A crystal structure of nonphosphorylated mitogen-activated protein kinase kinase 1 (MEK1) complexed with ATP-γS is reported in which MEK1 adopts the DFG-out conformation.
Keywords: MAP kinases, human mitogen-activated protein kinase kinase 1, MEK1, DFG motif, X-ray crystallography
Abstract
Eukaryotic protein kinases contain an Asp-Phe-Gly (DFG) motif, the conformation of which is involved in controlling the catalytic activity, at the N-terminus of the activation segment. The motif can be switched between active-state (DFG-in) and inactive-state (DFG-out) conformations: however, the mechanism of conformational change is poorly understood, partly because there are few reports of the DFG-out conformation. Here, a novel crystal structure of nonphosphorylated human mitogen-activated protein kinase kinase 1 (MEK1; amino acids 38–381) complexed with ATP-γS is reported in which MEK1 adopts the DFG-out conformation. The crystal structure revealed that the structural elements (the αC helix and HRD motif) surrounding the active site are involved in the formation/stabilization of the DFG-out conformation. The ATP-γS molecule was bound to the canonical ATP-binding site in a different binding mode that has never been found in previously determined crystal structures of MEK1. This novel ATP-γS binding mode provides a starting point for the design of high-affinity inhibitors of nonphosphorylated inactive MEK1 that adopts the DFG-out conformation.
1. Introduction
Protein kinases function as phosphorylation-dependent molecular switches that control many biological phenomena. The extracellular signal-regulated kinase pathway (also known as the Ras–Raf–MEK–ERK pathway) is an evolutionarily conserved signal-transduction cascade that regulates diverse cellular functions including cell proliferation, differentiation, migration and survival (Seger & Krebs, 1995 ▸; Kolch, 2000 ▸; McCubrey et al., 2007 ▸). The mitogen-activated protein kinase (MAPK) kinases (MEK1 and MEK2) are key components of the pathway that catalyse the phosphorylation of their only known physiological substrates, ERK1 and ERK2 (Wang et al., 2021 ▸). MEK1 and MEK2, the amino-acid sequences of which share over 80% identity (Zheng & Guan, 1993 ▸), are activated by Raf kinases through the dual phosphorylation of two neighboring serine residues in their activating segment (Ser218/Ser222 in human MEK1 and Ser222/Ser226 in human MEK2; Roskoski, 2012 ▸).
Phosphorylation of specific amino-acid residues in the activation segment is a common mechanism for regulating eukaryotic protein kinases. This event is accompanied by conformational changes in the key regulatory structural elements, which include the αC helix in the N-terminal lobe (N-lobe) and the conserved Asp-Phe-Gly (DFG) tripeptide motif at the amino-terminal base of the activation segment in the C-terminal lobe (C-lobe) (Johnson et al., 1996 ▸; Endicott et al., 2012 ▸). In the fully active state, the αC helix is rotated towards the active site and the conserved Glu in the αC helix forms a characteristic salt bridge with the conserved catalytic Lys in the β3 strand in the N-lobe. This conformation is called the αC-in conformation. The DFG motif is held in the DFG-in conformation, in which the Asp side chain chelates the catalytically important Mg2+ ion to orientate the ATP-binding site and the Phe side chain packs into a hydrophobic pocket in the groove between the two lobes of the kinase domain and contributes to the formation of a regulatory spine (Adams, 2001 ▸; Kornev et al., 2006 ▸; Taylor & Kornev, 2011 ▸).
To date, approximately 60 X-ray crystal structures of MEK1 in complexes with nucleotides, inhibitors and/or protein kinases have been reported in the Protein Data Bank (PDB; Berman et al., 2000 ▸). There is one report of a phosphorylation-site mutant S218E/S222E (Rice et al., 2012 ▸), but all others are reports of nonphosphorylated inactive MEK1: there are no structural reports of phosphorylated active MEK1. In all crystal structures of MEK1 the αC helix is displaced from its active-state position and the catalytically important salt bridge is broken, which is termed the αC-out conformation. In contrast, the DFG motif shows the DFG-in conformation, with two exceptions (PDB entries 5hze and 5yt3; N. A. Larsen, K. Bloudoff, V. Subramanian, K. Nomoto & J. Wang, unpublished work; S. Nakae, K. Doko, Y. Tada & T. Shirai, unpublished work). The catalytically active kinases adopt strikingly similar conformations, whereas inactive kinases show great conformational heterogeneity, as there are many ways to disrupt the arrangement of catalytic residues that has maximal activity.
