Abstract
Background and Aims
Current prediction models for early recurrence of hepatocellular carcinoma (HCC) after surgical resection remain unsatisfactory. The aim of this study was to develop evolutionary learning-derived prediction models with interpretability using both clinical and radiomic features to predict early recurrence of HCC after surgical resection.
Methods
Consecutive 517 HCC patients receiving surgical resection with available contrast-enhanced computed tomography (CECT) images before resection were retrospectively enrolled. Patients were randomly assigned to a training set (n = 362) and a test set (n = 155) in a ratio of 7:3. Tumor segmentation of all CECT images including noncontrast phase, arterial phase, and portal venous phase was manually performed for radiomic feature extraction. A novel evolutionary learning-derived method called genetic algorithm for predicting recurrence after surgery of liver cancer (GARSL) was proposed to design prediction models for early recurrence of HCC within 2 years after surgery.
Results
A total of 143 features, including 26 preoperative clinical features, 5 postoperative pathological features, and 112 radiomic features were used to develop GARSL preoperative and postoperative models. The area under the receiver operating characteristic curves (AUCs) for early recurrence of HCC within 2 years were 0.781 and 0.767, respectively, in the training set, and 0.739 and 0.741, respectively, in the test set. The accuracy of GARSL models derived from the evolutionary learning method was significantly better than models derived from other well-known machine learning methods or the early recurrence after surgery for liver tumor (ERASL) preoperative (AUC = 0.687, p < 0.001 vs. GARSL preoperative) and ERASL postoperative (AUC = 0.688, p < 0.001 vs. GARSL postoperative) models using clinical features only.
Conclusion
The GARSL models using both clinical and radiomic features significantly improved the accuracy to predict early recurrence of HCC after surgical resection, which was significantly better than other well-known machine learning-derived models and currently available clinical models.
Keywords: Hepatocellular carcinoma, Evolutionary learning, Machine learning, Recurrence, Surgery
Introduction
Hepatocellular carcinoma (HCC) is the sixth most commonly diagnosed cancer and the fourth leading cause of cancer death globally [1]. For patients with early stage HCC, surgical resection remains the most widely applied curative treatment for HCC [2, 3]. However, tumor recurrence may occur in nearly 70% of patients after resection, while early recurrence within 2 years of resection accounts for >70% of recurrence [4, 5]. Tumor burden, microvascular invasion, and liver function reserve have been reported to be risk factors associated with early HCC recurrence [4, 5, 6, 7].
To decrease the risk of recurrence after resection, there are several ongoing clinical trials of adjuvant immunotherapy but the inclusion criteria for those with high-risk of recurrence varied among the trials [8]. Improvement in the prediction of patients with high-risk of recurrence is required urgently for the development of future therapeutic strategies and may help to identify potential candidates who may benefit from adjuvant systemic therapies. Recently, the preoperative and postoperative early recurrence after surgery for liver tumor (ERASL) models have been proposed based on routine clinical parameters to classify patients with high-, moderate-, and low-risk of early recurrence [7]. Nevertheless, the discriminatory ability of ERASL models remains unsatisfactory, and radiomic information, which may provide crucial information [9], was not included in the model. For clinical decision-making, it is still necessary to further improve the prediction model.
Machine learning is a discipline that uses computational modeling to learn from data, meaning that performance at executing a specific task would be improved using experience [10]. Radiomics coupled with machine learning has been applied to HCC patients with promising results of predicting microvascular invasion before surgical resection [11]. The incorporation of radiomics and machine learning could improve the prediction accuracy of response to transarterial chemoembolization [12] and tumor recurrence in patients with solitary HCC using contrast-enhanced computed tomography (CECT) images [13]. Furthermore, various deep learning-based radiomics methods have been proposed to design prediction models in medical applications [14]. The deep learning-based fusion model used clinical features and CECT images to predict early HCC recurrence [15]. However, the high-level radiomic features extracted by the convolutional layers may suffer from low medical interpretability and high overfitting probability, especially when the training dataset is not large enough, not conducive to understanding, and making diagnostic decisions.
For clinical decision-making before surgical resection, it is desirable to accurately predict the early HCC recurrence. To design a personalized prediction model, it is necessary to identify the risk factors and biomarkers for improving interpretability and clinic practicality. Evolutionary learning means that the parameter values of machine learning methods are optimized using evolutionary algorithms. The aim of this study was to develop evolutionary learning-derived preoperative and postoperative models by combining clinical and radiomic features to predict early recurrence within 2 years after curative resection for HCC.
Materials and Methods
Patients
From October 1, 2007, to April 30, 2018, 1,352 consecutive HCC patients receiving surgical resection in Taipei Veterans General Hospital were retrospectively screened. Patients were excluded by the following criteria: (1) without CECT images within 3 months prior to surgery (n = 569), (2) lost to follow-up within 2 years after surgery (n = 144), (3) without curative surgery (n = 25), and (4) without complete dynamic CECT images including noncontrast phase, arterial phase, and portal venous phase (n = 97). Finally, 517 HCC patients with complete dynamic CECT images were enrolled in this study. The diagnosis of HCC and resectability was assessed before surgery by CECT images or magnetic resonance imaging (MRI), which fulfilled the diagnostic criteria of the American Association for the Study of Liver Diseases treatment guidelines for HCC [16], and was confirmed pathologically after surgery. Curative surgical resection was confirmed by CECT images or MRI after surgery. Patients were followed every 2–3 months with measurement of serum alpha-fetoprotein (AFP), ultrasound, CECT images, or MRI after the surgery. Recurrence-free survival was defined as the time from the date of curative surgery to the time of recurrence. Early recurrence was defined as tumor recurrence within 2 years after the surgery [4, 5, 6, 7].
