Abstract
Mesenchymal stem cell (MSC) exosomes promote tissue regeneration and repair, and thus might be used to treat many diseases; however, the influence of microenvironmental conditions on exosomes remains unclear. The present study aimed to analyze the effect of osteogenic induction on the functions of human umbilical cord MSC (HucMSC)-derived exosomes. Exosomes from standardized stem cell culture (Exo1) and osteogenic differentiation-exosomes (Exo2) were co-cultured with osteoblasts, separately. Cell counting kit-8 assays, alkaline phosphatase and alizarin red staining were used to observe the exosomes’ effects on osteoblast proliferation and differentiation. The levels of osteogenic differentiation-related proteins were analyzed using western blotting. Estrogen-deficient osteoporosis model mice were established, and treated with the two exosome preparations. Micro-computed tomography and hematoxylin and eosin staining were performed after 6 weeks. MicroRNAs in Exo1 and Exo2 were sequenced and analyzed using bioinformatic analyses. Compared with Exo1 group, Exo2 had a stronger osteogenic differentiation promoting effect, but a weaker proliferation promoting effect. In ovariectomy-induced osteoporosis mice, both Exo1 and Exo2 improved the tibial density and reversed osteoporosis in vivo. High-throughput microRNA sequencing identified 221 differentially expressed microRNAs in HucMSC-derived exosomes upon osteogenic induction as compared with the untreated control group. Importantly, we found that 41 of these microRNAs are potentially critical for MSC-secreted exosomes during osteogenic induction. Mechanistically, exosomal miRNAs derived from osteogenic induced-HucMSCs are involved in bone development and differentiation, such as osteoclast differentiation and the MAPK signaling pathway. The expression of hsa-mir-2110 and hsa-mir-328-3p gradually increased with prolonged osteogenic differentiation and regulated target genes associated with bone differentiation, suggesting that they are probably the most important osteogenesis regulatory microRNAs in exosomes. In conclusion, we examined the contribution of osteogenic induction to the function of exosomes secreted by HucMSCs following osteogenic differentiation in vitro and in vivo, and reveal the underlying molecular mechanisms of exosome action during osteoporosis.
Keywords: umbilical cord mesenchymal stem cells, exosomes, microenvironment, osteoporosis, MicroRNA, osteogenic differentiation
Introduction
Mesenchymal stem cells (MSCs) have the potential for multidirectional differentiation, immune regulation, and tissue repair and regeneration, and have therapeutic effects on a variety of orthopedic diseases 1 . However, after MSC transplantation, the number of cells remains very small, and the substitution effect of local cell differentiation is insufficient to support complex pathological changes 1,2 , suggesting that local proliferation and differentiation of MSCs might not be the mechanism by which MSCs exert their therapeutic effects. Recent studies have found that exosomes play an important role in intercellular communication 3,4 . MSC-derived exosomes contain a variety of bioactive substances, have low immunogenicity, regulate immunology, enhance cell proliferation, and promote tissue repair. For example, exosomes secreted by MSCs have therapeutic effects on diabetes, renal disease, and ischemic stroke 5,6 . MicroRNAs (miRNAs) are one of the main bioactive substances in exosomes, and comprise endogenous non-coding single-stranded small RNAs (18–25 nucleotides in length) that regulate gene expression, such as by binding to the 3’ untranslated region (UTR) region of target mRNAs 7 . Experimental studies have confirmed that miRNAs in MSC-derived exosomes play an important role in regulating cell function and treating various diseases 8 . High expression of exosome-derived miR-92a-3p in MSCs could promote cartilage regeneration by targeting WNT5A (encoding Wnt family member 5A) 9 . However, the secretion and function of exosomes are greatly influenced by the cell state and microenvironment. For example, MSC-derived exosomes in the tumor microenvironment cause the disappearance of normal functions of tissue repair, and then changes to the exosome cargoes, thereby promoting tumor cell proliferation and invasion, and eventually leading to the transformation of normal stromal cells to tumor fibroblasts 10 .
Human umbilical cord MSCs (HucMSCs) are usually collected in a non-invasive manner, have strong regenerative ability 11 , and have broad application prospects in regenerative medicine. However, the contribution of osteogenic induction to the function of exosomes remains unclear. In the present study, we explored the effects of exosomes secreted by HucMSCs in the microenvironment of osteogenic differentiation on the proliferation and differentiation of osteoblasts, and examined the molecular function of exosomes in both osteoblasts and ovariectomy-induced osteoporosis mice. High-throughput gene sequencing and bioinformatic techniques were utilized to analyze the differential expression of exosomal miRNAs, and to reveal the underlying molecular mechanisms of exosome action during osteoporosis.
Methods
Isolation and Characterization of HucMSCs
Human umbilical cords were taken from a healthy cesarean section of fetuses in the Second Affiliated Hospital of Shantou University. The experimental study was approved by the ethics Committee of the Second Affiliated Hospital of Shantou University Medical College and the informed consent of the pregnant women and their families. Umbilical cords were obtained from healthy newborn fetuses, and Wharton’s jelly tissues were isolated under sterile conditions. Tissues were cut to 1-2 mm3 size tissue pieces and inoculated in culture flasks containing 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) in a cell incubator at 37°C and 5% CO2 saturated humidity. When cells grew to 80% confluence, tissue blocks were removed, digested, and subcultured with 0.25% trypsin (Gibco, Grand Island, NY, USA) containing EDTA. HucMSCs after three passages (P3) were harvested and the cell concentration was adjusted to 1 × 106/mL. Antibodies labeled with fluorescein isothiocyanate (FITC), Peridinin chlorophyll protein complex (PerCP)-cyanine (Cy)™5.5, Allophycocyanin (APC), and Phycoerythrin (PE) were used (FITC-CD90, PerCP-Cy™5.5-CD105, APC-CD73, PE-CD45, PE-CD34, PECD11b, PE-CD19, and PE-HLA-DR) (BD Stemflow hMSC Analysis Kit, BD Biosciences, San Jose, CA, USA). The cells were incubated with the antibodies at room temperature for 30 min, washed with phosphate-buffered saline (PBS, Beyotime, Shanghai, China), centrifuged at 300 × g for 5 min, and the supernatant discarded. The cells were resuspended in PBS and analyzed using flow cytometry (BD Biosciences, San Jose, CA, USA). HucMSCs from P3 were seeded in 24-well cell culture plates (Corning, NY, USA). When the cells grew to 80-90% confluence, they were replaced with osteogenic differentiation induction conditioned medium, adipose differentiation conditioned medium, or cartilage differentiation conditioned medium (Cyagen Biosciences, Guangzhou, China). After 21 days of induced differentiation, osteogenic differentiation was detected using Alizarin red staining, and adipose differentiation was detected using oil red staining.
Isolation and Characterization of Exosomes Derived from HucMSCs
HucMSC-Exosome (Exo1): HucMSCs after P3 were cultured with Dulbecco’s modified Eagle’s medium (DMEM)/F12 conditioned medium containing 10% exosome-free FBS (Gibco, Grand Island, NY, USA), and the cell supernatant was collected after 48 h of culture. Osteogenic differentiation-Exosome (Exo2): HucMSCs after P3 were cultured to 80% confluence, washed twice with PBS, and replaced with osteogenic induction differentiation conditioned medium containing 10% exosome-free FBS, with 50 M vitamin C, 10 mM beta-phosphoglycerol, and 0.1 M dexamethasone (Sigma, St. louis, MO, USA). Cell supernatants were collected after 48 h of culture.