To improve our understanding of the regulatory dynamics of MEK1 activity, we searched for structural factors that affect the conformation of the DFG motif in MEK1. During a crystallographic study, we found that nonphosphorylated MEK1 adopts the DFG-out conformation in the crystal structure of its complex with ATP-γS when Mg2+ ions are not present. Here, we report this novel crystal structure. Inhibitors of MEK1 and MEK2 are being developed because aberrant activation of the Ras–Raf–MEK–ERK signaling pathway leads to diseases such as cancer (Caunt et al., 2015 ▸). In particular, MEK1 and MEK2 are attractive targets because their only known substrates are ERK1 and ERK2. This structure is expected to provide a basis for further exploration of MEK1/2 inhibitors.
2. Materials and methods
2.1. Macromolecular production
The MEK1 gene was amplified by PCR using a cDNA fragment encoding human Δ38MEK1 (residues 39–382) as a template. The PCR fragment was inserted into the NdeI–XhoI sites of the pET-30a vector (Novagen) incorporating a C-terminal hexahistidine tag. The Δ38MEK1 T292A/S298A mutant was constructed by amino-acid substitutions of Δ38MEK1, with the aim of increasing the chance of obtaining crystals which diffract to high resolution. Site-directed mutagenesis was performed using PfuTurbo polymerase (Stratagene) with pET-30a containing the Δ38MEK1 gene as the template. DNA sequences were verified by sequencing using a dye terminator cycle sequencing kit (Beckman Coulter) and a CEQ2000 fragment-analysis system (Beckman Coulter). The Δ38MEK1 T292A/S298A mutant was overexpressed in Escherichia coli BL21(DE3) cells (Novagen) for 24 h at 25°C by induction with 0.4 mM isopropyl β-d-1-thiogalactopyranoside. The cell pellets were harvested and sonicated in lysis buffer consisting of 50 mM Tris–HCl pH 8.0, 300 mM NaCl. After the cellular debris had been removed by centrifugation, the supernatant was loaded onto an Ni–NTA agarose column (Qiagen) and the protein was eluted with 200 mM imidazole. The Δ38MEK1 T292A/S298A mutant was further purified by anion-exchange chromatography using a Mono Q column (GE Healthcare) followed by size-exclusion chromatography using Superdex 200 (GE Healthcare). The purity of the expressed mutant proteins was confirmed by SDS–PAGE.
2.2. Crystallization
Prior to crystallization trials, the purified mutant protein was concentrated to 5 mg ml−1 in 25 mM Tris–HCl buffer pH 8.0 containing 5 mM MgCl2, 100 mM NaCl and 5 mM DTT using an ultrafiltration membrane (Millipore) with a molecular-weight cutoff of 10 kDa, and Li4-ATP-γS was added to the concentrated protein solution to a final concentration of 5 mM. Conditions for crystallization were initially searched for using the Crystal Screen and Index sparse-matrix screening kits (Hampton Research). After several optimization steps, well formed crystals were obtained in two weeks using the sitting-drop vapor-diffusion technique, in which 1.5 µl protein sample was mixed with 1.0 µl crystallization solution consisting of 19%(w/v) PEG 3350, 100 mM ammonium citrate buffer pH 6.0 at 4°C.
2.3. Data collection and processing
A diffraction data set was collected to 2.2 Å resolution from a flash-cooled crystal at 100 K on an ADSC Q315 CCD detector using synchrotron radiation of wavelength 1.0 Å at the BL38B1 station at SPring-8, Harima, Japan. The data set was processed and scaled using the HKL-2000 package (Otwinowski & Minor, 1997 ▸). Details of data collection and processing are provided in Table 1 ▸.
