. doi: 10.1242/dmm.006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palka M., Tomczak A., Grabowska K., Machowska M., Piekarowicz K., Rzepecka D., Rzepecki R. Laminopathies: what can humans learn from fruit flies. Cell. Mol. Biol. Lett. 2018;23:32. doi: 10.1186/s11658-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann H., Bar H., Kreplak L., Strelkov S.V., Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat. Rev. Mol. Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- 15.de Leeuw R., Gruenbaum Y., Medalia O. Nuclear lamins: thin filaments with major functions. Trends Cell Biol. 2018;28:34–45. doi: 10.1016/j.tcb.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Samson C., Petitalot A., Celli F., Herrada I., Ropars V., Le Du M.H., Nhiri N., Jacquet E., Arteni A.A., Buendia B., Zinn-Justin S. Structural analysis of the ternary complex between lamin A/C, BAF and emerin identifies an interface disrupted in autosomal recessive progeroid diseases. Nucleic Acids Res. 2018;46:10460–10473. doi: 10.1093/nar/gky736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turgay Y., Medalia O. The structure of lamin filaments in somatic cells as revealed by cryo-electron tomography. Nucleus. 2017;8:475–481. doi: 10.1080/19491034.2017.1337622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puckelwartz M.J., Depreux F.F., McNally E.M. Gene expression, chromosome position and lamin A/C mutations. Nucleus. 2011;2:162–167. doi: 10.4161/nucl.2.3.16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holaska J.M., Wilson K.L., Mansharamani M. The nuclear envelope, lamins and nuclear assembly. Curr. Opin. Cell Biol. 2002;14:357–364. doi: 10.1016/s0955-0674(02)00329-0. [DOI] [PubMed] [Google Scholar]
- 20.Dialynas G., Flannery K.M., Zirbel L.N., Nagy P.L., Mathews K.D., Moore S.A., Wallrath L.L. LMNA variants cause cytoplasmic distribution of nuclear pore proteins in Drosophila and human muscle. Hum. Mol. Genet. 2012;21:1544–1556. doi: 10.1093/hmg/ddr592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwerger M., Jaalouk D.E., Lombardi M.L., Isermann P., Mauermann M., Dialynas G., Herrmann H., Wallrath L.L., Lammerding J. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum. Mol. Genet. 2013;22:2335–2349. doi: 10.1093/hmg/ddt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuleesha Y., Puah W.C., Wasser M. Live imaging of muscle histolysis in Drosophila metamorphosis. BMC Dev. Biol. 2016;16:12. doi: 10.1186/s12861-016-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dialynas G., Shrestha O.K., Ponce J.M., Zwerger M., Thiemann D.A., Young G.H., Moore S.A., Yu L., Lammerding J., Wallrath L.L. Myopathic lamin mutations cause reductive stress and activate the nrf2/keap-1 pathway. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baird L., Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 2020;40 doi: 10.1128/MCB.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narasimhan M., Rajasekaran N.S. Reductive potential - a savior turns stressor in protein aggregation cardiomyopathy. Biochim. Biophys. Acta. 2015;1852:53–60. doi: 10.1016/j.bbadis.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajasekaran N.S., Connell P., Christians E.S., Yan L.J., Taylor R.P., Orosz A., Zhang X.Q., Stevenson T.J., Peshock R.M., Leopold J.A., Barry W.H., Loscalzo J., Odelberg S.J., Benjamin I.J. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandran S., Suggs J.A., Wang B.J., Han A., Bhide S., Cryderman D.E., Moore S.A., Bernstein S.I., Wallrath L.L., Melkani G.C. Suppression of myopathic lamin mutations by muscle-specific activation of AMPK and modulation of downstream signaling. Hum. Mol. Genet. 2019;28:351–371. doi: 10.1093/hmg/ddy332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caygill E.E., Brand A.H. The GAL4 system: a versatile system for the manipulation and analysis of gene expression. Methods Mol. Biol. 2016;1478:33–52. doi: 10.1007/978-1-4939-6371-3_2. [DOI] [PubMed] [Google Scholar]
