Abstract
Introduction
Breast cancer (BC) has become the malignant tumor with the highest incidence worldwide. As a critical components of epigenetic regulation complexes, chromobox (CBX) family members inhibit the transcription of target genes through chromatin modification, leading to the progression of various human diseases and cancers. So far, little is known about the role of different CBX members in BC, especially their association with immune cells.
Methods
We conducted the analysis of differential expression of CBXs using Oncomine and GEPIA, prognostic value of CBXs using GEPIA and Kaplan-Meier, genetic interaction of CBXs using cBioPortal and GeneMANIA, and immune cell infiltration of CBXs in BC patients using TIMER.
Results
The CBX2/3/4/8 expression levels were increased significantly, while the CBX6/7 expression levels were decreased. We found that CBX3 was significantly correlated with clinicopathological staging and short DFS in BC patients. High CBX3/5 expression was correlated with short OS in BC patients, while high expression of CBX4 was correlated with long OS in BC patients. In addition, the functions of CBXs family members mainly focus on methylated histone residue binding and chromatin organization. The CBXs expressions were closely related to the infiltration level of a variety of immune cells, including CD4/8+ T cells, B cells, neutrophils, macrophages and dendritic cells in BC cancers. The correlation between CBXs and immune cell infiltration was more common in Luminal BC than in Basal and Her-2 type.
Conclusion
This study may provide a new understanding for selection of molecular typing, therapeutic and prognostic biomarkers of CBX family in BC.
Keywords: comprehensive analysis, chromobox family members, breast cancer, prognosis, genetic interaction, immune infiltration
Introduction
CBX family members, typical components of polycomb repressive complexes 1 (PRC1), regulate heterochromatin,1 gene expression,2 and developmental programs, which involved in stem cell maintenance, cell differentiation, cell cycle regulation, cell aging, X chromosome inactivation, gene imprinting, occurrence and development of cancer. Up to now, the imbalance of CBXs is related to the occurrence, development, and recurrence of tumor by blocking differentiation of tumor stem cells and promoting their self-renewal in many cancers,3 such as BC,4 pancreatic adenocarcinoma,5 osteosarcoma,6 glioma7 and colon cancer.8
To date, eight CBX protein family in human genome have been identified, with conserved structure that contains a single N-terminal chromodomain. CBX proteins are further split into two groups based on molecular structure: HP1 group (includes CBX1/3/5) and Pc group (includes CBX2/4/6/7/8).9 Previous studies have found aberrant expressions and their prognostic values in some members of CBXs family. High expression of CBX4 was related to tumor size, pathological differentiation and poor survival, while the down-regulation of CBX7 was correlated with shorter OS in hepatocellular carcinoma (HCC) patients.10 High CBX3 expression was correlated with tumor size, vascular invasion, tumor recurrence, and poor survival in HCC patients. Moreover, CBX3 and other CBX family members (CBX1/5) are regulated by upstream miR-30a and target downstream genes (such as p21, CDK6, and CD44) to inhibit cell cycle and apoptosis, thereby promoting the growth, development, and invasion of liver cancer.11 However, the roles of different CBX members in the tumorigenesis and development of BC remain largely unknown and elusive.
In this study, our purpose was to determine the functional significance of CBX family members in BC by a variety of large databases. We analyzed different expression profiles, clinicopathological characteristics, prognostic values (including OS and DFS), CBXs gene alteration and interaction. We further explored the association between CBXs expression and immune cell infiltration. Our findings indicate that CBX may have complex and unique functions in the progression of BC.
Materials and Methods
Oncomine Database
Oncomine is a data platform that provides powerful genome expression analysis.12 It is used to analyze the mRNA level expression of CBX family members in BC and compare with normal breast tissue. In our study, the significance threshold was set as p < 0.01, a fold change ≥2 between cancer and normal tissues, and gene rank ≥ top 10%.
GEPIA
GEPIA is a new analytical tool based on TCGA and GTEx data,13 which can provide RNA expression data of thousands of tumors and normal tissue samples. In our study, GEPIA was used to analyze the expression of CBXs members and pathological staging in BC patients, and to determine the relationship between the expression and prognosis of BC. Student’s t test was utilized for statistical difference analysis of expression or pathological stage. Kaplan Meier curve was used to draw survival map. p < 0.05 was considered statistically significant.
The Kaplan-Meier Plotter
The Kaplan-Meier Plotter is a tool that uses GEO, EGA and TCGA data to analyze the association between gene expression and prognosis of tumor patients, and finally draw survival maps.14 In this study, we evaluated the correlation between CBXs mRNA expression and clinical endings of BC. All results (the best cut-off value, log-rank P value, HR, and 95% CIs) are automatically analyzed and presented by the website. p < 0.05 was considered statistically significant.
cBioPortal
cBioPortal is a platform based on TCGA database, which provides multi-dimensional and visual interactive exploration of genomics.15 We used it to obtain the information of genetic alterations and distribution of CBXs.