This study was approved by the Institutional Review Board, Taipei Veterans General Hospital, which complied with standards of the Declaration of Helsinki and current ethical guidelines. Due to the retrospective nature of the study, the Institutional Review Board waived the need for written informed consent. The identifying information of the enrolled subjects has been delinked and therefore authors could not access the information.
Biochemistry, Virological Tests, and Histological Features
The following clinical features and biochemistry were collected for analysis: age, sex, the body mass index, the Barcelona Clinic Liver Cancer (BCLC) stage, the Child-Pugh score, serum AFP, alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatinine, albumin, total bilirubin levels, and platelet count. Serum AFP was measured by chemiluminescent microparticle immunoassay (ARCHITECT AFP assay, Abbott Ireland Diagnostics Division, Sligo, Ireland). Serum biochemistry tests were measured by systemic multi-autoanalyzer (Technicon SMAC; Technicon Instruments Corp., Tarrytown, NY, USA). An albumin-bilirubin grade was calculated as previously described [17]. The histological features including the tumor size, tumor number, Edmonson histological grade, microvascular invasion, hepatic steatosis, and Ishak hepatic inflammation and fibrosis scores [18, 19] were added into the set of clinical features. A total of 26 preoperative clinical features and 5 postoperative pathological features were recorded (Table 1). The evolutionary method established 2 prediction models, preoperative and postoperative models adopting 26 and 31 clinical features, respectively.
Table 1.
Baseline characteristics for the 517 HCC patients undergoing surgical resection
| Feature | Training set (n = 362) | Test set (n = 155) | p value |
|---|---|---|---|
| Age, years | 62±12.3 | 61±13.1 | 0.676 |
| Male gender, n (%) | 294 (81.2) | 123 (79.4) | 0.628 |
| BMI, kg/m2 | 24.5±3.50 | 25.0±3.82 | 0.185 |
| Anti-HCV-positive, n (%) | 67 (18.5) | 19 (12.3) | 0.921 |
| HBsAg-positive, n (%) | 256 (70.7) | 111 (71.6) | 0.105 |
| BCLC stage 0/A/B/C, n (%) | 35/253/39/35 (9.7/69.9/10.8/9.7) | 14/117/18/6 (9/75.5/11.6/3.9) | 0.119 |
| Tumor size, cm | 5.7±3.72 | 6.2±4.31 | 0.703 |
| Tumor number 1/2/>2, n (%) | 294/44/24 (81.2/12.2/6.6) | 126/21/32 (81.3/13.5/5.2) | 0.640 |
| WBC count, /µL | 6,162±1,878 | 6,312±2,138 | 0.603 |
| Hemoglobin, g/dL | 13.5±1.95 | 13.3±1.82 | 0.296 |
| Platelet count, ×109/L | 184±76 | 189±82 | 0.607 |
| Prothrombin time-INR | 1.06±0.83 | 1.06±0.07 | 0.742 |
| ALB, g/dL | 4.0±0.43 | 4.0±2.67 | 0.969 |
| Na, mmol/L | 140±2.6 | 140±2.7 | 0.508 |
| K, mmol/L | 4.14±0.47 | 4.24±0.48 | 0.021 |
| BUN, mg/dL | 16±7.0 | 15±5.9 | 0.267 |
| Creatinine, mg/dL | 1.02±0.88 | 1.02±0.89 | 0.958 |
| Total bilirubin, mg/dL | 0.79±0.41 | 0.84±0.46 | 0.348 |
| ALT, U/L | 48±42 | 51±38 | 0.128 |
| AST, U/L | 46±31 | 54±45 | 0.073 |
| AFP, ng/mL | 20.72 (5.17–409.00) | 24.37 (6.14–978.84) | 0.276 |
| ALK-P, U/L | 87±49 | 96±59 | 0.157 |
| GGT, U/L | 43 (10–702) | 41.5 (13–626) | 0.518 |
| AAR | 1.14±0.79 | 1.20±0.83 | 0.662 |
| APRI | 0.64±0.52 | 0.84±1.26 | 0.491 |
| Tumor grade 1/2/>2, n (%) | 39/173/146 (10.9/48.3/40.8) | 19/70/65 (12.3/45.5/42.2) | 0.473 |
| Microvascular invasion, n (%) | 277 (76.5) | 105 (67.7) | 0.043 |
| Inflammation, n (%) | |||
| Mild | 16 (4.6) | 5 (3.4) | 0.825 |
| Moderate | 303 (86.8) | 129 (88.4) | |
| Severe | 30 (8.6) | 12 (8.2) | |
| Ishak fibrosis stage, n (%) | |||
| 0–2 | 157 (44) | 68 (44.1) | 0.339 |
| 3–4 | 96 (26.9) | 34 (22.1) | |
| 5–6 | 104 (29.1) | 52 (33.7) | |
| Hepatic steatosis, n (%) | |||
| <5% | 146 (48.3) | 69 (51.9) | 0.404 |
| 5–33% | 132 (43.7) | 58 (43.6) | |
| 34–66% | 23 (7.6) | 5 (3.8) | |
| >66% | 1 (0.3) | 1 (0.8) | |
| 2-year recurrence, n (%) | 168 (46.4) | 71 (45.8) | 0.976 |
HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein; BMI, body mass index; BCLC, Barcelona Clinic Liver Cancer; ALT, alanine aminotransferase; AST, aspartate aminotransferase; AAR; AST/ALT ratio; APRI, AST to platelet ratio index.
