Abstract
Introduction
Myocardial infarction is coronary artery-related heart disease, and the leading cause of mortality globally. Circular RNAs (circRNAs) are a new type of regulatory RNAs and participate in multiple pathological cardiac progression.
Methods
However, the function of circFoxo3 in MI-induced myocardial injury remains obscure.
Results
Significantly, we identified that circFoxo3 was downregulated in the MI rat model and the overexpression of circFoxo3 ameliorated MI-induced cardiac dysfunction and attenuated MI-induced autophagy in rat model. Meanwhile, the overexpression of circFoxo3 repressed oxygen–glucose deprivation (OGD)-induced autophagy, apoptosis, inflammation, and injury of cardiomyocyte in vitro. Mechanically, we identified that the expression of KAT7 was reduced by circFoxo3 overexpression in cardiomyocytes. Meanwhile, the expression of HMGB1 was repressed by the depletion of KAT7 in cardiomyocytes. The enrichment of histone H3 lysine 14 acetylation (H3K14ac) and RNA polymerase II (RNA pol II) on HMGB1 promoter was inhibited by the knockdown of KAT7. Moreover, the overexpression of circFoxo3 suppressed HMGB1 expression and KAT7 overexpression rescued the expression of HMGB1 in cardiomyocytes. The enrichment of KAT7, H3K14ac, and RNA poly II on HMGB1 promoter was decreased by circFoxo3 overexpression, while the overexpression of KAT7 could reverse the effect. The overexpression of KAT7 or HMGB1 could reverse circFoxo3-attenuated cardiomyocyte injury and autophagy in vitro. Thus, we conclude that circular RNA circFoxo3 relieved myocardial ischemia/reperfusion injury by suppressing autophagy via inhibiting HMGB1 by repressing KAT7 in MI.
Discussion
Our finding provides new insight into the mechanism by which circFoxo3 regulates MI-related cardiac dysfunction by targeting KAT7/HMGB1 axis.
Keywords: myocardial infarction, autophagy, circFoxo3, KAT7, HMGB1
Introduction
Myocardial infarction1 is a coronary artery-related cardiac disease, which ranks the leading cause of mortality globally2 and is commonly caused by an insufficient blood supply in heart, which causes ischemia and hypoxia of myocardial tissues, inflammatory response, death of cardiomyocytes, finally the myocardial injury.3,4 Among the MI-induced cardiomyocytes reactions, cell death plays a central role during the pathogenesis of MI and could be achieved by three major mechanisms including cell apoptosis, necrosis, and autophagy.5 Although accumulating evidence have unraveled the pathogenesis of MI, the incidence of MI remains steadily increasing, and exploration of effective therapeutic manners is urgent.6
CircRNAs are covalently closed noncoding RNA loops that are highly conserved and abundant, featured by higher stability in physical conditions in comparison with the linear RNAs.7 Accumulating studies have disclosed the participation of circRNAs in pathogenesis of various diseases, including MI.8,9 CircFoxo3 is a newly discovered circRNA that derives from FoxO3 gene, and functions as tumor activator in cancers including gastric cancer and glioblastoma.10,11 Recent studies also disclosed the participation of circFoxo3 in progression of coronary artery diseases.12,13 CircFoxo3 was also reported to protect cardiomyocytes against radiation-induced cardiotoxicity.14 Nevertheless, the function of circFoxo3 in MI remains unclear. Noteworthy, circRNAs mainly function through sponging microRNA (miRNA) or directly with targeted proteins.7 For example, circRHOT1 epigenetically regulates lung cancer progression via recruiting KAT5.15 Importantly, the previous studies have identified the critical function of circRNAs in the regulation of MI progression, including circTtc3, circCDYL, and circFndc3b.8,16,17 Moreover, high mobility group box 1 (HMGB1) belongs to non-histone DNA-binding protein that participates in regulation of gene expression, including DNA replication, gene transcription, as well as nucleosome assembly.18 Studies have indicated the functions of KAT7 in various diseases, such as cancers, sepsis, atherosclerosis, collagen disease, acute lung injury arthritis, epilepsy, as well as myocardial infarction.19–21
In this work, we evaluated the role of circFoxo3 during MI progression, found a cardioprotective function of circFoxo3 in both in vitro and in vivo MI model, deciphered the molecular mechanism that circFoxo3 suppressed the expression of HMGB1 and cardiomyocytes autophagy via KAT7/HMGB1 axis. Our findings may provide a novel therapeutic method for MI.
Materials and Methods
Cell Culture and Transfection
Rat cardiomyocytes H9c2 was purchased from the American Type Culture Collection (ATCC, USA), and cultured in DMEM (Thermo, USA) supplemented with 10% FBS (Gibco, USA) plus 1% penicillin/streptomycin at a 37°C humidified incubator that filled with 5% CO2. The overexpressing vectors of circFOXO3, KAT7 and HMGB1, the small interfering RNA targeting KAT7 and HMGB1, and the control oligonucleotides were purchased from Gene Pharma (Shanghai, China). The oligonucleotides (50 nM) were mixed with Lipofectamine 2000 (Invitrogen, USA), and cultured with cells for 48 hours following the manufacturer’s protocol.
