Abstract
In this study, we utilized retroviral transfer of cDNA libraries in order to identify oncogenes that are expressed in acute myeloid leukemia (AML). From screens using two different cell types as targets for cellular transformation, a single cDNA encoding a variant of the TrkA protooncogene was isolated. The protein product of this protooncogene, TrkA, is a receptor tyrosine kinase for nerve growth factor. The isolated transforming cDNA encoded a TrkA protein that contains a 75-amino-acid deletion in the extracellular domain of the receptor and was named ΔTrkA. ΔTrkA readily transformed fibroblast and epithelial cell lines. The deletion resulted in activation of the tyrosine kinase domain leading to constitutive tyrosine phosphorylation of the protein. Expression of ΔTrkA in cells led to the constitutive activation of intracellular signaling pathways that include Ras, extracellular signal-regulated kinase/mitogen-activated protein kinase, and Akt. Importantly, ΔTrkA altered the apoptotic and growth properties of 32D myeloid progenitor cells, suggesting ΔTrkA may have contributed to the development and/or maintenance of the myeloid leukemia from which it was isolated. Unlike Bcr-Abl, expression of ΔTrkA did not activate Stat5 in these cells. We have detected expression of ΔTrkA in the original AML sample by reverse transcriptase PCR and by Western blot analysis. While previous TrkA mutations identified from human tumors involved fusion to other proteins, this report is the initial demonstration that deletions within TrkA may play a role in human cancers. Finally, this report is the first to indicate mutations in TrkA may contribute to leukemogenesis.
The identification of cellular oncogenes has played an important role in understanding the molecular basis of cancer. Additionally, these studies have also provided the foundation for understanding various fundamental cellular processes. Studying mutated proteins or proteins expressed in an aberrant manner can unmask the role these proteins play in normal cell physiology. For example, the study of tumor-associated and mutated forms of the ras oncogenes has developed both mechanistic and functional descriptions of the role Ras proteins play in signaling pathways that control normal and neoplastic cell growth and differentiation. The identification of activated versions of tyrosine kinases (e.g., Abl and Src) as well as transcription factors (e.g., Fos, Jun, and Myc) has aided in the understanding of how these proteins normally function, as well as how they are regulated and interact with other signaling pathways in the cell.
DNA transfer screens for transformation have uncovered the oncogenic or cellular transforming potential of many proteins. The original gene transfer studies to identify oncogenes utilized genomic DNA isolated from a wide spectrum of human tumor cell lines and patient-derived tumor tissue. A significant outcome of these studies was the identification of mutated ras genes in 30% of all human cancers (6, 19, 24, 29, 56, 59, 62, 70). Other important oncogenes identified in this manner include vav, neu, met, ret, and trk, among others (14, 36, 45, 60, 65).
The use of genomic DNA for oncogene screening studies had several significant technical limitations that restricted efficient detection of transforming oncogenes. In particular, it is difficult to efficiently recover transforming sequences from the genomic DNA of the transformed recipient cells. Furthermore, the complexity of the entire human genome made it quite labor-intensive to adequately screen for activated oncogenes from a particular cell source. Advances in recombinant DNA technology have allowed a more efficient analysis using cDNA. Utilizing expression plasmids as a means to deliver, express, and recover cDNA sequences offers many advantages over genomic DNA transfer. In addition, cDNA represents the genes that are actually expressed in a given sample. Studies by Aaronson and colleagues using expression plasmid-based cDNA libraries identified a variety of oncogenes and included genes that encode for the heterotrimeric G alpha 12 subunit, the TC21 small GTPase, and the Ect2 and Ost Dbl family proteins (8–10, 48).
Whitehead et al., as well as Tsukamoto et al., utilized cDNA library screening that employed retrovirus-based expression vectors (69, 72). The greater efficiency of delivery of cDNA libraries provided by this method offers several advantages over traditional methods for screening for oncogenes. First, in contrast to DNA transfection, the efficiency of retroviral infection enhances the effective screening of the entire repertoire of genes expressed in a particular cell source. Second, it allows the use of cell lines that are not efficiently transfected for biological screens to detect growth-promoting genes. Genes identified by retroviral screening of cDNA libraries include highly transforming genes that encode the Lsc and Lfc Dbl family proteins, the RasGRP Ras guanine nucleotide exchange factor, and G2A-XGR G protein-coupled receptor (68, 73, 74, 76).
To date, cDNA expression library screens for novel oncogenes have primarily utilized immortalized or transformed cell lines as sources of cDNA. A potential concern in using cell lines is the fact that many tumor cell lines have been propagated in culture for extended periods and may not adequately represent the tumors from which they were derived. Another concern is that the gene expression profile may be altered when the cell line is propagated in vitro under artificial cell culture conditions. Thus, the utilization of patient-derived tumors as sources of cDNA may overcome these limitations and afford additional improvements in efforts to identify novel oncogenes important for the development of specific human malignancies. We chose to utilize the easy and highly efficient method developed by Whitehead et al. (72) to identify genes that may contribute to the development of acute myeloid leukemia (AML).
AML is a deadly disorder that is characterized by an aberrant accumulation of immature myeloid cells in the bone marrow and blood (49). A variety of genetic mutations have been found in AML, including point mutations in the N-ras gene and a variety of chromosomal translocations such as the promyelocytic leukemia-retinoic acid receptor (7, 20, 32, 52). Some mutations have been repeatedly identified because they are specifically analyzed by methods based on previous knowledge to look for them. However, any novel genetic mutation that could lead to the development of AML may be overlooked. Identification of additional oncogenes expressed in AML could provide great insight into how these leukemias develop and/or are maintained as well as help characterize how certain types of AML may respond to chemotherapeutic treatment. We were interested in expanding these studies on oncogenes expressed in AML by screening AML cDNA libraries by retrovirus-mediated transfer, which provides a more efficient approach to identifying novel genetic mutations in patients with AML. Here we describe the identification of a novel activating mutation in the TrkA protooncogene in a patient with AML. This is the first report of a TrkA mutation found in leukemia and the first demonstration of a deletion within TrkA in human cancer.
MATERIALS AND METHODS
cDNA library construction.
Blood from an AML patient, who had yet to undergo therapeutic treatment for the disease, was diluted in half with phosphate-buffered saline (PBS), and blood components were separated by spinning through an equal volume of Histopaque-1077 (Sigma Chemical Co.). Leukocytes were collected and washed with PBS. Total RNA was obtained using Trizol reagent (GIBCO-BRL) per the manufacturer's instructions. mRNA was purified from this total RNA using an mRNA purification kit (Amersham Pharmacia Biotech). cDNA was synthesized using a cDNA synthesis kit (GIBCO-BRL). The cDNA library was constructed essentially as described (72). Briefly, cDNA was treated with T4 DNA polymerase. BstXI adapters were then ligated to the blunt-ended cDNA, which was then size fractionated by agarose gel electrophoresis. cDNA was isolated and ligated into the pCTV1B retroviral vector (72) that had been cut with BstXI. This ligation was transformed into electrocompetent DH5α/P3 bacteria. Pooled bacteria were propagated and plasmid DNA was extracted using a plasmid midiprep kit (Bio-Rad Laboratories). The library used in this study contained about 4.6 × 106 independently isolated cDNA clones with an average size of approximately 1.5 kilobases.
Cell culture, retrovirus production, and retroviral infection.
Rat-1 fibroblasts, rat intestinal epithelial-1 (RIE-1) cells, and Bosc23 cells were grown in Dulbecco's modified Eagles medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (GIBCO-BRL). NIH 3T3 mouse fibroblasts were grown in DMEM supplemented with 10% calf serum (Hyclone Laboratories). 32D (clone 3) mouse myeloid progenitor cells were grown in RPMI supplemented with 10% FBS and 10% WEHI3B-conditioned medium (WEHI-cm) as a source of interleukin-3 (IL-3) (43). Penicillin and streptomycin were included in all media.
The AML cDNA library contained in the pCTV1B retroviral vector and all other retroviral vectors were converted to retrovirus using Bosc23 cells essentially as described (57). For library screening, the following number of cells were plated in 100-mm-diameter tissue culture dishes the day prior to infection: 5 × 105 Rat1 and NIH 3T3 cells and 8 × 105 RIE-1 cells. Infections were done using 1.5 to 3 ml of retrovirus, 1 to 1.5 ml of growth medium, and Polybrene (8 μg/ml) in a final volume of 3 or 4 ml per 100-mm-diameter dish. Retrovirus was removed 6 h later and replaced with growth medium. Infected cells were replenished with fresh growth medium every 2 to 3 days until primary foci appeared. Individual transformed foci were trypsinized and independently propagated.
To construct cell lines that expressed ΔTrkA, cells were infected with retrovirus made from the pBabepuro vector. For infections, 4 × 104 to 1 × 105 cells were plated in a well of a six-well plate. For 32D cells, 106 cells were infected. Following infection and two days of incubation, cells were passed into 100-mm-diameter dishes and selected in puromycin (1 μg/ml).