CECT Image Segmentation and Radiomic Feature Extraction
Interpretation and tumor segmentation of all CECT images were performed by 3 radiologists who were blinded to the clinical and pathological data. The 3 radiologists had read >2,000 liver CT studies per year. When contouring the tumor, the edge of the observed focal lesion within the liver was defined as an imaging appearance that is distinctive from the background according to the Liver Reporting and Data System [20, 21]. The image processing and semiautomatic tumor segmentation were performed using IntelliSpace Discovery (Philips, Eindhoven, The Netherlands).
For each patient's CT images of noncontrast phase, arterial phase, and portal venous phase, the images with the largest tumor diameter for each tumor in 3 phases were used for extracting radiomic features, including morphology, tumor edge, intensity, Haralick, invariant moment, and discrete wavelet transformed features for each phase. The detailed method of radiomic feature extraction is described in online suppl. Methods (for all online suppl. material, see www.karger.com/doi/10.1159/000518728).
The coarse-to-fine feature selection from a large number of radiomic features was done as follows. First, coarse feature selection was performed by the student's t test that the features with no significant difference among recurrence and nonrecurrence patients in 2 years were eliminated. Second, one of a group of features with high Pearson correlation coefficients (e.g., r > 0.9 in this study) was kept for the fine feature selection. Finally, the remaining features were selected using the optimal feature selection of the evolutionary method.
Evolutionary Learning-Derived Method Genetic Algorithm for Predicting Recurrence after Surgery of Liver Cancer
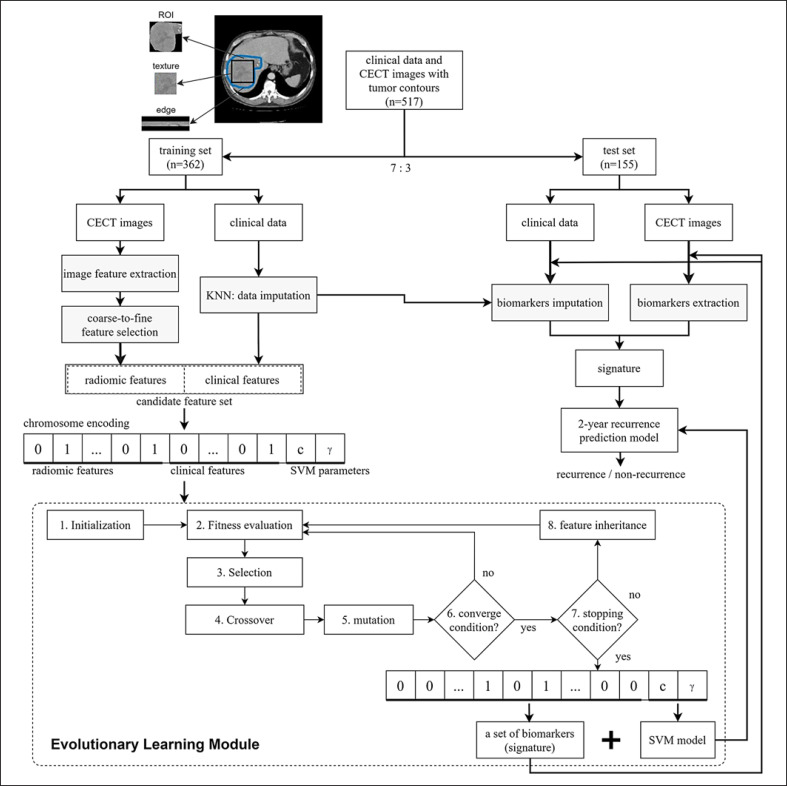
In this study, a novel evolutionary learning method called genetic algorithm for predicting recurrence after surgery of liver cancer (GARSL) is proposed to predict early HCC recurrence within 2 years after surgery. The illustrated flow chart of the GARSL method for predicting the 2-year HCC recurrence is shown in Figure 1. The whole dataset was divided into training and test datasets with the ratio 7:3. The radiomic and clinical features with imputation using a k-nearest neighbor method were cascaded into a candidate feature set. The prediction method GARSL used a well-known support vector machine (SVM) classifier which is a statistics-based supervised learning model. SVM performs classification or regression by mapping data into higher dimension feature space using a kernel function.
Fig. 1.
Illustrated flowchart of developing the evolutionary learning-derived method GARSL. GARSL, genetic algorithm for predicting recurrence after surgery of liver cancer.
The determination of both cost (c) and kernel (γ) parameters of SVM plays a vital role in modeling. All the optimal feature selection and parameter settings of SVM were conducted based on an intelligent evolutionary algorithm (IEA) [22]. The inheritable bi-objective combinatorial genetic algorithm (IBCGA) [23] with IEA was used to identify a small set of features (risk factors and radiomic features) and determine SVM parameter values while maximizing the fitness function. The fitness function is to maximize the prediction accuracy of Matthews correlation coefficient (MCC) for 10 fold cross-validation (CV) on the training dataset. The MCC measurement was used for dealing with the imbalanced dataset, in which 46.4% of HCC patients had the 2-year recurrence. The prediction model for the independent test can be trained using the output of IBCGA and the whole training dataset.
The optimal feature selection problem C (n, m) is to select a small number m from a large number n of candidate features in which the interaction among features exist. IBCGA aims to effectively solve the large-scale combinatorial optimization problem for delivering the value of m, the identified m features, and the values of c and γ. For applying IBCGA, all the candidate features were encoded into binary variables for optimal feature selection. The parameters (c, γ) were also encoded into the chromosome to be optimized at the same time. Based on the main effect difference (MED), the m features can be ranked according to the prediction contribution. The detailed description of the IBCGA and SVM algorithms can be found in supplementary methods. Some applications of IBCGA and IEA in designing prediction models for biomedicine research can refer the studies [24, 25, 26, 27, 28].
