Abstract
Background
The Stephania tetrandra S. Moore (S. tetrandra) is a medicinal plant belonging to the family Menispermaceae that has high medicinal value and is well worth doing further exploration. The wild resources of S. tetrandra were widely distributed in tropical and subtropical regions of China, generating potential genetic diversity and unique population structures. The geographical origin of S. tetrandra is an important factor influencing its quality and price in the market. In addition, the species relationship within Stephania genus still remains uncertain due to high morphological similarity and low support values of molecular analysis approach. The complete chloroplast (cp) genome data has become a promising strategy to determine geographical origin and understand species evolution for closely related plant species. Herein, we sequenced the complete cp genome of S. tetrandra from Zhejiang Province and conducted a comparative analysis within Stephania plants to reveal the structural variations, informative markers and phylogenetic relationship of Stephania species.
Results
The cp genome of S. tetrandra voucher ZJ was 157,725 bp, consisting of a large single copy region (89,468 bp), a small single copy region (19,685 bp) and a pair of inverted repeat regions (24,286 bp each). A total of 134 genes were identified in the cp genome of S. tetrandra, including 87 protein-coding genes, 8 rRNA genes, 37 tRNA genes and 2 pseudogene copies (ycf1 and rps19). The gene order and GC content were highly consistent in the Stephania species according to the comparative analysis results, with the highest RSCU value in arginine (1.79) and lowest RSCU value in serine of S. tetrandra, respectively. A total of 90 SSRs have been identified in the cp genome of S. tetrandra, where repeats that consisting of A or T bases were much higher than that of G or C bases. In addition, 92 potential RNA editing sites were identified in 25 protein-coding genes, with the most predicted RNA editing sites in ndhB gene. The variations on length and expansion extent to the junction of ycf1 gene were observed between S. tetrandra vouchers from different regions, indicating potential markers for further geographical origin discrimination. Moreover, the values of transition to transversion ratio (Ts/Tv) in the Stephania species were significantly higher than 1 using Pericampylus glaucus as reference. Comparative analysis of the Stephania cp genomes revealed 5 highly variable regions, including 3 intergenic regions (trnH-psbA, trnD-trnY, trnP) and two protein coding genes (rps16 and ndhA). The identified mutational hotspots of Stephania plants exhibited multiple SNP sites and Gaps, as well as different Ka/Ks ratio values. In addition, five pairs of specific primers targeting the divergence regions were accordingly designed, which could be utilized as potential molecular markers for species identification, population genetic and phylogenetic analysis in Stephania species. Phylogenetic tree analysis based on the conserved chloroplast protein coding genes indicated a sister relationship between S. tetrandra and the monophyletic group of S. japonica and S. kwangsiensis with high support values, suggesting a close genetic relationship within Stephania plants. However, two S. tetrandra vouches from different regions failed to cluster into one clade, confirming the occurrences of genetic diversities and requiring further investigation for geographical tracing strategy.
Conclusions
Overall, we provided comprehensive and detailed information on the complete chloroplast genome and identified nucleotide diversity hotspots of Stephania species. The obtained genetic resource of S. tetrandra from Zhejiang Province would facilitate future studies in DNA barcode, species discrimination, the intraspecific and interspecific variability and the phylogenetic relationships of Stephania plants.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-08193-x.
Keywords: Stephania tetrandra, Chloroplast genome, Phylogenetic relationship, Comparative analysis, Mutational hotspots
Background
The Stephania tetrandra S. Moore (S. tetrandra) is a kind of Stephania plants belonging to the tribe Menispermea of family Menispermaceae. It is a perennial herb that grows in the bush near the village, wilderness and roadside, and is widely distributed in the southern part of China [1]. The dried root of S. tetrandra is used as the traditional Chinese medicine Fangji in China with various pharmacological effects including diuretic, anti-inflammatory and an antirheumatic treatment [2]. Up to data, a total of 67 alkaloids and several other active compounds have been isolated and characterized from the roots and aerial parts of S. tetrandra [2]. The typical bisbenzylisoquinoline alkaloidhas tetrandrine derived from S. tetrandra have shown multiple biological activities, like anti-tumor, anti-inflammatory, prevention and treatment of various fibrotic diseases [3–5]. Tetrandrine was also reported to inhibit migration and invasion of human nasopharyngeal carcinoma dependent of MAPK and RhoA signaling pathways, suggesting its potential development in the application of anticancer agents for nasopharyngeal carcinoma therapy [6]. In addition, Fangji Huangqi Decoction was reported to protect the kidney directly and reduce the level of uric acid indirectly [7]. Therefore, the S. tetrandra has high medicinal value and is worthwhile to do further exploration.
According to the Chinese Pharmacopoeia, S. tetrandra is the only authentic source of Fangji [8]. However, due to the similarity in name and morphology, S. tetrandra is often mistakenly substituted and adulterated with roots from other plants, which contained different chemical compositions and therapeutic effects, such as Aristolochia fangchi [9]. In addition, the closely related Menispermaceae species of Cocculus orbiculatus, Menispermum dauricum and Sinomenium acutum also provided potential counterfeits for S. tetrandra in the market. The problem of wrong substitutions brings potential safety hazard and might leads damage to health. In addition, the classification of Stephania species has been conducted for many years using different methods. The traditional classification methods divided genus Stephania into three subgenera based on the comparison of appearance characters of plants [10]. With the development of molecular biology and DNA sequencing technology, molecular systematics research methods have also been used for taxonomic studies of the genus Stephania [11]. The DNA markers ITS and trnL-F have been applied to construct the phylogenetic tree of Stephania species. However, the analysis results gave controversial phylogenetic relationship among the three subgenera of the genus Stephania [11, 12]. Compared with the nuclear gene and partial chloroplast genes, the complete chloroplast genome with a large set of genes is generally considered as a useful tool for phylogenetic analysis and further development on species identification and restoration strategies [13]. The chloroplast genome of plants contains a lot of molecular information that is a good resource for plant systematics, population genomics and phylogenetic studies [14]. Thus, it is important to sequence the complete chloroplast genome and develop highly variable and informative markers to study the reticulate evolution history of the genus Stephania and to identify S. tetrandra and other adulterants accurately to ensure the clinical safety of S. tetrandra.
The chloroplast, which performs photosynthesis, is an essential organelle in plants and is generally non-recombinant and uniparentally inherited. In recent years, chloroplast genomic information has been widely used for developing molecular markers to classify medicinal plants and used for evolutionary studies [15, 16]. In most angiosperms, chloroplast genome is generally double stranded and circular [17]. It consists of a small single copy region (SSC), a large single copy region (LSC) and a pair of inverted repeat regions (IR) which separate SSC and LSC [18]. On average, the size of chloroplast genome varies from 115 kb to 165 kb [17]. There are many studies have reported that the chloroplast genome is expected to serve as a super-barcode to distinguish closely related species [19]. Comparative and phylogenetic analysis of six Ligularia species observed a close relationship between L. fischeri and L. jaluensis and the whole cp genome was expected to become a super-barcode to identify Ligularia species [20]. The ycf1b region from the complete chloroplast genome of Pterocarpus, was identified as a potential molecular marker for species identification of Pterocarpus wood [21]. The complete chloroplast genome sequence of S. tetrandra from Jiangxi Province has been determined previously [22]. However, the chloroplast genome information of S. tetrandra from Zhejiang Province has not been characterized and a comprehensive analysis of the chloroplast structures of Stephania species is still lacking. The aims of this study were to (i) conduct comparative research on the Stephania chloroplast genome, generating information on basic genome structure, codon usage bias, repetitive structure characteristics, and RNA editing sites; (ii) identify hotspot regions and develop potential DNA markers through high nucleotide diversity, and (iii) reconstruct the phylogenetic relationships of Menispermaceae species and determine the taxonomic status of Stephania based on the conserved chloroplast protein-coding genes data. Our results will be important gene basic data for the marker development, species discrimination, and the inference of phylogenetic relationships for family Menispermaceae based on these comprehensive analyses of chloroplast genomes.
Results
Chloroplast genome features of Stephania tetrandra
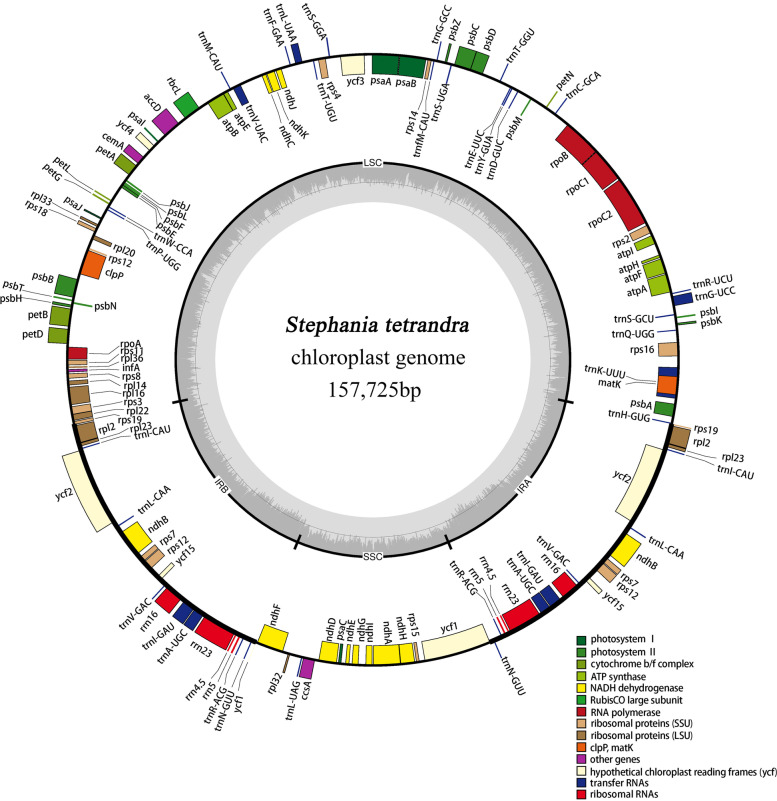
In total, 21,016,486 Illumina reads and 3,085,951,100 bases were obtained for Stephania tetrandra voucher ZJ by Illumina Hiseq 2500 platform. After cutting, selecting and assembling reads, the complete cp genome of S. tetrandra with a size of 157,725 bp was generated. It was a circular structure conserved 4 regions: a small single copy region (SSC; 19,685 bp), a large single copy region (LSC; 89,468 bp) and a pair of inverted repeat regions (IR; 24,286 bp of each) which separated SSC and LSC (Fig. 1). As shown in Table 1, the total number of genes annotated in the cp genome of S. tetrandra was 134, including 87 protein-coding genes, 8 rRNA genes, 37 tRNA genes and 2 pseudogene copies (ycf1 and rps19). Among the 134 genes, 18 genes were duplicated in IR region and 23 genes contained introns (Table 1). In addition, 11 protein-coding genes (rps16, petB, petD, atpF, ndhB (× 2), ndhA, rpoC1, rpl16 and rpl2 (× 2)) and 8 tRNA genes (trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU (× 2) and trnA-UGC (× 2)) contained only one intron, while 4 protein-coding genes (rps12(× 2), clpP and ycf3) had two introns.
Fig. 1.
