Abstract
The classical view of oxidative phosphorylation is that a proton motive force (PMF) generated by the respiratory chain complexes fuels ATP synthesis via ATP synthase. Yet, under glycolytic conditions, ATP synthase in its reverse mode also can contribute to the PMF. Here, we dissected these two functions of ATP synthase and the role of its inhibitory factor 1 (IF1) under different metabolic conditions. pH profiles of mitochondrial sub‐compartments were recorded with high spatial resolution in live mammalian cells by positioning a pH sensor directly at ATP synthase’s F1 and FO subunits, complex IV and in the matrix. Our results clearly show that ATP synthase activity substantially controls the PMF and that IF1 is essential under OXPHOS conditions to prevent reverse ATP synthase activity due to an almost negligible ΔpH. In addition, we show how this changes lateral, transmembrane, and radial pH gradients in glycolytic and respiratory cells.
Keywords: Mitochondrial F1FO ATP synthase, ΔpH, IF1, local pH measurements, proton motive force
Subject Categories: Membranes & Trafficking
Mitochondrial F1FO ATP synthase synthesizes or hydrolyses ATP depending on the metabolic conditions and controlled by IF1. Analysis of pH values using local pH sensors at ATP synthase and ATP synthase subcomplexes discloses these activities, which essentially contribute to the establishment of radial, transmembrane, and lateral pH gradients in cristae.
Introduction
Mitochondria are cellular power plants that are the major source of ATP under respiratory conditions. However, while the mechanisms by which ATP is produced have long been known, questions remain about a key regulatory step in the ATP synthesis process. Recent insights into the proton motive force (PMF) that drives ATP synthesis reveal a local and highly dynamic heterogeneity of PMF (Rieger et al, 2014; Sjoholm et al, 2017; Wolf et al, 2019; Toth et al, 2020). The extent to which ATP synthase activity contributes to this heterogeneity is poorly understood.
Under respiratory conditions, ATP is synthesized in process known as oxidative phosphorylation (OXPHOS). During this process, the respiratory complexes CI (NADH dehydrogenase, NADH : ubiquinone oxidoreductase) and CII (succinate dehydrogenase) deliver electrons to CIII (cytochrome c reductase, coenzyme Q: cytochrome c–oxidoreductase). From there, cytochrome c delivers the electrons to CIV (cytochrome c oxidase), the final electron acceptor of the electron transport chain (ETC). The redox activity of CI, CIII, and CIV is coupled to proton pumping from the matrix to the intracristal space (ICS). Thereby, this pumping generates a proton motive force (PMF), which is the transmembrane difference of the electrochemical potential of protons (ΔμH+). The PMF then drives ATP synthesis by ATP synthase (CV).
The PMF, as defined by Peter Mitchell (Mitchell, 1961, 1966; Mitchell & Moyle, 1967) in his chemiosmotic theory, is Δp (Δp = ΔµH+/F; F = Faraday constant), where Δp consists of an electric (ΔΨm) and a chemical part (ΔpHm): Δp = ΔΨm −2.3RT/F ΔpH (mV). Foremost, the Δp drives ATP synthesis, coupling the oxidative steps of the respiratory complexes to ADP phosphorylation by F1FO ATP synthase in a process known as oxidative phosphorylation (OXPHOS).
While the primary proton pumps of the respiratory chain are localized in cristae sheets, rows of dimers of F1FO ATP synthase line up along the cristae rims (Gilkerson et al, 2003; Vogel et al, 2006; Davies et al, 2011, 2012; Blum et al, 2019). F1FO ATP synthase consists of subcomplex FO, which is the proton pump located in the membrane, and subcomplex F1, which is hydrophilic and extends into the matrix. Proton flow from the intracristal space (ICS) into the matrix drives ADP phosphorylation (ATP synthase activity), but this reaction is reversible, and ATP synthase can function as an ATP hydrolase.
Thus, to block futile ATP hydrolysis, for example, by unassembled F1, many cells express inhibitory factor 1 (IF1) (Gledhill et al, 2007; Campanella et al, 2008; Fujikawa et al, 2012). IF1 activity is regulated by phosphorylation (Garcia‐Bermudez et al, 2015), Ca2+, and protons, making IF1 responsive to key functional parameters inside mitochondria (Boreikaite et al, 2019) and, thus, a master regulator of bioenergetics (Barbato et al, 2015; Garcia‐Bermudez & Cuezva, 2016). Yet, while IF1 is known to modulate inadvertent hydrolysis activity in cellulo, it is still debated whether it also modulates ATP synthase (Gledhill et al, 2007; Garcia‐Aguilar & Cuezva, 2018). In order to dissect ATP synthase/ase activity in situ and its impact on Δp, high‐resolution determination of local pH values is necessary.
Indeed, with progressive developments in high‐resolution imaging using ΔΨm and pH indicators, it has been possible to determine local Δp in mitochondrial sub‐compartments (Kondadi et al, 2020a; Wolf et al, 2020). So, it was recently possible to uncover that individual cristae can have disparate ΔΨm and that depolarization events might affect some cristae but not all (Wolf et al, 2019). In addition, work by our group and others has shown that a lateral pH exists between primary proton pumps and ATP synthase that downscales Δp to an intracristal Δp heterogeneity (Rieger et al, 2014; Sjoholm et al, 2017; Wolf et al, 2019; Kondadi et al, 2020b; Toth et al, 2020).
Here, we determined local pH values directly at mitochondrial ATP synthase to collect information about the forward and reverse activity of ATP synthase in situ and to link it to the Δp. To do this work, a pH‐sensitive GFP was introduced to the F1 subcomplex and the FO subcomplex via genetic fusion. This allowed us to determine pH values in the IMS (p‐side) and on the matrix side (n‐side) of the ATP synthase. To understand IF1’s role in controlling ATP synthase activity, IF1‐knockout cells and cells expressing constitutively active IF1‐H49K (Schnizer et al, 1996; Garcia‐Aguilar & Cuezva, 2018) were generated. For on‐side pH determination, the pH‐sensitive GFP derivative sEcGFP (Orij et al, 2009), also known as pHluorin, was used as a ratiometric pH sensor (Gao et al, 2004; Rieger et al, 2014). The generated pH profiles revealed that the local Δp was unexpectedly low under OXPHOS, which are ATP synthesis conditions. Moreover, our data clearly show that IF1 is needed to block ATP hydrolysis under OXPHOS conditions. Since the local Δp at sites of active ATP synthase is low under steady‐state OXPHOS, initiation of the reverse ATPase mode must be prevented. Accordingly, the Δp was maximal in respiring cells with a constitutively active IF1 variant (IF1‐H49K).
Results
Generation of IF1‐overexpressing and IF1‐knockout cell lines
To study the effect of F1FO ATP synthase on local pH values and dissect the ATP synthase and ATPase activity of CV, we generated different HeLa cell lines that either overexpressed the ATPase inhibitory factor IF1 (IF1‐OE) or had it knocked out (IF1‐KO) (Fig EV1A). To enhance IF1, an IF1‐H49K‐HA expressing cell line was generated, where IF1H49K is constitutively active (Schnizer et al, 1996; Garcia‐Aguilar & Cuezva, 2018). The total level of IF1 was doubled in the IF1H49K‐expressing cells (Fig EV1B). In both of these cell lines, the ATP synthase level was not changed (Fig EV1C). Repetitive determination of IF1 levels in hyperglycemic and respiratory cells indicated an increase of endogenous IF1 in the latter in two samples, no change in one sample, and a decrease of IF1 related to SU β in another sample (Fig EV1D).
Figure EV1. IF1 levels in cell lines used in this study.

- Relative IF1 levels of cells used in this study. N = 3 technical replicates, mean ± SD.
- Relative ATP synthase levels in IF1 OE and KO cells under glycolytic and respiratory conditions. Loading control: actin. Cells were either treated with 10 mM glucose (Glc) or 10 mM galactose (Gal) for 3 weeks.
- IF1 levels in relation to subunit B under glycolytic and respiratory conditions. Left: four individual blots are shown. Right: variability of IF1/SU β in respiratory cells compared to glycolytic cells. Cells were either treated with 25 mM glucose (HGlc) or 10 mM galactose (Gal) for 3 weeks.
Galactose induces a respiratory phenotype
In addition, we aimed to investigate the degree to which ATP synthase contributes to the local PMF under different metabolic conditions. A study in yeast reported dependency of intramitochondrial pH on the carbon source and thus fermentative versus respiratory metabolism (Orij et al, 2009). Similar, HeLa cells, which are derived from a cervix cancer, are responsive to the carbon source: a metabolic switch from glycolysis to OXPHOS is easily induced by changing from glucose to galactose (Rossignol et al, 2004). Therefore, to find out how different levels of respiratory activity (low and high) affects PMF and ATP synthase activity, we changed the sugar supply of HeLa cells, each for a longer period of time.
By comparing the metabolic profile of cells grown with high glucose (HGlc: 25 mM) or galactose (10 mM) for 3 weeks, we found that supplying cells with galactose resulted in increased respiration and less glycolysis, as determined with an automatic flux analyzer that records oxygen consumption rates (OCR) as a proxy for OXPHOS activity and extracellular acidification rates (ECAR) as a proxy for glycolytic activity. Galactose supplied cells also had a significantly higher basal respiration, respiration after inhibiting ATP synthase with oligomycin, and maximal respiration after uncoupling with FCCP (trifluoromethoxy carbonylcyanide phenylhydrazone) (Fig EV2A). Cells grown on (high or low) glucose were characterized by a low OCR/ECAR ratio, while cells grown on galactose‐containing medium had a high OCR/ECAR ratio (Fig 1A). As such, cells with a high OCR/ECAR ratio were called OXPHOS or respiratory cells. When comparing basal, CV‐linked, and maximal respiration rates between OXPHOS WT, IF‐KO, and IF‐OE cells, the IF‐OE cells displayed the lowest respiratory rates (Fig EV2B), suggesting that ATP synthase activity feeds back on the respiratory chain activity.
Figure EV2. Respiration rates of investigated cells under different metabolic settings.

- Determination of OXPHOS activity by monitoring oxygen consumption rates (OCR) and extracellular acidification rates (ECAR). After monitoring basal respiration, oligomycin (1 µM), uncoupler FCCP (trifluoromethoxy carbonylcyanide phenylhydrazone; 0.5 µM), rotenone, and antimycin A (Rot, 0.5 µM; AA, 0.5 µM) were added at the indicated times to determine CV‐linked respiration, maximum respiration, and OXPHOS‐specific oxygen consumption, respectively. OCR and ECAR were determined with an automatic flux analyzer (96XF, Seahorse/Agilent). ˜30,000 cells per well were seeded the day before measurement. Cells were supplied with high glucose (25 mM, HGlc), low glucose (5.6 mM), or galactose (10 mM, Gal) for 3 weeks. OCRs were in addition normalized on the relative mitochondrial mass. The error bars denote SEM.
- Basal, CV‐linked, and maximal respiration rates in respiratory cells with different IF1 levels. Three technical replicates from same clones, each technical replicate with 5 parallel measurements. Spare = reserve capacity, difference between basal and maximal respiration; Proton leak = respiration that is not linked to CV (proton leakage as a result of incomplete coupling); Non‐mito = Respiration that is not blocked upon addition of electron transport chain inhibitors. The error bars denote SEM.
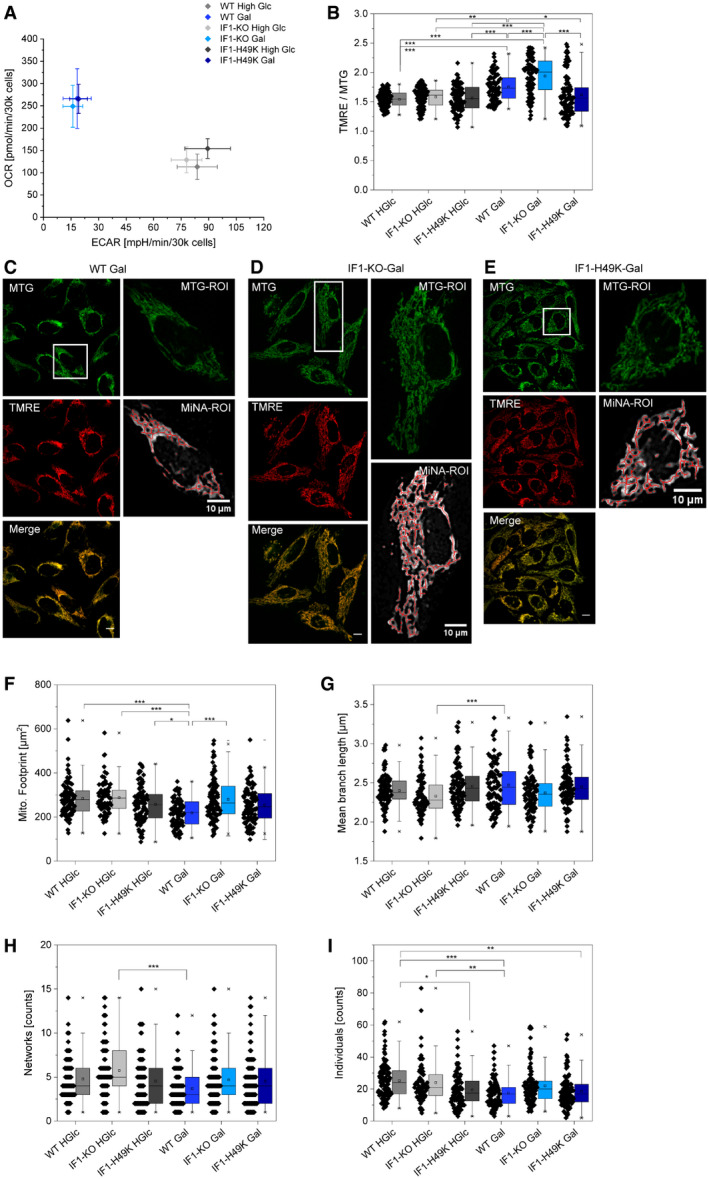
Figure 1. Characterization of cell lines used: metabolic profile, mitochondrial membrane potential, and morphology.
-
AMetabolic profiles of HeLa cells in dependence on IF1 levels under glycolytic and OXPHOS conditions generated by an automatic flux analyzer measuring oxygen consumption rates and extracellular acidification rates. n = 3 independent experiments (error bars = SD).
-
BMitochondrial membrane potential (Δψm) under the influence of overexpressed IF1‐H49K and the absence of IF1 in IF1‐KO cells. To estimate Δψm, HeLa cells were stained with TMRE (7 nM). In addition, HeLa cells were stained with the membrane potential‐independent dye MitoTracker™Green (MTG, 100 nM) for normalization to the mitochondrial area. Calculated TMRE/MTG ratios from n = 4 independent experiments are displayed for each cell. Statistics: one‐way ANOVA with post hoc Scheffé test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
-
CMiNA analysis of HeLa cells stained with the membrane potential‐independent dye MitoTracker™Green (MTG, 100 nM) to visualize and determine the mitochondrial area (= mitochondrial footprint). Left: HeLa WT cells grown in 10 mM galactose stained with 7 nM TMRE (red) plus 100 nM MTG (green). Co‐localization shown in yellow. Right: mitochondrial footprint (white) and mitochondrial skeleton (red) generated from MiNA analysis of the MTG channel (green).