GeneMANIA
GeneMANIA is a database used to generate hypotheses about gene function, analyze gene lists and determine gene priorities for functional analysis.16 It can be used to discover the interaction between two or more genes and the interaction between proteins, including protein-protein, protein-DNA, and genetic interaction, pathways, and genes physiological and biochemical translation, gene and protein expression, protein domain and phenotype screening. The gene interaction networks were identified using the GeneMANIA web server.
STRING
STRING is a database searching for protein-protein interactions,17 which includes the direct physical interaction between proteins and the indirect functional correlation between proteins. We used the unique computing method of STRING to obtain the different expression and interaction analysis of CBX through PPI network analysis.
TIMER
TIMER is a database of immune invasion, which uses RNA-seq expression profile to detect the infiltration of immune cells in tumor tissues.18 It provides the infiltration of six kinds of immune cells (CD4+ T cells, CD8+ T cells, B cells, neutrophils, macrophages and dendritic cells). We used TIMER to generate scatter plots to visualize the correlation between CBXs expression and BC immune infiltration.
Results
Differences in mRNA Expression of CBX Members in BC Patients
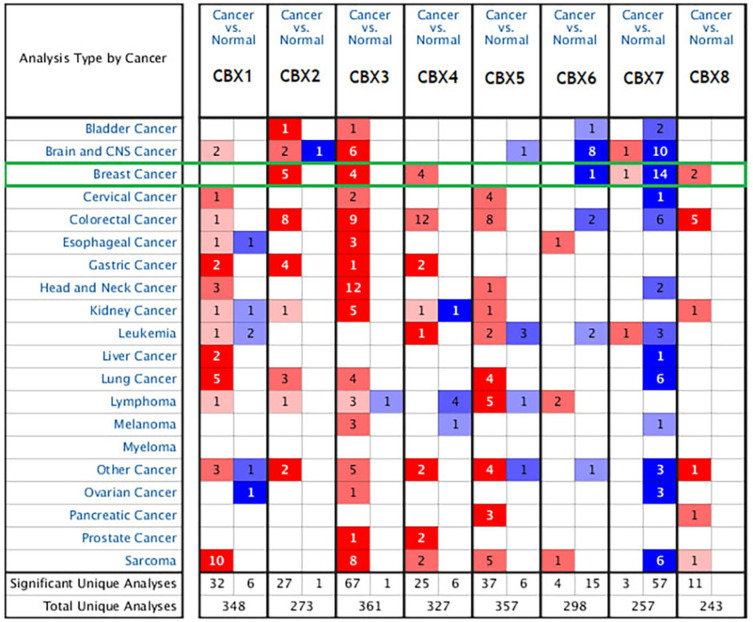
In order to study the expression differences of CBX members in BC patients, we used Oncomine data platform to analyze their transcription level expression (Figure 1). The data analysis results showed that the mRNA expression of CBX2/3/4/8 in BC tissues was increased significantly, while the mRNA expression of CBX6/7 was decreased than that in normal tissues. In detail, compared with normal tissues, the mRNA levels of CBX2/3/4/8 were overexpressed in BC tissues, with a fold change of 9.378 (p = 1.25E-15), 2.328 (p = 5.73E-04), 2.253 (p = 1.62E-14) and 2.048 (p = 1.10E-16), respectively. (Supplementary Table 1).
Figure 1.
The mRNA expression of CBX family proteins in different cancers in Oncomine. Red indicates increased expression and blue indicates decreased expression.
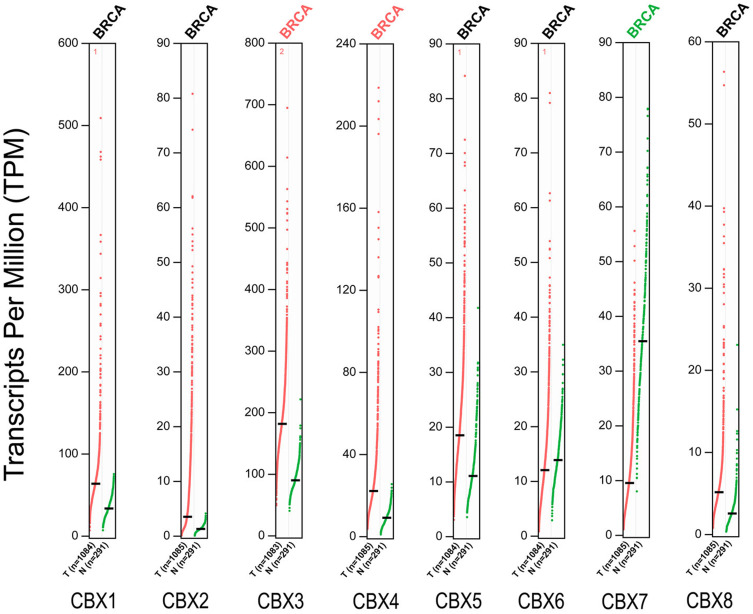
To further verify the expression differences of CBX family members, we used GEPIA data platform to compare mRNA expression of CBXs in BC and normal tissues. Our results showed that the expression levels of CBX3/4 in BC tissues were higher than those in normal tissues, while the expression level of CBX7 was lower than that in normal breast tissues (Figure 2). Together, these results were similar to those of Oncomine.