Circular chloroplast (CP) genome map of Stephania tetrandra. Genes drawn outside the circle are transcribed anti-clockwise, while those inside the circle are transcribed clockwise. Large single copy (LSC) region, inverted repeat (IRA, IRB) regions and small single copy (SSC) region are shown in the figure. The darker gray in the inner circle corresponds GC content whereas the lighter gray corresponds AT content. Different colors of genes represent their different functions
Table 1.
List of genes annotated in the chloroplast genomes of Staphania tetrandra voucher ZJ
| No. | Group of genes | Gene names | Amount |
|---|---|---|---|
| 1 | Photosystem I | psaJ, psaC, psaB, psaA, psaI | 5 |
| 2 | Photosystem II | psbE, psbB, psbT, psbN, psbH, psbJ, psbL, psbF, psbA, psbK, psbI, psbM, psbD, psbZ | 14 |
| 3 | Cytochrome b/f complex | petL, petG, petB*, petD*, petA, petN | 6 |
| 4 | ATP synthase | atpE, atpB, atpA, atpF*, atpH, atpI | 6 |
| 5 | NADH dehydrogense | ndhB*(× 2), ndhF, ndhD, ndhE, ndhJ, ndhK, ndhC, ndhA*, ndhG, ndhI, ndhH | 12 |
| 6 | RubisCO large subunit | rbcL | 1 |
| 7 | RNA polymerase | rpoA, rpoC2, rpoC1*, rpoB, | 4 |
| 8 | Ribosomal proteins (SSU) | rps11, rps8, rps3, rps19, rps7, rps14, rps4, rps18, rps12**(×2), rps16*, rps2, rps15 | 13 |
| 9 | Ribosomal proteins (LSU) | rpl33, rpl20, rpl36, rpl14, rpl16*, rpl22, rpl2*(×2), rpl23(× 2), rpl32, rpl33, rpl20 | 13 |
| 10 | Other genes | clpP**, infA, ccsA, accD, cemA, matK | 6 |
| 11 | Proteins of unknown function | ycf2(×2), ycf3**, ycf4, ycf1, ycf15(× 2) | 7 |
| 12 | Transfer RNAs | 37 tRNAs (8 contain an intron,7 in the IRs) | 37 |
| 13 | Ribosomal RNAs | rrn4.5(×2), rrn5(× 2), rrn16(× 2), rrn23(× 2) | 8 |
| 14 | pseudo genes | ycf1, rps19 | 2 |
One or two asterisks after a gene indicate that the gene contains one or two introns, respectively
The complete cp genomes of three Stephania species showed similarity in terms of the gene content, total length of complete cp genome and composition of GC content. The overall GC contents in the cp genomes of S. tetrandra and other Stephania species were calculated using MEGA 7.0. The total GC contents of the cp genomes were 38.18, 38.23 and 38.44% in S. tetrandra, S. japonica and S. kwangsiensis, respectively (Table 2). The GC content varied significantly in different regions of the cp genome. IR regions had the highest GC content in all the three cp genomes, followed by LSC region and SSC region. In addition, ycf1 and rps19 pseudogenes could be found in all of these three plants. Among their protein-coding genes, the initiation codons of psbC, rps19, rpl2 and ndhD genes were different from the standard initiation codon ATG. The psbC and rps19 genes in the three plants started with GTG, while rpl2 and ndhD in S. tetrandra, S. japonica started with ACG but in S. kwangsiensis started with ATA and GTG respectively. Similar results of rps19 started with GTG also reported in other Menispermaceae plants [23, 24]. Interestingly, the cp genome of S. tetrandra contains two ycf15 genes in the IR regions, which exhibited the most difference between S. tetrandra and the other two Stephania species.
Table 2.
Statistics on the basic features of the chloroplast genomes of the three Stephania species
| Characteristics | Stephania tetrandra | Stephania japonica | Stephania kwangsiensis |
|---|---|---|---|
| Accession number | MT849286 | KU204903.1 | MN654112.1 |
| Total length (bp) | 157,725 | 157,719 | 156,624 |
| LSC length (bp) | 89,468 | 88,693 | 87,759 |
| SSC length (bp) | 19,685 | 20,346 | 20,169 |
| IR length (bp) | 48,572 | 48,680 | 48,696 |
| Total Number of Genes | 134 | 132 | 132 |
| Coding Genes | 87 | 85 | 85 |
| rRNA Genes | 8 | 8 | 8 |
| tRNA Genes | 37 | 37 | 37 |
| Pesudo genes | 2 | 2 | 2 |
| GC content | |||
| Total (%) | 38.18 | 38.23 | 38.44 |
| LSC (%) | 36.31 | 36.42 | 36.66 |
| SSC (%) | 33.00 | 32.94 | 33.40 |
| IR (%) | 43.73 | 43.75 | 43.76 |
LSC Large single copy, SSC Small single copy, IR Inverted repeat, GC Guanine and Cytosine
Analysis of codon usage bias
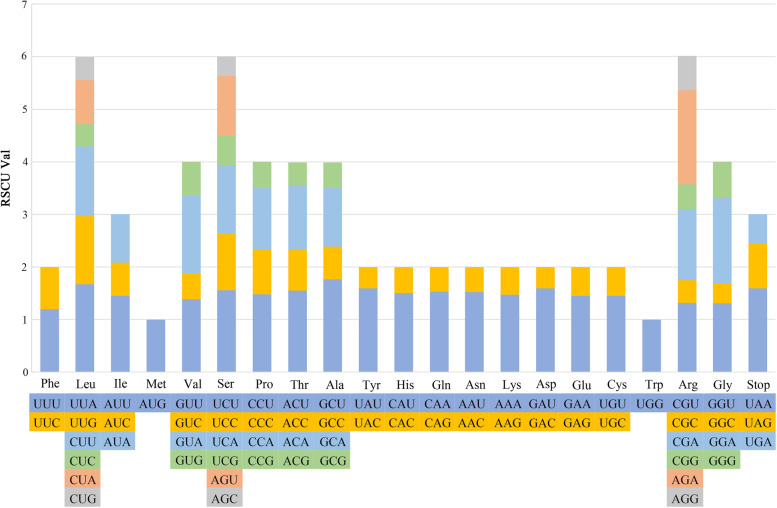
In the cp genome of S. tetrandra, it contained 78,750 bp of coding sequences consisting of 26,250 codons. Leucine was the most abundant amino acid in the cp genome of S. tetrandra, whereas cysteine was the least (Table 3). Regardless of stop codons, the most used codon was AUU, encoding isoleucine (Ile), with a number of 1065, while the least used codon was UGC, encoding cysteine (Cys), with a number of 84 in S. tetrandra. Relative synonymous codon usage (RSCU) reflected the ratio of the frequency of usage of a codon to the expected frequency [25]. Most of the amino acid have codon preferences in the cp genome of S. tetrandra, however, methionine (AUG) and tryptophan (UGG) were encoded by only one codon and exhibited no codon bias (Fig. 2). AGA (1.79) in arginine showed the highest RSCU value, and the lowest was AGC (0.37) in serine. Comparative analysis indicated the coded amino acids used in S. japonica and S. kwangsiensis were identical with that of S. tetrandra, with the number of codons of 26,005 and 26,019 in S. japonica and S. kwangsiensis, respectively. Moreover, similar trends on codon usage preference were observed in the three Stephaniae plants (Table 3).
Table 3.
Codon usage and codon-anticodon recognition patterns of three Stephania plants
| Codon | tRNA | Numbers and RSCU | ||
|---|---|---|---|---|
| S. tetrandra. | S. japonica | S. kwangsiensis | ||
| UUU(F) | 870/1.20 | 882/1.23 | 889/1.22 | |
| UUC(F) | trnF-GAA | 576/0.80 | 557/0.77 | 568/0.78 |
| UUA(L) | 749/1.67 | 768/1.72 | 757/1.69 | |
| UUG(L) | trnL-CAA | 581/1.30 | 556/1.24 | 567/1.27 |
| CUU(L) | 594/1.32 | 601/1.35 | 590/1.32 | |
| CUC(L) | 193/0.43 | 185/0.41 | 189/0.42 | |
| CUA(L) | 374/0.84 | 368/0.82 | 371/0.83 | |
| CUG(L) | 199/0.44 | 203/0.45 | 207/0.46 | |
| AUU(I) | 1065/1.45 | 1057/1.45 | 1056/1.46 | |
| AUC(I) | trnI-GAU | 464/0.63 | 447/0.61 | 460/0.63 |
| AUA(I) | 674/0.92 | 677/0.93 | 659/0.91 | |
| AUG(M) | trnM-CAU | 625/1.00 | 613/1.00 | 613/1.00 |
| GUU(V) | 506/1.39 | 510/1.41 | 506/1.39 | |
| GUC(V) | 179/0.49 | 175/0.48 | 181/0.50 | |
| GUA(V) | 537/1.48 | 534/1.47 | 535/1.46 | |
| GUG(V) | 233/0.64 | 231/0.64 | 239/0.65 | |
| UCU(S) | 540/1.56 | 526/1.56 | 524/1.55 | |
| UCC(S) | trnS-GGA | 371/1.07 | 362/1.07 | 370/1.09 |
| UCA(S) | trnS-UGA | 451/1.31 | 432/1.28 | 428/1.26 |
| UCG(S) | 199/0.57 | 191/0.57 | 196/0.58 | |
| CCU(P) | 417/1.48 | 409/1.46 | 397/1.42 | |
| CCC(P) | 236/0.84 | 234/0.84 | 253/0.90 | |
| CCA(P) | trnP-UGG | 331/1.17 | 318/1.14 | 321/1.15 |
| CCG(P) | 142/0.50 | 156/0.56 | 150/0.54 | |
| ACU(T) | 531/1.55 | 518/1.54 | 517/1.54 | |
| ACC(T) | trnT-GGU | 264/0.77 | 264/0.78 | 259/0.77 |
| ACA(T) | trnT-UGU | 417/1.22 | 409/1.21 | 405/1.21 |
| ACG(T) | 155/0.45 | 158/0.47 | 159/0.47 | |
| GCU(A) | 623/1.77 | 606/1.74 | 601/1.72 | |
| GCC(A) | 223/0.63 | 222/0.64 | 223/0.64 | |
| GCA(A) | 392/1.11 | 376/1.08 | 386/1.11 | |
| GCG(A) | 174/0.49 | 191/0.55 | 186/0.53 | |
| UAU(Y) | 747/1.59 | 749/1.57 | 731/1.57 | |
| UAC(Y) | trnY-GUA | 192/0.41 | 205/0.43 | 201/0.43 |
| UAA(*) | 46/1.59 | 41/1.45 | 43/1.52 | |
| UAG(*) | 25/0.86 | 26/0.92 | 26/0.92 | |
| CAU(H) | 490/1.50 | 475/1.50 | 468/1.49 | |
| CAC(H) | trnH-GUG | 163/0.50 | 158/0.50 | 162/0.51 |
| CAA(Q) | trnQ-UUG | 731/1.53 | 718/1.53 | 724/1.52 |
| CAG(Q) | 225/0.47 | 223/0.47 | 226/0.48 | |
| AAU(N) | 961/1.52 | 940/1.51 | 948/1.51 | |
| AAC(N) | 301/0.48 | 307/0.49 | 305/0.49 | |
| AAA(K) | 994/1.47 | 992/1.46 | 976/1.46 | |
| AAG(K) | 362/0.53 | 366/0.54 | 364/0.54 | |
| GAU(D) | 872/1.59 | 856/1.58 | 859/1.58 | |
| GAC(D) | trnD-GUC | 227/0.41 | 226/0.42 | 229/0.42 |
| GAA(E) | trnE-UUC | 958/1.45 | 953/1.46 | 953/1.45 |
| GAG(E) | 361/0.55 | 351/0.54 | 358/0.55 | |
| UGU(C) | trnC-GCA | 221/1.45 | 223/1.46 | 224/1.49 |
| UGC(C) | 84/0.55 | 82/0.54 | 77/0.51 | |
| UGA(*) | 16/0.55 | 18/0.64 | 16/0.56 | |
| UGG(W) | trnW-CCA | 470/1.00 | 465/1.00 | 465/1.00 |
| CGU(R) | trnR-ACG | 352/1.32 | 352/1.33 | 358/1.35 |
| CGC(R) | 115/0.43 | 112/0.42 | 111/0.42 | |
| CGA(R) | 360/1.35 | 343/1.30 | 352/1.32 | |
| CGG(R) | 125/0.47 | 132/0.50 | 124/0.47 | |
| AGU(S) | 390/1.13 | 386/1.14 | 384/1.13 | |
| AGC(S) | trnS-GCU | 127/0.37 | 130/0.38 | 132/0.39 |
| AGA(R) | trnR-UCU | 477.1.79 | 468/1.77 | 472/1.77 |
| AGG(R) | 174/0.64 | 176/0.67 | 179/0.67 | |
| GGU(G) | 588/1.31 | 584/1.30 | 583/1.30 | |
| GGC(G) | trnG-GCC | 168/0.38 | 176/0.39 | 171/0.38 |
| GGA(G) | 723/1.62 | 712/1.59 | 713/1.59 | |
| GGG(G) | 311/0.69 | 324/0.72 | 323/0.72 | |
Fig. 2.