-
DLeft: HeLa IF1‐KO cells grown in 10 mM galactose stained with 7 nM TMRE (red) plus 100 nM MTG (green). Right: mitochondrial footprint (white) and mitochondrial skeleton (red) generated from MiNA analysis of the MTG channel (green).
-
ELeft: HeLa IF1‐H49K cells grown in 10 mM galactose stained with 7 nM TMRE (red) plus 100 nM MTG (green). Right: mitochondrial footprint (white) and mitochondrial skeleton (red) generated from MiNA analysis of the MTG channel (green). Co‐localization shown in yellow. Scale bars = 10 µm.
-
FMitochondrial footprint (µm2) determined via MiNa analysis of HeLa cells. Cells were stained with the membrane potential‐independent dye MitoTracker™Green (MTG, 100 nM).
-
GMean branch length in µm determined via MiNA analysis.
-
HNumber of networks determined via MiNA analysis.
-
INumber of individuals determined via MiNA analysis.
Data information: Statistics: one‐way ANOVA with post hoc Scheffé test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Parameters from n = 4 independent experiments are displayed for each cell. (F‐I). Single dots indicate data from single cells (B, F–I). The error bars denote SD; the boxes represent the 25th to 75th percentiles. The vertical lines in the boxes represent the median values, whereas the square symbols in the boxes denote the respective mean values. The minimum and maximum values are denoted by x. Whiskers include outliners, coefficient 1.5.
ΔΨm is generally increased in respiratory cells and influenced by ATP synthase/IF1
To characterize mitochondrial PMF, we first determined the mitochondrial membrane potential ΔΨm by staining living cells with the membrane potential‐sensitive dye TMRE (tetramethylrhodamine ethyl ester). TMRE is a cell‐permeant, positively charged dye with excitation/emission maxima of ˜549/575 nm, and it accumulates in active mitochondria due to its relative negative charge. In addition, mitochondria were stained with MTG (MitoTracker™ Green FM), which was used as reference for mitochondrial mass (Wikstrom et al, 2007). The TMRE/MTG ratio was taken as a measure for the ΔΨm (Fig EV3A: glycolytic conditions, Fig EV3B: respiratory conditions).
Figure EV3. Determination of relative mitochondrial membrane potential.
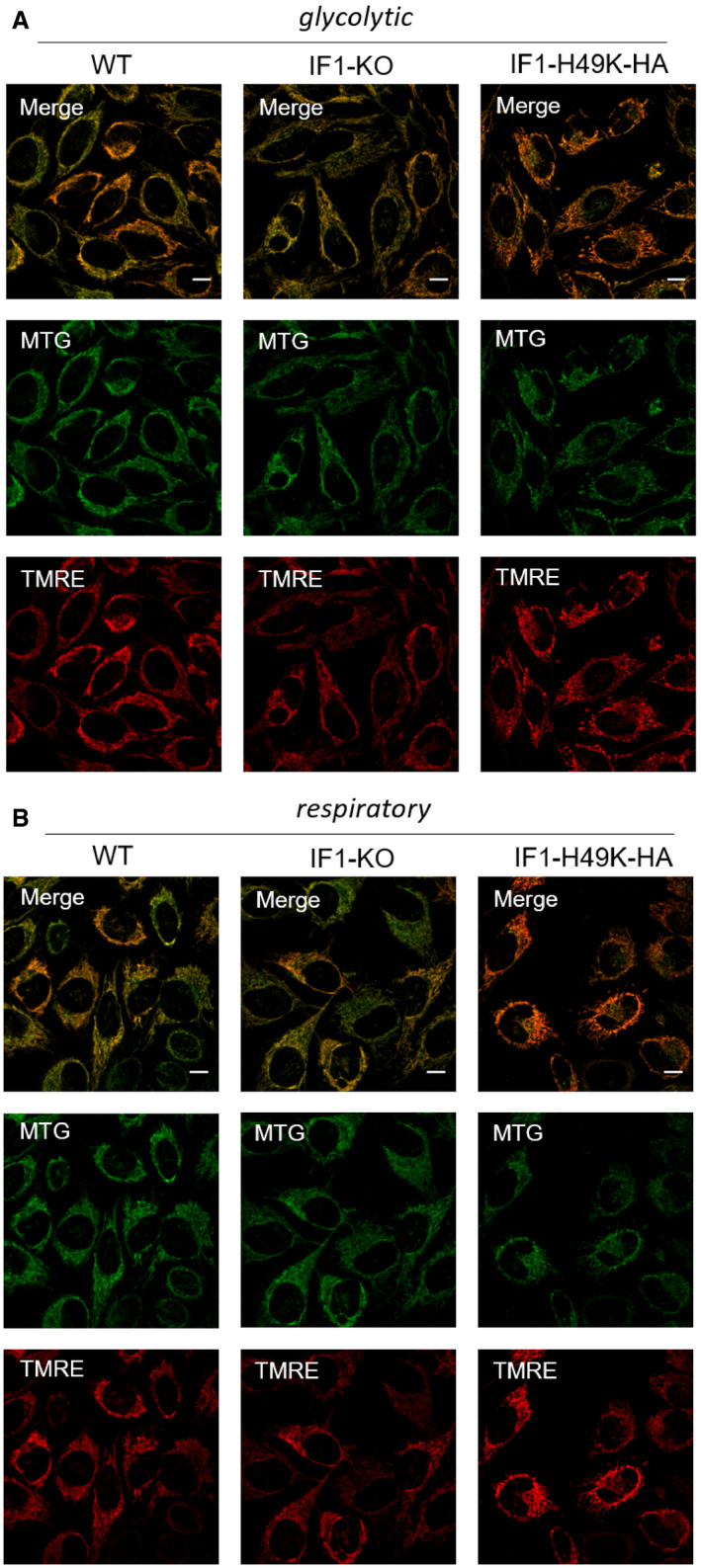
- ΔΨm under glycolytic conditions in the different HeLa cell lines with endogenous IF1 levels, IF1‐KO, and OE of IF1. MTG: MitoTracker™Green; TMRE: Tetramethylrhodamine ethyl ester.
- ΔΨm under respiratory conditions in the different cell lines with normal, KO‐, and OE of IF1.
Data information: Scale bars: 10 µm (A–B).
The MTG‐stained images were further used to determine mitochondrial morphology and networks. A cell‐by‐cell analysis was conducted by first generating a skeleton of the mitochondria using the MiNA plugin running in ImageJ (Valente et al, 2017). Under high glucose conditions, mitochondrial membrane potential and morphology were not affected by IF1 expression (Fig 1B–I). Specifically, in IF1‐KO and IF1H49K‐expressing cells, mitochondrial footprint (area), mean branch length, and network number were comparable. These findings are similar to those of another study, in which IF1 overexpression in glucose‐fed INS‐1E cells did not alter mitochondrial network morphology (Kahancova et al, 2020). Yet, IF1‐OE cells had fewer individual mitochondria (P < 0.05). Under galactose conditions, however, ΔΨm was significantly higher in IF1‐KO compared to WT cells, suggesting that ATPase activity contributed to ΔΨm in the IF1‐KO cells by reverse proton pumping. To confirm this, local pH measurements at ATP synthase on the p‐side of the IMM were required (see below). Also, the mitochondrial mass (footprint) was higher in IF1‐KO than in WT cells, probably representing a rescue mechanism to fulfill ATP demands (Fig 1F). Interestingly, although respiratory IF1‐OE cells had a lower ΔΨm than WT cells, the mitochondrial mass was not affected.
The clearest difference in mitochondrial morphology was observed when respiratory WT cells were compared with glycolytic IF1‐KO cells. Respiratory WT cells displayed a clear shift toward a fusion phenotype (greater networking): The mean branch length in respiratory WT cells increased (Fig 1G), and single networks and individuals decreased (Fig 1H and I). Respiratory WT cells (Gal) had a lower number of individual mitochondria compared to WT cells fed with high glucose (HGlc); this corresponds with previous studies indicating that respiratory HeLa cells showed greater networking (Rossignol et al, 2004). IF1‐OE cells (Gal and HGlc) had fewer individual mitochondria compared to WT HGlc (Fig 1I), but otherwise the mitochondrial network was unchanged. A similar observation was recently made in pancreatic beta cells overexpressing native IF1 (Kahancova et al, 2020). IF1‐OE was not correlated with higher respiration rates. In respiratory IF1‐KO cells, the increase of ΔΨm was not correlated with an increased mitochondrial fusion phenotype. Thus, we conclude that the activity of the respiratory chain is not the only factor that determines the morpho‐function of mitochondria (Bulthuis et al, 2019); the ATP synthase activity must also be considered.
Respiratory cells have higher oligomeric forms of ATP synthase
Next, we assessed how the metabolic conditions influence the assembly of ATP synthase (CV) into supercomplexes. Therefore, mitochondrial protein was separated by native gel electrophoresis, and we compared CV from WT HGlc cells, WT Gal cells, and IF1H49K‐OE cells; subunit β and IF1 were immunoblotted. We identified monomers, dimers, and oligomers of CV (Fig 2A), and IF1 associated with all assembly forms of ATP synthase. We found that respiratory cells tended to contain higher CV assembly states (dimers and oligomers) compared to glycolytic cells.
Figure 2. Assembly of OXPHOS complexes.

- BN‐PAGE and subsequent immuno‐blotting of mitochondrial extracts from indicated HeLa cell lines showing the assembly of F1FO ATP synthase and binding of IF1. Monomers (M), dimers (D), and oligomers (O) were detected. Left panel: Coomassie‐stained gel. Right panels: Immunoblotting and detection of ATP synthase and IF1. WT: wild‐type HeLa cells; IF*: cells expressing IF1‐H49K‐HA. Stars indicate oligomeric IF1 forms. I: complex I; III: complex III.
- Relative levels of ATP synthase monomers, dimers, and oligomers. Semi‐quantification of immunoblot shown in (A).
- Assembly of F1FO ATP synthase SU e‐sEcGFP (Sue#) into monomeric, dimeric, and oligomeric ATP synthase (CV). Proteins were separated by BN‐PAGE and immunoblotted as indicated. Left panel: detection of subunit β (α‐ATP5B); right panel: detection of sEcGFP with α‐GFP; O: oligomeric ATP synthase; D: dimeric (blue **); M: monomeric F1F0 ATP synthase (blue*).
- Assembly of sEcGFP‐tagged subunits CoxVIIIa‐sEcGFP (CIV#) into complex IV, and assembly of subunits γ‐sEcGFP (SU γ #) and SU e‐sEcGFP (Su e#) into ATP synthase. Proteins were separated by BN‐PAGE and immunoblotted. Lanes #1‐#10 were cut into three pieces for further evaluation. Lanes 1–2: complex IV assembly in WT and CoxVIIIa‐sEcGFP‐expressing cells, detection of CIV with anti‐Cox1. Lanes 3–4: complex V assembly in WT and SU γ‐sEcGFP‐expressing cells, detection with anti‐SU γ. Lanes 8–10: assembly of CIV# into CIV, SU γ‐sEcGFP (SUγ#) into CV, and SU e‐sEcGFP (SUe#) into CV. At lane #6 in the Coomassie‐stained gel, the positions of molecular weight markers (M) are shown. SC: respiratory supercomplexes; CIV2: complex IV dimers; CIV: complex IV monomer; s1: F1‐c8 subcomplexes; s2: subcomplexes of CV containing subunits b, e, and g (He et al, 2018). Blue **: dimeric F1F0 ATP synthase; blue*: monomeric F1F0 ATP synthase. Red asterisks: respiratory supercomplexes.
Source data are available online for this figure.
Moreover, the overexpression of IF1‐H49K stimulated an increase in oligomers in glycolytic cells as well as in respiratory cells (Fig 2B). This is in line with structural data showing that IF1 stabilizes CV tetramers (Pinke et al, 2020). Contrary, high glucose levels (glycolytic conditions) were correlated with lower IF1 levels (relative to subunit α of CV) (Kahancova et al, 2020), indicating that IF1 levels naturally change in dependency on metabolic conditions.
Implementation of local pH probes at OXPHOS complex CIV and at subunit e and γ in CV
To measure local pH, we genetically fused the pH probe sEcGFP (pHluorin, superecliptic GFP) (Miesenbock et al, 1998; Orij et al, 2009) to subunits of CIV, CV, and to the matrix protein mitochondrial processing peptidase (MPP), resulting in MPP‐sEcGFP, which was successfully imported into mitochondria (Appendix Fig S1). For attaching the probe to CIV and CV, sEcGFP was genetically fused to subunit γ (SU γ) of the F1 subcomplex of CV at the n‐side of the IMM, to subunit e (SU e) of FO subcomplex of CV at the p‐side of the cristae membrane, and to subunit CoxVIIIa of CIV at the p‐side of the IMM.
Protein complexes of cell lines expressing CV SU e‐sEcGFP, CV SU γ‐sEcGFP, and CoxVIIIa‐sEcGFP were separated by BN‐PAGE and immunoblotted. SU e‐sEcGFP (SUe#) assembled into the monomeric, dimeric, and oligomeric CV complexes (Fig 2C), and the assembly was not biased by the fusion of SU e to sEcGFP. CoxVIIIa‐sEcGFP (CIV#) was found in dimeric CIV and respiratory supercomplexes but, interestingly, not in monomeric CIV (Fig 2D). The assembly of CIV was also not affected by fusion of subunit CoxVIIIa to sEcGFP. Labeled SU γ was found in subcomplexes of CV (indicated as s1 and s2 in Fig 2D), which are not identical with F1 and FO. A minor fraction of SU γ‐sEcGFP was found in the monomeric holo CV (Fig 2D), which was similar to the assembly pattern we found for SU γ ‐Dendra in an earlier study (Appendix Fig S1 (Muster et al, 2010)). Expression of SU γ ‐sEcGFP did not affect the assembly of CV into monomers, as shown by comparing it with anti‐SU γ testing on the WT sample (Fig 2D, lanes #3 and #4).
According to a previous study from the Walker group (He et al, 2018), s1 is an F1‐c8 subcomplex, and s2 is a subcomplex containing subunits b, e, and g. However, the subcomplexes s1 and s2 were not detected when probed with anti‐ATP5B against subunit β (Fig 2C and D), which is the main compound of F1. Since the reaction between anti‐ATP5B and F1 is rather weak (Bisetto et al, 2013) SU β might not have been detected here. The correct localizations of tagged subunits CoxVIIIa‐sEcGFP in the cristae and MPP‐sEcGFP in the matrix were also confirmed by immuno‐EM in a study by Rosselin et al (2017), whereas the correct location of CV SU γ ‐EGFP in cristae was confirmed by Wilkens et al (2013) and of CV‐SU e ‐EGFP by Kondadi et al (2020a). Previous work has also shown that the assembly of GFP‐tagged SU γ into ATP synthase maintained its functionality (Prescott et al, 2003). Further, fusions to CoxVIIIa have been used in several studies without interfering with mitochondrial function (Liu et al, 2017).
In sum, the positioning of a pH probe at complex IV and subunit e of complex V was successful and did not hamper complex assembly. The pH probe at SU γ was mainly found in F1‐c8 subcomplexes of ATP synthase, which are anchored in the membrane (He et al, 2018). Yet, in cells expressing SU e‐sEcGFP and SU γ‐sEcGFP, respectively, an additional band of the size of free GFP was detected (Fig 2D). We cannot say whether free GFP is a translational by‐product, a purification artifact or the product of proteolytic activity.
