Abstract
ARHGEF9 encodes collybistin, a brain-specific guanosine diphosphate-guanosine-5′-triphosphate exchange factor that plays an important role in clustering of gephyrin and γ-aminobutyric acid type A receptors in the postsynaptic membrane. Overwhelming evidence suggests that defects in this protein can cause X-linked intellectual disability, which comprises a series of clinical phenotypes, including autism spectrum disorder, behavior disorder, intellectual disability, and febrile seizures. Here, we report a boy with clinical symptoms of severe intellectual disability, epilepsy, and developmental delay and regression. Trio exome sequencing (trio-clinical exome sequencing) identified a novel hemizygous deletion, c.656_c.669delACTTCTTTGAGGCC (p. His219Leu fs*9), in exon 5 of ARHGEF9. This variant was not reported in either the Genome Aggregation Database or our database of 309 patients with neurodevelopmental disorders. Oxcarbazepine and levetiracetam reduced the frequency of the patient’s epileptic seizures to a certain extent, but psychomotor developmental delay and developmental regression became more obvious with age. This case study seeks to report a de novo loss-of-function mutation of ARHGEF9, aiming to emphasize the genetic diagnosis of X-linked intellectual disability and further improve knowledge of the ethnic distribution of ARHGEF9 mutations.
Keywords: ARHGEF9, epilepsy, X-linked intellectual disability, protein truncation, loss-of-function mutation, exome sequencing
Background
Intellectual disability (ID) is a developmental brain disorder commonly characterized by mental retardation and adaptive behavior. Approximately 1% of people worldwide suffer from various degrees of ID. 1 In particular, according to a consensus estimate, males are affected by ID approximately 30% to 40% more often than females, and a major portion of patients are likely to have a clear X-linked segregation. 2 Although only approximately 5% of the entire complement of human genes is located on the X chromosome, 15% of X-linked genes are identified as ID risk genes. 3
Previous studies have shown that X-linked intellectual disability (XLID) is highly heterogeneous in both genetic and clinical aspects. XLID can usually be divided into two groups: syndromic ID, with physical features and comorbid illnesses, and nonsyndromic ID, without obvious comorbidities.4,5 Although next-generation sequencing (NGS) and diagnostic techniques have improved, currently, the majority of ID genes are still uncharacterized or not well-described. Therefore, more specific research is needed to clarify the mutations causing XLID. In recent years, a series of large-scale clinical cohort studies has been conducted. Through high-throughput and high-resolution microarrays and NGS technology, 150 XLID genes have been identified. Moreover, it has been posited that most established XLID genes play a crucial role in brain development, particularly in synaptic function.5,6
ARHGEF9 is one such gene on chromosome X q11. It acts as a guanine nucleotide exchange factor (GEF) for CDC42. Defects in this gene can cause epilepsy, ID (moderate to severe), developmental delay, autism spectrum disorder, and several other types of disability. 7 Currently, 27 pathogenic variants have been recorded, corresponding to 9 female and 18 male patients.5,7–19 Six copy number variants (CNVs) and one missense and two loss-of-function variants have been identified in female patients. All of these female patients have estimated X-chromosome inactivation (XCI). Compared with affected females, most male patients present severe clinical symptoms. In this case, we report a novel de novo loss-of-function (LoF) mutation identified in a male patient presenting with ID, developmental delay, and epilepsy. During the course of treatment, developmental regression was also noted. This case report describes a de novo single nucleotide variant (SNV) that connects the ARHGEF9 gene with developmental regression.