Figure 2.
The mRNA expression of CBX family proteins in BC in GEPIA. Red indicates increased expression and green indicates decreased expression.
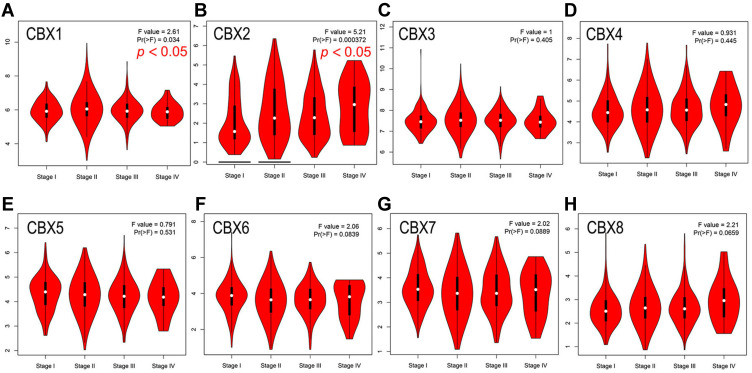
Then we evaluated the association between the differential expression of CBXs and the clinicopathological staging of BC patients. CBX2 group was highly correlated, followed by CBX1 group also has a high correlation, while the remaining six groups were not significantly different (Figure 3). All these suggest that CBX2, together with CBX1, may play an important role in the occurrence and development of BC.
Figure 3.
Correlation between CBXs expression and clinicopathological staging of BC in GEPIA. (A–H). Correlation between CBX1 (A), CBX2 (B), CBX3 (C), CBX4 (D), CBX5 (E), CBX6 (F), CBX7 (G), CBX8 (H) and clinicopathological staging of BC determined by GEPIA. CBX2 was highly correlated, followed by CBX1 also has a high correlation, while the remaining six groups were not significantly different.
Prognostic Evaluation of CBXs mRNA Expression in BC Patients
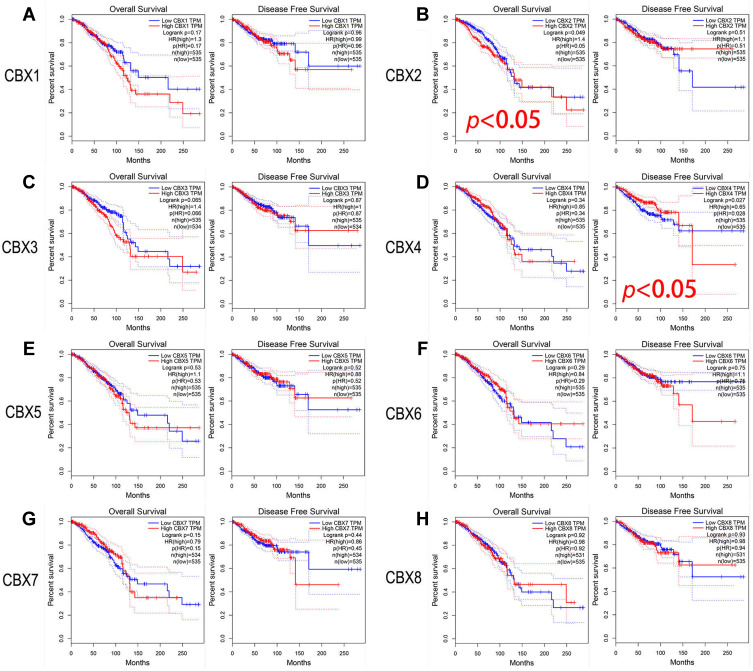
To assess the value of differential expression of CBXs in BC development, we used GEPIA to analyze the correlation between different CBXs and clinical outcomes. The curves of OS and DFS are shown in Figure 4. We found the high level of CBX2 mRNA expression in BC patients was significantly correlated with OS (p = 0.049), and the high level of CBX4 mRNA expression in BC patients was significantly correlated with DFS (p = 0.027). In addition, the remaining CBX members do not appear to have a significant impact on DFS or OS.
