Abstract

Subject Categories: Membranes & Trafficking; Microbiology, Virology & Host Pathogen Interaction; Structural Biology
We recently reported the first structures of the Plasmodium falciparum transporter PfFNT, both in the absence and presence of the inhibitor MMV007839 (Lyu et al, 2021). These structures indicated that PfFNT assembles as a pentamer. The bound MMV007839 was found in the middle of the elongated channel formed by each PfFNT protomer, adjacent to residue G107. MMV007839 exists in two tautomeric forms and can adopt either a cyclic hemiketal‐like structure or a linear vinylogous acid conformation (Fig 3A). Unfortunately, these two tautomeric forms could not be clearly distinguished based on the existing cryo‐EM data at 2.78 Å resolution. The bound MMV007839 inhibitor was reported as the cyclic hemiketal‐like form in the structure in Figs 3A and F, and 4C, Appendix Figs S10A and B, and S13 and in the online synopsis image.
Figure 3. Cryo‐EM structure of the PfFNT‐MMV007839 complex.
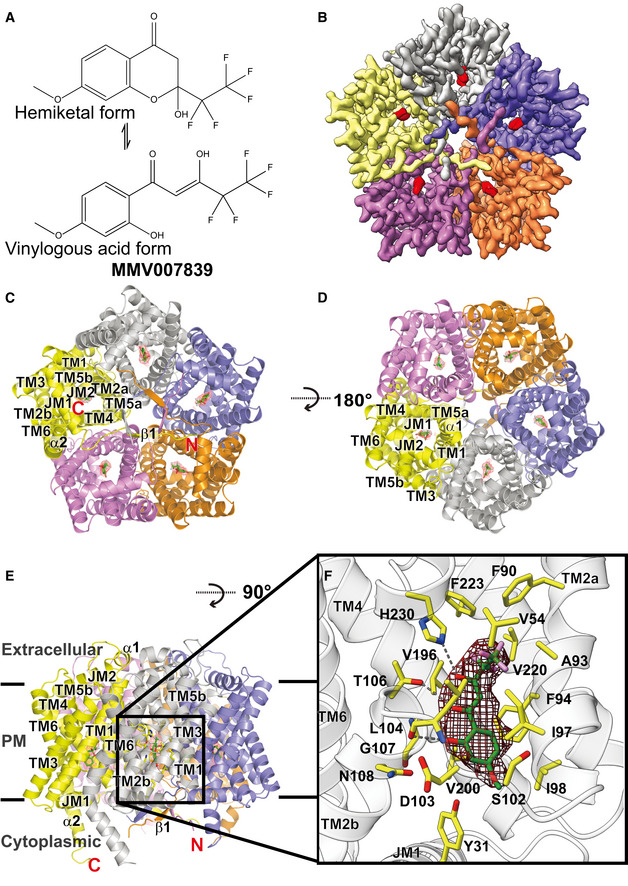
- Chemical structure of MMV007839. The compound can either be in cyclic hemiketal‐like or linear vinylogous acid tautomeric forms.
- Cryo‐EM density map of pentameric PfFNT viewed from the parasite’s cytoplasm. Densities of the five bound MMV007839 within the pentamer are colored red. The five protomers of pentameric PfFNT are colored yellow, slate, orange, purple, and gray.
- Ribbon diagram of the 2.18‐Å resolution structure of pentameric PfFNT‐MMV007839 viewed from the parasite’s cytoplasm. The five protomers of pentameric PfFNT are colored yellow, slate, orange, purple, and gray.
- Ribbon diagram of pentameric PfFNT‐MMV007839 viewed from the extracellular side of the parasite. The five protomers of pentameric PfFNT are colored yellow, slate, orange, purple, and gray.
- Ribbon diagram of pentameric PfFNT‐MMV007839 viewed from the parasite’s membrane plane. The five protomers of pentameric PfFNT are colored yellow, slate, orange, purple, and gray. Densities of the five bound MMV007839 are depicted as red meshes.
- The MMV007839‐binding site of PfFNT. The bound MMV007839 is colored green. Density of the bound MMV007839 is depicted as black mesh. Residues involved in forming the inhibitor binding site are colored yellow. The hydrogen bonds are highlighted with black dotted lines.