Establishment of MI Rat Model
All animal experiments were authorized by the Animals Ethics Committee of the First Affiliated Hospital of China Medical University, and were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. SD rats aged 8-weeks old (n = 6 for each group) were purchased from the Charles River laboratory (USA). To establish the in vivo MI model, we performed left anterior descending (LAD) ligation with 6–0 silk suture according to a previously reported protocol.22 Rats in treatment group were administrated with injection of indicated oligonucleotides (80 mg/kg in 100 μL PBS). While the rats in sham group received operation without ligation. One week after operation, the transthoracic two-dimensional (2D) echocardiography (Vevo 2100) ultrasound system was used to detect heart function. Left ventricular ejection fraction (LVEF), left ventricular fractional shortening (LVFS), left ventricle anterior wall thickness in diastole (LVAWd), and left ventricular posterior wall end diastole (LVPWd) were monitored. Then the rats were sacrificed, and heart tissues were collected for subsequent studies.
Enzyme Linked Immunosorbent Assay (ELISA)
The levels of inflammatory factors (IL-6 and IL-10) in rat serum, heart, and H9c2 cell culture medium were determined by ELISA assay (Invitrogen, USA) in accordance with manufacturer’s protocols.
Histological Analysis
The collected heart tissues were subjected to fixation in 4% paraformaldehyde (PFA), and made into paraffin-embedded 5-μm slices. To observe tissue damage, the samples were stained with hematoxylin and eosin (H&E), and photographed under a microscope (Leica, Germany). For triphenylterazolium chloride (TTC) staining, the aorta was isolated from rats and perfused with saline, followed by staining with Evans Blue solution (0.3%) (Sigma). Subsequently, the cardiac ventricle was sectioned and stained with TCC (1%).
Immunofluorescence
H9c2 cells were seeded on glass coverslips overnight, followed by fixation and permeabilization by ice-cold 100% methanol for 5 min at room temperature. After that, the cells were washed with PBS and blocked with 2% BSA in TBS buffer for 1 hour. The specific primary antibody against LC3 (Cell Signaling technology, USA) (1:100) in blocking solution was then used to hatch with the samples at 4°C overnight. Next day, the cells were incubated with Alexa fluor 633 secondary antibody for 1 hour at room temperature, followed by staining with DAPI for 10 minutes and mounted on slides. Cells were then imaged on a SP8 confocal microscope (Leica, Germany).
TdT-Mediated dUTP Nick End Labeling (TUNEL) Assay
The cell apoptosis in heart tissues was detected by One Step TUNEL Apoptosis Assay Kit (Beyotime, China) in accordance with the manufacturer’s description. In brief, the paraffin-embedded heart tissues were dewaxed, reacted with detective reagents in 37°C for 60 minutes. The nucleus was then stained with DAPI for 10 minutes. The fluorescence was observed under a microscope (Leica).
Statistical Analysis
The statistical analyses were realized by using SPSS software (USA). The statistical significance of the differences was determined by using Student’s t-test or one-way ANOVA method. The P value less than 0.05 was defined as statistically significant.
Results
The Overexpression of circFoxo3 Attenuates MI-Induced Cardiac Dysfunction in Rat Model
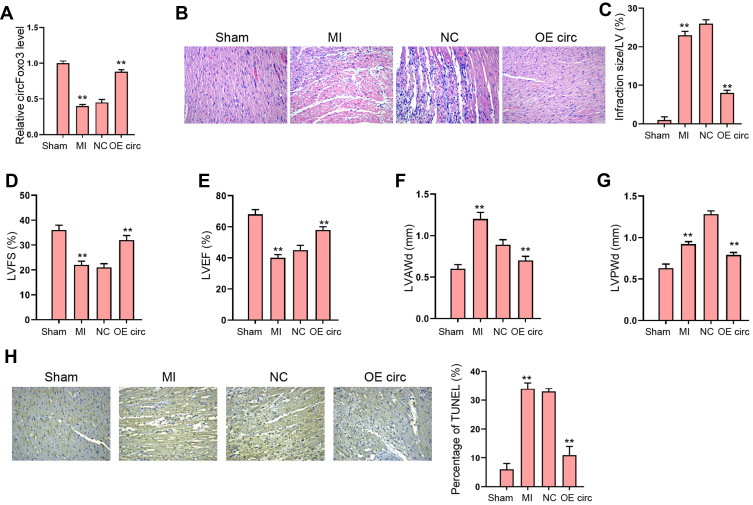
To identify the role of circFoxo3 in MI-induced abnormal cardiac function, we established a MI rat model and administrated circFoxo3 treatment in MI-infarcted regions. We revealed that circFoxo3 was reduced in MI rats and the effectiveness of circFoxo3 overexpression was validated in the model (Figure 1A). We observed inflammatory cell infiltration in MI heart tissues comparing with the sham group, and this infiltration was impeded by circFoxo3 overexpression (Figure 1B), along with decreased infarction size (Figure 1C). Echocardiography analysis showed that MI decreased the LVEF and LVFS, and upregulated LVAWd and LVPWd, whereas the administration of circFoxo3 overexpression could notably counteract the altered levels of LVEF, LVFS, LVAWd and LVPWd in MI rats (Figure 1D–G). Meanwhile, MI-induced apoptosis was attenuated by the overexpression of circFoxo3 in the rats (Figure 1H). These results demonstrated the successfully established MI rat model, and the protective role of circFoxo3 in MI (Supplementary Material).