Figure 4.
Prognostic evaluation of CBXs mRNA expression in BC patients (GEPIA). (B). High level of CBX2 mRNA expression in BC patients was significantly correlated with OS. (D). High level of CBX4 mRNA expression in BC patients was significantly correlated with DFS. CBX1 (A), CBX3 (C), CBX5 (E), CBX6 (F), CBX7 (G) and CBX8 (H) mRNA expression in BC patients were not associated with OS or DFS.
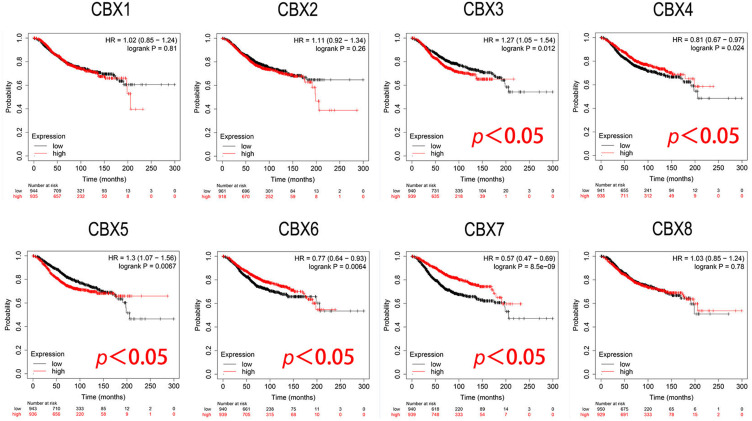
Furthermore, we conducted the analysis of prognostic value of CBXs in BC patients using Kaplan-Meier plotter. As shown in the subgroup analysis in Figure 5, the high expression of CBX3 (HR = 1.27, p = 0.012) and CBX5 (HR = 1.3, p = 0.0067) mRNA in BC patients was significantly correlated with short OS. However, the high expression of CBX4 (HR = 0.81, p = 0.024), CBX6 (HR = 0.77, p = 0.0064) and CBX7 (HR = 0.57, p = 8.5e-09) were significantly associated with long OS.
Figure 5.
Prognostic evaluation of CBXs mRNA expression in BC patients (Kaplan-Meier plotter). High expression of CBX3/5 mRNA was significantly correlated with short OS, and high expression of CBX4/6 and CBX7 were significantly associated with long OS in BC patients.
Analysis of CBXs Gene Alteration and Interaction in BC Patients
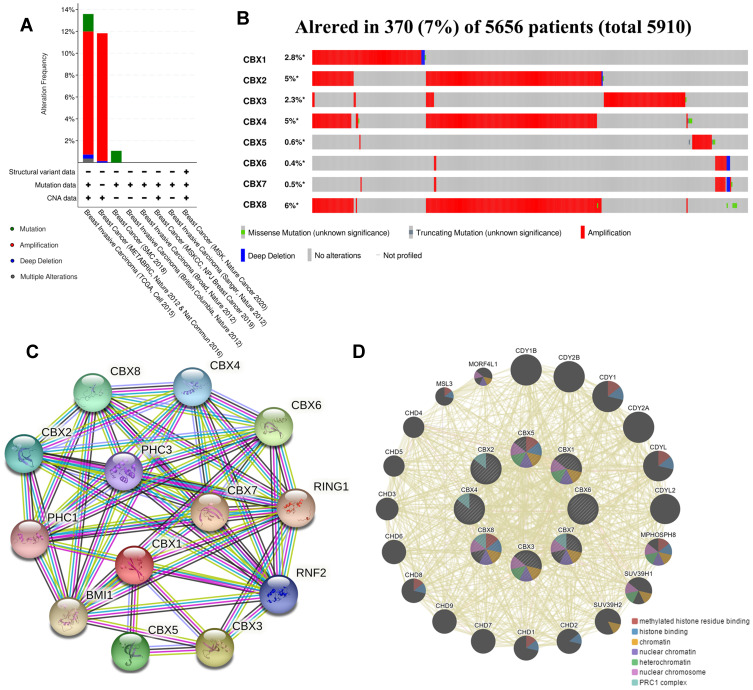
We used cBioPortal online website to analyze genetic alterations of CBXs in BC patients. In general, two or more changes were detected in BC of different groups. Gene amplification caused by various causes was more common in BC samples (Figure 6A). Among the 5656 cases of BC patients, 370 CBXs have altered, accounting for 7% (Figure 6B). Furthermore, in all BC samples, the incidence of CBX1/2/3/4/5/6/7/8 alterations were 2.8%, 5%, 2.3%, 5%, 0.6%, 0.4%, 0.5% and 6%, respectively.