Table 1. Summary of data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data-collection statistics | |
| X-ray source | BL38B1, SPring-8 |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 100 |
| Resolution (Å) | 34.65–2.20 (2.28–2.20) |
| Space group | C2221 |
| a, b, c (Å) | 47.815, 96.191, 138.617 |
| α, β, γ (°) | 90, 90, 90 |
| R merge † (%) | 7.7 (53.3) |
| 〈I/σ(I)〉 | 38.0 (4.7) |
| No. of observed reflections | 180567 |
| No. of unique reflections | 16503 |
| Completeness (%) | 99.0 (98.6) |
| Multiplicity | 10.9 (10.9) |
| Refinement statistics | |
| Resolution (Å) | 34.65–2.20 |
| Reflections (working/test) | 15654/839 |
| R work ‡ | 0.198 |
| R free § | 0.268 |
| R.m.s.d., bond lengths (Å) | 0.02 |
| R.m.s.d., bond angles (°) | 1.86 |
| Average B value (Å2) | 46.7 |
| Ramachandran plot¶ | |
| Most favored region (%) | 95.2 |
| Allowed regions (%) | 4.2 |
| Outliers (%) | 0.6 |
| PDB code | 3w8q |
R merge = \textstyle \sum_{hkl}\sum_{i}|I_{i}(hkl)- \langle I(hkl)\rangle|/\textstyle \sum_{hkl}\sum_{i}I_{i}(hkl), where Ii (hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity for multiple measurements.
R work = \textstyle \sum_{hkl}\big ||F_{\rm obs}|-|F_{\rm calc}|\big |/ \textstyle \sum_{hkl}|F_{\rm obs}|, where F obs is the observed structure factor and F calc is the calculated structure factor.
R free is the same as R work but calculated using 5% of the data that were not included in any refinement calculations.
Ramachandran plot values were given by Coot (Emsley et al., 2010 ▸).
2.4. Structure solution and refinement
The crystal structure was solved by the molecular-replacement method with MOLREP (Vagin & Teplyakov, 2010 ▸) from the CCP4 suite (Winn et al., 2011 ▸) using the structure of MEK1 (PDB entry 3eqd; Fischmann et al., 2009 ▸) as a search model. The quality of the model was gradually improved by alternating rounds of refinement using REFMAC5 (Murshudov et al., 2011 ▸) in the CCP4 suite and modification of the model using Coot (Emsley et al., 2010 ▸). The F o − F c difference map showed clear electron density corresponding to the ATP-γS molecule in the ATP-binding site (Fig. 1 ▸). Water molecules were added to the model at locations with 2F o − F c densities higher than 1.8σ and hydrogen-bonding stereochemistry using the water-picking function of Coot. The final refinement converged with R work and R free values of 0.198 and 0.268, respectively. The final refinement statistics are presented in Table 1 ▸. The stereochemistry of the final model was analyzed using Coot; two amino-acid residues, Gly116 and Gln243, fell into the outlier region of the Ramachandran plot. The coordinates and structure factors have been deposited in the Protein Data Bank under accession code 3w8q. Illustrations were prepared using PyMOL (DeLano, 2002 ▸).
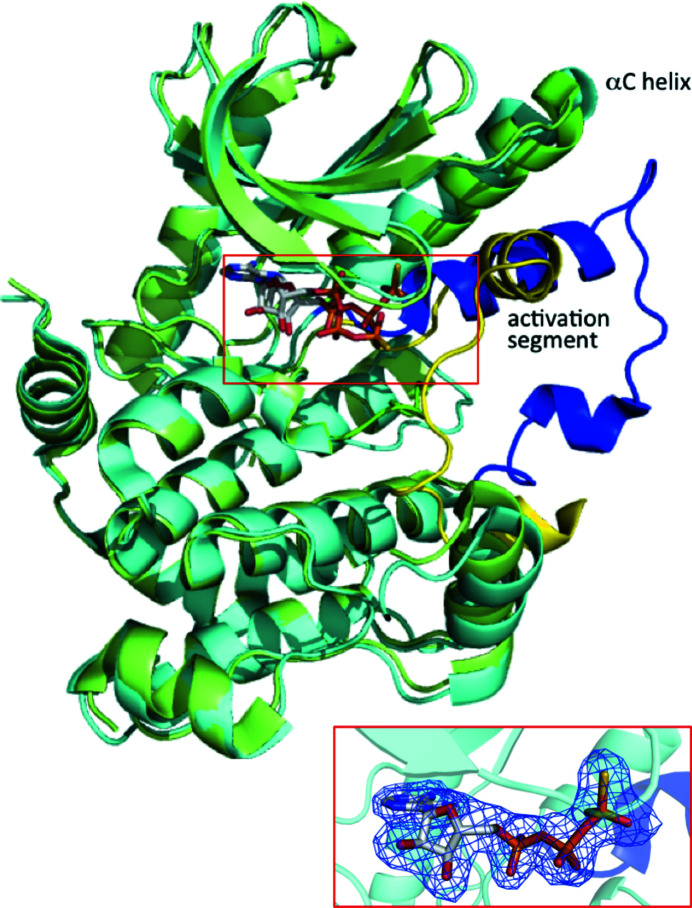
Figure 1.
