Dodiya et al. characterize an APPPS1-21 mouse model of Aβ amyloidosis with an antibiotic-perturbed microbiome. Fecal transplantation approaches and microglial depletion studies are employed to establish the causality between the gut microbiota, microglia, Aβ, and neurodegeneration.
Abstract
We previously demonstrated that lifelong antibiotic (ABX) perturbations of the gut microbiome in male APPPS1-21 mice lead to reductions in amyloid β (Aβ) plaque pathology and altered phenotypes of plaque-associated microglia. Here, we show that a short, 7-d treatment of preweaned male mice with high-dose ABX is associated with reductions of Aβ amyloidosis, plaque-localized microglia morphologies, and Aβ-associated degenerative changes at 9 wk of age in male mice only. More importantly, fecal microbiota transplantation (FMT) from transgenic (Tg) or WT male donors into ABX-treated male mice completely restored Aβ amyloidosis, plaque-localized microglia morphologies, and Aβ-associated degenerative changes. Transcriptomic studies revealed significant differences between vehicle versus ABX-treated male mice and FMT from Tg mice into ABX-treated mice largely restored the transcriptome profiles to that of the Tg donor animals. Finally, colony-stimulating factor 1 receptor (CSF1R) inhibitor-mediated depletion of microglia in ABX-treated male mice failed to reduce cerebral Aβ amyloidosis. Thus, microglia play a critical role in driving gut microbiome–mediated alterations of cerebral Aβ deposition.
Introduction
Increasing evidence from studies of the microbiota–brain axis has suggested that the gut microbiome plays a critical role in neurodevelopment, behavior, neuroinflammation, and potentially neurodegeneration (Diaz Heijtz et al., 2011; Braniste et al., 2014; Sharon et al., 2016). A variety of studies in preclinical animal models have offered support for the notion that a dysfunctional microbiota–brain axis might play an important role in brain disorders that include Alzheimer’s disease (AD; Dodiya et al., 2019; Minter et al., 2016; Vogt et al., 2017), Parkinson’s disease (Dodiya et al., 2020; Sampson et al., 2016), multiple sclerosis (Berer et al., 2011; Cekanaviciute et al., 2017), brain injury, stroke (Singh et al., 2016), and others. In preceding efforts, we provided substantial evidence supporting a strong connection between the gut microbiome and deposition of Aβ peptides and altered microglial phenotypes in mouse models that express familial AD-linked variants of APP and PS1 (Dodiya et al., 2019; Minter et al., 2017, 2016). Specifically, APPSWE/PS1ΔE9 (Jankowsky et al., 2001) and APPPS1-21 (Radde et al., 2006) mice treated with antibiotics (ABX) result in gut microbiome perturbations that are associated with reductions in amyloid β (Aβ) deposition and altered microglial transcriptional profiles that, surprisingly, are specific to male animals (Minter et al., 2016, 2017; Dodiya et al., 2019). Most importantly, when APPPS1-21 male mice treated with ABX throughout their lifetime were subjected to fecal microbiota transplantation (FMT) from naive age-matched APPPS1-21, and hence restored endogenous microbiota, we observed a partial restoration of Aβ deposition, thus establishing causality (Dodiya et al., 2019). The exact mechanism(s) by which the gut microbiome influences amyloidosis is not understood, but in view of our studies showing that gut microbiome changes parallel morphological and transcriptional alterations in microglia, it is not inconceivable that microglia play an important role in modulating AD-like phenotypes that are driven by the gut microbiota (Dodiya et al., 2019; Minter et al., 2017, 2016).
Microglia are brain-resident macrophages that originate from the yolk sac and enter the rudimentary brain before the formation of other neuroglial cells during embryonic stages (embryonic day 9.5 in mice). These cells expand and self-renew in adult life (Erny and Prinz, 2020). Besides their important immune functions in protection following infection or injury, these cells participate in embryonic wiring, synaptic transmission, and synaptic pruning and formation, as well as cell death (reviewed in Hong et al., 2016; Ransohoff and El Khoury, 2015; Reemst et al., 2016; Schafer and Stevens, 2015; Tay et al., 2017). Microglia use an array of cellular receptors and signaling molecules, collectively termed the “sensome,” to detect endogenous brain stimuli (such as protein aggregation or cellular injury) and exogenous peripheral stimuli (invasion of pathogenic microorganisms or their products) during adult life (Hickman et al., 2013). Some of these sensome genes (i.e., Clec7a, Itgb2, Icam4, Cd52, Cd79B, Cd101, Lgals9, Ptprc, and others) were also found to be expressed by microglia during the progenitor and embryonic phases, and the majority of the sensome genes showed highest expression in adults (Thion et al., 2018). Furthermore, compared with microglia from specific pathogen–free mice, germ-free (GF) microglia exhibit completely different transcriptome profiles and histological features that are reminiscent of immature microglia (Erny et al., 2015; Thion et al., 2018), findings that have been replicated in mice treated with ABX to generate “pseudo–GF” mice (Erny et al., 2015).
It is now well established that the microbiome can have a significant influence on Aβ deposition in mouse models (Dodiya et al., 2019; Harach et al., 2017; Minter et al., 2016), and studies have shown clear differences in microbiota profiles between normal healthy individuals and patients with AD (Vogt et al., 2017). Microglia, in conjunction with other glial cells, play a critical role in Aβ phagocytosis and clearance that influences amyloidosis and proinflammatory responses to Aβ deposits (Hansen et al., 2018). Recent evidence suggests that microglia under chronic activation phase (also termed disease-associated microglia [DAM] or neurodegenerative type microglia [MGnD]) can become detrimental and drive disease spread instead of disease clearance (Butovsky et al., 2014; Keren-Shaul et al., 2017; Mass et al., 2017). Most importantly, the common late-onset AD risk factors associated with rare variants of immune receptors are expressed by microglial cells (Guerreiro and Hardy, 2014; Tanzi, 2012). Additionally, depletion of microglia in several transgenic (Tg) and adeno-associated virus (AAV)–overexpresssion models (AAV-GFP/tau + clodronate and AAV-GFP/tau + colony-stimulating factor 1 receptor [CSF1R] inhibitor [PLX3397; Asai et al., 2015], APPPS1-21;CX3CR1−/− and R1.40;CXCR1−/− [Lee et al., 2010], APPSWE/PS1DE9+CSF1R inhibitor [GW2580; Olmos-Alonso et al., 2016], P301S+CSF1R-inhibitor [JNJ-527; Mancuso et al., 2019], 5XFAD+CSF1R inhibitor [PLX3397; Sosna et al., 2018], and 5XFAD+CSF1R-inhibitor [PLX5622; Spangenberg et al., 2019]) have been shown to reduce pathology and improve cognitive phenotypes. However, despite the significant evidence of microbiome–microglia interactions and microglial involvement in AD pathogenesis, the microbiome–microglia axis in disease remains poorly characterized.
In our earlier efforts, we demonstrated that FMT only leads to a partial restoration of Aβ amyloidosis in animals treated with long-term ABX (Dodiya et al., 2019), and this is likely due to persistent levels of ABX that could have eliminated critical bacterial species during daily FMT treatments. In view of our earlier demonstration that a short-term, postnatal exposure of ABX regimen was sufficient to reduce Aβ burden in APPSWE/PS1ΔE9 mice (Minter et al., 2017), we chose to repeat our studies in APPPS1-21 mice using the short-term ABX paradigm. The rationale for the current studies was the body of evidence suggesting that the commensal microbiome perturbation during the postnatal developmental time frame represents a crucial developmental window by which microbiota–host interactions mediate immuno- and neurodevelopment that may impact host physiology in later life (Cox et al., 2014; Hansen et al., 2013, 2012; Tognini, 2017). To investigate the role of alterations of the microbiome in early life on microgliosis and Aβ amyloidosis, we treated APPPS1-21 mice with ABX from postnatal day (PND) 14 to PND21, then housed the weaned mice with drinking water without any added ABX till the time of sacrifice.
We now report that early-life, postnatal ABX treatment results in sex-specific microbiome alterations that are associated with reduced extracellular deposition of Aβ, reduced levels of formic acid (FA)–insoluble Aβ peptides, alterations in the morphology of plaque-associated microglia, and Aβ-associated neurodegenerative changes that are specific to male APPPS1-21 mice. Similar to our previous findings in long-term ABX-treated APPPS1-21 mice (Dodiya et al., 2019), transcriptome analysis revealed profound changes in cortical mRNA levels in ABX-treated male mice only, suggestive of significant alterations in inflammation, microglial activation, and microglia development pathways. Moreover, FMT from age-matched Tg APPPS1-21 mice into short-term ABX-treated APPPS1-21 mice resulted in complete restoration of Aβ amyloidosis, microglial morphologies, and Aβ-associated neurodegenerative changes to those observed in vehicle-treated male mice. Importantly, cerebral cortex transcriptome profiles in short-term ABX-treated male mice that were exposed to FMT led to a near-complete restoration of the transcriptional signatures of vehicle-treated APPPS1-21 mice. Extending these findings, we now report that FMT from age-matched APPPS1-21 Tg mice (Tg-FMT) or WT (WT-FMT) C57Bl6 (non-Tg littermates) mice are equally competent in promoting Aβ amyloidosis in short-term ABX-treated APPPS1-21 mice at 9 wk. Finally, we now document that depletion of microglia with a CSF1R antagonist, PLX5622, in short-term ABX-treated APPPS1-21 mice fails to reduce Aβ amyloidosis compared with controls, thus arguing for an essential role of microglia in mediating microbiome-driven brain amyloidosis.
Results
Short-term ABX result in reduced Aβ amyloidosis in male mice only and Tg-FMT fully restores pathology
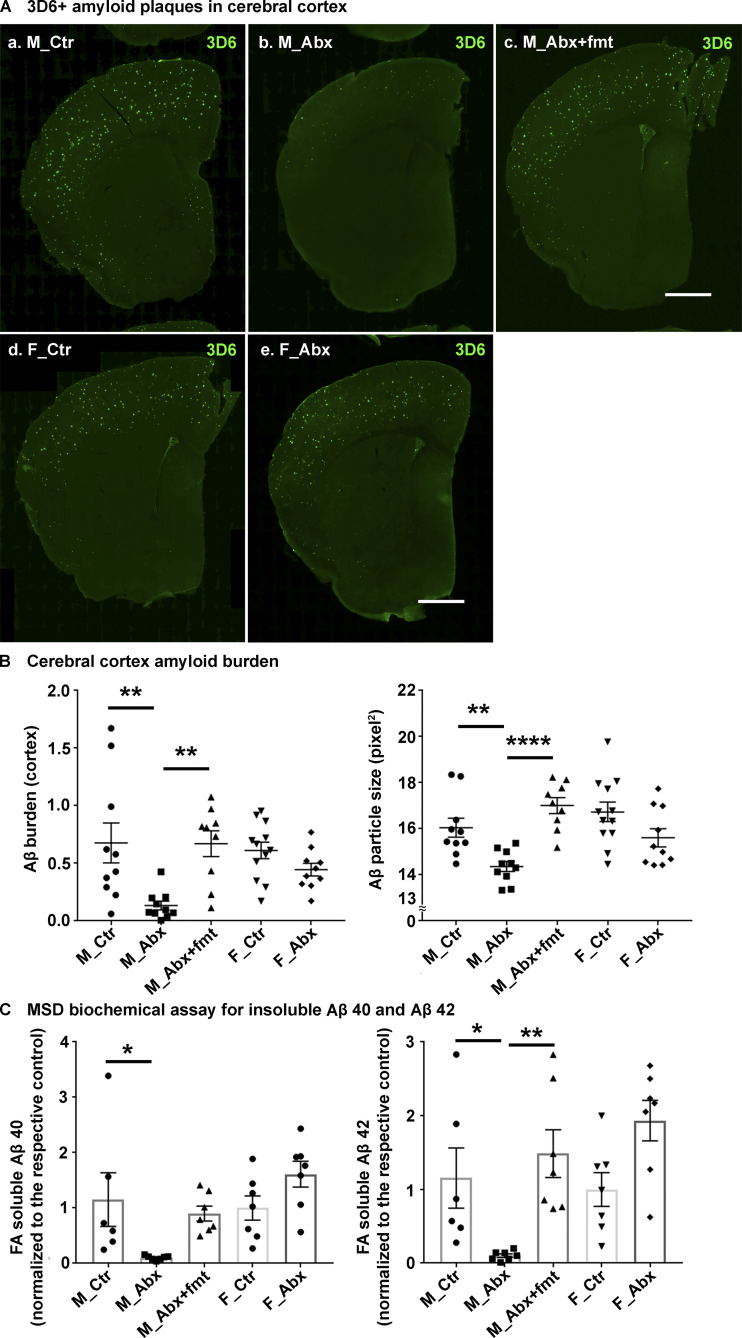
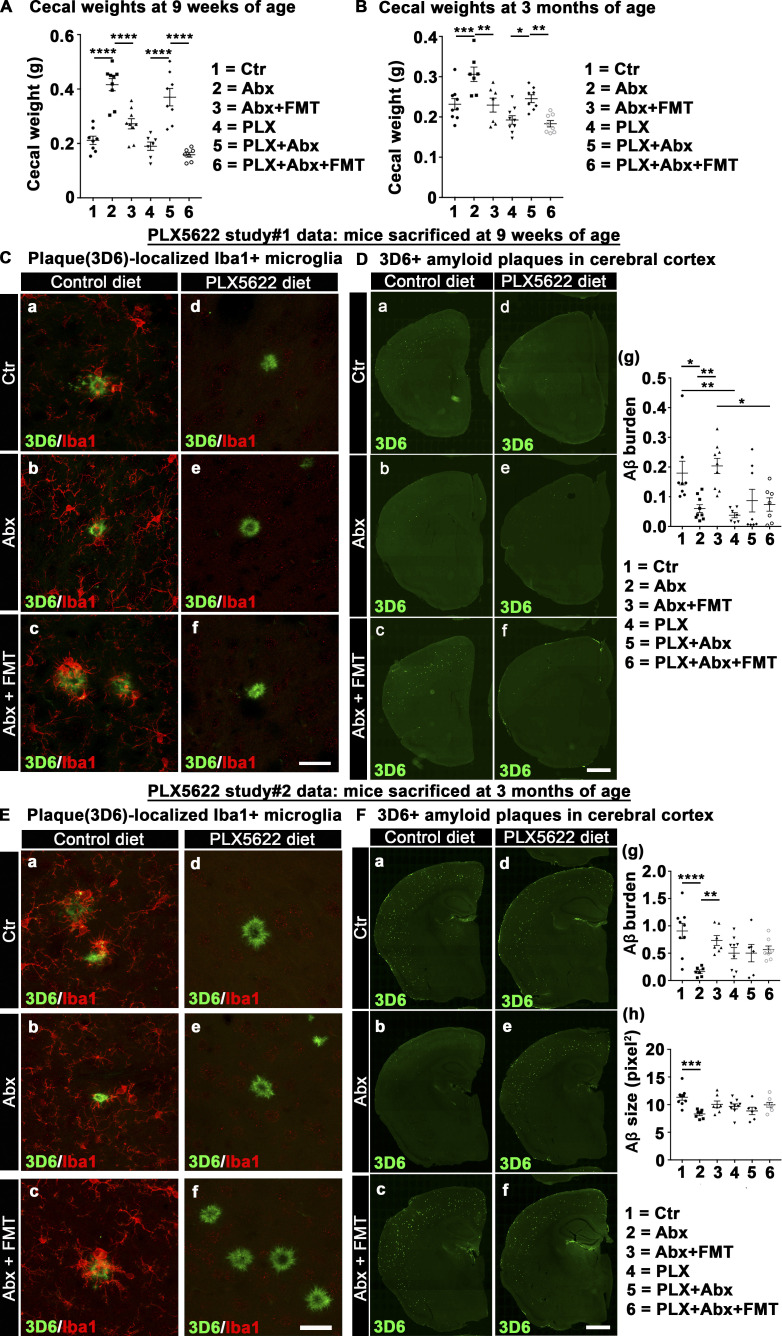
To investigate the effect of ABX on cerebral amyloidosis in APPPS1-21 mice, we treated male and female APPPS1-21 mice with ABX or vehicle from PND14 to PND21 and then provided regular drinking water till the time of sacrifice (9 wk). As lifelong ABX treatment only had a significant effect on Aβ deposition in male animals (Dodiya et al., 2019; Minter et al., 2016), we only performed FMT studies using age-matched APPPS1-21 Tg fecal slurries in ABX-treated male mice. Here, male mice treated with ABX postnatally were subjected to a regimen of daily FMT from age-matched Tg control mice (Tg-FMT) from PND24 till 9 wk of age. The levels of deposited Aβ were assessed by immunohistochemistry (IHC) using a well-established 3D6 antibody specific for the amino-terminus of Aβ (Dodiya et al., 2019) and by biochemical assays using the meso scale discovery (MSD) ELISA platform (Minter et al., 2016). At 9 wk of age, we show prominent Aβ deposition in the cortex of both male and female mice (Fig. 1 A, a–e), and short-term ABX resulted in significantly lower cortical Aβ burden and plaque size in male mice only (Fig. 1 A, a and b; quantified in Fig. 1 B), while female mice showed no significant changes in either Aβ burden or plaque size (Fig. 1 A, d and e; quantified in Fig. 1 B). Most importantly, Aβ burden and plaque size in ABX-treated male mice were restored to levels observed in vehicle-treated Tg male mice after treatment with fecal slurries from age-matched APPPS1-21 mice (Fig. 1 A, c; quantified in Fig. 1 B). Using ventral cerebral cortex tissue of the frozen half-hemisphere, we evaluated soluble and insoluble forms of Aβ1-40 and Aβ1-42 species (Fig. 1 C). While we observed no significant differences in soluble Aβ levels in ABX-treated male mice (Fig. S1 A), insoluble (FA–soluble) Aβ1-40 and Aβ1-42 levels were significantly reduced in extracts from ABX-treated male mice that were completely restored by introduction of Tg-FMT (Fig. 1 C). Similar to the histopathological studies, MSD analysis showed no significant differences in Aβ1-40 and Aβ1-42 species between ABX- and vehicle-treated female groups (Fig. 1 C). Taken together, these data indicate that short-term ABX treatment influences Aβ amyloidosis in the APPPS1-21 Tg mice in a sex-specific manner.
Figure 1.
Reduced Aβ-plaque pathology is only observed in short-term ABX-treated male mice, and Tg-donor FMT restores these changes. (A) Representative images of Aβ in the cortex of M_Ctr (a), M_Abx (b), M_Abx+FMT (c), F_Ctr (d), and F_Abx (e) using anti-Aβ monoclonal antibody. (B) Quantification of Aβ burden was performed using threshold-limited particle analysis of 3D6+ staining. One-way ANOVA showed significant changes in both Aβ burden (P = 0.002) and Aβ size (P < 0.0001). Specifically, M_Abx showed significant reduction in Aβ burden (P = 0.004) and Aβ size (P = 0.024) compared with M_Ctr. These changes were completely restored using FMT from age-matched Tg controls into ABX-treated male mice, similar to the levels of M_Ctr (P > 0.05). Female groups showed no significant differences in Aβ burden (P = 0.739) or Aβ size (P = 0.204). (C) MSD analysis of FA-soluble Aβ1-40 and Aβ1-42 levels in the right ventral cerebral cortex of vehicle-, ABX-, or ABX+FMT–treated mice using anti-Aβ mAb 4G8 (n = 6 or 7 mice/group). M_Abx showed significantly lower levels of detergent-insoluble, FA-soluble Aβ1-40 (P = 0.020) and Aβ1-42 (P = 0.038) compared with M_Ctr. FMT from age-matched Tg controls into ABX-treated male mice reverted these levels, similar to M_Ctr (P > 0.05). F_Abx showed no changes in FA-soluble Aβ1-40 (P = 0.243) but increased FA-soluble Aβ1-42 (P = 0.041) compared with F_Ctr. M_Ctr = vehicle-treated male, M_Abx = ABX-treated (PND14–PND21) male, M_Abx+FMT = ABX-treated (PND14–PND21) male, followed by FMT (PND24–PND63) from age-matched Tg-donor male, F_Ctr = vehicle-treated female, and F_Abx = ABX-treated (PND14–PND21) female. n = 9–12; mice per group unless otherwise mentioned. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Scale bar in panels A, c and A, e represents 1,000 µm and applies to all panels (A, a–e). Detailed statistics are listed in Table S9.
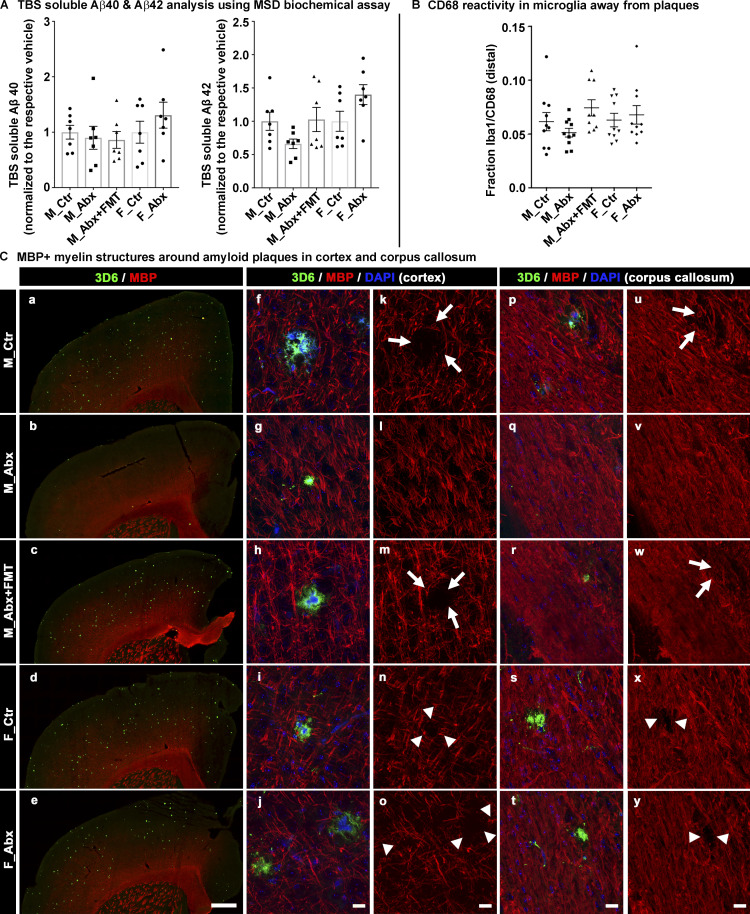
Figure S1.