Codon content and RSCU value of the 20 amino acid and stop codons in all protein-coding genes of Stephania tetrandra chloroplast genome. The color of the histogram corresponds to the color of codons
RNA editing sites
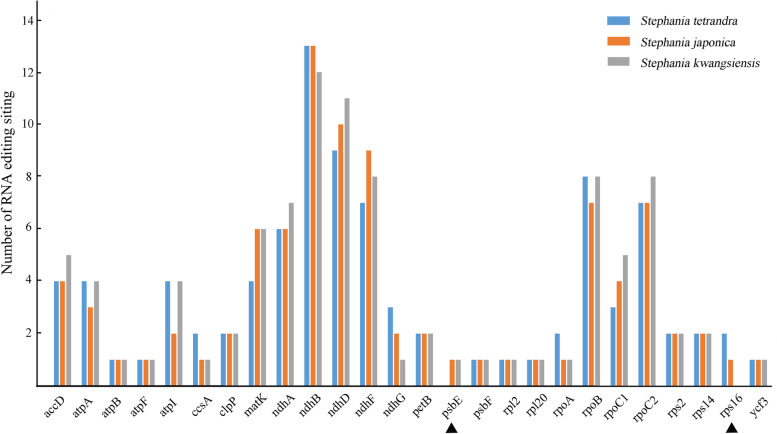
A total of 92 potential RNA editing sites have been predicted in 25 protein-coding genes of the cp genome of S. tetrandra (Fig. 3). Among 92 RNA editing sites, 24 codons were observed to be edited at the first nucleotide position whereas 66 codons were found to be edited at the second nucleotide position, and 2 codons edited both at the first and second nucleotide (Table 4). All of the detected codon changes in the cp genome of S. tetrandra showed C to T conversions. The ndhB gene had the largest number of RNA editing sites (13 editing sites), followed by ndhD (9 editing sites), while atpF, atpB, psbF, rpl20, ycf3 and rpl2 exhibited only one editing site in S. tetrandra. These editing sites resulted in 12 kinds of amino acid conversions (Table 4). The conversions of H to Y, P to S, L to F, R to W, R to C were due to codons edited at the first nucleotide position, while the S to L, P to L, S to F, T to M, A to V, T to I conversions were because of codons edited at the second nucleotide position. Only the conversion of P to F was due to codons edited at the first and second nucleotide positions. The conversion of serine to leucine (S to L) was the most abundant kind of conversion, accounting for 42.4%, followed by proline to leucine (P to L) and histidine to tyrosine (H to Y) accounted for 12.0% respectively. Furthermore, the predicted RNA editing sites in the cp genome of S. japonica and S. kwangsiensis showed similar results with that of S. tetrandra, with the number of 91 and 96 respectively (Fig. 3). It is important to note that the RNA editing site was lost in psbE gene of S. tetrandra and rps16 gene of S. kwangsiensis, which would bring crucial impacts on the translation and protein activity of these genes (Fig. 3). Since the close correlation between RNA editing sites and nucleotide substitution of protein coding genes, we performed further analysis to investigate the synonymous substitutions (Ks) and non-synonymous substitutions (Ka) of protein coding genes with abundant RNA editing sites (Table 5). The Ka/Ks ratios of most genes (23/25) in S. tetrandra were less than 0.5, suggesting an obvious purifying selection pattern. Particularly, both of the petB and psbF gene even exhibited a Ka/Ks value of 0, showing the two genes were possibly under strong purifying selection pressure (Table 5). However, atpF showed a Ka/Ks value greater than 1.00, which indicated it is under diversifying selection pattern and would play significant roles during pressure evolution in S. tetrandra (Table 5). The RNA editing site and nucleotide substitution of protein-coding genes provide valuable information for understanding of missense mutations in the cp genome of S. tetrandra.
Fig. 3.
Number of the RNA editing sites in the cp genome of Stephania tetrandra, Stephania japonica and Stephania kwangsiensis predicted by PREP-Cp program with a cutoff value 0.8
Table 4.
Amino acid conversion frequency of protein coding gene of three Stephania species
| Amino acid conversion | Edited position | Number and percentage | ||
|---|---|---|---|---|
| S. tetrandra | S. japonica | S. kwangsiensis | ||
| S-L | Second nucleotide | 39/42.4% | 34/37.8% | 34/36.2% |
| P-L | Second nucleotide | 11/12.0% | 13/14.4% | 14/14.9% |
| H-Y | First nucleotide | 11/12.0% | 10/11.1% | 11/11.7% |
| L-F | First nucleotide | 4/4.3% | 5/5.6% | 3/3.2% |
| S-F | Second nucleotide | 5/5.4% | 5/5.6% | 5/5.3% |
| T-M | Second nucleotide | 4/4.3% | 4/4.4% | 4/4.3% |
| A-V | Second nucleotide | 3/3.3% | 4/4.4% | 5/5.3% |
| P-S | First nucleotide | 2/2.2% | 4/4.4% | 4/4.3% |
| R-W | First nucleotide | 5/5.4% | 4/4.4% | 5/5.3% |
| T-I | Second nucleotide | 4/4.3% | 5/5.6% | 5/5.3% |
| R-C | First nucleotide | 2/2.2% | 0 | 2/2.1% |
| P-F | First and second nucleotide | 2/2.2% | 2/2.2% | 2/2.1% |
Table 5.
The value of Ka/Ks in 25 protein coding genes with RNA editing sites in S. tetrandra voucher ZJ
| Gene | Number of RNA editing sites | Non-synonymous substitutions (Ka) | Synonymous substitutions (Ks) | Ka/Ks |
|---|---|---|---|---|
| ndhB | 13 | 0.0035 | 0.0107 | 0.3271 |
| ndhD | 9 | 0.0106 | 0.0947 | 0.1119 |
| rpoB | 8 | 0.0045 | 0.0613 | 0.2887 |
| rpoC2 | 7 | 0.0177 | 0.0615 | 0.0732 |
| ndhF | 7 | 0.0202 | 0.0181 | 0.1869 |
| ndhA | 6 | 0.0074 | 0.0927 | 0.0798 |
| matK | 4 | 0.0373 | 0.0948 | 0.3936 |
| atpA | 4 | 0.0061 | 0.0617 | 0.0989 |
| accD | 4 | 00159 | 0.0444 | 0.3581 |
| atpI | 4 | 0.0054 | 0.0385 | 0.1403 |
| rpoC1 | 3 | 0.0071 | 0.0595 | 0.1193 |
| ndhG | 3 | 0.0101 | 0.0734 | 0.1376 |
| ccsA | 3 | 0.0239 | 0.1146 | 0.2086 |
| rpoA | 2 | 0.0207 | 0.0841 | 0.2461 |
| rps2 | 2 | 0.0037 | 0.0824 | 0.0449 |
| clpP | 2 | 0.0022 | 0.0554 | 0.0397 |
| rps14 | 2 | 0.0131 | 0.0453 | 0.2892 |
| rps16 | 2 | 0.0047 | 0.0295 | 0.1593 |
| petB | 2 | 0.08 | 0 | 0 |
| atpB | 1 | 0.0036 | 0.0643 | 0.0559 |
| atpF | 1 | 0.2122 | 0.1658 | 1.2799 |
| rpl2 | 1 | 0.0091 | 0.0108 | 0.8426 |
| rpl20 | 1 | 0.0116 | 0.0486 | 0.2387 |
| ycf3 | 1 | 0.0078 | 0.0423 | 0.1844 |
| psbF | 1 | 0 | 0 | 0 |
SSRs and long repeats analysis
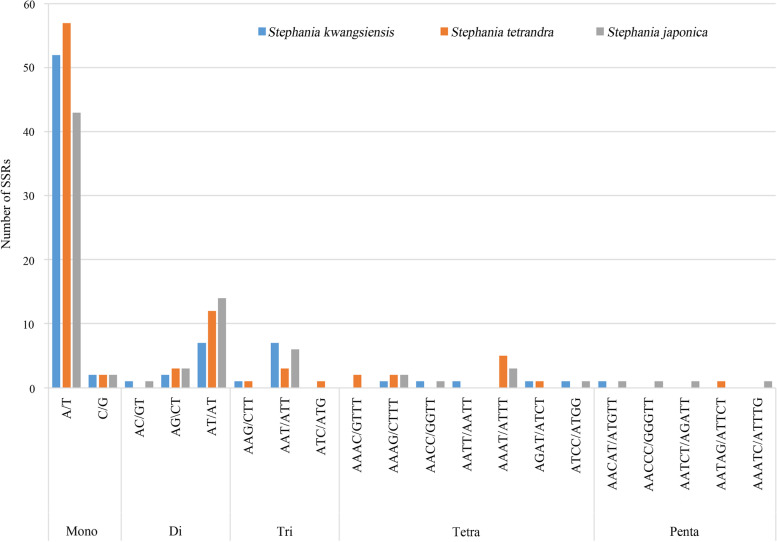
A total of 90, 80, 78 simple sequence repeats (SSRs) have been detected in the cp genomes of S. tetrandra, S. japonica and S. kwangsiensis, respectively (Fig. 4). In the cp genomes of the three Stephania plants, the mononucleotide repeats were consisted of 10–18 repeat units, dinucleotide repeats were consisted of 5–10 repeat units, trinucleotide repeats were consisted of 4–5 repeat units, tetranucleotide repeats were consisted of 3–4 repeat units, and pentanucleotide repeats contained 3 repeat units (Tables S1, S2, S3). SSRs composed of A/T were more abundant than those containing G or C in the cp genomes of the three Stephania plants. The mononucleotide repeat A/T was the most abundant, encountering 57 times in S. tetrandra, 52 times in S. kwangsiensis and 43 times in S. japonica, respectively (Table 6). And the second most repeat was the dinucleotide SSR AT/AT. The number and type of pentanucleotide SSRs in the cp genomes of the three plants exhibited some differences. The S. japonica had four different types of pentanucleotide SSRs (AACAT/ATGTT, AACCC/GGGTT, AATCT/AGATT, AAATC/ATTTG) whereas the S. tetrandra and S. kwangsiensis had only one type (AATAG/ATTCT in S. tetrandra, AACAT/ATGTT in S. kwangsiensis). The number of all types of pentanucleotide SSR was only one. No hexanucleotide SSR has been detected in the cp genomes of the three species. Compared with the IR region, SSRs were mainly distributed in LSC and SSC regions. These SSR markers would be useful for genetic diversity investigation and development of conservation strategy for Stephania plants.