Superposition of structures of MEK1 adopting the DFG-out conformation (this work; PDB entry 3w8q; cyan) and the DFG-in conformation (PDB entry 3eqd; Fischmann et al., 2009 ▸; green). The Cα atoms of both structures superimpose very well (r.m.s.d. of 1.0 Å) except for the αC helix and activation segment. MEK1 is represented as a ribbon diagram. The activation segments are shown in blue and yellow for the DFG-out and DFG-in conformations, respectively. The ATP-γS molecule is shown as a stick model. The ATP-γS binding site is shown in the box and an enlarged view corresponding to this region including an omit F o − F c map contoured at 3σ for the DFG-out conformation is shown in the inset.
2.5. Surface plasmon resonance measurement
Surface plasmon resonance (SPR) measurements were carried out using a Biacore T200 instrument at 25°C. In direct binding experiments, 20 µg ml−1 Δ38MEK1 T292A/S298A in 10 mM acetate buffer pH 5.0 was immobilized on a CM5 sensor chip and blocked with ethanolamine (7000 resonance units), and a reference cell was blocked with ethanolamine by the standard amine-coupling method using HBS-EP+ (10 mM HEPES pH 7.4, 150 mM NaCl, 0.05% Surfactant P20) as a running buffer. To investigate the binding activity of ATP-γS to Δ38MEK1 T292A/S298A, 25 mM Tris–HCl buffer pH 8.0 containing 120 mM NaCl and 1 mM DTT was used as the running buffer. The K d value of ATP-γS was determined by fitting the equilibrium responses at variable concentrations to a 1:1 binding model.
3. Results and discussion
3.1. Overall structure
The structure of Δ38MEK1 T292A/S298A in complex with ATP-γS was determined to 2.2 Å resolution (Fig. 1 ▸). The electron density corresponding to MEK1 was clearly interpreted except for the highly flexible region, the proline-rich loop (Val279–Ser304) and the C-terminal hexahistidine tag. The structure had a conserved typical kinase core comprising two lobes: a smaller N-lobe and a larger C-lobe. The crystal structure revealed that the ATP-γS molecule bound to the active site was located in a cleft between the two lobes in the absence of Mg2+ ions. However, Smith & Windsor (2007 ▸) reported that Mg2+ ions are required for nucleotide binding to both the phosphorylated and nonphosphorylated states of MEK1 based on the results of temperature-dependent circular-dichroism measurements using AMP-PNP as a nucleotide. Therefore, we attempted to determine the K d value for the binding of ATP-γS to nonphosphorylated MEK1 using SPR measurements. As a result, a dissociation constant of 5.8 µM was obtained, confirming that ATP-γS has a sufficient affinity for MEK1 even in the absence of Mg2+ ions.
The conformation of the activation segment (Asp208–Tyr240) was considerably different from that found in other crystal structures of MEK1 (Fig. 1 ▸). In most MEK1 crystal structures the activation segment is folded against the surface of the MEK1 core domain (Fig. 1 ▸), whereas in the present crystal structure it is folded into a unique conformation that extends outwards from the core domain (Fig. 1 ▸). The crystallographically equivalent subunit was created by the twofold symmetry operation to form a dimer. The accessible surface area buried in the dimer interface per subunit was calculated to be 5820 Å2, corresponding to 20.5% of the total subunit accessible surface area. The loop region (Ser222–Met230) in the central part of the activation segment was involved in the formation of the dimer interface; many residues in the loop formed hydrogen bonds and salt bridges at the dimer interface. This dimer formation probably helps the activation segment, especially its loop region, to adopt a particular conformation, even though the loop region is flexible and is not always visualized in crystal structures.