Figure 1.
The overexpression of circFoxo3 attenuates MI-induced cardiac dysfunction in rat model. (A–H) The MI Rat model was established and treated with injection of circFoxo3 vectors or control vectors surrounding the infarcted region. (A) Relative level of circFoxo3 RNA in heart tissues was detected by qRT-PCR. (B) HE staining of inflammatory cell infiltration in heat tissues three days after surgery. (C–G) Infarcted size,25 left ventricular fractional shortening (LVFS) (D), Left ventricular ejection fraction (LVEF) (E), left ventricle anterior wall thickness in diastole (LVAWd) (F), and left ventricular posterior wall end diastole (LVPWd) (G) were measured. (H) The apoptosis was detected by TUNEL staining. **p < 0.01.
The Overexpression of circFoxo3 Represses MI-Induced Autophagy and Inflammation in Rat Model
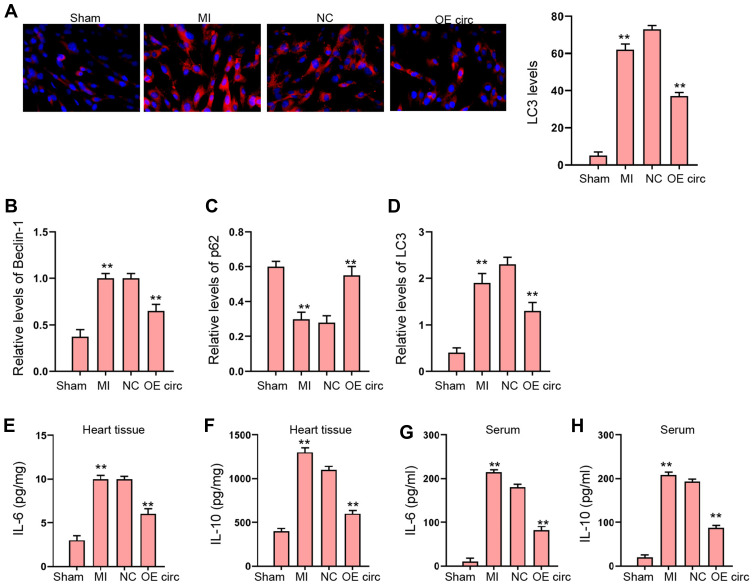
Then, we were interested in the function of circFoxo3 in autophagy and inflammation in MI rat model. We found that the expression of LC3 and Beclin-1 was induced and p62 expression was reduced in MI rats, in which the overexpression of circFoxo3 reversed the phenotype (Figure 2A–D). The levels of IL-6 and IL-10 were enhanced in MI rats, while the overexpression of circFoxo3 blocked the levels in the rats (Figure 2E–H).
Figure 2.
The overexpression of circFoxo3 represses MI-induced autophagy and inflammation in rat model. (A–H) The MI Rat model was established and treated with injection of circFoxo3 vectors or control vectors surrounding the infarcted region. (A) The levels of LC3 were measured by immunofluorescence. (B–D) The expression of Beclin-1, p62, and LC3 was detected by Western blot analysis. (E–H) The levels of IL-6 and IL-10 were analyzed by ELISA in heart tissues and serum. **p < 0.01.