Materials and methods
Clinical data
A Tibetan family was recruited at the Rehabilitation Medicine Center of the West China Second University Hospital. Clinical data were obtained using a standard assessment of neurodevelopmental disorders, including basic personal information, birth history, family history, physical examination (head circumference, muscle tone, facial appearance), system development (motor development, language development, autism, intelligence, sleep disorders, social interaction, seizures), and auxiliary examinations (head magnetic resonance imaging (MRI), electroencephalography, biochemical and metabolic indicators). All results were evaluated by two qualified pediatric neurologists. Signed informed consent was obtained from the patient’s parents, and the study was approved before the project began by the Ethical Committee of West China Second University Hospital of Sichuan University (K2017021). The reporting of this case conformed to CARE guidelines. 20
NGS
NGS was performed by a commercial vendor (Novogene, Tianjin, China) using a HiSeq 2000 platform (Illumina, San Diego, CA, USA). Sequencing libraries were generated using a SureSelect Human All Exon kit V6 (Agilent Technologies, Santa Clara, CA, USA). Processed reads were aligned to the human reference genome (GRCh38) using BWA v01.22 (https://github.com/lh3/bwa). 21 SNVs and insertion–deletion mutants were detected by SAMtools v1.0 (https://sourceforge.net/projects/samtools/files/samtools/1.0). 22 Functional annotation was performed by ANOVAR (https://annovar.openbioinformatics.org/en/latest/user-guide/download) and compared with the following databases: 1000 Genomes, dbSNP, Combined Annotation Dependent Depletion (CADD), Genome Aggregation Database (gnomAD), Exome Aggregation Consortium (ExAC), and Human Gene Mutation Database (HGMD). 23 Mutations were removed if the minor allele frequency of the mutation was >0.1% in more than one of the frequency databases, including the 1000 Genomes data (1000g_all), ExAC_all, and gnomAD (gnomAD_ALL and gnomAD_EAS). 24 De novo mutations were searched according to family-based mutation information using SAMtools. Sanger sequencing was used to validate the identified potential disease-causing variant.
Electroencephalography
Electroencephalography was used as a neurodiagnostic test to record the electrical activity of the brain. 25 Routine, scalp, awake, and interictal electroencephalogram (EEG) recordings were performed on the patient. Cap electrodes were applied with collodion, according to the 10–20 system, with linked mandibular references. The data were obtained on 16-channel Grass and Medtronic electroencephalography instruments (Shanghai, China). Common average referential, longitudinal, and transverse bipolar montages were used in all examinations. EEG recordings were filtered with 1 Hz high-pass, 35 Hz low-pass, and 60 Hz notch filters at a paper speed of 30 mm/s.
Case presentation
The Rehabilitation Medicine Center of the West China Second University Hospital has collected data on pediatric patients with neurodevelopmental disorders to better understand genetic neurodevelopmental conditions. Whole exome sequencing and CNV microarray technologies were used to establish a database of genetic samples from families with one proband and unaffected parents.
This male patient was the first child of healthy parents with no relevant family history. He was born at full term with spontaneous delivery after an uneventful pregnancy. His weight and length were both normal at birth. At 1 year of age, he suddenly presented with several episodes of neurological distress characterized by blank staring and no response to communication but without losing bowel or bladder control. The seizure episodes, which occurred once or twice per month, lasted 1 to 2 minutes each, and the patient would exhibit a period of lethargy. On admission to our hospital, the outpatient doctor prescribed levetiracetam. The seizures continued, but the frequency of seizures decreased to once every 1 to 2 months. At 3 years of age, the patient began to exhibit more frequent seizures, at 2 to 4 times per month. Oxcarbazepine and levetiracetam biotherapy led to a decrease in the seizure frequency to once per month. At the age of 3 years and 1 month, the patient’s weight was 11.9 kg (less than the fifth percentile), and his height was 93 cm (25th–50th percentile). The patient showed psychomotor developmental delays. However, he was able to sit and stand independently at 1 year of age and even walk alone at 23 months. Developmental regression was observed because of the frequent seizures. At the age of 3 years, he was suddenly unable to sit or stand without support. Currently, the patient cannot speak any words. Brain MRI did not show any morphological abnormality. Because the patient is young and lives in Tibet, he cannot attend the clinic regularly.