Figure 6.
Gene alteration and interaction of CBXs in BC patients. (A, B) Gene alteration and expression of CBXs in BC patients (cBioPortal). (C, D) Analysis of protein-protein interaction of CBXs (STRING and GeneMANIA).
In addition, we conducted PPI network analysis on differentially expressed CBX to explore their potential interactions using STRING. As shown in Figure 6C, a plurality of nodes and edges are obtained in the PPI network. CBXs, together with PHC1, PHC3, RING1, RNF2 and BMI1, are involved in regulating the development of embryos, pluripotent stem cells and tumors through various signaling pathways. Further GeneMANIA analysis showed that the functions of CBXs and their related molecules (such as CDY1, CDY1B, CDYL, CDY2B, CDYL2, CDY2A, SUV39H1 and SUV39H2) mainly focused on methylated histone residue binding, histone binding, chromatin, nuclear chromatin heterochromatin, PRC1 complex and nuclear chromosome (Figure 6D).
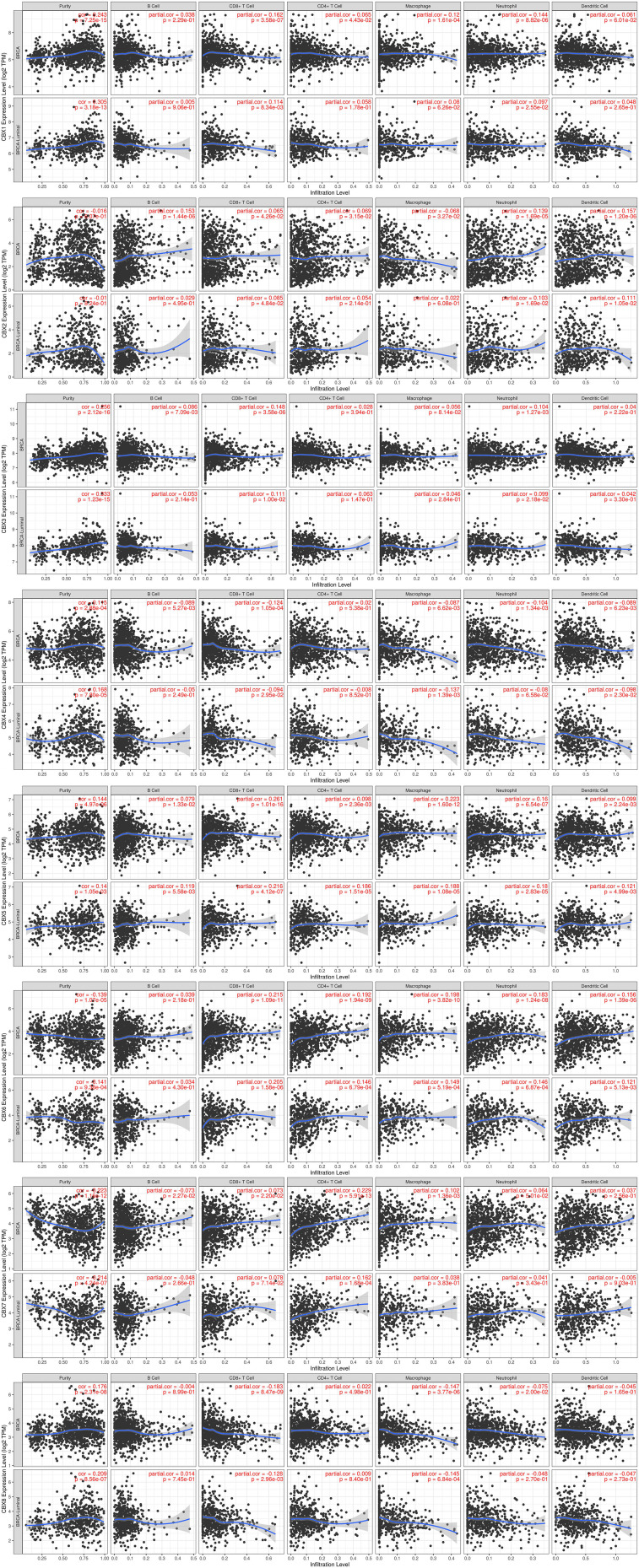
Correlation Between Immune Cell Infiltration and CBXs in BC Patients
The level of immune cell infiltration is considered to be closely related to the proliferation and progression of cancer cells. Thus, we further explored the association between CBXs expression and immune cell infiltration using TIMER (Figure 7 and Supplemental Figure S1). The CBX1 expression was related to the infiltration of CD4/8+ T cells, macrophages and neutrophils in BC. In detail, the expression of CBX1 was related to CD8+ T cells and neutrophils in Luminal type. Only neutrophil infiltration was related to the expression of CBX1 in Basal type, while no correlation was seen in the Her-2 type. The infiltration of all six host cells was closely correlated with the expression of CBX2, but not in Her-2 type. In Luminal type, the expression of CBX2 was associated with the infiltration of CD8+ T cells, dendritic cells and neutrophils. The expression of CBX2 was correlated with the infiltration of B cells and CD8+ T cells in Basal type. The CBX3 expression was positively correlated with B cell, CD8+ T cells and neutrophils in BC, while it was related to the infiltration of CD8+ T cells and neutrophils in Luminal type. No immunocyte infiltration was obviously related to CBX3 expression in Basal type and Her-2 type. CBX4 was negatively correlated with the infiltration of other immune cells except CD4+ T cells in BC, and it was also negatively correlated with the infiltration of CD8+ T cells, macrophages and dendritic cells in Luminal type. Similar to CBX3, CBX4 was not significantly associated with any immune cell infiltration in Basal and Her-2 subtypes. CBX5 expression was positively correlated with all six immunocytes in BC, which was also true in the Luminal type. Only CD8+ T cells was associated with CBX5 expression in the Basal type, while no relevance was found in Her-2 type. For both Luminal and BC, except for B cells, the infiltration of other immune cells was positively correlated with the CBX6 expression. The infiltration of CD4+ T cells, dendritic cells and neutrophils was positively correlated with CBX6 expression in Basal type, but there was no correlation in HER-2 type. With regard to CBX7, B cells, CD4/8+ T cells and macrophages had a statistical correlation