- 29.Gorczyca D., Ashley J., Speese S., Gherbesi N., Thomas U., Gundelfinger E., Gramates L.S., Budnik V. Postsynaptic membrane addition depends on the Discs-Large-interacting t-SNARE Gtaxin. J. Neurosci. 2007;27:1033–1044. doi: 10.1523/JNEUROSCI.3160-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 31.Pethig K., Genschel J., Peters T., Wilhelmi M., Flemming P., Lochs H., Haverich A., Schmidt H.H. LMNA mutations in cardiac transplant recipients. Cardiology. 2005;103:57–62. doi: 10.1159/000082048. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen D., Leistritz D.F., Turner L., MacGregor D., Ohson K., Dancey P., Martin G.M., Oshima J. Collagen expression in fibroblasts with a novel LMNA mutation. Biochem. Biophys. Res. Commun. 2007;352:603–608. doi: 10.1016/j.bbrc.2006.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPherson E., Turner L., Zador I., Reynolds K., Macgregor D., Giampietro P.F. Ovarian failure and dilated cardiomyopathy due to a novel lamin mutation. Am. J. Med. Genet. 2009;149A:567–572. doi: 10.1002/ajmg.a.32627. [DOI] [PubMed] [Google Scholar]
- 34.Budnik V., Koh Y.H., Guan B., Hartmann B., Hough C., Woods D., Gorczyca M. Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron. 1996;17:627–640. doi: 10.1016/s0896-6273(00)80196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weitkunat M., Schnorrer F. A guide to study Drosophila muscle biology. Methods. 2014;68:2–14. doi: 10.1016/j.ymeth.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 36.Sykiotis G.P., Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng H., Kerppola T.K. Visualization of the Drosophila dKeap1-CncC interaction on chromatin illumines cooperative, xenobiotic-specific gene activation. Development. 2014;141:3277–3288. doi: 10.1242/dev.110528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitoniak A., Bohmann D. Mechanisms and functions of Nrf2 signaling in Drosophila. Free Radic. Biol. Med. 2015;88:302–313. doi: 10.1016/j.freeradbiomed.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katsuoka F., Yamamoto M. Small Maf proteins (MafF, MafG, MafK): history, structure and function. Gene. 2016;586:197–205. doi: 10.1016/j.gene.2016.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forman H.J., Zhang H., Rinna A. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol. Aspect. Med. 2009;30:1–12. doi: 10.1016/j.mam.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schafer F.Q., Buettner G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 42.Winterbourn C.C., Metodiewa D. The reaction of superoxide with reduced glutathione. Arch. Biochem. Biophys. 1994;314:284–290. doi: 10.1006/abbi.1994.1444. [DOI] [PubMed] [Google Scholar]
- 43.Kanzok S.M., Fechner A., Bauer H., Ulschmid J.K., Muller H.M., Botella-Munoz J., Schneuwly S., Schirmer R., Becker K. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–646. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- 44.Sies H. Glutathione and its role in cellular functions. Free Radic. Biol. Med. 1999;27:916–921. doi: 10.1016/s0891-5849(99)00177-x. [DOI] [PubMed] [Google Scholar]
- 45.Handy D.E., Loscalzo J. Responses to reductive stress in the cardiovascular system. Free Radic. Biol. Med. 2017;109:114–124. doi: 10.1016/j.freeradbiomed.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lloret A., Fuchsberger T., Giraldo E., Vina J. Reductive stress: a new concept in alzheimer's disease. Curr. Alzheimer Res. 2016;13:206–211. doi: 10.2174/1567205012666150921101430. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H., Limphong P., Pieper J., Liu Q., Rodesch C.K., Christians E., Benjamin I.J. Glutathione-dependent reductive stress triggers mitochondrial oxidation and cytotoxicity. Faseb. J. 2012;26:1442–1451. doi: 10.1096/fj.11-199869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Limphong P., Zhang H., Christians E., Liu Q., Riedel M., Ivey K., Cheng P., Mitzelfelt K., Taylor G., Winge D., Srivastava D., Benjamin I. Modeling human protein aggregation cardiomyopathy using murine induced pluripotent stem cells. Stem Cells Transl Med. 2013;2:161–166. doi: 10.5966/sctm.2012-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez-Izquierdo A., Warren M., Riedel M., Cho S., Lai S., Lux R.L., Spitzer K.W., Benjamin I.J., Tristani-Firouzi M., Jou C.J. A near-infrared fluorescent voltage-sensitive dye allows for moderate-throughput electrophysiological analyses of human induced pluripotent stem cell-derived cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2014;307:H1370–H1377. doi: 10.1152/ajpheart.00344.