Subunit γ indicates a higher mobile complex of ATP synthase, while subunit g represents confined ATP synthase at cristae rims
Because respiratory cells showed a shift toward higher assembly states of active ATP synthase (reported above), we also expected to see different spatiotemporal behavior of CV in respiratory versus glycolytic cells. To compare the spatiotemporal organization of ATP synthase complexes labeled at F1 and FO, we performed tracking and localization microscopy (TALM) of individual ATP synthase particles. Therefore, as a subunit of F1, subunit γ was fused to the self‐labeling HaloTag, as recently described (Salewskij et al, 2020). Since we could not collect sufficient data for the localization of FO using CV Halo‐tagged SU e, we instead characterized the spatiotemporal behavior of SU g tagged with the HaloTag, because both of these FO subunits (e and g) closely interact, are co‐regulated, and stabilize oligomers of ATP synthase (Paumard et al, 2002; Wittig et al, 2008; Carraro et al, 2014; Pinke et al, 2020). Thus, we assume that holo‐complex CV labeled at SU g would exhibit the same behavior as when labeled at SU e.
In short, single ATP synthase particles were localized and tracked by a super‐resolution microscopy technique called tracking and localization microscopy (TALM) (Appelhans et al, 2012). HeLa cells stably transfected with the constructs were then stained by adding a fluorescent substrate, either tetramethylrhodamine‐HaloTag ligand (TMRHTL) or silicon rhodamine‐HTL (SiRHTL), and imaged with an inverted TIRF microscope (Nikon Eclipse TE2000 equipped with a 100× TIRF objective (N.A. 1.45, oil immersion) and an EMCCD camera (Evolve™512, Photometrics). Posttranslational labeling with low concentrations of fluorescent substrates (0.5–1 nM HTL‐TMR) for the HaloTag (Appelhans & Busch, 2017) allowed the dissection of single molecules. For further details, see Salewskij et al (2020). Image sequences of up to 1,000 frames (30 Hz) were recorded, and single emitters were identified and localized using a 2D Gaussian fit, taking into account the point spread function (PSF). Emitters that appeared in subsequent frames within a radius of 2 pixels (pixel size 107 nm) and that had similar intensity were treated as identical particles. The respective positions were then connected until the particle disappeared. Together, all connected particles of the same type resulted in a trajectory. Trajectories of one movie were superimposed to trajectory maps and further analyzed (Fig 3). Exemplary trajectory maps of ATP synthase labeled at SU γ (A) and SU g (A′) in single mitochondria are shown in Fig 3A and A′. The detailed views of the diffusion of CV‐SU γ and CV‐SU g clearly display a difference: Individual steps of CV‐SU γ appeared longer and differently oriented than the steps of CV‐SU g, which were shorter and more localized.
Figure 3. Single‐particle diffusion analysis of CV‐SU g reveals restrictive localization and diffusion of the oligomeric ATP synthase holo‐complex compared to ATP synthase monomers and subcomplexes.

-
A, A′Trajectory maps of ATP synthase labeled at SUγ (A) and SU g (A′) in single mitochondria. Insets: Detailed views of the diffusion of CV‐SU γ (A) and CV‐SU g (A′). Scale bars: 1 µm overview and 0.5 µm insets.
-
B, B′Generation of transformation matrices for linearization of mitochondria to analyze the directionality of the movement. The number of individual trajectories is shown in brackets.
-
C, C′Analysis of the step directions with respect to the longitudinal axis (±180° and 0°) and the orthogonal axis (from −90° and +90°) represented on a graduated disk. For the determination of directionality of CV‐SU γ and CV‐SU g, 5 successive steps were integrated. The number of analyzed steps is indicated in D and D′.
-
D, D′Heat maps showing the step magnitude in dependence on the step direction.
-
EDiffusion coefficients (‐log D) for CV labeled at SU γ and SU g in comparison. Left: probability density function (PDF) and cumulative density functions (CDF) for D from all trajectories. Right: box and whiskers plot. Mean diffusion coefficients D (open squares) calculated from all individual trajectories shown in A and A′, respectively.
-
FStep size diagram (PDF and CDF) for steps of SU γ and SU g in comparison.
Data information: Statistics: Mann–Whitney test (E, F). Diffusion coefficients of trajectories shown as single dots. The error bars denote SD; the boxes represent the 25th to 75th percentiles. The vertical lines in the boxes represent the median values, whereas the square symbols in the boxes denote the respective mean values. The minimum and maximum values are denoted by x. Whiskers include outliners, coefficient 1.5.
Next, we quantified the differences in detail. To detect possible movement in cristae membranes, trajectories were analyzed according to their orientation. To quantify the orientation with respect to the long axis of mitochondria, the direction of motion of a single molecule was described by a vector. Curved mitochondria were first linearized (Fig 3B and B′). For this purpose, the long axis of a curved mitochondrion was stretched using a transformation matrix to generate a linear mitochondrion. Then, the value of the direction vector (including five steps) was plotted in degrees [°] on a circular disk. The long axis of a mitochondrion was assigned ± 180° and 0°, and the orthogonal axis was assigned as −90° to 90°. CV‐SU γ displayed preferential movement along the orthogonal cristae (Fig 3C) with the majority of steps around ± 90° on the orientation disk (Fig 3D). This is in accordance with what was described earlier (Salewskij et al, 2020). In contrast, the trajectories of CV‐SU g had a different orientation, and cristae tracks were hardly observed (Fig 3C′ and D′). In addition, CV‐SU g had a significantly lower mobility (D median = 0.006 µm2/s) than CV‐SU γ with D = 0.015 µm2/s (Fig 3E). Also, the length of individual steps of CV‐SU g was, on average, significantly lower (86 nm) than for CV‐SU γ (182 nm) as displayed in Fig 3F.
In sum, ATP synthase subcomplexes labeled at F1‐SU γ were more mobile than ATP synthase labeled at FO‐SU g. CV‐SU γ trajectories display diffusion along the cristae membranes, while the holo‐complex tagged at SU g is quasi immobile. Since the holo‐complex assembles into oligomeric structures along the rims of cristae (Allen et al, 1989; Davies et al, 2011; Kuhlbrandt, 2019), a pH probe at SU e (equivalent to SU g), therefore, senses the pH at cristae on the p‐side of rims/tips. Furthermore, the pH probe at SU γ senses the pH mainly as F1‐SU γ or F1‐c8‐SU γ and in fully assembled ATP synthase at the n‐side of cristae membranes.
The spatiotemporal organization of ATP synthase is more restricted under OXPHOS conditions
We recently reported that the spatiotemporal organization of ATP synthase depends on its activity mode (Salewskij et al, 2020; Weissert et al, 2021). Namely, inhibiting the ATPase activity resulted in decreased mobility of ATP synthase complexes and subcomplexes (monomeric CV‐holo, s1, s2) labeled at subunit γ, while inhibiting ATP synthase activity by depleting the respiratory chain’s substrate increased the mobility of F1‐subunit γ. Here, we show that stimulation of the electron transport chain activity by galactose supply decreased the mobility of F1‐SU γ significantly in relation to increased ATP synthase activity. Specifically, CV particles in galactose showed mitochondrial localization and restricted mobility (Fig 4A). Further, the calculated diffusion coefficient's probability function (PDF) showed a clear shift toward lower mobility in galactose (Fig 4B), resulting in a significantly lower median diffusion coefficient D under OXPHOS conditions (D = 0.00898 µm2/s, Q1 = 0.0031, Q3 = 0.02967) than in glycolytic cells (D = 0.0177 µm2/s, Q1 = 0.00507, Q3 = 0.05386; Mann–Whitney; P < 0.001) (Fig 4C). The restriction could be due to an increase in dimeric and oligomeric forms of ATP synthase (Fig 2A and B).
Figure 4. Stimulation of ATP synthase activity is related to decreased mobility.
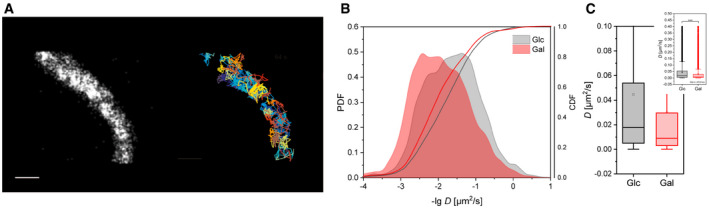
- Localizations and trajectory map of CV SU γ‐HaloTag/TMRHTL particles in a single mitochondrion. Scale bar: 1 µm. Cumulative images of 2,000 frames (recording 30 Hz). Each trajectory in a different color. Trajectories of the F1FO ATP synthase were generated by using the Multi‐Target‐Tracing‐Tool (MTT) (Sergé et al, 2008).
- Diffusion coefficients for ATP synthase under control conditions (5.6 mM glucose) and under OXPHOS conditions. The probability density function (PDF) and cumulative density function (CDF) of ‐log D are plotted. Number of trajectories analyzed: 8,622 (Glc) and 10,333 (Gal) from mitochondria in at least five different cells.
- Mean diffusion coefficients D calculated from all individual trajectories. Mann–Whitney test. ***P < 0.001; **P < 0.01; *P < 0.5. Cells analyzed > 5 per condition; number of trajectories: 4011 (Glc), 10333 (Gal); Q1: 0.00387 (Glc), 0.0031 (Gal); median D Glc: 0.0142 µm2/s, D Gal: 0.00898 µm2/s; Q3: 0.0504 (Glc); 0.02969 Gal; P < 10−31. The error bars denote SD; the boxes represent the 25th to 75th percentiles. The vertical lines in the boxes represent the median values, whereas the square symbols in the boxes denote the respective mean values. The minimum and maximum values are denoted by x. Whiskers include outliners, coefficient 1.5.
Calibration and characterization of local pH probes
Superecliptic GFP is a pH‐sensitive variant from the Aquorea victoria green fluorescent protein, with 9 mutations that lead to increased fluorescence and ratiometric pH sensitivity in comparison with the original ecliptic variant. Since the pH‐dependent emission spectra display an isosbestic point, sEcGFP can be used as a ratiometric pH sensor (Gao et al, 2004; Rieger et al, 2014) (Fig 5A). After excitation with 405 nm, the emission in two channels was simultaneously recorded, and the ratio was calculated (λ511/λ464). pH sensors MPP‐sEcGFP, CoxVIIIa‐sEcGFP, SU γ‐sEcGFP, and SU e‐sEcGFP were positioned as indicated in Fig 5B. First, pH calibration curves were generated by incubating cells expressing the different probes MPP‐sEcGFP, SU γ‐sEcGFP, and SU e‐sEcGFP in media with different pH values. For the equilibration with extracellular pH, ATP synthase inhibitor oligomycin and uncouplers FCCP (carbonyl cyanid‐p‐trifluoromethoxyphenylhydrazon) and nigericin were added (Fig 5C and D). Detergents were not required for pH equilibration, and the addition of triton or digitonin was not required to equilibrate with the external pH (Fig 5C and F). For each construct, a calibration curve was generated to identify and eliminate probe‐specific effects on the ratio, such as quenching or other effects by the specific nano‐environment. The calibration curves of cells expressing mt‐sEcGFP, SU γ‐sEcGFP, and SU e‐sEcGFP to different external pH values were similar, while the calibration for CoxVIIIa‐sEcGFP varied (Fig 5E). This was likely due to the specific nano‐environment of CoxVIIIa, as it is localized at the interface of respiratory supercomplexes (Rieger et al, 2017). Addition of FCCP, nigericin, and oligomycin resulted in pH equilibration between the local pH in the matrix, at CIV‐CoxVIIIa and CV SU e and SU γ, respectively, with the external pH (Fig 5G).
Figure 5. sEcGFP as a pH sensor.
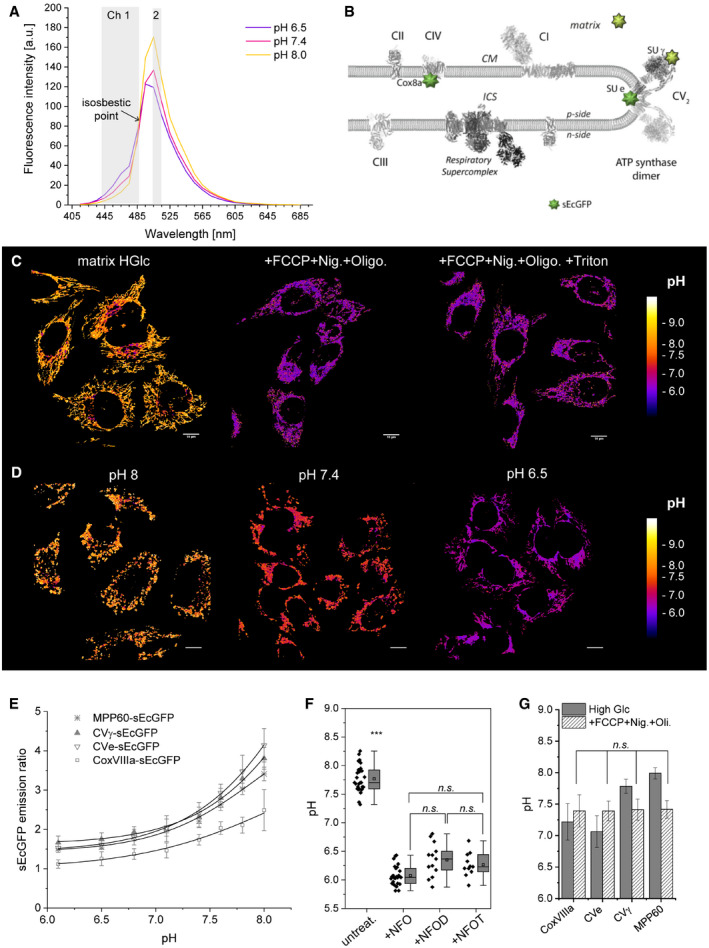
- Fluorescence emission spectrum of mt‐sEcGFP in HeLa cells in situ at different pH values (λex. = 405 nm, λem. = 415–705 nm; λ‐emission scan of transfected cells with 10 nm steps using a cLSM microscope). Before and after the isosbestic point (indicated), the pH dependence of the emission is reversed, allowing for a readout of pH‐dependent fluorescence emission in the two indicated channels and ratiometric imaging (λ511/λ464). The λ range for the first channel is set larger than the λ range for the second channel to allow for a better match of the signal intensities of the two different channels.
- Scheme showing the positions of the pH sensor fused to subunit of different OXPHOS complexes and MPP in the matrix. CIV: Cox8a (CoxVIIIa)‐sEcGFP; CV SU e‐sEcGFP, CV SU γ‐sEcGFP. Matrix: MPP‐sEcGFP. CI: complex I, CII: complex II, CIII: complex III, CIV: complex VI, CV2: dimeric complex V. ICS: intracristal space, CM: crista membrane.
- Images showing matrix pH equilibration with extracellular pH. Shown are ratiometric fluorescence images of MPP‐sEcGFP before and after addition of the uncouplers and inhibitors FCCP, nigericin, and oligomycin (plus 0.002% w/v Triton X‐100 where indicated). False color images (λ511/λ464) indicate ratio values.
