Abstract
Placental villous trophoblast mitochondrial respiratory function is critical for a successful pregnancy and environmental influences such as maternal obesity have been associated with respiratory impairment at term. More recently, a gestational high fat diet independent of maternal body composition, has been highlighted as a potential independent regulator of placental mitochondrial metabolism. The current study aimed to characterize the direct impact of a prolonged and isolated exposure to the dietary fatty acids Palmitate (PA) and Oleate (OA) upon placental cell mitochondrial respiratory function. BeWo cytotrophoblast (CT) and syncytiotrophoblast (SCT) cells were treated for 72 hours with 100 μM PA, OA or PA+OA (P/O). Live-cell metabolic function was analyzed via the Seahorse XF Mito and Glycolysis Stress tests. Immunoblots and spectrophotometric activity assays were utilized to examine the protein expression and function of electron transport chain (ETC) complexes and key mitochondrial regulatory enzymes. Syncytialization of BeWo cells resulted reduced respiratory activity in conjunction with altered complex I and II activity and decreased pyruvate dehydrogenase (PDH) protein expression and activity. PA and P/O treatments were associated with increased basal and maximal respiratory activities in BeWo CT cells without alterations in protein expression or activity of individual ETC complexes and mitochondrial substrate regulators. The metabolic suppression in BeWo SCTs was consistent with that previously observed in primary human trophoblast cell cultures, while the observed increases in respiratory activity in PA-treated BeWo CTs may be indicative of an early timepoint of specific dietary saturated fat-mediated placental cell mitochondrial dysfunction.
Keywords: Placenta, Trophoblast, BeWo, Mitochondria, Diet, Palmitate
Introduction
The prevalence of obesity within women of reproductive age has been increasing worldwide over the past several decades, and approximately 40 million pregnant women globally are obese [1–3]. Additional reports have highlighted that a majority of women of reproductive age have a hip-to-waist ratio (WHR) indicative of a high risk for metabolic disease development [4,5]. Increased body mass, obesity and elevated WHR prior to and during pregnancy have not only been associated with negative health impacts for the mother, but also with impaired metabolic health outcomes in the offspring [6–11]. For example, children born to mothers who were overweight or obese throughout the gestational period have an increased risk of developing metabolic complications such as type 2 diabetes mellitus and obesity compared to children born to mothers with lean body compositions [10]. Further, these children developed these “adult-associated” metabolic diseases as early as four years of age, and the prevalence of pediatric metabolic syndrome appears to be increasing [8,12,13].
The origin of early onset development of adult-associated diseases is the result of abberant in utero programming of fetal metabolism potentially induced by maternal obesity-mediated alterations to placental mitochondrial function [14–16]. Specifically, due to their close proximity to maternal blood, trophoblast cells within the placental chorionic villi may be important in facilitating the adverse effects observed with increased maternal adiposity [9,17]. In fact, the large multinuclear syncytiotrophoblast (SCT) cells, and underlying progenitor cytotrophoblast (CT) cells that comprise the chorionic villi in obese pregnancies have been found to display decreased mitochondrial respiratory activity (oxidative phosphorylation) as well as reduced cellular ATP content [17–19]. Obesity-associated impairments in placental mitochondrial function have also been identified through analysis of enzymes within mitochondrial energy production pathways. Specifically, the activity of citrate synthase (a proxy measurement of total cellular mitochondrial content) as well as the protein abundance and enzyme activity of electron transport chain (ETC) complex I (NADH Dehydrogenase) and complex II (Succinate Dehydrogenase) are decreased in villous trophoblast cells from term obese placentae, [19,20]. Since progenitor CT and differentiated SCT cells form the materno-fetal interface and regulate nutrient and gaseous exchange between mother and developing fetus, obesity-mediated mitochondrial dysfunction within these cell types can potentially alter transplacental nutrient transfer such that fetoplacental development becomes pathological [21].
Recently, analysis of dietary patterns in industrialized nations have discussed a link between poor diet and metabolic health risks, independent from measures of body composition [22,23]. Studies analyzing gestational diet composition have demonstrated that many pregnant women consume diets that are calorie-dense but nutrient lacking and especially high in saturated and monounsaturated fatty acids [24–27]. Further studies examined fasting blood lipid levels in pregnancy and discussed hyperlipidemia in obese women as well as that the saturated FA palmitate (C16:0, PA) and the monounsaturated FA oleate (C18:1 cis 9, OA) are the most abundant circulating NEFAs in pregnancy [28–30]. Circulating PA and OA levels have previously been found to be impacted by diet composition [31,32]. Increased levels of PA alone have been demonstrated in induce mitochondrial damage and impair mitochondrial oxidative function leading to decreased ATP content in a number of cell culture systems [33–35]. Additionally, maternal gestational diet has been identified as a regulator of placental function independent from maternal body composition, and a pre-gestational diet-reversal in obese non-human primates was associated with improved placental vascular function as well as improvements in fetal metabolic health [36–39]. As these diet reversal studies altered maternal fat intake to improve placental and fetal metabolic health, dietary fatty acids (FA) themselves may be important in mediating adverse outcomes in obese pregnancies. Collectively these data potentially highlight that the mitochondrial impairments observed in obese placentae could be due to dietary FA overabundance, and that changes in diet and fat content alone may have effects on placental function and ultimately offspring metabolic health.