The Overexpression of circFoxo3 Attenuates OGD-Induced Cardiomyocyte Injury in vitro
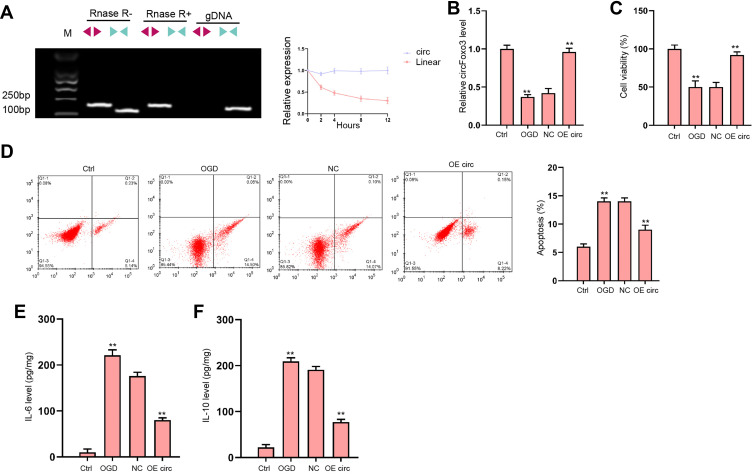
We evaluated the expression of canonical and back-spliced forms of Foxo3 by agarose gel electrophoresis assays and PCR in H9c2 cells in the presence or absence of RNase R supplementation. The circFoxo3 in cDNA, but not gDNA, was detectable using divergent primers with the treatment of RNase R (Figure 3A). We next explored the protective role of circFoxo3 in vitro. The expression of circFoxo3 was repressed in OGD-treated H9c2 cells and the effectiveness of circFoxo3 overexpression was validated in the cells (Figure 3B). CircFoxo3 overexpression facilitated cell viability (Figure 3C) and suppressed cell apoptosis (Figure 3D) under OGD stimulation. Besides, OGD treatment led to downregulated secretion of inflammatory cytokine IL-6 and IL-10 and the overexpression of circFoxo3 reversed the phenotypes (Figure 3E and F). These findings testified the in vitro protective effects of circFoxo3 in ischemic cardiomyocytes.
Figure 3.
The overexpression of circFoxo3 attenuates OGD-induced cardiomyocyte injury in vitro. (A) The back-spliced and canonical forms of Foxo3 expression was detected by agarose gel electrophoresis assays and PCR in H9c2 cells in the presence or absence of RNase R supplementation. (B–F) H9c2 cells were treated with OGD along with transfection of circFoxo3 overexpressing vectors (OE circ) or empty vectors (NC) for 48 hours. (B) Relative level of circFoxo3 RNA was detected by qRT-PCR.25 Cell viability was determined by CCK-8 assay. (D) Cell apoptosis was determined by flow cytometry. (E) The levels of IL-6 and IL-10 in culture medium were checked by ELISA assay. **p < 0.01.
The Overexpression of circFoxo3 Relieves OGD-Induced Autophagy in vitro
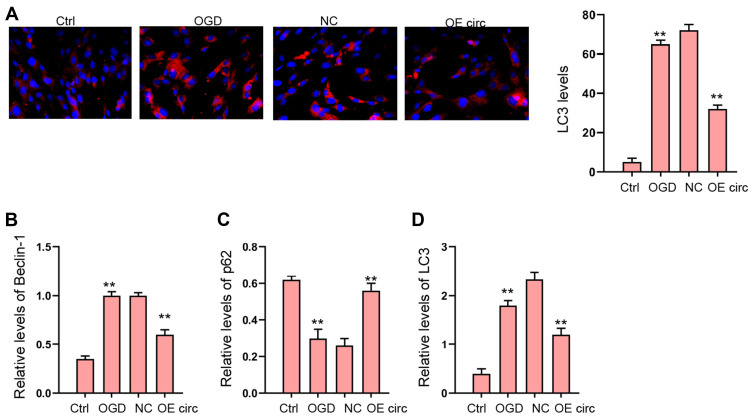
Next, we explored the effect of circFoxo3 on autophagy in OGD-treated H9c2 cells. Our data revealed that the expression of LC3 and Beclin-1 was induced and p62 expression was reduced in OGD-treated H9c2 cells, in which the overexpression of circFoxo3 reversed the phenotype (Figure 4A–D).
Figure 4.
The overexpression of circFoxo3 relieves OGD-induced autophagy in vitro. (A–D) H9c2 cells were treated with OGD along with transfection of circFoxo3 overexpressing vectors (OE circ) or empty vectors (NC) for 48 hours. (A) The levels of LC3 were measured by immunofluorescence. (B–D) The expression of Beclin-1, p62, and LC3 was detected by Western blot analysis. **p < 0.01.
The Overexpression of circFoxo3 Relieves OGD-Induced Autophagy in vitro
Next, we explored the effect of circFoxo3 on autophagy in OGD-treated H9c2 cells. Our data revealed that the expression of LC3 and Beclin-1 was induced and p62 expression was reduced in OGD-treated H9c2 cells, in which the overexpression of circFoxo3 reversed the phenotype (Figure 4A–D).