Results
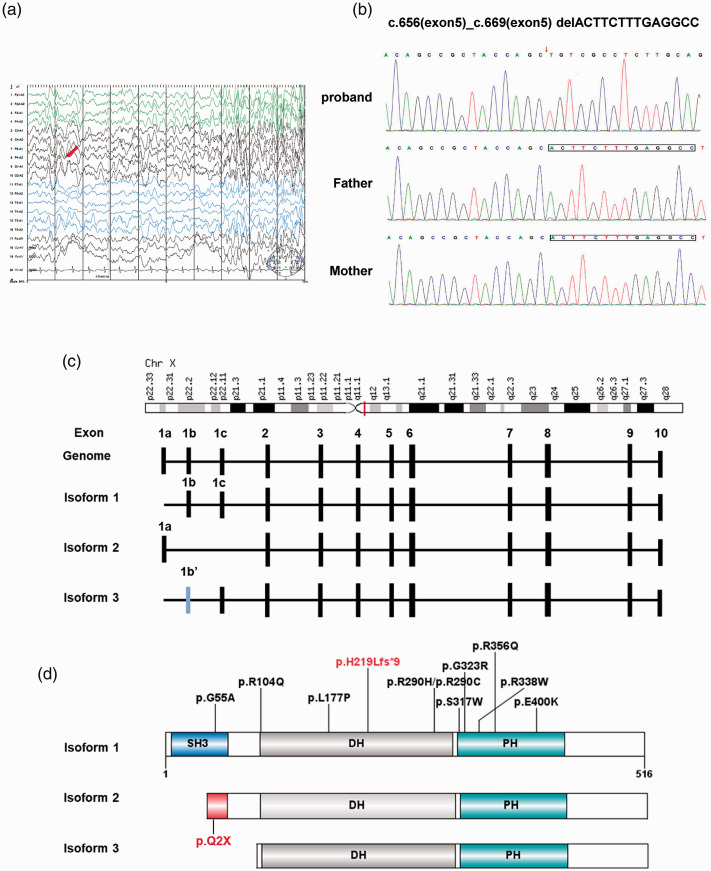
EEG results commonly provide strong evidence for underlying focal or diffuse cerebral dysfunction. This patient was instructed to remain relaxed during the EEG recordings. Electroencephalographic abnormalities occurred in this case, with bursts of generalized slow waves and focal slowing (red arrow) (Figure 1a). This slow wave activity was considered pathologic, indicating corresponding generalized cerebral dysfunction. No clinical seizures were observed during EEG recordings.
Figure 1.
Identification of a de novo mutation in ARHGEF9. (a) Electroencephalogram (EEG) of the proband in this study. The EEG recording was filtered with 1 Hz high-pass, 35 Hz low-pass, and 60 Hz notch filters at a paper speed of 30 mm/s. The red arrow indicates the location of slow waves. (b) Sanger sequencing of the ARHGEF9 gene mutation. DNA sequence traces of ARHGEF9 from the proband indicated a p.H219Lfs*9 variant. (c) ARHGEF9 is located on X chromosome (q11.1) and composed of 11 exons with three isoforms. (d) Schematic of ARHGEF9 protein with Src homology 3 (SH3), Dbl homology (DH), and pleckstrin homology (PH) domains. All reported missense and frameshift variants are indicated. The mutation identified in this proband was predicted to cause a truncated protein (red).
Because CNV microarrays cannot identify known or candidate pathogenic variations, whole exome sequencing was performed and subsequently used to predict two candidate genes (ARHGEF9 and SRPX2) that were X-linked, consistent with the clinical pattern of X-linked recessive inheritance. SRPX2 has been associated with XLID, but evidence supporting a role of SRPX2 in epilepsy and speech and cognitive defects is still questionable. 26 The missense SRPX2 variant c.257G>A/p.R86H was observed with an allele frequency of 0.0035 in the gnomAD database and was considered as a single nucleotide polymorphism. 24
Furthermore, a de novo frameshift mutation located in exon 5 of ARHGEF9 (p. His219Leu fs*9) was identified. ARHGEF9 has previously been reported to be an XLID gene, which is consistent with the phenotypic characteristics of the proband in this study. Table 1 summarizes all of the genetic findings and associated clinical phenotypes of patients with sequence or structural variants of ARHGEF9, including nine cases of CNVs and 18 cases of SNVs/insertion–deletion mutants. In this case, a deletion caused a frameshift mutation (p. His219Leu fs*9) and was proposed to encode a premature stop codon, thus producing a truncated protein lacking two important functional domains. This ARHGEF9 mutation identified by trio-clinical exome sequencing was confirmed by Sanger sequencing in the proband and his unaffected parents (Figure 1b). This deletion was only present in the proband and not either of his parents, indicating that this is a de novo protein-truncating mutation.