3.2. Conformation of the DFG motif and αC helix
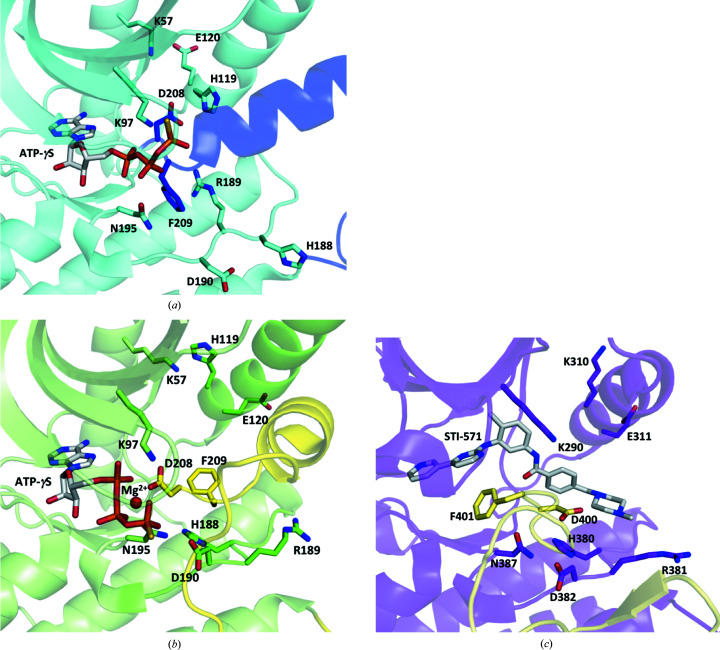
The most remarkable feature of the present structure is that the DFG motif (Asp208-Phe209-Gly210) adopts a unique ‘out’ conformation in which the Asp208 side chain faces away from the ATP-binding site and the Phe209 side chain moves from its canonical hydrophobic pocket to a solvent-exposed position (Fig. 2 ▸ a). This contrasts with the ‘in’ conformation, in which the Asp208 side chain points towards the ATP-binding site and coordinates to the Mg2+ ion, and the Phe209 side chain packs into the hydrophobic pocket (Fig. 2 ▸ b). This DFG-out conformation also differs from the typical ‘out’ conformation, called the flipped type, which has been observed in many kinases in complex with type II inhibitors (Zuccotto et al., 2010 ▸). In the flipped DFG-out conformation, the DFG Phe and Asp side chains swap positions, with the Phe side chain pointing towards the ATP-binding site (Fig. 2 ▸ c).
Figure 2.
A structural comparison of the DFG motif and αC helix of (a) MEK1 in the DFG-out conformation (PDB entry 3w8q), (b) MEK1 in the DFG-in conformation (PDB entry 3eqd) and (c) c-Abl in the flipped DFG-out conformation (PDB entry 1opj; Nagar et al., 2003 ▸). The ATP-γS molecule, the STI-571 inhibitor molecule and notable residues involved in structural changes are shown as stick models. The activation segments are shown in blue in (a), yellow in (b) and pale yellow in (c). The Mg2+ ion is indicated as a sphere colored magenta in (b).
In the present DFG-out conformation, the Asp208 side chain points towards the αC helix and forms a short contact with the His119 side chain: the distance between the Asp208 Oδ2 atom and His119 Nδ1 atom is 2.7 Å, suggesting the formation of a hydrogen bond or a salt bridge. Interestingly, the Phe209 side-chain phenyl ring was sandwiched between the side chains of two residues: Arg189 of the catalytic HRD motif and Asn195, a highly conserved metal-coordinating residue. The closest distance measured between the phenyl ring of Phe209 and the amide-bearing side chain of Asn195 was 3.4 Å. Of particular importance was the cation–π/π–π interaction between the phenyl ring of Phe209 and the guanidyl moiety of Arg189, in which the Arg189 side chain moved by about 8 Å to a new position through a crankshaft-like motion of the peptide backbone of the HRD motif: the average distance between the phenyl ring and the guanidyl moiety was 3.6 Å. This parallel orientation seems to stabilize the DFG-out conformation by minimizing the surface of the Phe side chain that is in contact with water.