- Ratiometric imaging of mitochondrial pH after equilibration with extracellular pH values pH = 8, pH = 7.4, and pH = 6.5. Scale bars = 10 µm.
- Calibration curves for the different sensor‐fusion proteins. Ratiometric response of sEcGFP to the local pH. Extracellular buffers with different pH values used: MES (2‐(N‐morpholino)ethanesulfonic acid, 20 mM, pH 5.6–6.1), BES (N,N‐bis(2‐hydroxyethyl)‐2‐aminoethanesulfonic acid, 20 mM, pH 6.1–7.6), or HEPPSO (4‐(2‐hydroxyethyl)‐piperazine‐1‐(2‐hydroxy)‐propanesulfonic acid, 20 mM, pH 7.1–8.0) in 125 mM KCl, 20 mM NaCl, 0.5 mM CaCl2, 0.5 mM MgSO4 plus FCCP (10 µM), nigericin (1 µM), and oligomycin (5 µg/ml). Each data point represents n = 2 independent experiments and mean values ± SD of at least 15 cells. NFO: nigericin + FCCP + oligomycin; NFOD: nigericin + FCCP + oligomycin + digitonin; NFOT: nigericin + FCCP + oligomycin + Triton X‐100.
- One‐way ANOVA comparison of pH values at SU γ of CV at the matrix site after treating cells with the cocktail (FCCP, nigericin, and oligomycin) for pH equilibration. Box (75%) and whisker plots, median (horizontal line in box), mean (box in box). ***P < 0.001; **P < 0.01; *P < 0.5, n.s.: non‐significant (N = 3 technical replicates, ≥ 10 cells analyzed). NFO: nigericin + FCCP + oligomycin; NFOD: nigericin + FCCP + oligomycin + digitonin (0.006% w/v); NFOT: nigericin + FCCP + oligomycin + Triton X‐100 (0.002% w/v).
- Local pH values at CoxVIIIa, SU e, SU γ, and in the matrix bulk in cells supplied with high glucose (High Glc, 25 mM) before and after the addition of FCCP, nigericin, and oligomycin, demonstrating successful pH equilibration with extracellular pH 7.3. Three technical replicates from same clones. Bars = mean, error bars ± SD. Statistics: one‐way ANOVA with post hoc Scheffé test.
pH measurements directly at ATP synthase allow for the discrimination of ATP synthase and ATPase activity in situ
Next, we determined local pH values under different metabolic conditions. The pH at SU e, at SU γ, and in the matrix was determined in glycolytic and respiratory cells (Figs 6A and EV4A–C). Under glycolytic conditions, the pH at SU e was pH = 7.06 ± 0.25 (SD). In the matrix, the pH at SU γ of CV and in the bulk were both alkaline (pHmatrix = 7.99 ± 0.09) (Fig 6B–D). We explain the low pH at SU e by reverse ATP synthase (ATPase) activity, which pumps protons from the matrix into the intracristal space. Under respiratory conditions, the proton concentration at SU e was lower (pH = 7.28 ± 0.24 (SD)) than under glycolytic conditions (pH = 7.06 ± 0.25) (Fig 6C). These values are similar to what was observed before (Rosselin et al, 2017). The pH at SU γ and in the matrix were more acidic under OXPHOS conditions, which is in line with proton pumping by ATP synthase into the matrix. At CIV, the local proton concentration was significantly lower in glycolytic (pH = 7.21 ± 0.26) than in respiratory conditions (pH = 6.88 ± 0.15, P = 8.6*10−11) due to increased proton pumping activity of CIV under respiratory (OXPHOS) conditions (Fig 6D).
Figure 6. Local pH values in glycolytic and oxidative mitochondria.

- Ratiometric imaging of pH in living HeLa cells at different OXPHOS complexes and in the matrix. WT matrix: MPP‐sEcGFP; WT SU γ: SU γ fused to sEcGFP; WT SU e: SU e fused to sEcGFP; WT CoxVIIIa: subunit CoxVIIIa fused to sEcGFP (all C‐terminally). HGlc: high glucose; Gal: galactose. Ratio images for IF1‐H49K OE cells are displayed in Fig EV4. Scale bars: 10 µm.
- Mitochondrial pH values at SU e, SU γ, and in the matrix bulk in hyper‐glycolytic cells (high glucose: HGlc, 25 mM).
- pH at SU e, SU γ, and in the matrix in respiring cells (Gal, 10 mM).
- pH at CoxVIIIa at complex IV in the ICS before and after stimulation of the respiratory chain.
Data information: Box (75%) and whiskers plots. Outlier included. The error bars denote SD; the boxes represent the 25th to 75th percentiles. The vertical lines in the boxes represent the median values, whereas the square symbols in the boxes denote the respective mean values. The minimum and maximum values are denoted by x. Statistics: one‐way ANOVA with post hoc Scheffé test: ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05. Three technical replicates from same clones (B–D).
Source data are available online for this figure.
Figure EV4. Mitochondrial pH mapping in HeLa WT, IF1‐KO, and IF1‐H49K under glycolytic and respiratory conditions.

- Local pH values in the matrix bulk in HeLa WT and IF1‐H49K cells supplied with high glucose or galactose, as indicated. Scale bars: 10 µm. WT data are also shown in Fig 6.
- Local pH values at CV SU g in WT and IF1‐H49K cells supplied with high glucose or galactose, as indicated. Scale bars: 10 µm. WT data are also shown in Fig 6.
- Local pH values at CV SU e in WT and IF1‐H49K cells supplied with high glucose or galactose, as indicated. Scale bars: 10 µm. WT data are also shown in Fig 6.
- Local pH values at CoxVIIIa (Cox8a) in WT and IF1‐H49K cells supplied with high glucose or galactose, as indicated. Scale bars: 10 µm. WT data are also shown in Fig 6.
- Influence of IF1 levels on the transmembrane, radial, and lateral pH gradients in mitochondria under glycolytic (High Glc) and oxidative (Gal) conditions. Solid black arrow = proton flow, meaning either ATP hydrolysis or ATP synthesis.
Data information: High glucose (High Glc, 25 mM); galactose (Gal, 10 mM); pH values for color scales correspond to ratio values of 1/1.5/2/2.5/3/3.5/4/4.5; conversion was performed using parameters from the according to calibration curves.
Source data are available online for this figure.
Inhibition of OXPHOS discloses forward and reverse activity of ATP Synthase
We next studied the effects of inhibitors that either targeted CV or the respiratory chain. We expected the following: Under glycolytic conditions when ATP is hydrolyzed and protons are pumped into the ICS, the pH at SU e should increase after the addition of oligomycin. In contrast, we anticipated that under respiratory conditions, a decrease of the pH at SU e would occur when ATP synthesis activity is blocked by oligomycin addition. This is exactly what we found: When we applied oligomycin under glycolytic conditions, the pH at SU e increased significantly (from pH = 7.06 ± 0.25 to pH = 7.32 ± 0.16; P = 4.9*10−5), while it decreased after oligomycin application under respiratory (OXPHOS) conditions (from pH = 7.28 ± 0.23 to pH = 6.8 ± 0.32; P = 1.3*10−13)) (Fig 7A). Next, we repeated the same experiments in IF1‐KO cells. In glycolytic IF1‐KO cells, oligomycin had the same effect as in WT cells, where the pH increased from pH = 6.95 ± 0.39 to pH = 7.31 ± 0.36 (P = 2.8 *10−6) (Fig 7B). Before oligomycin addition though, the pH at SU e in IF1‐KO in glycolytic conditions was lower than in WT cells. Together, this confirms that the acidic pH before oligomycin addition was due to reverse proton pumping by ATPase activity. Under OXPHOS conditions, the pH at SU e was higher in IF1‐KO cells under OXPHOS conditions (IF1‐KO: pH = 7.39 ± 0.28; WT: pH = 7.28 ± 0.24; P = 0.038). Surprisingly though, oligomycin had no effect in respiring IF1‐KO cells, and we currently cannot explain this without knowing additional parameters. In this context, it should be mentioned that oligomycin increased the heterogeneity of ΔΨm, as recent high‐resolution imaging data suggest (Wolf et al, 2019).
Figure 7. Inhibitor effects on the local pH value at SU e in the intracristal space.

- Mitochondrial pH at CV SU e measured in HeLa WT in glycolytic (HGlc) and respiring (Gal) conditions. Where indicated, 5 µg/ml oligomycin was added to inhibit CV activity.
- Mitochondrial pH at CV SU e in IF1‐KO cells. Conditions: glycolytic (HGlc) and respiring (Gal) with and without oligomycin.
- Mitochondrial pH at CV SU e in IF1‐OE cells. Conditions: glycolytic (HGlc) and respiring (Gal) with and without oligomycin.
- Fluorescence image showing the localization of SU e‐sEcGFP in WT cells. Scale bar: 10 µm.
- pH calibration curve for testing the effect of antimycin A on local pH at SU e.
- Effect of inhibiting the respiratory chain at complex III (CIII) with antimycin A (AA) on the local pH at SU e in WT cells (glycolytic conditions).
- Local pH at SU e in IF1‐KO cells before and after inhibiting CIII.
- Local pH at SU e in IF1‐OE cells before and after inhibiting CIII.
Data information: Box (75%) and whiskers plots. Whiskers: outlier included. The error bars denote SD; the boxes represent the 25th to 75th percentiles. The vertical lines in the boxes represent the median values, whereas the square symbols in the boxes denote the respective mean values. The minimum and maximum values are denoted by x. Statistics: one‐way ANOVA with post hoc Scheffé test: ***P ≤ 0.001; n.s.: non‐significant. Technical replicates from same clones: 3 (A–C), 2 (F–H).
Finally, we determined pH values at SU e in WT and IF1‐OE cells and the effect of oligomycin. Oligomycin addition had no further effect on local pH at SU e in IF1‐H49K, indicating that previous ATP hydrolysis was already blocked by IF1‐H49K (Fig 7C). The pH at SU e under OXPHOS conditions was similar to the pH in high glucose conditions. Oligomycin addition resulted in acidification at SU e, since protons were no longer translocated from the ICS to the matrix and, thus, the PMF generated by the primary proton pumps was not depleted.
Inhibition of respiratory chain results in stimulation of ATP hydrolysis
Inhibition of the respiratory chain resulted in acidification at SU e. Since this was only observed in WT and IF1‐KO cells, we conclude that this acidification is the result of stimulated ATP hydrolysis, which is suppressed in IF1‐H49K OE cells (Fig 7F–H).
IF1 inhibition of ATPase results in increased pH at SU e
Here, we discuss and compare in more detail the local pH values at ATP synthase subunit e in WT cells, cells stably expressing IF1‐H49K and IF‐KO cells. IF1 inhibits the reverse ATPase activity. If ATPase is mainly active under glycolytic conditions, IF1‐OE should increase the pH at SU e. Indeed, we found that the pH was significantly higher in IF1‐H49K cells compared to control cells (Fig EV4C) and IF1‐KO cells (Fig 8A), since ATPase was now inhibited and no protons were pumped into the ICS by reverse ATP synthase activity. When IF1‐KO and IF1‐H49K cells were compared, the difference in the pH at SU e was significant: The pH value of SU e in IF1‐KO cells was pH = 7.39 ± 0.28, whereas the value in IF1‐H49K‐expressing cells was pH = 7.19 ± 0.25 (P < 0.0001). This demonstrates that the presence or absence of IF1 significantly affects the pH at SU e of ATP synthase also under OXPHOS conditions. Moreover, a positive effect of IF1‐H49K in the presence of endogenous IF1 indicates that endogenous IF1 is not maximally activated at basic pH and when the phosphorylation potential is high.
Figure 8. The activity of ATP synthase determines lateral, radial, and transmembrane pH gradients.

- Mitochondrial pH at CV SU e is compared for HeLa WT, IF1‐KO, and IF1‐H49K cells under glycolytic and respiring conditions.
- Mitochondrial pH at CV SU γ measured in HeLa WT, IF1‐KO, and IF1‐H49K cells under glycolytic and respiring conditions.
- pH in the matrix bulk (MPP60 as mitochondrial targeting sequence) measured in HeLa WT, IF1‐KO, and IF1‐H49K cells under glycolytic and respiring conditions.
- pH at respiratory complex IV at SU CoxVIIIa in the ICS measured in HeLa WT, IF1‐KO, and IF1‐H49K cells under glycolytic and respiring conditions.
- pH differences between SU e and SU γ of ATP synthase SU e are part of FO, SU γ of F1. Triple stars in columns indicate that the difference between the compared pH values constituting the ΔpH was significant; P ≤ 0.001.
- Radial pH gradient between matrix bulk and subunit γ of ATP synthase.
- pH difference between SU e of ATP synthase and matrix bulk.
- Transmembrane pH difference between local pH at CIV (CoxVIIIa) and matrix pH.
- Lateral pH gradient between CIV SU CoxVIIIa and CV SU e. Be aware that the pH under glycolytic conditions at SU e was lower than the pH at CIV in the ICS, resulting in a “negative” ΔpH. (A–I) One‐way ANOVA comparison of sets of pH values at the indicated localizations. Box (75%) and whisker plots, median (horizontal line in box), mean (box in box). P ≤ 0.001: ***P ≤ 0.01: **P ≤ 0.05: *; n.s.: non‐significant (N = 3). Statistics: one‐way ANOVA with post hoc Schéffe test for normal distribution.
- pH measurements in mitochondria in live cells expressing sEcGFP as the pH sensor at SU γ. The pH was color‐coded according to respective emission ratio. The pH at SU γ in IF1‐H49K‐HA expressing cells is compared with WT cells. (Scale bar: 1 µm).
- Schematic drawing showing the resolved pH values and resulting gradients under respiratory and glycolytic conditions. The color gradient was generated following the ratiometric pH code used throughout the study.
Data information: (A–D) One‐way ANOVA comparison of sets of pH values at the indicated localizations. Box (75%) and whiskers plots (outlier included, coefficient 1.5). The error bars denote SD, the boxes represent the 25th to 75th percentiles. The vertical lines in the boxes represent the median values, whereas the square symbols in the boxes denote the respective mean values. The minimum and maximum values are denoted by x. error bars = SD. P ≤ 0.001: ***P ≤ 0.01: **P ≤ 0.05: *; n.s.: non‐significant (N = 3, technical replicates from same cell clones, data from individual cells are shown). Statistics: one‐way ANOVA with post hoc Schéffe test for normal distribution. (E‐I) One‐way ANOVA comparison of measured pH values; shown is ΔpH as the calculated difference between the mean pH values from data shown in A‐D. P ≤ 0.001: ***P ≤ 0.01: **, P ≤ 0.05: *; n.s.: non‐significant.