Collected cell supernatants were subjected to 2000 × g gradient centrifugation (Eppendorf, Hamburg, Germany) at 4°C for 20 min and the pellet was discarded. The supernatant was removed to a new centrifuge tube and centrifuged at 10,000 × g at 4°C for 40 min. The pellet was discarded and the supernatant was removed to a new centrifuge tube. The pellet was collected by centrifugation at 100,000 × g for 60 min, resuspended by PBS and centrifuged at 100,000 × g (Beckman Coulter, Indianapolis, IN, USA) for 60 min at 4°C. The pellets were resuspended in PBS, and then filtered and sterilized through 0.22 µm sterile filter membrane, and stored in a freezer (Panasonic, Osaka, Japan) at −80°C.
After adjusting the exosome suspension to the appropriate concentration with PBS, the exosomes were dripped on special carbon-film copper mesh for electron microscopy observation. The exosomes on the mesh were stained with 2% phosphotungstic acid for 2-3 min, air-dried naturally, and photographed using transmission electron microscopy (Hitachi, Tokyo, Japan). Western blotting was used to detect three exosome-derived marker proteins, CD9, CD81, and HSP70 (ProteinTech, Rosemont, IL, USA). Vesicle diameter in the exosome suspensions was measured using a particle size analyzer (Malvern Nanosight NS300, Malvern, UK).
Isolation of Osteoblasts
Osteoblasts were derived from the skulls of suckling SD rats. Osteoblasts in the skull were isolated using an appropriate amount of 0.2% type II collagenase (Gibco, Grand Island, NY, USA)) and inoculated into T75 culture flasks (Corning, NY, USA)) containing DMEM medium (Gibco, Grand Island, NY, USA) (containing 10% FBS, penicillin 100 U/mL, streptomycin 0.1 mg/mL).
Cell Counting Kit-8 (CCK-8)
The isolated osteoblasts were seeded into 96-well plates at a concentration of 3 × 103/mL, and 100 µL of cell suspension was added into each well. After 12 h, Exo1 and Exo2 were co-cultured with the osteoblasts at concentrations of 0, 0.05, 0.1, and 0.2 mg/mL. Four control duplicate wells were set at each concentration. After incubation for 36 and 60 h, 10 µL of CCK-8 (MCE, Monmouth Junction, NJ, USA) reagent was added to each well, and after incubation at 37°C for 2 h in the dark, the absorbance at 450 nm wavelength was detected using a microplate reader (Tecan, Männedorf, Switzerland). For each concentration group, the experiment was repeated three times, and statistical analysis was performed.
Alkaline Phosphatase (ALP) and Alizarin Red Staining
The isolated osteoblasts were seeded in 24-well plates, and after 24–48 h, when the cells had grown to 80% confluence, osteogenic induction was performed using osteogenic induction medium. Co-cultures with Exo1 and Exo2, at 0.2, 0.1, and 0 mg/mL were set, respectively. Each concentration gradient had four duplex holes, and the solution was changed every 3 days. An alkaline phosphatase kit (Beyotime, Shanghai, China) was used for analysis after 10 days. After 21 days, alizarin red (Solarbio, Beijing, China) staining was performed according to the manufacturer’s instructions. The results for the groups were compared and analyzed according to the depth of staining.
Western Blotting
Osteoblasts were co-cultured with Exo1 and Exo2 in osteogenic induction medium conditions, respectively. Three concentration of 0.1, 0.2, and 0 mg/mL were set. After 7 days of culture, the cells were collected and 150 µL of Radioimmunoprecipitation assay (RIPA) Lysis Buffer (Beyotime, Shanghai, China) was added to each well of the 6-well plate for lysis. All protein concentrations were determined using a bicinchoninic acid (BCA) protein concentration determination kit (Beyotime, Shanghai, China). The protein concentrations in the samples were adjusted so that the protein content was the same in the same volume. The SDS-PAGE gel preparation kit (Beyotime, Shanghai, China) was used for constant voltage protein electrophoresis at 80 V for 40 min, followed by 120 V for 30 min. The separated proteins were transferred to PVDF membranes (0.2 μm; Thermo Scientific, Rockford, IL, USA) at 300 mA for 80 min. The membranes were the incubated in 10% skimmed milk (BD Biosciences, San Jose, CA, USA) for 1 h at room temperature, incubated overnight with primary antibodies at 4°C, followed by incubation with horseradish peroxidase-labeled secondary antibody (Beyotime, Shanghai, China) at room temperature for 1 h. Immunoreactive proteins on the membranes were visualized using a chemiluminescent imager (Bio-Rad, Hercules, CA, USA) using extremely hypersensitive ECL luminescent reagents. The mouse-derived primary antibodies recognized several typical osteogenic differentiation-related proteins, including RUNX family transcription factor 2 (RUNX2), osteopontin (OST), collagen type I alpha 1 Chain (COL1A1), and ALP (Abcam, Cambridge, MA, USA), and the internal reference was detected using mouse-derived anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies (ProteinTech, Rosemont, IL, USA). The protein expression intensity was compared and analyzed according to the ratio of the gray value of indicated protein to the gray value of the internal reference protein.
Animal Model of Osteoporosis
All C57BL/6 J mice were purchased from Beijing Weitong Lihua Laboratory Animal Technology Co., Ltd (Beijing, China). Forty 10-week-old C57 female mice were divided into five groups with eight mice in each group: Sham group, ovariectomized (OVX) group, OVX+Exo1 group, OVX+Exo2 group, and OVX+E2 (estradiol) group. The experiments were performed as described previously 12 . All mice were anesthetized with 2% pentobarbital sodium. Except for the sham group, ovariectomy was performed to simulate the osteoporotic disease model caused by estrogen deficiency. The sham group underwent a sham operation as the control group. Ovariectomized mice treated with estradiol (OVX+E2 group) are included in our in vivo experiments and served as a positive control to evaluate the efficiency of different treatments in osteoporosis therapy. A small incision was made on the back of mice to remove the ovaries, including part of the fallopian tube, and the incision was sutured with 5-0 artificial absorbable sutures. After 1 week of recovery time, the experiment was carried out. The OVX+Exo1 group, OVX+Exo2 group, and OVX+E2 group were intraperitoneally injected with Exo1 (0.5 mg/kg), Exo2 (0.5 mg/kg), and E2 (0.15 mg/kg), respectively. The OVX group was injected with the same volume of PBS. Injection every 3 days for 6 weeks.
Over time, ovariectomized mice will develop osteoporosis because of estrogen deficiency. Specifically, the effects of exosome therapy (OVX+Exo1 and OVX+Exo2) on osteoporosis were compared with ovariectomization (OVX) and estrogen therapy (OVX+E2). Six weeks later, the tibia of mice were taken, and five groups were subjected to micro computed tomography (microCT) scanning, to detect and analysis various data indicators: Bone volume (BV), relative bone volume (BV/trabecular volume (TV)), cortical bone area (Ct.Ar), cortical bone thickness (Ct.Th), bone surface (BS), ratio of bone surface area to bone volume (BS/TV), bone mineral content (BMC), bone mineral density (BMD), trabecular number (Tb.N), trabecular separation/Spacing (Tb.Sp), trabecular thickness (Tb.Th), and trabecular structure pattern index (Structure model index (SMI)). After micro-CT (SCANCO, Wangen-Brüttisellen, Switzerland) scanning, mouse tibias were fixed with 4% paraformaldehyde and decalcified with 10% EDTA for 4 weeks, and paraffin-embedded sectioning was performed. After hematoxylin and eosin (HE) staining, the sections were observed and analyzed under a microscope.