CircFoxo3 Reduces HMGB1 Expression by Inhibiting KAT7
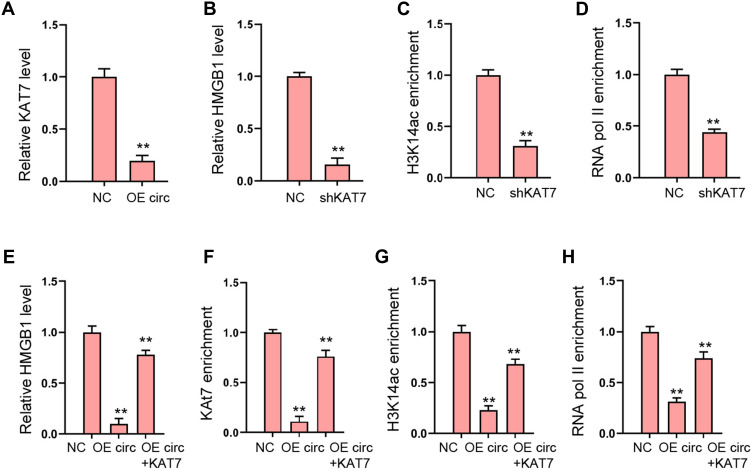
Then, we explored the potential mechanism underlying circFoxo3-mediated cardiomyocyte injury. We observed that the expression of KAT7 was reduced by circFoxo3 overexpression in H9c2 cells (Figure 5A). Meanwhile, the expression of HMGB1 was repressed by the depletion of KAT7 in H9c2 cells (Figure 5B). The enrichment of histone H3 lysine 14 acetylation (H3K14ac) and RNA polymerase II (RNA pol II) on HMGB1 promoter was inhibited by the knockdown of KAT7 (Figure 5C and D). Moreover, the overexpression of circFoxo3 suppressed HMGB1 expression and KAT7 overexpression rescued the expression of HMGB1 in H9c2 cells (Figure 5E). The enrichment of KAT7, H3K14ac, and RNA poly II on HMGB1 promoter was decreased by circFoxo3 overexpression, while the overexpression of KAT7 could reverse the effect (Figure 5F–H).
Figure 5.
CircFoxo3 reduces HMGB1 expression by inhibiting KAT7. (A) The expression of KAT7 was measured by qPCR in H9c2 cells treated with circFoxo3 overexpressing vectors. (B–D) The H9c2 cells were treated with KAT7 shRNA. (B) The expression of HMGB1 was measured by qPCR.25 The enrichment of H3K14ac on HMGB1 promoter was detected by ChIP assays. (D) The enrichment of RNA poly II on HMGB1 promoter was analyzed by ChIP assays. (E–H) The H9c2 cells were treated circFoxo3 overexpressing vectors, or co-treated with circFoxo3 overexpressing vectors and KAT7 overexpressing vectors. (E) The expression of HMGB1 was measured by qPCR. (F–H) The enrichment of KAT7, H3K14ac, and RNA poly II was measured by ChIP assays. **p < 0.01.
The Depletion of KAT7 Attenuates OGD-Induced Cardiomyocyte Injury in vitro
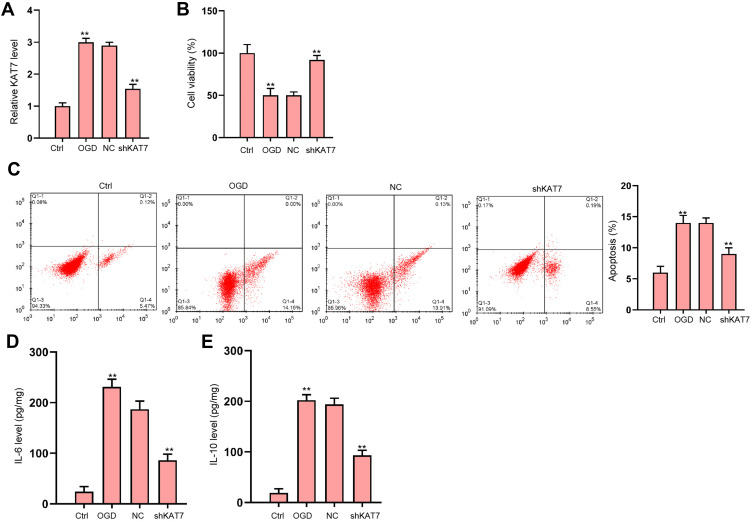
Next, we analyzed the role of KAT7 in vitro. The expression of KAT7 was increased in OGD-treated H9c2 cells and the effectiveness of KAT7 depletion was validated in the cells (Figure 6A). KAT7 depletion promoted cell viability (Figure 6B) and suppressed cell apoptosis (Figure 6C) under OGD stimulation. Besides, OGD treatment led to reduced secretion of inflammatory cytokine IL-6 and IL-10 (Figure 6D and E).
Figure 6.
The depletion of KAT7 attenuates OGD-induced cardiomyocyte injury in vitro. (A–E) H9c2 cells were treated with OGD along with transfection of KAT7 shRNA for 48 hours. (A) Relative level of KAT7 RNA in heart tissues was detected by qRT-PCR. (B) Cell viability was determined by CCK-8 assay.25 Cell apoptosis was determined by flow cytometry. (D and E) The levels of IL-6 and IL-10 in culture medium were checked by ELISA assay. **p < 0.01.