Table 1.
Genetic findings and phenotypic features of ARHGEF9 variants.
| Variant | Sex | Inheritance | Symptoms | ID | MRI | Epilepsy | EEG findings | AEDs | Response to AEDs | Ref | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CNV | |||||||||||
| #1 | 46,XX, t(X;20) (q12; P13) | F | De novo | DD, SZ | Severe | Normal | Yes | Abnormal | VPA, PHT, PGB | Improved | (Marco et al. 2009)7 |
| #2 | 46, X, t(X;18) (q11.1; q11.21) | F | De novo | DD, SZ | Severe | Normal | Yes | Abnormal | CZP, VPA | Improved | (Kalscheuer et al. 2009)9 |
| #3 | 46, X, inv(X) (q11.1; q27.3) | F | De novo | DD | Moderate | NR | No | N/A | N/A | N/A | (Alber et al. 2017)13 |
| #4 | 1290 kb Xq11.1 deletion | M | De novo | DD, SZ | Severe | Normal | Yes | Abnormal | PB, CBZ, CLB, TPM, OXC, LEV | Controlled | (Lesca et al. 2011)10 |
| #5 | 737 kb Xq11.1 deletion | M | De novo | DD, SZ | Severe | Normal | Yes | Abnormal | VPA | Controlled | (Shimojima et al. 2011)5 |
| #6 | 217 kb Xq11.1 deletion | M | De novo | DD, SZ | Severe | NR | Yes | Abnormal | VPA | Controlled | (Machado et al. 2016)11 |
| #7 | 82 kb Xq11.1 deletion | F | De novo | DD | Mild | NR | No | N/A | N/A | N/A | (Bhat et al. 2016)14 |
| #8 | 27 kb Xq11.1 deletion | F | De novo | DD | Moderate | Normal | No | N/A | N/A | N/A | (Alber et al. 2017)13 |
| #9 | 7.5 kb Xq11.1 deletion | F | De novo | DD, SZ | Moderate | NR | Yes | Normal | NR | Seizure free | (Alber et al. 2017)13 |
| Frameshift and Nonsense | |||||||||||
| #10 | c.656_c.669 del (p.H219L fs*9) | M | De novo | DD, SZ | Severe | Normal | Yes | Abnormal | LEX, OXC | Improved | This study |
| #11 | c.381 + 3A>G | M | De novo | DD, SZ | Severe | NR | Yes | Abnormal | VPA, LEX | N/A | (Yao et al. 2020)17 |
| #12 | c.4C>T (p.Q2*) | M | Maternal | DD, SZ | Severe | Abnormal | Yes | Abnormal | Multiple AEDs | Refractory | (Shimojima et al. 2011)5 |
| #13 | c.899G>A (p.W300*) | M | De novo | SZ | Severe | Abnormal | Yes | Abnormal | VPA, LEX, CZP | Refractory | (Ghesh et al. 2019)18 |
| #14 | c.865C>T (p.R289*) | F | Paternal | DD | Moderate | Normal | Yes | Normal | N/A | N/A | (Ghesh et al. 2019)18 |
| #15 | c.1056G>A (E7 skipping) | F | De novo | DD, SZ | Moderate | Normal | Yes | Abnormal | VPA, LEX | Partially controlled | (Ghesh et al. 2019)18 |
| Missense | |||||||||||
| #16 | c.164G>C (p. G55A) | M | De novo | DD, SZ | Severe | Abnormal | Yes | NR | PB, LTG | Refractory | (Harvey et al. 2004)8 |
| #17 | c.311G>A (p. R104Q) | M | De novo | DD, SZ | Severe | Abnormal | Yes | Abnormal | Multiple AEDs | Refractory | (Alber et al. 2017)13 |
| #18 | c.530T>C (p. L177P) | M | De novo | DD, SZ | Severe | Normal | Yes | Abnormal | VPA, LEV, LTG | Improved | (Alber et al. 2017)13 |
| #19 | c.535G>A (p. E179K) | F | De novo | DD, SZ | Moderate | N/A | Yes | N/A | N/A | N/A | (Scala et al. 2021)19 |
| #20 | c.868C>T (p. R290C) | M | Maternal | DD, SZ | Severe | N/A | Yes | Abnormal | Multiple AEDs | Refractory | (J. Y. Wang et al. 2018)12 |
| #21 | c.869G>A (p. R290H) | M | De novo | DD, SZ | Moderate | Abnormal | Yes | Abnormal | Multiple AEDs | Refractory | (Alber et al. 2017)13 |