MicroRNA Sequencing Analysis
When the P3 generation HucMSCs grew to 80% confluence, they were divided into three groups, and each group had three samples. The control group was replaced with conditioned medium without exosome serum, and the supernatant was collected after 48 h of culture; osteogenic group 3 and osteogenic group 7 were replaced with osteogenic induction differentiation medium containing 10% exosome-free serum, and the supernatant was collected after 48 h and 7 days of culture, respectively. The cell supernatants of the three groups were separated by ultracentrifugation. RNA sequencing uses next generation sequencing (NGS) technology was used to obtain the sequences of miRNAs (18–30 nt or 18–40 nucleotides) in exosomes. Sequencing data were then compared with databases to identify and analyze the small RNA sequences.
The sequencing results of three groups of miRNAs analyzed statistically for differentially expressed miRNAs. The p -value was corrected by multiple hypothesis tests using the Q value. Genes with two or more coincidence differences and a Q-value less than or equal to 0.001 were considered as significant differentially expressed genes. The domain value of p -value was determined by controlling the FDR (False Discovery Rate). The FDR value of the difference test was obtained, and the multiple of differential expression of a gene between different samples was calculated according to the expression amount of the gene, as calculated using the FPKM value (Fragments per Kilobase of transcript per Million mapped reads). The smaller the FDR value, the greater the difference multiples, indicating more significant expression differences. Differentially expressed genes were defined as those with an FDR < 0.001 and > 2-fold expression difference.
RNAhybrid 13 , miRanda 14 , and TargetScan 15 were used to predict the potential target genes of the miRNAs, and then the intersections of the results predicted by three software were noted. According to the results of differential miRNA detection, hierarchical clustering analysis was performed using the pheatmap function in the R software 16 to form a clustering heat map of differentially expressed miRNAs between the groups. The biological functions of genes were investigated using Kyoto Encyclopedia of Genes and Genome (KEGG) Pathway analysis. According to KEGG Pathway 17 public database, pathway significance enrichment analysis was carried out to identify those pathways that were significantly enriched in candidate genes compared with the whole genome background. Pathways with a Q value < 0.05 were defined as pathways that were significantly enriched in differentially expressed genes. Pathway significant enrichment can identify the most important biochemical metabolic pathways and signal transduction pathways in which the candidate genes participate.
Statistical Analysis
Statistical analysis was performed using the software Statistical Product and Service Solutions (SPSS)19.0 (IBM Corp., Armonk, NY, USA). GraphPad Prism 8.0 (GraphPad. Inc, La Jolla, CA, USA) and ImageJ Launcher software (NIH, Bethesda, MD, USA) were used for image editing and gray value analysis, respectively. The two groups were compared and analyzed using t-tests. Differences were considered significant at P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Results
Typical Features of HucMSCs and HucMSC-Derived Exosomes
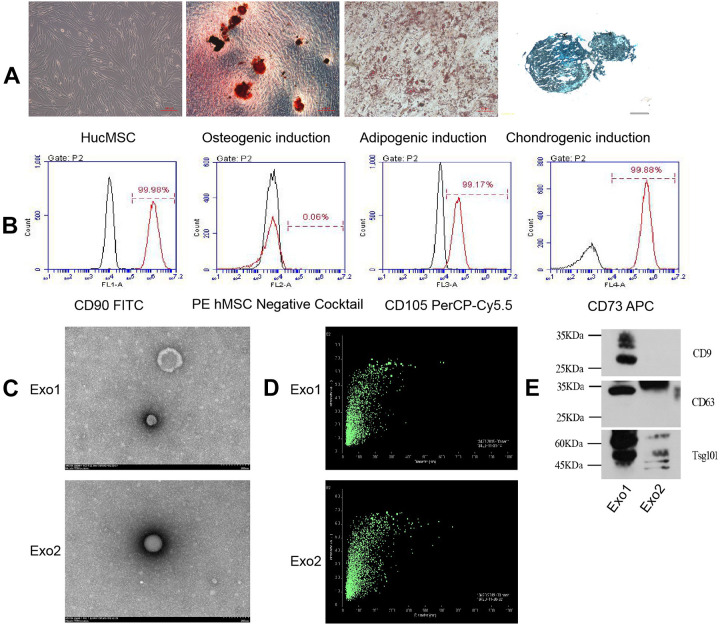
HucMSCs isolated from Wharton’s jelly tissue of the umbilical cord are typically fusiform, triangular, or fibrous. When they approach maturity, they appear to from a whirlpool or broom shape. We first monitored and analyzed the differentiated states of HucMSCs induced with different conditional media. When HucMSCs differentiated into osteoblasts, Alizarin red could stain the mineralized nodules red or orange red; when they differentiated into adipocytes, oil red dye could dye the fat drops in adipocytes orange red; after 30 days of induction, chondrocytes were formalin-fixed and paraffin-embedded, and stained with alixin blue, which partly showed the endo-acidic mucopolysaccharides in cartilage tissue (Fig. 1A). Moreover, the HucMSCs were confirmed by high percentage of positive staining for CD73 (99.88%), CD90 (99.98%), and CD105 (99.17%) as well as negative staining for CD45, CD34, CD11b, CD19, and HLA-DR (Fig. 1B). During the induction of the osteogenic differentiation microenvironment, the cell proliferative capacity of HucMSCs was inhibited within the first three days, their cell morphology changed, and aggregated nodules without refractive properties gradually appeared after another four days. Following 21 days of osteogenic induction, mineralized nodules with strong refraction existed in the culture dish and were stained red by Alizarin red. To explore the contribution of osteogenic induction to exosomes, we concentrated on the comparison between exosomes secreted by HucMSCs from P3 cultured with normal conditional medium (Exo1) and with osteogenic induction differentiation conditioned medium exosomes (Exo2). The morphology of the both types of HucMSC-derived exosomes were confirmed using transmission electron microscopy, which revealed that the exosomes were typical vesicles Fig. 1C) and the particle size range of the exosomes in suspension was 20–200 µm (Fig. 1D). Furthermore, western blotting analysis also indicated that the HucMSC-derived exosomes expressed the characteristic surface markers, such as CD9, CD81, and tumor susceptibility 101 (TSG101) (Fig. 1E).
Figure 1.
Structure and appearance of HucMSCs and their exosomes. (A) HucMSCs have a whirlpool-like or broom-like appearance. Mineralized nodules induced by HucMSC osteogenic differentiation are stained red by Alizarin red (shown at original magnification, × 100). Fat droplets induced by HucMSC adipose differentiation are stained orange with oil red stain (shown at original magnification, × 200). Endoacid mucopolysaccharides formed by HucMSC cartilage differentiation are stained blue by alixin blue (shown at original magnification, × 100). (B) Flow cytometry detected positive expression of HucMSC CD73, CD90, and CD105, and negative expression of CD45, CD34, CD11b, CD19, and HLA-DR. Black histograms represent the isotype controls, and the red peak represents the marker indicated. (C) Under the electron microscope, the exosomes showed a double-layer membrane vesicle structure. (D) The vesicle diameter in the sample suspension was distributed in the range of 20–200 μm. (E) Western blotting confirmed that the exosomes expressed CD9, CD63, and TSG101 proteins.