To gain more insights on the local pH at ATP synthase, we also determined pH values at subunit γ of ATP synthase in WT, IF1‐KO, and IF1‐H49K cells. The C terminus of SU γ fused to sEcGFP extruded from the F1‐head (F1‐SU γ or F1‐c8‐SU γ) such that the sensor can detect pH values close on the cristae membrane. In contrast to the pH at SU e, the pH at F1‐SU γ in WT and IF1‐H49K cells was higher under glycolytic than under respiratory conditions (Fig EV4B). In IF1‐KO cells, the pH was the same under glycolytic and OXPHOS conditions (Fig 8B). Under glycolytic conditions, the presence of IF1‐H49K resulted in a pH decrease at SU γ. Under OXPHOS conditions, IF1 had no effect on the local pH at SU γ; the pH was the same in IF1‐KO and IF1‐H49K‐overexpressing cells. To better understand the differences in local pH at F1‐CV under glycolytic conditions, we complementarily determined the matrix pH in the different cell lines in both metabolic states (glycolytic and OXPHOS) (Fig 8C). No difference was found between WT and IF1‐OE under glycolytic conditions. Respiration decreased the matrix bulk pH compared to glycolytic conditions, but IF1‐H49K expression had no influence on the matrix pH in OXPHOS mitochondria. This was expected when only ATP synthase had been active. In IF1‐KO cells, matrix pH was lowest under glycolytic and OXPHOS conditions. At the current state, we cannot interpret the drop of pH at the n‐side and in the matrix in IF1‐KO cells without further knowledge.
The local pH at complex IV is not affected by active ATPase
Next, we compared the pH values measured at respiratory complex IV (CIV) in the different IF1 cell lines under glycolytic and OXPHOS conditions. Therefore, sEcGFP was fused to subunit CoxVIIIa at the C terminus localized at the p‐side (Fig 8D). Under glycolytic conditions, the pH at CoxVIIIa was significantly higher than under OXPHOS conditions, since fewer protons were pumped into the ICS. Higher OXPHOS activity linked with proton pumping resulted in local acidic pH values below pH< 7. IF1 had no effect on pH values at CIV in glycolytic cells, since neither IF‐KO nor IF1‐H49K‐expressing cells exhibited pH differences at CIV. Surprisingly, however, IF1‐H49K‐expressing cells exhibited the most acidic pH at CIV under OXPHOS conditions (pH = 6.49 ± 0.17), which was significantly lower than pH = 6.88 (± 0.15; P < 10−19) in WT cells (Figs 8D and EV4D). This suggests either an accelerated cytochrome c oxidase proton pumping activity in cells expressing IF1‐H49K or a retarded diffusion of protons from the primary proton pump CIV (proton source) to CV (proton sink) (Rieger et al, 2014). Since the ultrastructure of cristae is changed in IF1‐H49K‐expressing cells (Weissert et al, 2021), a different coupling between proton source and sink (Toth et al, 2020) is also likely.
ATP synthesis is possible at low Δp
The determined pH values at ATP synthase subunits SU e and SU γ were then used to calculate the corresponding ΔpH across the crista membrane. It has to be kept in mind, though, that due to assembly of SU γ into ATP synthase subcomplexes, this was not necessarily the ΔpH across single ATP synthase molecules. Whether a difference was significant was determined by statistical analysis (one‐way ANOVA test; P < 0.001: ***). In mitochondria with reverse ATP synthase (=ATPase) activity, the pH difference was highest with ΔpH = 0.75 under glycolytic conditions. IF1‐KO and IF1‐H49K‐expressing cells had a lower ΔpH (Fig 8E). In OXPHOS cells, the pH difference was ΔpH ˜0.13 under ATP synthesis conditions in WT, IF‐KO, and IF1‐H49K‐expressing cells. Thus, IF1‐H49K reduced the transmembrane ΔpH at ATP synthase under glycolytic conditions, while it did not significantly affect the ΔpH under OPXHOS conditions. Together, these data clearly show that ATPase builds up a local ΔpH at ATP synthase under glycolytic conditions due to reverse proton pumping. In contrast, and quite surprisingly, the transmembrane ΔpH, and thus, the actual Δp was low under steady‐state OXPHOS conditions at the ATP synthase. This suggests that ATP synthesis is possible at very low ΔpH values: a ΔpH = 0.13 means that the proton concentration at the p‐side is only 1.1x higher than at the n‐side!
The local pH at SU γ of F1FO ATP synthase is generally lower than in the matrix bulk
Next, we compared the pH between the matrix bulk and the sensor at SU γ (Fig 8F). Indeed, the pH was lower at SU γ than in the matrix bulk, indicating a radial gradient from the membrane surface toward the bulk. This suggests that proton concentrations near the surface and in the bulk are not in equilibrium, e.g., due to a delay in proton exchange. Such a scenario was proposed before (Heberle et al, 1994; Mulkidjanian et al, 2006) and has been recently intensively reviewed (Lee, 2019). Obviously, diffusion is not sufficient to maintain a homogenous pH in a compartment (Mulkidjanian, 2006) under the conditions in which primary and secondary proton pumps are active (Strauss et al, 2008; Rieger et al, 2014; Sjoholm et al, 2017; Toth et al, 2020). When the matrix bulk pH was used to calculate ΔpH, the nominal ΔpH was higher (Fig 8F and G). Regardless of the reference, though, the ΔpH was lower under OXPHOS conditions compared to glycolytic conditions. Strikingly, in IF‐KO cells, the ΔpH was the smallest under OXPHOS conditions, suggesting that IF1 has an important role in maintaining Δp.
Next, we calculated the ΔpH between CIV and the matrix. Irrespective of the metabolic condition, the calculated ΔpH > 0.6. In IF1‐H49K‐expressing cells, the calculated pH difference was ΔpH > 1.2 under OXPHOS conditions. This was due to the exceptionally low pH at CIV (Fig 8H).
Lateral pH gradients between primary proton pumps and ATP synthase reverse under glycolytic conditions
Finally, we compared pH values at subunit CoxVIIIa of CIV and subunit SU e of ATP synthase. Previously, we and others had reported a lateral pH gradient in the IMS (Rieger et al, 2014; Toth et al, 2020). As observed before, the pH difference was ΔpH > 0.4 under OXPHOS conditions with a higher proton concentration at CIV. This corresponds to a > 3× higher proton concentration at CIV, the primary proton pump (Fig 8I). Under glycolytic conditions, however, the calculated ΔpHCoxVIIIa‐SU e was negative, indicating an inverse pH gradient with a lower pH (higher proton concentration) at SU e of ATP synthase. The inverse lateral pH gradient was also found in IF1‐KO cells. This can only be explained by the reverse activity of ATP synthase, pumping protons into the IMS while hydrolyzing ATP. As examples, we show the pH maps of WT and IF1‐H49K‐OE mitochondria in detail in Fig 8J. Together, the experimentally obtained pH profiles of the mitochondrial sub‐compartments reveal lateral and radial pH gradients (Fig 8K). The pH profiles for all conditions are schematically depicted in Fig EV4E. In IF1‐ OE cells, local pH levels might be additionally determined by ultrastructural changes in cristae that result in a different distribution pattern of OXPHOS complexes (Fig EV5A–C).
Figure EV5. The spatiotemporal organization of ATP synthase is affected by IF1‐H49K.

- Mobility pattern of F1FO ATP synthase F1‐SU γ under control conditions. Recording: 59 Hz, 3,000 frames, total recording time 51 s (Movies EV1 and EV2). The panels show trajectory maps of the F1FO ATP synthase in two individual mitochondria. Inset: selected trajectories from a third mitochondrion showing orthogonal trajectories (arrowheads). Trajectories of the F1FO ATP synthase were generated by using the Multi‐Target‐Tracing‐Tool (MTT). Every single trajectory is in one color.
- Mobility pattern of F‐SU γ in cells expressing IF1‐HA, (Movie EV3) in expanded information. Recording rate: 59 Hz, 3,000 frames.
Data information: Scale bars: 1 µm (A, B, C), 0.5 µm (inset A). Reprinted under the terms of the Creative Commons Attribution License (Weissert et al, 2021).
IF1 is required to optimize mitochondrial ATP production rates under all conditions
Finally, we correlated mitochondrial and glycolytic ATP synthesis rates and mitochondrial ATP level in living cells with the ΔpH data. To obtain ATP synthesis rates, OCR and ECAR were recorded before and after serial addition of mitochondrial inhibitors (oligomycin and rotenone/antimycin A) and pathway‐specific ATP production rates were calculated (Mookerjee et al, 2017). Under glycolytic conditions, HeLa WT cells showed low mitochondrial ATP production rates (< 400 pmol/min*30 K cells) but high cytosolic ATP production rates (> 600 pmol/min*30 K cells) (Fig 9A and B). Under OXPHOS conditions, ATP synthesis rates increased > 2×, while cytosolic ATP synthesis rates decreased significantly. However, the increase in ATP synthesis rates under OXPHOS conditions was not observed in IF‐KO cells, emphasizing IF1’s role in blocking ATP hydrolysis under OXPHOS conditions due to low ΔpH/Δp. Also, ATP levels were lower in IF1‐KO cells under OXPHOS conditions (Fig 9C).
Figure 9. IF1 is beneficial for ATP production even under OXPHOS conditions.

- Mitochondrial ATP production rates in HeLa cells the presence and absence of IF1. Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were determined with an automatic flux analyzer (Seahorse/Agilent). After monitoring basal respiration, oligomycin (1 µM), and rotenone plus antimycin A (Rot; 0.5 µM; AA, 0.5 µM) were added sequentially to determine glycolytic and mitochondrial ATP production rates. ˜12 technical replicates per measurement, N = 4 independent assays.
- Cytosolic ATP production rates. Each data point represents the mean ATP production rate of one well (˜12 technical replicates per measurement, N = 4 independent assays.).
- Determination of mitochondrial ATP levels with the fluorogenic dye ATP‐red™ (5 µM, prestaining for 15 min). To normalize on mitochondrial mass, mitochondria were stained with MitoTracker™ Green (MTG, 100 nM for 30 min). Two technical replicates from same clones.
Data information: (A–C) Statistics: One‐way ANOVA with post hoc Scheffé test. The error bars denote SD; the boxes represent the 25th to 75th percentiles. Whiskers: outliners include coefficient 1.5. The vertical lines in the boxes represent the median values, whereas the square symbols in the boxes denote the respective mean values. The minimum and maximum values are denoted by x.***P ≤ 0.001; **P ≤ 0.01; n.s.: non‐significant.
Under glycolytic but not OXPHOS conditions, IF1‐H49K had a positive effect on ATP production rates. We conclude that the endogenous IF1 was not fully active under glycolytic conditions, likely because the matrix pH was basic (pH > 7.5), and IF1 is activated at pH < 7.5 (Zanotti et al, 2009; Boreikaite et al, 2019). The pH‐independently active IF1 (IF1‐H49K) obviously blocked ATPase more efficiently. Finally, ATP levels were determined with the fluorogenic BioTracker™ ATP‐Red™dye (Millipore), which locates to mitochondria. ATP‐Red fluorescence was normalized on the mitochondrial mass (MitoTracker™Green signal). Generally, mitochondria with high respiration rates (OXPHOS conditions) had a significantly higher ATP content than mitochondria with lower respiration (glycolytic conditions), irrespective of IF1 levels (Fig 9C). IF1‐H49K expressing glycolytic cells had more mitochondrial ATP than WT cells, in accordance with higher mitochondrial ATP synthesis rates and inhibition of ATP hydrolysis. IF1‐KO cells had lower ATP levels than WT cells, which can be explained by higher ATP hydrolysis rates and/or export of ATP to supplement the cytosol with ATP, as suggested above. Thus, these data suggest that in WT HeLa cells, a subpopulation of ATP synthase is in the reverse mode. Under OXPHOS conditions, IF1 is required to block this. IF1‐H49K‐expressing cells also had lower mitochondrial ATP levels than WT cells (but higher levels than IF1‐KO cells). The same observation was made with pancreatic beta cells, where mitochondrial and cytosolic ATP substantially dropped in IF1‐OE (Kahancova et al, 2020). Whether this is due to hampered coupling of ATP synthase and respiration could be tested by determining P/O values. A decreased coupling could arise due to the recently observed altered cristae structure in IF1‐H49K‐expressing cells (Weissert et al, 2021).
Together, these data clearly demonstrate the importance of IF1 for blocking futile ATP hydrolysis under OXPHOS conditions.
Discussion
Although PMF has been intensively studied since the 1960s, much of the work has remained at a descriptive level due to methodological limitations. Only recently has the development of functional imaging and biosensing techniques made it possible to visualize mitochondrial PMF in living cells in situ. The use of membrane potential‐sensitive dyes has revealed new fundamental features, such as the occurrence of rapid changes in PMF of individual mitochondria (i); differential PMF in individual cristae; and cristae remodeling capable of tuning PMF (ii) (Wolf et al, 2019, 2020; Kondadi et al, 2020a; Segawa et al, 2020). To this end, readout of local pH sensors revealed that lateral pH gradients exist between H+ source and sink in individual cristae (iii) and thus PMF is nonhomogeneous along the intermembrane (iv) (Rieger et al, 2014; Sjoholm et al, 2017; Toth et al, 2020). Apparently, local separation of respiratory supercomplexes and ATP synthase is an essential prerequisite for tuning electron transfer and PMF formation as well as its utilization (Strauss et al, 2008; Cogliati et al, 2013; Rieger et al, 2014; Wolf et al, 2019).
Here, we specialized in how PMF is shaped around ATP synthase F1 and FO and correlated this with the activity as well as spatiotemporal organization of ATP synthase. To do this, we performed high‐resolution pH measurements on the F1 and F0 part of ATP synthase, determined the mobility and distribution of ATP synthase under different metabolic conditions in the cristae membrane using single molecule tracking and localization microscopy, and deciphered the fraction of ATP synthase or ATPase activity via altering the levels of inhibitory factor IF1. The inhibitory factor IF1 is a key peptide for the regulation of the activity of the mitochondrial ATP synthase enzyme, which further influences the bioenergetics and structure of mitochondria and eventually cell fate (Campanella et al, 2008; Fujikawa et al, 2012; Barbato et al, 2015; Garcia‐Bermudez & Cuezva, 2016). The pH maps we generated by fusing the pH sensor sEcGFP to subunit CoxVIIIa of CIV, subunit e at FO, and SU γ at F1 of CV and mitochondrial processing peptidase in the matrix and revealed significant differences between glycolytic and respiratory cells. Moreover, we succeeded in elucidating the impact of IF1 to the local pH at ATP synthase under different metabolic conditions. Tumor cell lines can easily be reset to different metabolic states by changing the sugar supply (Rossignol et al, 2004). High glucose sugar supply (25 mM) was used to induce a glycolytic phenotype, which was characterized by a low OCR with no spare capacity and a high cytosolic ATP synthesis rate. Alternatively, a respiratory phenotype was induced by supplying galactose/pyruvate showing significant higher OCR rates and an almost twofold spare capacity (Fig EV2). We assumed that IF1 levels mainly would affect pH under glycolytic conditions, as it plays a role in glucose‐modulated adaptation in tumor cells (Domenis et al, 2012). Contrary, we did not except much influence of IF1 on pH under oxidative conditions. We assumed that the role of IF1 would be restricted to inhibit ATP hydrolysis during ATP synthase assembly (Song et al, 2018) but would not affect ATP synthase activity. In order to determine the effects of IF1, we generated an IF1‐KO cell line and a cell line stably expressing the constitutively active IF1‐H49K mutant (Schnizer et al, 1996; Garcia‐Aguilar & Cuezva, 2018).