Statistical Analysis
The descriptive values were expressed as mean ± standard deviation or as a median (ranges) when appropriate. The Mann-Whitney U test was used to compare continuous variables. Pearson χ2 analysis or the Fisher's exact test was used to compare categorical variables. The Kaplan-Meier method was used to estimate survival rates. A 2-tailed p < 0.05 was considered statistically significant. The statistical analyses for the descriptive data were performed using the IBM SPSS Statistics V22 (IBM, Armonk, NY, USA).
Patient and Public Involvement
Patients or the public were not involved in the design, or conduct, or reporting, or dissemination plans of our research.
Results
A total of 517 HCC patients with available CECT images and 2-year recurrence outcomes were finally enrolled in this study. Patients were randomly assigned to the training set (n = 362) and the test set (n = 155) in a ratio of 7:3. The baseline characteristics of the patients in the training and test sets are shown in Table 1. The patient characteristics between the 2 groups were generally comparable, except that patients in the test set had slightly higher serum potassium levels and lower percentage of microvascular invasion. During a median follow-up period of 47.4 months (range 2.2–162.2 months), 239 (46.2%) patients developed early recurrence after curative surgical resection.
Coarse-to-Fine Feature Selection and Analysis
A total of 6,284 images in the noncontrast phase, 5,919 images in the arterial phase, and 6,190 images in the portal venous phase were considered. For each of the 517 patients, there were 1,451 radiomic features extracted for 1 phase, including 24 morphology features, 6 edge features, 11 intensity features, 440 Haralick features, 7 Hu moment invariant features, and 963 discrete wavelet transformed features. Among the 4,353 (= 1,451 × 3) radiomic features, a total of 112 features after coarse feature selection were retained. A total of 138 and 143 features were served as the input candidate features of IBCGA to design the preoperative and postoperative models, respectively.
There were 41 features selected for designing the preoperative model with the MCC and accuracy of 0.476 and 69.89% of 10-CV, respectively. The preoperative model achieved the test area under the receiver operating characteristic curve (AUC), accuracy, and MCC of 0.739, 72.90%, and 0.453, respectively (Table 2). The top 20 features in the preoperative model ranked by the MED score and statistical significance of each feature were shown in online suppl. Table 1. The distribution of top 20 features in positive prediction and negative prediction and statistical significance of each feature within whole data were given in online suppl. Figure 1.
Table 2.
Performance comparison of GARSL with other well-known machine learning models to predict early recurrence using both clinical and radiomic features
| Method | 10-CV (n = 362) |
Test set (n = 155) |
|||
|---|---|---|---|---|---|
| accuracy, % | MCC | accuracy, % | MCC | AUC | |
| GARSL | 69.89 | 0.476 | 72.90 | 0.453 | 0.739 |
| C4.5 | 56.35 | 0.119 | 57.42 | 0.172 | 0.610 |
| Random tree | 51.93 | 0.038 | 61.29 | 0.228 | 0.615 |
| Hoeffding tree | 61.33 | 0.216 | 68.39 | 0.361 | 0.718 |
| Logistic model tree | 63.54 | 0.262 | 67.74 | 0.346 | 0.716 |
| Logistic regression | 54.14 | 0.085 | 60.65 | 0.212 | 0.642 |
| Naive Bayes | 61.05 | 0.211 | 67.10 | 0.335 | 0.716 |
Formulas, indexes, and modeling details of every method are described in the online suppl. File. CV, cross-validation; MCC, Matthews correlation coefficient; AUC, area under the receiver operating characteristic curve; GARSL, genetic algorithm for predicting recurrence after surgery of liver cancer.
From online suppl. Table 1, there were 5 statistically significant clinical features that the tumor size, BCLC stage, tumor number, GGT, and AST were ranked at 1, 3, 9, 12, and 15, respectively, based on the MED score. There were 4, 8, and 3 radiomic features from the images of noncontrast phase, arterial phase, and portal venous phase, respectively. The wavelet features with ranks 2 and 4 came from the tumor texture of the arterial phase in the presentation of frequency domain. The Haralick feature rank 5 came from the tumor texture of the noncontrast phase in the presentation of spatiality domain. The morphology and edge features of tumors ranked 6 and 7 also play an important role. It is noted that although some features were not significant, they were important in classifying samples in the subset of training datasets. Note that the feature of tumor intensity was not selected. The experimental results reveal that the clinical and radiomic features including texture, morphology, tumor edge features in the CECT images of 3 phases are informative. The features selected for GARSL preclinical, GARSL radiomics, GARSL preoperative, and GARSL postoperative models are shown in online suppl. Tables 2–5.
Comparison of the GARSL Model with Machine Learning Methods
The evolutionary learning-derived method GARSL designed prediction models with interpretable features using an optimization approach to feature selection. The same training dataset and 138 features (26 preclinical features and 112 radiomic features) were used to design preoperative models for performance comparison between GARSL and other well-known machine learning methods using the WEKA software [29] (Table 2). The 4 kinds of decision tree methods were compared. C4.5 is a statistical classifier and is probably the most widely used machine learning method to date. Random tree is a decision tree with stochastic process. Hoeffding tree is an incremental decision tree designed for large dataset. Logistic model tree is a combination of logistic regression and decision tree. Logistic regression is a statistical model for predicting a binary dependent variable. The naïve Bayes classifier is a probabilistic machine learning method based on the Bayes theorem. Among the 7 prediction models, the GARSL model has the highest accuracy (69.89%) and MCC (0.476) of 10-CV, and test AUC (0.739), accuracy (72.90%), and MCC (0.453), and is significantly better than other 6 prediction models.