| #22 | c.881T>C (p.I294T) | M | Maternal | DD, SZ | Severe | Normal | Yes | Normal | VPA | Refractory | (Zhang et al. 2020)17 |
| #23 | c.950C>G (p. S317W) | M | Maternal | SZ | Severe | Abnormal | Yes | Abnormal | VPA, CBZ, CLB | Improved | (Alber et al. 2017)13 |
| #24 | c.967G>A (p. G323R) | M | Maternal | DD, SZ | Severe | Abnormal | Yes | Abnormal | VPA | Seizure free | (Klein et al. 2017)33 |
| #25 | c.1012C>T (p. R338W) | M | Maternal | SZ | Moderate | N/A | Yes | N/A | N/A | N/A | (Long et al. 2015)15 |
| #26 | c.1067G>A (p. R356Q) | M | Maternal | - | Mild | N/A | No | N/A | N/A | N/A | (Alber et al. 2017)13 |
| #27 | c.1070G>T (p. R357I) | M | Maternal | DD, SZ | Mild | Normal | Yes | Normal | N/A | N/A | (Zhang et al. 2020)17 |
| #28 | c.1198G>A (p. E400K) | M | De novo | DD | Moderate | Normal | No | N/A | N/A | N/A | (de Ligt et al. 2012)16 |
VPA, valproic acid; PHT, phenytoin; PGB, pregabalin; CLB, clobazam; CBZ, carbamazepine; PB, phenobarbitone; TPM, topiramate; LEV, levobupivacaine; OXC, oxcarbazepine; CZP, clonazepam; LTG, lamotrigine; N/A, not applicable; NR, not reported, ID, intellectual disability; MRI, magnetic resonance imaging; EEG, electroencephalogram; AED, anti-epileptic drug; DD, developmental delay; SZ, seizure; CNV, copy number variant; LEX, levetiracetam; M, male; F, female.
Discussion
ARHGEF9, which encodes collybistin (also known as hPEM2 in humans), can modulate cognitive function through its structural and functional activities. 27 Collybistin serves as a synaptic Rho GEF (for Rho/Rac/Cdc42-like GTPases) because it contains a Dbl homology (DH) domain. Rho GEFs have been shown to activate small GTPases of the Rho family by rapidly catalyzing the dissociation of GDP, allowing binding of GTP to induce an active state. 28 Additionally, a DH domain composed of approximately 180 amino acids is conserved among all Rho GEF family members and is responsible for the interactions and activation of Rho GTPase. The DH domain of collybistin activates Cdc42 but does not activate Rac1 or RhoA. 29 This domain is also believed to be involved in an interaction with gephyrin, which is an extensively studied scaffold protein responsible for anchoring, clustering, and stabilizing glycine and γ-aminobutyric acid type A receptors. 30 Moreover, it is critical for inhibitory synapse formation and function. However, the role of DH domain activity on Cdc42 in gephyrin clustering is unclear. Experiments in mice have shown that gephyrin and γ-aminobutyric acid type A receptor clustering were not affected by hippocampal deletion of Cdc42. 29 In our study, this frameshift mutation deleted 14 nucleotides in the center of the DH domain, leading to a functionally defective isoform with a disrupted DH domain and total loss of the pleckstrin homology domain and the C terminus. To our knowledge, nine CNVs and 18 frameshift/missense mutations have currently been identified in patients with ARHGEF9-related disorders (Table 1). However, the genotype–phenotype relationship has not been well correlated.