Isolation of transforming cDNAs and cloning into retroviral vectors.
The genomic DNA of cells propagated from transformed foci was isolated by proteinase K treatment and extraction (13). In order to obtain the integrated cDNA from retroviral infection, PCR was performed using primers for regions just outside of the cDNA cloning site in pCTV1B (72). These primers were pCTV-5′, 5′-CCTCACTCCTTCTCTAGCTC-3′, and pCTV1-3′, 5′-AACAAATTGGACTAATCGATACG-3′. PCRs contained 200 to 400 ng of genomic DNA, a 10 μM concentration of each primer, 1× cloned Pfu buffer, a 0.2 mM concentration of each deoxynucleoside triphosphate, 10% dimethyl sulfoxide, and 2.5 U of cloned Pfu polymerase (Stratagene) in 50-μl reaction mixtures. PCR products were gel purified, digested with MluI and BsiWI (New England Biolabs), and cloned into the pCTV3 retroviral vector (72). Retrovirus was made using this vector, and cells were infected to verify that transformation was caused by the rescued cDNA.
The ΔTrkA cDNA was cloned into the SalI site of the pBabepuro retroviral vector (51). The TrkA cDNA was cloned from pMexTrkA (a gift from Mariano Barbacid) into the EcoRI site of pBabepuro. The H-Ras61L cDNA was cloned from the pZIP-NeoSV(x)1 vector into pBabepuro.
Western blot analysis.
Primary antibodies that were used in this study include anti-Trk (sc7268), anti-extracellular signal-regulated kinase (anti-ERK) (sc93G), and anti-Stat5 (sc1656) (Santa Cruz Biotechnology), anti-phospho-ERK, anti-phospho-Akt, anti-Akt, anti-phospho-Stat5, and anti-phospho-TrkA(Tyr490) (New England Biolabs, Inc./Cell Signaling Technology) and anti-Ras (OP40; Oncogene Research Products-Calbiochem). Western blotting was performed per the manufacturer's instructions, and primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia Biotech). Blots were developed using enhanced chemilluminescence (Amersham Pharmacia Biotech). For Western blot analysis, equal number of cells were lysed in 2× sample buffer (20 mM NaPO4 [pH 7.0], 20% glycerol, 10% β-mercaptoethanol, 0.2 M dithiothreitol, and 0.02% bromophenol blue) prior to electrophoresis. Western blot analyses on the AML patient samples were performed following protein extraction using Trizol reagent (GIBCO-BRL) per the manufacturer's instructions.
32D cell apoptosis and growth analyses.
To assay 32D cell response to IL-3 deprivation, cells were washed twice in RPMI containing 10% FBS in order to remove the WEHI-cm containing IL-3. Cells were cultured in RPMI containing 10% FBS at a density of 5 × 105 cells per ml. Cell viability following IL-3 removal was monitored daily using trypan blue exclusion.
For experiments analyzing 32D cell growth in low-IL-3 conditions, parental 32D cells were first tested with varying concentrations of WEHI-cm to determine a level of WEHI-cm that would not support continued proliferation. This level was 0.5% for the batch of WEHI-cm that was utilized. Cells were washed twice in RPMI containing 10% FBS. Cells were placed in RPMI containing 10% FBS and 0.5% WEHI-cm at a density of 2 × 105 per ml. Cell growth and viability were monitored daily by trypan blue exclusion.
Measurement of Ras, ERK, Akt, and Stat5 activation and NGF treatment.
Prior to analyzing the relative levels of active Ras in 32D cells, cells were washed twice in RPMI only and starved in conical tubes in RPMI only at a density of 106 per ml for 3 h. Cells were then analyzed for active Ras by utilizing an activated Ras pull-down assay as previously described (67). Approximately 1 mg of lysate protein was used for this assay. For NIH 3T3 cells, cells were placed in DMEM containing 0.5% calf serum for 20 h prior to assaying for the relative amounts of active Ras or Western blotting for activated ERK and Akt. For the analysis of ERK and Stat5 activity in 32D cells, cells were cultured at a concentration of 2.5 × 105/ml in the absence of WEHI-cm for 3 h before Western blotting. For nerve growth factor (NGF) treatment, cells were plated at a density of 4 × 105 cells per well in six-well plates and treated the next day for various times with 100 ng of NGF (Boehringer Mannheim, Inc.) per ml. Cells were washed in PBS containing 100 μM sodium vanadate and analyzed by Western blotting.
PCR from cDNA library.
The TrkA cDNA, ΔTrkA cDNA, and the AML cDNA libraries were analyzed by PCR in order to detect the deletion of ΔTrkA. Primers were designed that would detect wild-type TrkA as a 326-bp PCR product and ΔTrkA as a 101-bp product. These primers were: 5′, 5′-TCCCGGCCAGTGTGCAGCTG-3′, and 3′, 5′-AGGGATGGGGTCCTCGGGGTTGAA-3′. PCRs contained 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 75 mM KCl, a 0.2 mM concentration of each deoxynucleoside triphosphate, 200 ng of each primer, 10 ng of plasmid DNA or 100 ng of cDNA library, and 2.5 U of Pfu polymerase.
RESULTS
Isolation of a novel mutation in the TrkA protooncogene in a patient with AML.
The efficiency of screening for oncogenes has increased with the advent of using retroviruses to deliver DNA into cells. We were interested in utilizing retroviruses to efficiently screen patient samples for oncogenes expressed in AML. Several myeloid leukemia samples were obtained, and cDNA libraries were constructed within the pCTV1B retroviral vector (72). One library contained 4.6 × 106 independently isolated cDNAs and was used to screen a variety of cell types for transformation. These cell lines included NIH 3T3 mouse fibroblasts, Rat1 fibroblasts, RIE-1 cells, and 32D myeloid progenitor cells. NIH 3T3 cells have been classically used as a target cell type for the isolation of new oncogenes because it is well known that these cells can easily be transformed by a single oncogene. Rat1 fibroblasts and RIE-1 cells were also used because of their very low rate of spontaneous transformation, high efficiency of infection by retroviruses, and because epithelial cells are the cellular origin of the majority of human cancers. In addition, 32D myeloid cells were chosen as a unique cell line to isolate AML-associated oncogenes whose expression could deregulate the growth of cells of the myeloid lineage of the hematopoietic system. While it has been documented that a wide range of oncogenes can transform NIH 3T3 cells, some of these oncogenes cannot fully transform 32D cells (12, 47) (data not shown). This suggests that 32D cells may require activation of cell-type specific signaling pathways that lead to transformation. Therefore, screening multiple cell types increases the likelihood of identifying expressed genes that may have transforming potential.
From both the Rat1 and RIE-1 screens, a 2.3-kb cDNA was isolated from a population of cells derived from a transformed focus. Since it is possible that multiple cDNAs can be introduced into a cell simultaneously and given the fact that these cells were not a clonal population, it had to be determined if the isolated cDNA was sufficient to cause cellular transformation. NIH 3T3, Rat1, and RIE-1 cells that were infected with virus containing the isolated cDNA readily became morphologically transformed (data not shown), confirming that expression of this cDNA was sufficient to cause transformation.
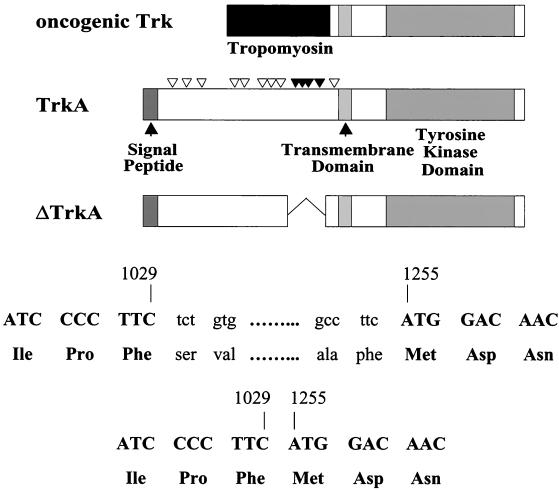
Sequence analysis of this cDNA indicated that this expressed gene was a variant of the TrkA protooncogene, which encodes the receptor for NGF, TrkA (38). The protein encoded by this cDNA contained an in-frame deletion of 225 nucleotides encoding 75 amino acids of TrkA (Fig. 1). This deletion corresponds to a region just outside of the transmembrane domain of the receptor. Our designation for this truncated version of human TrkA is ΔTrkA. The Trk oncogene was originally discovered as a transforming gene from a colon carcinoma biopsy specimen (45). This gene was the result of a fusion of sequences of the gene for tropomyosin to sequences of an unknown gene that was later identified as trkA (Fig. 1).
FIG. 1.