HucMSC-Derived Exosomes Promote Cell Proliferation and Differentiation of Osteoblasts
To determine the effects of exosomes on osteoblasts, exosomes derived from standardized stem cell culture (Exo1) and from osteogenic differentiation (Exo2) were co-cultured individually with osteoblasts for 36 h and 60 h. The absorbance of OD450 was measured using CCK-8 assays, and it was found that Exo1 significantly promoted osteoblast proliferation in a concentration-dependent manner (Fig. 2A). However, the proliferative ability of osteoblasts under concentration gradients of Exo2 were similar to those of the control group (Fig. 2B). When the concentration of exosomes was 100 μg/mL, staining for osteoblast differentiation was deepest in the Exo2 group, which indicated that exosomes produced following induction of osteogenic differentiation promote osteogenesis better than exosomes produced under normal culture (Fig. 2C, D). Compared with the control group, the cells in the Exo1 and Exo2 groups were stained strongly, accompanied by large amounts of mineralized deposits. When the concentration of exosomes increased to 200 μg/mL, the staining intensity and mineralized deposits were enhanced (Fig. 2E–H), which showed that exosomes could promote osteogenic differentiation in a concentration-dependent manner. Furthermore, the protein levels of osteogenic differentiation-related proteins (including RUNX2, COL1A1, ALP, and OST) in the cells co-cultured with either Exo1 or Exo2 were higher than those without treatment, especially the levels of RUNX2, ALP, and OST in Exo2 group (Fig. 3A,B).
Figure 2.
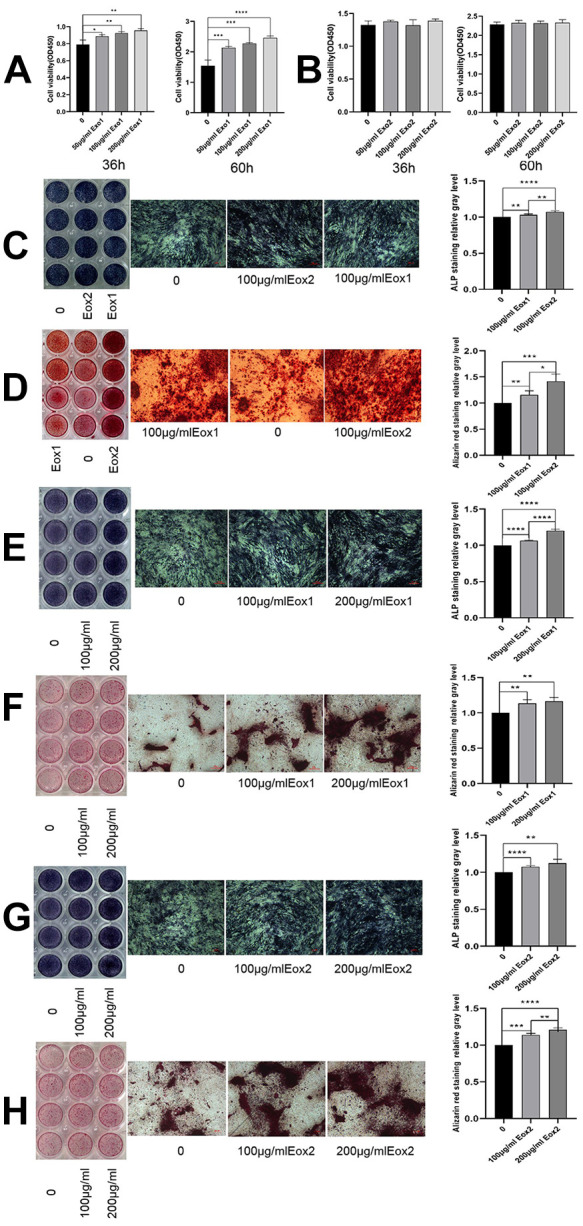
HucMSC exosomes promote osteogenic proliferation and differentiation. (A) After co-culture of Exo1 with osteoblasts, the results of CCK-8 assays showed a significant promoting of proliferation in a concentration-dependent manner. (B) Exo2 did not promote proliferation after co-culture with osteoblasts. Exo1 and Exo2 concentrations were 100 μg/mL. (C) Images show ALP staining, in which the color in the Exo2 image was darker than that for Exo1. (D) Images show alizarin red staining, in which Exo2 produced the most mineralized nodules, indicating that Exo2 promotes stronger osteogenic differentiation than Exo1. The concentration of Exo1 was set at 200, 100, and 0 μg/mL, and the concentration of Exo2 was set at 200, 100, and 0μg/mL. E and G show ALP staining, and F and H Alizarin red staining of exosomes. The staining increased with increasing concentration, indicating that Exo1 and Exo2 promoted osteogenic differentiation in a concentration-dependent manner (shown at the original magnification, × 100).
Figure 3.
HucMSC exosomes promote osteoblast differentiation, affect osteoblast differentiation-related protein levels, and reduce osteoporosis. (A) Western blotting validation of the effects of Exo1 and Exo2 on the levels of RUNX2, COL1A1, ALP, and OST proteins in osteoblasts. The exosomes promoted the differentiation of osteoblasts in a concentration-dependent manner, and Exo2 showed a stronger effect on promoting osteogenic differentiation than Exo1. (B) Statistical analysis of the expression of osteoblast differentiation-related proteins showed that the levels of RUNX2, COL1A1, ALP, and OST in the Exo1 group and Exo2 group were significantly higher than that in the control group. (C) HE staining of bone tissue sections of mice, compared with OVX group, showed that osteoporosis was reduced in the Exo1 group and Exo2 group (shown at original magnification, × 50).
HucMSC-Derived Exosomes Mitigate Osteoporosis in vivo
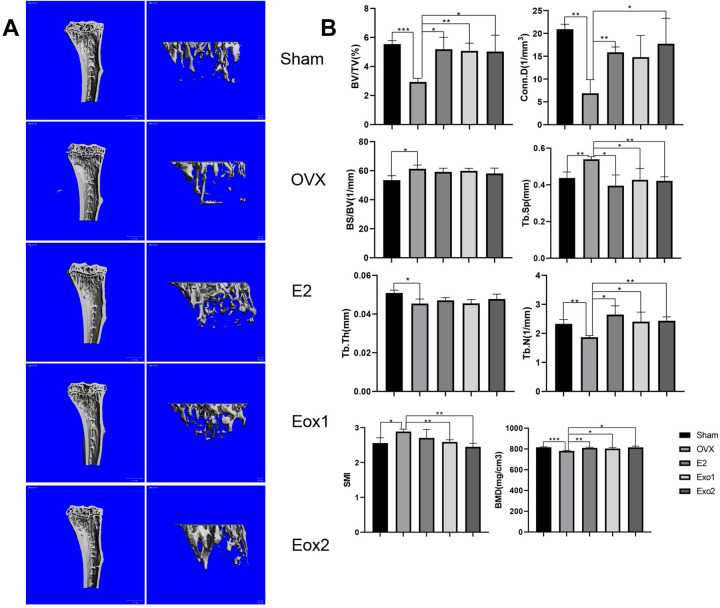
Estrogen deficiency leads to osteoporosis after ovariectomization, while estrogen supplementation is well known and widely used to reverse ovariectomy-induced osteoporosis in mice. To examine whether exosomes derived from MSCs are able to prevent bone loss, we established an ovariectomy-induced osteoporosis mouse model. The mice were divided into five groups: Sham group (Sham), ovariectomized group (OVX), ovariectomized group supplemented with estradiol (OVX+E2), and ovariectomized groups treated with either Exo1 (OVX+Exo1) or Exo2 (OVX+Exo2). Six weeks after the application of exosomes, mouse tibia and cancellous bone were harvested, and analyzed by H&E staining, micro-CT scanning, and using various parameters of bone structure. H&E staining of paraffin sections of mouse tibia and cancellous bone indicated that the degree of osteoporosis is significantly reduced in Exo1 and Exo2 groups, compared with that in the OVX group (Fig. 3C). These results were further validated in micro-CT 3D reconstruction images of tibia and cancellous bone (Fig. 4A). Compared with the OVX group, the values of BV/TV, Conn. D, TB. Sp, TB. N, and BMD were all significantly increased in both the Exo1 and Exo2 groups, whereas there was no statistically remarkable change of BS/BV value and Tb.Th value among these groups (Fig. 4B). Moreover, the SMI value of either the Exo1 or Exo2 group was lower than that of the OVX group (Fig. 4B). These findings suggested that both Exo1 and Exo2 are able to mitigate the symptoms of osteoporosis in mice.