Fig. 4.
Number of different types of SSRs in the cp genomes of Stephania tetrandra, Stephania japonica and Stephania kwangsiensis, setting parameters as 10 for mononucleotide SSRs (Mono), 5 for dinucleotide SSRs (Di), 4 for trinucleotide SSRs (Tri), 3 each for tetranucleotide (Tetra), pentanucleotide (Penta) and hexanucleotide (Hexa) SSRs
Table 6.
The SSR types of the three Stephania plants
| SSR type | Repeat unit | Amount | ||
|---|---|---|---|---|
| Stephania kwangsiensis | Stephania tetrandra | Stephania japonica | ||
| Mono | A/T | 52 | 57 | 43 |
| C/G | 2 | 2 | 2 | |
| Di | AC/GT | 1 | 0 | 1 |
| AG/CT | 2 | 3 | 3 | |
| AT/AT | 7 | 12 | 14 | |
| Tri | AAG/CTT | 1 | 1 | 0 |
| AAT/ATT | 7 | 3 | 6 | |
| ATC/ATG | 0 | 1 | 0 | |
| Tera | AAAC/GTTT | 0 | 2 | 0 |
| AAAG/CTTT | 1 | 2 | 2 | |
| AACC/GGTT | 1 | 0 | 1 | |
| AATT/AATT | 1 | 0 | 0 | |
| AAAT/ATTT | 0 | 5 | 3 | |
| AGAT/ATCT | 1 | 1 | 0 | |
| ATCC/ATGG | 1 | 0 | 1 | |
| Penta | AACAT/ATGTT | 1 | 0 | 1 |
| AACCC/GGGTT | 0 | 0 | 1 | |
| AATCT/AGATT | 0 | 0 | 1 | |
| AATAG/ATTCT | 0 | 1 | 0 | |
| AAATC/ATTTG | 0 | 0 | 1 | |
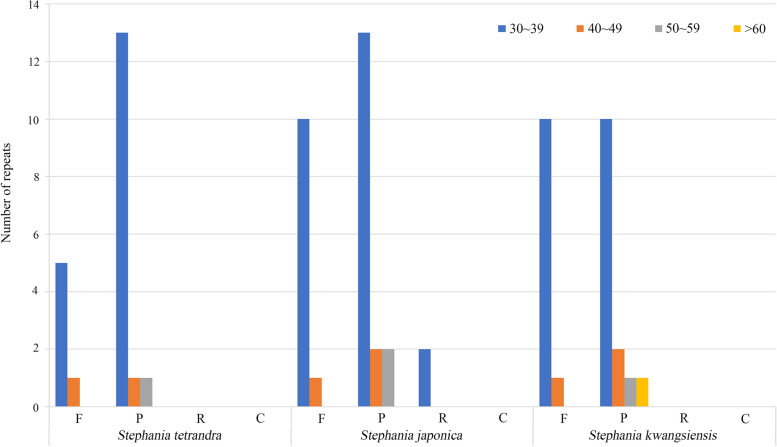
REPuter program was used to analyze long repeat sequences in the cp genomes of the three Stephania plants. The results indicated three types of repetition (forward, reverse and palindrome), with a length of exceeding 30 bp. After removing the two IR regions, which were the largest palindromic repeats in the chloroplast genome, we identified a total of 21 long repeat sequences in S. tetrandra cp genome, including 6 forward and 15 palindromic repeats (Fig. 5). S. japonica have been identified 11 forward, 17 palindromic and 2 reverse repeats in cp genome, while S. kwangsiensis contained 11 forward, 14 palindromic repeats in cp genome. Most of the long repeats are 30–39 bp in length. The palindromic repeat was the most abundant long repeat type and no complimentary repeat has been found in the cp genomes of the three Stephania plants. Furthermore, these detected repeats were mainly distributed in LSC region of Stephania species.
Fig. 5.
The analysis of the number and length of the long repeats identified from Stephania tetrandra, Stephania japonica and Stephania kwangsiensis chloroplast genomes. The type of long repeats contains forward (F), palindromic (P), reverse (R) and complement (C). The hamming distance of 3, the minimal repeats of 30 and the maximum repeats of 50 were applied during the calculating process
IR expansion and contraction
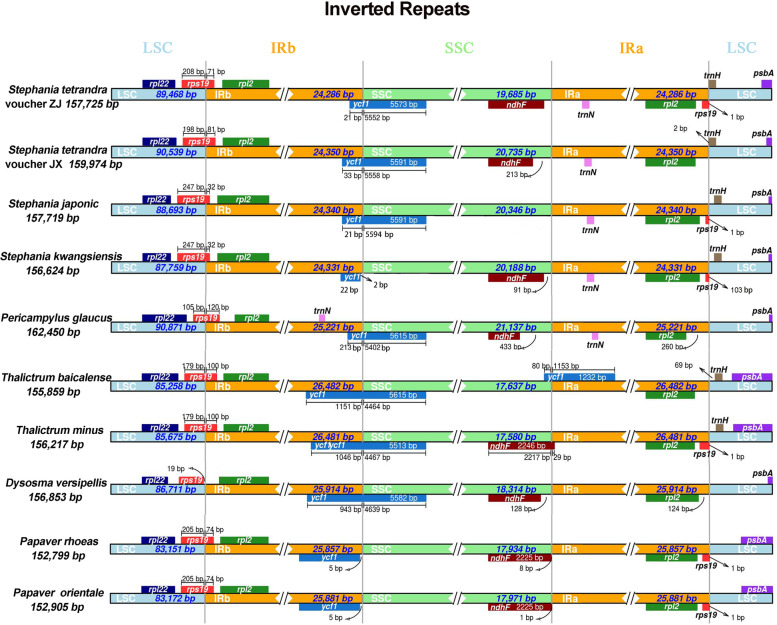
The expansion and contraction of IR region is the most common evolutionary event in genome evolution, which is considered to be one of the reasons for the length change between different genomes [26]. Therefore, the junctions of LSC/IR and IR/SSC are sometimes regarded as an index of chloroplast genome evolution. To evaluate the potential impact of the junction changes in the chloroplast genomes of Stephania species, we compared the LSC/IRb/SSC/IRa boundary regions of four Stephania plants and other closely related species from Menispermaceae, Berberidaceae, Ranunculaceae and Papaveraceae to determine the unique and common cp genome features (Fig. 6). The length of the IR regions was similar within the Stephania plants, ranging from 24,286 bp in S. tetrandra voucher ZJ to 24,350 bp in S. tetrandra voucher JX (Fig. 6). However, the Stephania species harbored the shortest IR regions compared with other plants from Ranunculales with the longest IR region of 26,482 bp in Thalictrum baicalense. Our results indicated that the locating position of ycf1 gene was highly variable at the boundary of IRB-SSC region in Stephania species. The length of ycf1 gene in S. tetrandra voucher ZJ was 18 bp shorter than that of S. tetrandra specimen from Jiangxi Province, with embedded length in IRB region of 21 and 33 bp, respectively (Fig. 6). The variation in gene length and expansion extent of ycf1 from two S. tetrandra vouchers indicated potential markers for further geographical origin discrimination. However, the contraction of IR region in S. kwangsiensis led to the formation of pseudogene ycf1, which was located completely in the IRB region with 2 bp away from the SSC-IRB border. It is worth noting that the overlap between ycf1 gene and the IRB region varied from 213 to 1151 bp in the cp genome from Menispermaceae, Berberidaceae and Ranunculaceae plants, which was significantly longer than that of Stephania species (Fig. 6). The junction between LSC and IRB region was located in the intergenic rps19 in most species of Ranunculales except Dysosma delavayi, in which the rps19 gene was completely encoded in LSC region and exhibited a 19 bp distance to the junction of the LSC/IRB region (Fig. 6). The length of IRB expansion to rps19 varied among the Ranunculales species ranging from 32 bp to 120 bp. However, a highly conserved length of rps19 gene (279 bp) was predicted in most of these Ranunculales plants except Pericampylus glaucus encoded a rps19 gene of 225 bp (Fig. 6). Beside ycf1 and rps19, other genes of ndhF, rpl2, rpl22 and trnH(GUG) were also found in the the LSC/IR and SSC/IR borders among the cp genomes from Ranunculales species.
Fig. 6.
Comparison of junctions between the large single-copy (LSC), small single-copy (SSC) and inverted repeat (IR) regions among 10 chloroplast genomes, including four Stephania plants, one closely related species in family of Menispermaceae, and five species from Berberidaceae, Ranunculaceae and Papaveraceae. The numbers of above the gene features indicate the distance between the ends of genes and border sites
Substitutions and InDels analysis
SNP (single nucleotide polymorphism) is a genetic marker formed by single nucleotide variation in the genome, which generally refers to single nucleotide variation with mutation frequency greater than 1% [27]. Using Pericampylus glaucus as reference, SNPs and InDels variations in cp genomes of the four Stephania plants have been analyzed. The analysis results revealed that transition substitution was more than transversion substitution in the cp genome of the four Stephania plants, which caused the values of transition to transversion ratio (Ts/Tv) much higher than 1 (Table 7). Moreover, the SNP sites mainly distributed in LSC region, followed by SSC region and IR region.
Table 7.