Schematic diagrams of oncogenic Trk, TrkA, and ΔTrkA. Oncogenic Trk contains sequences of tropomyosin in place of most of the extracellular domain of TrkA. ΔTrkA contains an in-frame 75-amino-acid deletion of the extracellular domain of TrkA. The deletion is the result of a loss of 225 nucleotides from the trkA gene (nucleotides 1030 to 1254). Nucleotide numbering is based on the TrkA GenBank submission (accession number M23102). Nucleotides shown in lowercase type are not present in the ΔTrkA cDNA. The triangles above TrkA indicate sites of glycosylation. Four glycosylation sites (solid triangles) are deleted in ΔTrkA. The signal peptide, transmembrane domain, and tyrosine kinase domain in TrkA are indicated.
ΔTrkA causes growth transformation of NIH 3T3 fibroblasts, Rat1 fibroblasts, RIE-1 epithelial cells, and 32D myeloid cells.
TrkA is a receptor tyrosine kinase that binds and is activated by NGF (33, 34, 38). It is possible that overexpression of the TrkA tyrosine kinase domain leads to its constitutive activation, resulting in aberrant activation of intracellular cascades, which results in cellular transformation. Utilizing the pBabepuro retroviral vector, wild-type TrkA and ΔTrkA were stably expressed in NIH 3T3, Rat1, and RIE-1 cells (Fig. 2 and data not shown). Unlike expression of ΔTrkA, TrkA did not induce cellular transformation in RIE-1 cells or other cell types (Fig. 2 and data not shown). RIE-1 cell transformation induced by ΔTrkA was essentially indistinguishable from that caused by the activated Ras protein, H-Ras61L (Fig. 2). These transformed cells were highly refractile and spindle shaped. In addition, NIH 3T3, Rat1, and RIE-1 cells expressing ΔTrkA readily grew in soft agar, indicating this activated TrkA protein transformed these cells to a state of anchorage-independent growth (data not shown).
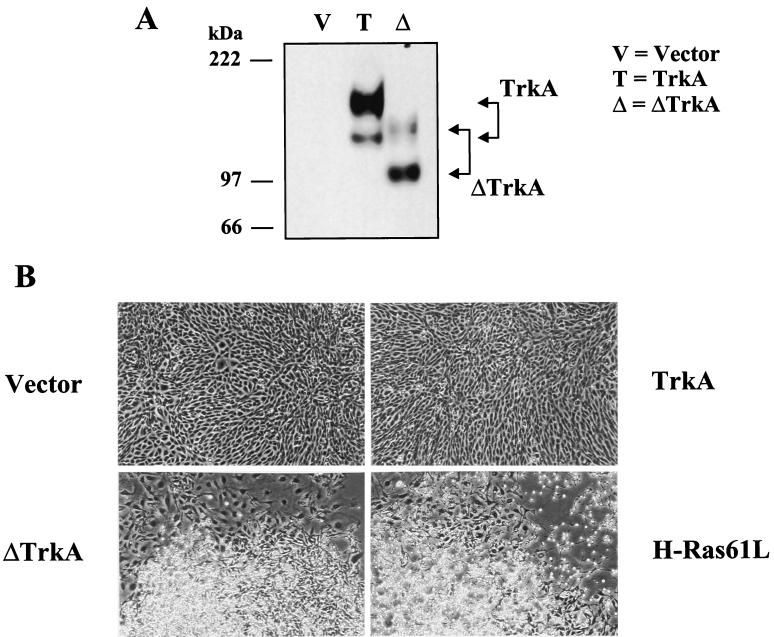
FIG. 2.
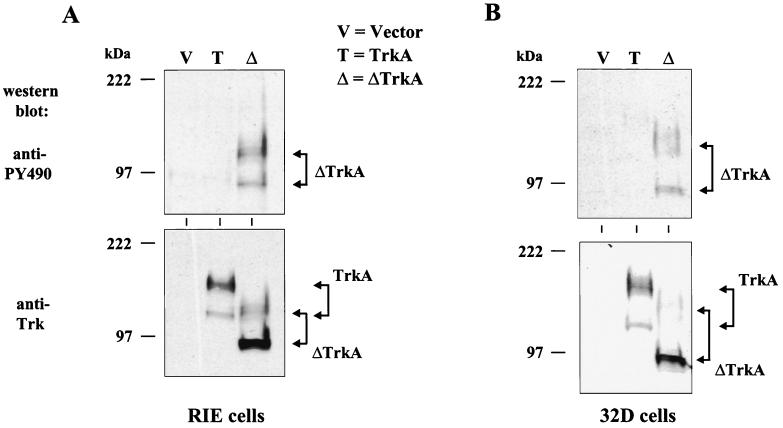
ΔTrkA transforms RIE-1 cells. RIE-1 cell lines were made that stably express empty vector, TrkA, ΔTrkA, and H-Ras61L. (A) Expression of Trk proteins was analyzed by Western blotting with anti-Trk antibodies. The position of TrkA and ΔTrkA proteins are indicated, and the molecular masses of standards are indicated at left. (B) ΔTrkA morphologically transforms RIE-1 cells, while wild-type TrkA does not. The highly refractile and morphologically transformed ΔTrkA cells are essentially indistinguishable from those caused by oncogenic H-Ras61L.
Western blot analysis of cells expressing TrkA or ΔTrkA indicated that TrkA migrated as a doublet at about 140 and 110 kDa, whereas ΔTrkA migrated as proteins of about 115 and 98 kDa (Fig. 2). While the predicted molecular mass of TrkA is about 86 kDa, its apparent molecular mass has been previously characterized as a result of glycosylation (46). The 140-kDa protein is fully processed TrkA, while the 110-kDa protein is only partially glycosylated. The difference in molecular weight between ΔTrkA and TrkA is due in part to the loss of 75 amino acids in ΔTrkA. The 75-amino-acid deletion in ΔTrkA removes four glycosylation sites from the protein, which also likely contributes to the lower molecular weight of ΔTrkA. While the fully processed form of TrkA is more abundant than the partially glycosylated form, the partially glycosylated form of ΔTrkA is more abundant than its fully processed form (Fig. 2). Again, this may be explained by the loss of glycosylation sites in ΔTrkA.
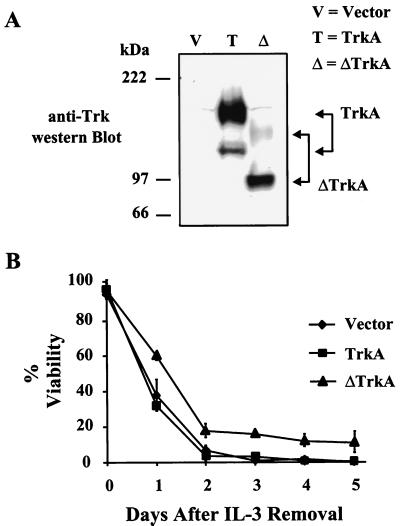
While it was clear that ΔTrkA was capable of functioning as an oncogene in fibroblasts and epithelial cells, its effect on a myeloid cell line would be more relevant to the disease in which it was derived from. 32D cells are murine, nontransformed, myeloid progenitor cells that depend on IL-3 for viability and growth (28). These cells have been shown to be transformed by oncogenes that cause human leukemias (e.g., Bcr-Abl) and therefore represent a cell type more relevant to the study of an oncogene of myeloid origin (42). 32D cells were established to stabily express TrkA and ΔTrkA. Comparable levels of TrkA and ΔTrkA protein were expressed in mass populations of 32D cells following retroviral infection and drug selection (Fig. 3A).
FIG. 3.
Expression of ΔTrkA in 32D myeloid cells delays apoptosis in response to IL-3 deprivation. (A) 32D cells stably expressing empty vector, TrkA, or ΔTrkA were analyzed by Western blotting with anti-Trk antibodies. (B) 32D cells expressing vector, TrkA, or ΔTrkA were deprived of IL-3, and cell viability over time was analyzed by trypan blue exclusion. Error bars represent standard deviation within a single experiment. Essentially identical results were observed with three independently derived sets of cell lines.
32D cells undergo apoptosis when cultured in the absence of IL-3 (1). These cells are considered fully transformed, or growth factor independent, when they do not die and continue to proliferate in the absence of IL-3. The Bcr-Abl protein, which is believed to be the causative agent of Philadelphia chromosome-positive human leukemias, is an example of a protein that can fully transform these cells (42). 32D cells expressing either empty vector, TrkA, ΔTrkA, or Bcr-Abl were cultured in the absence of IL-3, and cell growth and viability were measured by trypan blue exclusion. 32D cells expressing either empty vector or TrkA died rapidly in the absence of IL-3 (Fig. 3). This rate of death was similar to uninfected parental cells (data not shown). Expression of Bcr-Abl in these cells prevented cell death, and these cells continued to proliferate in the absence of IL-3 (data not shown). Cells expressing ΔTrkA died at a slower rate than vector and TrkA cells (Fig. 3). A significant fraction of these cells remained viable for 10 days or even longer, a result that was reproducible with the three stable ΔTrkA 32D cells lines that were created (data not shown). In addition, in one cell line, cells expressing ΔTrkA eventually became growth factor independent and could proliferate indefinitely in the absence of IL-3 (data not shown). However, this was not reproducibly observed when additional independently derived cell lines were analyzed. The fact that ΔTrkA could delay apoptosis in response to IL-3 but could not support the continued proliferation of these cells explains why ΔTrkA was not cloned out of a 32D cell screen for IL-3 independence, following infection with the AML cDNA library.