Comparison between GARSL and ERASL Models
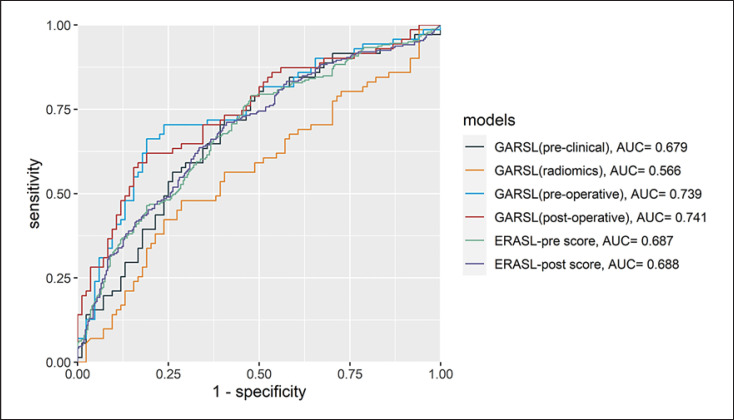
The performance comparison between the GARSL and ERASL models for prediction of early recurrence on the test set is shown in Table 3 and Figure 2. The ERASL-pre model composed of gender, ABLI grade, AFP, tumor size, and tumor number, while the ERASL-post model had the addition of microvascular invasion. Compared to other preoperative prediction models, the GARSL preoperative model using both preclinical features and radiomic features from CECT images had the best C-index of 0.695 and AUC of 0.739, which was significantly better than the GARSL using preclinical features (p < 0.001), GARSL using radiomic features (p < 0.001), and ERASL using preclinical features (p < 0.001). After incorporation of 5 postoperative pathological features, the GARSL postoperative model had further improved C-index of 0.710 and AUC of 0.741, which was significantly better than the GARSL preoperative model (p < 0.001) and the ERASL-post model (p < 0.001).
Table 3.
Performance of the GARSL and ERASL models for prediction of early recurrence
| Method | Feature type | Training set (n = 362) |
Test set (n = 155) |
|||
|---|---|---|---|---|---|---|
| AUC | C-index (SE) | AUC | C-index (SE) | p value | ||
| GARSL | Preclinical | 0.863 | 0.790 (0.016) | 0.679 | 0.647 (0.034) | <0.001* |
| GARSL | Radiomics | 0.740 | 0.518 (0.023) | 0.566 | 0.533 (0.035) | <0.001* |
| GARSL | Preoperative | 0.781 | 0.738 (0.018) | 0.739 | 0.695 (0.032) | REF |
| GARSL | Postoperative | 0.767 | 0.723 (0.019) | 0.741 | 0.710 (0.031) | <0.001* |
| ERASL-pre | Preoperative | 0.667 | 0.659 (0.021) | 0.687 | 0.672 (0.017) | <0.001* |
| ERASL-post | Postoperative | 0.672 | 0.656 (0.022) | 0.688 | 0.666 (0.018) | <0.001† |
C-index, concordance index; AUC, area under the receiver operating characteristic curve; SE, standard error; ERASL, early recurrence after surgery for liver tumor; GARSL, genetic algorithm for predicting recurrence after surgery of liver cancer.
p value compared to the GARSL preoperative score. † p value compared to the GARSL postoperative score.
Fig. 2.
Receiver operating characteristic curves of the GARSL and ERASL models in predicting early recurrence in the test set. AUC, area under the receiver operating characteristic curve; ERASL, early recurrence after surgery for liver tumor; GARSL, genetic algorithm for predicting recurrence after surgery of liver cancer.
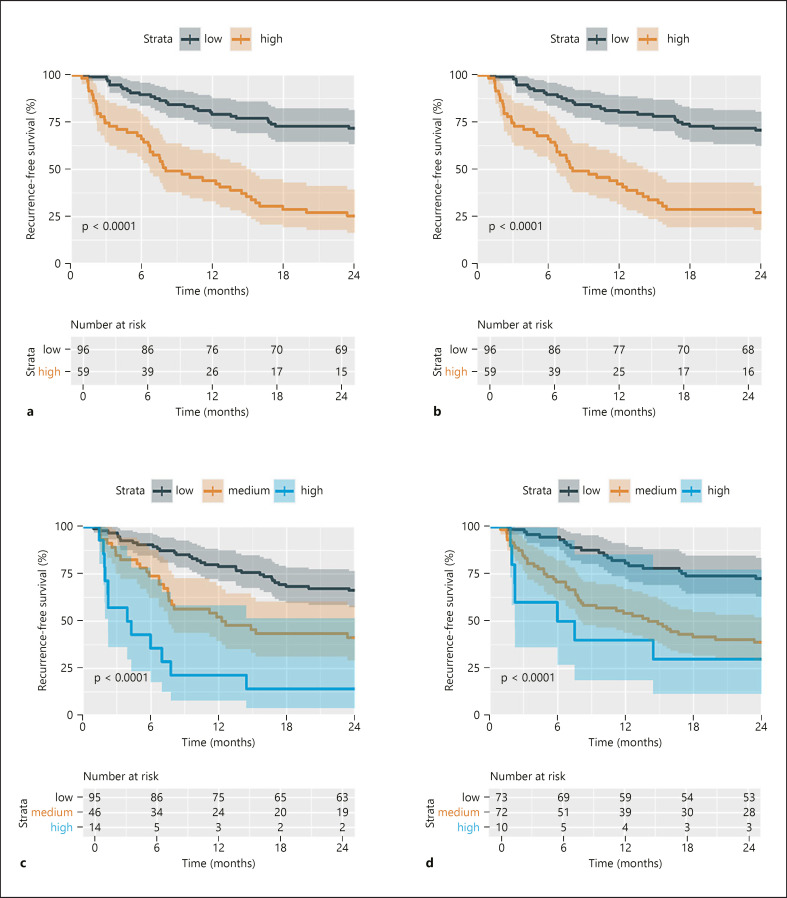
The 2 year recurrence rates in the high- and low-risk groups on the test set were 75% and 38% for GARSL preoperative model (Fig. 3a) and 73 and 29% for GARSL postoperative model (Fig. 3b), whereas the 2-year recurrence rate in the high-, medium-, and low-risk groups were 86, 59, and 34% for the ERASL pre model (Fig. 3c), and 70, 61, and 27% for ERASL-post model (Fig. 3d).