Functionally-deficient ARHGEF9 caused by chromosomal abnormalities has been previously found in nine patients, including six female patients who suffered from complete or extreme skewed X inactivation in favor of the abnormal X chromosome, and their conditions were similar to those of hemizygous male patients. 13 However, despite chromosome-wide silencing, approximately 15% of X-linked genes escape X inactivation, thus some XLID genes can be partially expressed because of another inactive X chromosome.3,31 To date, three male patients with inactivation of the ARHGEF9 gene caused by chromosome abnormalities have been reported. The lengths of the missing fragments were 1290 kb, 737 kb, and 217 kb. These three fragment deletions were all located immediately below the centromere and included the entire ARHGEF9 gene as well as a micro-RNA gene (mIR1468). A 1290-kb Xq11.11 microdeletion disrupting ARHGEF9 was found in a 6-year-old male patient with severe ID, epilepsy (onset: 5 months), developmental delay, macrosomia, and mild dysmorphism. The patient was hyperactive with attention deficit disorder, and his brain MRI results were normal. 10 The second reported microdeletion mutation, a 737-kb Xq11.1 deletion containing ARHGEF9, was identified by Shimojima et al. in a 5-year-old boy who also suffered from severe ID, epilepsy (onset: 24 months), development delay, and macrosomia. Brain MRI (Table 1) showed a relatively hypoplastic frontal lobe, and dysmorphic features were not reported in this patient. 5 The third 217-kb deletion was reported by Machado et al. in an 18-year-old male patient with severe mental impairment, developmental delay, epilepsy, and no dysmorphic features. 11 Similarly, the most significant phenotypes in the present patient were developmental delay and epilepsy, which are consistent with the above three patients.
Affected patients with ARHGEF9 variants include nine female patients exhibiting six chromosomal abnormalities, two LoF variants, and one missense variant. Seven of these patients presented with moderate ID and a less severe phenotype. Most affected females present skewed XCI patterns in the peripheral blood. Moreover, the mothers of the affected male patients were all healthy, indicating that the random skewing of X inactivation protects females from X-linked recessive disorders. Although Ghesh et al. reported two female patients with random XCI patterns, the XCI patterns in the peripheral blood may be different from those in other tissues, including the brain.18,32
According to the gnomAD database, the probability of being loss-of-function intolerant (pLI) score of ARHGEF9 is 1, reflecting that this gene is extremely LoF intolerant. In total, only five LoF variants of ARHGEF9 have been reported in the literature. Shimojima et al. identified the first nonsense variant in a cohort study. 5 Twenty-three male patients with severe ID and epilepsy were recruited, and their genotypes were verified by Sanger sequencing. A p.Q2 variant, which was located in the first exon, was identified in a 5.5-year-old boy with severe ID, epilepsy (onset: 20 months), and developmental delay. 5 Exon 1a encodes a long N terminus (MQWIRGGSGM) that is only present in isoform 2 of ARHGEF9. However, isoform 1 and isoform 3 begin with a shorter sequence (MTL) encoded by exon 1b (Figure 1c), indicating that p.Q2 only affects the expression of isoform 2 and does not affect the other two isoforms. Because this mutation occurred in the second codon of the variants, it represents total LoF of the encoding protein (Figure 1d). Another protein truncation mutation was recently reported by Yao et al. 17 It was a splice variant that could cause a truncated protein, which was shown by a transcriptional experiment. 17 Ghesh et al. recently reported three novel LoF variants (p.K352/E7 skipping, p.R289*, and p.W300*), which is the first report of affected females with random XCI patterns, even when taking missense variants into account. 18 The present study reported a novel nonsense variant in a male patient. Similar to other males with LoF variants of ARHGEF9, severe ID and epilepsy were observed. Moreover, this report provides further evidence of developmental regression in affected patients with pathogenic ARHGEF9 variants. However, as this patient could not visit the clinic often, we could not determine the precise time that developmental regression occurred.