Soluble Aβ1-40 Aβ1-42 levels measured by MSD biochemical assay, graphical presentation of CD68 reactivity in Iba1 away from plaques using ImageJ, and MBP expression in cerebral cortex and corpus callosum using qualitative comparison. (A) TBS soluble levels of Aβ1-40 and Aβ1-42 in cerebral cortex homogenates are shown using the MSD biochemical assay. n = 6–7 mice per group. (B) Graphical presentation of the colocalization measure of CD68-reactive Iba1+ microglia cells in microglia away from the plaques is shown. n = 8–10 per group. (C) Representative low-magnification (a–e) and high-magnification images (f–y) of 3D6+ plaque (green)-localized, MBP (red) from vehicle-treated male (a, f, k, p, and u), ABX-treated male (b, g, l, q, and v), ABX+FMT–treated male (c, h, m, r, and w), vehicle-treated female (d, i, n, s, and x), and ABX-treated female (e, j, o, t, and y) mice. Higher-magnification images of single-plaque microenvironments were taken from either cerebral cortex (f–o) or corpus callosum (p–y) in a random manner. Note that the arrows in k, m, u, and w indicate loss of MBP in control and ABX+FMT–treated male mice. Arrowheads in n, o, x, and y represent loss of MBP in F_Ctr and F_Abx mice. Note the loss of MBP either by arrows or arrowheads is more prominent in cortex compared with corpus callosum. ABX-treated male mice showed very minimal, if any, loss of MBP-positive structures around Aβ plaques. Scale bar in e represents 500 µm and applies to a–e. Scale bars in j, o, t, and y represent 15 µm and apply to f–y. M_Ctr = vehicle-treated male, M_Abx = ABX-treated male from PND14–PND21 only, M_Abx+FMT = ABX-treated male (PND14–PND21), followed by fecal microbiota transfer (PND24–PND63) from age-matched Tg donor male mice, F_Ctr = vehicle-treated female, and F_Abx = ABX-treated female from PND14–PND21 only. Error bars are mean ± SEM.
Short-term ABX mediate alterations in fecal gut microbiota profiles in males and females, taxa restoration with FMT in male group
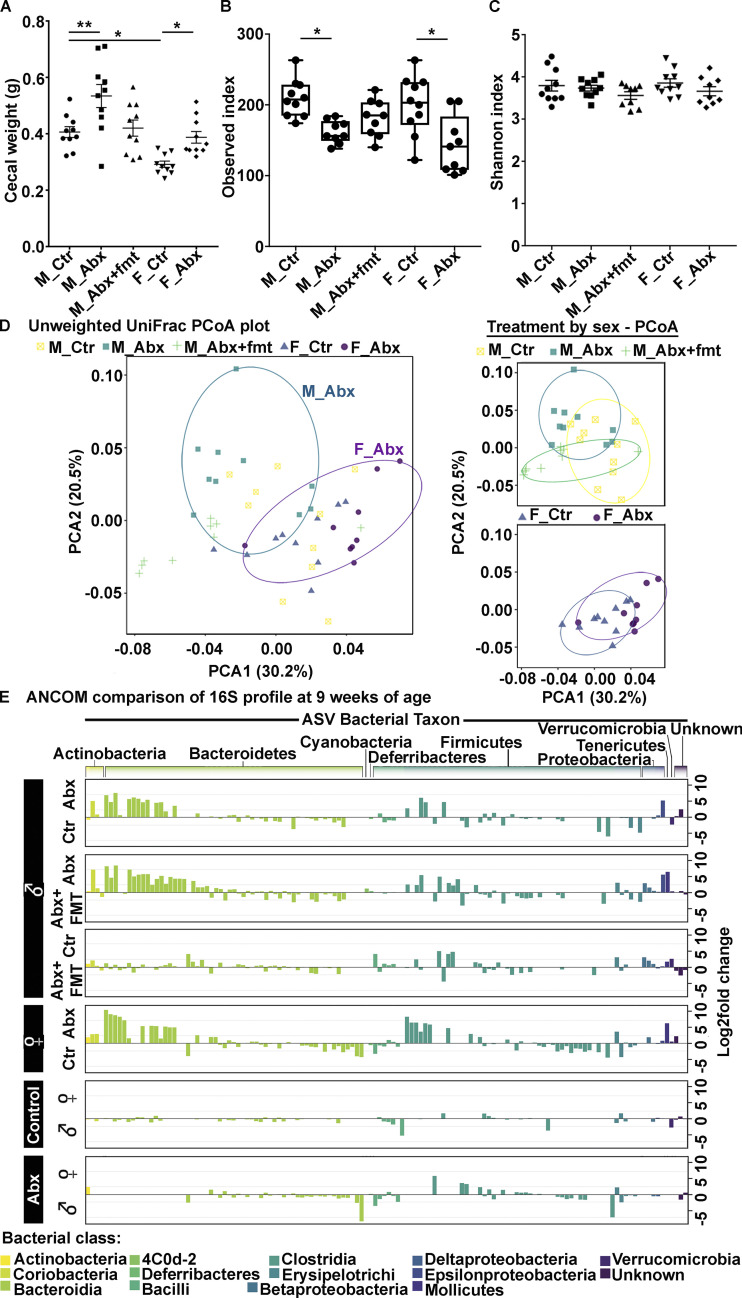
Having established that short-term ABX treatment influences Aβ amyloidosis in the APPPS1-21 Tg mice in a sex-specific manner, it was critical to determine the impact of ABX on gut microbiota profiles. For this, ceca from male and female APPPS1-21 mice that were subjected to postnatal ABX or vehicle gavages were collected and weighed at the time of sacrifice (9 wk of age). As expected, the ceca of ABX-treated mice showed significantly higher weights compared with their vehicle-treated counterparts (Fig. 2 A), as described previously (Dodiya et al., 2019; Minter et al., 2017, 2016). Additionally, the cecal weights of ABX-treated male mice that were subjected to Tg-FMT were restored to the cecal weights of Tg male controls.
Figure 2.
Short-term ABX changes cecal weight and fecal microbiota profile in male and female APPPS1-21 mice at 9 wk of age. (A) Cecal weights from vehicle- or ABX-treated male and female mice. ABX resulted in cecal enlargement in both sexes. One-way ANOVA: F(4, 45) = 11.02, P < 0.0001. Sidak post hoc analysis showed significantly larger cecal size in ABX-treated male (0.534 ± 0.041) and ABX-treated female (0.388 ± 0.021) mice compared with their vehicle-treated counterparts (M_Ctr = 0.406 ± 0.019, F_Ctr = 0.292 ± 0.0113; P = 0.004, P = 0.013, respectively). FMT treatment in ABX-treated male mice restored cecal weights (0.420 ± 0.028), similar to vehicle-treated male mice (0.406 ± 0.019; P = 0.992). (B and C) The α-diversity was measured by using observed index (B) and Shannon index (C). Observed index showed reduced diversity in ABX-treated male and ABX-treated female mice compared with their vehicle-treated counterparts (Kruskal-Wallis: P = 0.03, P = 0.01, respectively). Shannon index comparison showed no significant differences between groups (P > 0.05). (D) PCoA plot generated using unweighted version of the UniFrac distance metric. The two components explained 30.2% of the variance. ABX treatment of male and female mice resulted in separate clusters at the time of sacrifice. (E) Log2 fold change of taxa presentation from ANCOM comparison of 16S profile of fecal microbiota. ABX treatment resulted in sex-specific taxa changes. Observe the flat line for comparison between groups M_Ctr and M_Abx+FMT and M_Ctr and F_Ctr, suggesting no major changes between these groups. M_Ctr = vehicle-treated male, M_Abx = ABX-treated (PND14–PND21) male, M_Abx+FMT = ABX-treated (PND14–PND21) male followed by FMT (PND24–PND63) from age-matched Tg-donor male, F_Ctr = vehicle-treated female, and F_Abx = ABX-treated (PND14–PND21) female. n = 9 or 10 mice per group. Data are mean ± SEM. *, P < 0.05; **, P < 0.01. PCoA, principal coordinate analysis.
16S ribosomal RNA (rRNA) amplicon sequencing was performed on fresh fecal pellets collected from individual mice at 9 wk of age, and subsequent statistical analyses were performed at 100% nucleotide identity (amplicon sequence variants [ASVs]). Analysis of microbial α-diversity revealed significantly lower bacterial richness in ABX-treated male and female groups compared with their vehicle-treated counterparts (Fig. 2 B), while differences in Shannon diversity were not significant (Fig. 2 C). Analysis of β-diversity using the unweighted UniFrac metric (Lozupone and Knight, 2005) identified no differences between vehicle-treated male and female groups (PERMANOVA, P = 0.164), while ABX-treated male and female groups did differ significantly from each other (PERMANOVA, P = 0.001; Fig. 2 D). Similar results were observed using the weighted UniFrac metric, with vehicle-treated mice showing no significant differences between the sexes (PERMANOVA, P = 0.05) and ABX-treated mice differing significantly by sex (PERMANOVA, P = 0.001).
Analysis of composition of microbiomes (ANCOM; Mandal et al., 2015) identified a number of ASVs whose proportion differed significantly between ABX-treated mice and their vehicle-treated counterparts (Fig. 2 E and Table 1). Relative to vehicle-treated males, ABX-treated males exhibited a greater proportion of ASVs in the order Bacteroidales (phylum Bacteroidetes), including Parabacteroides distasonis, Bacteroides sp., Odoribacter sp., Prevotella spp., as well as ASVs in the families Paraprevotellaceae, Rikenellaceae, and S24-7. Other bacteria found to exhibit a greater proportion in ABX-treated males included Olsenella profus (phylum Actinobacteria) and ASVs in the phylum Firmicutes including Lactobacillus sp., Allobaculum sp., and family Lachnospiraceae. Helicobacter sp. (phylum Proteobacteria) was also in greater proportion in ABX-treated males compared with vehicle-treated males (Table 1). Bacteria exhibiting a decrease in ABX-treated males relative to vehicle-treated males included Bifidobacterium pseudolongum (phylum Actinobacteria), Lactobacillus reuteri, Coprococcus sp., and Allobaculum sp. (phylum Firmicutes), Sutterella (phylum Proteobacteria), and Akkermansia muciniphila (phylum Verrucomicrobia; Table 1). The identification of ASVs in the families Lachnospiraceae and S24-7 as differing significantly between vehicle-treated and ABX-treated males supports similar findings from our previous investigations (Dodiya et al., 2019; Minter et al., 2017, 2016).
Table 1. ANCOM analysis (90% threshold of detection) of fecal microbiota at 9 wk from short-term ABX and FMT study.
| Comparison | ||
| F_Vehicle vs. F_ABX | Higher in vehicle | Higher in Abx |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__; g__; s__ | p__Actinobacteria; c__Actinobacteria; o__Bifidobacteriales; f__Bifidobacteriaceae; g__Bifidobacterium; s__pseudolongum | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; | p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; s__ | p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Olsenella; s__profusa | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__distasonis | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Paraprevotellaceae; g__Paraprevotella; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotella; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Paraprevotellaceae;; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae;; | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotella; s__ | |
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | |
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__reuteri | p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__ | |
| p__Firmicutes; c__Bacilli; o__Turicibacterales; f__Turicibacteraceae; g__Turicibacter; s__ | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__; s__ | |
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Ruminococcus; s__gnavus | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Coprococcus; s__ | |
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae;; | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae;; | |
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcus; s__ | p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | |
| p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | p__Proteobacteria; c__Betaproteobacteria; o__Burkholderiales; f__Alcaligenaceae; g__Sutterella; s__ | |
| p__Proteobacteria; c__Deltaproteobacteria; o__Desulfovibrionales; f__Desulfovibrionaceae; g__Desulfovibrio; s__C21_c20 | p__Proteobacteria; c__Epsilonproteobacteria; o__Campylobacterales; f__Helicobacteraceae; g__Helicobacter; s__ | |
| p__Proteobacteria;;;;; | ||
| p__Verrucomicrobia; c__Verrucomicrobiae; o__Verrucomicrobiales; f__Verrucomicrobiaceae; g__Akkermansia; s__muciniphila | ||
| M_ABX vs. F_ABX | Higher in males | Higher in females |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__; g__; s__ | p__Actinobacteria; c__Actinobacteria; o__Bifidobacteriales; f__Bifidobacteriaceae; g__Bifidobacterium; s__pseudolongum | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; s__ | p__Firmicutes; c__Clostridia; o__Clostridiales;;; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Paraprevotellaceae;; | p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__distasonis | ||
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotella; s__ | ||
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae;; | ||
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | ||
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; | ||
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__ | ||
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__reuteri | ||
| p__Firmicutes; c__Bacilli; o__Turicibacterales; f__Turicibacteraceae; g__Turicibacter; s__ | ||
| p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | ||
| p__Proteobacteria; c__Betaproteobacteria; o__Burkholderiales; f__Alcaligenaceae; g__Sutterella; s__ | ||
| p__Verrucomicrobia; c__Verrucomicrobiae; o__Verrucomicrobiales; f__Verrucomicrobiaceae; g__Akkermansia; s__muciniphila | ||
| M_Vehicle vs. F_Vehicle | Higher in males | Higher in females |
| p__Actinobacteria; c__Actinobacteria; o__Bifidobacteriales; f__Bifidobacteriaceae; g__Bifidobacterium; s__pseudolongum | p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Olsenella; s__profusa | |
| p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; | p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Paraprevotellaceae; g__Paraprevotella; s__ | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae;; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Paraprevotellaceae;; | p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__ | p__Proteobacteria; c__Deltaproteobacteria; o__Desulfovibrionales; f__Desulfovibrionaceae; g__Desulfovibrio; s__C21_c20 | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__distasonis | p__Proteobacteria; c__Epsilonproteobacteria; o__Campylobacterales; f__Helicobacteraceae; g__Helicobacter; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotella; s__ | ||
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | ||
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__ | ||
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__reuteri | ||
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Coprococcus; s__ | ||
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Peptostreptococcaceae; g__; s__ | ||
| p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | ||
| p__Proteobacteria; c__Betaproteobacteria; o__Burkholderiales; f__Alcaligenaceae; g__Sutterella; s__ | ||
| p__Proteobacteria;;;;; | ||
| p__Verrucomicrobia; c__Verrucomicrobiae; o__Verrucomicrobiales; f__Verrucomicrobiaceae; g__Akkermansia; s__muciniphila | ||
| M_Vehicle vs. M_ABX | Higher in vehicle | Higher in Abx |
| p__Actinobacteria; c__Actinobacteria; o__Bifidobacteriales; f__Bifidobacteriaceae; g__Bifidobacterium; s__pseudolongum | p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Olsenella; s__profusa | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; s__uniformis | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotella; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Odoribacteraceae; g__Odoribacter; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Paraprevotellaceae;; | |
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__ | |
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__reuteri | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__distasonis | |
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Coprococcus; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotella; s__ | |
| p__Firmicutes; c__Clostridia; o__Clostridiales;;; | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__; s__ | |
| p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | |
| p__Proteobacteria; c__Betaproteobacteria; o__Burkholderiales; f__Alcaligenaceae; g__Sutterella; s__ | p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; | |
| p__Verrucomicrobia; c__Verrucomicrobiae; o__Verrucomicrobiales; f__Verrucomicrobiaceae; g__Akkermansia; s__muciniphila | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__; s__ | |
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae;; | ||
| p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | ||
| p__Proteobacteria; c__Epsilonproteobacteria; o__Campylobacterales; f__Helicobacteraceae; g__Helicobacter; s__ | ||
| M_ABX vs. FMT | Higher in ABX | Higher in FMT |
| p__Actinobacteria; c__Actinobacteria; o__Bifidobacteriales; f__Bifidobacteriaceae; g__Bifidobacterium; s__pseudolongum | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; | |
| p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotella; s__ | |
| p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Olsenella; s__profusa | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae;; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Odoribacteraceae; g__Odoribacter; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Paraprevotellaceae; g__Paraprevotella; s__ | p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Paraprevotellaceae;; | p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__ | p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__reuteri | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__distasonis | p__Firmicutes; c__Clostridia; o__Clostridiales; f__; g__; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotella; s__ | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__; s__ | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Coprococcus; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__AF12; s__ | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae;; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae;; | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Oscillospira; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | p__Firmicutes; c__Clostridia; o__Clostridiales;;; | |
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__ | p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | |
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Clostridiaceae;; | ||
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__; s__ | ||
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae;; | ||
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcus; s__flavefaciens | ||
| p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | ||
| p__Proteobacteria; c__Betaproteobacteria; o__Burkholderiales; f__Alcaligenaceae; g__Sutterella; s__ | ||
| p__Proteobacteria; c__Betaproteobacteria;;;; | ||
| p__Proteobacteria; c__Deltaproteobacteria; o__Desulfovibrionales; f__Desulfovibrionaceae; g__Desulfovibrio; s__C21_c20 | ||
| p__Proteobacteria; c__Epsilonproteobacteria; o__Campylobacterales; f__Helicobacteraceae; g__Helicobacter; s__ | ||
| p__Tenericutes; c__Mollicutes; o__RF39; f__; g__; s__ | ||
| p__Verrucomicrobia; c__Verrucomicrobiae; o__Verrucomicrobiales; f__Verrucomicrobiaceae; g__Akkermansia; s__muciniphila | ||
| M_Vehicle vs. FMT | Higher in vehicle | Higher in FMT |
| p__Actinobacteria; c__Actinobacteria; o__Bifidobacteriales; f__Bifidobacteriaceae; g__Bifidobacterium; s__pseudolongum | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Bacteroidaceae; g__Bacteroides; | |
| p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__; s__ | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Prevotellaceae; g__Prevotella; s__ | |
| p__Actinobacteria; c__Coriobacteriia; o__Coriobacteriales; f__Coriobacteriaceae; g__Olsenella; s__profusa | p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Odoribacteraceae; g__Odoribacter; s__ | p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Paraprevotellaceae;; | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae;; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__ | p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae;; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Porphyromonadaceae; g__Parabacteroides; s__distasonis | p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae; g__; s__ | p__Proteobacteria; c__Deltaproteobacteria; o__Desulfovibrionales; f__Desulfovibrionaceae; g__Desulfovibrio; s__C21_c20 | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae;; | p__Proteobacteria;;;;; | |
| p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__S24-7; g__; s__ | ||
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__ | ||
| p__Firmicutes; c__Bacilli; o__Lactobacillales; f__Lactobacillaceae; g__Lactobacillus; s__reuteri | ||
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Clostridiaceae;; | ||
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae; g__Anaerostipes; s__ | ||
| p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae; g__Ruminococcus; s__flavefaciens | ||
| p__Firmicutes; c__Erysipelotrichi; o__Erysipelotrichales; f__Erysipelotrichaceae; g__Allobaculum; s__ | ||
| p__Proteobacteria; c__Betaproteobacteria; o__Burkholderiales; f__Alcaligenaceae; g__Sutterella; s__ | ||
| p__Proteobacteria; c__Betaproteobacteria;;;; | ||
| p__Proteobacteria; c__Epsilonproteobacteria; o__Campylobacterales; f__Helicobacteraceae; g__Helicobacter; s__ | ||
| p__Tenericutes; c__Mollicutes; o__RF39; f__; g__; s__ | ||
| p__Verrucomicrobia; c__Verrucomicrobiae; o__Verrucomicrobiales; f__Verrucomicrobiaceae; g__Akkermansia; s__muciniphila | ||
p, phylum; c, class; o, order; f, family; g, genus, s, species.
Many, but not all, of the ABX-associated microbial changes in male mice were restored to the profiles observed in Tg male mice following Tg-FMT. In particular, both vehicle and FMT-treated males had a similar increased proportion of several Firmicutes, including Lactobacillus reuteri, Allobaculum, and Oscillospira, as well as the family Lachnospiraceae. Similarly, ABX-treated female mice showed many changes in taxa compared with vehicle-treated female mice, but most importantly, ABX-male and ABX-female groups showed significant differences in several taxa (Table 1), suggesting that early-life ABX treatment resulted in sex-specific taxa differences at the time of sacrifice.
Collectively, these data indicate that early-life ABX treatment results in sex-specific microbiota differences at 9 wk of age and that male Tg FMT restores these changes in ABX-treated male mice.
Short-term ABX-mediated microbiota perturbations result in sex-specific alterations in the morphology and activation status of plaque-localized microglia
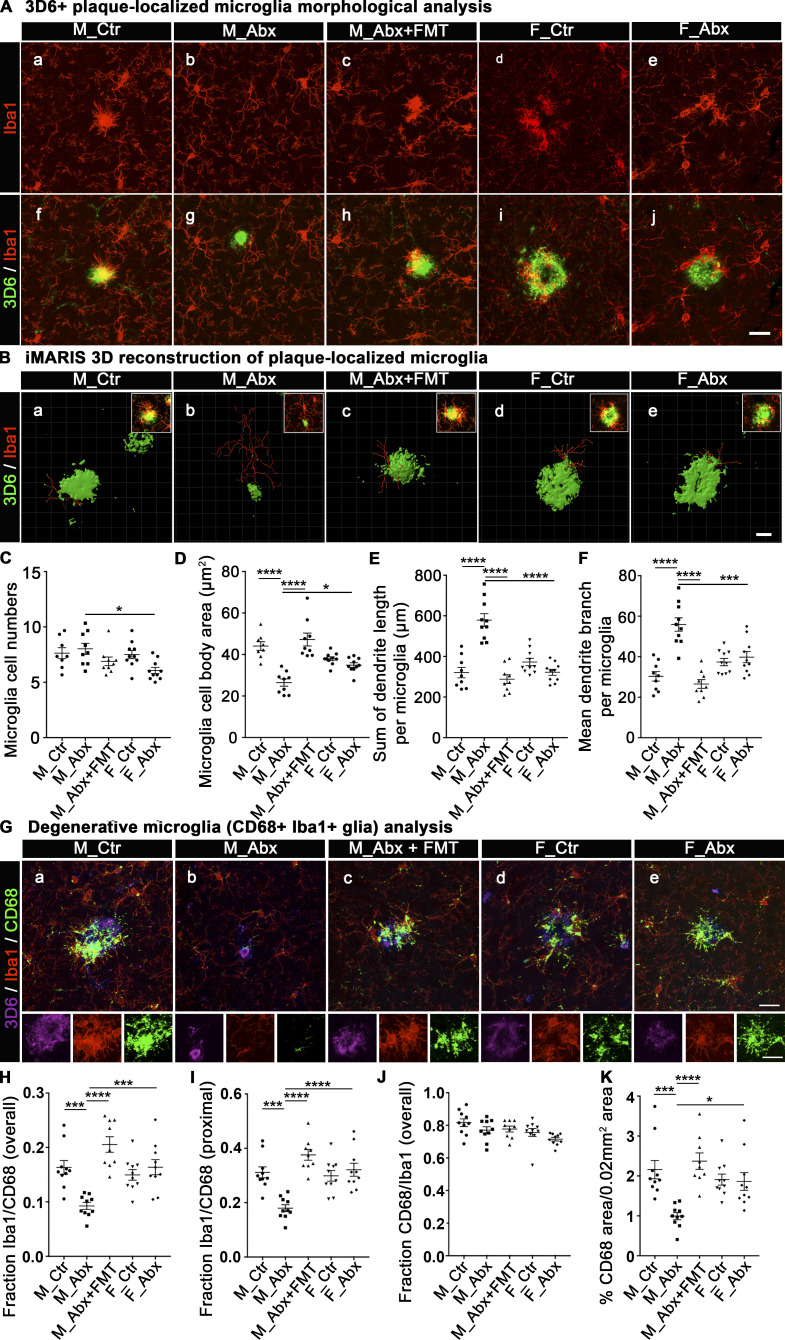
In our earlier report using lifelong ABX, we showed that gut microbiota perturbations influenced microglial morphology and transcriptome profiles in male mice only (Dodiya et al., 2019). To investigate the status of microglial morphology in our current model of early-life microbiota-perturbed APPPS1-21 mice, we performed IHC studies to evaluate 3D6+amyloid plaque–localized Iba1+microglial cells in the cerebral cortex (Fig. 3). We collected three-dimensional (3D) Z-stacks (30 planes of Z-stack covering 8.66-µm thickness) of high-magnification images that allowed us to evaluate plaque-localized microglia within a 0.02-mm2 area that contains a 3D6+amyloid plaque (Fig. 3, A and B). Using ImageJ software (National Institutes of Health [NIH]), we evaluated microglial cell numbers (Fig. 3 C) and microglial cell body areas (Fig. 3 D). ABX treatment did not affect microglial cell numbers within 0.2 mm2 of plaque microenvironment (Fig. 3 C), but we observed significant differences in microglial cell body areas in brains of mice treated with ABX versus vehicle (Fig. 3 D); ABX treatment resulted in smaller microglial cell body sizes only in ABX-treated male mice compared with the vehicle-treated male cohort. Importantly, Tg-FMT introduced into ABX-treated male mice reversed these changes to that observed in vehicle-treated mice. No significant differences in microglial cell body areas were observed in ABX-treated female mice compared with vehicle-treated females.