Fig. 3.
Kaplan-Meier curves of the early RFS stratified by the GARSL and ERASL models. GARSL preoperative model (a), GARSL postoperative model (b), ERASL-pre model (c), ERASL-post model (d). ERASL, early recurrence after surgery for liver tumor; RFS, recurrence-free survival; GARSL, genetic algorithm for predicting recurrence after surgery of liver cancer.
Discussion
In this study, we developed evolutionary learning-derived GARSL models using both clinical and radiomic features for predicting early recurrence of HCC after surgical resection. The experimental results showed that the GARSL models using additional radiomic features performed well and were significantly better than the current clinical model ERASL and several well-known machine learning models.
Our novel evolutionary learning-derived method GARSL aims to identify a minimal set of features with interaction while maximizing prediction accuracy by considering all the available candidate features. The feature selection of GARSL identifies a set of features with interaction at a time rather than individual features one by one, which was different from traditional feature selection methods based on the p value of individual features or human domain knowledge without considering feature interaction. Therefore, some related features such as ALT and AST, or tumor factors and BCLC stage, would be selected if they could improve the prediction accuracy. The situation might occur that the addition of some significant features such as AFP (p value = 6.89 × 10−4 using the Mann-Whitney U test) could not increase the prediction accuracy. Some features which were not selected did not necessarily mean that they were not significant but that they did not improve the accuracy of the final model. The features usefulness in the prediction of recurrence is somewhat different from the aspects of human knowledge.
Most researchers used 70/30 of the training/test ratio in separating small datasets [30]. The influence of data splitting depends on the dataset and prediction model. Generally, a small training dataset (or ratio) would frequently result in overtraining, while the performance may be not representative if the test dataset is too small in evaluating prediction models. If the dataset is large enough, 80/20 is commonly used. If the performance of the training set and test set is too different, overtraining may exist. When the training dataset is not large enough, there is always the possibility of overtraining if the method aims to maximize the prediction accuracy. Generally, the 10 fold CV or 5-CV is utilized in training the model to reduce the overtraining degree, which was used in our proposed method GARSL. Another approach to reducing the possibility of overtraining is to increase the size of the training dataset. Learning from the feedback of the prediction model, the test accuracy would be increased and close to the training accuracy.
Although many factors have been identified to be associated with early recurrence of HCC in the past decades, currently there are only few prediction models. The recently proposed ERASL-pre and ERASL-post models, which stratified patients into high-, medium-, and low-risk of early recurrence, were derived from independent predictors of early recurrence by using the conventional Cox regression hazard model. Experimental results showed that the GARSL models, which only stratify patients into high- and low-risk groups of 2-year early recurrence, had better prediction accuracy than the ERASL-pre and ERASL-post models. Our data suggest that evolutionary learning-derived prediction models may significantly improve the predictive accuracy than conventional regression models and by input additional radiomic features further improves the accuracy than using clinical features only. Of note, although the accuracy of the GARSL postoperative model was significantly better than the GARSL preoperative model, the AUC was close and recurrence rates between high- and low-risk groups of the 2 GARSL models were similar (Table 3; Fig. 2). A recent study has shown that some pathological features, such as microvascular invasion, might be predicted by radiomic features [11], which may explain why adding postoperative pathological features only slightly improve the predictive accuracy.
Currently, several machine learning methods have been developed but which method has better performance was unclear. In this study, we compared the performance of the GARSL method and other well-known machine learning methods using the same feature set, including 26 clinical features and 112 radiomic features. Our results showed that GARSL achieved the best performance in terms of accuracy, MCC, and AUC on both training and test datasets (Table 2), suggesting that the evolutionary learning method GARSL have the advantages over the compared machine learning methods: (1) IBCGA is good at feature selection using the IEA and (2) the optimal feature selection and parameter values of SVM were simultaneously optimized by maximizing the 10-CV accuracy.
The machine learning approach has been introduced to many aspects of HCC, including the radiomic analysis using MRI [31], digital pathology [32], incorporation of genomic data [33], and microRNA signature [34]. MRI, which includes T1, T2, diffusion-weighted imaging, as well as dynamic contrast phases and hepatobiliary phase, may provide more radiomic information as compared to CECT images. Although tumor segmentation using MRI is more complicated, whether MRI radiomics have more prognostic value than CT radiomics warrants further research. By incorporation of more comprehensive data in machine learning models, such as other imaging modalities, digital pathology, and genomics, would further improve precision medicine in the future.
Our GARSL models would assist physicians and surgeons on treatment decision and follow-up program for patients with resectable HCC. For patients with low-risk of HCC recurrence, surgical resection would be highly encouraged, whereas for patients with high-risk of early recurrence after surgical resection, a more stringent surveillance strategy might be given and clinical trials of adjuvant or neoadjuvant systemic therapies could be considered [8]. Nevertheless, the feasibility of applying this model in real-world practice remains challenging given the complexity of the model. The successful deployment of the artificial intelligence-driven health technologies, including auto-segmentation for liver tumor, requires investment to strengthen the underlying health system in the future [35].