Conclusion
We report a patient with XLID with severe mental retardation, developmental regression, and epilepsy. Sequencing analysis identified a novel ARHGEF9 mutation (c.656_c.669delACTTCTTTGAGGCC). This deletion is a frameshift mutation (p. His219Leu fs*9) and is proposed to encode a premature stop codon, thus producing a truncated protein. The lack of pleckstrin homology and DH domains led to LoF of collybistin. More importantly, this is one of the few clinical cases in which ARHGEF9 has been associated with developmental regression, and thus it is crucial for extending the knowledge of XLID and NGS analysis.
Acknowledgements
We gratefully thank the patient and his parents for their support and the Department of Rehabilitation Medicine, West China Second University Hospital for the clinical diagnosis and technical support.
Footnotes
Availability of data and materials: The annotated mutations of this patient and the raw NGS data are available upon request from the corresponding author.
Author contributions: T. Q. conceived and designed the experiments and analyzed data. Q. D. and Q. W. wrote the manuscript.
Ethics approval and consent to participate: The study was approved by the Ethical Committee of West China Second University Hospital of Sichuan University (K2017021). Written informed consent was obtained from the parents of this patient at the time of diagnosis.
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and publication of this article: This study was supported by the National Natural Science Foundation of China (81520108020).
ORCID iD: Qiu Wang https://orcid.org/0000-0001-5615-463X
References
- 1.Ellison JW Rosenfeld JA andShaffer LG.. Genetic basis of intellectual disability. Annu Rev Med 2013; 64: 441–450. [DOI] [PubMed] [Google Scholar]
- 2.Gecz J Shoubridge C andCorbett M.. The genetic landscape of intellectual disability arising from chromosome X. Trends Genet 2009; 25: 308–316. [DOI] [PubMed] [Google Scholar]
- 3.Neri G, Schwartz CE, Lubs HA, et al. X-linked intellectual disability update 2017. Am J Med Genet A 2018; 176: 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ropers HH andHamel BC.. X-linked mental retardation. Nat Rev Genet 2005; 6: 46–57. [DOI] [PubMed] [Google Scholar]
- 5.Shimojima K, Sugawara M, Shichiji M, et al. Loss-of-function mutation of collybistin is responsible for X-linked mental retardation associated with epilepsy. J Hum Genet 2011; 56: 561–565. [DOI] [PubMed] [Google Scholar]
- 6.Raymond FL. X linked mental retardation: a clinical guide. J Med Genet 2006; 43: 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marco EJ, Abidi FE, Bristow J, et al. ARHGEF9 disruption in a female patient is associated with X linked mental retardation and sensory hyperarousal. BMJ Case Rep 2009; 2009: bcr06.2009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harvey K, Duguid IC, Alldred MJ, et al. The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J Neurosci 2004; 24: 5816–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalscheuer VM, Musante L, Fang C, et al. A balanced chromosomal translocation disrupting ARHGEF9 is associated with epilepsy, anxiety, aggression, and mental retardation. Hum Mutat 2009; 30: 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesca G, Till M, Labalme A, et al. De novo Xq11.11 microdeletion including ARHGEF9 in a boy with mental retardation, epilepsy, macrosomia, and dysmorphic features. Am J Med Genet A 2011; 155A: 1706–1711. [DOI] [PubMed] [Google Scholar]
- 11.Machado CO, Griesi-Oliveira K, Rosenberg C, et al. Collybistin binds and inhibits mTORC1 signaling: a potential novel mechanism contributing to intellectual disability and autism. Eur J Hum Genet 2016; 24: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang JY, Zhou P, Wang J, et al. ARHGEF9 mutations in epileptic encephalopathy/intellectual disability: toward understanding the mechanism underlying phenotypic variation. Neurogenetics 2018; 19: 9–16. [DOI] [PubMed] [Google Scholar]