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDermott-Roe C., Lv W., Maximova T., Wada S., Bukowy J., Marquez M., Lai S., Shehu A., Benjamin I., Geurts A., Musunuru K. Investigation of a dilated cardiomyopathy-associated variant in BAG3 using genome-edited iPSC-derived cardiomyocytes. JCI Insight. 2019;4 doi: 10.1172/jci.insight.128799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tondeur S., Assou S., Nadal L., Hamamah S., De Vos J. [Biology and potential of human embryonic stem cells] Ann. Biol. Clin. 2008;66:241–247. doi: 10.1684/abc.2008.0224. [DOI] [PubMed] [Google Scholar]
- 52.Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R., Collins F.S. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lammerding J., Schulze P.C., Takahashi T., Kozlov S., Sullivan T., Kamm R.D., Stewart C.L., Lee R.T. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohrl J.M., Arnold R., Djabali K. Nuclear pore complexes cluster in dysmorphic nuclei of normal and progeria cells during replicative senescence. Cells. 2021;10 doi: 10.3390/cells10010153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steele-Stallard H.B., Pinton L., Sarcar S., Ozdemir T., Maffioletti S.M., Zammit P.S., Tedesco F.S. Modeling skeletal muscle laminopathies using human induced pluripotent stem cells carrying pathogenic LMNA mutations. Front. Physiol. 2018;9:1332. doi: 10.3389/fphys.2018.01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh F., Charles A.L., Schlagowski A.I., Bouitbir J., Bonifacio A., Piquard F., Krahenbuhl S., Geny B., Zoll J. Reductive stress impairs myoblasts mitochondrial function and triggers mitochondrial hormesis. Biochim. Biophys. Acta. 2015;1853:1574–1585. doi: 10.1016/j.bbamcr.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 57.Kk S.N., Devarajan A., Karan G., Sundaram S., Wang Q., van Groen T., Monte F.D., Rajasekaran N.S. Reductive stress promotes protein aggregation and impairs neurogenesis. Redox Biol. 2020;37:101739. doi: 10.1016/j.redox.2020.101739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parzych K.R., Klionsky D.J. An overview of autophagy: morphology, mechanism, and regulation. Antioxidants Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burke B. Lamins and apoptosis: a two-way street? J. Cell Biol. 2001;153:F5–F7. doi: 10.1083/jcb.153.3.f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lamark T., Svenning S., Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61:609–624. doi: 10.1042/EBC20170035. [DOI] [PubMed] [Google Scholar]
- 61.Jeong S.J., Zhang X., Rodriguez-Velez A., Evans T.D., Razani B. p62/SQSTM1 and selective autophagy in cardiometabolic diseases. Antioxidants Redox Signal. 2019;31:458–471. doi: 10.1089/ars.2018.7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katsuragi Y., Ichimura Y., Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 63.Carre-Mlouka A., Gaumer S., Gay P., Petitjean A.M., Coulondre C., Dru P., Bras F., Dezelee S., Contamine D. Control of sigma virus multiplication by the ref(2)P gene of Drosophila melanogaster: an in vivo study of the PB1 domain of Ref(2)P. Genetics. 2007;176:409–419. doi: 10.1534/genetics.106.063826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avila A., Silverman N., Diaz-Meco M.T., Moscat J. The Drosophila atypical protein kinase C-ref(2)p complex constitutes a conserved module for signaling in the toll pathway. Mol. Cell Biol. 2002;22:8787–8795. doi: 10.1128/mcb.22.24.8787-8795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ichimura Y., Kominami E., Tanaka K., Komatsu M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy. 2008;4:1063–1066. doi: 10.4161/auto.6826. [DOI] [PubMed] [Google Scholar]