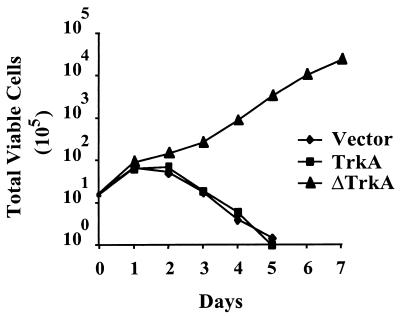
To determine if ΔTrkA could alter the growth properties of 32D cells, cells expressing ΔTrkA were plated in low-IL-3 conditions. The level of IL-3 used in these experiments did not support the growth of parental 32D cells (data not shown). Under these conditions, 32D cells expressing ΔTrkA maintained a high level of viability and were able to proliferate indefinitely while cells expressing vector control or TrkA underwent cell death (Fig. 4 and data not shown). Together, these data suggest that expression of ΔTrkA in myeloid cells has an inhibitory effect on apoptotic mechanisms and promotes reduced growth factor dependence for proliferation. Therefore, ΔTrkA can alter the cell survival and growth properties of myeloid progenitor cells.
FIG. 4.
Expression of ΔTrkA in 32D myeloid cells promotes growth in low concentrations of IL-3. 32D cells stably expressing empty vector, TrkA, or ΔTrkA were plated under suboptimal IL-3 conditions (0.5% WEHI-cm). Total viable cells were determined over time by trypan blue exclusion. Nearly identical results were obtained from the two independently derived sets of cell lines that were analyzed.
ΔTrkA is constitutively hyperphosphorylated on tyrosines and causes sustained activation of Ras, ERK, and Akt but not Stat5.
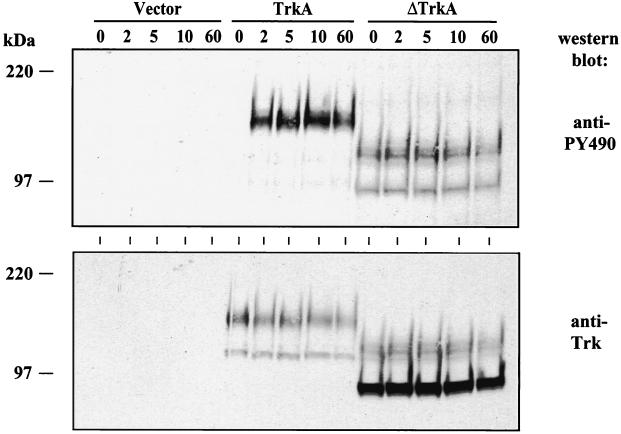
The oncogenic form of Trk has been shown to have increased intrinsic tyrosine kinase activity (5, 50). Activation of TrkA leads to the tyrosine phosphorylation of multiple tyrosines on the cytoplasmic portion of the receptor (34, 38). Two of these sites of phosphorylation (tyrosines 674 and 675) have been shown to be correlated with tyrosine kinase activity and another one (tyrosine 490) with the binding of the Shc adapter protein (21, 53, 61). To analyze the tyrosine phosphorylation state of ΔTrkA, an antibody (PY490) that specifically recognizes TrkA tyrosine 490 when it is phosphorylated was utilized. In both RIE-1 cells and 32D cells, this antibody only recognized ΔTrkA and not wild-type TrkA, suggesting that ΔTrkA is constitutively tyrosine phosphorylated and chronically stimulates downstream signaling (Fig. 5). Another antibody that recognizes tyrosines 674 and 675 when phosphorylated also only recognized ΔTrkA (data not shown). Treatment of RIE-1 cells expressing vector, TrkA, or ΔTrkA with NGF rapidly induced the tyrosine phosphorylation of TrkA but did not affect the phosphorylation of ΔTrkA, as measured by antibodies that recognize tyrosine 490 as well as antibodies that recognize tyrosines 674 and 675 (Fig. 6 and data not shown). Thus, constitutively upregulated, ligand-independent tyrosine kinase activity could explain the transforming properties of ΔTrkA.
FIG. 5.
ΔTrkA is constitutively tyrosine phosphorylated in RIE-1 cells and 32D myeloid progenitor cells. RIE-1 cells (A) and 32D cells (B) stably expressing empty vector, TrkA, and ΔTrkA were analyzed by Western blotting with antibodies that recognize phosphorylated tyrosine 490 of TrkA (PY490) (top panels). The blots were stripped and reprobed with anti-Trk antibodies (bottom panels).
FIG. 6.
NGF does not increase the tyrosine phosphorylation of ΔTrkA. RIE cells expressing either vector, TrkA, or ΔTrkA were treated with NGF for the times indicated (in minutes). Total cell lysates were collected and analyzed by Western blotting with anti-PY490, which specifically recognizes phosphorylation of tyrosine 490 of TrkA (top panel). The blot was stripped and western blotted with anti-Trk antibodies (bottom panel).
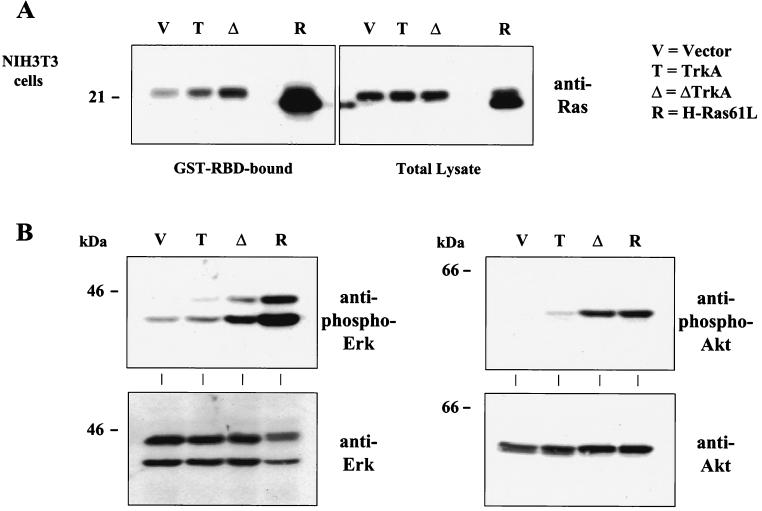
Transient activation of TrkA by NGF leads to recruitment of the Shc adapter protein and subsequent activation of the Ras GTPase (3, 21, 35, 39, 53). To date, whether oncogenic Trk transformation leads to constitutive activation of Ras has not been shown. To determine if the constitutive activation of ΔTrkA signals to components known to be downstream of activated TrkA, the level of activated Ras was measured in NIH 3T3 cells expressing ΔTrkA. Utilizing an activated Ras pull-down assay, it was shown that expression of ΔTrkA leads to a constitutive elevation in the amount of activated Ras (Ras-GTP) (Fig. 7A). In addition, expression of ΔTrkA led to the constitutive activation of ERK mitogen-activated protein (MAP) kinases as well as the Akt serine kinase, as measured by Western blot analyses with phospho-specific antibodies that recognize the activated forms of these kinases (Fig. 7B). A chemical inhibitor (U0126) of the ERK activator, MEK, blocked transformation by ΔTrkA (data not shown). This confirms that ΔTrkA constitutively elicits intracellular signals.
FIG. 7.
Cells expressing ΔTrkA contain elevated levels of Ras-GTP and activated ERK MAP kinases and Akt. (A) The level of Ras activation was measured by the Ras glutathione S-transferase–Ras binding domain (GST-RBD) pull-down assay. Bound (active) Ras (Ras-GTP) and total cell lysates of NIH 3T3 cells expressing vector, TrkA, ΔTrkA, and H-Ras61L were analyzed by Western blotting with anti-pan (non-isoform-specific) Ras antibodies. H-Ras61L migrates faster than wild-type endogenous Ras. (B) NIH 3T3 cells were analyzed for activation of ERK (left, top panel) and Akt (right, top panel) by Western blotting total cell lysates with antibodies that recognize the activated, phosphorylated forms of these kinases. Blots were also probed with anti-ERK (left, bottom panel) and anti-Akt (right, bottom panel) antibodies as controls.