Figure 4. Structure of the central channel in the PfFNT‐MMV007839 protomer.
-
CA cartoon of the central channel formed within a PfFNT protomer. The channel contains one constriction site in this conformational state. Residues forming the constriction and the K35‐D103‐N108 and K177‐E229‐N234 triads are illustrated as sticks. Residues F94, I97, and L104, which form the first constriction site in the apo‐PfFNT structure, are also included in the figure.
Eric Beitz alerted us to the findings reported by his group that the linear vinylogous acid tautomer of MMV007839 constitutes the binding and inhibitory entity of PfFNT (Golldack et al, 2017).
Improved resolution of the PfFNT‐MMV007839 complex shows binding of the vinylogous acid form
We have re‐purified PfFNT and made cryo‐EM samples for high‐resolution structural determination (see Additional Methods). Extensive data collection and multiple rounds of refinements using these new data allowed us to improve the resolution of the PfFNT‐MMV007839 complex structure to 2.18 Å (Fig 3B–E and Table 1). Based on these new cryo‐EM data, we unambiguously identified that MMV007839 is in the form of linear vinylogous acid, confirming the earlier functional studies (Golldack et al, 2017). Residues Y31, V54, A93, F90, F94, I97, I98, S102, D103, L104, T106, G107, N108, V196, V200, V220, F223, and H230, all within 4.5 Å of MMV007839, are responsible for forming the inhibitor‐binding site (Fig 3F).
Table 1.
Cryo‐EM data collection, processing, and refinement statistics.
| Data set | PfFNT reconstituted in nanodiscs (1E3D1) |
|---|---|
| PfFNT‐MMV007839 | |
| Data collection and processing | |
| Magnification | 81,000 |
| Voltage (kV) | 300 |
| Electron Microscope | Krios‐GIF‐K3 |
| Defocus range (μm) | −1.0 to −2.5 |
| Total exposure time (s) | 2 |
| Energy filter width (eV) | 20 |
| Pixel size (Å) | 1.08 |
| Total dose (e−/ Å2) | 38 |
| Number of frames | 52 |
| Does rate (e−/ phys. Pixel/s) | 12.24 |
| No. of initial micrographs | 4,345 |
| No. of initial particles | 7,917,294 |
| No. of final particles | 1,044,196 |
| Symmetry | C1 |
| Resolution after density modification | 2.18 |
| Refinement | |
| Model resolution cut‐off (Å) | 2.18 |
| Model composition | |
| No. of Protein residues | 1,406 |
| No. ligands | 5 |
| RMSD a | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.540 |
| Validation | |
| MolProbity score | 2.19 |
| Ramachandran plot (%) | |
| Favored (%) | 96.78 |
| Allowed (%) | 3.22 |
| Disallowed (%) | 0 |
| CC b Mask | 0.66 |
| CC box | 0.50 |
| CC Vol | 0.65 |
Root mean square deviation.
Correlation coefficient.
We note the following changes regarding the interaction of PfFNT with MMV007839: The linear vinylogous acid form of MMV007839 contacts residues in the inhibitor‐binding site that were not reported in Lyu et al (2021). These are residues V54, S102, D103, and N108.
Correction to the manuscript:
We therefore correct Figs 3 and 4C, Appendix Figs S10 and S13. A complete Appendix with corrected Appendix Figs S10 and S13 is provided. The synopsis image has been corrected in the online version of the manuscript. We further correct the text describing inhibitor binding related to Fig 3F, Appendix Figs S10 and S13 as follows.
Figure 3. Original.