Comparison of substitutions and InDels in four Stephania species
| Comparative analyses of substitutions | |||||||||
| Species | Region | Transition Substitutions | Transversion Substitutions | Total Number of SNP sites | Ts/Tv | ||||
| A/G | C/T | A/T | A/C | C/G | G/T | ||||
| S. tetrandra voucher ZJ | Large single copy | 158 | 173 | 13 | 61 | 15 | 35 | 455 | 2.6 |
| S. tetrandra voucher JX | 177 | 207 | 20 | 64 | 19 | 40 | 527 | 2.7 | |
| S. japonica | 242 | 245 | 28 | 69 | 26 | 60 | 670 | 2.5 | |
| S. kwangsiensis | 228 | 239 | 39 | 87 | 25 | 61 | 679 | 2.2 | |
| S. tetrandra voucher ZJ | Inverted repeat | 8 | 9 | 0 | 3 | 2 | 1 | 23 | 2.1 |
| S. tetrandra voucher JX | 10 | 7 | 1 | 5 | 1 | 3 | 27 | 1.7 | |
| S. japonica | 10 | 11 | 1 | 4 | 2 | 5 | 33 | 1.9 | |
| S. kwangsiensis | 9 | 13 | 1 | 5 | 1 | 5 | 34 | 1.8 | |
| S. tetrandra voucher ZJ | Small single copy | 43 | 38 | 5 | 19 | 4 | 18 | 127 | 2.0 |
| S. tetrandra voucher JX | 64 | 61 | 15 | 21 | 9 | 10 | 180 | 2.2 | |
| S. japonica | 68 | 92 | 21 | 25 | 11 | 21 | 238 | 2.2 | |
| S. kwangsiensis | 73 | 66 | 13 | 27 | 7 | 24 | 210 | 1.9 | |
| Comparative analyses of InDels sites | |||||||||
| Species | Large single copy | Inverted repeat | Small single copy | ||||||
| No’s of Indels | InDel average length | No’s of Indels | InDel average length | No’s of Indels | InDel average length | ||||
| S. tetrandra voucher ZJ | 11 | 88.636 | 5 | 104.400 | 5 | 28.200 | |||
| S. tetrandra voucher JX | 13 | 42.581 | 2 | 87.300 | 5 | 28.200 | |||
| S. japonica | 19 | 37.421 | 6 | 89.500 | 5 | 28.200 | |||
| S. kwangsiensis | 21 | 34.571 | 4 | 134.250 | 5 | 23.400 | |||
A total of 21 InDels in the protein-coding sequences of the cp genome from S. tetrandra voucher ZJ were identified, including 11 InDels locating in LSC region, 5 InDels locating in SSC region, and 5 InDels locating in one IR region. However, only 20 InDel substitutions were revealed in the cp genome coding regions of S. tetrandra voucher JX, with the number of 13, 2 and 5 distributing in the LSC, SSC and IR region, respectively (Table 7). In addition, a total of 101 InDels have been identified in four Stephania plants, including 64 in the LSC region, 17 in the SSC region and 20 the in the IR region, respectively. The highest number of InDels substitutions was observed in S. japonica and S. kwangsiensis (30), while the lowest number was detected in S. tetrandra voucher JX (20). The LSC region of cp genome harbored the largest number of InDel sites in Stephania species. Furthermore, S. tetrandra voucher ZJ showed the largest InDel average length (88.636 bp) in LSC region, while S. kwangsiensis exhibited the largest InDel average length in IR region (134.250 bp). The Indel average length in SSC region ranged from 23.4 bp to 28.2 bp in the cp genome of four Stephania species, which represented the shortest one (Table 7).
Identification and analysis of divergence regions
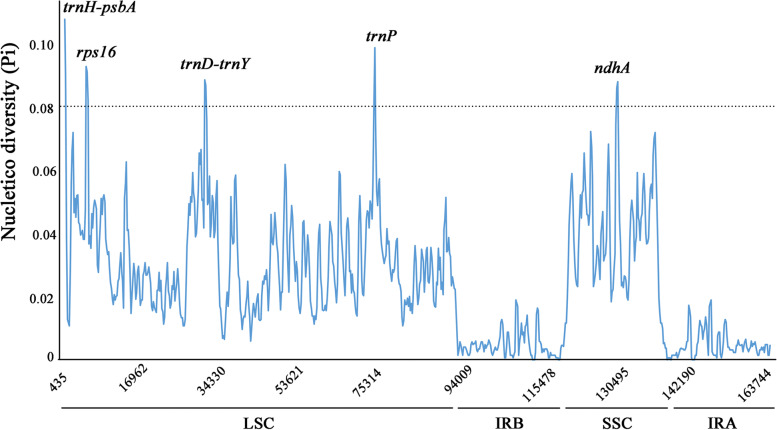
In order to evaluate the sequences divergence level, the complete cp genomes of the four Stephania plants have been multiple aligned and used DnaSP software to calculate nucleotide variability (Pi). A total of 5 mutational hotspot loci with high Pi value (> 0.08) have been screened out in the Stephania species, including 3 intergenic regions (trnH-psbA, trnD-trnY, trnP) and two protein coding genes (rps16 and ndhA) (Fig. 7). These high Pi value regions suggested to be divergence regions in the complete cp genome of the four Stephania plants. The highest Pi value was identified in the region of trnH-psbA (0.10639), followed by that from trnP region (0.0975). The most conserved region with the lowest pi value was found in the IR regions, further confirming that the IR regions were highly conserved in the chloroplast genomes of Stephania plants.
Fig. 7.
Nucleotide diversity (Pi) analysis for chloroplast genomes from the Staphania plants. Sliding window length was 800 bp and step size was selected as 200 bp
To further evaluate the diversities of 5 mutational hotspots among Stephania plants, we carried out a comparative analysis to determine the numbers of SNP sites and Gaps, as well as the values of non-synonymous to synonymous substitution (Ka/Ks) ratio. Multiple SNP sites were identified in the 5 mutational hotspots, with the highest SNP number of 136 in rps16 region (Table 8). In addition, a large number of Gaps were detected in the 5 mutational hotspots with the length ranging from 7 to 83 bp, which led to different mutational hotspot length in the four Stephania plants. The Ka/Ks ratio analysis demonstrated that the 5 mutational hotspots were suffering different selection pressures in the Stephania species. Most of the mutational hotspots exhibited a Ka/Ks ratio value much smaller than 1.00, indicating a purifying selection pattern. However, the Ka/Ks values of trnH-psbA in S. tetrandra voucher ZJ (1.0892), trnD-trnY in S. japonica (1.4076), S. kwangsiensis (1.1759), trnP in S. tetrandra voucher ZJ (1.1528), S. japonica (1.1237), S. kwangsiensis (1.5410) and ndhA in S. tetrandra voucher ZJ (1.3464), S. japonica (1.0783) were greater than 1.00, demonstrating that these genes were under diversifying selection pattern and more sensitive to the environment pressure. It is interesting to note that the Ka/Ks values of these identified mutational hotspots were all less than 1.00 in S. tetrandra voucher JX, suggesting potential population genetic variations between the two S. tetrandra species from Jiangxi and Zhejiang Province (Table 8). These results indicated the mutational hotspots could provide potential molecular markers to resolve the difficulties in species identification of Stephania species. According to the conserved regions surrounding the variable sties of the mutational hotspots, 5 pairs of PCR primers were designed to amplify the potential molecular markers (trnH-psbA, rps16, trnP, ndhA and trnD-trnY), which provided an effective tool for species identification and population genetic investigation in Stephania plants (Table 9). Furthermore, the significant SNP and Gap differences on five mutational regions observed between S. tetrandra voucher ZJ and voucher JX demonstrated high genetic differentiations among S. tetrandra populations from different regions, suggesting the potential of developing informative interspecifically variable sites as markers to verify its geographic origin.
Table 8.
Multiple analysis of the mutational hotspots in four Stephania plants
| Mutational hotspots | Species | Length | Number of SNP sites | Total length of Gaps | Ka/ks |
|---|---|---|---|---|---|
| trnH-psbA | S. tetrandra voucher ZJ | 543 bp | / | / | 1.0892 |
| S. tetrandra voucher JX | 597 bp | 62 | 30 | 0.9363 | |
| S. japonica | 594 bp | 83 | 29 | 0.7585 | |
| S. kwangsiensis | 556 bp | 121 | 31 | 0.8556 | |
| rps16 | S. tetrandra voucher ZJ | 1146 bp | / | / | 0.5510 |
| S. tetrandra voucher JX | 1206 bp | 97 | 26 | 0.5689 | |
| S. japonica | 1124 bp | 83 | 29 | 0.7639 | |
| S. kwangsiensis | 827 bp | 136 | 83 | 0.7030 | |
| trnD-trnY | S. tetrandra voucher ZJ | 404 bp | / | / | 0.7224 |
| S. tetrandra voucher JX | 475 bp | 65 | 14 | 0.8217 | |
| S. japonica | 481 bp | 70 | 8 | 1.4076 | |
| S. kwangsiensis | 481 bp | 57 | 9 | 1.1759 | |
| trnP | S. tetrandra voucher ZJ | 662 bp | / | / | 1.1528 |
| S. tetrandra voucher JX | 687 bp | 77 | 28 | 0.9143 | |
| S. japonica | 673 bp | 91 | 18 | 1.1237 | |
| S. kwangsiensis | 616 bp | 106 | 32 | 1.5410 | |
| ndhA | S. tetrandra voucher ZJ | 531 bp | / | / | 1.3464 |
| S. tetrandra voucher JX | 492 bp | 97 | 8 | 0.7692 | |
| S. japonica | 507 bp | 84 | 7 | 1.0783 | |
| S. kwangsiensis | 492 bp | 91 | 21 | 0.9638 |
Gaps consists of the insertion and deletion of single bases and fragments
Table 9.
PCR primers designed according to the mutational hotspots within four Stephania species
| Mutational hotspots | PCR primers | Expected length |
|---|---|---|
| trnH-psbA | F:CGCCGTAGTAAATAGGAGA | 860 bp |
| R:TCATCAACCGYGCTAACCT | ||
| rps16 | F:RATACAATAAGCAAGCTC | 957 bp |
| R:TCCCRAAACAAGAAAACG | ||
| trnD-trnY | F:GTGCTCTGACCGATTGAACT | 525 bp |
| R:GGCAATATGTCTACGCTGGT | ||
| trnP | F:TAGGTAGGGATGACAGGA | 918 bp |
| R:GACCCGAACCATAGAGTA | ||
| ndhA | F:CRAATCCMAAATTAGACCA | 668 bp |
| R:GACGCTTAGGAACACCAA |
Phylogenetic analysis
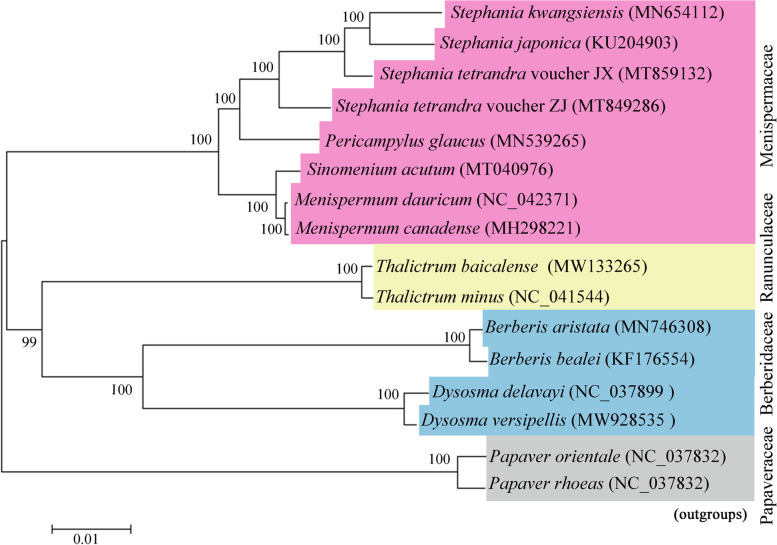
The conserved chloroplast genomes have been indicated as effective approach for the phylogenetic relationship studies of plants from different taxa [28]. The phylogenetic tree was constructed using maximum likelihood (ML) method for Ranunculales plants based on coding sequences of 76 protein-coding genes, including 8 species from family Menispermaceae, 2 species from family Ranunculaceae and 4 species from family Berberidaceae. Papaver orientale and P. rhoeas were chosen as the outgroups for phylogenetic analysis. As shown in Fig. 8, all nodes received high support values in the ML tree, and it was congruent with those obtained from the previous study. The phylogeny analysis showed S. japonica and S. kwangsiensis consisted into a stable monophyletic group with high bootstrap values, which exhibited a stable sister relationship with S. tetrandra voucher JX and voucher ZJ, indicating a close genetic relationship among the four Stephania plants. Interestingly, S. tetrandra voucher ZJ and voucher JX failed to clustered into the same branch, revealing obvious discrepancies on cp genomes of S. tetrandra from different regions (Fig. 8). In addition, the Stephania species formed a clade with Pericampylus glaucus with strong statistical support, indicated a relatively close relationship between the genus Stephania and Pericampylus in Trib. Menispermeae. Moreover, the species from the genus of Menispermum and Sinomenium combined together to form a robust monophyletic clade, which showed sister relationship to the group of Stephania and Pericampylus plants. The entire subclade comprising of species from family of Berberidaceae and Ranunculaceae grouped with the clade of family Menispermaceae, which were clustered together to form the monophyletic group of core Ranunculales (Fig. 8). The complete chloroplast sequence of S. tetrandra voucher ZJ could provide useful information for clarifying the phylogenetic relationship of family Menispermaceae as well as the classification of subgenera and species groups in Stephania.