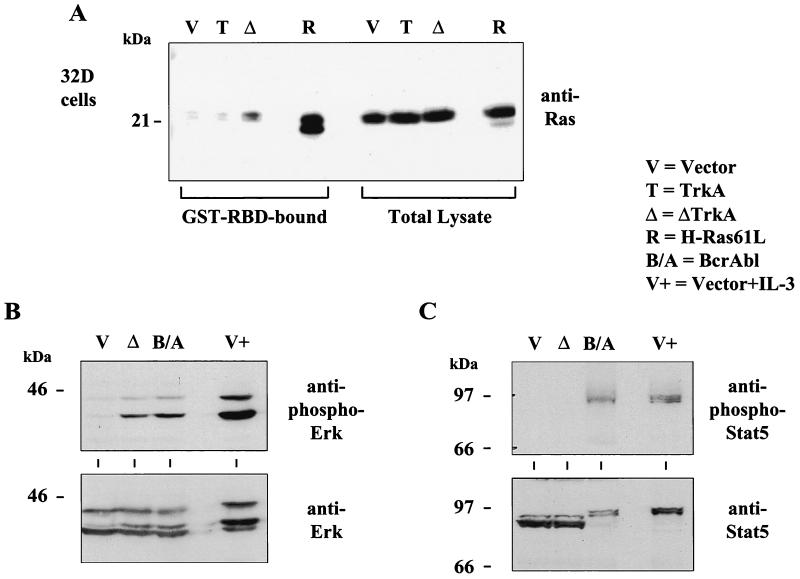
Similar experiments were performed in 32D cells expressing ΔTrkA. These cells also contained constitutively elevated levels of active Ras and ERK MAP kinases (Fig. 8A and B). We were unable to determine if ΔTrkA activates Akt in these cells. The phospho-Akt antibodies used did not clearly detect activated Akt even under conditions of IL-3 stimulation or Bcr-Abl expression, two signals that are known to activate Akt. In addition, we analyzed the activation state of Stat5 in these cells. The activation state of Stat5 can be monitored by the phosphorylation status of tyrosine 694 (26). Using this phospho-specific antibody, we observed Stat5 activation by both Bcr-Abl and IL-3 stimulation but not ΔTrkA expression (Fig. 8C).
FIG. 8.
32D cells expressing ΔTrkA contain elevated levels of activated Ras and ERK MAP kinases but do not contain activated Stat5. (A) The level of Ras activation was measured by the Ras glutathione S-transferase–Ras binding domain (GST-RBD) pull-down assay. Bound (active) Ras (Ras-GTP) and total cell lysates of 32D cells expressing vector, TrkA, ΔTrkA, and H-Ras61L were analyzed by Western blotting with anti-pan Ras antibodies. H-Ras61L migrates faster than wild-type endogenous Ras. (B) Cell lysates of 32D cells expressing vector, ΔTrkA, Bcr-Abl, or vector cells stimulated with IL-3 were analyzed by Western blotting with antibodies that recognize activated, phosphorylated ERK MAP kinases. Blots were also analyzed with anti-ERK antibodies. (C) Cell lysates of 32D cells expressing vector, ΔTrkA, Bcr-Abl, or vector cells stimulated with IL-3 were analyzed by Western blotting with antibodies that recognize activated, phosphorylated Stat5. Blots were also probed with anti-Stat5 antibodies. A mobility shift caused by Stat5 phosphorylation can be seen.
ΔTrkA was expressed in the AML patient.
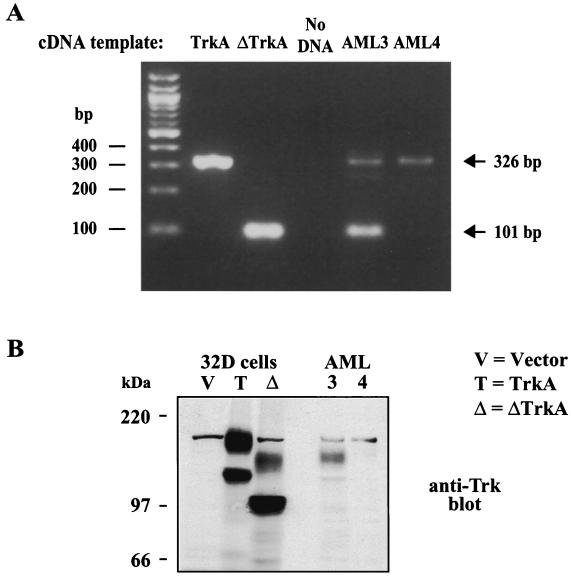
Finally, it was possible that the deletion in ΔTrkA was a result of the transfection during the retroviral production or a result of the retroviral integration process. It has been previously documented that the trkA cDNA undergoes frequent rearrangements during standard transfection techniques and that these alterations generate transforming versions of TrkA (54). To determine if the deletion in ΔTrkA was present before transfection and therefore originating from the patient, the cDNA library that was generated from the patient was analyzed by PCR. Based on the ΔTrkA sequence, primers were designed that would detect ΔTrkA cDNA as a 101-bp PCR product and wild-type TrkA cDNA as a 326-bp product. PCR analysis of the AML patient-derived cDNA library (AML3) that was used to clone ΔTrkA generated both the 101-bp and 326-bp PCR products, indicating that ΔTrkA was present as an expressed gene in the patient (Fig. 9A). Only the wild-type TrkA PCR product was present after PCR analysis of cDNA generated from mRNA isolated from an unrelated AML patient (AML4). Sequence analysis of the 101-bp PCR product confirmed the deletion of the same Trk nucleotides that are deleted in the ΔTrkA cDNA sequence. In addition, just upstream of the deletion there is a point mutation in the ΔTrkA cDNA that changes the serine at amino acid 300 to a cysteine, in comparison to the published TrkA sequence (GenBank accession number M23102). This mutation was also present in the 326-bp product, indicating that the cDNA matches the wild-type trkA allele at this position. We analyzed the sequence of this region of trkA from two additional AML samples as well as from a normal donor. In all cases this codon encodes for a cysteine and not a serine (data not shown). This suggests that the sequence of TrkA deposited in GenBank may contain an error at this position.
FIG. 9.
ΔTrkA was expressed in the AML patient. (A) Primers were designed based on the ΔTrkA cDNA sequence in order to discriminate between wild-type TrkA and ΔTrkA. These primers were used to PCR amplify TrkA cDNA, ΔTrkA cDNA, AML3 library cDNA (the library screened in this study), and AML4 library cDNA (an unrelated AML patient sample). The 326-bp PCR fragment indicates the presence of the wild-type TrkA cDNA, while the 101-bp PCR product indicates the presence of the ΔTrkA cDNA. (B) Total cell lysates of 32D cells expressing vector, TrkA, ΔTrkA, and protein extracts from the AML3 patient and the unrelated AML4 patient were analyzed by Western blotting with anti-Trk antibodies.
Finally, to confirm the PCR analysis that indicated ΔTrkA was expressed in the patient, we analyzed protein from the patient-derived leukemic cells by Western blotting. Antibodies that recognize Trk detected a protein that was smaller than wild-type TrkA and that migrated closely with ΔTrkA expressed in 32D cells (Fig. 9B). This suggests that ΔTrkA was expressed in the AML patient.
DISCUSSION
The Trk oncogene was identified from a colon carcinoma (45). Sequences of the gene for nonmuscle tropomyosin were found fused upstream of sequences that encoded a protein that had homology to tyrosine kinases. This latter gene was subsequently cloned as the TrkA protooncogene, and its product, TrkA, is a receptor tyrosine kinase that binds to, is activated by, and elicits the biological properties of the neurotrophin, NGF (33–35, 38, 39). TrkA is a member of a family of neurotrophin receptors (2). In addition to being present in the original colon carcinoma, TrkA has been found mutated in papillary thyroid carcinomas (4). In all of these cases, the gene for TrkA is found rearranged with another gene, such as those encoding tropomyosin and TPR among others (27, 55). This results in the replacement of sequences at the TrkA amino terminus with amino acids encoded by the other gene. It is believed that fusion of TrkA to these proteins results in constitutive activation of the tyrosine kinase activity of TrkA.
Activation of TrkA by NGF leads to activation of the Ras-Raf-ERK pathway as well as the phosphatidylinositol (PI) 3-kinase and phospholipase C-γ (reviewed in references 35 and 39). Activation of Ras is mediated through a protein complex formation involving tyrosine-phosphorylated TrkA, Shc, and Grb2, which in turn binds to the Ras activator Sos. While it is believed that Ras activation plays an important role in signaling by TrkA, the specific roles for Ras are inconclusive. TrkA signaling, as a result of NGF treatment, induces differentiation of the pheochromocytoma cell line PC12 and inhibits apoptosis induced by serum removal. Differentiation is believed to be mediated by activation of the Ras-ERK pathway (35, 39). However, NGF-mediated survival of these cells does not require Ras (75). Cell survival is believed to be mediated through a PI 3-kinase-dependent mechanism that signals to the antiapoptotic kinase Akt (18, 25, 75). Interestingly, Ras is required for NGF-mediated survival signaling in primary neurons (35, 39). While NGF and Trk studies have focused primarily on PC12 cells, the Trk oncogene has been shown to transform a variety of other cell types, including fibroblasts and hematopoietic cells (11, 37, 66).