Figure 3.
Short-term ABX alter microglial morphologies and activation status in male mice, and FMT reverts these changes. (A) Representative images of 3D6+ plaque (green)-localized, Iba1+ microglia (red) from M_Ctr, M_Abx, M_ABX+FMT, F_Ctr, and F_Abx groups. (B) Representative images of iMARIS 3D reconstruction and inserts of compressed 3D Z-stacks from M_Ctr, M_Abx, M_Abx+FMT, F_Ctr, and F_Abx. (C) ImageJ quantification of cell numbers showed no differences in plaque-localized microglial cell numbers in M_Abx (P = 0.964) and F_Abx (P = 0.088) compared with their controls. (D) Cell body areas measured using ImageJ showed significantly reduced Iba1+ cell body size in M_Abx compared with M_Ctr (P < 0.0001), which was restored in M_Abx+FMT group that was similar to M_Ctr (P = 0.7926). Female mice showed no significant changes in microglial cell body area (P = 0.755). (E and F) The dendrite branch length (E) and dendrite branch point measures (F) using iMARIS 3D reconstruction showed significantly higher mean branch length and branch points per microglia in M_Abx compared with M_Ctr (P < 0.0001), which was restored in M_Abx+FMT, similar to M_Ctr (P = 0.8717, P = 0.8472). Female mice showed no significant differences in both measures (P = 0.522, P = 0.958). (G) Representative images of 3D6+ plaque (purple)-localized CD68 (lysosome marker: green) reactive Iba1+ microglial cells (red) from M_Ctr, M_Abx, M_Abx+FMT, F_Ctr, and F_Abx in the top panels and cropped inserts of each channel in the bottom panels. (H) The fraction of CD68-reactive microglia was significantly lower in M_Abx compared with M_Ctr (P = 0.0005) and reverted in M_Abx+FMT, similar to M_Ctr (P = 0.11). Female mice showed no significant differences (P = 0.908). (I) Microglia in close proximity to plaques showed significantly lower CD68 reactivity in M_Abx compared with M_Ctr (P = 0.0002) and reverted in M_Abx+FMT, similar to M_Ctr (P = 0.1674). Female mice showed no differences (P = 0.921). (J) Majority (∼80%) of CD68-reactive structures colocalized with Iba1+ cells, and there were no differences among any groups (P > 0.05). (K) Overall CD68 reactivity was significantly diminished in M_Abx compared with M_Ctr (P = 0.0003) and reverted in M_Abx+FMT, similar to M_Ctr (P = 0.93). Female mice showed no differences (P = 0.999). M_Ctr = vehicle-treated male, M_Abx = ABX-treated (PND14–PND21) male, M_Abx+FMT = ABX-treated (PND14–PND21) male, followed by FMT (PND24–PND63) from age-matched Tg-donor male, F_Ctr = vehicle-treated female, and F_Abx = ABX-treated (PND14–PND21) female. n = 8–10 mice per group. Data are mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Scale bars in A, j, B, e, and G, e represent 20 µm, 10 µm, and 20 µm and apply to all panels in A, a–j, B, a–e, and G, a–e, respectively. Detailed statistics are listed in Table S9.
To further access the morphology of plaque-localized microglia, we employed Imaris software to generate 3D reconstructions as established previously (Dodiya et al., 2019). The total “dendrite” branch length and total number of branch points of plaque-localized microglia were evaluated (Fig. 3 B, E, and F). ABX treatment resulted in significantly longer dendritic branch lengths in male, but not in female, mice compared with their vehicle-treated counterparts. Similarly, dendritic branch points were significantly higher in ABX-treated male, but not in female, mice compared with their vehicle-treated cohorts. Furthermore, Tg-FMT into ABX-treated male mice resulted in complete restoration of the morphological parameters to those observed in vehicle-treated male Tg mice.
Finally, to assess the activated phagocytic state of plaque-localized microglia, we performed IHC with antibodies specific for myeloid cell-specific cluster of differentiation 68 (CD68) antigen. CD68 is primarily expressed in microglial cells within the brain parenchyma (Fiala et al., 2002) and is commonly used as a marker of activated phagocytic microglia (Walker and Lue, 2015); microglia in the resting phase express very low levels of this antigen (Lee et al., 2002). For confocal microscopy, we evaluated plaque-localized microglia within a 0.02-mm2 area that contains a 3D6+amyloid plaque, as indicated above (Fig. 3 G). The area covered by CD68 was quantified using the “Threshold” and “Analyze Particles” (inclusion size of 1-Infinity) functions from ImageJ within the region of interest (0.02 mm2). Clustering of Iba1+ microglia around 3D6+ Aβ plaques was accompanied by higher CD68 immunoreactivity in vehicle-treated APPPS1-21 male mice that was significantly reduced in ABX-treated male mice (Fig. 3 H). In addition, microglia in close proximity to plaques showed significant changes in CD68 reactivity (Fig. 3 I), while microglia away from plaques showed no significant changes in CD68 expression following ABX treatment (Fig. S1, B). More importantly, Tg-FMT into ABX-treated male mice restored CD68 immunoreactivity to levels seen in vehicle-treated mice (Fig. 3, G–I and K). The vast majority of CD68 immunoreactive cells were microglia as evaluated by the fraction of CD68/Iba1 colocalization measures (Fig. 3 J). Moreover, ABX did not alter CD68 immunoreactive plaque-localized microglia in female mice compared with the vehicle-treated group (Fig. 3, G–K).
Collectively, these data indicate that in male mice, short-term ABX-perturbed microbiome resulted in lower numbers of degenerative microglia (aka DAM or MGnD, as defined by larger cell bodies, reduced dendritic branch lengths and branch points, and higher CD68 reactivity). These morphological phenotypes were fully restored with Tg-FMT in ABX-treated male mice. On the other hand, short-term ABX-perturbed microbiome had no impact on either cell body size or dendritic morphology of plaque-localized microglia in female mice.
Short-term ABX-mediated microbiota perturbations result in sex-specific alterations in Aβ-associated degenerative changes
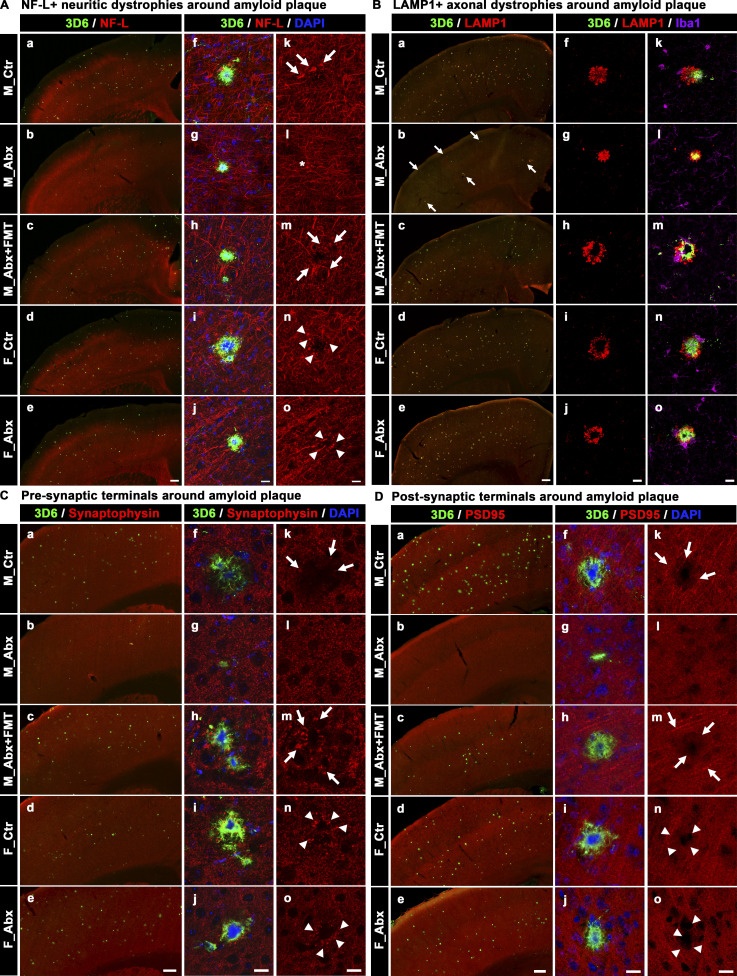
To further characterize the role of microbiome-perturbed amyloidosis and its impact on cortical degenerative changes, we performed IHC with antibodies specific for neurons (neurofilament light chain [NF-L]), lysosomes (LAMP1), synapses (Synaptophysin, PSD95), and myelin (myelin basic protein [MBP]). The qualitative measures were evaluated by comparing lower-magnification images of entire cerebral cortex scanned using a slide scanner, followed by a comparison of higher-magnification images using an SP8 confocal microscope of a randomly selected amyloid plaque area.
First, we evaluated dystrophic axons using NF-L. NF-L, a cytoskeleton protein, is exclusively expressed in neurons and located primarily in axons (Bridel et al., 2019). NF-L is a marker for active axonal injury and neuritic damage, possibly including degenerating fibers (Bacioglu et al., 2016; Masliah et al., 1993). Axonal pathology in AD brains can be visible in the form of dystrophic neurites, mainly axonal swellings or dilatations, in close proximity to Aβ plaques. Similarly, we observed NF-L immunoreactive swollen or dilated axons in close proximity to Aβ plaques in vehicle-treated mice that were completely absent in ABX-treated male mice; FMT restored the neuritic damage in ABX-treated male mice to levels seen in vehicle-treated mice (Fig. 4 A). In contrast, female mice showed no differences in levels of neuritic dystrophy between vehicle- or ABX-treated mice.
Figure 4.
Short-term ABX reduces Aβ-associated degenerative changes in male mice, and FMT reverts these changes in ABX-treated male mice. (A) Representative low (a–e) and high (f–o) magnification images of 3D6+ plaque (green)-localized, NF-L (red) from M_Ctr (a, f, and k), M_Abx (b, g, and l), M_Abx+FMT (c, h, and m), F_Ctr (d, i, and n), and F_Abx (e, j, and o) mice. Note the arrows in k and m indicate swollen neurites in M_Ctr and M_Abx+FMT, while asterisk in l represents lack of swollen neurites in M_Abx. Arrowheads in n and o represent swollen dystrophic neurites in F_Ctr and F_Abx mice. (B) Representative low (a–e) and high (f–o) magnification images of LAMP1 immunolabeled (red) dystrophic axons in close proximity to 3D6+ plaque (green) from M_Ctr (a, f, and k), M_Abx (b, g, and l), M_Abx+FMT (c, h, and m), F_Ctr (d, i, and n), and F_Abx (e, j, and o) mice. Note the arrows in b indicate lower levels of Aβ (green) plaques and associated LAMP1 reactivity (red) in M_Abx compared with M_Ctr (a). Majority of LAMP1 reactivity was observed around the plaques, and Iba1+ microglia (purple) cells did not colocalize with LAMP1 (red). (C) Representative low (a–e) and high (f–o) magnification of 3D6+ plaque-localized synaptophysin (a presynaptic terminal marker: red) from M_Ctr (a, f, and k), M_Abx (b, g, and l), M_Abx+FMT (c, h, and m), F_Ctr (d, i, and n), and F_Abx (e, j, and o) mice. Note the arrows in k and m indicate swollen dystrophic presynaptic terminals and lack of synaptophysin boutons in close proximity to Aβ plaques in M_Ctr and M_Abx+FMT. Arrowheads in n and o represent swollen dystrophic presynaptic terminals in F_Ctr and F_Abx mice. (D) Representative low (a–e) and high (f–o) magnification of 3D6+ plaque-localized PSD95 (a postsynaptic terminal marker: red) from M_Ctr (a, f, and k), M_Abx (b, g, and l), M_Abx+FMT (c, h, and m), F_Ctr (d, i, and n), and F_Abx (e, j, and o) mice. Note the arrows in k and m indicate the absence of PSD95 immunoreactive postsynaptic terminals in close proximity to Aβ plaques in M_Ctr and M_Abx+FMT. Arrowheads in n and o represent the absence of PSD95 immunoreactive postsynaptic terminals in close proximity to Aβ plaques in F_Ctr and F_Abx mice. M_Ctr = vehicle-treated male, M_Abx = ABX-treated (PND14–PND21) male, M_Abx+FMT = ABX-treated (PND14–PND21) male, followed by FMT (PND24–PND63) from age-matched Tg-donor male, F_Ctr = vehicle-treated female, and F_Abx = ABX-treated (PND14–PND21) female. Scale bars in A, e, B, e, C, e, and D, e represent 200 µm and apply to all panels in A, a–e, B, a–e, C, a–e, and D, a–e. Scale bars in A, j, A, o, B, j, B, o, C, j, C, o, D, j, and D, o represent 15 µm and apply to all panels in A, f–o, B, f–o, C, f–o, and D, f–o.
Second, we evaluated lysosome-associated membrane protein-1 (LAMP1), a selective marker of late endosomes and lysosomes that accumulates intensively around Aβ plaques (Gowrishankar et al., 2015) and represents dystrophic axons around Aβ plaques (Meilandt et al., 2020). Similarly, we observed higher LAMP1 immunolabeling, present in all Aβ plaques in vehicle-treated male mice, while ABX treatment showed reductions in LAMP1 immunoreactivity that paralleled ABX-mediated reductions in Aβ amyloidosis (Fig. 4 B). FMT into ABX-treated male mice restored LAMP1 immunolabeling similar to that observed in vehicle-treated male mice. Again, female mice showed no obvious differences in LAMP1 immunoreactivity between vehicle- and ABX-treated animals.
To investigate Aβ-related synaptic alterations as a function of Aβ deposition and/or oligomeric Aβ species, we examined pre- and postsynaptic elements with antibodies specific for synaptophysin (Fig. 4 C) and PSD-95 (Fig. 4 D), respectively, using qualitative IHC comparisons. Under high magnification, vehicle-treated male mice showed high levels of synaptophysin-reactive swollen tortuous dystrophic neurites in close proximity to Aβ, suggestive of Aβ-driven synaptic alterations. Interestingly, ABX-treated male mice showed intact synaptophysin-labeled structures in close proximity to Aβ plaques, while FMT into ABX-treated animals resulted in restoration of dystrophic synaptophysin-labeled neurites in close proximity to Aβ plaques. Similar to the observations with the presynaptic marker, PSD95-immunolabeling was diminished in close proximity to Aβ plaques suggestive of Aβ-related synaptic alterations. ABX-treated male mice showed minimal, if any, loss in the levels of PSD95-immunoreactive structures around Aβ plaques while FMT treatment reverted these changes in ABX-treated male mice. Female mice showed no changes in either synaptophysin or PSD95-labeled structures irrespective of vehicle or ABX treatments.
Finally, we investigated myelin abnormalities using antibodies to MBP (Fig. S1 C). We observed visible loss of myelin in close proximity to Aβ plaques (largely in the cerebral cortex and occasionally in the corpus callosum) of the vehicle-treated male mice, and these alterations were completely absent in ABX-treated male mice. FMT into ABX-treated male mice resulted in myelin abnormalities similar to vehicle-treated male mice. In contrast, we did not observe any difference in plaque-mediated myelin abnormalities in female mice with vehicle or ABX treatments.
Short-term ABX treatment results in sex-specific alterations in circulating cytokines and chemokines and Tg-FMT restores these changes in male APPPS1-21 mice
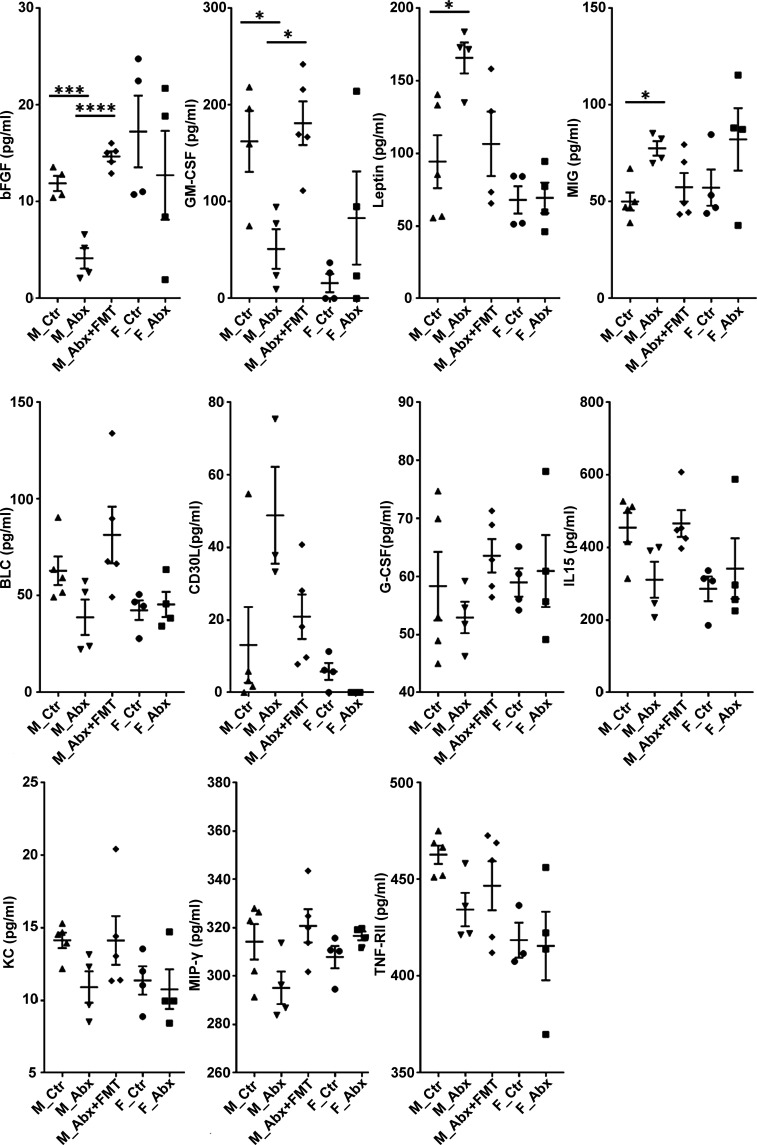
It is well documented that the gut microbiota can influence peripheral immune cell function and the production of plasma levels of cytokine and chemokines (Schirmer et al., 2016). Using a commercially available cytokine array quantibody kit (RayBiotech; QAM-CYT-5), we quantified 40 cytokines and chemokines in the plasma samples of mice treated with vehicle or ABX. At 9 wk of age, numerous cytokines and chemokines showed changes in ABX-treated male mice compared with vehicle-treated controls, and these factors were completely restored to levels seen in vehicle-treated Tg male mice following administration of Tg-FMT (Fig. S2). Prominent reductions of basic fibroblast growth factor (bFGF) and GM-CSF in ABX-treated males, as well as significant elevations of leptin and MIG, were observed in plasma of ABX-treated males compared with control cohorts. Interestingly, Tg-FMT fully restored these cytokines/chemokines to levels observed in Tg control mice. Female mice showed either no differences or opposite differences in levels of these factors with ABX treatment compared with the male groups. Additional factors also showed a trend toward increased or decreased levels with ABX that were restored by Tg-FMT. Remaining cytokines/chemokines in this cytokine array quantibody kit (Eotaxin-1 [CCL11], Eotaxin-2 [MPIF-2/CCL24], Fas Ligand [TNFSF6], ICAM-1 [CD54], IFN-γ, IL-1 α [IL-1 F1], IL-1 β [IL-1 F2], IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-10, IL-12 p40, IL-13, IL-17A, IL-21, LIX, MCP-1 [CCL2], MCP-5, M-CSF, MIP-1 α [CCL3], Platelet Factor 4 [CXCL4], RANTES [CCL5], TARC [CCL17], I-309 [TCA-3/CCL1], TNF-α, and TNF RII [TNFRSF1B]) did not show any significant differences or trends among male or female groups. These findings suggest that short-term ABX-perturbed gut microbiome influences circulating chemokines and cytokines in a sex-specific manner and Tg-FMT results in complete restoration of these levels in ABX-treated male mice.
Figure S2.
Plasma cytokines and chemokines measured by RayBiotech cytokine array. Using serial standard concentrations provided in the QAM-CYT-5 kit manual, intensity values were plotted against cytokine concentrations for standard curve preparations and for cytokine calculations. The values were then recorded in picogram per milliliters and plotted using GraphPad Prism for comparison analysis among groups. bFGF, GM-CSF, leptin, and MIG showed significant differences among male groups only. Cytokines/chemokines such as BLC, G-CSF, IL-15, KC, MIP-γ, and TNF-Rll showed trends of reduction with ABX treatment in male mice compared with controls, whose levels were restored with FMT in ABX-treated male mice. Compared with those, CD30L showed a trend of increase in ABX-treated male mice compared with control male mice whose levels were restored with FMT in ABX-treated male mice. n = 4 or 5 mice per group. Data are mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. M_Ctr = vehicle-treated male, M_Abx = ABX-treated male from PND14–PND21 only, M_Abx+FMT = ABX-treated male (PND14–PND21), followed by FMT (PND24–PND63) from age-matched Tg donor male mice, F_Ctr = vehicle-treated female, and F_Abx = ABX-treated female from PND14–PND21 only. BLC, B lymphocyte chemoattractant; G-CSF, granulocyte CSF; KC, keratinocytes-derived chemokine.