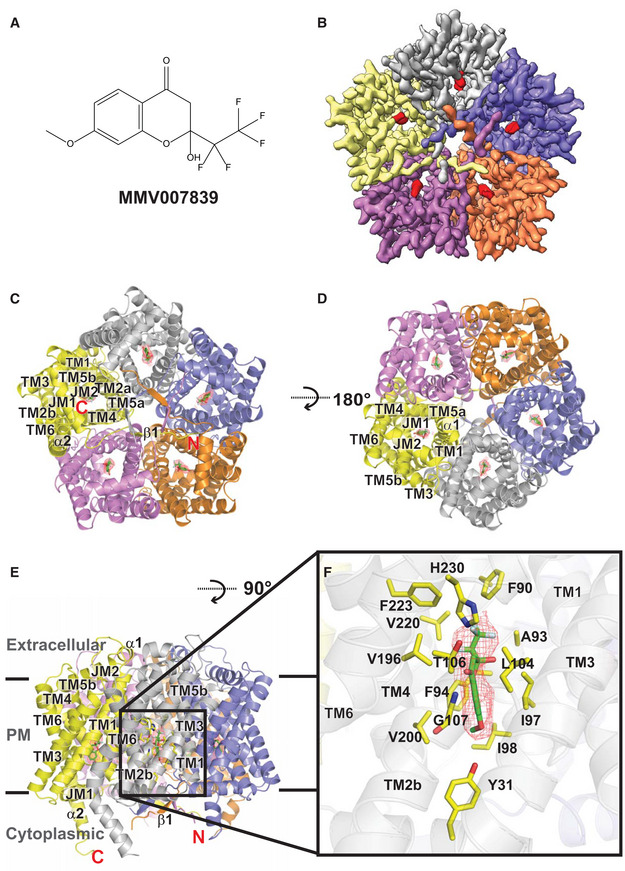
Figure 4. Original.
Page 4, last paragraph in Lyu et al:
“Our high quality cryo‐EM map allows us to unambiguously depict the location of bound MMV007839. The bound inhibitor localized right next to G107 and is closely attached to this residue (Fig 3F). It appears that the backbone nitrogen of G107 forms a hydrogen bond with an oxygen atom from the carbonyl group of the MMV007839 inhibitor. Apparently, the space between G107 and MMV007839 is so tight that there is no room to accommodate an amino acid side chain in this region”.
It is corrected to:
“Our high‐quality cryo‐EM map allows us to unambiguously depict the location of bound MMV007839. The bound inhibitor localized right next to G107 and is closely attached to this residue (Fig 3F). Apparently, the space between G107 and MMV007839 is so tight that there is no room to accommodate an amino acid side chain in this region”.
Page 6, first paragraph in Lyu et al:
“Residues Y31, F90, A93, F94, I97, I98, L104, T106, G107, V196, V200, V220, F223, and H230, which are within 4.5 A from MMV007839, form the inhibitor binding site (Fig 3F). Surprisingly, most of these residues are hydrophobic in nature. Therefore, the primary association between PfFNT and the inhibitor should be via hydrophobic interaction. In addition, the two nitrogen atoms of the imidazole ring of H230 are 4.6 and 4.7 A away from the oxygen atoms of the hydroxyl and carbonyl groups of MMV007839, respectively. We postulate that these two nitrogen atoms participate in electrostatic contributions to secure inhibitor binding”.
Is corrected to:
“Residues Y31, V54, F90, A93, F94, I97, I98, S102, D103, L104, T106, G107, N108, V196, V200, V220, F223, and H230, which are within 4.5 Å from MMV007839, form the inhibitor‐binding site (Fig 3F). Surprisingly, many of these residues are hydrophobic in nature, interacting with this inhibitor via hydrophobic interaction. In addition, the side‐chain oxygen of T106, side‐chain nitrogen of H230, and backbone oxygen of L104 specifically interact with the hydroxyl and oxo groups of MMV007839, forming three hydrogen bonds to further secure the binding”.
Page 6, third paragraph in Lyu et al:
“In comparison with the apo‐PfFNT and PfFNT‐MMV007839 structures, the side chains of several amino acids that participate in forming the channel have significantly shifted their location and orientation to accommodate for inhibitor binding. These residues include Y31, F90, F94, I97, I98, L104, T106, V196, V200, F223, and H230, in which many of them are involved in forming the binding site (Fig 3F and Appendix Fig S10). In addition, G107 is found to shift the backbone location to expand the size of the pocket for housing the MMV007839 molecule”.
Is corrected to:
“In comparison with the apo‐PfFNT and PfFNT‐MMV007839 structures, the side chains of several amino acids that participate in forming the channel have significantly shifted their location and orientation to accommodate for inhibitor binding. These residues include Y31, F90, F94, I97, I98, S102, D103, L104, T106, G107, V196, V200, F223, and H230, in which many of them are involved in forming the binding site (Fig 3F and Appendix Fig S10)”.