in BC, while the infiltration of CD4+ T cells, B cells, dendritic cells and neutrophils have a positive correlation with CBX7 in Basal type. Only the CD4+ cells infiltration was positively correlated with the expression of CBX7 in the other two subtypes. For CBX8, it was negatively correlated with the infiltration of B cells in Her-2 type, the infiltration of macrophages and CD8+ T cells in Luminal type, the infiltration of CD+8 T cells, macrophages and neutrophils in BC. Obviously, the correlation between CBXs and immune cell infiltration was more common in Luminal BC than in Basal and Her-2 type.
Figure 7.
Correlation between immune cell infiltration and CBXs in BC patients and Luminal BC determined by TIMER.
Discussion
BC is a complex disease due to the accumulation of different epigenetic changes and gene mutations,19 which considered to be one of the important hallmarks of tumors.20 In BC, the alterations of oncogene and tumor suppressor gene expression levels are mainly controlled by epigenetic mechanisms, especially histone modification and DNA methylation.21 CBX family members, a critical components of epigenetic regulation complexes, inhibit the transcription of target genes through chromatin modification, leading to the progression of various human diseases and cancers. CBXs have been shown to play critical roles in colorectal cancer22 and HCC.23 However, the roles of different CBX members in the tumorigenesis and development of BC need to be clarified. We comprehensively analyzed CBXs in BC from the aspects of expression, gene alteration, prognostic value and immune cell infiltration.
CBX3, a member of the CBX family proteins, is proved to be involved in human tumorigenesis and patient prognosis, but the exact function of CBX3 in tumor is not clear. CBX3 is proven to promote tumor proliferation and predict poor survival in HCC.11 Similarly, CBX3 was overexpressed in tongue squamous cell carcinoma (TSCC), which play a key role in tongue tumorigenesis by regulating GS/S phase via p21 pathway,24 was an independent prognostic factor in TSCC patients.25 In colorectal adenocarcinoma (COAD), CBX3 was validated as a target for miR-375, and its overexpression could promote the malignant behaviors of COAD cells by lncRNA-SNHG17/miR-375/CBX3 axis.26 Similar to the results of the TCGA data, CBX3 was detected to be elevated in non-small cell lung cancer (NSCLC). Interestingly, there was a highly significant association between the expression of CBX3 and EGFR mutations in NSCLC patients.27 Inhibition of EGFR signal did not change the expression level of CBX3 in NSCLC cell, suggesting that the expression of CBX3 may be independent of EGFR downstream signals. Overexpression of CBX3 in gastric cancer (GC) cells significantly promoted its malignant phenotype and participated in the regulation of genes related to cell cycle, mismatch repair and immune related pathways. The CBX3 expression was negatively associated with the abundance of tumor infiltrating lymphocytes and the response to immunotherapy.28 So far, there is little literature on the role of CBX3 in BC. Our findings suggest that CBX3 contributes to BC progression and is expected to serve as a potential prognostic biomarker and therapeutic target in BC.