First, we found that the pH at SU e was acidic under glycolytic conditions (pHWT = 7.06 and pHIF1‐KO = 6.95). This can be explained by reverse proton pumping in HeLa WT and IF1‐KO cells linked to ATP hydrolysis. Under respiratory conditions, the pH was significantly higher (pHWT = 7.28 and pHIF1‐KO = 7.39) due to proton pumping from the ICS into the matrix by active ATP synthase. In IF1‐H49K cells, the pH at subunit e was significantly higher under glycolytic conditions (pH = 7.28), indicating an inhibition of ATP hydrolysis. Under respiratory conditions, the pH was similar as in WT cells and as in IF1‐H49K expressing glycolytic cells. If IF1 had inhibited ATP synthesis under OXPHOS conditions as suggested elsewhere (Sanchez‐Cenizo et al, 2010), we would have expected a lower pH at SU e in IF1‐H49K cells compared to control cells. However, the data showed only a tendency (P = 0.083) for pH decrease at SU e (Fig 8A). The comparison of IF1‐KO and IF1‐H49K cells showed a significant difference between the pH at SU e: in IF1‐KO cells with a pH = 7.39 ± 0.28 and in IF1‐H40K cells with a pH = 7.19 ± 0.25. This clearly shows that IF1 influences the pH at SU e of ATP synthase even under OXPHOS conditions. Accordingly, the decisive factor for the pH at SU e is whether the ATP synthase acts as a proton source (reverse mode) or as a sink (ATP synthesis mode). Additional pH measurements at CV SU γ in the F1 subcomplex at the n‐side of the membrane confirmed the influence of IF1 on ATP hydrolase activity in glycolytic cells. The pH in WT cells at SU γ was clearly higher than in IF1‐H49K where ATPase activity was blocked. In respiring cells, no difference was found between the pH at SU γ in WT and IF1‐H49K expressing cells. This is in accordance with the model that IF1‐H49K only blocks reverse proton pumping by ATPase activity (Gledhill et al, 2007).
Most interestingly was the result that in WT cells the overall ΔpH at CV (SU γ ‐ SU e) was lower (ΔpHOXPHOS˜0.13) in respiring mitochondria with forward ATP synthase activity than in mitochondria with reverse ATP synthase (=ATPase) activity (ΔpHglycolytic˜0.72). This finding has two implications: The feasibility of ATP hydrolase to generate a significant PMF (i) and that for mitochondrial ATP synthesis obviously a high ΔpH is not a prerequisite (ii). A ΔpH = 0.13 is equivalent to a 1.1‐fold difference in the proton concentration! This resembles the situation of specific bacteria that are able to produce ATP even with unfavorable pH gradients (Preiss et al, 2010; Krulwich et al, 2011). Overall, the importance of a high PMF for ATP synthesis seems to have been overestimated. In the steady state, it is obviously sufficient that protons are continuously delivered, similar as it was recently shown in an in vitro model (Toth et al, 2020). The low PMF under ATP synthesis conditions is a tricky situation, since it would in principle stimulate reverse ATP synthase activity. In order to prevent futile activity, IF1 obviously is essential under OXPHOS conditions! However, how can IF1 be activated when the phosphorylation potential is in principle high? Indeed, phosphorylation of IF1 would prevent its binding to ATP synthase (Garcia‐Bermudez et al, 2015). A possible explanation of IF1 activation despite a high phosphorylation potential is provided by the observation that the pH at SU γ was lowered under OXPHOS conditions (pHOXPHOS = 7.41 ± 0.20; pHglycolytic = 7.78 ± 0.11). This would likely shift the equilibration toward protonated IF1, which is the active form as recently reported (Boreikaite et al, 2019).
Furthermore, we showed here that the local pH is important for the calculation of PMF. For example, the bulk matrix pH was always higher than the local pH at SU γ, only in IF1‐KO cells no difference was found. This means that the calculated PMF when related to the matrix pH is higher than when related to the local pH at F1‐SU γ. The finding that the pH measured at the ATP synthase subunit differs from the pH in the matrix bulk may be explained if the release of protons from the membrane into the bulk is delayed, as suggested previously (Heberle et al, 1994), or if the matrix itself is sub‐compartmented where ATP synthase (sub‐) complexes are spatially separated from MPP. Because super‐resolved localization of in nascendi CV subcomplexes in yeast did not provide evidence for spatial orchestration of CV assembly, but CV is assembled throughout the crista membrane (Stoldt et al, 2018), we conclude that diffusion of protons from the surface into the bulk is delayed as previously suggested (Mulkidjanian et al, 2006). To verify this, we would need to test whether other surface sensors on the n‐side would also report a lower pH than in the matrix or whether this is a specific property of the sensor on the proton pump ATP synthase.
Moreover, we confirmed that the physical separation shown between the respiratory complexes as proton sources and ATP synthase as proton sink results in a lateral pH gradient (Strauss et al, 2008; Rieger et al, 2014; Toth et al, 2020). This lateral pH gradient was reversed under glycolytic conditions, another evidence for active proton pumping of reverse ATP synthase/ATPase. Under respiratory conditions, the proton concentration at CIV, the primary proton pump, was higher than in glycolytic conditions. Notably, this pH drop at CIV was very prominent in IF1‐H49K cells under respiratory conditions, indicating a stimulation of CIV activity. These cells also exhibited the highest mitochondrial ATP synthesis rate. With regard to the current model of respiratory control for the regulation of cytochrome c oxidase, which suggests that CIV is negatively controlled by the ATP/ADP ratio and a high ΔΨm (Ludwig et al, 2001), this is at the first glance perplexing. The data suggest that despite a considerable PMF (ΔpH > 1.0) and high mitochondrial ATP synthesis rate, complex IV obviously was not inhibited but rather stimulated. A possible explanation is that respiration and ATP synthesis were partially uncoupled in IF1‐H49K cells. Since overexpression of IF1‐H49K can evoke structural changes of the inner membrane architecture (Weissert et al, 2021), the kinetic coupling between CIV and CV (Toth et al, 2020) could be partially abandoned. This hypothesis finds support in the observation that the lateral pH difference between CIV and CV was also higher in IF1‐H49K cells than in control cells (Fig 8l). Figure EV4E summarizes the findings of this paper and shows differences in lateral, radial, and transmembrane pH gradients under the influence of IF1 under different metabolic conditions.
In sum, we here elucidated the contribution of ATP synthase to local PMF. We found that IF1 is a key physiological instrument to prevent futile ATP hydrolysis (Fujikawa et al, 2012), even under OXPHOS conditions, where obviously the steady‐state ΔpH at the ATP synthase is very low. An additional mechanism to prevent futile ATP synthase/hydrolase at the spot, is a possible spatial separation of ATP synthase and ATPase as recent findings of our group indicate (Salewskij et al, 2020).
The local pH differences and diverse pH gradients dissected here may partially explain recent observations of local ΔΨm differences within mitochondria and between individual cristae (Wolf et al, 2019, 2020; Kondadi et al, 2020a; Segawa et al, 2020). By combining these new high‐resolution noninvasive methods for determining pH values and ΔΨm, our understanding of the PMF is currently actively advancing.
Conclusion
ATP synthase activity substantially determines the local Δp in mammalian cells, as revealed by local pH measurement presented here. Strikingly, the Δp at ATP synthase is almost negligible under OXPHOS conditions. Therefore, to prevent futile ATP synthesis/hydrolysis cycles, IF1 is essential, especially under OXPHOS conditions. Furthermore, pH profiling of mitochondrial sub‐compartments disclosed that the pH is not laterally or radially balanced in mitochondria in living cells. This is due to the physical separation of primary proton pumps and ATP synthase. In this context, diffusion of protons is apparently not sufficient to ensure proton equilibration under steady‐state conditions. These findings, in combination with the observed heterogeneity of the mitochondrial membrane potential (Wolf et al, 2019), are important for understanding the generation and modulation of the proton motive force (PMF).
Materials and Methods
Cell culture
HeLa cells were purchased from the Leibniz Institute DSMZ‐German Collection of Microorganisms and Cell Cultures). The HeLa cells (wildtype, IF1‐KO, and IF1‐H49K‐HA) were cultivated in XF Base Medium Minimal DMEM (Agilent) supplemented with 10% FBS (Fetal bovine serum), 1% HEPES (4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid), 1% NEAA (non‐essential amino acids), 2% NaHCO3 and either 10 mM D‐Galactose (plus 4 mM alanyl‐l‐glutamine) or 25 mM d‐Glucose (plus 2 mM alanyl‐l‐glutamine) at 37°C and 5% CO2. When confluency was reached, cells were split using 1 ml Trypsin/EDTA for 5 min at 37°C (and 5% CO2).
Single molecule tracking and localization microscopy (from Salewskij et al, 2020)
Single molecule tracking and localization was performed at a room temperature with an inverted microscope (Nikon, Eclipse TE2000) equipped with a TIRF condenser, an oil immersion objective designed for TIRF (100× oil/N.A. 1.49/0.12 Apochromat, Nikon), an EMCCD camera (Evolve™512, Photometrics, 512 × 512 pixel imaging array, 16 μm × 16 μm pixel area). To fulfill the Nyquist oversampling, a 1.5× magnitude focusing lens was used. A quad line beam splitter (zt405/ 488/561/633–640) was mounted in combination with an emission quadband filter (HC 446 nm/523 nm/600 nm/677 nm). A 561 nm laser (Melles Griot, 20 mW, DPSS) was coupled into the device via a single‐mode glass fiber (FC/PC). The measurements were performed as described before (Weissert et al, 2021). In short, HeLa cells stable transfected with F1FO ATP synthase subunit ɣ fused C‐terminally to a HaloTag were posttranslationally labeled with TMRHTL (1 nM) for 20 min, washed twice in PBS and once in medium without phenol red before imaging. Assembly and correct intramitochondrial localization were shown earlier with constructs where similar sized EGFP and pHluorin, respectively, were genetically fused to the same position in SU γ as exchange of by the HaloTag7 (Muster et al, 2010; Rosselin et al, 2017). The TMRHTL concentration was set for sub‐stoichiometric labeling and low enough to visualize single molecule signals. The incident angle was set to achieve a highly inclined laminated optical light sheet (Tokunaga et al, 2008). Usually, 3,000 frames were recorded. Frame rates were 33 and 59 Hz, respectively. Individual signals with a signal‐to‐noise ratio above 5 were fitted with a 2D Gaussian fit for localizing molecules. From all frames, a cumulative localization map was generated after movement of mitochondria during the recording time was excluded. For generating trajectory maps, single molecules in subsequent frames were connected when the following criteria were matched: they must have been within a radius of maximal 2 px (pixel size 107 nm) and the intensity distribution profile must have been identical. Only trajectories with a minimum duration of 160 ms were analyzed.
Theoretical background (from Weissert et al, 2021)
Single fluorescent molecules, whose size is below the diffraction limit, project a three‐dimensional diffraction pattern onto the image plane, which is defined as the point spread function (PSF). The PSF can be calculated by Abbe’s law:
| (1) |
with d being the diameter of the PSF, λ the wavelength of the emitted light, and NA being the numerical aperture of the microscope’s objective. The numerical aperture describes the light gathering capability of an objective and depends on the refractive index n of the medium in which the lens is used and the half angular aperture of the objective α. The “full width at half maximum” (FWHM) criterion is used to describe a PSF, whereby FWHM is the width at 50% of the intensity maximum. For TMR (λem. = 582 nm) and an objective with NA of 1.49, the PSF diameter is approximately 390 nm.
According to the Rayleigh criterion, the resolution of the system is:
| (2) |
where r is the minimal distance between the two objects. n is the refractive index of the medium between the objective and sample, λ is the wavelength of the emitted light, α is the half angular aperture of the objective, and NA is the numerical aperture of the microscope objective. Practically, this means that two separate light points can just be distinguished if the intensity maximum of the first point coincides with the first intensity minimum of the second point. Here, the theoretical resolution is ˜240 nm. The pixel size was 106.7 nm for an EMCCD camera (512 × 512 px chip) with a pixel area of 16 µm × 16 µm, thus fulfilling the Nyquist–Shannon sampling theorem, which demands a twofold oversampling rate for the desired spatial resolution. In order to probe the spatiotemporal behavior of F1FO ATP synthase, the multi‐target tracer was used (Sergé et al, 2008). First, the particle was localized by applying a 2D Gaussian fit. The accuracy of the localization highly depends on the brightness of the signal (corresponding to the number of emitted photons N), the pixel size of the EMCCD/CMOS camera, and the background noise:
| (3) |
where σ is the standard error of the mean value of photon distribution, N is the number of gathered photons, a is the pixel size of the EMCCD/CMOS detector, b is the standard deviation of the background, and si is the standard deviation in dimension i, with i being either x or y dimension.
Single molecule localization and tracking
Single molecules were recorded over several frames. The determined PSF was used to localize single emitters by a 2D Gaussian fit (Betzig et al, 2006; Hess et al, 2006; Gould et al, 2009). The multiple‐target tracing algorithm (MTT) (Sergé et al, 2008) was used to generate trajectories. To obtain diffusion data, the mean square displacement (MSD, r 2) was determined and the diffusion coefficient was calculated according to:
| (4) |
| (5) |
where r2 is the MSD, D is the lateral diffusion coefficient, Δt is the lag time, n is the dimension, and ε is the localization error. The exponent α is a factor that distinguishes modes of diffusion, with α < 1 for anomalous diffusion. Anomalous diffusion is expected when the movement of particles is restricted by structural constraints such as physical obstacles or inclusion in micro‐compartments. Further details are found in Appelhans and Busch (2017) (Appelhans et al, 2018).
Statistics
For statistical analysis, we used ANOVA with post hoc Mann–Whitney or Scheffé tests, as indicated, for. Details are mentioned at the figure legends.
Generation of IF1‐KO and IF1‐H49K‐HA cell lines
The IF1‐knockout cells (IF1‐KO) were manufactured using the CRISPR/Cas technique. The first step was to find the appropriate targeting sequence for crRNA in the vicinity of the START codon. The crRNA was cloned into the pSpCas9(BB)‐2A‐GFP (PX458) vector (Addgene). Thus, the resulting vector contained both sgRNA and Cas9 nuclease. To enable selection with puromycin, the pac gene was inserted at the cutting site. Therefore, a construct was cloned with homologous arms to the right and left of the place where Cas9 is to be cut with the pac gene in between. HeLa cells were co‐transfected with both constructs and transformed cells were selected by adding 0.5 µg/ml puromycin. Positive clones were isolated and the IF1 knockdown tested by qPCR and WESTERN. For cloning of IF1‐H49K‐HA, the forward primer GAAGAAGAAATCGTTCATCATAAG with an EcoRV restriction site and the reverse primer CTTGTGTTTTTTCAAAGCTGCCAGTTGTTC with an EcoRI restriction site was used. The template was integrated into the pSems26 vector after cutting out the sequence encoding for CoxVIIIa‐snap‐tag purchased from NEB Biosciences™ (formerly Covalys™). The plasmid backbone of pSems26 encodes for ampicillin resistance and has a CMV promoter. For generation of a stable cell line expressing IF1‐H49K‐HA, transfected HeLa cells were selected for stable neomycin resistance by growth in the presence of 0.8 mg/ml G418 (Calbiochem #345810). Positive clones were tested by WESTERN.