In this study, we identified a mutation in the TrkA gene in a patient with AML. This is the first example of a TrkA mutation identified from a leukemia patient. While previously described mutations in this gene have involved rearrangements with other genes, the mutation described here is an internal, in-frame deletion (Fig. 1). The deleted sequences are from within exon 8 but are not inclusive of the entire exon. A direct repeat of CCTTC, present just before the deleted sequences and at the end of the deleted sequences, may have been involved in a recombination event that removed these sequences. This deletion removes 75 amino acids in the extracellular domain of the TrkA receptor tyrosine kinase.
The resulting protein product, which we have named ΔTrkA, is highly transforming in Rat1 and NIH 3T3 fibroblasts and also in RIE-1 epithelial cells (Fig. 2 and data not shown). This transformation is seen both morphologically and by anchorage-independent growth. ΔTrkA, unlike overexpressed wild-type TrkA, is constitutively phosphorylated on multiple tyrosines (Fig. 5 and data not shown). These include tyrosine 490, which is an important regulator of TrkA signaling to downstream targets, including both Ras and PI 3-kinase, as well as tyrosines 674 and 675 whose phosphorylation has been shown to correlate with kinase activity (30, 61). Constitutive activation of Ras and downstream pathways containing ERK and Akt have been observed in cells expressing ΔTrkA (Fig. 7 and 8). These data indicate that the tyrosine kinase activity of ΔTrkA is deregulated, resulting in its constitutive activation and chronic stimulation of downstream signaling pathways.
It is likely that the deletion in the extracellular domain confers a conformational change in the protein that deregulates the kinase domain. These data are in agreement with in vitro analyses using the trkA cDNA where a spontaneous deletion of a portion of the extracellular domain resulted in a transforming protein (17). Further analyses indicated that mutation of cysteine 345 to serine resulted in a weakly transforming form of TrkA (17). Many cysteines, including cysteine 345 of TrkA, are conserved in Trk family members and therefore may be important determinants of the structure of Trk proteins. This cysteine is deleted in ΔTrkA. Therefore, it appears that subtle changes in the extracellular region of TrkA, which may alter the tertiary structure of the protein, can alter the activation state of the intracellular tyrosine kinase domain. In addition, the deletion that created ΔTrkA removed several glycosylation sites that may affect protein function. Glycosylation of TrkA has been shown to inhibit TrkA kinase activity (71). It was shown that deglycosylated TrkA was constitutively activated but did not signal to downstream targets like the ERK pathway. It is speculated that glycosylation may prevent spontaneous homo-interactions of TrkA molecules that may result in activation of the tyrosine kinase domain (71). While this suggests a mechanism by which ΔTrkA may exhibit elevated tyrosine kinase activity, it should be noted that the hyperglycosylated form of ΔTrkA contains more tyrosine phosphorylation than the underglycosylated form (Fig. 5).
While ΔTrkA transforms both fibroblasts and epithelial cells, its identification from an AML patient suggests it could alter the growth properties of myeloid cells. Expression of ΔTrkA in 32D myeloid cells slowed the rate of apoptosis in response to IL-3 withdrawal (Fig. 3). In addition, it allowed the proliferation of 32D myeloid cells in concentrations of IL-3 that could not support the growth of control cells (Fig. 4). The original Trk oncogene isolated from a colon carcinoma has also been shown to transform hematopoietic cells (37). 32D cells expressing ΔTrkA had elevated levels of activated Ras and ERK MAP kinases, but did not contain constitutively activated Stat5 (Fig. 8). This is unlike the expression of Bcr-Abl which activates all of these pathways and renders 32D cells independent of IL-3 for viability and growth. Stat5 has been shown to regulate the expression of Bcl-X to inhibit apoptosis (22, 63). The lack of Stat5 activation by ΔTrkA may explain why these cells require low levels of IL-3 to retain viability. These levels of IL-3 were enough to activate Stat5 (G. W. Reuther, Q. T. Lambert, and C. J. Der, unpublished data). Interestingly, the single cell line that did become IL-3 independent after ΔTrkA expression had elevated levels of activated Stat5, suggesting that a second mutation occurred that led to activation of this pathway and that this may have cooperated with other ΔTrkA-induced signals to transform these cells (Reuther et al., unpublished data). Importantly, Stat5 has been shown to cooperate with the PI 3-kinase pathway to transform an IL-3-dependent hematopoietic cell line (58). Chemical inhibitors of both the MEK and PI 3-kinase pathways completely blocked the ability of ΔTrkA to inhibit apoptosis upon IL-3 deprivation (Reuther et al., unpublished data). These results are consistent with previous work suggesting that both of these pathways contribute to blocking cell death and rendering cells IL-3 independent (12, 64). Mutational or signal-induced activation of Ras has been shown to alter the apoptotic properties of 32D cells, and mutations in N-Ras are frequently found in AML (Reuther et al., unpublished data and (7, 15, 16, 52)). Thus, ΔTrkA may contribute to the development and/or maintenance of leukemia through the constitutive upregulation of Ras activity.
We were able to detect a Trk protein smaller than TrkA and similar in size to ΔTrkA in the patient from which ΔTrkA was cloned (Fig. 9B). ΔTrkA was highly expressed in these cells, suggesting the majority of the leukemic cells expressed this mutated form of TrkA. This is consistent with ΔTrkA providing a growth advantage and enrichment of these cells. It would have been interesting to analyze the activation states of various signaling pathways in these cells. Unfortunately, this type of analysis is complicated by several factors, including, and most importantly, the lack of appropriate negative control cells to compare the sample to.
While we have so far been unable to transform mouse primary hematopoietic cells with ΔTrkA, two recent reports describe an ETV6-TrkC fusion protein and a TEL-TrkC fusion protein that were isolated from patients with AML (23, 44). These proteins were able to induce a myeloproliferative disorder in mice. Thus, it is likely that activation of Trk family members may play a role in the development of various leukemias. We analyzed 11 additional AML samples and did not identify a deletion in this region of TrkA in any samples. Based on this analysis, it is likely that ΔTrkA is not a common mutation but rather a sporadic event. However, our analysis does not exclude the possibility that TrkA may be activated by other mechanisms in leukemia.
While TrkA expression was thought to be specific to neuronal cells, it is expressed in a wide range of tissues and its expression has been identified in AML patient samples (31, 40, 41, 55). Therefore, a mutation in the trkA gene has the potential to contribute to the development of myeloid leukemias. ΔTrkA alters the growth and apoptotic properties of myeloid cells (Fig. 3 and 4). ΔTrkA may therefore provide hematopoietic cells with a growth advantage by altering mitogenic signaling. Additionally, it may prevent the normal turnover of these cells by altering the apoptotic signals that help define the makeup of the hematopoietic system. Alteration of mitogenic and/or apoptotic signals by ΔTrkA could have contributed to the expansion and accumulation of white blood cells, leading to a leukemic state.
ACKNOWLEDGMENTS
We thank Beverly S. Mitchell for assistance in obtaining AML blood samples, Aylin S. Ulku for construction of the pBabePuro H-Ras61L expression plasmid, Mariano Barbacid for the pMexTrkA expression plasmid, and Warren S. Pear for Bosc23 cells and helpful discussions.
G.W.R. is a recipient of the Cancer Research Institute/Merrill Lynch Fellowship. This work was supported by grants from the National Institutes of Health to C.J.D. (CA42978, CA55008, and CA63071).