The αC helix (Lys104–Glu120) showed the ‘out’ conformation, in which the αC helix was rotated outwards towards the β-strands in the N-lobe and the catalytically important salt bridge between Lys97 and Glu114 was broken. This feature is common to all nonphosphorylated MEK1 structures reported to date. However, there was a slight difference in the orientation of the αC helix between the MEK1 structure in the DFG-in state and that in the DFG-out state: His119 points towards the αA helix and interacts with Lys57 in the DFG-in state (Fig. 2 ▸ b), whereas His119 of the αC helix interacts with Asp208 in the DFG-out state (Fig. 2 ▸ a). This finding suggests that the αC helix is involved in the formation/stabilization of the DFG-out structure in MEK1.
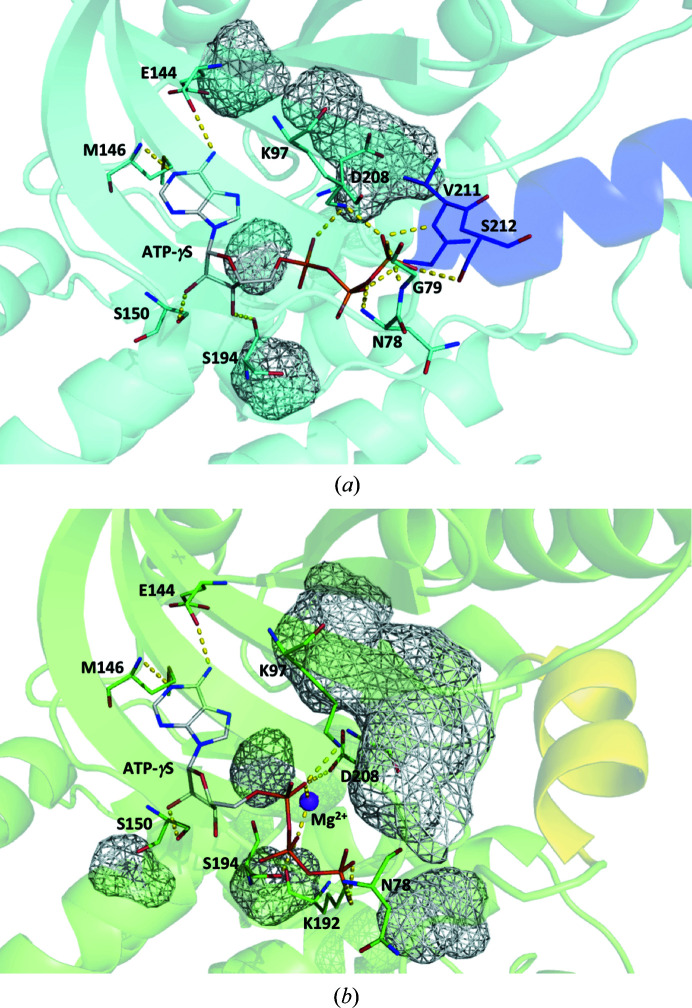
3.3. Binding mode of ATP-γS
The intermolecular distances between the ATP-γS molecule and MEK1 are summarized in Table 2 ▸. The adenine moiety fitted into the adenine-binding pocket and formed hydrogen bonds to Glu144 and Met146 in the hinge region connecting the N- and C-lobes (Fig. 3 ▸ a). This binding mode was identical to that observed in the DFG-in state (Fig. 3 ▸ b). The ribose moiety was oriented slightly towards the inside of the protein, and a hydrogen bond was newly formed between the C3′ OH group and the Ser194 carbonyl O atom in the DFG-out state.
Table 2. Interactions of ATP-γS with MEK1(out) adopting the DFG-out conformation.