The Depletion of KAT7 Attenuates OGD-Induced Autophagy in vitro
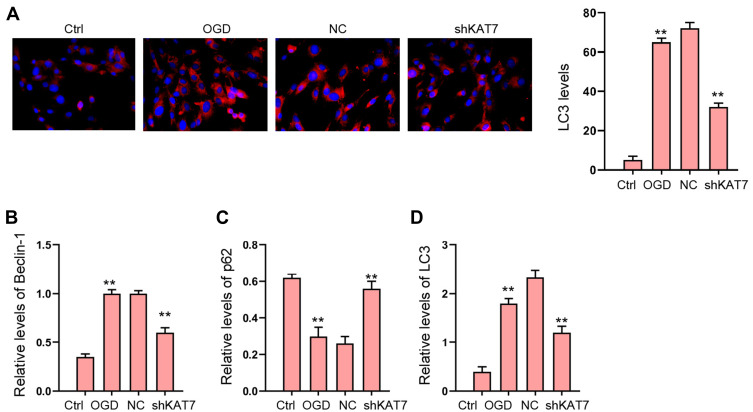
Next, we explored the effect of KAT7 on autophagy in OGD-treated H9c2 cells. Our data revealed that the expression of LC3 and Beclin-1 was induced and p62 expression was reduced in OGD-treated H9c2 cells, in which the knockdown of KAT7 reversed the phenotype (Figure 7A–D).
Figure 7.
The depletion of KAT7 attenuates OGD-induced autophagy in vitro. (A–D) H9c2 cells were treated with OGD along with transfection of KAT7 shRNA for 48 hours. (A) The levels of LC3 were measured by immunofluorescence. (B–D) The expression of Beclin-1, p62, and LC3 was detected by Western blot analysis. **p < 0.01.
CircFoxo3 Attenuates OGD-Induced Cardiomyocyte Injury and Autophagy by Targeting KAT7/HMGB1 Axis in vitro
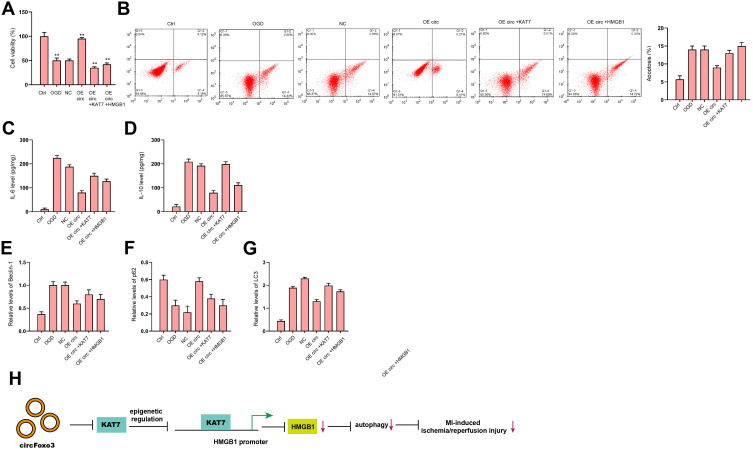
We next tried to validate the role of circFoxo3/KAT7/HMGB1 axis in the regulation of cardiomyocyte injury and autophagy. The overexpression of circFoxo3 enhanced cell viability (Figure 8A) and suppressed cell apoptosis (Figure 8B) under OGD stimulation, while the overexpression of KAT7 or HMGB1 could reverse this effect. The IL-6 and IL-10 secretions were repressed by the overexpression of circFoxo3 in OGD-treated H9c2 cells, in which the overexpression of KAT7 or HMGB1 rescued the levels of IL-6 and IL-10 (Figure 8C and D). The expression of LC3 and Beclin-1 was reduced and p62 expression was enhanced by circFoxo3 overexpression in OGD-treated H9c2 cells, in which the overexpression of KAT7 or HMGB1reversed the phenotypes (Figure 8E–G).
Figure 8.
CircFoxo3 attenuates OGD-induced cardiomyocyte injury and autophagy by targeting KAT7/HMGB1 axis in vitro. (A–G) H9c2 cells were treated with OGD along with transfection of circFoxo3 overexpressing vectors, or co-treated with circFoxo3 overexpressing vectors and KAT7 or HMGB1 overexpressing vectors. (A) Cell viability was determined by CCK-8 assay. (B) Cell apoptosis was determined by flow cytometry. (C and D) The levels of IL-6 and IL-10 in culture medium were checked by ELISA assay. (E–G) The expression of Beclin-1, p62, and LC3 was detected by Western blot analysis. (H) A diagram model of this study was shown. **p < 0.01.
Discussion
MI serves as a coronary artery-related heart disease, and the leading cause of mortality globally.1 CircRNAs are a new type of regulatory RNAs and participate in multiple pathological cardiac progression. Nevertheless, the role of circFoxo3 in MI-induced myocardial injury remains obscure. In the current study, we discovered the novel function of circFoxo3 in the modulation of myocardial ischemia/reperfusion injury.