Figure 4.
Therapeutics effects of HucMSC exosomes in osteoporosis model mice. (A) MicroCT scans of tibia and cancellous bone of mice after 6 weeks, showing that Exo1 and Exo2 induced increased bone mass relative to the Sham group in three-dimensional images. (B) Compared with the control group, after statistical analysis, the relative bone volume (BV/TV), bone density (BMD), trabecular Connectivity density (Conn.D), trabecular number (Tb.N), trabecular separation (Tb.Sp), and trabecular structure pattern index (SMI) were significantly different, and the results demonstrated that Exo1 and Exo2 had therapeutic effects on osteoporosis.
Identification of microRNAs in Exosomes using Bioinformatics Analysis
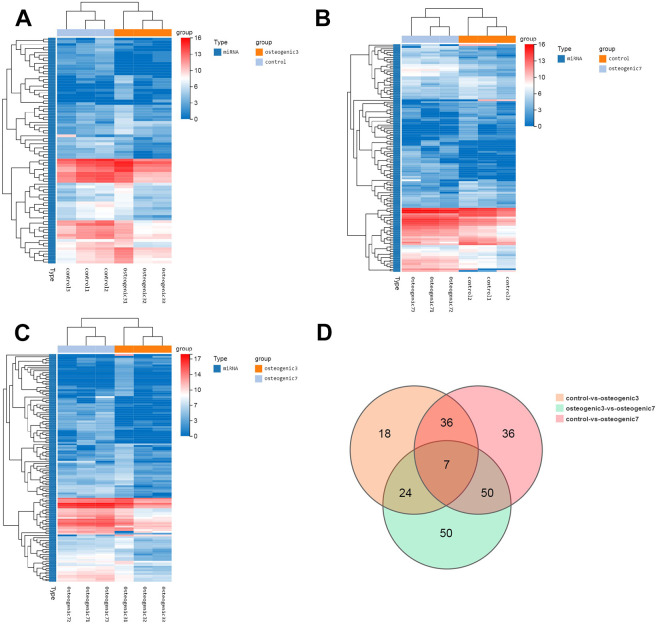
To identify which microRNAs participate in MSC-secreted exosomes after osteogenic induction, HucMSCs were firstly maintained in conditioned medium without exosome serum (control group) or osteogenic induction differentiation medium for 48 h (osteogenic group 3) and 7 days (osteogenic group 7). Exosomes were isolated from the supernatant derived from these HucMSCs and were further analyzed by microRNA sequencing. We identified 85 microRNAs that were differentially expressed by greater than a 2-fold change in HucMSC-derived exosomes of osteogenic group 3 relative to control samples without induction (Fig. 5A). Interestingly, the number and the extent of change of differentially expressed miRNAs gradually increased with prolonged osteogenic differentiation (Fig. 5B). Compared with HucMSC-derived exosomes of osteogenic group 3, enhanced osteogenic differentiation induced the upregulation of 67 microRNAs and the downregulation of 64 microRNAs by greater than 2-fold in the exosomes of osteogenic group 7 (Fig. 5C). Importantly, prolonged osteogenic differentiation confirmed that 41 microRNAs are potentially critical for MSC-secreted exosomes during osteogenic induction, because the alteration of these microRNAs were consistent and similar between osteogenic group 3 and osteogenic group 7 (Fig. 5D and Table 1). Furthermore, the predicted target genes of the differentially expressed miRNAs were analyzed using TargetScan and miRBase databases. KEGG pathway enrichment analysis indicated that miRNAs derived from MSC-secreted exosomes after osteogenic induction indeed regulated genes involved in human diseases (such as cancers and endocrine diseases), organismal systems, metabolic pathways, and environmental information processing (Fig. 6A, B). Specifically, miRNAs derived from MSC-secreted exosomes following osteogenic induction participated in osteoclast differentiation, the mitogen-activated protein kinase (MAPK) signaling pathway, and cytokine-cytokine receptor interaction in both osteogenic group 3 and osteogenic group 7. Moreover, with longer osteogenic differentiation culture time, the target genes of miRNAs within MSC-secreted exosomes were significantly related to bone development and differentiation, such as osteoclast differentiation, phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3 K)-protein kinase B (AKT) and mammalian target of rapamycin (mTOR) signaling pathways (Fig. 6C). Consistently, the potentially important microRNAs of MSC-secreted exosomes following osteogenic induction also regulate genes involved in the MAPK signaling pathway (Fig. 6D). These findings suggested that the regulation by miRNAs of the MAPK signaling pathway might be one of the mechanisms by which exosomes regulate cell differentiation and treat osteoporosis. Two miRNAs related to the above signaling pathways were screened out, hsa-mir-328-3p and hsa-mir-2110. The target gene of hsa-mir-328-3p was CHRD (encoding chordin), while hsa-mir-2110 was predicted to target TNF (also known as TNF-alpha, encoding tumor necrosis factor; Fig. 6E).
Figure 5.
The osteogenic microenvironment affects the miRNA profile of HucMSC exosomes. (A) Clustering heatmap of control vs. osteogenic 3 differentially expressed miRNAs. (B) Clustering heatmap of control vs. osteogenic 7 differentially expressed miRNAs. (C) Clustering heatmap of osteogenic 3 vs. osteogenic 7 differentially expressed miRNAs, showing the expression level of differentially expressed miRNAs by color, with log2 (expression value + 1) of the sample on the horizontal axis and genes on the vertical axis. The redder the color of the block, the higher the expression level; the bluer the color, the lower the expression level. (D) VENN/UpSetR plots of miRNA expression differences among the control group, osteogenic group 3, and osteogenic group 7.
Table 1.
Critical microRNAs in MSC-Secreted Exosomes During Osteogenic Induction.