Real‐Time ATP Rate Assay via oxygen consumption measurements
Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) of intact HeLa cells were recorded with the Seahorse XFe96 Extracellular Flux Analyzer (Agilent Technologies). 30,000 cells were sown per well of a 96‐well XF cell culture microplate 24 h before the experiment. 60 min before the Agilent Seahorse XF Real‐Time ATP Rate Assay, cells were washed with XF DMEM medium pH 7.4 (with 5 mM HEPES, without phenol red, glucose, pyruvate, and l‐glutamine, 103575‐100 from Agilent/Seahorse Technologies) with supplements (1 mM pyruvate, 2 mM L‐glutamine and 5.6 or 25 mM D‐glucose, respectively, 10 mM D‐galactose—adjusted to pH 7.4), added to fresh XF assay medium and incubated at 37°C before loading into the XFe Analyzer. Supplements were from Roth. Injections were as follows: (i) 1 µM oligomycin, (ii) 0.5 μM rotenone/0.5 µM antimycin. Seahorse XF technology measures the flux of both H+ production (ECAR) and O2 consumption (OCR), simultaneously. By obtaining OCR and ECAR data under basal conditions and after serial addition of mitochondrial inhibitors (oligomycin and rotenone/antimycin A), total cellular ATP production rates and pathway‐specific mitochondrial ATP and glycolytic ATP production rates can be measured (Mookerjee et al, 2017). The series of calculations used to transform the OCR and ECAR data to ATP production rates is performed using the Seahorse XF Real‐Time ATP Rate Assay Report Generator from Agilent. Briefly, glycolytic ATP production rate (pmol ATP/min) = glycolytic proton efflux rate (pmol H+/min) is associated with the conversion of glucose to lactate in the glycolytic pathway. Mitochondrial ATP production rate is associated with oxidative phosphorylation in the mitochondria and defined as: OCRATP (pmol O2/min) = OCR (pmol O2/min) ‐ OCROligo (pmol O2/min).
SDS–PAGE and BN‐PAGE
For SDS–PAGE, cell lysates from samples of confluent T‐25 flasks (1 flask would last for 10–15 gels) were heated for 5 min (95°C) and then separated on 12% Tricine–SDS–PAGEs and transferred to PVDF membranes.
For blue native PAGE, cell lysates from 2 confluent T175 flasks were processed as described recently (Salewskij et al, 2020). In short, harvested cells were lysed, mitochondria enriched by differential centrifugation and solubilized via Digitonin ˜50% (TLC) (Sigma‐Aldrich, #D5628) 6 g/g mitochondria (Acin‐Perez et al, 2008) as described in Wittig et al (2007). First, the concentration of crude mitochondria was determined via the Bradford method using BSA as a standard. For BN‐PAGE analysis, 50 μg protein per pocket was loaded and separated via the vertical native gel 3–12% SERVAGel™ (Serva, #43251.01). In addition, a NativeMark™ unstained protein standard (Thermo Fisher, #LC0725) was used for molecular weight estimation. However, it should be taken into consideration that in gradient gels, native membrane proteins move slightly different than free protein standard, especially in the high molecular weight range. Cathode and anode buffers were prepared as described in Wittig et al (2006).
Immunoblotting
Briefly, cell lysates from samples of confluent T‐25 flasks (1 flask would last for 10–15 gels) were heated for 5 min (95°C). 20 µg of denatured protein was then resolved by SDS–PAGE using self‐casted 4–12% Tricine–Bis–Acrylamide gels (Bio‐Rad). Gel contents were electrotransferred to PVDF membranes (Millipore) using a semidry TransferApparatus (Trans‐Blot SD Bio‐Rad). Equal loading was evaluated by Ponceau S staining. Membranes were blocked with 10% nonfat dry milk in TBS‐T (200 mM Tris, 1.37 M NaCl, + 0.1% Tween‐20) for 1 h. Membranes were incubated with primary antibodies overnight at 4°C. IF1 was detected with anti‐ATPIF1 antibody (CD6P1Q) purchased from Cell Signaling (#13268), VDAC was detected with an anti‐VDAC antibody (Cell signaling #4661), subunit β was detected with an anti‐ATP5B antibody (Proteintech #17247‐1‐AP), subunit e with an anti‐ATP5I antibody (Abcam #122241), and polyclonal anti‐SU γ (ATP5C1) was a gift from Ilka Wittig. GFP/sEcGFP was detected with polyclonal anti‐GFP antibody (host: rabbit, #AB10145, purchased from Merck Millipore.) To detect CIV, anti‐MTOC1 (a‐Cox1, host mouse, monoclonal) from Invitrogen antibodies (#459600) was used. Monoclonal anti‐actin was purchased from Sigma‐Aldrich (#A1978, host mouse). For the secondary antibody, Peroxidase AffiniPure Goat Anti‐Mouse IgG + IgM (H + L) (111‐035‐068) or Goat Anti‐Rabbit IgG (H + L) (111‐035‐045) from Jackson Immuno Research was used. After washes in TBS‐T, detection was performed with HRP‐conjugated secondary antibodies (1:2,000). Membranes were washed in TBS‐T (Tris‐buffered saline (TBS) and Tween 20) and developed using standard chemiluminescence with ECL (SuperSignal WestPico Thermo Fisher) and imaged by ChemiDoc BioRaD.
Fluorescence microscopy
Fluorescence imaging was carried out with confocal laser scanning microscope (Leica TCS SP8 SMD) equipped with a 63× water objective (N.A. 1.2) and a tunable white light laser. HyDs with GaASP photocathodes were used as detectors. Measurements were performed in a humidified chamber at 37°C and 5% CO2.
Local pH measurements
For in situ pH determination, HeLa cells transfected with sEcGFP (Miesenbock et al, 1998) (or superecliptic pHluorin = pHl, derivate of green fluorescent protein) fusion constructs (calcium phosphate method) were used. sEcGFP can be used as a ratio‐metric pH sensor (Rieger et al, 2014), when emission at 470 and 510 nm is recorded (Gao et al, 2004). The original template for sEcGFP was a gift from Prof. Jürgen Klingauf. The local pH sensors were generated by genetically fusing sEcGFP to subunits of CV, CIV, and targeting it to the matrix, respectively (Fig 1D). In detail, sEcGFP was fused to subunit γ (SU γ) of the F1 subcomplex of CV at the n‐side of the IMM, to subunit e (SU e) of FO subcomplex of CV at the p‐side of the cristae membrane and to subunit CoxVIIIa of CIV at the p‐side of the IMM (Rosselin et al, 2017). To generate CoxVIIIa‐sEcGFP, the sEcGFP template was integrated into the pSems26 CoxVIIIa‐snap vector purchased from NEB Biosciences™ (formerly Covalys™) after cutting out the sequence encoding for the snap‐tag. The plasmid backbone of pSems26 encodes for ampicillin resistance for bacterial selection and contained the neomycin resistance for selection of stable clones. The promoter was from CMV. To generate SU e‐sEcGFP and SU γ‐sEcGFP, respectively, CoxVIIIa was cut out and the coding sequences for SU e and SU γ, respectively, inserted at the N terminus of sEcGFP. The correct localizations of the fusion constructs have been confirmed by Immuno‐EM in a different study (Rosselin et al, 2017). An additional pH sensor was generated for the matrix. The mitochondrial targeting sequence of the mitochondrial processing peptidase MPP was fused to sEcGFP to obtain a soluble matrix pH sensor (mt‐sEcGFP). Stable cell lines were generated by addition of 0.5 µg/ml puromycin or 0.8 mg/ml G418 and selection and testing of positive clones. CoxVIIIa‐sEcGFP and CV SU e‐sEcGFP were used to determine ICS pH and CV, SU γ‐sEcGFP, and mt‐sEcGFP to determine matrix pH (Rieger et al, 2014; Sohnel et al, 2016). Local pH was recorded in situ by confocal fluorescence emission ratio imaging by an inverse confocal laser scanning microscope (Leica TCS SP8 SMD). All measurements were performed at 37°C in fresh medium or buffer. Ratio images were obtained after excitation with a single wavelength (λex. = 405 nm) by recording in two wavelength regimes (λem. = 440–490 nm and 505–517 nm). For pH calibration, cells were perfused for 3–5 min with PBS (PAA, pH 7.0–7.5 with CaCl2 and MgCl2). PBS was exchanged for MES (25 mM, pH 6.1), BES (25 mM, pH 6.5, 7.1, and 7.4) or HEPPSO (25 mM, pH 7.8, 8.0, and 8.2) buffer adjusted to the desired pH by 1 M HCl or NaOH and supplemented with the following compounds: 125 mM KCl, 20 mM NaCl, 0.5 mM CaCl2, 0.5 mM MgSO4, 10 µM FCCP, 1 µM nigericin, and 5 µg/ml oligomycin. FCCP, nigericin, and oligomycin were purchased from Enzo Life. MES was purchased from Biomol and BES and HEPPSO from Sigma. Other common chemicals were sourced from Roth. Images were taken after 10‐30 min of incubation.
Membrane potential measurements
The mitochondrial membrane potential, Δψm, was determined using the membrane potential‐sensitive dye TMRE (tetramethylrhodamine ethyl ester). TMRE is a cell‐permeant, positively charged dye with excitation/emission maxima ˜549/575 nm, which accumulates in active mitochondria due to its relative negative charge. In addition, mitochondria were stained with MTG (MitoTracker™ Green FM) which was used as reference for mitochondrial mass. Cells grown on glass coverslips were incubated in medium with MTG (100 nM) and TMRE (7 nM) for 30 min. Cells were washed twice with PBS. Cells in fresh medium plus 7 nM TMRE were kept at 37°C and 5% CO2 during the measurement. Fluorescence was recorded with a cLSM (Leica SP8, equipped with a white light laser, WLL, a 63× water objective, N.A., and special hybrid detectors HyD). Z‐stacks (three slices, step size 360 nm) were recorded simultaneously for MTG and TMRE signals. MTG was excited with the laser line 488 nm (WLL), and emission was recorded between 500 and 510 nm. TMRE was excited with laser line 561 nm (WLL), and emission between 620 and 750 nm was recorded. TMRE/MTG ratios were created by using ImageJ® software (NIH Image). Therefore, the Otsu filter mask was used as a mask for mitochondria and the background was set to NaN. The mean respective fluorescence intensities (all mitochondria/cell) were determined.
Mitochondrial ATP via fluorescence imaging of ATP‐red
The BioTracker™ ATP‐red dye (Millipore) is a fluorogenic indicator for ATP in mitochondria (Wang et al, 2016). To load the dye, the cells were incubated in medium with 5 μM ATP‐red for 15 min at 37°C (5% CO2). Before measurement, the cells were washed twice with medium before fresh medium was added. The fluorescence was recorded with a cLSM (Leica SP8, 60× water objective) at 37°C and 5% CO2. ATP‐red was excited at 561 nm (white light laser), and the emission from Z‐stacks (three slices, step size 360 nm) was recorded between 580 and 650 nm. The subtracted background images for intensity analysis were created with ImageJ® software (NIH Image). The filter mask Otsu was used as mask for the cells, and the background was set to NaN.
Statistics
Statistical analysis was performed using OriginPro version 9.6, 2019 (OriginLab Cooperation, Northampton, MA). After testing for normal distribution (Kolmogorov–Smirnov), one‐way ANOVA test with post hoc Schéffe test was selected for comparative measurements. At least three independent measurements on different days with different cell culture samples were the basis.
pH calibration
sEcGFP has two emission peaks (λem1 = 511 nm and λem2 = 464 nm, λexc = 405 nm) that reversely respond to pH changes. The emission spectrum obtained by a fluorescence scan (10 nm steps) of cells stably expressing matrix‐targeted sEcGFP shows the existence of an isosbestic point at 484 nm Thus, the fluorescence intensity below and after 484 nm shows an opposite dependency on pH and sEcGFP can be used as ratio‐metric pH probe. The fluorescence emission ratio of λem1/λem2 reports pH changes (Rieger et al, 2014).
The ratio of Em.511/Em.464 was plotted in dependence on the pH. For all constructs, the λem1/λem2 value increased. The dependency was fitted with a Boltzmann fit to obtain the respective conversion from emission ratio to pH.
MiNA analysis
To determine the morphology under different conditions, the cells were stained with 100 nM.
MTG (MitoTracker® Green FM). After 30‐min incubation, cells were washed twice with PBS. Cells in fresh medium were kept at 37°C and 5% CO2 during the measurement. Fluorescence was recorded with a cLSM (Leica SP8, equipped with a white light laser, WLL, a 63× water objective, N.A., and special hybrid detectors HyD). MTG was excited with the laser line 488 nm (WLL), and emission was recorded between 500 and 510 nm (single slices). Finally, the mitochondrial morphology (mean branch length, mitochondrial footprint, networks, and individuals) was analyzed with the MiNA plugin (Valente et al, 2017) for ImageJ® software (NIH Image).
Author contributions
KBB, BR, and TA conceptualized the study; KBB, TA, BR, JV, and M‐TB contributed to methodology; KBB, BR, TA, JV, and M‐TB validated the data; KBB, BR, TA, JV, and M‐TB made formal analysis; KBB, BR, TA, JV, and M‐TB investigated the data; KBB contributed to resources; KBB and BR contributed to writing—original draft preparation; KBB and BR contributed to writing—review and editing; KBB, BR, TA, and M‐TB visualized the study; KBB and BR supervised the data; KBB contributed to project administration; KBB acquired funding. All authors have read and agreed to the published version of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Source Data for Expanded View
Source Data for Figure 2
Source Data for Figure 6
Acknowledgements
The study was supported by grants from the DFG (Bu2288/1‐2), (Bu2288/1‐3; FOR 2848/1), and the CRC SFB944 (INST 190/1672 and the z‐Project). Part of the data were acquired in the iBiOS facility in Osnabrück. We thank Wladislaw Kohl and Patrick Duwe for technical assistance and Giulia Nebel for important preliminary studies. We also would like to thank Michael Hippler and Armen Mulkidjanian for stimulating discussions. Open Access funding enabled and organized by Projekt DEAL.
EMBO reports (2021) 22: e52727.
Data availability
No data were deposited in a public database.