Both Tg- and WT-FMT revert gut microbiota profiles in short-term ABX-treated male APPPS1 mice and impact both Aβ amyloidosis and microglial phenotypes in a similar manner
In the experiments presented above and our earlier efforts, we employed FMT from age-matched Tg donor males (Tg-FMT) into ABX-treated male mice in order to restore the ABX-perturbed microbiome. While successful (Fig. 1, Fig. 2, and Fig. 3), it was unclear whether expression of FAD-linked human PS1 or APP transgenes and potentially the presence of oligomeric Aβ species (see below) in the APPPS1-21 mice might have affected peripheral or central physiology in a manner that, in turn, influenced production of specific metabolites that drive the observed phenotypes. To rule out this possibility, we used the same short-term ABX paradigm in Tg male mice followed by FMT from age-matched non-Tg mice (WT-FMT) or Tg-FMT.
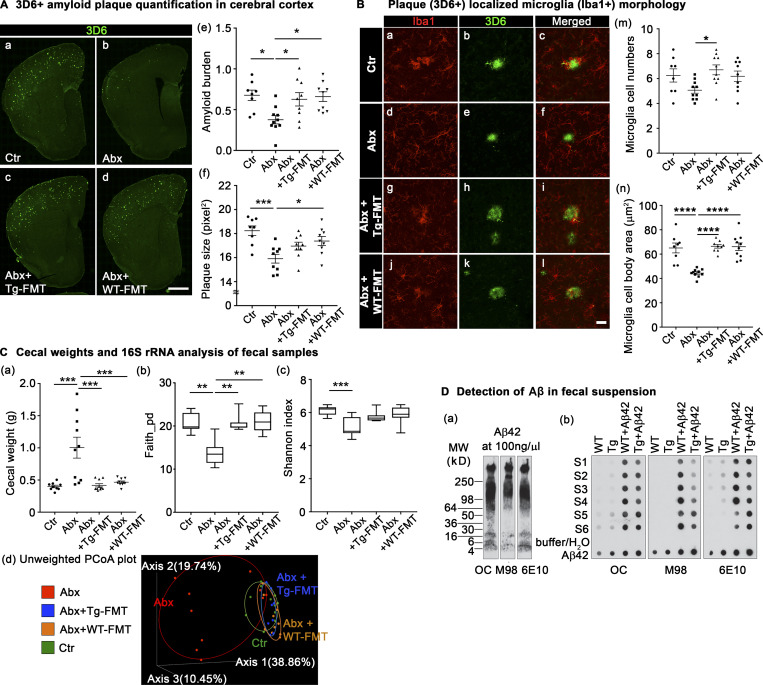
As expected, IHC using the 3D6 antibody revealed significantly reduced Aβ amyloidosis in ABX-treated male Tg mice compared with vehicle-treated controls (Fig. 5 A, a and b; quantified in e), but most importantly, both WT-FMT and Tg-FMT resulted in complete restoration of cerebral Aβ amyloidosis in ABX-treated APPPS1-21 male mice to levels observed in vehicle-treated mice (Fig. 5 A, a, c, and d; quantified in e). Amyloid plaque size analysis also showed reductions with ABX treatment and reversal with FMT treatment, independent of the donor (Fig. 5 A, f).
Figure 5.
FMT from either Tg (APPPS1-21) or WT (non-Tg littermates) male mice restores cerebral Aβ amyloidosis and microglial morphologies in ABX-treated male mice. (A) Representative immunofluorescence images of Aβ plaque burden in the cortex of Ctr (a), Abx (b), Abx+Tg-FMT (c), and Abx+WT-FMT (d) using anti–Aβ-specific antibody, 3D6. Quantification of Aβ burden (e) and amyloid size (f) was performed using threshold-limited particle analysis of 3D6 staining. Aβ burden (P = 0.013) and Aβ size (P = 0.0007) analyses showed a significant reduction in Abx compared with Ctr. These changes were reverted by FMT from either Tg or non-TG (WT) age-matched donor mice into ABX-treated male mice, similar to Ctr group (P > 0.05). (B) Representative images of 3D6+ plaque (green)-localized Iba1+ microglia cells (red) from Ctr (a–c), Abx (d–f), Abx+Tg-FMT (g–i), and Abx+WT-FMT (j–l). ImageJ quantification of plaque-localized microglial cell numbers (m) showed changes between Abx+Tg-FMT and Abx groups (P = 0.0234). Other groups showed no differences compared with controls (P > 0.05). Cell body area measures (n) showed reduced size in Abx compared with Ctr (P < 0.0001), which was restored with FMT from either Tg or WT donors. (C) Cecal weight analysis (a): ABX resulted in cecal enlargement (P = 0.0001), and both WT-FMT (P = 0.0004) and Tg-FMT (P = 0.0001) in ABX-treated male mice showed significantly lower cecal weights. The α-diversity was measured using Faith phylogenetic diversity (b) and Shannon index (c). ABX resulted in reduced Faith diversity in Abx compared with Ctr (b; Kruskal-Wallis [KW]: P = 0.0012). Both WT- and Tg-FMT showed significantly higher diversity compared with Abx (KW: P = 0.0017, P = 0.0033), similar to Ctr (KW: P > 0.99). Shannon index showed significantly lower diversity in Abx (KW: P = 0.0009) but no changes among other groups (c). Unweighted UniFrac PCoA plot (d): The two components explained 38.86% of the variance. Abx resulted in a separate cluster compared with Ctr, and both WT- and Tg-FMT–transplanted Abx groups overlapped with Ctr group. (D) OC, M98, and 6E10 antibodies were used to detect Aβ42 species. Western blot analysis of Aβ oligomers and fibers in Aβ42 solution incubated at RT for 8 d (a). Dot blot analysis of Aβ42 in fecal suspension and Aβ42-spiked fecal suspension (b). Buffer and MilliQ water were used as negative controls, and Aβ42 (10 ng, 20 ng, 50 ng, and 100 ng) was used as positive controls. S1–S6 represents n = 6 samples in both groups. Ctr = vehicle-treated male, Abx = ABX-treated (PND14–PND21) male, Abx+Tg-FMT = ABX-treated (PND14–PND21) male, followed by FMT (PND24–PND63) from age-matched Tg-donor male, Abx+WT-FMT = ABX-treated (PND14–PND21) male, followed by FMT (PND24–PND63) from age-matched non-Tg C57Bl6 donor male mice. n = 8–10 mice per group unless otherwise noted. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Scale bar in A, d represents 1,000 µm and applies to A, a–d. Scale bar in B, l represents 20 µm and applies to B, a–l. Detailed statistics are listed in Table S9. PCoA, principal coordinate analysis.
Evaluation of microglia cell numbers in a 0.02-mm2 area containing Aβ plaque showed no significant change in microglial cell numbers around Aβ plaques with ABX treatment compared with controls. However, the Tg-FMT group showed significantly higher microglial cell numbers compared with ABX-treated mice. Most importantly, both Tg- or WT-FMT groups showed no significant differences in microglial cell numbers compared with vehicle-treated Tg mice (Fig. 5 B, m). Similar to the results above (Fig. 3 D), we observed that microglial cell size was significantly reduced in ABX-treated mice compared with their vehicle-treated cohorts, but ABX-treated male mice subjected to either WT-FMT or Tg-FMT restored microglial cell body size to that observed in controls (Fig. 5 B, n).
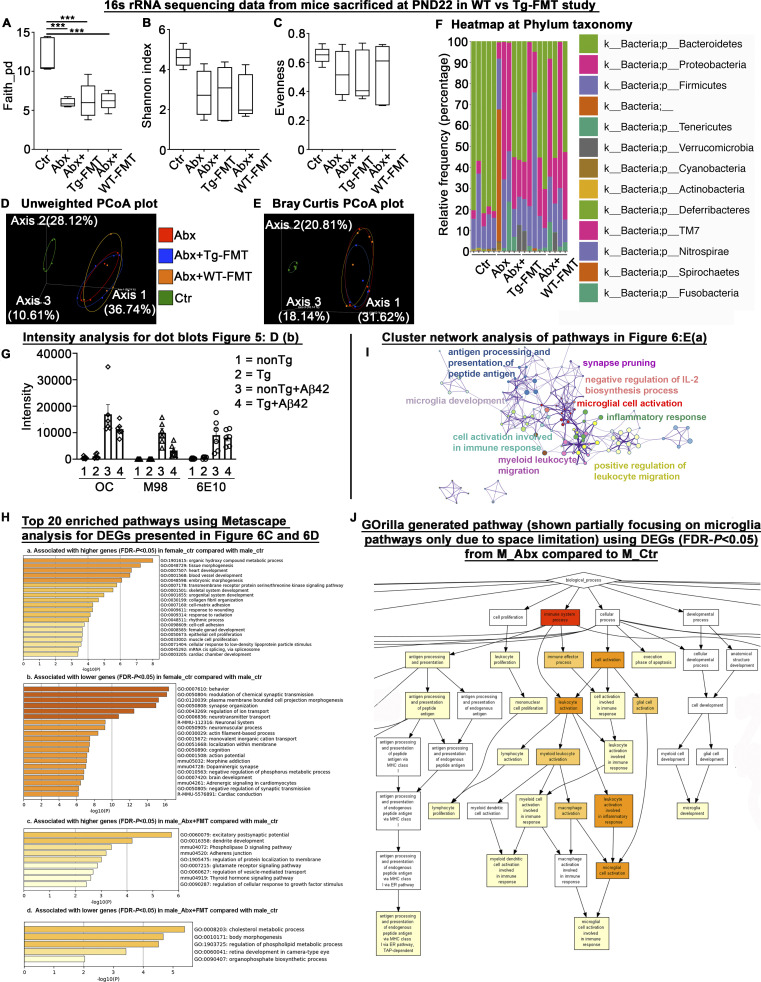
To investigate the microbiota profile differences between ABX and WT- or Tg-FMT mice groups, we performed 16S rRNA amplicon sequencing on fresh fecal pellets collected from individual mice 1 d after completion of ABX-treatments (PND22) and at the time of sacrifice (9 wk of age). At PND22, analysis of microbial α-diversity revealed significantly lower bacterial richness in all ABX groups compared with vehicle-treated male mice (Fig. S3 A). Shannon diversity indices and evenness indices showed no significant differences between vehicle-treated mice compared with all ABX-treated groups (Fig. S3, B and C). Analysis of β-diversity using the unweighted UniFrac metric (Fig. S3 D) showed significant differences between vehicle-treated mice and all ABX-treated groups (PERMANOVA, vehicle vs. ABX: P = 0.013; vehicle vs. ABX+Tg-FMT: P = 0.012; vehicle vs. ABX+WT-FMT: P = 0.006). As expected, no differences were observed between all ABX-treated groups at PND22 (PERMANOVA, ABX vs. ABX+Tg-FMT: P = 0.90; ABX vs. ABX+WT-FMT: P = 0.60). Bray-Curtis dissimilarities showed similar differences among all groups (Fig. S3 E). ANCOM (as mentioned above) identified differences between control and all ABX groups at phylum (Fig. S3 F) and species (Table S1) taxonomy. Altogether, these data suggest that ABX administration during PND14–PND21 resulted in microbiota perturbations at PND22 to all ABX groups and in a similar manner.
Figure S3.
16S rRNA sequencing analysis of fecal microbiota profile at PND22 in WT- versus Tg-FMT study groups, intensity measurements for dot blots presented in Fig. 5 D, b; cluster network analysis of lower DEGs in M_Abx group presented in Fig. 6 E, a; and RNA-seq data analysis from short-term ABX male and female mice sacrificed at 9 wk of age. The 16s rRNA was employed to confirm the ABX-related fecal microbiota perturbations in ABX-, ABX+WT-FMT, and ABX+Tg-FMT groups compared with the vehicle-treated male group a day after cessation of ABX (PND22). (A) Faith’s diversity showed significant differences among groups (one-way ANOVA: F(3,15) = 13.93, P = 0.0001). Compared with vehicle-treated male mice, ABX-treated mice in ABX, ABX+Tg-FMT, and ABX+WT-FMT groups showed significantly lower diversity (Ctr vs. ABX, Ctr vs. ABX+Tg-FMT, Ctr vs. WT+FMT; P = 0.0006 for each comparison). (B and C) Shannon index (B) and evenness (C) showed no significant differences among groups (P > 0.05). (D) PCoA plot generated using unweighted version of the UniFrac distance metric. The two components explained 36.74% of the variance. ABX treatments resulted in separate clusters compared with Ctr group, and ABX, ABX+Tg-FMT, and ABX+WT-FMT group clusters overlapped with each other at PND22. (E) Bray Curtis PCoA plot showed similar results as unweighted PCoA plot. (F) Relative frequency percentage was plotted at the phylum taxonomy. Ctr = vehicle-treated male, Abx = ABX-treated male, Abx+Tg-FMT = ABX-treated male with FMT from age-matched Tg donor male mice, and Abx+WT-FMT = ABX-treated male with FMT from age-matched non-Tg C57Bl6 donor male mice. n = 4 or 5 mice per group for A–F. Data are mean ± SEM for A–F. ***, P < 0.001. (G) Graphical presentation of intensity measurements for dot blots presented in Fig. 5 D, b. Error bars are mean ± SEM. (H, a) Heatmap associated with higher genes (FDR-P < 0.05) in vehicle-treated female compared with vehicle-treated male mice. (H, b) Heatmap associated with lower genes (FDR-P < 0.05) in vehicle-treated female compared with vehicle-treated male mice. (H, c) Heatmap associated with higher genes (FDR-P < 0.05) in FMT-transplanted ABX-treated male mice compared with vehicle-treated male mice. (H, d) Heatmap associated with lower genes (FDR-P < 0.05) in FMT-transplanted ABX-treated male mice compared with vehicle-treated male mice. (I) cluster network analysis of lower DEG-associated pathways in M_Abx compared with M_Ctr group presented in Fig. 6 E, a. (J) GO biological process analysis using GOrilla. Identified microglial activation as a major impacted pathway similar to topmost GO biological pathways analyzed by Metascape depicting a crucial involvement of microglia in our established model. PCoA, principal coordinate analysis.
To further validate the efficacy of ABX and ABX+FMT treatments, we measured cecal weights at the time of sacrifice at 9 wk of age (Fig. 5 C, a). As expected, ABX-treated mice showed significantly higher cecal weights compared with the vehicle-treated mice, and both WT- and Tg-FMT resulted in complete restoration of cecal enlargements. At 9 wk of age, analysis of microbial α-diversity revealed significantly lower bacterial richness in ABX-treated males compared with their vehicle-treated counterparts (Fig. 5 C, b), and both WT- and Tg-FMT treated groups showed higher bacterial richness compared with ABX-treated mice. In contrast, Shannon index showed significantly lower diversity in ABX-treated mice only and no changes among other groups (Fig. 5 C, c). Analysis of β-diversity using the unweighted UniFrac metric identified no differences between vehicle-treated male, ABX+Tg-FMT, and ABX+WT-FMT groups (PERMANOVA, vehicle vs. ABX+Tg-FMT: P = 0.053; vehicle vs. ABX+WT-FMT: P = 0.062; ABX+Tg-FMT vs. ABX+WT-FMT: P = 0.08), while ABX-treated mice were significantly different from vehicle-treated mice (PERMANOVA, P = 0.001; Fig. 5 C, d). ANCOM identified several taxa differences between ABX and control groups (Table S2). Interestingly, both WT- and Tg-FMT–treated groups showed a majority of taxa restoration compared with ABX-treated males and similar to those observed in vehicle-treated groups.
Absence of Aβ oligomers in Tg-FMT
Having demonstrated that age-matched Tg-FMT introduced into ABX-treated male mice restores Aβ amyloidosis and microglial phenotypes to levels seen in vehicle-treated Tg mice, we asked whether these phenotypes might be the result of inadvertent introduction of Aβ seeds that might be present in the fecal slurries from Tg mice fecal pellets that could transit from the gut to the brain and drive amyloidosis. To investigate the presence of Aβ oligomers and fibrils in fecal samples of Tg mice at 9 wk of age, we performed dot blot analysis using three Aβ-specific antibodies (OC, Kayed et al., 2007; M98, Nussbaum et al., 2012; McLean et al., 2013; and 6E10, Thinakaran et al., 1996). We first prepared a mixture of oligomerized Aβ42 peptides and performed Western blot assays using the three antibodies. On 4–20% gradient gels, OC and 6E10 recognize monomeric, oligomeric, and high-molecular-weight Aβ42 aggregates, while M98 mainly recognizes high-molecular-weight Aβ42 aggregates (Fig. 5 D, a). Fecal suspensions from six (S1–S6) non-Tg (WT) and APPPS1-21 (Tg) animals as well as suspensions spiked with aged Aβ42 solution (WT+Aβ42, Tg+Aβ42, Aβ42 final concentration = 100 ng/µl) were dot blotted in triplicate on nitrocellulose membranes, and aged Aβ42 solutions (10 ng, 20 ng, 50 ng, and 100 ng) were used as positive controls and PBS or H2O was used as negative control (Fig. 5 D, b). OC and human-specific 6E10 detected very low signals, if any, in fecal suspensions but strong signals in Aβ42-spiked suspensions (Fig. S3 G). As mAb 6E10 is human specific, any signals detected in the slurry from WT animals must be nonspecific. Finally, M98, specific for Aβ fibrils, only detected signals in spiked suspensions and with pure Aβ42.
These data suggest that the FMT from age-matched WT or Tg male mice are, at least in this semi-quantitative assessment, devoid of Aβ oligomers/fibrils; hence, it is highly unlikely that the FMT-mediated restoration of Aβ amyloidosis in ABX-treated APPPS1-21 mice is driven by nucleation by Aβ seeds present in the fecal slurries.
Transcript levels of cortical mRNAs are altered in short-term ABX-treated male mice and largely restored with Tg-FMT
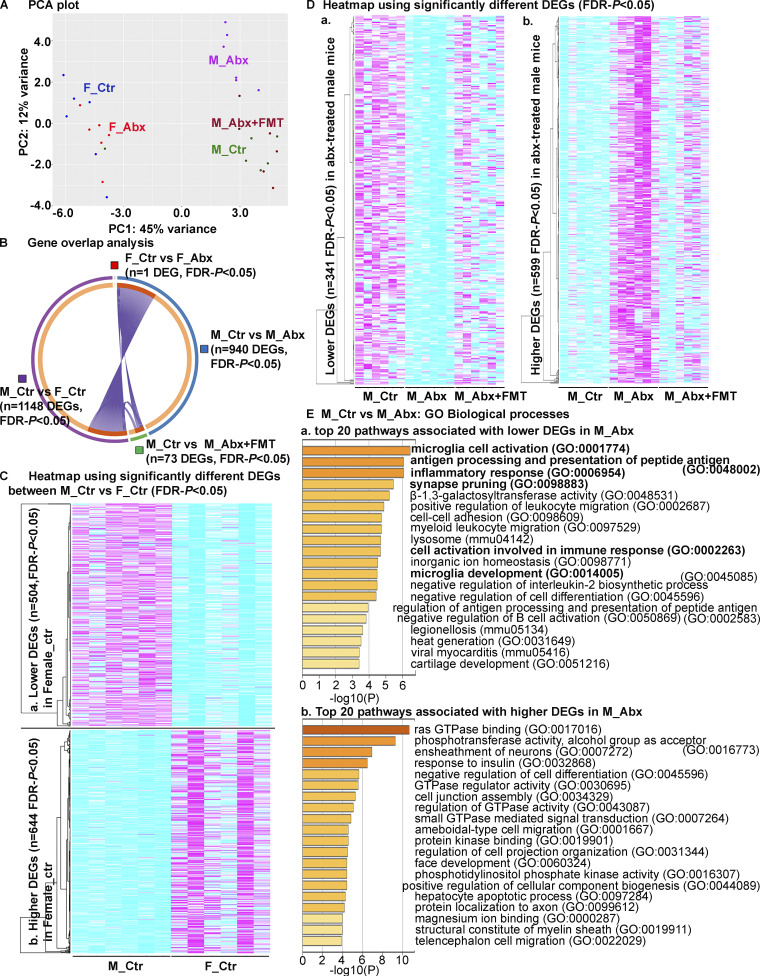
We extracted RNA from the dorsal cerebral cortex of cohorts of male and female APPPS1-21 Tg mice subject to vehicle or short-term ABX and then analyzed transcriptome changes using RNA sequencing (RNA-seq) at the time of sacrifice. Principal component analysis (PCA) revealed sex-specific differences in vehicle- and ABX-treated APPPS1-21 groups (Fig. 6 A). Here, we observed marked differences in gene expression between vehicle-treated male and female groups (n = 1,148 differentially expressed genes [DEGs]; false discovery rate [FDR]–P < 0.05; Fig. 6 B). We used a metascape gene ontology (GO) analyses platform to assign biological relevance to a set of higher (Fig. 6 C, a) and lower (Fig. 6 C, b) transcripts. Compared with the male group, female mice showed higher expression of genes associated with metabolic process (GO1901615), tissue morphogenesis (GO0048729), and many others (Fig. S3 H) and lower expression of genes associated with behavior (GO0007610), chemical synaptic transmission (GO0050804), synapse organization (GO0050808), and other neuronal- or neurotransmitter-related pathways (Fig. S3 H). ABX treatment resulted in significant alterations in the male cohort (n = 940 DEGs; 599 were up-regulated and 341 were down-regulated in ABX-treated male mice; FDR-P < 0.05; Fig. 6 B), while the female mice did not show major changes between groups (n = 1 DEG; FDR-P < 0.05; Fig. 6 B). Most importantly, Tg-FMT in ABX-treated male mice restored the majority of transcriptome changes to those seen in vehicle-treated males (n = 73 DEGs; 21 significantly lower and 52 significantly higher compared with male control; FDR-P < 0.05; Fig. 6, B, and D). Interestingly, when compared with vehicle-treated males, ABX-treated males showed lower expression of genes associated with multiple pathways related to the immune system. Specifically, microglial cell activation (GO0001774), antigen processing and presentation of peptide antigen (GO0048002), inflammatory response (GO0006954), positive regulation of leukocyte migration (GO0002687), myeloid leukocyte migration (GO0097529), cell activation involved in immune response (GO0002263), microglia development (GO0014005), negative regulation of IL-2 biosynthesis (GO0045085), negative regulation of B cell activation (GO0050869), and others (Fig. 6 E, a; and Fig. S3 I). Moreover, genes associated with microglia activation, including Aif1, App, C1qa, Cst7, Cx3cr1, Tyrobp, Tlr2, and Trem2, were significantly down-regulated (Fig. 6 E, a; Table S3). Upon expansion of this highly ranked category, we detected several cytokine production pathways (IL-1B, IL-10, IL-6, TNF-superfamily, IFN-γ: GO:0032652, GO:0032675, GO:0001817, GO:0032731, GO:0032635, GO:0001819, GO:0032612, GO:0032732, GO:0032755, GO:0001816, GO:0032653, GO:0042533, GO:0032651, GO:0032680, GO:1903555, GO:0042035, GO:0071706, GO:0042535, GO:0042089, GO:0032760, GO:1903557, GO:0042107, GO:0032640), macrophage activation (GO:0002281, GO:0042116), leukocyte activation (GO:0002269, GO:0002274), and detection of external biotic stimulus (GO:0098581, GO:0009595) associated with lower expression of genes in ABX-treated male mice (Table S3). These pathways were also among the highly ranked pathways associated with lower gene expression in ABX-treated male mice when this dataset was evaluated using GO enrichment analysis and visualization tool (GOrilla; Eden et al., 2009) online services (Fig. S3 J). Molecular functions associated with lower expression of lysosomal genes (mmu04142), including Abca2, Cd68, Ctsd, Ctss, Hexb, Idua, Laptm5, Atp6v0a2, Naglu, Ctsz, Gusb, Slc17a5, and Gnptab, were highly ranked. Additionally, ABX-treated male mice showed higher expression of genes associated with many pathways, including Ras GTPase-binding (GO0017016), ensheathment of neurons (GO0007272), subgroups of gliogenesis (GO0042063), glial cell differentiation (GO0010001), oligodendrocyte differentiation (GO0048709), and astrocyte differentiation (GO0048708; Table S4). Most importantly, these pathways were specific to the ABX-treated male versus vehicle-treated male comparisons and were not enriched in the vehicle-treated female versus vehicle-treated male or ABX+FMT–treated male versus vehicle-treated male groups (Fig. S3 H).