| Gene ID | Type | log2 (osteogenic3/control) | Q value (control-vs-osteogenic3) | log2 (osteogenic7/control) | Q value (control-vs-osteogenic7) |
|---|---|---|---|---|---|
| hsa-miR-429 | miRNA | 5.559943827 | 5.94E-06 | 4.576757059 | 8.55E-05 |
| hsa-miR-34b-3p | miRNA | 4.835578269 | 7.58E-07 | 3.576757059 | 2.85E-04 |
| hsa-miR-370-5p | miRNA | 4.521469679 | 3.38E-10 | 3.924680362 | 6.08E-10 |
| hsa-miR-1270 | miRNA | 4.312016313 | 7.52E-05 | 4.11732544 | 3.04E-06 |
| hsa-miR-4454 | miRNA | 3.106012853 | 2.03E-42 | 1.924680362 | 1.13E-17 |
| hsa-miR-619-5p | miRNA | 2.651866316 | 1.29E-27 | 3.309618254 | 4.71E-83 |
| hsa-miR-150-5p | miRNA | 2.549055511 | 1.45E-16 | 1.232802657 | 1.27E-04 |
| hsa-miR-365b-3p | miRNA | 2.192869048 | 6.99E-21 | 2.844581823 | 3.93E-67 |
| hsa-miR-365a-3p | miRNA | 2.192869048 | 6.99E-21 | 2.844581823 | 3.93E-67 |
| hsa-miR-10a-5p | miRNA | 1.9682471 | 0 | 1.770096322 | 0 |
| hsa-miR-615-3p | miRNA | 1.880959829 | 0 | 2.250443714 | 0 |
| hsa-miR-328-3p | miRNA | 1.867586949 | 0 | 2.19413482 | 0 |
| hsa-let-7d-3p | miRNA | 1.741556772 | 0 | 1.18681054 | 3.65E-253 |
| hsa-miR-675-5p | miRNA | 1.732348112 | 1.01E-09 | 4.150100476 | 2.57E-184 |
| hsa-miR-10b-5p | miRNA | 1.618077307 | 0 | 2.21915872 | 0 |
| hsa-miR-574-3p | miRNA | 1.417150974 | 1.01E-202 | 1.573633359 | 0 |
| hsa-miR-204-5p | miRNA | 1.140409225 | 2.75E-162 | 1.272320854 | 0 |
| hsa-miR-433-3p | miRNA | 1.099339047 | 1.31E-22 | 1.482060103 | 1.00E-71 |
| hsa-miR-2110 | miRNA | 1.087768336 | 1.11E-32 | 1.348170145 | 7.06E-85 |
| hsa-miR-382-5p | miRNA | -1.020675274 | 1.68E-191 | -1.127515849 | 0 |
| hsa-miR-25-3p | miRNA | -1.056251171 | 9.69E-233 | -1.885178647 | 0 |
| hsa-miR-345-5p | miRNA | -1.244377035 | 7.53E-13 | -1.739934388 | 7.69E-32 |
| hsa-miR-146a-5p | miRNA | -1.349331158 | 0 | -2.411734531 | 0 |
| hsa-miR-629-5p | miRNA | -1.366055592 | 1.89E-04 | -1.210249218 | 1.44E-05 |
| hsa-miR-590-3p | miRNA | -1.383129105 | 1.46E-08 | -1.112156406 | 1.58E-09 |
| hsa-miR-21-5p | miRNA | -1.422721873 | 0 | -1.495275091 | 0 |
| hsa-miR-377-5p | miRNA | -1.470392251 | 3.23E-06 | -2.023537096 | 5.11E-14 |
| hsa-miR-1246 | miRNA | -1.526516068 | 0 | -1.174642613 | 0 |
| hsa-miR-188-5p | miRNA | -1.857908688 | 5.08E-04 | -3.052599562 | 1.13E-09 |
| hsa-miR-329-3p | miRNA | -2.299418399 | 3.91E-07 | -1.856679352 | 2.02E-08 |
| hsa-miR-3074-5p | miRNA | -2.625922718 | 0 | -1.817295416 | 0 |
| hsa-miR-136-3p | miRNA | -2.633121752 | 8.20E-69 | -1.679582695 | 1.66E-61 |
| hsa-miR-27a-3p | miRNA | -2.827688894 | 0 | -3.067134097 | 0 |
| hsa-miR-598-3p | miRNA | -2.857908688 | 8.33E-05 | -3.052599562 | 1.65E-07 |
| hsa-miR-30d-5p | miRNA | -2.904639738 | 1.57E-37 | -1.203166423 | 7.61E-19 |
| hsa-miR-299-3p | miRNA | -2.946717955 | 7.58E-11 | -2.090782755 | 1.40E-11 |
| hsa-miR-337-5p | miRNA | -4.024267074 | 2.79E-39 | -2.785998541 | 1.89E-45 |
| hsa-miR-549a-5p | miRNA | -4.147415305 | 3.53E-05 | -4.342106179 | 7.27E-08 |
| hsa-miR-655-3p | miRNA | -5.147415305 | 1.46E-05 | -2.342106179 | 5.61E-05 |
| hsa-miR-410-5p | miRNA | -5.360409029 | 9.24E-12 | -2.555099902 | 3.08E-11 |
| hsa-miR-4423-5p | miRNA | -5.388423405 | 1.88E-06 | -2.998151778 | 3.73E-07 |
Figure 6.
KEGG pathway enrichment analyses of target genes of miRNA in HucMSC-derived exosomes. (A) KEGG enrichment bubble map of target genes predicted by differentially expressed miRNA between osteogenic group 3 and control group. (B) Bubble map of target genes predicted by differentially expressed miRNA between osteogenic group 7 and control group. (C) KEGG enrichment analysis of target genes predicted by differentially expressed miRNA between osteogenic group 3 and 7. The X-axis is the enrichment ratio (the ratio of the number of genes annotated to an item in a selected gene set to the total number of genes annotated to that item by the species), and the Y-axis is KEGG Pathway; the size of the bubble represents the number of genes annotated to a KEGG Pathway. The color represents the enrichment P value. (D) KEGG enrichment result of target genes regulated by critical microRNAs in MSC-secreted exosomes during osteogenic induction. (E) TargetScan and miRBase databases were applied to analyze the target genes of hsa-mir-328-3p and hsa-mir-2110. Shown are complementary base pairing sequences of the miRNA 3´-UTR region.
Discussion
Exosomes are carriers and transfer agents of intercellular information, and the in-depth study of exosomes is important to reveal the mechanism of intercellular information exchange. As physiological vesicles, exosomes can be targeted to alter specific regulatory cell function materials that they contain, thereby improving the accuracy and effectiveness of disease treatment. The application of cell-derived vesicles can reduce the risks associated with transplantation and minimize the possible immune response and ectopic tissue development problems caused by MSC transplantation 1,9,18 . Experimental studies showed that exosomes derived from MSCs have therapeutic effects on many diseases, and their cargoes are important to regulate cell function and play a therapeutic role 18 . Therefore, the present study aimed to explore the mechanism of exosomes-related osteogenic differentiation, and identify potential molecular targets related to the proliferation and differentiation of osteoblasts.
Previous studies have proved that the exosomes from HucMSCs have therapeutic effects in a variety of diseases, such as immune diseases, Alzheimer’s disease, inflammatory bowel disease, spinal cord nerve injury, cancer, and ischemia, which are similar to the therapeutic effects of HucMSCs 5,19 . To further explore the impact of exosomes in osteogenic differentiation, we isolated the exosomes produced by HucMSCs. The osteogenic differentiation of cultured human umbilical cord mesenchymal stem cells in lineage-specific culture condition was performed according to an established method 7 –9 . During the initial differentiation of mesenchymal stem cells, the expression levels of important genes usually change within 48 h 20 . In this study, HucMSC cell proliferation was inhibited during the first 3 days of growth in the osteogenic differentiation microenvironment; after 7 days of culture, cell morphology changed and osteogenesis-related proteins were expressed, as evidenced by western blotting detection. Indeed, alkaline phosphatase staining and alizarin red staining also confirmed that exosomes secreted by HucMSCs induced with osteogenic differentiation conditioned medium for 48 h promoted osteogenic differentiation 20–21 .
Exosomes derived from standardized stem cell culture (Exo1) and from osteogenic differentiation (Exo2) promoted osteogenic differentiation, as observed using ALP and alizarin red staining; however, the effect of Exo2 was more obvious. Western blotting showed that the levels of osteogenic differentiation-related proteins RUNX2, COL1A1, ALP, and OST were higher in the two exosome preparations than that in the blank control group, and was concentration dependent. The results of CCK-8 assays showed that Exo1 promoted the proliferation of osteoblasts, while Exo2 did not. These results showed that in the microenvironment of osteogenic differentiation, the regulatory function of exosomes had changed, the effect of exosomes on osteogenic differentiation was enhanced, and the microenvironment reduced the promoting effect on osteoblast proliferation.