- 66.Jain A., Rusten T.E., Katheder N., Elvenes J., Bruun J.A., Sjottem E., Lamark T., Johansen T. p62/Sequestosome-1, autophagy-related gene 8, and autophagy in Drosophila are regulated by nuclear factor erythroid 2-related factor 2 (NRF2), independent of transcription factor TFEB. J. Biol. Chem. 2015;290:14945–14962. doi: 10.1074/jbc.M115.656116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jain A., Lamark T., Sjottem E., Larsen K.B., Awuh J.A., Overvatn A., McMahon M., Hayes J.D., Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 69.Kim Y.C., Guan K.L. mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y., Zhang H. Regulation of autophagy by mTOR signaling pathway. Adv. Exp. Med. Biol. 2019;1206:67–83. doi: 10.1007/978-981-15-0602-4_3. [DOI] [PubMed] [Google Scholar]
- 71.Munson M.J., Ganley I.G. MTOR, PIK3C3, and autophagy: signaling the beginning from the end. Autophagy. 2015;11:2375–2376. doi: 10.1080/15548627.2015.1106668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scott R.C., Juhasz G., Neufeld T.P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tonelli C., Chio I.I.C., Tuveson D.A. Transcriptional regulation by Nrf2. Antioxidants Redox Signal. 2018;29:1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsieh J.Y., Li S.Y., Tsai W.C., Liu J.H., Lin C.L., Liu G.Y., Hung H.C. A small-molecule inhibitor suppresses the tumor-associated mitochondrial NAD(P)+-dependent malic enzyme (ME2) and induces cellular senescence. Oncotarget. 2015;6:20084–20098. doi: 10.18632/oncotarget.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Meo S., Napolitano G., Venditti P. Mediators of physical activity protection against ROS-linked skeletal muscle damage. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20123024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bellezza I., Riuzzi F., Chiappalupi S., Arcuri C., Giambanco I., Sorci G., Donato R. Reductive stress in striated muscle cells. Cell. Mol. Life Sci. 2020;77:3547–3565. doi: 10.1007/s00018-020-03476-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wendel A. Measurement of in vivo lipid peroxidation and toxicological significance. Free Radic. Biol. Med. 1987;3:355–358. doi: 10.1016/s0891-5849(87)80047-3. [DOI] [PubMed] [Google Scholar]
- 78.Jaeschke H., Kleinwaechter C., Wendel A. NADH-dependent reductive stress and ferritin-bound iron in allyl alcohol-induced lipid peroxidation in vivo: the protective effect of vitamin E. Chem. Biol. Interact. 1992;81:57–68. doi: 10.1016/0009-2797(92)90026-h. [DOI] [PubMed] [Google Scholar]
- 79.Ghyczy M., Boros M. Electrophilic methyl groups present in the diet ameliorate pathological states induced by reductive and oxidative stress: a hypothesis. Br. J. Nutr. 2001;85:409–414. doi: 10.1079/bjn2000274. [DOI] [PubMed] [Google Scholar]
- 80.Xie H.B., Cammarato A., Rajasekaran N.S., Zhang H., Suggs J.A., Lin H.C., Bernstein S.I., Benjamin I.J., Golic K.G. The NADPH metabolic network regulates human alphaB-crystallin cardiomyopathy and reductive stress in Drosophila melanogaster. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Korge P., Calmettes G., Weiss J.N. Increased reactive oxygen species production during reductive stress: the roles of mitochondrial glutathione and thioredoxin reductases. Biochim. Biophys. Acta. 2015;1847:514–525. doi: 10.1016/j.bbabio.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pimentel D., Haeussler D.J., Matsui R., Burgoyne J.R., Cohen R.A., Bachschmid M.M. Regulation of cell physiology and pathology by protein S-glutathionylation: lessons learned from the cardiovascular system. Antioxidants Redox Signal. 2012;16:524–542. doi: 10.1089/ars.2011.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao P., Sharir H., Kapur A., Cowan A., Geller E.B., Adler M.W., Seltzman H.H., Reggio P.H., Heynen-Genel S., Sauer M., Chung T.D., Bai Y., Chen W., Caron M.G., Barak L.S., Abood M.E. Targeting of the orphan receptor GPR35 by pamoic acid: a potent activator of extracellular signal-regulated kinase and beta-arrestin2 with antinociceptive activity. Mol. Pharmacol. 2010;78:560–568. doi: 10.1124/mol.110.066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rajasekaran N.S., Varadharaj S., Khanderao G.D., Davidson C.J., Kannan S., Firpo M.A., Zweier J.L., Benjamin I.J. Sustained activation of nuclear erythroid 2-related factor 2/antioxidant response element signaling promotes reductive stress in the human mutant protein aggregation cardiomyopathy in mice. Antioxidants Redox Signal. 