Figure 6.
Short-term ABX results in sex-specific changes of cortical transcriptome profiles. (A) PCA plot of cerebral cortex transcriptomes. n = 6 mice per group. (B) Gene overlap analysis showing number of significantly different genes. Note the numbers of significant DEGs were the highest between sexes (n = 1,148 DEGs: M_Ctr vs. F_Ctr). ABX treatment in male groups showed higher number of DEGs (n = 940, M_Ctr vs. M_Abx) compared with the female groups (n = 1 DEG, F_Ctr vs. F_Abx). Only 73 DEGs were significantly different between M_Ctr vs. M_Abx+FMT, suggesting that FMT restored the transcriptome profiles in M_Abx mice to those of M_Ctr group. (C) Heatmap of DEGs (FDR-P < 0.05) between M_Ctr and F_Ctr with clusters (left). 504 DEGs were significantly lower (a), and 644 DEGs were significantly higher (b) in F_Ctr compared with M_Ctr. Each column is an individual animal, n = 6 mice per group. (D) Heatmap of DEGs (FDR-P < 0.05) between M_Ctr and M_Abx with clusters (left). 341 DEGs were significantly lower (a), and 599 DEGs were significantly higher (b) in M_Abx compared with M_Ctr. Compared with M_Ctr, M_Abx+FMT showed 73 DEGs (55 DEGs were higher, and 21 DEGs were lower) that were significantly different. Each column is an individual animal, n = 6 mice per group. (E) GO biological processes, molecular functions, and KEGG pathways analysis based on the DEGs (FDR-P < 0.05) between M_Ctr and M_Abx groups. Panels show heatmap of top 20 down-regulated pathways (a), while b shows top 20 down-regulated pathways in M_Abx compared with M_Ctr using Metascape. M_Ctr = vehicle-treated male, M_Abx = ABX-treated (PND14–PND21) male, M_Abx+FMT = ABX-treated (PND14–PND21) male, followed by FMT (PND24–PND63) from age-matched Tg-donor male, F_Ctr = vehicle-treated female, and F_Abx = ABX-treated (PND14–PND21) female. GTPase, guanosine triphosphatase.
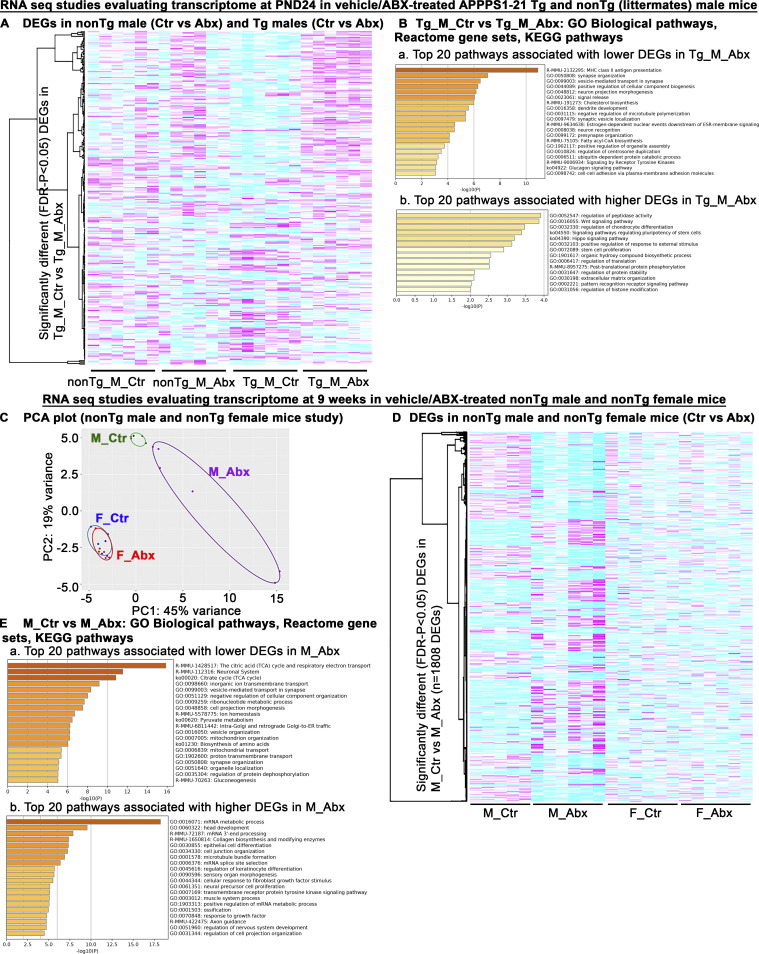
In addition to the 9 wk of ABX and vehicle comparisons in Tg male mice (Fig. 6), we investigated the transcriptome profiles in Tg male and non-Tg male littermates 3 d after cessation of treatment (PND24). In this study, animals (non-Tg littermates and Tg male mice) were administered ABX or vehicle (PND14–PND21) and were sacrificed at PND24. RNA-seq analysis was performed on RNA from the dorsal cerebral cortex as above. ABX treatment showed changes in DEGs in a genotype-dependent manner (Tg_M_Ctr vs. Tg_M_Abx: n = 208 DEGs; FDR-P < 0.05; Table S5; and nonTg_M_Ctr vs. nonTg_M_Abx: n = 79 DEGs; FDR-P < 0.05; Fig. S4 A and Table S6). Metascape pathway analyses of significantly lower and higher DEGs revealed several pathways that were affected in ABX-treated Tg male mice compared with vehicle-treated Tg male mice at PND24 (Fig. S4 B). MHC class ll antigen presentation (R-MMU-2132295) was the topmost pathway associated with reduced DEGs in ABX-Tg male mice, suggestive of reduced microglial or astrocytic priming at PND24 age (Das and Chinnathambi, 2019) and could be related to reduced microglial cell activation observed in ABX-Tg male mice at 9 wk of age (GO:0001774; Fig. 6 E). Up-regulation of MHC class II is an early sign of microglia activation in response to central nervous system (CNS) injury (Aloisi et al., 2000).
Figure S4.
Short-term ABX treatment alters cerebral cortex transcriptome profiles at PND24 in a genotype-dependent manner, and short-term ABX treatment in non-Tg male and female littermates alters cerebral cortex transcriptome profiles in a sex-specific manner. In this study, animals (non-Tg littermates and Tg male mice) were administered ABX or vehicle (PND14–PND21) and were sacrificed at PND24, 3 d after the cessation of ABX/vehicle treatments. RNA-seq analysis was performed on RNA from the dorsal cerebral cortex. (A) ABX treatment showed changes in DEGs in a genotype-dependent manner (Tg_M_Ctr vs. Tg_M_Abx: n = 208 DEGs; FDR-P < 0.05; Table S5; and non-Tg_M_Ctr vs. non-Tg_M_Abx: n = 79 DEGs; FDR-P < 0.05; Table S6). (B, a) Heatmap associated with lower genes (FDR-P < 0.05) in ABX-treated APPPS1-21 Tg male compared with vehicle-treated APPPS1-21 Tg male mice at PND24. (B, b) Heatmap associated with higher genes (FDR-P < 0.05) in ABX-treated APPPS1-21 Tg male compared with vehicle-treated APPPS1-21 Tg male mice at PND24. (C–E) Dorsal cortical transcriptome of cohorts of non-Tg male and non-Tg female mice subject to ABX or vehicle treatment from PND14 to PND21 and sacrificed at 9 wk of age. (C) PCA plot. (D) Significantly different DEGs from non-Tg M_Ctr compared with non-Tg M_Abx groups are plotted using a heatmap to visualize the sex-specific alterations in the cortical transcriptome. ABX- and vehicle-treated non-Tg male mice showed profound differences in cerebral cortex transcripts (n = 1,808 DEGs; FDR-P < 0.05), while non-Tg female mice showed no major changes between ABX- and vehicle-treated groups (n = 1 DEG; FDR-P < 0.05). (E, a) Heatmap associated with lower genes (FDR-P < 0.05) in ABX-treated male mice compared with vehicle-treated male mice. (E, b) Heatmap associated with higher genes (FDR-P < 0.05) in ABX-treated male mice compared with vehicle-treated male mice. acyl-CoA, acyl Coenzyme A; ESR, estrogen receptors.
We then asked whether the sex-specific transcriptome differences (Fig. 6) observed herein, and as reported previously (Dodiya et al., 2019), are specific to the APPPS1-21 Tg line by examining the dorsal cortical transcriptome of cohorts of non-Tg male and female mice subject to ABX or vehicle treatment from PND14 to PND21 and sacrificed at 9 wk of age. As observed above, ABX treatments resulted in sex-specific group differences as shown in a PCA plot (Fig. S4 C). Marked differences in gene expression were observed between ABX- and vehicle-treated non-Tg male mice (n = 1,808 DEGs; FDR-P < 0.05; Fig. S4, D and E; and Table S7), while non-Tg female mice showed no major changes between ABX- and vehicle-treated groups (n = 1 DEG; FDR-P < 0.05; Fig. S4 D and Table S8).
Collectively, our RNA-seq data from Tg and non-Tg studies suggest that early life perturbations of the gut microbiome alter the cortical transcriptome profile in a sex-specific manner. Specific to Tg APPPS1-21 male mice, ABX treatment results in alterations in cortical transcripts that are associated with microglia priming or activation at both PND24 and 9 wk of age. Most importantly, Tg-FMT restored the majority of transcriptional readouts observed in the ABX-treated Tg-male mice group to levels observed in vehicle-treated animals at 9 wk of age. These transcriptome changes related to microglial physiology suggest that microglia play a central role in driving the pathological and physiological phenotypes that are observed in the short-term ABX treatment paradigm used herein.
Microglia play an essential role in mediating short-term ABX-perturbed microbiome’s effect on Aβ amyloidosis
The mechanism(s) by which Tg- or WT-FMT restores Aβ amyloidosis in ABX-treated male mice is unclear. However, we have clearly demonstrated that ABX-treated male mice show significant effects on microglial gene expression and morphology. To determine the role of microglia in modulation of Aβ amyloidosis, we used an established strategy to deplete microglia in the brain (Dagher et al., 2015). Microglia originate from the yolk sac and colonize the CNS during early development through the action of CSF1, which regulates proliferation, differentiation, and function of macrophage lineage cells through binding to its specific receptor, CSF1R (Erblich et al., 2011). Under physiological conditions, microglia are the principal cells in the CNS that express CSF1R (Erblich et al., 2011), and PLX5622, a potent inhibitor of CSF1R tyrosine kinase activity (inhibitory constant = 5.9 nM) with more than 50-fold selectivity over a panel of 230 kinases, depletes over 98% of microglia in mice treated for 7 d (1,200 mg/kg in chow; Dagher et al., 2015). Additionally, microglia elimination leads to no discernable deficits in behavior or learning and memory in the tasks tested (Dagher et al., 2015; Elmore et al., 2014). We used our established short-term ABX paradigm, wherein ABX groups received a high-dose ABX cocktail from PND14 to PND21. ABX+FMT groups were gavaged with ABX in a similar fashion, followed by Tg-FMT from PND24 till the time of sacrifice. Microglial depletion groups received PLX5622 chow starting 3 d after vehicle/ABX treatments (PND24), with or without FMT gavages (PND24 till time of sacrifice). Mice were sacrificed at 9 wk or 3 mo of age to evaluate the role of microglia in microbiome-mediated cerebral amyloidosis. Efficacy of the ABX treatment was confirmed using cecal weights at the time of sacrifice at both 9 wk (Fig. 7 A) and 3 mo of age (Fig. 7 B). As expected, ABX-treated mice showed significantly larger ceca compared with their vehicle-treated counterparts at both 9 wk and 3 mo of age. Compared with ABX-treated mice, FMT into ABX-treated mice resulted in a reduction of cecal weight to that observed in vehicle-treated mice.
Figure 7.
PLX5622-induced microglia depletion fails to reduce amyloidosis in ABX-treated male mice. (A–F) Male mice were assigned to vehicle (PND14–PND21), ABX (PND14–PND21), or ABX+FMT (ABX [PND14–PND21]+FMT [PND24–until sacrifice]) groups that were subjected to control or PLX5622 diet (PND24–until sacrifice) and sacrificed at 9 wk of age (A, C, and D) or 3 mo of age (B, E, and F). At both 9 wk (A) and 3 mo (B) of age, ABX resulted in cecal enlargement in control and PLX groups, while Abx+FMT showed cecal weights similar to controls. (C) Representative high magnification (a–f) images of 3D6+ plaque (green)-localized Iba1+ microglia (red) from Ctr (a and d), Abx (b and e), and Abx+FMT (c and f) male mice that were assigned to a control (a–c) or PLX5622 (d–f) diet. Note the complete absence of microglia and altered plaque morphology in PLX5622-treated groups (d–f). (D) Representative images of 3D6+ Aβ plaques in Ctr (a and d), Abx (b and e), and Abx+FMT (c and f) groups assigned to a control (a–c) or PLX5622 (d–f) diet. Quantification of Aβ plaque burden using threshold-limited particle analysis of 3D6+ staining is shown in g. ABX treatment resulted in significant reduction of amyloid burden only in control diet–treated mice, and FMT reverted these changes similar to control group. No significant differences in amyloid burden were observed among PLX5622 groups. (E) Representative high-magnification (a–f) images of 3D6+ plaque (green)-localized Iba1+ microglia (red) from Ctr (a and d), Abx (b and e), and Abx+FMT (c and f) male mice that were assigned to a control (a–c) or PLX5622 (d–f) diet. Note the complete absence of microglia and altered plaque morphology in PLX5622-treated groups (d–f). (F) Representative images of 3D6+ Aβ plaques in Ctr (a and d), Abx (b and e), and Abx+FMT (c and f) male mice that were assigned to a control (a–c) or PLX5622 (d–f) diet. Quantification of Aβ plaque burden using threshold-limited particle analysis of 3D6+ staining is shown in g. ABX-treatment showed significant reduction in amyloid burden only in control diet–treated groups, and FMT reverted these changes similar to control group. No differences in amyloid burden were observed among PLX5622 diet–treated groups (P > 0.99). Plaque size quantification (h) showed significant differences within control diet–treated groups. Ctr = vehicle-treated male, Abx = ABX-treated male, Abx+FMT = ABX-treated male with FMT from age-matched Tg donor male mice, PLX = PLX5622 diet–treated vehicle-gavaged mice, PLX+Abx = ABX-treated male mice on PLX5622 diet, PLX+Abx+FMT = ABX-treated male with FMT from age-matched Tg donor male mice on PLX5622 diet. n = 8–10 mice per group. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Scale bars in C, f and E, f represent 15 µm and apply to C, a–f and E, a–f, while scale bars in D, f and F, f represent 1,000 µm and apply to D, a–f and F, a–f. Detailed statistics are listed in Table S9.
PLX5622 treatment of APPPS1-21 mice for 9 wk resulted in >98% elimination of microglia throughout the cerebral cortex compared with control diet–treated groups (Fig. 7 C, a–f). As expected, IHC revealed significantly lower levels of 3D6+ amyloid plaques in ABX-treated male mice (Fig. 7 D, b; quantified in g), and Tg-FMT significantly elevated Aβ amyloidosis in the ABX-treated animals (Fig. 7 D, c; quantified in g). Importantly, PLX5622-mediated elimination of microglia resulted in significantly lower Aβ plaque burden (Fig. 7 D, d; quantified in g). These levels were similar to the levels observed in the control diet–treated ABX group (Fig. 7 D, b; quantified in g). All PLX5622 diet–treated groups showed no differences in Aβ deposition (Fig. 7 D, d–f; quantified in g), with amyloid burden closer to zero compared with the control diet–treated vehicle group. The absence of Aβ burden (floor effect) among all PLX groups impeded our investigations of a potential microglial role in established microbiome-driven Aβ amyloidosis in this 9-wk-old cohort. Hence, we chose to perform a longer-term PLX study wherein mice received similar treatments as indicated above until the time of sacrifice at 3 mo of age. Similar to PLX5622-treated male mice at 9 wk of age (Fig. 7 C, d–f), PLX5622-treated male mice at 3 mo of age also showed depletion of microglia around the majority of Aβ plaques in the cerebral cortex compared with control diet–treated mice (Fig. 7 E, a–f). We continuously observed similar beneficial effects of ABX at 3 mo of age, and FMT showed complete reversal of amyloidosis (Fig. 7 F, a–c; quantified in g). Interestingly and in contrast to the results observed in mice treated with PLX5622 for 9 wk (Fig. 7 D), PLX5622 diet–treated mice showed no significant differences in amyloid burden compared with control diet–treated mice at 3 mo of age (Fig. 7 F, a and d; quantified in g). Most importantly, ABX administration (Fig. 7 F, e) showed no significant changes in amyloidosis after PLX5622 treatment compared with either control diet–treated (Fig. 7 F, a) or PLX5622 diet–treated (Fig. 7 F, d) vehicle groups (Fig. 7 F, quantified in g). It is noteworthy that the remaining plaques in PLX5622-treated mice at 9 wk (Fig. 7 C) and 3 mo of age (Fig. 7 E) are reminiscent of the plaque morphologies observed in TREM2-deficient mice (Ulland et al., 2017) and in 5XFAD mice treated with PLX5622 (Spangenberg et al., 2019), settings in which plaques exhibit a spiculed, less-compacted structure. These data suggest that microglial cells play a critical role in mediating the ABX-driven lowering of cerebral Aβ amyloidosis.
Discussion
A series of preceding efforts from our group has demonstrated that male APPPS1-21 mice treated with ABX throughout life had reductions in levels of deposited Aβ peptides and alterations in the morphology and transcriptional readouts of cortical microglia, suggestive of an M0, homeostatic signature, compared with male APPPS1-21 mice treated with vehicle (Dodiya et al., 2019). Moreover, FMT from male APPPS1-21 mice into long-term ABX-treated male APPPS1-21 mice leads to a partial restoration of Aβ deposition and microglial transcriptional signatures associated with a neurodegenerative MGnD phenotype that is a feature of control APPPS1-21 mice at the time of sacrifice (Dodiya et al., 2019). In view of the demonstration that FMT only leads to partial restoration of Aβ deposition in long-term ABX-treated animals, we surmised that ABX in the drinking water likely eliminated specific bacterial species and/or their metabolites in the FMT that might be critical for driving Aβ pathology. To address this issue, we chose to employ a paradigm that we previously demonstrated as resulting in lowering Aβ burden in male APPSWE/PS1DE9 mice (Minter et al., 2017). Using this short-term ABX paradigm, wherein APPPS1-21 mice were subjected to oral gavage only at PND14–21, then housed with regular drinking water till the time of sacrifice, we now demonstrate that short-term ABX treatment results in reduced Aβ amyloidosis and altered microglial phenotypes in APPPS1-21 male mice. To these findings, we now offer several additional insights that strengthen our hypothesis that the gut microbiota play an important, albeit presently mechanistically silent, role in the regulation of Aβ amyloidosis and microglial physiology. First, short-term ABX treatment leads to reductions in levels of FA–soluble and deposited Aβ peptides only in male animals, and FMT, devoid of Aβ fibrils, from either Tg or WT age-matched donor male mice leads to complete restoration of these levels. Second, sex-specific differences in bacterial taxa are readily apparent following short-term ABX treatment at 9 wk of age. Third, at the time of sacrifice, we observed that the morphology of plaque-associated microglia in male, but not female, APPPS1-21 mice treated with short-term ABX is markedly altered, suggestive of a homeostatic M0 phenotype, and that FMT from male APPPS1-21 mice fully reverts these phenotypes to an MGnD/DAM state that is observed in mice treated with vehicle. Fourth, Aβ amyloidosis–related apparent degenerative marker changes (neuritic loss, axonal dysfunction, synaptic loss, and myelin loss) were drastically reduced in ABX-treated male mice, and FMT reverted these changes in ABX-treated male mice. Fifth, transcriptomic analysis of total cortical RNA revealed that short-term ABX treatment of male mice leads to modulation of microglial activation and development pathways, as described earlier (Erny et al., 2015), and that ABX-treated mice provided with FMT leads to a restoration of these transcript levels to those seen in control mice. Finally, we assessed the contribution of microglia to ABX-mediated, reduced Aβ amyloidosis in APPPS1-21 mice at 9 wk and 3 mo of age. We now report that short-term ABX fails to reduce Aβ amyloidosis following CSF1R antagonist–mediated elimination of microglia. Collectively, our studies strengthen our previous findings that the microbiome plays an important role in sex-specific modulation of Aβ amyloidosis in the APPPS1-21 model and that microglia play an essential role in mediating the effects of ABX-perturbed microbiome on cerebral amyloid deposition in male mice.
Our current findings and those reported earlier (Dodiya et al., 2019) document sex-specific differences in ABX-perturbed microbiome-mediated alterations of Aβ deposition and microglial phenotypes. Absent mechanistic explanations for these observations, we have shown considerable differences in the microbiome of male versus female animals treated with ABX, and it is tempting to speculate that these differences might underlie the sex-specific modulation of cortical Aβ pathology and microglial phenotypes. One such taxon, Akkermansia muciniphila, consistently resulted in higher abundance in ABX-treated female mice compared with vehicle-treated female animals, while at lower abundance in ABX-treated male compared with vehicle-treated male mice in the current and previous reports (Dodiya et al., 2019), suggesting a potential involvement of this taxon in the observed differences in amyloidosis. Akkermansia has been shown to be associated with various neurodegenerative disorders (Keshavarzian et al., 2015; Jangi et al., 2016; Berer et al., 2017; Engen et al., 2017). However, the exact role(s) of A. muciniphila (and other significantly different taxa) on Aβ deposition and microglial phenotypes can only be determined using transplantation studies using selected bacteria or a combination of strains in well-characterized GF or pseudo-GF mouse models of Aβ amyloidosis. Additionally, sex-specific gut microbiota differences have been reported due to hormonal effects (Org et al., 2016) that are strain dependent. In addition, the gut microbiome may mediate sex-differences in host physiology (Weger et al., 2019) and diseases, including autoimmunity (Yurkovetskiy et al., 2013), liver cancer (Xie et al., 2017), metabolic disorders (Peng et al., 2020), and potentially neurodegenerative diseases (Cox et al., 2019). It will be critical to assess the impact of ABX-mediated differences in the sex-specific microbiome on cortical Aβ pathology and microglial phenotypes using GF AD models as well as in ovariectomized or masculinized models to address bacterial strain effects on brain physiology in a sex-dependent manner.