Fig. 8.
Phylogenetic relationships based on the conserved chloroplast protein coding genes from four Stephania plants and other representative Ranunculales species using maximum likelihood (ML) method. The number on each node represents the bootstrap value from 500 replicates. Papaver orientale and Papaver rhoeas were set as the outgroups
Discussion
In this study, we sequenced and assembled the complete cp genome of S. tetrandra from Zhejiang province and analyzed its codon usage bias and RNA editing sites. We also conducted a comparative analysis on the RNA editing sites, codon usage bias, repeat sequences, substitutions and InDels events within the complete cp genomes of the Stephania plants and constructed the phylogenetic tree. Our results provide basic data for further studies on the identification and phylogenetic relationship of Menispermaceae plants. The genome database of S. tetrandra voucher ZJ would be also beneficial for the subsequent specific molecular species identification and population genetic analysis from different regions in Stephania plants.
The Stephania is a genus of Menispermaceae, with three subgenera of Subgen. Botryodiscia, Subgen. Stephania and Subgen. Tuberiphania [29]. S. tetrandra is the only plant that belonging to the Subgen. Botryodiscia and has attracted considerable interest owing to its significant medicinal values. The species of S. japonica and S. kwangsiensis were classified to the Subgen. Stephania and Subgen. Tuberiphania, respectively. The ML trees constructed based on ITS and trnL-F in previous reports showed conflicting results in the determination of the relationship among the three subgenera of the genus Stephania. The ML trees based on ITS showed that Subgen. Tuberiphania species formed a branch and S. tetrandra clustered with the Subgen. Tuberiphania species to form a monophyletic group [12]. However, the ML trees of trnL-F indicated that the clade of Subgen. Stephania species exhibited a sister relationship with that of Subgen. Tuberiphania species, and S. tetrandra clustered with Perichasma laetificata and the combined group of Subgen. Stephania and Subgen. Tuberiphania species [12]. Besides molecular investigation strategy, the chemical composition analysis could also provide valuable information for the relationship research within Stephania species. For instance, S. cepharantha belonging to the Subgen. Tuberiphania showed no chemical compounds of L-tetrahydropalmatine and tetrandrine, which exhibited similar chemical compositions with plants from Subgen. Stephania [30]. The chemical analysis indicated S. cepharantha may be a transitional type between Subgen. Tuberiphania and Subgen. Stephania [30]. In addition, hasubanan alkaloids were the unique alkaloids in plants of genus Stephania, which were considered to be of great significance in the study of classification and evolution of Stephania [31]. Most of plants classified to Subgen. Stephania and Subgen. Tuberiphania, which are rich sources of hasubanan alkaloids. However, there are no such chemical in S. tetrandra, indicating a closer relationship between Subgen. Stephania and Subgen. Tuberiphania. Our result showed a close phylogenetic relationship between Subgen. Stephania and Subgen. Tuberiphania, which was consistent with the previous ML analysis based on trnL-F marker. Furthermore, both of the S. tetrandra samples from Jiangxi and Zhejiang Province respectively clustered with combined clade of Subgen. Stephania and Subgen. Tuberiphania, further confirming the reasonability of dividing the S. tetrandra as a separate subgenus (Fig. 8). The chloroplast genome of S. tetrandra voucher ZJ provides a valuable reference for the study of phylogenetic relationship within Stephania genus, which required more accurate and abundant chloroplast genome information from Stephania plants.
Codon usage bias and RNA editing were important events for the protein-coding genes in plant cp genomes, which was closely related with the molecular evolutionary phenomena such as mutation, selection, and random genetic drift. Qin et al. (2013) suggested that the synonymous codon usage bias was correlated with intron number in plants, which may be due to DNA methylation [32]. The GC content in codon positions was also indicated as an important role during the evolution of genomic structure and one of the major factors in the codon usage biases shaping [33]. Our results demonstrated that among the 31 codons with RSCU value higher than 1, 29 codons ended with A or U. However, most of the codons (28) with RSCU value less than 1 ended with G or C. This result indicated that S. tetrandra preferred to use synonymous codons with a third base of A or U, which was similar with that of Fagopyrum dibotrys and Salix wilsonii [34, 35]. The RNA editing process is an essential maturation mechanism to avoid incorrect RNA mutations and is widespread in the chloroplast genome of plants [36]. The RNA editing sites analysis of the cp genome is vital to understand the correct translation process and mutations of genes [14]. In our study, 92 RNA editing sites were predicted in 25 protein-coding genes of the cp genome of S. tetrandra, which may affect the structure and function of proteins. Previous studies have shown that the deletion editing sites of psbF-26 in Arabidopsis would affect the assembly of PSII complex seriously [37]. In addition, the RNA editing of rpoA and clpP chloroplast transcripts by CLB19 played crucial roles for the s essential for correct chloroplast development and phenotype development [38]. Our results found the RNA editing sites of psbE and rps16 genes were lost in certain Stephania species (Fig. 3). However, the functional changes caused by the deletion of RNA editing sites in Stephania plants remained unknown and required further exploration.
Mutational hotspots from chloroplast genome have been demonstrated as potential molecular markers that might be developed for phylogenetic relationships analysis and identification among closely related species. Jiao et al. (2019) have found seven variable regions from the chloroplast genome of Pterocarpus species, and suggested ycf1b could be applied as a high-resolution DNA barcode for species identification of Pterocarpus wood [21]. In addition, Bi et al. (2018) selected ten regions with relatively high variability from the cp genome of Fritillaria species and demonstrated that the regions of ycf1a and ycf1b showed highest variability, providing valuable resources for the study of species identification, phyletic evolution, breeding direction and population genetics [39]. In this study, we also calculated nucleotide variability (Pi) using sliding window analysis and identified 5 high Pi value regions and designed five pairs of primers. These markers were believed to be potential divergence regions in the complete cp genome of the four Stephania plants, providing the basis for subsequent molecular identification. The trnH-psbA region had the highest Pi value and the trnP also exhibited high variability in the cp genomes which might be developed as a molecular marker for subsequent research on resolving the difficulties of identification and phylogenetic relationships analysis of Stephania plants. Furthermore, the divergence regions mainly distributed in LSC and SSC region, which was consist with the previous reports of Chaenomeles and Lancea species [40, 41]. Multiple SNP sites and Gaps were identified in the divergence regions, which provided basic information for the cost-effective, authentic and robust molecular markers design in Stephania plants. The Ka/Ks ratio analysis indicated four mutational hotspots (trnH-psbA, trnP, trnD-trnY and ndhA) in S. tetrandra voucher ZJ were suffering diversifying, which would be more sensitive to the environment changes (Table 8). It is interesting to note that S. tetrandra from different regions exhibited significant differences in the complete genome sequences. For instance, the cp genome size of S. tetrandra voucher JX is 2249 bp longer than that of S. tetrandra voucher ZJ in our study. Furthermore, the cp genome of S. tetrandra voucher JX exhibited insertion of a base A in ycf15 gene, which caused the base dislocation to move backward and led to the untranslatability of this gene [22]. It is well known that S. tetrandra is distributed widely in tropical and subtropical regions of Asia and Africa, generating potentials of great numbers of genetic diversities and unique population structures. Moreover, the geographical origin of herbal medicines is an important factor influencing the quality of the medicinal materials [42]. Our results have revealed the occurrence of genetic variation events of S. tetrandra from different regions, which might bring significant differences to the medicinal treatment efficacy and cause health benefits to consumers. Therefore, it is extremely important to develop effective strategy to accurately discriminate the geographical origin of S. tetrandra. These results indicated the mutational hotspots of cp genome would be valuable tools for species geographical diversity investigation of S. tetrandra, providing a potential approach to determine different original species and ensure its pharmaceutical activity in the market. However, the available complete chloroplast genomes of S. tetrandra from different regions are still dramatically insufficient at present, the development of new DNA barcodes based on the chloroplast genome needs further study.
Distribution of repeats in the genome could infer highly polymorphic regions in the genome [17]. SSRs could be developed as molecular markers for species identification, population genetic and phylogenetic relationship analysis [43], In addition, SSR have also been extensively used in genetic analysis, functional gene mapping and quantitative trait locus (QTL) mapping [44]. Chu et al. (2020) have selected 36 SSR primers pairs for identification of cauliflower and broccoli varieties and confirmed that SSR molecular markers could be used to identify cauliflower and broccoli varieties [45]. Huang et al. (2021) developed EST-SSR markers of Tetraena mongolica, suggesting that the SSRs might be useful for further study of genetic structure and adaptive evolutionary mechanism and utilization of the germplasm [46]. Moreover, the mononucleotide and dinucleotide repeats were the most and second most abundant SSRs in the cp genomes of two Caldesia species [47]. Here, we also found that the mononucleotide repeat A/T was the most abundant SSR and the second most was the dinucleotide SSR AT/AT, which was consist with the previous reports. Furthermore, SSRs have been demonstrated to be generally composed of A or T repeats and infrequently contain C and G repeats in the cp genomes of seven Populus species [48]. This result was consisted with our research and the abundance of AT base content in SSRs might be one of the reasons for the high AT content in the chloroplast genome. The chloroplast genomes of the Stephania species are rich in SSRs and were good resources to develop SSRs markers for its further studies.