2011;14:957–971. doi: 10.1089/ars.2010.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kannan S., Muthusamy V.R., Whitehead K.J., Wang L., Gomes A.V., Litwin S.E., Kensler T.W., Abel E.D., Hoidal J.R., Rajasekaran N.S. Nrf2 deficiency prevents reductive stress-induced hypertrophic cardiomyopathy. Cardiovasc. Res. 2013;100:63–73. doi: 10.1093/cvr/cvt150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bhide S., Trujillo A.S., O'Connor M.T., Young G.H., Cryderman D.E., Chandran S., Nikravesh M., Wallrath L.L., Melkani G.C. Increasing autophagy and blocking Nrf2 suppress laminopathy-induced age-dependent cardiac dysfunction and shortened lifespan. Aging Cell. 2018;17 doi: 10.1111/acel.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bartlett B.J., Isakson P., Lewerenz J., Sanchez H., Kotzebue R.W., Cumming R.C., Harris G.L., Nezis I.P., Schubert D.R., Simonsen A., Finley K.D. p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy. 2011;7:572–583. doi: 10.4161/auto.7.6.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramos F.J., Chen S.C., Garelick M.G., Dai D.F., Liao C.Y., Schreiber K.H., MacKay V.L., An E.H., Strong R., Ladiges W.C., Rabinovitch P.S., Kaeberlein M., Kennedy B.K. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3003802. 144ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liao C.Y., Anderson S.S., Chicoine N.H., Mayfield J.R., Academia E.C., Wilson J.A., Pongkietisak C., Thompson M.A., Lagmay E.P., Miller D.M., Hsu Y.M., McCormick M.A., O'Leary M.N., Kennedy B.K. Rapamycin reverses metabolic deficits in lamin A/C-deficient mice. Cell Rep. 2016;17:2542–2552. doi: 10.1016/j.celrep.2016.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang L.L., Zhang X.Y., Lu Y.Y., Bi Y.D., Liu X.L., Fang F. The role of autophagy in murine cytomegalovirus hepatitis. Viral Immunol. 2021 doi: 10.1089/vim.2020.0024. [DOI] [PubMed] [Google Scholar]
- 91.Fao L., Mota S.I., Rego A.C. Shaping the Nrf2-ARE-related pathways in Alzheimer's and Parkinson's diseases. Ageing Res. Rev. 2019;54:100942. doi: 10.1016/j.arr.2019.100942. [DOI] [PubMed] [Google Scholar]
- 92.Woerner A.C., Frottin F., Hornburg D., Feng L.R., Meissner F., Patra M., Tatzelt J., Mann M., Winklhofer K.F., Hartl F.U., Hipp M.S. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- 93.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 94.Dodson M., Redmann M., Rajasekaran N.S., Darley-Usmar V., Zhang J. KEAP1-NRF2 signalling and autophagy in protection against oxidative and reductive proteotoxicity. Biochem. J. 2015;469:347–355. doi: 10.1042/BJ20150568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Magracheva E., Kozlov S., Stewart C.L., Wlodawer A., Zdanov A. Structure of the lamin A/C R482W mutant responsible for dominant familial partial lipodystrophy (FPLD) Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:665–670. doi: 10.1107/S1744309109020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Petrillo S., Pelosi L., Piemonte F., Travaglini L., Forcina L., Catteruccia M., Petrini S., Verardo M., D'Amico A., Musaro A., Bertini E. Oxidative stress in Duchenne muscular dystrophy: focus on the NRF2 redox pathway. Hum. Mol. Genet. 2017;26:2781–2790. doi: 10.1093/hmg/ddx173. [DOI] [PubMed] [Google Scholar]
- 97.Schoenmakers E., Agostini M., Mitchell C., Schoenmakers N., Papp L., Rajanayagam O., Padidela R., Ceron-Gutierrez L., Doffinger R., Prevosto C., Luan J., Montano S., Lu J., Castanet M., Clemons N., Groeneveld M., Castets P., Karbaschi M., Aitken S., Dixon A., Williams J., Campi I., Blount M., Burton H., Muntoni F., O'Donovan D., Dean A., Warren A., Brierley C., Baguley D., Guicheney P., Fitzgerald R., Coles A., Gaston H., Todd P., Holmgren A., Khanna K.K., Cooke M., Semple R., Halsall D., Wareham N., Schwabe J., Grasso L., Beck-Peccoz P., Ogunko A., Dattani M., Gurnell M., Chatterjee K. Mutations in the selenocysteine insertion sequence-binding protein 2 gene lead to a multisystem selenoprotein deficiency disorder in humans. J. Clin. Invest. 