It is plausible that altered peripheral inflammation in ABX-treated male mice exerts effects on microglia, thus leading to reduced amyloidosis compared with ABX-treated female mice; several reports have invoked an involvement of peripheral inflammation in central inflammation (Perry et al., 2007; Wendeln et al., 2018). In this regard, either gut microbiota (Dorrestein et al., 2014; Ivanov et al., 2009) or microbial metabolites (Wikoff et al., 2009) could affect peripheral immune cells to modulate mucosal immune responses (Kamada et al., 2013; Mazmanian et al., 2005) and systemic immunity (Belkaid and Naik, 2013). Specifically, we herein report that ABX-treated male mice exhibited significant reductions in levels of bFGF and GM-CSF and increased levels of leptin and MIG in plasma. Most importantly, FMT into ABX-treated males restored the levels of these factors in the plasma to those observed in vehicle-treated male animals. In this regard, bFGF, a growth factor secreted by macrophages and brain resident glial cells, was shown to affect Aβ pathophysiology (Gómez-Pinilla et al., 1990), possibly via glial interactions (Quon et al., 1990). GM-CSF is a well-characterized pro-inflammatory factor that was shown to have direct effects on microglial activity/proliferation (Giulian et al., 1994; Zaheer et al., 2007), and anti–GM-CSF antibody delivered into the brains of Tg2576 mice resulted in altered microglial activity and reduced levels of Aβ1-42 (Manczak et al., 2009). Thus, it is not inconceivable that reduction in levels of bFGF and GM-CSF in the periphery could influence microglial activity that, in turn, might affect amyloidosis outcomes in ABX- and ABX+FMT–treated mice. MIG (CXCL9) is another chemokine that is considered to play a role in the interplay between neurons and glial cells, but very few studies (Lee et al., 2008) have explored the role of this chemokine in Alzheimer’s subjects or mouse models. Leptin, an adipocytokine produced in both the periphery and centrally, is proposed to have beneficial effects on Aβ amyloidosis (reviewed in Marwarha and Ghribi, 2012); delivery of leptin into the brains of APPSWEPS1ΔE9 mice reduced Aβ pathology, and it was suggested that the mechanism involved enhanced microglial phagocytosis (Pérez-González et al., 2014). Future studies using brain-impermeant CSF1R-antagonists, such as PLX73086, that deplete peripheral myeloid cells (Bellver-Landete et al., 2019) may allow us to delineate the potential role of peripheral CSF1R-expressing cells on microglial function in our model.
It should be noted that Sun et al. (2019) recently reported that 6-mo-old APPSWEPS1ΔE9 mice treated with ABX for 3 d and subsequently provided with WT-FMT daily for a month resulted in lower levels of Aβ pathology and decreased τ phosphorylation compared with APPSWEPS1ΔE9 at 7 mo of age. In addition, this study also showed that WT-FMT into 6-mo-old APPSWEPS1ΔE9 mice also increased levels of PSD95 and synapsin l. In contrast, we show that ABX (PND14–PND21) results in reduced amyloidosis in APPPS1-21 mice. Additionally, in our efforts to restore microbiota by either WT or Tg-FMT, we observed a restoration of pathology with both WT-FMT and Tg-FMT groups, which is contradictory to observed reduction in pathology with WT-FMT in APPSWEPS1ΔE9 at 7 mo of age. There are several important differences between the two studies that explain the discrepant results: (1) difference in ABX regimens; ABX administration at PND14–PND21 versus 6 mo (Sun et al., 2019); this is important to emphasize, as it is well established that the commensal microbiome perturbation during the postnatal developmental time frame represents a crucial developmental window by which microbiota–host interactions mediate immuno- and neurodevelopment that may impact host physiology in later life (Cox et al., 2014; Hansen et al., 2013, 2012; Tognini 2017); (2) the APPSWEPS1ΔE9 mice develop pathology between 6 and 7 mo, while the APPPS1-21 mice develop pathology by 6–7 wk; (3) the transgenes in the APPSWEPS1ΔE9 are driven by a mouse prion protein promoter that is expressed widely in the periphery and in the CNS, while the transgenes expressed in the APPPS1-21 mice are driven by the neuronal-specific Thy1 promoter; it is conceivable that expression of the transgenes in peripheral cells might influence the overall outcomes; and (4) the APPSWEPS1ΔE9 mice exhibit age-dependent changes in microbiota with decreased diversity as a function of age (Chen et al., 2020), while we do not observe any differences in microbiota profiles between WT (non-Tg littermates) and Tg–APPPS1-21 mice at PND22 or 7 wk of age (Fig. S5), thus making it very difficult to compare the two studies.
Figure S5.
16S rRNA analysis of fecal microbiota profile at PND22 and 7 wk of age in WT (non-Tg littermates) versus Tg APPPS1-21 mice. (A and B) 16S rRNA analysis of fecal microbiota profile was evaluated in freshly collected fecal pellet at PND22 (A) and 7 wk of age (B) in WT (non-Tg littermates) compared with Tg APPPS1-21 male mice. (A) PCoA plot generated using unweighted version of the UniFrac distance metric. The two components explained 17.2% of the variance. Both non-Tg and Tg group clusters overlapped at PND22. (B and C) The α-diversity was measured by using Faith phylogenetic diversity (B) and evenness (C). Both indices showed no changes among non-Tg and Tg groups (Kruskal-Wallis [KW]: P > 0.05 for both). (D) Heatmap of raw abundance shows comparable species profile between non-Tg and Tg mice at PND22. (E–H) Similar to the microbiota diversity readouts at PND22, microbiota profiles between non-Tg and Tg were also similar at 7 wk of age. (E) PCoA plot generated using unweighted version of the UniFrac distance metric. The two components explained 31.8% of the variance. (F and G) Faith phylogenetic diversity (F) and evenness (G) indices showed no changes among non-Tg and Tg groups (KW: P > 0.05 for both). (H) Heatmap of raw abundance shows similar species profiles between non-Tg and Tg mice at 7 wk of age. KW, Kruskal-Wallis; PCoA, principal coordinate analysis.
The mechanism(s) by which ABX modifies microglial transcriptional signatures (Dodiya et al., 2019) and morphology (Dodiya et al., 2019; Minter et al., 2017, 2016) and reduces Aβ amyloidosis specifically in male animals is not entirely clear. It is well established that the postnatal developmental time frame is where the commensal microbiome perturbations can affect host immune responses (Hansen et al., 2013) and neurodevelopment (Borre et al., 2014). In this regard, the microbiota have been shown to affect microglia maturation (Erny et al., 2015), and it has been established that microglia have an innate immune memory (Wendeln et al., 2018). Taken together with the finding that the median half-life of microglia in the mouse brain is ∼15–18 mo (Füger et al., 2017), it is highly conceivable that the microglial response to amyloid through alterations in the microbiome that occur in short-term ABX-treated male mice can be sustained for almost the entire lifetime of the animal. It is clear that microglia exhibit a unique M0 homeostatic molecular and functional signature in the healthy brain (Butovsky et al., 2014; Gautier et al., 2012; Hickman et al., 2013) and become neurotoxic or neurodegenerative (MGnD type) in disease conditions (aka DAM). Hence, it is plausible that perturbations of the microbiome in early life could affect microglial priming and maturation. Supporting this notion, our unbiased RNA-seq analysis (Table S3) revealed significant reductions in expression of microglial sensome genes (P2ry12, P2ry13, Gpr34, Cx3cr1, Ccr5, C3ar1, Ly86, Cd68, Trem2, Cd180, Tlr2, Clec7a, Itgam, Siglech, Cd33, Cd52, Cd84, Lag-3, Ptprc, Tnfrsf13b, Cd53, and Slamf9; Table S3) compared with the top 100 microglia sensome genes reported by Hickman et al. (2013). Most importantly, none of these microglia sensome genes were highly expressed in ABX-treated male mice (Table S4), suggesting that microglial maturation could be impacted in our pseudo-GF APPPS1-21 male mice. Among these, 22% of the down-regulated sensome genes were genes sensing apoptotic neurons (Trem2); substances released following neuronal injury, such as nucleotides and adenosine, and molecules expressed in the cell surface (Siglech) and soluble chemokine and related receptors (Ccr5, Cx3cr1, C3ar1), and proteins involved in sensing bacterial ligands (Ly86, Cd68, Tlr2, Cd180, Clec7a; Hickman et al., 2013). Additionally, caspase-3 and caspase-7 genes were significantly lower in ABX-treated male mice. Interestingly, inhibition of the caspase-3/7 pathway was shown to effectively block microglial activation (Burguillos et al., 2011). Importantly, GO term analysis revealed significantly reduced immune activity–related pathways (i.e., microglial cell activation [GO0001774], antigen processing and presentation of peptide antigen [GO0048002], inflammatory response [GO0006954], positive regulation of leukocyte migration [GO0002687], myeloid leukocyte migration [GO0097529], cell activation involved in immune response [GO0002263], microglia development [GO0014005], negative regulation of IL-2 biosynthesis [GO0045085], and negative regulation of B cell activation [GO0050869] pathways) in our ABX-treated APPPS1-21 male mice. While comparing these gene expression patterns would support the notion that microglia in ABX-treated male mice exhibit immature microglial phenotypes that lead to enhanced phagocytosis and clearance of extracellular Aβ oligomers or deposited Aβ, these conclusions need to be tempered by the fact that our transcriptional and morphological readouts are biased to analyses made at the time of sacrifice, where Aβ deposition is significantly diminished and microglia appear to have adopted an M0 phenotype. Unfortunately, we currently have no information pertaining to the temporal dynamics of microglia–Aβ interactions, and it will be essential to extend these studies using two-photon imaging in living animals that will allow us to visualize, at a temporal level, microglial maturation, migration, microglial–plaque interactions, and dynamics and to ask critical questions pertaining to plaque seeding, growth, and clearance.
The most provocative finding of our current studies is the demonstration that microglia play an important role in driving ABX-mediated reductions of Aβ in male APPPS1-21 mice. The mechanism(s) underlying this fascinating result remains to be established, but we offer several scenarios. Spangenberg et al. (2019) have reported that CSF1R-mediated depletion of microglia impairs parenchymal plaque deposition in the 5XFAD mouse model with rare ThioflavinS+ core plaques only observed in areas in the brain (i.e, the subiculum, where a subpopulation of CSF1R-resistant microglia remain). Microglial repopulation following removal of PLX5622 leads to robust levels of ThioS+ plaques, suggesting that microglia are critical for plaque seeding. Consistent with the view shared by Spangenberg et al. (2019), it is not inconceivable that microglia ingest low levels of Aβ (or oligomeric Aβ species that are “invisible” by IHC approaches) and concentrate these in acidic vesicles, including lysosomes, leading to conformational changes that drive nucleation. These seeds are subsequently released into the extracellular space via exocytic vesicles or by Aβ-induced death of microglial cells, as has been demonstrated by Baik et al. (2016). Alternatively, and has been suggested by Venegas et al. (2017), Aβ that is sensed by microglial pattern-recognition receptors leads to pathological innate immune activation of the NLRP3 inflammasome that recruits the adaptor protein ASC, which triggers helical fibrillar assembly of ASC. These ASC fibrils then recruit the effector caspase-1, leading to autoproteolytic activation and subsequent assembly of ASC fibrils into large ASC specks. In mouse macrophages, ASC specks recruit and activate caspase-1, which induces maturation of the cytokine IL-1β and pyroptotic cell death (Franklin et al., 2014). The presence of extracellular ASC specks can then cross-seed Aβ (Venegas et al., 2017).
How does the presence of microglia reduce Aβ amyloidosis in ABX-treated male mice? We now offer a highly speculative model: microglia ingest Aβ to generate highly aggregated Aβ species that impose an enormous cellular metabolic burden as the microglia attempt to degrade these recalcitrant protein aggregates. This ultimately leads to the demise of the microglial cell. The extracellular, highly aggregated Aβ peptides are then reingested by microglia that migrate to these deposits, and the process continues. This “capture-release-recapture” model was originally posited to explain the observation that tattoo pigment particles persist for indefinite periods of time and are resistant to clearance (Baranska et al., 2018). Tattoo pigments are primarily metal salts in the submicrometer range, and those that remain at the site of injection in the epidermis are exclusively found within dermal macrophages. These cells are continuously replenished from circulating monocytes to compensate for the loss of dying macrophages. According to the pigment capture–release–recapture model, when tattoo pigment–laden macrophages die during the course of adult life, neighboring macrophages recapture the released pigments and ensure the macroscopic stability and long-term persistence of tattoos. By analogy, and in view of the findings from the laboratories of Heneka (Venegas et al., 2017) and Mook-Jung (Baik et al., 2016), the capture–release–recapture model of tattoo persistence might very well be the explanation of the findings provided herein. Future studies using long-term in vivo single-cell imaging will be critical to document microglial ingestion of extracellular Aβ aggregates, accumulation within lysosomes, and subsequent release of nonhydrolyzable Aβ species from dying cells in order to validate the capture-release-recapture hypothesis. In this regard, Füger et al. (2017) used real-time two-photon imaging to demonstrate that while the loss of nonplaque-associated microglia in APPPS1-21 mice is similar to that seen in non-Tg mice, there is a threefold increase in the numbers of dividing nonplaque-associated microglia in the APPPS1-21 mice, and these newly divided cells migrate to the plaques. These studies indirectly argue that newly dividing microglial cells are replacing dead microglial cells that are plaque associated.
In conclusion, our findings fully support the conclusion that ABX-mediated perturbations of the microbiome have sex-specific influences on brain Aβ amyloidosis and microglial homeostasis and that microglia play a central role in modulation of gut microbiota–mediated modulation of Aβ deposition.
Materials and methods
Animal housing and handling
APPPS1-21 mice (obtained from M. Jucker, University of Tubingen, Tubingen, Germany) on a C57BL6Cj background were housed in an animal research center (ARC) facility at The University of Chicago under 12-h light/dark conditions with an ad libitum access to chow and water unless otherwise noted. The experimental procedures were approved by the institutional animal care and use committee at The University of Chicago.
Experimental models
Short-term ABX and FMT study
Short-term ABX treatment was performed using established protocol (Minter et al., 2017) using both male and female APPPS1-21 mice. In brief, pups receiving the antibiotic cocktail were gastric gavaged with 200 µl of antibiotic cocktail (4 mg/ml kanamycin [Sigma-Aldrich; K4000-5 g], 0.35 mg/ml gentamicin [Sigma-Aldrich; G1914-250 mg], 8,500 U/ml colistin [Sigma-Aldrich; C4461-1 g], 2.15 mg/ml metronidazole [Sigma-Aldrich; M1547-25 g], and 0.45 mg/ml vancomycin [Sigma-Aldrich; V2002-1 g]) from PND14 to PND21, followed by an ad libitum access to regular drinking water until the time of sacrifice at 9 wk of age. During the gastric gavage delivery, all mice were transferred to a new sterile cage to avoid microbial contamination from accumulated fecal pellets in cages. Parents from the same cage as pups receiving ABX treatment were euthanized after weaning the pups and were not used for future breedings. The vehicle-treated group received 200 µl of sterile ARC water in a similar fashion. ABX/vehicle treatment for this study was performed by H.B. Dodiya.
To evaluate the causality between gut microbiome and Alzheimer’s pathogenesis, FMT was performed from age-matched Tg APPPS1-21 donor male mice into ABX-treated male mice. For preparation of the fresh fecal slurry (Dodiya et al., 2019) for short-term ABX+FMT experiments, fresh fecal pellets from age-matched Tg APPPS1-21 donor male mice were collected and immediately mixed in autoclaved water (5 mg fecal content dissolved in 1 ml sterile water). The feces was homogenized, and the supernatant was collected after allowing the suspension to settle down by using gravity for 5 min. Mice in the ABX+FMT group received 200 µl of freshly prepared fecal slurry from the age-matched donor mouse by gastric gavage daily, starting on PND24 until the time of sacrifice. The ABX or vehicle-treated group received 200 µl of sterile water in a similar fashion to nullify the everyday gavage effect.
WT-FMT versus Tg-FMT study
To compare the pathogenicity between WT and Tg microbiomes, another set of ABX-treated APPPS1-21 mice was employed. For this study, we only used male mice, as ABX-treated female mice do not exhibit reduced amyloidosis. ABX/vehicle treatment (PND14–PND21) for all pups pertaining to WT versus Tg study was performed by I.Q. Weigle under H.B. Dodiya’s supervision. FMT from age-matched male Tg APPPS1-21 or non-Tg littermates was prepared as per above method by C.J. Roman-Santiago under H.B. Dodiya’s supervision and used to implant Tg or WT microbiota into ABX-treated APPPS1-21 male mice. Respective controls were gavaged with ARC water as mentioned above.
PLX treatment study
To determine the role of microglia in modulation of Aβ amyloidosis in our model, we used an established strategy to deplete microglia in the brain (Dagher et al., 2015). We only used male mice for PLX studies to investigate the role of microglial cells in our established microbiome–brain axis, as female mice showed no beneficial effects of ABX-perturbed microbiome on amyloidosis. ABX/vehicle gavages (PND14–PND21) were performed by HBD and age-matched Tg-FMT (PND24–9 wk or PND24–3 mo) gavages were performed by either HBD or IQW. Mice receiving ABX+Tg-FMT were randomly assigned to CSF-1 inhibitor compound (PLX5622)–infused chow or control chow from PND24 until the time of sacrifice. PLX5622 was provided by Plexxikon and formulated in AIN-76A chow diet by Research Diets facility at the 1,200-ppm dose as used previously (Dagher et al., 2015).
Necropsy and tissue harvesting
Approved protocols by The University of Chicago Animal Care and Use Protocols were used to harvest different organs from mice at the time of sacrifice as published (Dodiya et al., 2019). On the day of sacrifice, mice received an intraperitoneal injection of ketamine and xylazine mixture. Upon confirmation of deep anesthesia, blood samples were collected using cardiac puncture using a 25-gauge needle and were stored in buffered sodium citrate blood collection tubes (BD Vacutainer; 366393) on ice. Right after the blood collection, the descending aorta was clamped, and the mice were perfused using physiological cold saline (pH 7.4) for 3 min. Brains were then extracted from skulls and dissected into two hemispheres (one hemisphere for histology: postfixed by 4% paraformaldehyde, another for transcriptome analysis: frozen immediately). Cecum was collected and weighed to evaluate the efficacy of the antibiotic treatments. Upon completion, plasma was collected by centrifugation at 2,000 rpm for 10 min at 4°C by using a Beckman Coulter centrifuge and stored at −80 for future use.
Fecal microbiota analysis
DNA extractions were performed on fecal pellets using the MoBio PowerSoil 96 Well Soil DNA Isolation Kit (catalog no. 12955) following the standard DNA extraction protocol outlined by the Earth Microbiome Project (http://www.earthmicrobiome.org/). We amplified the V4 region of the 16S rRNA gene using the 515f and 806r primers and mitochondrial blockers to reduce amplification of host mitochondrial DNA (Caporaso et al., 2011, 2012; Kozich et al., 2013). Sequencing was performed using paired-end 150 base reads on an Illumina HiSeq sequencing platform through the University of Illinois at Chicago Sequencing Core, following standardized sequencing protocols described in Caporaso et al. (2012). Full DNA extraction, amplification, and sequencing protocols and standards are available at http://www.earthmicrobiome.org/protocols-and-standards. We used the Quantitative Insights Into Microbial Ecology pipeline (Caporaso et al., 2010) and vsearch8.1 (Rognes et al., 2016) to perform standard demultiplexing and quality filtering. We identified unique ASVs using the Deblur method (Amir et al., 2017), and taxonomy was assigned using the Greengenes Database (May 2013 release; http://greengenes.lbl.gov). Statistical analyses were performed in R, relying primarily on vegan and phyloseq libraries. For a complete list of libraries and code used for data analyses and microbiome figure generation, visit http://www.github.com/hollylutz/AlzheimersMP.
Fecal slurry Aβ oligomer measurement
Preparation of Aβ42 solution and formation of Aβ42 oligomers and fibers
Aβ powder was purchased from Biolegend (catalog no. 932501). The powder was first dissolved in 0.1 M NaOH and incubated at room temperature for 10 min. Then the dissolved Aβ42 was diluted to 100 ng/µl in PBS. The Aβ42 solution was then allowed to aggregate at room temperature for 8 d. The formation of Aβ42 oligomers and fibers was confirmed by Western blot using a 4–20% gradient gel.
Preparation of the mixture of fecal suspension and Aβ42
Fecal suspensions from 6 non-Tg and 6 Tg mice at 50-mg/ml concentrations were centrifuged at 10,000 rpm for 10 min at 4°C, and supernatant of each sample was saved. For the spiking experiment, aged Aβ42 solution was mixed with the supernatant so that the final concentration of Aβ42 was 100 ng/µl.
Dot blot analysis
20 μl of the supernatant of fecal suspension or the mixture of fecal suspension and Aβ42 was loaded onto a nitrocellulose membrane using a dot blot apparatus (Bio-Rad) per the manufacturer’s instruction. The blot was blocked in a blocking buffer containing 5% blocking grade milk (Bio-Rad) and 1× PBS, 0.1% Tween-20 detergent at room temperature for 1.5 h and incubated with primary antibody in a cold room overnight. The blot was washed and incubated with secondary antibody for 1 h at room temperature. After washing, the blot was developed with enhanced chemiluminescence reagents (Perkin-Elmer). Anti-amyloid fibrils OC (catalog no. AB2286), anti–Aβ fibrils, clone M98 (catalog no. MABN640) were purchased from EMD Millipore. 6E10 was purchased from Biolegend.