This study has some limitations. First, this is a single-center study. The accuracy of the GARSL model needs further external validation from other institutions in the future. Second, the impact of viral load or antiviral therapy in patients with HBV or HCV infection was not analyzed in the current model. The input of more clinical parameters might further improve the accuracy of the model. Third, the input of CECT radiomics needs manual segmentation of HCC by experienced radiologists. More efforts are required for the development of automated tumor segmentation to facilitate future application of radiomics analysis.
In conclusion, we developed an evolutionary learning-derived method GARSL for designing prediction models with interpretable features by incorporation of clinical and radiomic features of CECT images to predict early recurrence of HCC after surgical resection. The experimental results showed that the well-known clinical features (tumor size, BCLC stage, and tumor number) and radiomic feature types (texture, morphology, and edge of tumors) in 3 phases of CECT images play important roles with quantitative assessment in predicting early recurrence of HCC through an optimal feature selection. The GARSL models had significantly better prediction performance than some typical machine learning models and currently available clinical models. Our study shows potential for use of this approach in designing and guiding patient care in the future.
Statement of Ethics
This study gained consent to Institutional Review Board in Taipei Veterans General Hospital (IRB number: 2020-05-008BC) and was conducted ethically in accordance with the World Medical Association Declaration of Helsinki. Due to the retrospective nature of the study, the Institutional Review Board waived the need for written informed consent. The identifying information of the enrolled subjects has been delinked and therefore authors could not access the information.
Conflict of Interest Statement
Y.-H.H. has received research grants from Gilead Sciences and Bristol-Meyers Squibb, and honoraria from Abbvie, Gilead Sciences, Bristol-Meyers Squibb, Ono Pharmaceutical, Merck Sharp & Dohme, Eisai, Eli Lilly, Ipsen, and Roche and has served in an advisory role for Abbvie, Gilead Sciences, Bristol-Meyers Squibb, Ono Pharmaceuticals, Eisai, Eli Lilly, Ipsen, Merck Sharp & Dohme, and Roche. The other authors declare no conflicts of interest.
Funding Sources
The study was supported by grants from Taipei Veterans General Hospital, Taipei, Taiwan (V109E-008-2, V109E-008-2[110]) and Ministry of Science and Technology, Taiwan (MOST 106-2634-F-075-001-, MOST 107-2634-F-075-001-, MOST 108-3011-F-075-001-, MOST 108-2221-E-009-127-, MOST 108-2218-E-029-004-, MOST 109-2221-E-009-129-, MOST 109-2740-B-400-002-) and was financially supported by the “Center For Intelligent Drug Systems and Smart Bio-devices (IDS2B)” from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Author Contributions
I.-C. Lee contributed to data acquisition; study concept and design; analysis and interpretation of data; drafting of the manuscript; and statistical analysis. J.-Y. Huang, T.-C. Chen, and C.-H. Yen contributed to implementation and analysis of all machine learning methods. N.-C. Chiu, H.-E. Hwang, C.-A. Liu, R.-C., and Lee contributed to image interpretation and segmentation. G.-Y. Chau, J.-G. Huang, Y.-P. Hung, and Y. Chao contributed to data acquisition. S.-Y. Ho and Y.-H. Huang contributed to the study concept and design; obtained funding; critical revision of the manuscript for important intellectual content; and study supervision.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Supplementary data
Acknowledgements
The authors thank the Clinical Research Core Laboratory, Taipei Veterans General Hospital for providing their facilities to conduct this study.
References
- 1.Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, et al. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology. 2020 Jul;159((1)):335–49.e15. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Association for the Study of the Liver EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018 Jul;69((1)):182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Vibert E, Schwartz M, Olthoff KM. Advances in resection and transplantation for hepatocellular carcinoma. J Hepatol. 2020 Feb;72((2)):262–76. doi: 10.1016/j.jhep.2019.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Imamura H, Matsuyama Y, Tanaka E, Ohkubo T, Hasegawa K, Miyagawa S, et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003 Feb;38((2)):200–7. doi: 10.1016/s0168-8278(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 5.Portolani N, Coniglio A, Ghidoni S, Giovanelli M, Benetti A, Tiberio GA, et al. Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg. 2006 Feb;243((2)):229–35. doi: 10.1097/01.sla.0000197706.21803.a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu JC, Huang YH, Chau GY, Su CW, Lai CR, Lee PC, et al. Risk factors for early and late recurrence in hepatitis B-related hepatocellular carcinoma. J Hepatol. 2009 Nov;51((5)):890–7. doi: 10.1016/j.jhep.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Chan AWH, Zhong J, Berhane S, Toyoda H, Cucchetti A, Shi K, et al. Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J Hepatol. 2018 Dec;69((6)):1284–93. doi: 10.1016/j.jhep.2018.08.027. [DOI] [PubMed] [Google Scholar]
- 8.Kudo M. A paradigm change in the treatment strategy for hepatocellular carcinoma. Liver Cancer. 2020 Aug;9((4)):367–77. doi: 10.1159/000507934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017 Dec;14((12)):749–62. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 10.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019 Apr 4;380((14)):1347–58. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 11.Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, et al. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019 Jun;70((6)):1133–44. doi: 10.1016/j.jhep.2019.02.023. [DOI] [PubMed] [Google Scholar]