2010;120:4220–4235. doi: 10.1172/JCI43653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sasaki-Honda M., Jonouchi T., Arai M., Hotta A., Mitsuhashi S., Nishino I., Matsuda R., Sakurai H. A patient-derived iPSC model revealed oxidative stress increases facioscapulohumeral muscular dystrophy-causative DUX4. Hum. Mol. Genet. 2018;27:4024–4035. doi: 10.1093/hmg/ddy293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mateos J., De la Fuente A., Lesende-Rodriguez I., Fernandez-Pernas P., Arufe M.C., Blanco F.J. Lamin A deregulation in human mesenchymal stem cells promotes an impairment in their chondrogenic potential and imbalance in their response to oxidative stress. Stem Cell Res. 2013;11:1137–1148. doi: 10.1016/j.scr.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 100.Caron M., Auclair M., Donadille B., Bereziat V., Guerci B., Laville M., Narbonne H., Bodemer C., Lascols O., Capeau J., Vigouroux C. Human lipodystrophies linked to mutations in A-type lamins and to HIV protease inhibitor therapy are both associated with prelamin A accumulation, oxidative stress and premature cellular senescence. Cell Death Differ. 2007;14:1759–1767. doi: 10.1038/sj.cdd.4402197. [DOI] [PubMed] [Google Scholar]
- 101.Pekovic V., Gibbs-Seymour I., Markiewicz E., Alzoghaibi F., Benham A.M., Edwards R., Wenhert M., von Zglinicki T., Hutchison C.J. Conserved cysteine residues in the mammalian lamin A tail are essential for cellular responses to ROS generation. Aging Cell. 2011;10:1067–1079. doi: 10.1111/j.1474-9726.2011.00750.x. [DOI] [PubMed] [Google Scholar]
- 102.Fabrini R., Bocedi A., Pallottini V., Canuti L., De Canio M., Urbani A., Marzano V., Cornetta T., Stano P., Giovanetti A., Stella L., Canini A., Federici G., Ricci G. Nuclear shield: a multi-enzyme task-force for nucleus protection. PLoS One. 2010;5:e14125. doi: 10.1371/journal.pone.0014125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Padmakumar V.C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A.A., Gotzmann J., Foisner R., Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 104.Libotte T., Zaim H., Abraham S., Padmakumar V.C., Schneider M., Lu W., Munck M., Hutchison C., Wehnert M., Fahrenkrog B., Sauder U., Aebi U., Noegel A.A., Karakesisoglou I. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol. Biol. Cell. 2005;16:3411–3424. doi: 10.1091/mbc.e04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Puckelwartz M.J., Kessler E., Zhang Y., Hodzic D., Randles K.N., Morris G., Ju Earley, Hadhazy M., Holaska J.M., Mewborn S.K., Pytel P., McNally E.M. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum. Mol. Genet. 2009;18:607–620. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Atalaia A., Ben Yaou R., Wahbi K., De Sandre-Giovannoli A., Vigouroux C., Bonne G. Laminopathies' treatments systematic review: a contribution towards a 'treatabolome. J. Neuromuscul. Dis. 2021;8:419–439. doi: 10.3233/JND-200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee J., Termglinchan V., Diecke S., Itzhaki I., Lam C.K., Garg P., Lau E., Greenhaw M., Seeger T., Wu H., Zhang J.Z., Chen X., Gil I.P., Ameen M., Sallam K., Rhee J.W., Churko J.M., Chaudhary R., Chour T., Wang P.J., Snyder M.P., Chang H.Y., Karakikes I., Wu J.C. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature. 2019;572:335–340. doi: 10.1038/s41586-019-1406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oh J., Lee S.H., Choi J., Choi J.R., Kim S., Cha Y.J., Choi H.K., Won D., Yoon H.G., Park S.W., Kang S.M., Lee S.T., Lee S.H. Establishment of a novel human iPSC line (YCMi003-A) from a patient with dilated cardiomyopathy carrying genetic variant LMNA p.Asp364His. Stem Cell Res. 2021;56:102508. doi: 10.1016/j.scr.2021.102508. [DOI] [PubMed] [Google Scholar]
- 109.Shemer Y., Mekies L.N., Ben Jehuda R., Baskin P., Shulman R., Eisen B., Regev D., Arbustini E., Gerull B., Gherghiceanu M., Gottlieb E., Arad M., Binah O. Investigating LMNA-related dilated cardiomyopathy using human induced pluripotent stem cell-derived cardiomyocytes. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22157874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.