Conclusion
In summary, in this study we have determined the complete cp genome sequence of S. tetrandra voucher ZJ from Zhejiang Province and analyzed its features. Comparative analysis between S. tetrandra voucher ZJ and other Stephania species revealed that the available complete cp genomes from this genus were highly conserved in terms of overall structure, genome size, GC contents, and gene numbers, orders and functions. Comparative analysis involving cp genomes of Stephania plants revealed 5 highly variable regions (trnH-psbA, rps16, trnD-trnY, trnP and ndhA), with multiple SNP sites and Gaps, as well as different non-synonymous to synonymous substitution (Ka/Ks) ratio values. Moreover, five pairs of specific primers targeting the mutational hotspots were accordingly designed, which could be utilized as potential molecular markers for resolving the difficulties in studies regarding species identification, population genetic and phylogenetic analysis in Stephania species. The number and types of SSRs, long repeat sequences and the value of Ts/Tv were also detected for developing potential and effective molecular markers. The Maximum likelihood (ML) phylogenetic analysis showed that S. japonica and S. kwangsiensis consisted into a monophyletic group and exhibited a stable sister relationship with both of the S. tetrandra plants from different regions, indicating a close genetic relationship among the four Stephania species. The cp genome of S. tetrandra voucher ZJ provides valuable information for the development of accurate molecular approaches for its geographical origin determination. Our comprehensive analysis of these complete chloroplast genomes will contribute to medicinal resource conservation, genetic diversity, genome evolution and adaptation history, and phylogenetic relationship investigation of Stephania plants.
Methods
Plant material and DNA extraction
The S. tetrandra plant was collected from the mountain in Fuyang District of Zhejiang Province (30°05′38.4″N 119°53′24″E) are identified by Dr. Guanghui Liao of Zhejiang Chinese Medical University. The specimen was deposited in Medicinal Herbarium Center of Zhejiang Chinese Medical University (Herbarium Code: MHCZCMU; Collector and Identifier: Guanghui Liao: voucher number: FJZJ-190826). Modified cetyltrimethylammonium brofmide (CTAB) method was used to extracted total genomic DNA from dried leaves ground with liquid nitrogen [49]. Final DNA quality was assessed by a Nanodrop spectrophotometer (Thermo Fisher Scientific, USA), and DNA integrity was examined by electrophoresis on a 1.0% agarose gel.
Genome sequencing, assembly and annotation
Total genomic DNA was sequenced using the Illumina Hiseq Platform. The quality of paired-end Illumina reads was assessed with FastQC, and the low-quality reads were removed using Fastp. Then the filtered reads were assembled de novo using metaSPAdes with the complete cp genome of S. japonica (NC_029432) as reference and the protein-coding genes, mRNA genes, tRNA genes were annotated by GeSeq annotation tool [50]. The CPGAVAS2 software also used to annotate protein-coding genes [51]. Then the annotation of chloroplast genome was further manually corrected by BLAST. OrganellarGenomeDRAW (OGDRAW) tool was used to draw the circular chloroplast genome map of S. tetrandra [52]. The fully annotated cp genome was finally deposited at the GenBank database (Accession Number: MT849286).
Comparative analysis of cp genomes
MEGA 7.0 [53] was used to analyze the genome feature and Codon W software was used to investigate the distribution of codon usage using the RSCU value [54]. To predict the number of RNA editing site, the PREP-Cp program was employed with a cutoff value 0.8 [55]. The IR scope was further applied to analyze the LSC/IRb/SSC/IRa boundary locations in ten Ranunculales species cp genomes [56]. As for the repeats analysis, simple sequence repeats markers in the three Stephania species were detected by MISA, setting parameters as 10 for mononucleotide SSRs, 5 for dinucleotide SSRs, 4 for trinucleotide SSRs, 3 each for tetranucleotide, pentanucleotide and hexanucleotide SSRs [57]. Long repeats of four different type (forward (F), palindromic (P), reverse (R), and complementary (C)) were calculated by REPuter, with hamming distance 3, minimal repeats 30 and maximum computed repeats 50 [58].
Analysis of substitutions and InDels, as well as adaptive evaluation
Using pericampylus glaucus (MN539265) as reference sequence, the protein-coding sequences of LSC, SSC and IR regions of the four Stephania plants (including S. tetrandra voucher JX) were pairwise aligned with corresponding parts of the reference sequence using MAFFT v7.037b [59]. MEGA7.0 was used to determined substitutions numbers and types of the sequences [53], and numbers of InDels events and their average lengths were detected by Dnasp v6 [60]. In order to analyzed the Ka and Ks substitution rates and Ka/Ks ratio, Pericampylus glaucus was compared with S. tetrandra in 25 protein coding genes. Besides, the Ka/Ks value of 5 mutation hotspots in four Stephania plants were also evaluate separately. The alignment was carried out by MAFFT v7.037b [58], and the calculation of the value of Ka/Ks was implemented by DnaSP v6 [60]. The number of mutation sites of the other three Stephania species with S. tetrandra voucher ZJ as the reference in five mutation regions was analyzed respectively by MEGA 7.0 [53].
Identification of divergence regions and PCR primers designing
The complete cp genomes of the four Stephania plants were multiple aligned by MAFFT v7.037b to evaluate sequence divergence [59], and nucleotide variability (Pi) was calculated by sliding window analysis of DnaSP v6 setting parameters as window length for 800 sites and step size for 200 sites [14]. Specific primers were designed in conserved nucleotide sequences at both ends of mutation hotspots by primer premier 5, and met the conditions of GC content ranging from 40 to 60%, as well as primer length ranging from 15 bp to 30 bp.
Phylogenetic analysis
Stephania kwangsiensis (MN654112), Stephania japonica (KU204903), Sinomenium acutum (MT040976), Menispermum canadense (MH298221), Menispermum dauricum (NC_042371), Pericampylus glaucus (MN539265), Stephania tetrandra (MT859132), Thalictrum baicalense (MW133265), Thalictrum minus (NC_041544), Berberis aristata (MN746308), Berberis bealei (KF176554), Dysosma delavayi (NC_037899), Dysosma versipellis (MW928535), Papaver orientale (NC_037832) and Papaver rhoeas (NC_037832) were downloaded from NCBI. Seventy-six protein-coding gene sequences that existed in S. tetrandra voucher ZJ and other 15 representative species were multiple aligned by MAFFT v7.037b [59] and then analyzed using MEGA 7.0 [53] by maximum-likelihood (ML) method to constructed the phylogenetic tree. ML trees were constructed based on the Kimura 2-parameter model with 500 bootstrap replications. Papaver orientale and Papaver rhoeas were set as the outgroups.
Supplementary Information
Additional file 1: Table S1. SSRs identified in the cp genome of Stephania tetrandra. Table S2. SSRs identified in the cp genome of Stephania japonica. Table S3. SSRs identified in the cp genome of Stephania kwangsiensis.
Acknowledgements
We are grateful for the help provided by Genesky Biotechnology Co., Ltd. (Shanghai, China) in the analysis of raw illumina data in this study.
Conflict of interest
No potential conflict of interest was reported by the authors.
Abbreviations
- BLAST
Basic local alignment search tool
- cp
Chloroplast
- DNA
Deoxyribonucleic acid
- IR
Inverted repeat
- LSC
Large single copy region
- SSC
Small single copy region
- SSR
Simple sequence repeats
- SNP
Single nucleotide polymorphism
- InDel
Insertion and deletion
Authors’ contributions
SD, YG and RC designed the experiments and organized the manuscript. GL, SY and ZY contributed to the resource sampling and species identification. SD and QW performed the experiments. SD, ZY and YG analyzed the data and wrote the main manuscript, prepared figures and tables, RC and YG supervised the project, all authors have reviewed and approved the manuscript.
Funding
This work was financially supported by the Opening Project of Zhejiang Provincial Preponderant and Characteristic Subject of Key University (Traditional Chinese Pharmacology), Zhejiang Chinese Medical University (No. ZYAOX2018033).
Availability of data and materials
The chloroplast genome of Stephania tetrandra assembled in this study have been deposited in the National Center for Biotechnology and Information (NCBI) under the accession number of MT849286. The raw sequence data reported in this study have been deposited in NCBI with the BioProject, SRA, and BioSample numbers of PRJNA735711, SRR14748481 and SAMN19594056, respectively (https://submit.ncbi.nlm.nih.gov/subs/sra/SUB9808741/overview). The other cp genomes used in this study were downloaded from the NCBI.
Declarations
Ethics approval and consent to participate
Since the project of Survey of Chinese Material Medical Resources of Zhejiang Province, Dr. Guanghui Liao has obtained the permission by Zhejiang Chinese Medical University to collect plant species from mountain of Fuyang District. Because of the important medical value of Stephania tetrandra, Dr. Liao provided us the plant specimen for further molecular study. The plant material collection and experimental research were conducted according to the Plant Protection and Regulation of Zhejiang Chinese Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shujie Dong and Zhiqi Ying contributed equally to this work.
Contributor Information
Yuqing Ge, Email: geyuqing@hotmail.com.
Rubin Cheng, Email: biothcheng@hotmail.com.
References
- 1.Moore S. Flora Reipublicae Popularis Sinicae. Beijing: Science Press; 1996. Stephania tetrandra; p. 52. [Google Scholar]
- 2.Jiang YP, Liu M, Liu HT, Liu S. A critical review: traditional uses, phytochemistry, pharmacology and toxicology of Stephania tetrandra S. Moore (Fen Fang Ji) Phytochem Rev. 2020;24:1–41. doi: 10.1007/s11101-020-09673-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xi Y, Zhang HJ, Ye ZG, Zhang GP. Research development on modern pharmacological effect of tetrandrine. China J Chin Mater Med. 2020;45:20–28. doi: 10.19540/j.cnki.cjcmm.20190830.401. [DOI] [PubMed] [Google Scholar]
- 4.Zhong ZY, Qian Z, Zhang X, Chen FC, Ni S, Kang ZR, et al. Tetrandrine prevents bone loss in ovariectomized mice by inhibiting RANKL-induced osteoclastogenesis. Front Pharmacol. 2020;10:1530. doi: 10.3389/fphar.2019.01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thawabteh A, Juma S, Bader M, Karaman D, Scrano L, Bufo SA, et al. The biological activity of natural alkaloids against herbivores, cancerous cells and pathogens. Toxins. 2019;11(11):656. doi: 10.3390/toxins11110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu SH, Chueh FS, Chou YC, Ma YS, Peng SF, Lin CC, et al. Tetrandrine inhibits cell migration and invasion in human nasopharyngeal carcinoma NPC-TW 039 cells through inhibiting MAPK and RhoA signaling pathways. J Food Biochem. 2020;27:e13387. doi: 10.1111/jfbc.13387. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Xue N, Li HL, Chen Z, Yu W. Study on mechanism of Fangji Huangqi Decoction on hypouricemic effect and renal protection in hyperuricemia mice. China J Chin Mater Med. 2020;45(21):5248–5255. doi: 10.19540/j.cnki.cjcmm.20200630.401. [DOI] [PubMed] [Google Scholar]
- 8.Chinese Pharmacopoeia Commission . Pharmacopoeia of the People’s Republic of China. Beijing: China Medical Science Press; 2020. Fangji; pp. 155–157. [Google Scholar]
- 9.Chen JF, Zhao Q, Si DD, Nie AZ, Wang YY, Deng ZF, et al. Comprehensive profiling of Stephania tetrandra (Fangji) by stepwise DFI and NL-dependent structure annotation algorithm-based UHPLC-Q-TOF-MS and direct authentication by LMJ-HRMS. J Pharm Biomed Anal. 2020;185:113225. doi: 10.1016/j.jpba.2020.113225. [DOI] [PubMed] [Google Scholar]
- 10.Luo XR. A systematic notes on the genus stephania of China. Bull Bot Res. 1982;(01):33–59.