Immunocytochemistry
The immunofluorescence staining was performed to evaluate Aβ amyloidosis and microglial morphologies as published previously (Dodiya et al., 2019). In brief, a full series of 40-µm-thick brain sections was used for Aβ (3D6, 1:10,000) staining, and a half series of sections was used for microglia staining (Iba1, 1:500). Briefly, free-floating 40-µm level-matched sections were washed with dilution media for 60 min (10 min/wash). After that, sections were incubated in serum blocking solution for 1 h at room temperature, followed by primary antibody incubations for overnight in a 4°C refrigerator. The next day, sections were washed with dilution media for 60 min (10 min/wash), followed by secondary antibody incubation at room temperature for 1 h. We used donkey anti-mouse 488 (Invitrogen; 1:500) and donkey anti-rabbit 594 (Invitrogen; 1:500) as secondary antibodies for Aβ and microglia staining, respectively. Sections were then washed with dilution media and mounted on glass slides. Sections were then coverslipped by using Fluoromount aqueous mounting medium (Sigma-Aldrich; F4680). 3D6+ Aβ plaque images were captured by using a 3D Histech Pannoramic MIDI whole-slide scanner with a Zeiss AxioCam MRM CCD camera by The University of Chicago Integrated Light Microscopy Facility personnel. Microglia images were captured by using a Leica SP8 3D STED laser-scanning confocal microscope under 40×1.5 magnification. Similarly, we performed IHC with antibodies specific for activated/degenerative microglia (CD68; catalog no. 137002; FA-11 clone; 1:250), neurons (NF-L; catalog no. ab223343; 1:200), lysosomes (LAMP1; catalog no. ab25245, 1:200), synapses (Synaptophysin; catalog no. ab32594; 1:200; PSD95; catalog no. ab238135; 1:200), and myelin (MBP; catalog no. ab218011; 1:200) followed by a combination of host-specific secondary antibodies: donkey anti-mouse 488 (Invitrogen; catalog no. A21202, 1:500), goat anti-rat 555 (Invitrogen; catalog no. A21434, 1:500), donkey anti-rabbit 647 (Invitrogen; catalog no. A31573,1:500), and donkey anti-rabbit 555 (Invitrogen; catalog no. A32794). When needed, nuclear counterstaining was performed using DAPI (Thermo Fisher Scientific; catalog no. 62248) at the concentration of 1 µg/ml by incubating samples for 3 min followed by 10-min washes (3×) using 1× Tris-buffered saline (TBS) solution.
Aβ burden analysis
Aβ burden and amyloid plaque size were evaluated per the established protocol (Dodiya et al., 2019). In brief, six sections (past olfactory bulb to the mid-hippocampal level) at an equidistance of 480 µm were used for 3D6+ amyloid plaque analyses. Using Fiji ImageJ software (NIH; ImageJ 1.51n), average plaque size and Aβ fraction area (amyloid burden) were generated by two independent observers in a blind manner. Individual images were created with the 3D Histech Pannoramic viewer software (3DHistech Kft). Images were then normalized, and an automated thresholding based on entropy of the histogram was used to identify the amyloid plaques. After images were converted into 8-bit format, specific threshold number was applied, which was then followed by “fill holes” and “watershed” algorithms for a binary conversion. Finally, plaque number, plaque size area, and total area occupied by plaque (i.e., Aβ burden) were calculated using “analyze particles.” Graphs were plotted using data on amyloid burden and amyloid plaque size.
Microglia morphology and activation status marker (CD68) analysis
Microglia morphology was evaluated in the same manner as published previously (Dodiya et al., 2019). Microglial cell counts and cell body area were measured using Fiji ImageJ software (NIH; ImageJ 1.51n), while 3D reconstruction to measure microglial cell branch points and microglial cell branch length parameters were recorded using Bitplane Imaris software v.9.1.2 (Andor Technology PLC). Using a Leica SP8 3D STED laser-scanning confocal microscope, Z-stack images of individual plaques from 40-µm-thick sections were obtained under 40 × 1.5 magnification with 0.30-µm step increments in the z-plane. A total of five microenvironments containing microglia surrounding an Aβ plaque were collected in a 3D Z-stack manner. These 3D Z-stacks were then imported into ImageJ software where microglia (Iba1 channel) containing Z-stack was processed using Z-project probe for maximum-intensity projections. These compressed images were used to measure the number of microglial cells surrounding plaques and cell body area per microglial cell. The average number of microglia/plaques in an 0.02-µm2 area was collected for each case. By using freehand selection, each microglial cell body was outlined, and the area was measured.
3D reconstruction was performed to measure microglial cell branch length and branch point numbers per the published protocol (Dodiya et al., 2019). The Z-stack images of microglia surrounding individual plaques were obtained under 40×1.5 magnification from 40-µm-thick sections with 0.30-µm step increments in the z-plane. These Z-stacks were then imported into Bitplane Imaris software v.9.1.2. Using the filament tool, all microglial dendrite-like processes were mapped and recorded. The surface tool was used to establish the realm of the Aβ plaque. Quantification of average dendrite length and the total number of branch points were collected using an in-built analysis program.
Microglial activation status was evaluated using immunofluorescence staining followed by confocal microscope imaging and ImageJ analysis for combination of markers CD68/Iba1/3D6. CD68 (lysosomal marker) and Iba1 (microglial marker) colocalization fraction around 3D6+ plaques was measured using Fiji ImageJ software (NIH; ImageJ 1.51n). 3D Z-stacks for Iba1/CD68/3D6 channels were captured as mentioned above using a Leica SP8 3D STED laser-scanning confocal microscope. A total of six to seven microenvironments containing microglia surrounding Aβ plaques were collected and used to evaluate the fraction colocalization of Iba1 with CD68 reactivity. The area covered by CD68 was quantified using the “Otsu Dark” threshold and Analyze Particles (inclusion size of 1-Infinity) functions from ImageJ software (NIH) within the region of interest (0.02 mm2).
Brain protein extraction
To quantify soluble and insoluble Aβ levels, we chose the ventral half of each frozen brain. Briefly, the −80°C stored frozen brains were dissected into dorsal and ventral halves on dry ice. The ventral halves were weighed and homogenized with 5× (wt/vol) volume of TBS containing 1× Halt Protease Inhibitor cocktail (Thermo Fisher Scientific) and 5 mM EDTA. After sonication, the homogenized samples were subjected to ultracentrifugation at 100,000×g for 60 min at 4°C. The supernatant fraction was collected to detect TBS soluble Aβ levels. The remaining pellet fraction was further extracted in 10× (wt/vol) volume of 70% FA followed by homogenization. The homogenized samples were then subjected to ultracentrifugation at 100,000×g for 60 min at 4°C to collect the supernatant fraction that contains TBS-insoluble and FA-soluble Aβ levels. The supernatants were frozen immediately and shipped on dry ice to Harvard University for MesoScale Aβ analysis.
MesoScale Aβ analysis
Levels of Aβ peptides were analyzed through protocols previously reported (Minter et al., 2016). Specifically, the assay was performed on an electrochemiluminescence-based multiarray method using the Quickplex SQ 120 system from MSD Meso Scale Diagnostics LLC. The MesoScale Aβ 4G8 kits were used to detect Aβ peptides in a 96-well–based assay. First, 96-well plates were blocked with diluents provided by the manufacturer with shaking for 1 h at room temperature. The experimental samples and MesoScale protein standards were resuspended in the manufacturer-supplied detection antibodies. The mixed solutions were placed on a shaker for 2 h at RT, followed by washing and adding of the reading buffer. The electrochemiluminescence signals were captured, signals were obtained for all samples and standard proteins, and the sample Aβ levels were analyzed using the MesoScale protein standards.
RayBiotech cytokine/chemokine analysis
Plasma collected at the time of sacrifice (9 wk of age) was used to perform the cytokine/chemokine measurements. Quantibody membrane mouse cytokine array Q5 kit (RayBiotech; QAM-CYT-5) was used to detect levels of 40 different cytokines (bFGF, BLC [CXCL13], CD30 Ligand [TNFSF8], Eotaxin-1 [CCL11], Eotaxin-2 [MPIF-2/CCL24], Fas Ligand [TNFSF6], GCSF, GM-CSF, ICAM-1 [CD54], IFN-γ, IL-1 α [IL-1 F1], IL-1 β [IL-1 F2], IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-10, IL-12 p40, IL-13, IL-15, IL-17A, IL-21, KC [CXCL1], Leptin, LIX, MCP-1 [CCL2], MCP-5, M-CSF, MIG [CXCL9], MIP-1 α [CCL3], MIP-1 γ, Platelet Factor 4 [CXCL4], RANTES [CCL5], TARC [CCL17], I-309 [TCA- 3/CCL1], TNF-α, TNF RI [TNFRSF1A], and TNF RII [TNFRSF1B] per the manufacturer’s instructions. The Quantibody array is a multiplexed sandwich ELISA-based quantitative array platform that allows us to determine the concentration of multiple cytokines simultaneously. Briefly, plasma samples were diluted (2×) using sample diluent provided in the kit. A glass microarray slide, which is divided into 16 wells of identical cytokine antibody arrays, was dried, and blocking was performed using the sample diluent followed by an incubation of 100 µl standard cytokines (the array-specific cytokine standards whose concentrations have been predetermined) or unknown plasma samples in properly labeled wells for 2 h. After the microarray slides were washed using wash buffers I and II, slides were incubated into detection antibody cocktail for 2 h. Wash buffers I and II were then applied, and slides were brought into Cy3 equivalent dye-conjugated streptavidin to incubate in the dark at room temperature for 1 h. Following last steps of washes, the glass microarray slide setup was disassembled, washed, and dried in the dark at room temperature. The slides were sent to RayBiotech Life for free scanning and data extraction service. Upon receiving the extracted data, manufacturer-recommended median fluorescence minus background numbers for recorded intensities was used for all standards and individual samples. Intensity data were normalized relative to the blanks for each sample. Using serial standard concentrations provided in the QAM-CYT-5 kit manual, intensity values were plotted against cytokine concentrations for standard curve preparations and for cytokine calculations. The values were then recorded in picogram per milliliters and plotted using GraphPad Prism for comparison analysis among groups.
Cerebral cortex RNA extraction
Total RNA was isolated from the dorsal cerebral cortex per published protocol (Dodiya et al., 2019). Briefly, TRIzol reagent was used to isolate total RNA followed by RNAeasy Micro cleanup (Qiagen) procedure according to the manufacturer’s instructions. The quality of the total RNA was evaluated using the Agilent Bioanalyzer. RNA-seq library preparations and Illumina HiSeq4000 were performed at The University of Chicago Genomics Core Facility, and data files were collected in FASTQ format for the bioinformatic analysis.
RNA-seq bioinformatics analysis
The quality of DNA reads, in FASTQ format, was evaluated using FastQC (Andrews, 2010). Adapters were trimmed, and reads of poor quality or aligning to rRNA sequences were filtered using Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The cleaned reads were aligned to the mouse genome (mm10) using STAR (Dobin et al., 2013). Read counts for each gene were calculated using HTSeq-Counts (Anders et al., 2015) in conjunction with a gene annotation file for mm10 obtained from Ensembl (http://useast.ensembl.org/index.html). A comprehensive quality control report was generated using MultiQC (Ewels et al., 2016). Differential expression was determined using DESeq2 (Love et al., 2014). The cutoff for determining significant DEGs was an FDR-adjusted P value <0.05. The data discussed in this publication have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus and are accessible through GEO Series accession no. GSE185407 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185407). GO analysis and identification of DEGs belonging to specific pathways were performed using Metascape (Zhou et al., 2019), and the results were confirmed using DAVID online service (Huang et al., 2009) and GOrilla online services (Eden et al., 2009).
Statistical analysis
GraphPad Prism software (version 7.0e) was used to run statistical analyses. Kruskal-Wallis (nonparametric) analysis was performed to compare α- and β-diversity indices in 16S rRNA microbiota analysis with multiple groups comparison correction. One-way ANOVA was used to compare multiple groups (short-term ABX studies and WT- vs. Tg-FMT studies) unless otherwise stated, followed by a post hoc comparison. Two-way ANOVA was used (PLX 9-wk and PLX 3-mo studies) to compare the effect of a control or PLX diet and ABX or ABX-FMT treatments followed by Sidak post hoc comparison. Statistical P value below 0.05 was considered for significant differences. Detailed statistical information is provided in the figure legends or Table S9.
Online supplemental material
Fig. S1 shows soluble Aβ1-40 Aβ1-42 levels measured by MSD biochemical assay (A), graphical presentation of CD68 reactivity in Iba1 away from plaques using ImageJ (B), and MBP expression in cerebral cortex and corpus callosum using qualitative comparison (C). Fig. S2 shows plasma cytokines and chemokines measured by RayBiotech cytokine array. Using serial standard concentrations provided in the QAM-CYT-5 kit manual, intensity values were plotted against cytokine concentrations for standard curve preparations and for cytokine calculations. The values were then recorded in picogram per milliliters and plotted using GraphPad Prism for comparison analysis among groups. bFGF, GM-CSF, leptin, and MIG showed significant differences among male groups only. Cytokines/chemokines such as BLC, G-CSF, IL-15, KC, MIP-γ, and TNF-Rll showed a trend of reduction with ABX treatment in male mice compared with controls, whose levels were restored with FMT in ABX-treated male mice. Compared with those, CD30L showed a trend of increase in ABX-treated male mice compared with control male mice, whose levels were restored with FMT in ABX-treated male mice. Fig. S3 shows 16S rRNA sequencing analysis of fecal microbiota profile at PND22 in WT- versus Tg-FMT study groups (A–F), intensity measurements for dot blots presented in Fig. 5 D, b (G), cluster network analysis of lower DEGs in M_Abx group presented in Fig. 6 E, a (I), and additional RNAseq data from short-term ABX male and female mice sacrificed at 9 wk of age (H–J). (A–F) The 16s rRNA was employed to confirm the ABX-related fecal microbiota perturbations in ABX-, ABX+WT-FMT, and ABX+Tg-FMT groups compared with vehicle-treated male group a day after cessation of ABX (PND22). Data in panels G and H–J are supplemental data to Fig. 5 D, b and Fig. 6, respectively. Fig. S4 shows that short-term ABX treatment alters cerebral cortex transcriptome profiles at PND24 in a genotype-dependent manner (A and B) and short-term ABX treatment in non-Tg male and female littermates alters cerebral cortex transcriptome profiles at 9 wk in a sex-specific manner (C–E). In these studies, animals (non-Tg and Tg littermates) were administered ABX or vehicle (PND14–PND21) and sacrificed at either PND24 (3 d after the cessation of ABX/vehicle treatments; A and B) or 9 wk of age (C–E). RNA-seq analysis was performed on RNA from the dorsal cerebral cortex. Fig. S5 shows 16S rRNA analysis of fecal microbiota profile at PND22 and 7 wk of age in WT (non-Tg littermates) versus Tg APPPS1-21 mice. Table S1 shows ANCOM analysis of fecal microbiota at PND22 from the WT- versus Tg-FMT study. Table S2 shows ANCOM analysis of fecal microbiota at 9 wk from the WT- versus Tg-FMT study. Table S3 shows lower DEGs in ABX-treated APPPS1-21 male mice associated pathways. Table S4 shows higher DEGs in ABX-treated APPPS1-21 male mice associated pathways. Table S5 compares DEGs between Tg_M_Ctr (group 9) and Tg_M_Abx (group 7) at PND24. Table S6 compares DEGs between non-Tg_M_Ctr (group 5) and non-Tg_M_Abx (group 6) at PND24. Table S7 compares DEGs between non-Tg_M_Ctr (group 3) and non-Tg_M_Abx (group 4) at 9 wk of age. Table S8 compares DEGs between non-Tg_F_Ctr (group 2) and non-Tg_F_Abx (group 1) at 9 wk of age. Table S9 details statistical information for mentioned comparisons in each figure legend.
Supplementary Material
shows ANCOM analysis of fecal microbiota at PND22 from WT- versus Tg-FMT study.
shows ANCOM analysis of fecal microbiota at 9 wk from WT- versus Tg-FMT study.
shows lower DEGs in ABX-treated APPPS1-21 male mice associated pathways.
shows higher DEGs in ABX-treated APPPS1-21 male mice associated pathways.
compares DEGs between Tg_M_Ctr (group 9) and Tg_M_Abx (group 7) at PND24.
compares DEGs between non-Tg_M_Ctr (group 5) and non-Tg_M_Abx (group 6) at PND24.
compares DEGs between non-Tg_M_Ctr (group 3) and non-Tg_M_Abx (group 4) at 9 wk of age.
compares DEGs between non-Tg_F_Ctr (group 2) and non-Tg_F_Abx (group 1) at 9 wk of age.
details statistical information for mentioned comparisons in each figure legend.
Acknowledgments
We thank Ms. Shirley Bond and Dr. Christine Labno for slide scanner image processing at The University of Chicago Integrated Light Microscopic Core Facility. We thank Ms. Mikayla Marchuk, Dr. Shou Siming, and Dr. Pieter W. Faber for their help with bioanalyzer and next-generation sequencing services at The University of Chicago Genomics Core Facility.
These studies were supported by the Cure Alzheimer’s Fund (to S.S. Sisodia and R.E. Tanzi), Open Philanthropy Project and Good Ventures Foundation (to S.S. Sisodia and R.E. Tanzi), Bright Focus Foundation Research Fellowship grant ID# A2019032F (to H.B. Dodiya), Alzheimer’s Association Research Fellowship award (to H.B. Dodiya), and the Luminescence Foundation (to S.S. Sisodia).
Author contributions: H.B. Dodiya conceived and performed short-term ABX experiments, WT- versus Tg-FMT experiments, PLX5622 experiments, and non-Tg littermates; assessed the gut microbiota profile and designed and performed bioinformatics analysis for WT- versus Tg-FMT study, cytokine assessments, immunocytochemistry experiments, SP8 laser confocal microscope imaging, Imaris evaluation, RNA-seq experiments, and data interpretation, preparation of brain homogenates for MSD biochemical assay, final figures preparation, and manuscript preparation. H.L. Lutz assessed the gut microbiota profile, designed and performed bioinformatics analysis for short-term ABX study, and reviewed and critiqued the manuscript. I.Q. Weigle generated mice and performed ABX to WT- versus Tg-FMT study mice, fecal DNA extraction for the WT- versus Tg-FMT study, and IHC using Aβ-related degenerative markers and performed FMT in PLX5622 studies and brain sectioning for PLX5622 studies. P. Patel and M.J. Schipma performed RNA-seq–related bioinformatics and assisted in generating RNA-seq figures. J. Michalkiewicz generated mice pertaining to PLX studies and short-term ABX to non-Tg mice studies as well as performed IHC for PLX studies. C.J. Roman-Santiago performed FMT oral gavage, brain sectioning, and amyloidosis evaluation in the WT- versus Tg-FMT study under H.B. Dodiya’s supervision. C. Zhang and Y. Liang performed MSD biochemical assays using TBS and FA-soluble brain homogenates. A. Srinath performed ImageJ analysis of CD68 reactivity in microglia under H.B. Dodiya’s supervision. Xulun Zhang performed dot blot analysis of Aβ oligomers in fecal slurries of WT compared with Tg mice. J. Xia and M. Olszewski quantified amyloidosis and microglia for short-term ABX studies using ImageJ as second blind observers under H.B. Dodiya’s supervision. Xiaoqiong Zhang maintained and genotyped APPPS1-21 mice for short-term ABX and FMT experiments. E. Chang, R.E. Tanzi, and J.A. Gilbert reviewed and critiqued the manuscript. S.S. Sisodia developed the microbiota-related hypothesis, co-designed the experimental study, and reviewed and critiqued the manuscript.