- 12.Chen M, Cao J, Hu J, Topatana W, Li S, Juengpanich S, et al. Clinical-radiomic analysis for pretreatment prediction of objective response to first transarterial chemoembolization in hepatocellular carcinoma. Liver Cancer. 2021 Feb;10((1)):38–51. doi: 10.1159/000512028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji GW, Zhu FP, Xu Q, Wang K, Wu MY, Tang WW, et al. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: a multi-institutional study. EBioMedicine. 2019 Dec;50:156–65. doi: 10.1016/j.ebiom.2019.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lao J, Chen Y, Li ZC, Li Q, Zhang J, Liu J, et al. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep. 2017 Sep 4;7((1)):10353. doi: 10.1038/s41598-017-10649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Chen Q, Iwamoto Y, Aonpong P, Lin L, Hu H, et al. Deep fusion models of multi-phase CT and selected clinical data for preoperative prediction of early recurrence in hepatocellular carcinoma. IEEE Access. 2020;8:139212–20. [Google Scholar]
- 16.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011 Mar;53((3)):1020–2. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson PJ, Berhane S, Kagebayashi C, Satomura S, Teng M, Reeves HL, et al. Assessment of liver function in patients with hepatocellular carcinoma: a new evidence-based approach-the ALBI grade. J Clin Oncol. 2015 Feb 20;33((6)):550–8. doi: 10.1200/JCO.2014.57.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995 Jun;22((6)):696–9. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 19.Lee IC, Huang YH, Chan CC, Huo TI, Chu CJ, Lai CR, et al. Correlation between clinical indication for treatment and liver histology in HBeAg-negative chronic hepatitis B: a novel role of alpha-fetoprotein. Liver Int. 2010 Sep;30((8)):1161–8. doi: 10.1111/j.1478-3231.2010.02301.x. [DOI] [PubMed] [Google Scholar]
- 20.Moura Cunha G, Chernyak V, Fowler KJ, Sirlin CB. Up-to-date role of CT/MRI LI-RADS in hepatocellular carcinoma. J Hepatocell Carcinoma. 2021;8:513–27. doi: 10.2147/JHC.S268288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spieler B, Sabottke C, Moawad AW, Gabr AM, Bashir MR, Do RKG, et al. Artificial intelligence in assessment of hepatocellular carcinoma treatment response. Abdom Radiol. 2021 Mar 31;46((8)):3660–71. doi: 10.1007/s00261-021-03056-1. [DOI] [PubMed] [Google Scholar]
- 22.Ho S-Y, Shu L-S, Chen J-H. Intelligent evolutionary algorithms for large parameter optimization problems. IEEE Trans Evol Computat. 2004 Dec;8((6)):522–41. [Google Scholar]
- 23.Ho SY, Chen JH, Huang MH. Inheritable genetic algorithm for biobjective 0/1 combinatorial optimization problems and its applications. IEEE Trans Syst Man Cybern B Cybern. 2004 Feb;34((1)):609–20. doi: 10.1109/tsmcb.2003.817090. [DOI] [PubMed] [Google Scholar]
- 24.Yerukala Sathipati S, Ho SY. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Sci Rep. 2017 Aug 8;7((1)):7507. doi: 10.1038/s41598-017-07739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai MJ, Wang JR, Yang CD, Kao KC, Huang WL, Huang HY, et al. PredCRP: predicting and analysing the regulatory roles of CRP from its binding sites in Escherichia coli. Sci Rep. 2018 Jan 17;8((1)):951. doi: 10.1038/s41598-017-18648-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yerukala Sathipati S, Ho SY. Identifying a miRNA signature for predicting the stage of breast cancer. Sci Rep. 2018 Oct 31;8((1)):16138. doi: 10.1038/s41598-018-34604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yerukala Sathipati S, Sahu D, Huang HC, Lin Y, Ho SY. Identification and characterization of the lncRNA signature associated with overall survival in patients with neuroblastoma. Sci Rep. 2019 Mar 26;9((1)):5125. doi: 10.1038/s41598-019-41553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai MJ, Wang JR, Ho SJ, Shu LS, Huang WL, Ho SY. GREMA: modelling of emulated gene regulatory networks with confidence levels based on evolutionary intelligence to cope with the underdetermined problem. Bioinformatics. 2020 Jun 1;36((12)):3833–40. doi: 10.1093/bioinformatics/btaa267. [DOI] [PubMed] [Google Scholar]
- 29.Frank E, Hall M, Trigg L, Holmes G, Witten IH. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20((15)):2479–81. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen QH, Ly H-B, Ho LS, Al-Ansari N, Le HV, Tran VQ, et al. Influence of data splitting on performance of machine learning models in prediction of shear strength of soil. Math Probl Eng. 2021 Feb 8;2021((4832864)):1–15. [Google Scholar]
- 31.Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY. Radiomics on gadoxetic acid-enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin Cancer Res. 2019 Jul 1;25((13)):3847–55. doi: 10.1158/1078-0432.CCR-18-2861. [DOI] [PubMed] [Google Scholar]
- 32.Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, et al. Predicting survival after hepatocellular carcinoma resection using deep-learning on histological slides. Hepatology. 2020;72((6)):2000–13. doi: 10.1002/hep.31207. [DOI] [PubMed] [Google Scholar]
- 33.Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018 Mar 15;24((6)):1248–59. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yerukala Sathipati S, Ho SY. Novel miRNA signature for predicting the stage of hepatocellular carcinoma. Sci Rep. 2020 Sep 2;10((1)):14452. doi: 10.1038/s41598-020-71324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwalbe N, Wahl B. Artificial intelligence and the future of global health. Lancet. 2020 May 16;395((10236)):1579–86. doi: 10.1016/S0140-6736(20)30226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.