While obesity has been well associated with functional impairments in villous trophoblast mitochondria, the underlying independent effects of dietary saturated and monounsaturated NEFAs in mediating these phenotypes remains uncertain. The objective of this study was to examine the direct prolonged effects of PA and OA on CT and SCT mitochondrial metabolism in isolation from the effects of maternal body composition and obesity. It was postulated that a prolonged exposure to the dietary NEFAs PA and OA both independently and in combination would impair mitochondrial function in BeWo CT and SCT cells.
Materials and Methods:
Materials
All chemicals were purchased from Millipore Sigma (Oakville, Canada) unless otherwise indicated.
Cell culture conditions
BeWo cells are a choriocarcinoma-derived immortalized cell line that are representative system of villous trophoblast cells of the human placenta, and have been previously utilized to examine human villous trophoblast metabolic function [40,41]. Under basal conditions these cells are proliferative and CT-like, and when cultured with a cAMP analogue (8-Br-cAMP) they differentiate into SCT-like cells that human chorionic gonadotropin (hCG) [42,43]. In the current study, BeWo cells were utilized as a model to examine the independent effects of dietary NEFAs as these cells are isolated from effects of maternal body composition and gestational diet.
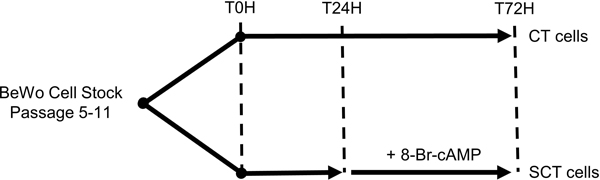
BeWo cells (CCL-98) were purchased from the American Type Culture Collection (ATCC; Cedarlane Labs, Burlington, Canada) (CCL-98™) and cultured in F12K media (Gibco, ThermoFisher, Missasauga, Canada) supplemented with 1% Penicillin-Streptomycin (Invitrogen, ThermoFisher, Missasauga, Canada) and 10% Fetal Bovine Serum (Gibco). Cells were maintained at 37 °C in 5% CO2/95% atmospheric air and used between passages 5–15 for all experiments. For each experiment, BeWo CT cells were plated at stated specific experimental densities and allowed to adhere to cell culture plates overnight prior to NEFA treatment. For all NEFA treatments, fats were conjugated 2:1 (molar ratio) to fatty acid free bovine serum albumin (BSA). NEFA medias were heated at 37°C for at least 30 mins prior to media changes to ensure equilibrium state of FA-BSA complexes [44]. Media supplemented with 0.3% BSA were used for control cultures in all experiments (BSA-ctrl). For SCT differentiation, 250 μM 8-Br-cAMP was added to experimental cell culture media at 24H and 48H. Media in all experimental conditions were replenished every 24H. A schematic showing the experimental timeline is shown in Figure 1.
Figure 1. Cell culture timeline for BeWo cell NEFA-treatments.
BeWo cells were plated and allowed to adhere overnight prior to NEFA-treatment initiation at T0H. At T24H 250 μM 8-Br-cAMP or a vehicle control was added to the cell culture plates to allocate cells to either CT or SCT cell type. All metabolic activity assays and cell collections were performed at T72H.
Cell viability
Cell viability in response to increasing concentrations (0–200μM) of PA, OA, and a 1:1 ratio of PA and OA (P/O) conjugated 2:1 (molar ratio) to BSA was assessed via the CellTiter-Fluor live-cell protease activity assay (Promega Corporation, Madison, WI, USA). BeWo cells were plated at 1×104 cells/well in 96-well opaque walled plates, and viability was assessed at 72H for both CT and SCT cell types as per the manufacturer’s instructions, using a Spectromax M5 plate reader (Molecular Devices, San Jose, CA, USA).
Immunofluorescent analysis of BeWo syncytialization
To assess the syncytialization of NEFA-treated BeWo cultures, percent fusion was of each treatment group was determined via zona occuldens-1 (ZO-1) immunofluorescent cell imaging [45]. In brief, BeWo cells were plated at a density of 1.4×105 cells/well on glass coverslips coated with laminin (2 μg/cm2) in 6-well cell culture plates and cultured for 72H with 100 μM of PA, OA or P/O conjugated 2:1 (molar ratio) to BSA. At 72H cells were washed once with Phosphate Buffered Saline (PBS) and fixed with 1:1 methanol:acetone for 30 mins at −20°C. Fixed cells were washed with PBS and permeabilized with 0.3% Triton-X 100 in PBS for 5 mins. Coverslips were then washed three times with PBS and blocked with 10% goat serum (abcam, Toronto, Canada) in PBS (blocking buffer). Coverslips were then incubated with ZO-1 antibody (1:200 dilution in blocking buffer; catalog No. 33–9100, Invitrogen) for 1 hour at room temperature. After washing with PBS three times coverslips were incubated with donkey anti-mouse IgG secondary antibody, Alexa Fluor 594 (1:2000 dilution in blocking buffer; Catalog No. R37115, Invitrogen) for 1 hour at room temperature. Coverslips were then washed 3 more times in PBS and mounted on slides using Fluoroshield with DAPI. Immunofluorescent images were then captured using the Zeiss Axioimager Z1 microscope (Carl Zeiss AG, Oberkochen, Germany) at the Biotron Integrated Microscopy Facility, Biotron Research Centre, Western University, London, Ontario, Canada. Nuclei and fused BeWo cells were then counted using ZEN software (Carl Zeiss AG), and percent fusion calculated as the percentage of total counted cells lacking ZO-1 expression.