References
- Aloisi, F., Ria F., and Adorini L.. 2000. Regulation of T-cell responses by CNS antigen-presenting cells: different roles for microglia and astrocytes. Immunol. Today. 21:141–147. 10.1016/S0167-5699(99)01512-1 [DOI] [PubMed] [Google Scholar]
- Amir, A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z., Kightley E.P., Thompson L.R., Hyde E.R., Gonzalez A., and Knight R.. 2017. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems. 2:e00191. 10.1128/mSystems.00191-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed June 1, 2021)
- Asai, H., Ikezu S., Tsunoda S., Medalla M., Luebke J., Haydar T., Wolozin B., Butovsky O., Kügler S., and Ikezu T.. 2015. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 18:1584–1593. 10.1038/nn.4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacioglu, M., Maia L.F., Preische O., Schelle J., Apel A., Kaeser S.A., Schweighauser M., Eninger T., Lambert M., Pilotto A., et al. 2016. Neurofilament Light Chain in Blood and CSF as Marker of Disease Progression in Mouse Models and in Neurodegenerative Diseases. Neuron. 91:56–66. 10.1016/j.neuron.2016.05.018 [DOI] [PubMed] [Google Scholar]
- Baik, S.H., Kang S., Son S.M., and Mook-Jung I.. 2016. Microglia contributes to plaque growth by cell death due to uptake of amyloid β in the brain of Alzheimer’s disease mouse model. Glia. 64:2274–2290. 10.1002/glia.23074 [DOI] [PubMed] [Google Scholar]
- Baranska, A., Shawket A., Jouve M., Baratin M., Malosse C., Voluzan O., Vu Manh T.P., Fiore F., Bajénoff M., Benaroch P., et al. 2018. Unveiling skin macrophage dynamics explains both tattoo persistence and strenuous removal. J. Exp. Med. 215:1115–1133. 10.1084/jem.20171608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid, Y., and Naik S.. 2013. Compartmentalized and systemic control of tissue immunity by commensals. Nat. Immunol. 14:646–653. 10.1038/ni.2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellver-Landete, V., Bretheau F., Mailhot B., Vallières N., Lessard M., Janelle M.E., Vernoux N., Tremblay M.E., Fuehrmann T., Shoichet M.S., and Lacroix S.. 2019. Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat. Commun. 10:518. 10.1038/s41467-019-08446-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berer, K., Mues M., Koutrolos M., Rasbi Z.A., Boziki M., Johner C., Wekerle H., and Krishnamoorthy G.. 2011. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 479:538–541. 10.1038/nature10554 [DOI] [PubMed] [Google Scholar]
- Berer, K., Gerdes L.A., Cekanaviciute E., Jia X., Xiao L., Xia Z., Liu C., Klotz L., Stauffer U., Baranzini S.E., et al. 2017. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. USA. 114:10719–10724. 10.1073/pnas.1711233114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borre, Y.E., O’Keeffe G.W., Clarke G., Stanton C., Dinan T.G., and Cryan J.F.. 2014. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol. Med. 20:509–518. 10.1016/j.molmed.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Braniste, V., Al-Asmakh M., Kowal C., Anuar F., Abbaspour A., Tóth M., Korecka A., Bakocevic N., Ng L.G., Kundu P., et al. 2014. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 6:263ra158. 10.1126/scitranslmed.3009759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridel, C., van Wieringen W.N., Zetterberg H., Tijms B.M., Teunissen C.E., Alvarez-Cermeño J.C., Andreasson U., Axelsson M., Bäckström D.C., Bartos A., et al. and the NFL Group . 2019. Diagnostic Value of Cerebrospinal Fluid Neurofilament Light Protein in Neurology: A Systematic Review and Meta-analysis. JAMA Neurol. 76:1035–1048. 10.1001/jamaneurol.2019.1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguillos, M.A., Deierborg T., Kavanagh E., Persson A., Hajji N., Garcia-Quintanilla A., Cano J., Brundin P., Englund E., Venero J.L., and Joseph B.. 2011. Caspase signalling controls microglia activation and neurotoxicity. Nature. 472:319–324. 10.1038/nature09788 [DOI] [PubMed] [Google Scholar]
- Butovsky, O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E., et al. 2014. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17:131–143. 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., and Knight R.. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 108(Suppl 1):4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekanaviciute, E., Yoo B.B., Runia T.F., Debelius J.W., Singh S., Nelson C.A., Kanner R., Bencosme Y., Lee Y.K., Hauser S.L., et al. 2017. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA. 114:10713–10718. 10.1073/pnas.1711235114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Fang L., Chen S., Zhou H., Fan Y., Lin L., Li J., Xu J., Chen Y., Ma Y., and Chen Y.. 2020. Gut Microbiome Alterations Precede Cerebral Amyloidosis and Microglial Pathology in a Mouse Model of Alzheimer’s Disease. BioMed Res. Int. 2020:8456596. 10.1155/2020/8456596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, L.M., Yamanishi S., Sohn J., Alekseyenko A.V., Leung J.M., Cho I., Kim S.G., Li H., Gao Z., Mahana D., et al. 2014. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 158:705–721. 10.1016/j.cell.2014.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, L.M., Abou-El-Hassan H., Maghzi A.H., Vincentini J., and Weiner H.L.. 2019. The sex-specific interaction of the microbiome in neurodegenerative diseases. Brain Res. 1724:146385. 10.1016/j.brainres.2019.146385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagher, N.N., Najafi A.R., Kayala K.M., Elmore M.R., White T.E., Medeiros R., West B.L., and Green K.N.. 2015. Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice. J. Neuroinflammation. 12:139. 10.1186/s12974-015-0366-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, R., and Chinnathambi S.. 2019. Microglial priming of antigen presentation and adaptive stimulation in Alzheimer’s disease. Cell. Mol. Life Sci. 76:3681–3694. 10.1007/s00018-019-03132-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz, R., Wang S., Anuar F., Qian Y., Björkholm B., Samuelsson A., Hibberd M.L., Forssberg H., and Pettersson S.. 2011. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA. 108:3047–3052. 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodiya, H.B., Kuntz T., Shaik S.M., Baufeld C., Leibowitz J., Zhang X., Gottel N., Zhang X., Butovsky O., Gilbert J.A., and Sisodia S.S.. 2019. Sex-specific effects of microbiome perturbations on cerebral Aβ amyloidosis and microglia phenotypes. J. Exp. Med. 216:1542–1560. 10.1084/jem.20182386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodiya, H.B., Forsyth C.B., Voigt R.M., Engen P.A., Patel J., Shaikh M., Green S.J., Naqib A., Roy A., Kordower J.H., et al. 2020. Chronic stress-induced gut dysfunction exacerbates Parkinson’s disease phenotype and pathology in a rotenone-induced mouse model of Parkinson’s disease. Neurobiol. Dis. 135:104352. 10.1016/j.nbd.2018.12.012 [DOI] [PubMed] [Google Scholar]
- Dorrestein, P.C., Mazmanian S.K., and Knight R.. 2014. Finding the missing links among metabolites, microbes, and the host. Immunity. 40:824–832. 10.1016/j.immuni.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden, E., Navon R., Steinfeld I., Lipson D., and Yakhini Z.. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 10:48. 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore, M.R., Najafi A.R., Koike M.A., Dagher N.N., Spangenberg E.E., Rice R.A., Kitazawa M., Matusow B., Nguyen H., West B.L., and Green K.N.. 2014. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 82:380–397. 10.1016/j.neuron.2014.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen, P.A., Dodiya H.B., Naqib A., Forsyth C.B., Green S.J., Voigt R.M., Kordower J.H., Mutlu E.A., Shannon K.M., and Keshavarzian A.. 2017. The Potential Role of Gut-Derived Inflammation in Multiple System Atrophy. J. Parkinsons Dis. 7:331–346. 10.3233/JPD-160991 [DOI] [PubMed] [Google Scholar]
- Erblich, B., Zhu L., Etgen A.M., Dobrenis K., and Pollard J.W.. 2011. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One. 6:e26317. 10.1371/journal.pone.0026317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny, D., and Prinz M.. 2020. How microbiota shape microglial phenotypes and epigenetics. Glia. 68:1655–1672. 10.1002/glia.23822 [DOI] [PubMed] [Google Scholar]
- Erny, D., Hrabě de Angelis A.L., Jaitin D., Wieghofer P., Staszewski O., David E., Keren-Shaul H., Mahlakoiv T., Jakobshagen K., Buch T., et al. 2015. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18:965–977. 10.1038/nn.4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels, P., Magnusson M., Lundin S., and Käller M.. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 32:3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala, M., Liu Q.N., Sayre J., Pop V., Brahmandam V., Graves M.C., and Vinters H.V.. 2002. Cyclooxygenase-2-positive macrophages infiltrate the Alzheimer’s disease brain and damage the blood-brain barrier. Eur. J. Clin. Invest. 32:360–371. 10.1046/j.1365-2362.2002.00994.x [DOI] [PubMed] [Google Scholar]
- Franklin, B.S., Bossaller L., De Nardo D., Ratter J.M., Stutz A., Engels G., Brenker C., Nordhoff M., Mirandola S.R., Al-Amoudi A., et al. 2014. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat. Immunol. 15:727–737. 10.1038/ni.2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füger, P., Hefendehl J.K., Veeraraghavalu K., Wendeln A.C., Schlosser C., Obermüller U., Wegenast-Braun B.M., Neher J.J., Martus P., Kohsaka S., et al. 2017. Microglia turnover with aging and in an Alzheimer’s model via long-term in vivo single-cell imaging. Nat. Neurosci. 20:1371–1376. 10.1038/nn.4631 [DOI] [PubMed] [Google Scholar]
- Gautier, E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., et al. Immunological Genome Consortium . 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118–1128. 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulian, D., Li J., Li X., George J., and Rutecki P.A.. 1994. The impact of microglia-derived cytokines upon gliosis in the CNS. Dev. Neurosci. 16:128–136. 10.1159/000112099 [DOI] [PubMed] [Google Scholar]
- Gómez-Pinilla, F., Cummings B.J., and Cotman C.W.. 1990. Induction of basic fibroblast growth factor in Alzheimer’s disease pathology. Neuroreport. 1:211–214. 10.1097/00001756-199011000-00009 [DOI] [PubMed] [Google Scholar]
- Gowrishankar, S., Yuan P., Wu Y., Schrag M., Paradise S., Grutzendler J., De Camilli P., and Ferguson S.M.. 2015. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc. Natl. Acad. Sci. USA. 112:E3699–E3708. 10.1073/pnas.1510329112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro, R., and Hardy J.. 2014. Genetics of Alzheimer’s disease. Neurotherapeutics. 11:732–737. 10.1007/s13311-014-0295-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, C.H., Nielsen D.S., Kverka M., Zakostelska Z., Klimesova K., Hudcovic T., Tlaskalova-Hogenova H., and Hansen A.K.. 2012. Patterns of early gut colonization shape future immune responses of the host. PLoS One. 7:e34043. 10.1371/journal.pone.0034043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, C.H., Metzdorff S.B., and Hansen A.K.. 2013. Customizing laboratory mice by modifying gut microbiota and host immunity in an early “window of opportunity”. Gut Microbes. 4:241–245. 10.4161/gmic.23999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, D.V., Hanson J.E., and Sheng M.. 2018. Microglia in Alzheimer’s disease. J. Cell Biol. 217:459–472. 10.1083/jcb.201709069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harach, T., Marungruang N., Duthilleul N., Cheatham V., Mc Coy K.D., Frisoni G., Neher J.J., Fåk F., Jucker M., Lasser T., and Bolmont T.. 2017. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci. Rep. 7:41802. 10.1038/srep41802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman, S.E., Kingery N.D., Ohsumi T.K., Borowsky M.L., Wang L.C., Means T.K., and El Khoury J.. 2013. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16:1896–1905. 10.1038/nn.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S., Dissing-Olesen L., and Stevens B.. 2016. New insights on the role of microglia in synaptic pruning in health and disease. Curr. Opin. Neurobiol. 36:128–134. 10.1016/j.conb.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Ivanov, I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi, S., Gandhi R., Cox L.M., Li N., von Glehn F., Yan R., Patel B., Mazzola M.A., Liu S., Glanz B.L., et al. 2016. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7:12015. 10.1038/ncomms12015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky, J.L., Slunt H.H., Ratovitski T., Jenkins N.A., Copeland N.G., and Borchelt D.R.. 2001. Co-expression of multiple transgenes in mouse CNS: a comparison of strategies. Biomol. Eng. 17:157–165. 10.1016/S1389-0344(01)00067-3 [DOI] [PubMed] [Google Scholar]
- Kamada, N., Seo S.U., Chen G.Y., and Núñez G.. 2013. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13:321–335. 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- Kayed, R., Head E., Sarsoza F., Saing T., Cotman C.W., Necula M., Margol L., Wu J., Breydo L., Thompson J.L., et al. 2007. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2:18. 10.1186/1750-1326-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul, H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169:1276–1290.e17. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Keshavarzian, A., Green S.J., Engen P.A., Voigt R.M., Naqib A., Forsyth C.B., Mutlu E., and Shannon K.M.. 2015. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 30:1351–1360. 10.1002/mds.26307 [DOI] [PubMed] [Google Scholar]
- Kozich, J.J., Westcott S.L., Baxter N.T., Highlander S.K., and Schloss P.D.. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79:5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.B., Nagai A., and Kim S.U.. 2002. Cytokines, chemokines, and cytokine receptors in human microglia. J. Neurosci. Res. 69:94–103. 10.1002/jnr.10253 [DOI] [PubMed] [Google Scholar]
- Lee, K.S., Chung J.H., Lee K.H., Shin M.J., Oh B.H., and Hong C.H.. 2008. Bioplex analysis of plasma cytokines in Alzheimer’s disease and mild cognitive impairment. Immunol. Lett. 121:105–109. 10.1016/j.imlet.2008.09.004 [DOI] [PubMed] [Google Scholar]
- Lee, S., Varvel N.H., Konerth M.E., Xu G., Cardona A.E., Ransohoff R.M., and Lamb B.T.. 2010. CX3CR1 deficiency alters microglial activation and reduces beta-amyloid deposition in two Alzheimer’s disease mouse models. Am. J. Pathol. 177:2549–2562. 10.2353/ajpath.2010.100265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone, C., and Knight R.. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso, R., Fryatt G., Cleal M., Obst J., Pipi E., Monzón-Sandoval J., Ribe E., Winchester L., Webber C., Nevado A., et al. NIMA Consortium . 2019. CSF1R inhibitor JNJ-40346527 attenuates microglial proliferation and neurodegeneration in P301S mice. Brain. 142:3243–3264. 10.1093/brain/awz241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak, M., Mao P., Nakamura K., Bebbington C., Park B., and Reddy P.H.. 2009. Neutralization of granulocyte macrophage colony-stimulating factor decreases amyloid beta 1-42 and suppresses microglial activity in a transgenic mouse model of Alzheimer’s disease. Hum. Mol. Genet. 18:3876–3893. 10.1093/hmg/ddp331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, S., Van Treuren W., White R.A., Eggesbø M., Knight R., and Peddada S.D.. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. 10.3402/mehd.v26.27663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwarha, G., and Ghribi O.. 2012. Leptin signaling and Alzheimer’s disease. Am. J. Neurodegener. Dis. 1:245–265. [PMC free article] [PubMed] [Google Scholar]
- Masliah, E., Mallory M., Deerinck T., DeTeresa R., Lamont S., Miller A., Terry R.D., Carragher B., and Ellisman M.. 1993. Re-evaluation of the structural organization of neuritic plaques in Alzheimer’s disease. J. Neuropathol. Exp. Neurol. 52:619–632. 10.1097/00005072-199311000-00009 [DOI] [PubMed] [Google Scholar]
- Mass, E., Jacome-Galarza C.E., Blank T., Lazarov T., Durham B.H., Ozkaya N., Pastore A., Schwabenland M., Chung Y.R., Rosenblum M.K., et al. 2017. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature. 549:389–393. 10.1038/nature23672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian, S.K., Liu C.H., Tzianabos A.O., and Kasper D.L.. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 122:107–118. 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- McLean, D., Cooke M.J., Albay R. III, Glabe C., and Shoichet M.S.. 2013. Positron emission tomography imaging of fibrillar parenchymal and vascular amyloid-β in TgCRND8 mice. ACS Chem. Neurosci. 4:613–623. 10.1021/cn300226q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilandt, W.J., Ngu H., Gogineni A., Lalehzadeh G., Lee S.H., Srinivasan K., Imperio J., Wu T., Weber M., Kruse A.J., et al. 2020. Trem2 Deletion Reduces Late-Stage Amyloid Plaque Accumulation, Elevates the Aβ42:Aβ40 Ratio, and Exacerbates Axonal Dystrophy and Dendritic Spine Loss in the PS2APP Alzheimer’s Mouse Model. J. Neurosci. 40:1956–1974. 10.1523/JNEUROSCI.1871-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter, M.R., Zhang C., Leone V., Ringus D.L., Zhang X., Oyler-Castrillo P., Musch M.W., Liao F., Ward J.F., Holtzman D.M., et al. 2016. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci. Rep. 6:30028. 10.1038/srep30028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter, M.R., Hinterleitner R., Meisel M., Zhang C., Leone V., Zhang X., Oyler-Castrillo P., Zhang X., Musch M.W., Shen X., et al. 2017. Antibiotic-induced perturbations in microbial diversity during post-natal development alters amyloid pathology in an aged APPSWE/PS1ΔE9 murine model of Alzheimer’s disease. Sci. Rep. 7:10411. 10.1038/s41598-017-11047-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum, J.M., Schilling S., Cynis H., Silva A., Swanson E., Wangsanut T., Tayler K., Wiltgen B., Hatami A., Rönicke R., et al. 2012. Prion-like behaviour and tau-dependent cytotoxicity of pyroglutamylated amyloid-β. Nature. 485:651–655. 10.1038/nature11060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos-Alonso, A., Schetters S.T., Sri S., Askew K., Mancuso R., Vargas-Caballero M., Holscher C., Perry V.H., and Gomez-Nicola D.. 2016. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer’s-like pathology. Brain. 139:891–907. 10.1093/brain/awv379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org, E., Mehrabian M., Parks B.W., Shipkova P., Liu X., Drake T.A., and Lusis A.J.. 2016. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 7:313–322. 10.1080/19490976.2016.1203502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, C., Xu X., Li Y., Li X., Yang X., Chen H., Zhu Y., Lu N., and He C.. 2020. Sex-specific association between the gut microbiome and high-fat diet-induced metabolic disorders in mice. Biol. Sex Differ. 11:5. 10.1186/s13293-020-0281-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-González, R., Alvira-Botero M.X., Robayo O., Antequera D., Garzón M., Martín-Moreno A.M., Brera B., de Ceballos M.L., and Carro E.. 2014. Leptin gene therapy attenuates neuronal damages evoked by amyloid-β and rescues memory deficits in APP/PS1 mice. Gene Ther. 21:298–308. 10.1038/gt.2013.85 [DOI] [PubMed] [Google Scholar]
- Perry, V.H., Cunningham C., and Holmes C.. 2007. Systemic infections and inflammation affect chronic neurodegeneration. Nat. Rev. Immunol. 7:161–167. 10.1038/nri2015 [DOI] [PubMed] [Google Scholar]
- Quon, D., Catalano R., and Cordell B.. 1990. Fibroblast growth factor induces beta-amyloid precursor mRNA in glial but not neuronal cultured cells. Biochem. Biophys. Res. Commun. 167:96–102. 10.1016/0006-291X(90)91735-B [DOI] [PubMed] [Google Scholar]
- Radde, R., Bolmont T., Kaeser S.A., Coomaraswamy J., Lindau D., Stoltze L., Calhoun M.E., Jäggi F., Wolburg H., Gengler S., et al. 2006. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 7:940–946. 10.1038/sj.embor.7400784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff, R.M., and El Khoury J.. 2015. Microglia in Health and Disease. Cold Spring Harb. Perspect. Biol. 8:a020560. 10.1101/cshperspect.a020560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reemst, K., Noctor S.C., Lucassen P.J., and Hol E.M.. 2016. The Indispensable Roles of Microglia and Astrocytes during Brain Development. Front. Hum. Neurosci. 10:566. 10.3389/fnhum.2016.00566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes, T., Flouri T., Nichols B., Quince C., and Mahé F.. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 4:e2584. 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, T.R., Debelius J.W., Thron T., Janssen S., Shastri G.G., Ilhan Z.E., Challis C., Schretter C.E., Rocha S., Gradinaru V., et al. 2016. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell. 167:1469–1480.e12. 10.1016/j.cell.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, D.P., and Stevens B.. 2015. Microglia Function in Central Nervous System Development and Plasticity. Cold Spring Harb. Perspect. Biol. 7:a020545. 10.1101/cshperspect.a020545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A., Ter Horst R., Jansen T., Jacobs L., Bonder M.J., et al. 2016. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 167:1125–1136.e8. 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon, G., Sampson T.R., Geschwind D.H., and Mazmanian S.K.. 2016. The Central Nervous System and the Gut Microbiome. Cell. 167:915–932. 10.1016/j.cell.2016.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, V., Roth S., Llovera G., Sadler R., Garzetti D., Stecher B., Dichgans M., and Liesz A.. 2016. Microbiota Dysbiosis Controls the Neuroinflammatory Response after Stroke. J. Neurosci. 36:7428–7440. 10.1523/JNEUROSCI.1114-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosna, J., Philipp S., Albay R. III, Reyes-Ruiz J.M., Baglietto-Vargas D., LaFerla F.M., and Glabe C.G.. 2018. Early long-term administration of the CSF1R inhibitor PLX3397 ablates microglia and reduces accumulation of intraneuronal amyloid, neuritic plaque deposition and pre-fibrillar oligomers in 5XFAD mouse model of Alzheimer’s disease. Mol. Neurodegener. 13:11. 10.1186/s13024-018-0244-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg, E., Severson P.L., Hohsfield L.A., Crapser J., Zhang J., Burton E.A., Zhang Y., Spevak W., Lin J., Phan N.Y., et al. 2019. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun. 10:3758. 10.1038/s41467-019-11674-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., Xu J., Ling Y., Wang F., Gong T., Yang C., Ye S., Ye K., Wei D., Song Z., et al. 2019. Fecal microbiota transplantation alleviated Alzheimer’s disease-like pathogenesis in APP/PS1 transgenic mice. Transl. Psychiatry. 9:189. 10.1038/s41398-019-0525-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi, R.E. 2012. The genetics of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2:a006296. 10.1101/cshperspect.a006296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, T.L., Savage J.C., Hui C.W., Bisht K., and Tremblay M.E.. 2017. Microglia across the lifespan: from origin to function in brain development, plasticity and cognition. J. Physiol. 595:1929–1945. 10.1113/JP272134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinakaran, G., Teplow D.B., Siman R., Greenberg B., and Sisodia S.S.. 1996. Metabolism of the “Swedish” amyloid precursor protein variant in neuro2a (N2a) cells. Evidence that cleavage at the “beta-secretase” site occurs in the golgi apparatus. J. Biol. Chem. 271:9390–9397. 10.1074/jbc.271.16.9390 [DOI] [PubMed] [Google Scholar]
- Thion, M.S., Low D., Silvin A., Chen J., Grisel P., Schulte-Schrepping J., Blecher R., Ulas T., Squarzoni P., Hoeffel G., et al. 2018. Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell. 172:500–516.e16. 10.1016/j.cell.2017.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini, P. 2017. Gut Microbiota: A Potential Regulator of Neurodevelopment. Front. Cell. Neurosci. 11:25. 10.3389/fncel.2017.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulland, T.K., Song W.M., Huang S.C., Ulrich J.D., Sergushichev A., Beatty W.L., Loboda A.A., Zhou Y., Cairns N.J., Kambal A., et al. 2017. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell. 170:649–663.e13. 10.1016/j.cell.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas, C., Kumar S., Franklin B.S., Dierkes T., Brinkschulte R., Tejera D., Vieira-Saecker A., Schwartz S., Santarelli F., Kummer M.P., et al. 2017. Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature. 552:355–361. 10.1038/nature25158 [DOI] [PubMed] [Google Scholar]
- Vogt, N.M., Kerby R.L., Dill-McFarland K.A., Harding S.J., Merluzzi A.P., Johnson S.C., Carlsson C.M., Asthana S., Zetterberg H., Blennow K., et al. 2017. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 7:13537. 10.1038/s41598-017-13601-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, D.G., and Lue L.F.. 2015. Immune phenotypes of microglia in human neurodegenerative disease: challenges to detecting microglial polarization in human brains. Alzheimers Res. Ther. 7:56. 10.1186/s13195-015-0139-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger, B.D., Gobet C., Yeung J., Martin E., Jimenez S., Betrisey B., Foata F., Berger B., Balvay A., Foussier A., et al. 2019. The Mouse Microbiome Is Required for Sex-Specific Diurnal Rhythms of Gene Expression and Metabolism. Cell Metab. 29:362–382.e8. 10.1016/j.cmet.2018.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendeln, A.C., Degenhardt K., Kaurani L., Gertig M., Ulas T., Jain G., Wagner J., Häsler L.M., Wild K., Skodras A., et al. 2018. Innate immune memory in the brain shapes neurological disease hallmarks. Nature. 556:332–338. 10.1038/s41586-018-0023-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff, W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., and Siuzdak G.. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA. 106:3698–3703. 10.1073/pnas.0812874106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, G., Wang X., Zhao A., Yan J., Chen W., Jiang R., Ji J., Huang F., Zhang Y., Lei S., et al. 2017. Sex-dependent effects on gut microbiota regulate hepatic carcinogenic outcomes. Sci. Rep. 7:45232. 10.1038/srep45232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy, L., Burrows M., Khan A.A., Graham L., Volchkov P., Becker L., Antonopoulos D., Umesaki Y., and Chervonsky A.V.. 2013. Gender bias in autoimmunity is influenced by microbiota. Immunity. 39:400–412. 10.1016/j.immuni.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaheer, A., Zaheer S., Sahu S.K., Knight S., Khosravi H., Mathur S.N., and Lim R.. 2007. A novel role of glia maturation factor: induction of granulocyte-macrophage colony-stimulating factor and pro-inflammatory cytokines. J. Neurochem. 101:364–376. 10.1111/j.1471-4159.2006.04385.x [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., and Chanda S.K.. 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10:1523. 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows ANCOM analysis of fecal microbiota at PND22 from WT- versus Tg-FMT study.
shows ANCOM analysis of fecal microbiota at 9 wk from WT- versus Tg-FMT study.
shows lower DEGs in ABX-treated APPPS1-21 male mice associated pathways.
shows higher DEGs in ABX-treated APPPS1-21 male mice associated pathways.
compares DEGs between Tg_M_Ctr (group 9) and Tg_M_Abx (group 7) at PND24.
compares DEGs between non-Tg_M_Ctr (group 5) and non-Tg_M_Abx (group 6) at PND24.
compares DEGs between non-Tg_M_Ctr (group 3) and non-Tg_M_Abx (group 4) at 9 wk of age.
compares DEGs between non-Tg_F_Ctr (group 2) and non-Tg_F_Abx (group 1) at 9 wk of age.
details statistical information for mentioned comparisons in each figure legend.