- 13.Alber M, Kalscheuer VM, Marco E, et al. ARHGEF9 disease: phenotype clarification and genotype-phenotype correlation. Neurol Genet 2017; 3: e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhat G, LaGrave D, Millson A, et al. Xq11.1-11.2 deletion involving ARHGEF9 in a girl with autism spectrum disorder. Eur J Med Genet 2016; 59: 470–473. [DOI] [PubMed] [Google Scholar]
- 15.Long P, May MM, James VM, et al. Missense mutation R338W in ARHGEF9 in a family with X-linked intellectual disability with variable macrocephaly and macro-orchidism. Front Mol Neurosci 2015; 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Ligt J, Willemsen MH, Van Bon BWM, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- 17.Yao R, Zhang Y, Liu J, et al. Clinical and molecular characterization of three novel ARHGEF9 mutations in patients with developmental delay and epilepsy. J Mol Neurosci 2020; 70: 908–915. [DOI] [PubMed] [Google Scholar]
- 18.Ghesh L, Vincent M, Delemazure AS, et al. Autosomal recessive Treacher Collins syndrome due to POLR1C mutations: report of a new family and review of the literature. Am J Med Genet A 2019; 179: 1390–1394. [DOI] [PubMed] [Google Scholar]
- 19.Scala M, Zonneveld-Huijssoon E, Brienza M, et al. De novo ARHGEF9 missense variants associated with neurodevelopmental disorder in females: expanding the genotypic and phenotypic spectrum of ARHGEF9 disease in females. Neurogenetics 2021; 22: 87–94. [DOI] [PubMed] [Google Scholar]
- 20.Gagnier JJ, Kienle G, Altman DG, et al. The CARE guidelines: consensus-based clinical case reporting guideline development. Headache 2013; 53: 1541–1547. [DOI] [PubMed] [Google Scholar]
- 21.Li H andDurbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K Li M andHakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020; 581: 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller-Putz GR. Electroencephalography. Handb Clin Neurol 2020; 168: 249–262. [DOI] [PubMed] [Google Scholar]
- 26.Piton A Redin C andMandel JL.. XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am J Hum Genet 2013; 93: 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid T, Bathoorn A, Ahmadian MR, et al. Identification and characterization of hPEM-2, a guanine nucleotide exchange factor specific for Cdc42. J Biol Chem 1999; 274: 33587–33593. [DOI] [PubMed] [Google Scholar]
- 28.Miller MB, Yan Y, Eipper BA, et al. Neuronal Rho GEFs in synaptic physiology and behavior. Neuroscientist 2013; 19: 255–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy-Alla S, Schmitt B, Birkenfeld J, et al. PH-domain-driven targeting of collybistin but not Cdc42 activation is required for synaptic gephyrin clustering. Eur J Neurosci 2010; 31: 1173–1184. [DOI] [PubMed] [Google Scholar]
- 30.Saiepour L, Fuchs C, Patrizi A, et al. Complex role of collybistin and gephyrin in GABAA receptor clustering. J Biol Chem 2010; 285: 29623–29631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berletch JB, Yang F, Xu J, et al. Genes that escape from X inactivation. Hum Genet 2011; 130: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musalkova D, Sticova E, Reboun M, et al. Variable X-chromosome inactivation and enlargement of pericentral glutamine synthetase zones in the liver of heterozygous females with OTC deficiency. Virchows Arch 2018; 472: 1029–1039. [DOI] [PubMed] [Google Scholar]
- 33.Klein KM, Pendziwiat M, Eilam A, et al. The phenotypic spectrum of ARHGEF9 includes intellectual disability, focal epilepsy and febrile seizures. J Neurol 2017; 264: 1421–1425. [DOI] [PubMed] [Google Scholar]