As a critical pathological modulator, circRNAs are found to participate in the modulation of MI-related myocardial injury. It has been reported that Circular RNA CircFndc3b regulates heart repair by FUS/VEGF-A signaling in MI model.8 Circular RNA Ttc3 modulates cardiac dysfunction induced by MI by targeting miR-15b.16 Circular RNA ACR relieves cardiac ischemia/reperfusion damage by inhibiting autophagy through modulating the Pink1/FAM65B signaling.23 These reports demonstrate the crucial roles of circRNAs in the regulation of cardiac ischemia/reperfusion injury. But the understanding of the function of circRNAs in MI-induced cardiac ischemia/reperfusion injury is still elusive. In this work, we identified that circFoxo3 was downregulated in the MI rat model and the overexpression of circFoxo3 ameliorated MI-induced cardiac dysfunction and attenuated MI-induced autophagy in rat model. Meanwhile, the overexpression of circFoxo3 represses oxygen–glucose deprivation (OGD)-induced autophagy, apoptosis, inflammation, and injury of cardiomyocyte in vitro. These data present an innovative role of circFoxo3 in the inhibition of MI-induced cardiac dysfunction, providing crucial evidence for the important function of circRNAs in MI. Meanwhile, our study is consistent with the previous investigation that circRNAs play important roles in the progression of MI-induced cardiac ischemia/reperfusion injury.
The previous studies have identified the critical function of HMGB1 in the modulation of MI progression. It has been identified the inhibition of lncRNA TUG1 enhances miR-142-3p to attenuate myocardial injury during ischemia/reperfusion via targeting Rac1- and HMGB1-induced autophagy.24 Resolvin D1 represses MI by targeting HMGB1 pathway in a rodent model.25 TLR9 is required for HMGB1-regulated post-MI tissue repair by regulating angiogenesis, cardiac healing, and apoptosis.20 These reports reveal the important effect of HMGB1 on MI. In this investigation, we found that the expression of KAT7 was reduced by circFoxo3 overexpression in cardiomyocytes. Meanwhile, the expression of HMGB1 was repressed by the depletion of KAT7 in cardiomyocytes. The enrichment of histone H3 lysine 14 acetylation (H3K14ac) and RNA polymerase II (RNA pol II) on HMGB1 promoter was inhibited by the knockdown of KAT7. Moreover, the overexpression of circFoxo3 suppressed HMGB1 expression and KAT7 overexpression rescued the expression of HMGB1 in cardiomyocytes. The enrichment of KAT7, H3K14ac, and RNA poly II on HMGB1 promoter was decreased by circFoxo3 overexpression, while the overexpression of KAT7 could reverse the effect. We validated that the depletion of KAT7 attenuates OGD-induced cardiomyocyte injury and autophagy in vitro. The overexpression of KAT7 or HMGB1 could reverse circFoxo3-attenuated cardiomyocyte injury and autophagy in vitro. Our mechanism investigation reveals an unreported role of KAT7 in the MI progression, providing new mechanism of circFoxo3-mediated cardiomyocyte injury and autophagy involving KAT7/HMGB1 axis. There are some limitations of current study. We evaluated the effect of circFoxo3 on myocardial ischemia/reperfusion injury in vitro and in vivo but the clinical significance of circFoxo3 in myocardial ischemia/reperfusion injury should be confirmed in future investigations. Meanwhile, the KAT7/HMGB1 axis may be just one of the downstream mechanisms of circFoxo3-mediated myocardial ischemia/reperfusion injury and other potential mechanisms should be explored in the future.