- 11.Wang HC. Floral organogenesis and molecular phylogeny of Menispermeae (Menispermaceae): Ph.D. Thesis. Wuhan: Chinese Academy of Sciences; 2005.
- 12.Xie DT. A taxonomic study of the genus Stephania Lour. from China. Shanghai: Fudan University; 2014. [Google Scholar]
- 13.Gao CS, Wang QR, Ying ZQ, Ge YQ, Cheng RB. Molecular structure and phylogenetic analysis of complete chloroplast genomes of medicinal species Paeonia lactiflora from Zhejiang Province. Mitochondrial DNA Part B. 2020;5(1):1077–1078. doi: 10.1080/23802359.2020.1721372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somaratne Y, Guan DL, Wang WQ, Zhao L, Xu SQ. The complete chloroplast genomes of two Lespedeza species: insights into codon usage bias, RNA editing sites, and phylogenetic relationships in Desmodieae (Fabaceae: Papilionoideae) Plants. 2019;9(1):51. doi: 10.3390/plants9010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JH, Jung JY, Choi HI, Kim NH, Park JY, Lee Y, et al. Diversity and evolution of major Panax species revealed by scanning the entire chloroplast intergenic spacer sequences. Genet Resour Crop Evol. 2013;60(2):413–425. doi: 10.1007/s10722-012-9844-4. [DOI] [Google Scholar]
- 16.Sun JL, Han Y, Cui XM, Liu Y. Development and application of chloroplast molecular markers in Panax notoginseng. China J Chin Mater Med. 2020;45(6):1342–1349. doi: 10.19540/j.cnki.cjcmm.20200104.105. [DOI] [PubMed] [Google Scholar]
- 17.Lee SR, Kim K, Lee BY, Lim CE. Complete chloroplast genomes of all six Hosta species occurring in Korea: molecular structures, comparative, and phylogenetic analyses. BMC Genomics. 2019;20(1):833. doi: 10.1186/s12864-019-6215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iram S, Hayat MQ, Tahir M, Gul A, Abdullah AI. Chloroplast genome sequence of Artemisia scoparia: comparative analyses and screening of mutational hotspots. Plants. 2019;8(11):476. doi: 10.3390/plants8110476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang AS, Wu HW, Zhu XC, Lin JM. Species identification of Conyza bonariensis assisted by chloroplast genome sequencing. Front Genet. 2018;9:374. doi: 10.3389/fgene.2018.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen XL, Zhou JG, Cui YX, Wang Y, Duan BZ, Yao H. Identification of Ligularia herbs using the complete chloroplast genome as a super-barcode. Front Pharmacol. 2018;9:695. doi: 10.3389/fphar.2018.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao LC, Lu Y, He T, Li JN, Yin YF. A strategy for developing high-resolution DNA barcodes for species discrimination of wood specimens using the complete chloroplast genome of three Pterocarpus species. Planta. 2019;250(1):95–104. doi: 10.1007/s00425-019-03150-1. [DOI] [PubMed] [Google Scholar]
- 22.Cao L, Mu ZJ, Sheng SS, Chen YY, Zhong GY, Du XL. The complete chloroplast genome of Stephania tetrandra (Menispermaceae) Mitochondrial DNA Part B. 2020;5(4):3819–3820. doi: 10.1080/23802359.2020.1840935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hina F, Jin ZY, Yang ZP, Li P, Fu CX. The complete chloroplast genome of Menispermum dauricum (Menispermaceae, Ranunculales) Mitochondrial DNA Part B. 2018;3(2):913–914. doi: 10.1080/23802359.2018.1501306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hina F, Li JK, Chen C, Fu CX. Characterization of the complete chloroplast genome of an important medicinal plant, Sinomenium acutum (Menispermaceae, Ranunculales) Mitochondrial DNA Part B. 2020;5(2):1305–1306. doi: 10.1080/23802359.2020.1732239. [DOI] [Google Scholar]
- 25.Nie LP, Cui YX, Wu LW, Zhou JG, Xu ZC, Li YH, et al. Gene losses and variations in chloroplast genome of parasitic plant macrosolen and phylogenetic relationships within santalales. Int J Mol Sci. 2019;20(22):5812. doi: 10.3390/ijms20225812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen XF, Wu ML, Liao BS, Liu ZX, Bai R, Xiao SM, et al. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 2017;22(8):1330. doi: 10.3390/molecules22081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J, Li WH, Zhu D, Yu QH, Zhang Z, Sun MH, et al. Association of single nucleotide polymorphisms in MTHFR and ABCG2 with the different efficacy of first-line chemotherapy in metastatic colorectal cancer. Med Oncol. 2014;31(1):802. doi: 10.1007/s12032-013-0802-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han TY, Li MM, Li JW, Lv H, Ren BR, Chen J, et al. Comparison of chloroplast genomes of Gynura species: sequence variation, genome rearrangement and divergence studies. BMC Genomics. 2019;20(1):791. doi: 10.1186/s12864-019-6196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lour. Fl. Cochinch . Flora Reipublicae Popularis Sinicae. Beijing: Science Press; 1996. Stephania; p. 40. [Google Scholar]
- 30.Zhu ZY, Feng YX, Ho LY, Wang YC. Utilization of medicinal plants of genus Stephania of Menispermaceae in China. Acta Pharm Sin. 1983;18(6):460–467. doi: 10.16438/j.0513-4870.1983.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Huang JM, Guo QX, Pan SL. A preliminary study on the phytochemical taxonomy of the genus Stephania. West China J Pharm Sci. 1999;02:39–41. [Google Scholar]
- 32.Qin Z, Cai ZQ, Xia GM, Wang MC. Synonymous codon usage bias is correlative to intron number and shows disequilibrium among exons in plants. BioMed Central. 2013;14:56. doi: 10.1186/1471-2164-14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang ZC, Dai W, Wang Y, Lu CP, Fan HJ. Analysis of synonymous codon usage patterns in torque Teno sus virus 1 (TTSuV1) Arch Virol. 2013;158(1):145–154. doi: 10.1007/s00705-012-1480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang XM, Zhou T, Bai GQ, Zhao YM. Complete chloroplast genome sequence of Fagopyrum dibotrys: genome features, comparative analysis and phylogenetic relationships. Sci Rep. 2018;8(1):12379. doi: 10.1038/s41598-018-30398-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen YN, Hu N, Wu HT. Analyzing and characterizing the chloroplast genome of Salix wilsonii. Biomed Res Int. 2019;5190425. 10.1155/2019/5190425. [DOI] [PMC free article] [PubMed]
- 36.Wang MX, Liu H, Ge LQ, Xing GW, Wang M, Song WN, et al. Identification and analysis of RNA editing sites in the chloroplast transcripts of Aegilops tauschii L. Genes. 2016;8(1):13. doi: 10.3390/genes8010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai WH, Ji DL, Peng LW, Guo JK, Ma JF, Zou MJ, et al. LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 2009;150(3):1260–1271. doi: 10.1104/pp.109.136812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, Gutiérrez-Nava MDLL, Cantero A, et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 2008;56(4):590–602. doi: 10.1111/j.1365-313X.2008.03634.x. [DOI] [PubMed] [Google Scholar]
- 39.Bi Y, Zhang MF, Xue J, Dong R, Du YP, Zhang XH. Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Sci Rep. 2018;8(1):1184. doi: 10.1038/s41598-018-19591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun JH, Wang YH, Liu YL, Xu C, Yuan QJ, Guo LP, et al. Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Sci Rep. 2020;10(1):11466. doi: 10.1038/s41598-020-67943-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chi XF, Wang JL, Gao QB, Zhang FQ, Chen SL. The complete chloroplast genomes of two Lancea species with comparative analysis. Molecules. 2018;23(3):602. doi: 10.3390/molecules23030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Q, Wu XB, Zhang DQ. Comparison of the abilities of universal, super, and specific DNA barcodes to discriminate among the original species of Fritillariae cirrhosae bulbus and its adulterants. PLoS One. 2020;15(2):e0229181. 10.1371/journal.pone.0229181. [DOI] [PMC free article] [PubMed]
- 43.Chen XL, Cui YX, Nie LP, Hu HY, Xu ZC, Sun W, et al. Identification and phylogenetic analysis of the complete chloroplast genomes of three Ephedra herbs containing ephedrine. Biomed Res Int. 2019;5921725. 10.1155/2019/5921725. [DOI] [PMC free article] [PubMed]
- 44.Yang JJ, Zhang J, Han RX, Zhang F, Mao AJ, Luo J, et al. Target SSR-Seq: a novel SSR genotyping technology associate with perfect SSRs in genetic analysis of cucumber varieties. Front Plant Sci. 2019;10:531. doi: 10.3389/fpls.2019.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu YX, Deng S, Li SG, Liu D, Chen HR, Ren L, et al. Validation of microsatellite markers and its application for varieties identification in cauliflower and broccoli. Mol Plant Breed. 2020. https://kns.cnki.net/kcms/detail/46.1068.s.20201113.1321.004.html.
- 46.Huang L, Jia YY, Zhang YN, Li JB, Dang ZH. Identification and development of the EST-SSR markers in the desert plant Tetraena Mongolica. J Plant Genet Resour. 2021;22(02):540–549. [Google Scholar]
- 47.Mwanzia VM, Nzei JM, Yan D-Y, Kamau PW, Chen JM, Li Z-Z. The complete chloroplast genomes of two species in threatened monocot genus Caldesia in China. Genetica. 2019;147(5–6):381–390. doi: 10.1007/s10709-019-00079-x. [DOI] [PubMed] [Google Scholar]
- 48.Zong D, Gan PH, Zhou AP, Li JY, Xie ZL, Duan AA, et al. Comparative analysis of the complete chloroplast genomes of seven Populus species: insights into alternative female parents of Populus tomentosa. PLoS One. 2019;14(6):e0218455. doi: 10.1371/journal.pone.0218455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doyle JJDJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 50.Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, et al. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45(W1):W6–11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, et al. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019;47(W1):W65–W73. doi: 10.1093/nar/gkz345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47(W1):W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharp PM, Li WH. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mower JP. The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37 Web Server:W253–W259. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinform. 2018;34(17):3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 57.Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurtz S, Schleiermacher C. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics. 1999;15(5):426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- 59.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34(12):3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. SSRs identified in the cp genome of Stephania tetrandra. Table S2. SSRs identified in the cp genome of Stephania japonica. Table S3. SSRs identified in the cp genome of Stephania kwangsiensis.
Data Availability Statement
The chloroplast genome of Stephania tetrandra assembled in this study have been deposited in the National Center for Biotechnology and Information (NCBI) under the accession number of MT849286. The raw sequence data reported in this study have been deposited in NCBI with the BioProject, SRA, and BioSample numbers of PRJNA735711, SRR14748481 and SAMN19594056, respectively (https://submit.ncbi.nlm.nih.gov/subs/sra/SUB9808741/overview). The other cp genomes used in this study were downloaded from the NCBI.