For the sake of comparison, the interactions of ATP-γS with MEK1(in) adopting the DFG-in conformation (PDB entry 3eqd; Fischmann et al., 2009 ▸) are also provided.
| MEK1(out) | MEK1(in) | |||
|---|---|---|---|---|
| ATP-γS atom | Atom | Distance (Å) | Atom | Distance (Å) |
| N1 | Met146 N | 3.2 | Met146 N | 3.1 |
| N6 | Glu144 O | 2.8 | Glu144 O | 2.7 |
| O2′ | Ser150 Oγ | 2.7 | Ser150 Oγ | 2.8 |
| O3′ | Ser194 O | 2.6 | ||
| O1α | Lys97 Nζ | 2.8 | Lys97 Nζ | 2.6 |
| Asp208 Oδ2 | 2.9 | |||
| Mg2+ | 2.5† | |||
| O1β | Gly210 N | 2.8 | Ser194 Oγ | 3.0 |
| Mg2+ | 2.1† | |||
| O3β | Asn78 N | 2.8 | ||
| S1γ ‡ | Gly79 N | 3.3 | Asn78 N | 3.5 |
| O2γ | Lys97 Nζ | 2.6 | Lys192 Nζ | 2.7 |
| Val211 N | 3.0 | |||
| O3γ | Ser212 Oγ | 2.7 | ||
Figure 3.
Structural comparison of the ATP-γS binding mode between (a) the DFG-out state (PDB entry 3w8q) and (b) the DFG-in state (PDB entry 3eqd). The ATP-γS molecule is shown as a wire model and residues interacting with ATP-γS are shown as stick models. Short helices are colored blue in (a) and yellow in (b). Mg2+ is indicated as a sphere colored magenta in (b). The cavities are shown in a mesh representation in gray. Short contacts are shown by dotted lines in yellow.
There was a significant difference in the binding mode of the triphosphate moiety between the DFG-out and DFG-in states. The triphosphate moiety formed a hydrogen bond to the Asp208 side chain in the DFG-in state, but did not interact with Asp208 in the DFG-out state. In the DFG-in state, there was a unique cavity adjacent to the ATP-binding site (Fig. 3 ▸ b). The DFG Asp208 and β3-strand Lys97 residues seem to function as walls that partition the cavity. The short helix, which is a characteristic of the activation segment of MEK1, also contributes to cavity formation. This cavity is the binding site for the so-called type III inhibitors (Ohren et al., 2004 ▸; Dar & Shokat, 2011 ▸). In the DFG-out state, the DFG Asp residue and the short helix at the N-terminus of the activation segment resulted in elimination of the target space for the type III inhibitor and the formation of a deep cleft continuous with the ATP-binding site (Fig. 3 ▸ a). The phosphate moiety of ATP-γS was able to penetrate the deep cleft and interact with the Gly210, Val211 and Ser212 residues of the short helix.
4. Conclusion
We determined the X-ray structure of nonphosphorylated MEK1 to obtain further insight into the structural features of inactive kinases. The structure revealed that MEK1 can adopt the DFG-out conformation with the following characteristics: (i) the DFG Asp residue points towards the αC helix and (ii) the DFG Phe residue moves out of its canonical hydrophobic pocket and interacts with the residues of the HRD motif, which generates conformational changes. The crystal structure revealed that the structural elements surrounding the active site, the αC helix and the HRD motif, are involved in the formation and stability of the DFG-out conformation of MEK1. The structure also revealed that the ATP analog ATP-γS can tightly bind to the ATP-binding cleft even though its shape is changed. Protein kinases are some of the most important molecular targets for therapeutic agents in various diseases. This structural information is expected to provide valuable insights into the design of inhibitor molecules because compounds that can stabilize the DFG-out conformation have potential as selective inhibitors of nonphosphorylated MEK1.
Supplementary Material
PDB reference: human mitogen-activated protein kinase kinase 1, 3w8q
Acknowledgments
X-ray data collections were performed on beamline BL38B1 at the SPring-8 synchrotron-radiation facility, Hyogo, Japan with the approval of the Japan Synchrotron Radiation Research Institute (Proposal Nos. 2014B1452, 2015A1018 and 2015B2092) and on beamline BL44XU under the Cooperative Research Program of the Institute for Protein Research, Osaka University (Proposal Nos. 2014A6929 and 2014B6929).
Funding Statement
This work was funded by Ministry of Education, Culture, Sports, Science and Technology grant 26440035 to Toshiji Tada; Japan Agency for Medical Research and Development grant JP21am0101111 .