The increased expression of CBX5 in GC tissue means its carcinogenic effect. It is considered to promote the proliferation, metastasis and infiltration of GC cells, and can be regulated by microRNA-758-3p,29 which also indicates that CBX5 may become a therapeutic target. In addition, CBX5 can also promote tumor proliferation and metastasis through LOXL1-AS1/miR-589-5p pathway in renal cell carcinoma.30 In glioblastoma, CBX5 can activate and enhance DNA damage repair pathway, promote DNA repair, thus repair DNA damage induced by temozolomide,31 eventually lead to chemotherapy resistance and reduce the effect of chemotherapy.32 Similar to our results, CBX5 was found to be associated with triple-negative breast cancer (TNBC). CBX5 silencing can inhibit cell proliferation and metastasis, while overexpression promotes the proliferation and metastasis of TNBC cells, and can be regulated by miR-2335-5p/lncRNA SNHG11 signaling pathway.33 These results together indicate that CBX5 plays a carcinogenic role in many cancers including BC.
We also conducted the analysis of gene alteration and interaction of the CBXs in BC patients. In addition, predicted functions and pathways of the mutations analysis demonstrated CBXs, together with PHC1, PHC3, RING1, RNF2 and BMI1, are involved in regulating the development of embryos, pluripotent stem cells and tumors through various signaling pathways. Further GeneMANIA analysis showed that the functions of CBXs family and their related molecules mainly focused on methylated histone residue binding, histone binding, chromatin, nuclear chromatin heterochromatin, nuclear chromosome and PRC1 complex. These together prove that the CBX family is involved in the development and progress of BC.
It is now clear that, in addition to cancer cells, the tumor microenvironment also contains a series of immune cells,20 which help cancer cells escape immune supervision through a series of mechanisms, and eventually lead to tumor. Tumor associated immune cells may have anti-tumor or tumor-promoting functions. The tumor-antagonizing immune cells mainly consist of CD4/8+ T cells, natural killer cells, macrophages and neutrophils. Our research demonstrated that differentially expressed CBXs might be significantly associated with infiltration of immune cell types, and there are differences in the correlation between CBXs and immune cells in different BC types, indicating CBXs may be a biomarker for molecular typing of BC. This study may provide more information for CBXs to become a therapeutic molecular target and biomarker.
Conclusion
In general, we comprehensively analyzed the expression difference, prognostic value and association with immune cell infiltration of CBX family members in BC. The CBX2/3/4/8 expression levels were increased significantly, while the CBX6/7 expression levels were decreased in BC tissues. We found that CBX3 was significantly correlated with clinicopathological staging and short DFS in BC patients. High mRNA expression of CBX3/5 was correlated with short OS in BC patients. While high mRNA expression of CBX4 was correlated with long OS in BC patients. In addition, the functions of differentially expressed CBXs mainly focus on methylated histone residue binding and chromatin organization. The differentially expression of CBXs were closely related to the infiltration level of a variety of immune cells, including CD4+ T cells, CD8+ T cells, B cells, neutrophils, macrophages and dendritic cells in BC cancers. The correlation between CBXs and immune cell infiltration was more common in Luminal BC than in Basal and Her-2 type. This study may provide a new understanding for selection of molecular typing, therapeutic and prognostic biomarkers of CBX family in BC.
Acknowledgments
We sincerely thank the contributors to the database.
Abbreviations
BC, breast cancer; CBX, Chromobox; GEPIA, gene expression profiling interactive analysis; DFS, disease-free survival; OS, overall survival; PRC1, polycomb repressive complexes 1; HCC, hepatocellular carcinoma; TSCC, tongue squamous cell carcinoma; COAD, Colorectal adenocarcinoma; NSCLC, non-small cell lung cancer; GC, gastric cancer; TNBC, triple-negative breast cancer.