Page 7, first paragraph of “Discussion”. The statement:
“Additionally, the side chains of residues Y31, K35, F90, I98, D103, L104, T106, N108, K177, F223, E229 and N234 are observed to switch their relative positions when compared with the conformations of these five protomers (Appendix Fig S13)”.
Is corrected to:
“Additionally, the side chains of residues Y31, K35, F90, F94, I98, D103, L104, T106, N108, K177, F223, E229, and N234 are observed to switch their relative positions when compared with the conformations of these five protomers (Appendix Fig S13)”.
We apologize for not choosing the vinylogous acid form of MMV007839 shown by Golldack et al as the binding entity (Golldack et al, 2017) as the correct conformation.
Additional methods
The procedures for protein purification, sample preparation, cryo‐EM data collection, data processing, and model refinement were the same as those described for the PfFNT‐MMV007839 complex in Lyu et al Initially, 2,567,008 particles were selected from the cryo‐EM data. After several rounds of 2D classifications, 1,044,196 particles were selected for ab initio 3D reconstruction heterogeneous refinement and further processed with non‐uniform and local CTF refinement (Fig EV1). A soft mask covering the PfFNT protein area was used for the final local focused refinement, resulting in a 2.43 Å global resolution map based on the gold standard Fourier shell correlation (FSC 0.143). The cryo‐EM map was then improved by density modification to a final resolution of 2.18 Å (gold‐standard FSC 0.143).
Figure EV1. Cryo‐EM structure of the PfFNT‐MMV007839 complex.
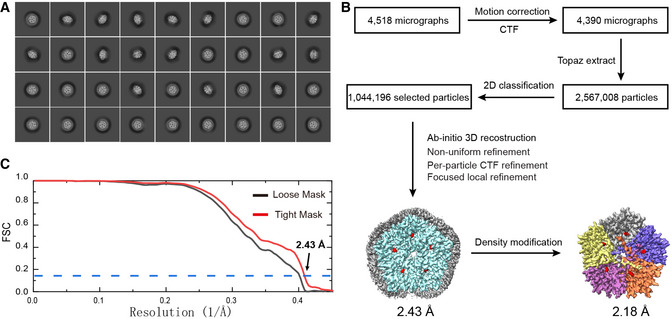
- Representative 2D classes.
- Data processing flowchart showing resolution of 2.43 Å. The resolution of the PfFNT‐MMV007839 pentamer was further enhanced to 2.18 Å after density modification, where the cryo‐EM density maps of the five protomers of PfFNT are colored yellow, slate, orange, purple and gray. The densities of bound MMV007839 are in red.
- Fourier shell correlation (FSC) curves showing resolution of 2.43 Å.
Supporting information
Appendix
Expanded View Figures PDF
Acknowledgements
This work was supported by NIH Grants R01AI145069 (E.W.Y.), R01AI130131 (J.W.K.) and U19AI129326 (J.W.K.).
Addendum to: EMBO Rep (2021) 22: e51628. DOI 10.15252/embr.202051628 | Published online 20 January 202133471955
Data availability
The cryo‐EM maps of the new PfFNT‐MMV007839 structure were deposited in the EMDB under ID code EMD‐24076 (https://www.ebi.ac.uk/emdb/search/EMD‐24076). Atomic coordinates have been deposited in the PDB with accession code 7MXY (https://www.rcsb.org/structure/unreleased/7MXY). Raw cryo‐EM data are available on request.
References
- Golldack A, Henke B, Bergmann B, Wiechert M, Erler H, Blancke Soares A, Spielmann T, Beitz E (2017) Substrate‐analogous inhibitors exert antimalarial action by targeting the Plasmodium lactate transporter PfFNT at nanomolar scale. PLoS Pathog 13: e1006172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu M, Su C‐C, Kazura JW, Yu EW (2021) Structural basis of transport and inhibition of the Plasmodium falciparum transporter PfFNT. EMBO Rep 22: e51628 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Data Availability Statement
The cryo‐EM maps of the new PfFNT‐MMV007839 structure were deposited in the EMDB under ID code EMD‐24076 (https://www.ebi.ac.uk/emdb/search/EMD‐24076). Atomic coordinates have been deposited in the PDB with accession code 7MXY (https://www.rcsb.org/structure/unreleased/7MXY). Raw cryo‐EM data are available on request.