References
- Acin‐Perez R, Fernandez‐Silva P, Peleato ML, Perez‐Martos A, Enriquez JA (2008) Respiratory active mitochondrial supercomplexes. Mol Cell 32: 529–539 [DOI] [PubMed] [Google Scholar]
- Allen RD, Schroeder CC, Fok AK (1989) An investigation of mitochondrial inner membranes by rapid‐freeze deep‐etch techniques. J Cell Biol 108: 2233–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhans T, Beinlich FR, Richter CP, Kurre R, Busch KB (2018) Multi‐color localization microscopy of single membrane proteins in organelles of live mammalian cells. J vis Exp 136: e57690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhans T, Busch K (2017) Single molecule tracking and localization of mitochondrial protein complexes in live cells. Methods Mol Biol 1567: 273–291 [DOI] [PubMed] [Google Scholar]
- Appelhans T, Richter CP, Wilkens V, Hess ST, Piehler J, Busch KB (2012) Nanoscale organization of mitochondrial microcompartments revealed by combining tracking and localization microscopy. Nano Lett 12: 610–616 [DOI] [PubMed] [Google Scholar]
- Barbato S, Sgarbi G, Gorini G, Baracca A, Solaini G (2015) The inhibitor protein (IF1) of the F1F0‐ATPase modulates human osteosarcoma cell bioenergetics. J Biol Chem 290: 6338–6348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott‐Schwartz J, Hess HF (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313: 1642–1645 [DOI] [PubMed] [Google Scholar]
- Bisetto E, Comelli M, Salzano AM, Picotti P, Scaloni A, Lippe G, Mavelli I (2013) Proteomic analysis of F1F0‐ATP synthase super‐assembly in mitochondria of cardiomyoblasts undergoing differentiation to the cardiac lineage. Biochim Biophys Acta 1827: 807–816 [DOI] [PubMed] [Google Scholar]
- Blum TB, Hahn A, Meier T, Davies KM, Kuhlbrandt W (2019) Dimers of mitochondrial ATP synthase induce membrane curvature and self‐assemble into rows. Proc Natl Acad Sci USA 116: 4250–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boreikaite V, Wicky BIM, Watt IN, Clarke J, Walker JE (2019) Extrinsic conditions influence the self‐association and structure of IF1, the regulatory protein of mitochondrial ATP synthase. Proc Natl Acad Sci USA 116: 10354–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulthuis EP, Adjobo‐Hermans MJW, Willems P, Koopman WJH (2019) Mitochondrial morphofunction in mammalian cells. Antioxid Redox Signal 30: 2066–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella M, Casswell E, Chong S, Farah Z, Wieckowski MR, Abramov AY, Tinker A, Duchen MR (2008) Regulation of mitochondrial structure and function by the F1Fo‐ATPase inhibitor protein, IF1. Cell Metab 8: 13–25 [DOI] [PubMed] [Google Scholar]
- Carraro M, Giorgio V, Sileikyte J, Sartori G, Forte M, Lippe G, Zoratti M, Szabo I, Bernardi P (2014) Channel formation by yeast F‐ATP synthase and the role of dimerization in the mitochondrial permeability transition. J Biol Chem 289: 15980–15985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana‐Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC et al (2013) Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155: 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Anselmi C, Wittig I, Faraldo‐Gomez JD, Kuhlbrandt W (2012) Structure of the yeast F1Fo‐ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci USA 109: 13602–13607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kuhlbrandt W (2011) Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA 108: 14121–14126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenis R, Bisetto E, Rossi D, Comelli M, Mavelli I (2012) Glucose‐modulated mitochondria adaptation in tumor cells: a focus on ATP synthase and inhibitor Factor 1. Int J Mol Sci 13: 1933–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa M, Imamura H, Nakamura J, Yoshida M (2012) Assessing actual contribution of IF1, inhibitor of mitochondrial FoF1, to ATP homeostasis, cell growth, mitochondrial morphology, and cell viability. J Biol Chem 287: 18781–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Knight MR, Trewavas AJ, Sattelmacher B, Plieth C (2004) Self‐reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol 134: 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Aguilar A, Cuezva JM (2018) A review of the inhibition of the mitochondrial ATP synthase by IF1 in vivo: reprogramming energy metabolism and inducing mitohormesis. Front Physiol 9: 1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Bermudez J, Cuezva JM (2016) The ATPase inhibitory factor 1 (IF1): a master regulator of energy metabolism and of cell survival. Biochim Biophys Acta 1857: 1167–1182 [DOI] [PubMed] [Google Scholar]
- Garcia‐Bermudez J, Sanchez‐Arago M, Soldevilla B, Del Arco A, Nuevo‐Tapioles C, Cuezva JM (2015) PKA Phosphorylates the ATPase inhibitory factor 1 and Inactivates its capacity to bind and inhibit the mitochondrial H(+)‐ATP synthase. Cell Rep 12: 2143–2155 [DOI] [PubMed] [Google Scholar]
- Gilkerson RW, Selker JM, Capaldi RA (2003) The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett 546: 355–358 [DOI] [PubMed] [Google Scholar]
- Gledhill JR, Montgomery MG, Leslie AG, Walker JE (2007) How the regulatory protein, IF(1), inhibits F(1)‐ATPase from bovine mitochondria. Proc Natl Acad Sci USA 104: 15671–15676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould TJ, Verkhusha VV, Hess ST (2009) Imaging biological structures with fluorescence photoactivation localization microscopy. Nat Protoc 4: 291–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Ford HC, Carroll J, Douglas C, Gonzales E, Ding S, Fearnley IM, Walker JE (2018) Assembly of the membrane domain of ATP synthase in human mitochondria. Proc Natl Acad Sci USA 115: 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle J, Riesle J, Thiedemann G, Oesterhelt D, Dencher NA (1994) Proton migration alone the membrane surface and retarded surface to bulk transfer. Nature 370: 379–382 [DOI] [PubMed] [Google Scholar]
- Hess ST, Girirajan TP, Mason MD (2006) Ultra‐high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J 91: 4258–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahancova A, Sklenar F, Jezek P, Dlaskova A (2020) Overexpression of native IF1 downregulates glucose‐stimulated insulin secretion by pancreatic INS‐1E cells. Sci Rep 10: 1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondadi AK, Anand R, Hansch S, Urbach J, Zobel T, Wolf DM, Segawa M, Liesa M, Shirihai OS, Weidtkamp‐Peters S et al (2020a) Cristae undergo continuous cycles of membrane remodelling in a MICOS‐dependent manner. EMBO Rep 21: e49776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondadi AK, Anand R, Reichert AS (2020b) Cristae membrane dynamics ‐ a paradigm change. Trends Cell Biol 30: 923–936 [DOI] [PubMed] [Google Scholar]
- Krulwich TA, Sachs G, Padan E (2011) Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9: 330–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W (2019) Structure and mechanisms of F‐type ATP synthases. Annu Rev Biochem 88: 515–549 [DOI] [PubMed] [Google Scholar]
- Lee JW (2019) Electrostatically localized proton bioenergetics: better understanding membrane potential. Heliyon 5: e01961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang L, Long Q, Weaver D, Hajnoczky G (2017) Choosing proper fluorescent dyes, proteins, and imaging techniques to study mitochondrial dynamics in mammalian cells. Biophys Rep 3: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig B, Bender E, Arnold S, Huttemann M, Lee I, Kadenbach B (2001) Cytochrome C oxidase and the regulation of oxidative phosphorylation. ChemBioChem 2: 392–403 [DOI] [PubMed] [Google Scholar]
- Miesenbock G, De Angelis DA, Rothman JE (1998) Visualizing secretion and synaptic transmission with pH‐sensitive green fluorescent proteins. Nature 394: 192–195 [DOI] [PubMed] [Google Scholar]
- Mitchell P (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi‐osmotic type of mechanism. Nature 191: 144–148 [DOI] [PubMed] [Google Scholar]
- Mitchell P (1966) Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol Rev Camb Philos Soc 41: 445–502 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Moyle J (1967) Chemiosmotic hypothesis of oxidative phosphorylation. Nature 213: 137–139 [DOI] [PubMed] [Google Scholar]
- Mookerjee SA, Gerencser AA, Nicholls DG, Brand MD (2017) Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J Biol Chem 292: 7189–7207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY (2006) Proton in the well and through the desolvation barrier. Biochem Biophys Acta 1757: 415–427 [DOI] [PubMed] [Google Scholar]
- Mulkidjanian AY, Heberle J, Cherepanov DA (2006) Protons @ interfaces: implications for biological energy conversion. Biochim Biophys Acta 1757: 913–930 [DOI] [PubMed] [Google Scholar]
- Muster B, Kohl W, Wittig I, Strecker V, Joos F, Haase W, Bereiter‐Hahn J, Busch K (2010) Respiratory chain complexes in dynamic mitochondria display a patchy distribution in life cells. PLoS One 5: e11910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orij R, Postmus J, Ter Beek A, Brul S, Smits GJ (2009) In vivo measurement of cytosolic and mitochondrial pH using a pH‐sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 155: 268–278 [DOI] [PubMed] [Google Scholar]
- Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J (2002) The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J 21: 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinke G, Zhou L, Sazanov LA (2020) Cryo‐EM structure of the entire mammalian F‐type ATP synthase. Nat Struct Mol Biol 27: 1077–1085 [DOI] [PubMed] [Google Scholar]
- Preiss L, Yildiz O, Hicks DB, Krulwich TA, Meier T (2010) A new type of proton coordination in an F(1)F(o)‐ATP synthase rotor ring. PLoS Biol 8: e1000443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott M, Nowakowski S, Gavin P, Nagley P, Whisstock JC, Devenish RJ (2003) Subunit gamma‐green fluorescent protein fusions are functionally incorporated into mitochondrial F1F0‐ATP synthase, arguing against a rigid cap structure at the top of F1. J Biol Chem 278: 251–256 [DOI] [PubMed] [Google Scholar]
- Rieger B, Junge W, Busch KB (2014) Lateral pH gradient between OXPHOS complex IV and F(0)F(1) ATP‐synthase in folded mitochondrial membranes. Nat Commun 5: 1–6 [DOI] [PubMed] [Google Scholar]
- Rieger B, Shalaeva DN, Sohnel AC, Kohl W, Duwe P, Mulkidjanian AY, Busch KB (2017) Lifetime imaging of GFP at CoxVIIIa reports respiratory supercomplex assembly in live cells. Sci Rep 7: 46055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselin M, Santo‐Domingo J, Bermont F, Giacomello M, Demaurex N (2017) L‐OPA1 regulates mitoflash biogenesis independently from membrane fusion. EMBO Rep 18: 451–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA (2004) Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 64: 985–993 [DOI] [PubMed] [Google Scholar]
- Salewskij K, Rieger B, Hager F, Arroum T, Duwe P, Villalta J, Colgiati S, Richter CP, Psathaki OE, Enriquez JA et al (2020) The spatio‐temporal organization of mitochondrial F1FO ATP synthase in cristae depends on its activity mode. Biochim Biophys Acta Bioenerg 1861: 148091 [DOI] [PubMed] [Google Scholar]
- Sanchez‐Cenizo L, Formentini L, Aldea M, Ortega AD, Garcia‐Huerta P, Sanchez‐Arago M, Cuezva JM (2010) Up‐regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+‐ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J Biol Chem 285: 25308–25313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnizer R, Van Heeke G, Amaturo D, Schuster SM (1996) Histidine‐49 is necessary for the pH‐dependent transition between active and inactive states of the bovine F1‐ATPase inhibitor protein. Biochim Biophys Acta 1292: 241–248 [DOI] [PubMed] [Google Scholar]
- Segawa M, Wolf DM, Hultgren NW, Williams DS, van der Bliek AM, Shackelford DB, Liesa M, Shirihai OS (2020) Quantification of cristae architecture reveals time‐dependent characteristics of individual mitochondria. Life Sci Alliance 3: e201900620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergé A, Bertaux N, Rigneault H, Marguet D (2008) Dynamic multiple‐target tracing to probe spatiotemporal cartography of cell membranes. Nat Methods 5: 687–694 [DOI] [PubMed] [Google Scholar]
- Sjoholm J, Bergstrand J, Nilsson T, Sachl R, Ballmoos CV, Widengren J, Brzezinski P (2017) The lateral distance between a proton pump and ATP synthase determines the ATP‐synthesis rate. Sci Rep 7: 2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohnel AC, Kohl W, Gregor I, Enderlein J, Rieger B, Busch KB (2016) Probing of protein localization and shuttling in mitochondrial microcompartments by FLIM with sub‐diffraction resolution. Biochim Biophys Acta 1857: 1290–1299 [DOI] [PubMed] [Google Scholar]
- Song J, Pfanner N, Becker T (2018) Assembling the mitochondrial ATP synthase. Proc Natl Acad Sci USA 115: 2850–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt S, Wenzel D, Kehrein K, Riedel D, Ott M, Jakobs S (2018) Spatial orchestration of mitochondrial translation and OXPHOS complex assembly. Nat Cell Biol 20: 528–534 [DOI] [PubMed] [Google Scholar]
- Strauss M, Hofhaus G, Schroder RR, Kuhlbrandt W (2008) Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J 27: 1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Imamotu N, Sakata‐Sogawa K (2008) Highly inclined thin illumination enables clear single‐molecule imaging in cells. Nat Methods 5: 159–161 [DOI] [PubMed] [Google Scholar]
- Toth A, Meyrat A, Stoldt S, Santiago R, Wenzel D, Jakobs S, von Ballmoos C, Ott M (2020) Kinetic coupling of the respiratory chain with ATP synthase, but not proton gradients, drives ATP production in cristae membranes. Proc Natl Acad Sci USA 117: 2412–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente AJ, Maddalena LA, Robb EL, Moradi F, Stuart JA (2017) A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem 119: 315–326 [DOI] [PubMed] [Google Scholar]
- Vogel F, Bornhovd C, Neupert W, Reichert AS (2006) Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol 175: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yuan L, Zeng X, Peng J, Ni Y, Er JC, Xu W, Agrawalla BK, Su D, Kim B et al (2016) A multisite‐binding switchable fluorescent probe for monitoring mitochondrial ATP level fluctuation in live cells. Angew Chem Int Ed Engl 55: 1773–1776 [DOI] [PubMed] [Google Scholar]
- Weissert V, Rieger B, Morris S, Arroum T, Psathaki OE, Zobel T, Perkins G, Busch KB (2021) Inhibition of the mitochondrial ATPase function by IF1 changes the spatiotemporal organization of ATP synthase. Biochim Biophys Acta Bioenerg 1862: 148322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom JD, Katzman SM, Mohamed H, Twig G, Graf SA, Heart E, Molina AJ, Corkey BE, de Vargas LM, Danial NN et al (2007) beta‐Cell mitochondria exhibit membrane potential heterogeneity that can be altered by stimulatory or toxic fuel levels. Diabetes 56: 2569–2578 [DOI] [PubMed] [Google Scholar]
- Wilkens V, Kohl W, Busch K (2013) Restricted diffusion of OXPHOS complexes in dynamic mitochondria delays their exchange between cristae and engenders a transitory mosaic distribution. J Cell Sci 126: 103–116 [DOI] [PubMed] [Google Scholar]
- Wittig I, Carrozzo R, Santorelli FM, Schagger H (2006) Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim Biophys Acta 1757: 1066–1072 [DOI] [PubMed] [Google Scholar]
- Wittig I, Karas M, Schagger H (2007) High resolution clear native electrophoresis for in‐gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics 6: 1215–1225 [DOI] [PubMed] [Google Scholar]
- Wittig I, Velours J, Stuart R, Schagger H (2008) Characterization of domain interfaces in monomeric and dimeric ATP synthase. Mol Cell Proteomics 7: 995–1004 [DOI] [PubMed] [Google Scholar]
- Wolf DM, Segawa M, Kondadi AK, Anand R, Bailey ST, Reichert AS, van der Bliek AM, Shackelford DB, Liesa M, Shirihai OS (2019) Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J 38: e101056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DM, Segawa M, Shirihai OS, Liesa M (2020) Method for live‐cell super‐resolution imaging of mitochondrial cristae and quantification of submitochondrial membrane potentials. Methods Cell Biol 155: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti F, Gnoni A, Mangiullo R, Papa S (2009) Effect of the ATPase inhibitor protein IF1 on H+ translocation in the mitochondrial ATP synthase complex. Biochem Biophys Res Commun 384: 43–48 [DOI] [PubMed] [Google Scholar]