REFERENCES
- 1.Askew D S, Ashmun R A, Simmons B C, Cleveland J L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 2.Barbacid M. Neurotrophic factors and their receptors. Curr Opin Cell Biol. 1995;7:148–155. doi: 10.1016/0955-0674(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 3.Blaikie P, Immanuel D, Wu J, Li N, Yajnik V, Margolis B. A region in Shc distinct from the SH2 domain can bind tyrosine-phosphorylated growth factor receptors. J Biol Chem. 1994;269:32031–32034. [PubMed] [Google Scholar]
- 4.Bongarzone I, Pierotti M A, Monzini N, Mondellini P, Manenti G, Donghi R, Pilotti S, Grieco M, Santoro M, Fusco A. High frequency of activation of tyrosine kinase oncogenes in human papillary thyroid carcinoma. Oncogene. 1989;4:1457–1462. [PubMed] [Google Scholar]
- 5.Borrello M G, Pelicci G, Arighi E, De Filippis L, Greco A, Bongarzone I, Rizzetti M, Pelicci P G, Pierotti M A. The oncogenic versions of the Ret and Trk tyrosine kinases bind Shc and Grb2 adaptor proteins. Oncogene. 1994;9:1661–1668. [PubMed] [Google Scholar]
- 6.Bos J L. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 7.Bos J L, Verlaan-de Vries V M, van der Eb A J, Janssen J W, Delwel R, Lowenberg B, Colly L P. Mutations in N-ras predominate in acute myeloid leukemia. Blood. 1987;69:1237–1241. [PubMed] [Google Scholar]
- 8.Chan A M, Fleming T P, McGovern E S, Chedid M, Miki T, Aaronson S A. Expression cDNA cloning of a transforming gene encoding the wild-type G alpha 12 gene product. Mol Cell Biol. 1993;13:762–768. doi: 10.1128/mcb.13.2.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan A M, McGovern E S, Catalano G, Fleming T P, Miki T. Expression cDNA cloning of a novel oncogene with sequence similarity to regulators of small GTP-binding proteins. Oncogene. 1994;9:1057–1063. [PubMed] [Google Scholar]
- 10.Chan A M, Miki T, Meyers K A, Aaronson S A. A human oncogene of the RAS superfamily unmasked by expression cDNA cloning. Proc Natl Acad Sci USA. 1994;91:7558–7562. doi: 10.1073/pnas.91.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleveland J L, Dean M, Rosenberg N, Wang J Y, Rapp U R. Tyrosine kinase oncogenes abrogate interleukin-3 dependence of murine myeloid cells through signaling pathways involving c-myc: conditional regulation of c-myc transcription by temperature-sensitive v-abl. Mol Cell Biol. 1989;9:5685–5695. doi: 10.1128/mcb.9.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleveland J L, Troppmair J, Packham G, Askew D S, Lloyd P, Gonzalez-Garcia M, Nunez G, Ihle J N, Rapp U R. v-raf suppresses apoptosis and promotes growth of interleukin-3-dependent myeloid cells. Oncogene. 1994;9:2217–2226. [PubMed] [Google Scholar]
- 13.Coen D M. Quantitation of rare cDNAs by PCR. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1992. pp. 1–2. [Google Scholar]
- 14.Cooper C S, Park M, Blair D G, Tainsky M A, Huebner K, Croce C M, Vande W G. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 15.Cortez D, Kadlec L, Pendergast A M. Structural and signaling requirements for BCR-ABL-mediated transformation and inhibition of apoptosis. Mol Cell Biol. 1995;15:5531–5541. doi: 10.1128/mcb.15.10.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortez D, Stoica G, Pierce J H, Pendergast A M. The BCR-ABL tyrosine kinase inhibits apoptosis by activating a Ras-dependent signaling pathway. Oncogene. 1996;13:2589–2594. [PubMed] [Google Scholar]
- 17.Coulier F, Kumar R, Ernst M, Klein R, Martin-Zanca D, Barbacid M. Human trk oncogenes activated by point mutation, in-frame deletion, and duplication of the tyrosine kinase domain. Mol Cell Biol. 1990;10:4202–4210. doi: 10.1128/mcb.10.8.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datta K, Bellacosa A, Chan T O, Tsichlis P N. Akt is a direct target of the phosphatidylinositol 3-kinase. Activation by growth factors, v-src and v-Ha-ras, in Sf9 and mammalian cells. J Biol Chem. 1996;271:30835–30839. doi: 10.1074/jbc.271.48.30835. [DOI] [PubMed] [Google Scholar]
- 19.Der C J, Krontiris T G, Cooper G M. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci USA. 1982;79:3637–3640. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 21.Dikic I, Batzer A G, Blaikie P, Obermeier A, Ullrich A, Schlessinger J, Margolis B. Shc binding to nerve growth factor receptor is mediated by the phosphotyrosine interaction domain. J Biol Chem. 1995;270:15125–15129. doi: 10.1074/jbc.270.25.15125. [DOI] [PubMed] [Google Scholar]
- 22.Dumon S, Santos S C, Debierre-Grockiego F, Gouilleux-Gruart V, Cocault L, Boucheron C, Mollat P, Gisselbrecht S, Gouilleux F. IL-3 dependent regulation of Bcl-xL gene expression by STAT5 in a bone marrow derived cell line. Oncogene. 1999;18:4191–4199. doi: 10.1038/sj.onc.1202796. [DOI] [PubMed] [Google Scholar]
- 23.Eguchi M, Eguchi-Ishimae M, Tojo A, Morishita K, Suzuki K, Sato Y, Kudoh S, Tanaka K, Setoyama M, Nagamura F, Asano S, Kamada N. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25) Blood. 1999;93:1355–1363. [PubMed] [Google Scholar]
- 24.Eva A, Tronick S R, Gol R A, Pierce J H, Aaronson S A. Transforming genes of human hematopoietic tumors: frequent detection of ras-related oncogenes whose activation appears to be independent of tumor phenotype. Proc Natl Acad Sci USA. 1983;80:4926–4930. doi: 10.1073/pnas.80.16.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke T F, Yang S I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 26.Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greco A, Pierotti M A, Bongarzone I, Pagliardini S, Lanzi C, Della P G. TRK-T1 is a novel oncogene formed by the fusion of TPR and TRK genes in human papillary thyroid carcinomas. Oncogene. 1992;7:237–242. [PubMed] [Google Scholar]
- 28.Greenberger J S, Sakakeeny M A, Humphries R K, Eaves C J, Eckner R J. Demonstration of permanent factor-dependent multipotential (erythroid/neutrophil/basophil) hematopoietic progenitor cell lines. Proc Natl Acad Sci USA. 1983;80:2931–2935. doi: 10.1073/pnas.80.10.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall A, Marshall C J, Spurr N K, Weiss R A. Identification of transforming gene in two human sarcoma cell lines as a new member of the ras gene family located on chromosome 1. Nature. 1983;303:396–400. doi: 10.1038/303396a0. [DOI] [PubMed] [Google Scholar]
- 30.Hallberg B, Ashcroft M, Loeb D M, Kaplan D R, Downward J. Nerve growth factor induced stimulation of Ras requires Trk interaction with Shc but does not involve phosphoinositide 3-OH kinase. Oncogene. 1998;17:691–697. doi: 10.1038/sj.onc.1201980. [DOI] [PubMed] [Google Scholar]
- 31.Kaebisch A, Brokt S, Seay U, Lohmeyer J, Jaeger U, Pralle H. Expression of the nerve growth factor receptor c-TRK in human myeloid leukaemia cells. Br J Haematol. 1996;95:102–109. doi: 10.1046/j.1365-2141.1996.d01-1874.x. [DOI] [PubMed] [Google Scholar]
- 32.Kakizuka A, Miller W H J, Umesono K, Warrell R P J, Frankel S R, Murty V V, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan D R, Hempstead B L, Martin-Zanca D, Chao M V, Parada L F. The trk proto-oncogene product: a signal transducing receptor for nerve growth factor. Science. 1991;252:554–558. doi: 10.1126/science.1850549. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan D R, Martin-Zanca D, Parada L F. Tyrosine phosphorylation and tyrosine kinase activity of the trk proto-oncogene product induced by NGF. Nature. 1991;350:158–160. doi: 10.1038/350158a0. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan D R, Miller F D. Signal transduction by the neurotrophin receptors. Curr Opin Cell Biol. 1997;9:213–221. doi: 10.1016/s0955-0674(97)80065-8. [DOI] [PubMed] [Google Scholar]
- 36.Katzav S, Martin-Zanca D, Barbacid M. vav, a novel human oncogene derived from a locus ubiquitously expressed in hematopoietic cells. EMBO J. 1989;8:2283–2290. doi: 10.1002/j.1460-2075.1989.tb08354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katzav S, Martin-Zanca D, Barbacid M, Hedge A M, Isfort R, Ihle J N. The trk oncogene abrogates growth factor requirements and transforms hematopoietic cells. Oncogene. 1989;4:1129–1135. [PubMed] [Google Scholar]
- 38.Klein R, Jing S Q, Nanduri V, O'Rourke E, Barbacid M. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell. 1991;65:189–197. doi: 10.1016/0092-8674(91)90419-y. [DOI] [PubMed] [Google Scholar]