References
- Adams, J. A. (2001). Chem. Rev. 101, 2271–2290. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Biswal, H. S. & Wategaonkar, S. (2009). J. Phys. Chem. A, 113, 12763–12773. [DOI] [PubMed]
- Caunt, C. J., Sale, M. J., Smith, D. P. & Cook, J. S. (2015). Nat. Rev. Cancer, 15, 577–592. [DOI] [PubMed]
- Dar, A. C. & Shokat, K. M. (2011). Annu. Rev. Biochem. 80, 769–795. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Endicott, J. A., Noble, M. E. M. & Johnson, L. N. (2012). Annu. Rev. Biochem. 81, 587–613. [DOI] [PubMed]
- Fischmann, T. O., Smith, C. K., Mayhood, T. W., Myers, J. E. Jr, Reichert, P., Mannarino, A., Carr, D., Zhu, H., Wong, J., Yang, R.-S., Le, H. V. & Madison, V. S. (2009). Biochemistry, 48, 2661–2674. [DOI] [PubMed]
- Johnson, L. N., Noble, M. E. M. & Owen, D. J. (1996). Cell, 85, 149–158. [DOI] [PubMed]
- Kolch, W. (2000). Biochem. J. 351, 289–305. [PMC free article] [PubMed]
- Kornev, A. P., Haste, N. M., Taylor, S. S. & Ten Eyck, L. F. (2006). Proc. Natl Acad. Sci. USA, 103, 17783–17788. [DOI] [PMC free article] [PubMed]
- Krepps, M. K., Parkin, S. & Atwood, D. A. (2001). Cryst. Growth Des. 1, 291–297.
- McCubrey, J. A., Steelman, L. S., Chappell, W. H., Abrams, S. L., Wong, E. W. T., Chang, F., Lehmann, B., Terrian, D. M., Milella, M., Tafuri, A., Stivala, F., Libra, M., Basecke, J., Evangelisti, C., Martelli, A. M. & Franklin, R. A. (2007). Biochim. Biophys. Acta, 1773, 1263–1284. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nagar, B., Hantschel, O., Young, M. A., Scheffzek, K., Veach, D., Bornmann, W., Clarkson, B., Superti-Furga, G. & Kuriyan, J. (2003). Cell, 112, 859–871. [DOI] [PubMed]
- Ohren, J. F., Chen, H., Pavlovsky, A., Whitehead, C., Zhang, E., Kuffa, P., Yan, C., McConnell, P., Spessard, C., Banotai, C., Mueller, W. T., Delaney, A., Omer, C., Sebolt-Leopold, J., Dudley, D. T., Leung, I. K., Flamme, C., Warmus, J., Kaufman, M., Barrett, S., Tecle, H. & Hasemann, C. A. (2004). Nat. Struct. Mol. Biol. 11, 1192–1197. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Rice, K. D., Aay, N., Anand, N. K., Blazey, C. M., Bowles, O. J., Bussenius, J., Costanzo, S., Curtis, J. K., Defina, S. C., Dubenko, L., Engst, S., Joshi, A. A., Kennedy, A. R., Kim, A. I., Koltun, E. S., Lougheed, J. C., Manalo, J.-C. L., Martini, J.-F., Nuss, J. M., Peto, C. J., Tsang, T. H., Yu, P. & Johnston, S. (2012). ACS Med. Chem. Lett. 3, 416–421. [DOI] [PMC free article] [PubMed]
- Roskoski, R. Jr (2012). Biochem. Biophys. Res. Commun. 417, 5–10.
- Seger, R. & Krebs, E. G. (1995). FASEB J. 9, 726–735. [PubMed]
- Smith, C. K. & Windsor, W. T. (2007). Biochemistry, 46, 1358–1367. [DOI] [PubMed]
- Taylor, S. S. & Kornev, A. P. (2011). Trends Biochem. Sci. 36, 65–77. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wang, C., Wang, H., Zheng, C., Liu, Z., Gao, X., Xu, F., Niu, Y., Zhang, L. & Xu, P. (2021). Eur. J. Med. Chem. 218, 113386–113397. [DOI] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Zheng, C.-F. & Guan, K.-L. (1993). J. Biol. Chem. 268, 11435–11439. [PubMed]
- Zuccotto, F., Ardini, E., Casale, E. & Angiolini, M. (2010). J. Med. Chem. 53, 2681–2694. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human mitogen-activated protein kinase kinase 1, 3w8q