In conclusion, we concluded that circular RNA circFoxo3 relieved myocardial ischemia/reperfusion injury by suppressing autophagy via inhibiting HMGB1 by repressing KAT7 in MI (Figure 8H). Our finding provides new insight into the mechanism by which circFoxo3 regulates MI-related cardiac dysfunction by targeting KAT7/HMGB1 axis.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Amani H, Habibey R, Hajmiresmail SJ, Latifi S, Pazoki-Toroudi H, Akhavan O. Antioxidant nanomaterials in advanced diagnoses and treatments of ischemia reperfusion injuries. J Mater Chem B. 2017;5:9452–9476. doi: 10.1039/C7TB01689A [DOI] [PubMed] [Google Scholar]
- 2.Anderson JL, Morrow DA. Acute myocardial infarction. N Engl J Med. 2017;376:2053–2064. doi: 10.1056/NEJMra1606915 [DOI] [PubMed] [Google Scholar]
- 3.Cabello JB, Burls A, Emparanza JI, Bayliss SE, Quinn T. Oxygen therapy for acute myocardial infarction. Cochrane Database Syst Rev. 2016;12:CD007160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed GW, Rossi JE, Cannon CP. Acute myocardial infarction. Lancet. 2017;389:197–210. doi: 10.1016/S0140-6736(16)30677-8 [DOI] [PubMed] [Google Scholar]
- 5.Whelan RS, Kaplinskiy V, Kitsis RN. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu Rev Physiol. 2010;72:19–44. doi: 10.1146/annurev.physiol.010908.163111 [DOI] [PubMed] [Google Scholar]
- 6.Chen WW, Gao RL, Liu LS, et al. China cardiovascular diseases report 2015: a summary. J Geriatr Cardiol. 2017;14:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garikipati VNS, Verma SK, Cheng Z, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. 2019;10:4317. doi: 10.1038/s41467-019-11777-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang S, Li X, Zheng H, et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S, Liao K, Miao Z, et al. CircFOXO3 promotes glioblastoma progression by acting as a competing endogenous RNA for NFAT5. Neuro Oncol. 2019;21:1284–1296. doi: 10.1093/neuonc/noz128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang T, Jiang HS, Zhang BT, Liu G. CircFOXO3 functions as a molecular sponge for miR-143-3p to promote the progression of gastric carcinoma via upregulating USP44. Gene. 2020;753:144798. doi: 10.1016/j.gene.2020.144798 [DOI] [PubMed] [Google Scholar]
- 12.Lin SP, Hu J, Wei JX, et al. Silencing of circFoxO3 protects HT22 cells from glutamate-induced oxidative injury via regulating the mitochondrial apoptosis pathway. Cell Mol Neurobiol. 2020;40:1231–1242. doi: 10.1007/s10571-020-00817-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou YL, Wu WP, Cheng J, et al. CircFOXO3 rs12196996, a polymorphism at the gene flanking intron, is associated with circFOXO3 levels and the risk of coronary artery disease. Aging. 2020;12:13076–13089. doi: 10.18632/aging.103398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu Y, Xie X, Lin L. circFOXO3 protects cardiomyocytes against radiation induced cardiotoxicity. Mol Med Rep. 2021;23:1. [DOI] [PubMed] [Google Scholar]
- 15.Ren X, Yu J, Guo L, Ma H. Circular RNA circRHOT1 contributes to pathogenesis of non-small cell lung cancer by epigenetically enhancing C-MYC expression through recruiting KAT5. Aging. 2021;13:20372–20382. doi: 10.18632/aging.203417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai L, Qi B, Wu X, et al. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol. 2019;130:10–22. doi: 10.1016/j.yjmcc.2019.03.007 [DOI] [PubMed] [Google Scholar]
- 17.Zhang M, Wang Z, Cheng Q, et al. Circular RNA (circRNA) CDYL induces myocardial regeneration by ceRNA after myocardial infarction. Med Sci Monit. 2020;26:e923188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson U, Yang H, Harris H. Extracellular HMGB1 as a therapeutic target in inflammatory diseases. Expert Opin Ther Targets. 2018;22:263–277. doi: 10.1080/14728222.2018.1439924 [DOI] [PubMed] [Google Scholar]
- 19.Vijayakumar EC, Bhatt LK, Prabhavalkar KS. High Mobility Group Box-1 (HMGB1): a potential target in therapeutics. Curr Drug Targets. 2019;20:1474–1485. doi: 10.2174/1389450120666190618125100 [DOI] [PubMed] [Google Scholar]
- 20.Liu FY, Fan D, Yang Z, et al. TLR9 is essential for HMGB1-mediated post-myocardial infarction tissue repair through affecting apoptosis, cardiac healing, and angiogenesis. Cell Death Dis. 2019;10:480. doi: 10.1038/s41419-019-1718-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng M, Tang Y, Li W, et al. The endotoxin delivery protein HMGB1 mediates caspase-11-dependent lethality in sepsis. Immunity. 2018;49:740–753 e7. doi: 10.1016/j.immuni.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang P, Wang L, Li Q, et al. Atorvastatin enhances the therapeutic efficacy of mesenchymal stem cells-derived exosomes in acute myocardial infarction via up-regulating long non-coding RNA H19. Cardiovasc Res. 2020;116:353–367. doi: 10.1093/cvr/cvz139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou LY, Zhai M, Huang Y, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2019;26:1299–1315. doi: 10.1038/s41418-018-0206-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu R, Li Z, Wang Q. Resolvin D1 attenuates myocardial infarction in a rodent model with the participation of the HMGB1 pathway. Cardiovasc Drugs Ther. 2019;33:399–406. doi: 10.1007/s10557-019-06884-y [DOI] [PubMed] [Google Scholar]
- 25.Bauza MDR, Gimenez CS, Locatelli P, et al. High-dose intramyocardial HMGB1 induces long-term cardioprotection in sheep with myocardial infarction. Drug Deliv Transl Res. 2019;9:935–944. doi: 10.1007/s13346-019-00628-z [DOI] [PubMed] [Google Scholar]