- 39.Klesse L J, Parada L F. Trks: signal transduction and intracellular pathways. Microsc Res Tech. 1999;45:210–216. doi: 10.1002/(SICI)1097-0029(19990515/01)45:4/5<210::AID-JEMT4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 40.Koizumi H, Morita M, Mikami S, Shibayama E, Uchikoshi T. Immunohistochemical analysis of TrkA neurotrophin receptor expression in human non-neuronal carcinomas. Pathol Int. 1998;48:93–101. doi: 10.1111/j.1440-1827.1998.tb03877.x. [DOI] [PubMed] [Google Scholar]
- 41.Labouyrie E, Parrens M, de Mascarel A, Bloch B, Merlio J P. Distribution of NGF receptors in normal and pathologic human lymphoid tissues. J Neuroimmunol. 1997;77:161–173. doi: 10.1016/s0165-5728(97)00055-6. [DOI] [PubMed] [Google Scholar]
- 42.Laneuville P, Heisterkamp N, Groffen J. Expression of the chronic myelogenous leukemia-associated p210bcr/abl oncoprotein in a murine IL-3 dependent myeloid cell line. Oncogene. 1991;6:275–282. [PubMed] [Google Scholar]
- 43.Lee J C, Hapel A J, Ihle J N. Constitutive production of a unique lymphokine (IL 3) by the WEHI-3 cell line. J Immunol. 1982;128:2393–2398. [PubMed] [Google Scholar]
- 44.Liu Q, Schwaller J, Kutok J, Cain D, Aster J C, Williams I R, Gilliland D G. Signal transduction and transforming properties of the TEL-TRKC fusions associated with t(12;15)(p13;q25) in congenital fibrosarcoma and acute myelogenous leukemia. EMBO J. 2000;19:1827–1838. doi: 10.1093/emboj/19.8.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin-Zanca D, Hughes S H, Barbacid M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature. 1986;319:743–748. doi: 10.1038/319743a0. [DOI] [PubMed] [Google Scholar]
- 46.Martin-Zanca D, Oskam R, Mitra G, Copeland T, Barbacid M. Molecular and biochemical characterization of the human trk proto-oncogene. Mol Cell Biol. 1989;9:24–33. doi: 10.1128/mcb.9.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mavilio F, Kreider B L, Valtieri M, Naso G, Shirsat N, Venturelli D, Reddy E P, Rovera G. Alteration of growth and differentiation factors response by Kirsten and Harvey sarcoma viruses in the IL-3-dependent murine hematopoietic cell line 32D C13(G) Oncogene. 1989;4:301–308. [PubMed] [Google Scholar]
- 48.Miki T, Smith C L, Long J E, Eva A, Fleming T P. Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature. 1993;362:462–465. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- 49.Miller K B. Clinical manifestations of acute myeloid leukemia. In: Hoffman R, Benz E J, Shattil S J, Furie B, Cohen H J, Silberstein L E, editors. Hematology: basic principles and practice. New York, N.Y: Churchill Livingstone, Inc.; 1995. pp. 993–1014. [Google Scholar]
- 50.Mitra G, Martin-Zanca D, Barbacid M. Identification and biochemical characterization of p70TRK, product of the human TRK oncogene. Proc Natl Acad Sci USA. 1987;84:6707–6711. doi: 10.1073/pnas.84.19.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Needleman S W, Kraus M H, Srivastava S K, Levine P H, Aaronson S A. High frequency of N-ras activation in acute myelogenous leukemia. Blood. 1986;67:753–757. [PubMed] [Google Scholar]
- 53.Obermeier A, Lammers R, Wiesmuller K H, Jung G, Schlessinger J, Ullrich A. Identification of Trk binding sites for SHC and phosphatidylinositol 3′-kinase and formation of a multimeric signaling complex. J Biol Chem. 1993;268:22963–22966. [PubMed] [Google Scholar]
- 54.Oskam R, Coulier F, Ernst M, Martin-Zanca D, Barbacid M. Frequent generation of oncogenes by in vitro recombination of TRK protooncogene sequences. Proc Natl Acad Sci USA. 1988;85:2964–2968. doi: 10.1073/pnas.85.9.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pahlman S, Hoehner J C. Neurotrophin receptors, tumor progression and tumor maturation. Mol Med Today. 1996;2:432–438. doi: 10.1016/1357-4310(96)84847-7. [DOI] [PubMed] [Google Scholar]
- 56.Parada L F, Tabin C J, Shih C, Weinberg R A. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 57.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosa S S, Dumon S, Mayeux P, Gisselbrecht S, Gouilleux F. Cooperation between STAT5 and phosphatidylinositol 3-kinase in the IL-3-dependent survival of a bone marrow derived cell line. Oncogene. 2000;19:1164–1172. doi: 10.1038/sj.onc.1203418. [DOI] [PubMed] [Google Scholar]
- 59.Santos E, Tronick S R, Aaronson S A, Pulciani S, Barbacid M. T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB- and Harvey-MSV transforming genes. Nature. 1982;298:343–347. doi: 10.1038/298343a0. [DOI] [PubMed] [Google Scholar]
- 60.Schechter A L, Hung M C, Vaidyanathan L, Weinberg R A, Yang-Feng T L, Francke U, Ullrich A, Coussens L. The neu gene: an erbB-homologous gene distinct from and unlinked to the gene encoding the EGF receptor. Science. 1985;229:976–978. doi: 10.1126/science.2992090. [DOI] [PubMed] [Google Scholar]
- 61.Segal R A, Bhattacharyya A, Rua L A, Alberta J A, Stephens R M, Kaplan D R, Stiles C D. Differential utilization of Trk autophosphorylation sites. J Biol Chem. 1996;271:20175–20181. doi: 10.1074/jbc.271.33.20175. [DOI] [PubMed] [Google Scholar]
- 62.Shimizu K, Goldfarb M, Suard Y, Perucho M, Li Y, Kamata T, Feramisco J, Stavnezer E, Fogh J, Wigler M H. Three human transforming genes are related to the viral ras oncogenes. Proc Natl Acad Sci USA. 1983;80:2112–2116. doi: 10.1073/pnas.80.8.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Socolovsky M, Fallon A E, Wang S, Brugnara C, Lodish H F. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction. Cell. 1999;98:181–191. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- 64.Songyang Z, Baltimore D, Cantley L C, Kaplan D R, Franke T F. Interleukin 3-dependent survival by the Akt protein kinase. Proc Natl Acad Sci USA. 1997;94:11345–11350. doi: 10.1073/pnas.94.21.11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takahashi M, Ritz J, Cooper G M. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell. 1985;42:581–588. doi: 10.1016/0092-8674(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 66.Tapley P, Lamballe F, Barbacid M. K252a is a selective inhibitor of the tyrosine protein kinase activity of the trk family of oncogenes and neurotrophin receptors. Oncogene. 1992;7:371–381. [PubMed] [Google Scholar]
- 67.Taylor S J, Shalloway D. Cell cycle-dependent activation of Ras. Curr Biol. 1996;6:1621–1627. doi: 10.1016/s0960-9822(02)70785-9. [DOI] [PubMed] [Google Scholar]
- 68.Tognon C E, Kirk H E, Passmore L A, Whitehead I P, Der C J, Kay R J. Regulation of RasGRP via a phorbol ester-responsive C1 domain. Mol Cell Biol. 1998;18:6995–7008. doi: 10.1128/mcb.18.12.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsukamoto T, Huang T, Guzman R C, Chen X, Pascual R V, Kitamura T, Nandi S. Isolation of oncogenes from rat mammary tumors by a highly efficient retrovirus expression cloning system. Biochem Biophys Res Commun. 1999;265:7–12. doi: 10.1006/bbrc.1999.1625. [DOI] [PubMed] [Google Scholar]
- 70.Vogt P K, Bos T J. jun: oncogene and transcription factor. Adv Cancer Res. 1990;55:1–35. doi: 10.1016/s0065-230x(08)60466-2. [DOI] [PubMed] [Google Scholar]
- 71.Watson F L, Porcionatto M A, Bhattacharyya A, Stiles C D, Segal R A. TrkA glycosylation regulates receptor localization and activity. J Neurobiol. 1999;39:323–336. doi: 10.1002/(sici)1097-4695(199905)39:2<323::aid-neu15>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 72.Whitehead I, Kirk H, Kay R. Expression cloning of oncogenes by retroviral transfer of cDNA libraries. Mol Cell Biol. 1995;15:704–710. doi: 10.1128/mcb.15.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitehead I, Kirk H, Tognon C, Trigo-Gonzalez G, Kay R. Expression cloning of lfc, a novel oncogene with structural similarities to guanine nucleotide exchange factors and to the regulatory region of protein kinase C. J Biol Chem. 1995;270:18388–18395. doi: 10.1074/jbc.270.31.18388. [DOI] [PubMed] [Google Scholar]
- 74.Whitehead I P, Khosravi-Far R, Kirk H, Trigo-Gonzalez G, Der C J, Kay R. Expression cloning of lsc, a novel oncogene with structural similarities to the Dbl family of guanine nucleotide exchange factors. J Biol Chem. 1996;271:18643–18650. doi: 10.1074/jbc.271.31.18643. [DOI] [PubMed] [Google Scholar]
- 75.Yao R, Cooper G M. Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science. 1995;267:2003–2006. doi: 10.1126/science.7701324. [DOI] [PubMed] [Google Scholar]
- 76.Zohn I E, Klinger M, Karp X, Kirk H, Symons M, Chrzanowska-Wodnicka M, Der C J, Kay R J. G2A is an oncogenic G protein-coupled receptor. Oncogene. 2000;19:3866–3877. doi: 10.1038/sj.onc.1203731. [DOI] [PubMed] [Google Scholar]