Osteoporosis is led by an imbalance of the differentiation ratio of MSCs to osteoblasts and adipocytes. Osteoporosis is characterized mainly by decreased numbers of osteoblasts, enhanced osteoclast activity, and the increase in bone absorption caused by changes of the cellular microenvironment, such as ageing and estrogen deficiency 22–23 . Estrogen deficiency causes osteoporosis after ovariectomization, while estrogen supplementation is traditionally used to reverse ovariectomy-induced osteoporosis. Thus, we used ovariectomized mice treated with estradiol as a positive control in our experiments to evaluate the outcome of the exosome treatment in vivo. Specifically, the effects of exosome therapy on osteoporosis were compared with ovariectomization and estrogen therapy. As indicated by micro-CT scanning, H&E staining, and analyses of multiple parameters of bone structure, we found that both Exo1 and Exo2 exerted positive preventive effects on osteoporosis in vivo. Furthermore, the effects of Eox1 and Eox2 were very similar, suggesting that exosomes derived from HucMSCs themselves, without any induction, are probably sufficient to reverse the symptoms of osteoporosis in mice. By contrast, the pathogenesis of osteoporosis is multifaceted, which is closely related to the proliferation and differentiation of mesenchymal stem cells. In this study, in vitro experiments revealed that Exo1 promoted the proliferation of osteoblasts, but Exo2 did not. These findings in turn imply that the molecular mechanisms responsible for the therapeutic effects on osteoporosis of Exo1 and Exo2 are probably different, which remain to be further studied.
The core mechanism of microRNAs is to induce mRNA degradation or inhibit the expression of their target genes through complementary pairing with mRNA base sequences, which is involved in biological processes such as cell differentiation, growth, migration, and apoptosis 7 . Exosomes are carriers of information between cells or tissues; therefore, microRNAs in exosomes play an important role in the regulation of cell function 24 . We found that the types and expression of miRNAs changed constantly changing with the extension osteogenic differentiation time, indicating that changes in the microenvironment can affect the cargoes of exosomes. Furthermore, KEGG pathway enrichment showed that target genes of miRNAs derived from exosomes upon osteogenic induction were enriched in osteoclast differentiation and MAPK signaling pathways, which are known to be closely related to the occurrence and development of osteoporosis 25–26 . Target genes of important MSC-secreted exosome-derived miRNAs that changed with osteogenic differentiation time were also significantly related to bone development and differentiation. These findings are highly consistent with the previous studies on the pathological changes observed in the process of osteoporosis 23 .
In the prediction of microRNA targeting genes, we found that CHRD is one of the targets of hsa-mir-328-3p. Chordin binds to bone morphogenetic protein (BMP) and inhibits the activation of BMP related signaling pathways 27 . The results suggested that the inhibition of CHRD expression led to the significant increase in ALP expression and extracellular mineral deposition. Chordin might represent a new target to promote bone regeneration 27–28 . In addition, we speculated that the target gene of hsa-mir-2110 is TNF. The TNF superfamily is mainly secreted by macrophages. It can bind to and act through its receptors TNFRSF1A/TNFR1 and TNFRSF1B/TNFBR. TNFs are involved in a wide range of biological processes, including cell proliferation, differentiation, and apoptosis 29 –31 . In the process of osteoclast differentiation, TNF plays an important role in the differentiation and formation of osteoclasts. It is a necessary inducting factor of almost all osteoclast differentiation signaling pathways 23 . Therefore, hsa-mir-2110 is a potentially crucial target to alleviate osteoporosis.
Studies on bone marrow MSCs in elderly patients with osteoporosis showed that the osteogenic differentiation ability of MSCs decreased, the potential of adipogenic differentiation in MSCs increased, and their osteoclast activity increased with aging 22 , 32 . The inhibition the by exosomal miRNAs on adipocyte differentiation of MSCs and osteoclasts probably contributes to the treatment of osteoporosis. Consistent with our findings, a large number of experimental studies have revealed that the inhibition of MSC adipose differentiation and osteoclast differentiation-related signaling pathways by miRNAs can promote osteogenic differentiation and can treat osteoporosis 7 , 33 , 34 . Similarly, our miRNA sequencing data revealed that the target genes of the miRNAs in HucMSC-derived exosomes induced with osteogenic induction medium are involved in the osteoclast differentiation and MAPK signaling pathways. Therefore, microRNAs in exosomes might target genes to inhibit adipose differentiation and osteoclast activity in the early stage of osteogenic differentiation. The molecular mechanism underlying the balance between osteoclastic and adipogenic differentiation regulated by exosomal miRNAs should be further explored in the future.
Conclusions
In the microenvironment of osteoblast differentiation culture, HucMSC-derived exosomes can significantly promote the differentiation of osteoblasts, and displayed certain therapeutic and preventative effects on an OVX mouse osteoporosis model. It is suggested that exosomes are influenced by the cells in the differentiation stage and carry related substances to promote osteoblast differentiation. Bioinformatic analysis showed that osteogenic differentiation changed the microRNA profile in exosomes, and the target genes of these miRNAs might not only act on osteogenic differentiation, but also on the pathways related to adipogenic differentiation and osteoclastic differentiation of MSCs. Therefore, the prevention and treatment of osteoporosis may be subjected to multifaceted regulation.
Acknowledgments
We thank the Central Laboratory of Medical College of Shantou University for its aid and technical support.
Abbreviations
- MSC
Mesenchymal stem cell
- HucMSC
Human umbilical cord MSCs
- Exo1
Exosomes from standardized stem cell culture
- Exo2
osteogenic differentiation-exosomes
- OB
Osteoblast
- RUNX2
RUNX family transcription factor 2
- OST
osteopontin
- COL1A1
collagen type I alpha 1 Chain
- ALP
Alkaline phosphatase
- OVX
ovariectomized
- E2
Estradiol
- BV/TV
Bone volume to total tissue volume ratio
- Ct.Ar
cortical bone area
- Ct.Th
cortical bone thickness
- BS/TV
ratio of bone surface area to bone volume
- BMD
bone mineral density
- Tb.N
trabecular number
- Tb.Sp
trabecular separation/Spacing
- Tb.Th
trabecular thickness
- Conn.D
Connective density
- SMI
Structure Model Index
Footnotes
Author Contributions: Ge Yahao performed the experiments, created the graphs, and wrote the manuscript. Wang Xinjia contributed to the conception and design, financial and administrative support, and manuscript reviewing and revising. Dong Mingming and Chang Qi collated analyzed data. Qiu Yisen, Lian Zhen and Han Yafei helped with statistical analysis.
Availability of Data and Materials: All data generated or analyzed during this study are included in this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Guangdong Science and Technology Special Fund Mayor Project (No: 20190301-65), 2020 Li Ka Shing Foundation Cross-Disciplinary Research Grant (2020LKSFG01D) and Shantou City Science and Technology Plan Project, Guangdong Province, China (No: 2018155, 2019-106-37). The funding bodies had no role in the design of the study, the collection, analysis, and interpretation of the data, and the writing of the manuscript. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
ORCID iD: Ge Yahao, MM  https://orcid.org/0000-0001-9691-5376
https://orcid.org/0000-0001-9691-5376
Ethics Approval: The experimental study was approved by the ethics Committee of the Second Affiliated Hospital of Shantou University Medical College and the informed consent of the pregnant women and their families. This experimental study was approved by the Medical Laboratory Animal Ethics Committee, and all experimental procedures were performed in accordance with the procedures of the Medical Laboratory Animal Center of Shantou University Medical College.