Ethics Approval and Consent to Participate
Our study was approved by the medical ethics committee of the Second Affiliated Hospital of Xi’an Jiaotong University and met the requirements of its medical ethics.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Strom AR, Biggs RJ, Banigan EJ, et al. HP1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. Elife. 2021;10:e36972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kloetgen A, Duggimpudi S, Schuschel K, et al. YBX1 indirectly targets heterochromatin-repressed inflammatory response-related apoptosis genes through regulating CBX5 mRNA. Int J Mol Sci. 2020;21:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Wijnen AJ, Bagheri L, Badreldin AA, et al. Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer and development. Bone. 2021;143:115659. [DOI] [PubMed] [Google Scholar]
- 4.Iqbal MA, Siddiqui S, Ur Rehman A, et al. Multiomics integrative analysis reveals antagonistic roles of CBX2 and CBX7 in metabolic reprogramming of breast cancer. Mol Oncol. 2021;15(5):1450–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen LY, Cheng CS, Qu C, et al. CBX3 promotes proliferation and regulates glycolysis via suppressing FBP1 in pancreatic cancer. Biochem Biophys Res Commun. 2018;500(3):691–697. [DOI] [PubMed] [Google Scholar]
- 6.Ma C, Nie XG, Wang YL, et al. CBX3 predicts an unfavorable prognosis and promotes tumorigenesis in osteosarcoma. Mol Med Rep. 2019;19(5):4205–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao SP, Wang F, Yang M, et al. CBX3 promotes glioma U87 cell proliferation and predicts an unfavorable prognosis. J Neurooncol. 2019;145(1):35–48. [DOI] [PubMed] [Google Scholar]
- 8.Fan Y, Li H, Liang X, Xiang Z. CBX3 promotes colon cancer cell proliferation by CDK6 kinase-independent function during cell cycle. Oncotarget. 2017;8(12):19934–19946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wotton D, Merrill JC. Pc2 and SUMOylation. Biochem Soc Trans. 2007;35(Pt 6):1401–1404. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Tang J, Liao D, et al. Chromobox homolog 4 is correlated with prognosis and tumor cell growth in hepatocellular carcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S684–692. [DOI] [PubMed] [Google Scholar]
- 11.Zhong X, Kan A, Zhang W, et al. CBX3/HP1γ promotes tumor proliferation and predicts poor survival in hepatocellular carcinoma. Aging (Albany NY). 2019;11(15):5483–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–w102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koboldt DC, Fulton R, McLellan Met al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 21.Selvakumar P, Badgeley A, Murphy P, et al. Flavonoids and other polyphenols act as epigenetic modifiers in breast cancer. Nutrients. 2020;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Pan Y, Cao Z, Zhao S. Comprehensive analysis of prognostic value and immune infiltration of chromobox family members in colorectal cancer. Front Oncol. 2020;10:582667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ning G, Huang YL, Zhen LM, et al. Transcriptional expressions of Chromobox 1/2/3/6/8 as independent indicators for survivals in hepatocellular carcinoma patients. Aging (Albany NY). 2018;10(11):3450–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Chen W, Fu X, Su X, Yang A. CBX3 promotes tumor proliferation by regulating G1/S phase via p21 downregulation and associates with poor prognosis in tongue squamous cell carcinoma. Gene. 2018;654:49–56. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Fu X, Su X, Yang A. CBX3/HP1γ is upregulated in tongue squamous cell carcinoma and is associated with an unfavorable prognosis. Exp Ther Med. 2018;15(5):4271–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Zhan Y, Wang J, Wang J, Guo J, Kong D. lncRNA-SNHG17 promotes colon adenocarcinoma progression and serves as a sponge for miR-375 to regulate CBX3 expression. Am J Transl Res. 2020;12(9):5283–5295. [PMC free article] [PubMed] [Google Scholar]
- 27.Chang SC, Lai YC, Chen YC, Wang NK, Wang WS, Lai JI. CBX3/heterochromatin protein 1 gamma is significantly upregulated in patients with non-small cell lung cancer. Asia Pac J Clin Oncol. 2018;14(5):e283–e288. [DOI] [PubMed] [Google Scholar]
- 28.Lin H, Lian J, Xia L, Guan G, You J. CBX3 promotes gastric cancer progression and affects factors related to immunotherapeutic responses. Cancer Manag Res. 2020;12:10113–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J, Zhang Z, Pan L, Zhou Y. Identification of miR-758-3p as potential modulator of CBX5 expression in gastric cancer. Technol Cancer Res Treat. 2018;17:1533033818816061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Wu C, Zhang J. Long non-conding RNA LOXL1-AS1 sponges miR-589-5p to up-regulate CBX5 expression in renal cell carcinoma. Biosci Rep. 2020;40:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YH, Kuo CY, Stark JM, Shih HM, Ann DK. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res. 2013;41(11):5784–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi GZ, Huang G, Guo M, et al. Acquired temozolomide resistance in MGMT-deficient glioblastoma cells is associated with regulation of DNA repair by DHC2. Brain. 2019;142(8):2352–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu L, Zhang W, Wang P, et al. LncRNA SNHG11 aggravates cell proliferation and migration in triple-negative breast cancer via sponging miR-2355-5p and targeting CBX5. Exp Ther Med. 2021;22(2):892. [DOI] [PMC free article] [PubMed] [Google Scholar]