Statement of Human and Animal Rights: All procedures in this study were conducted in accordance with the procedures of the Medical Laboratory Animal Center of Shantou University Medical College and approved by the Medical Laboratory Animal Ethics Committee.
Statement of Informed Consent: Written informed consent was obtained from the patients for their anonymized information to be published in this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1. Phinney DG, Pittenger MF. Concise review: MSC-derived exosomes for cell-free therapy. Stem Cells. 2017;35(4):851–858. [DOI] [PubMed] [Google Scholar]
- 2. Phan J, Kumar P, Hao D, Gao K, Farmer D, Wang A. Engineering mesenchymal stem cells to improve their exosome efficacy and yield for cell-free therapy. J Extracell Vesicles. 2018;7(1):1522236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gimona M, Pachler K, Laner-Plamberger S, Schallmoser K, Rohde E. Manufacturing of human extracellular vesicle-based therapeutics for clinical use. Int J Mol Sci. 2017;18(6):1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lai RC, Yeo RW, Lim SK. Mesenchymal stem cell exosomes. Semin Cell Dev Biol. 2015;40:82–88. [DOI] [PubMed] [Google Scholar]
- 5. Yaghoubi Y, Movassaghpour A, Zamani M, Talebi M, Mehdizadeh A, Yousefi M. Human umbilical cord mesenchymal stem cells derived-exosomes in diseases treatment. Life Sci. 2019;233:116733. [DOI] [PubMed] [Google Scholar]
- 6. Mendt M, Rezvani K, Shpall E. Mesenchymal stem cell-derived exosomes for clinical use. Bone Marrow Transplant. 2019;54(suppl 2):789–792. [DOI] [PubMed] [Google Scholar]
- 7. Seenprachawong K, Tawornsawutruk T, Nantasenamat C, Nuchnoi P, Hongeng S, Supokawej A. miR-130a and miR-27b enhance osteogenesis in human bone marrow mesenchymal stem cells via specific down-regulation of peroxisome proliferator-activated receptor gamma. Front Genet. 2018;9:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwong FN, Richardson SM, Evans CH. Chordin knockdown enhances the osteogenic differentiation of human mesenchymal stem cells. Arthritis Res Ther. 2008;10(3):R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang J, Liu X, Li H, Chen C, Hu B, Niu X, Li Q, Zhao B, Xie Z, Wang Y. Exosomes/tricalcium phosphate combination scaffolds can enhance bone regeneration by activating the PI3K/Akt signaling pathway. Stem Cell Res Ther. 2016;7(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma Z, Cui X, Lu L, Chen G, Yang Y, Hu Y, Lu Y, Cao Z, Wang Y, Wang X. Exosomes from glioma cells induce a tumor-like phenotype in mesenchymal stem cells by activating glycolysis. Stem Cell Res Ther. 2019;10(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Naji A, Eitoku M, Favier B, Deschaseaux F, Rouas-Freiss N, Suganuma N. Biological functions of mesenchymal stem cells and clinical implications. Cell Mol Life Sci. 2019;76(17):3323–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qi X, Zhang J, Yuan H, Xu Z, Li Q, Niu X, Hu B, Wang Y, Li X. Exosomes secreted by human-induced pluripotent stem cell-derived mesenchymal stem cells repair critical-sized bone defects through enhanced angiogenesis and osteogenesis in osteoporotic rats. Int J Biol Sci. 2016;12(7):836–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kruger J, Rehmsmeier M. RNAhybrid. microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;211:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. [DOI] [PubMed] [Google Scholar]
- 17. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elahi FM, Farwell DG, Nolta JA, Anderson JD. Preclinical translation of exosomes derived from mesenchymal stem/stromal cells. Stem Cells. 2020;38(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yin S, Ji C, Wu P, Jin C, Qian H. Human umbilical cord mesenchymal stem cells and exosomes: bioactive ways of tissue injury repair. Am J Transl Res. 2019;11(3):1230–1240. [PMC free article] [PubMed] [Google Scholar]
- 20. van de Peppel J, Strini T, Tilburg J, Westerhoff H, van Wijnen AJ, van Leeuwen JP. Identification of three early phases of cell-fate determination during osteogenic and adipogenic differentiation by transcription factor dynamics. Stem Cell Reports. 2017;8(4):947–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang W, Dong R, Diao S, Du J, Fan Z, Wang F. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Res Ther. 2017;8(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q, Cao J, Xie N, Velletri T, Zhang X, Xu C, et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23(7):1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park-Min KH. Metabolic reprogramming in osteoclasts. Semin Immunopathol. 2019;41(5):565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kusuma GD, Carthew J, Lim R, Frith JE. Effect of the microenvironment on mesenchymal stem cell paracrine signaling: opportunities to engineer the therapeutic effect. Stem Cells Dev. 2017;26(9):617–631. [DOI] [PubMed] [Google Scholar]
- 25. Wang W, Bai J, Zhang W, Ge G, Wang Q, Liang X, Li N, Gu Y, Li M, Xu W, Yang H, et al. Protective effects of punicalagin on osteoporosis by inhibiting osteoclastogenesis and inflammation via the NF-kappaB and MAPK pathways. Front Pharmacol. 2020;11:696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. An Y, Zhang H, Wang C, Jiao F, Xu H, Wang X, Luan W, Ma F, Ni L, Tang X, Liu M, et al. Activation of ROS/MAPKs/NF-kappaB/NLRP3 and inhibition of efferocytosis in osteoclast-mediated diabetic osteoporosis. FASEB J. 2019;33(11):12515–12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Troilo H, Barrett AL, Wohl AP, Jowitt TA, Collins RF, Bayley CP, Zuk AV, Sengle G, Baldock C. The role of chordin fragments generated by partial tolloid cleavage in regulating BMP activity. Biochem Soc Trans. 2015;43(5):795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vizoso FJ, Eiro N, Cid S, Schneider J, Perez-Fernandez R. Mesenchymal stem cell secretome: toward cell-free therapeutic strategies in regenerative medicine. Int J Mol Sci. 2017;18(9):1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao B. TNF and bone remodeling. Curr Osteoporos Rep. 2017;15(3):126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gaur U, Aggarwal BB. Regulation of proliferation, survival and apoptosis by members of the TNF superfamily. Biochem Pharmacol. 2003;66(8):1403–1408. [DOI] [PubMed] [Google Scholar]
- 31. Yang N, Wang G, Hu C, Shi Y, Liao L, Shi S, Cai Y, Cheng S, Wang X, Liu Y, Tang L, et al. Tumor necrosis factor alpha suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis. J Bone Miner Res. 2013;28(3):559–573. [DOI] [PubMed] [Google Scholar]
- 32. Sui BD, Hu CH, Zheng CX, Jin Y. Microenvironmental views on mesenchymal stem cell differentiation in aging. J Dent Res. 2016;95(12):1333–1340. [DOI] [PubMed] [Google Scholar]
- 33. You L, Pan L, Chen L, Gu W, Chen J. MiR-27a is essential for the shift from osteogenic differentiation to adipogenic differentiation of mesenchymal stem cells in postmenopausal osteoporosis. Cell Physiol Biochem. 2016;39(1):253–265. [DOI] [PubMed] [Google Scholar]
- 34. Li B. MicroRNA regulation in osteogenic and adipogenic differentiation of bone mesenchymal stem cells and its application in bone regeneration. Curr Stem Cell Res Ther. 2018;13(1):26–30. [DOI] [PubMed] [Google Scholar]