Quantitative-PCR analysis of BeWo syncytialization
Relative mRNA transcript abundance of the gene encoding the beta subunit of human chorionic gonadotropin beta (CGB) was used as an indirect quantification of BeWo syncytialization following NEFA-treatment. Cells were plated at a density of 2×105 cells/well 6-well plates and treated with 100 μM NEFAs as described above. Total RNA was then extracted via phenol-chloroform extraction using TRIzol reagent (Invitrogen) as per the manufacturer’s specifications. Isolated RNA was then reverse transcribed using random decamer primers and M-MLV reverse transcriptase (Invitrogen) and RT-qPCR was subsequently performed using the CFX384 Real Time System (BioRad, Mississauga, Canada).
Target CGB primer sequences were obtained from Malhotra et al., (2015); primer sequences for OVOL1 and ERVW1 were obtained from Kusama et al., (2018); and housekeeping reference genes (ATCB and PSMB6) were designed using the NCBI/Primer-BLAST tool (Supplementary Table 1). Amplification of target gene expression was reported as fold change relative to control CT cells using the 2-ΔΔCt method with the ΔCt of each treatment condition being assessed as Ct(target) – Geometric mean of Ct(ATCB) and Ct(PMSB6).
Quantification of apoptosis
The activation state of pro-apoptotic pathways in BeWo CT and SCT cells in response to 100 μM NEFA treatments was assessed via caspase-3/7 activity at 72H. Cells were plated at a density of 1×104 cells/well in opaque walled 96-well plates and treated with 100 μM NEFAs as described above. Live cell caspase 3/7 activity was determined at 72H utilizing the Caspase-Glo 3/7 Assay (Promega Corporation) as per manufacturer’s instructions. BeWo CT cells treated with 100 μM of each rotenone (ETC complex I inhibitor) and antimycin A (ETC complex II inhibitor) were utilized as a positive control for upregulation of pro-apoptotic pathways [48].
Optimization of Seahorse XF Assay Protocols
BeWo cells were plated at the determined optimal density of 7.5×103 cells/well in Seahorse XF24 V7PS cell culture microplates (Agilent Technologies, Santa Clara, CA, USA) coated with 9 μg/cm2 collagen I (Gibco). The optimal concentration of the ATP synthase inhibitor oligomycin (1.5 μg/mL) was determined as per manufacturer’s instructions. The mitochondrial uncoupler dinitrophenol (DNP; 50 μM) was utilized to measure maximal mitochondrial respiratory activity. Attempts to follow the manufacturer’s recommended uncoupler FCCP resulted in unstable and declining OCR measurements. Previous reports have identified that progesterone, which is secreted by BeWo cells, recouples FCCP uncoupled mitochondria, but not DNP uncoupled mitochondria, and the optimization of DNP in our system produced the required stable OCR increase to measure maximal respiratory rate [49,50]. The complex I inhibitor rotenone (0.5 μM), complex II inhibitor antimycin A (0.5 μM), D-glucose (10 mM), and 2-deoxy-D-glucose (50 mM) were used as per manufacturer’s recommendation.
BeWo cell oxidative function
The optimized Mitochondrial (Mito) Stress Test was utilized to quantify the oxidative function of NEFA-treated (100 μM) BeWo CTs and SCTs. At the 72H timepoint the culture medium exchanged with Mito Stress Test assay media (Seahorse XF base media (Agilent Technologies) supplemented with 10 mM D-glucose, 2 mM sodium pyruvate, and 2 mM L-glutamine (Gibco, ThermoFisher, Mississauga, Canada), pH 7.4), and cells were incubated at 37°C in 100% atmospheric air for 45 mins. Following basal OCR readings subsequent injections of oligomycin; DNP; and rotenone combined with antimycin A were utilized to measure parameters of mitochondrial respiratory function (Supplementary Fig. 1A/B).
Cells were lysed via freeze-thaw method in Seahorse assay media and total DNA was quantified using Hoescht dye fluorescence [51,52]. Briefly, cell lysates were mixed with 2x Hoescht Dye assay mix (0.0324 mM Hoescht 33342 Dye (Pierce, ThermoFisher Scientific, Mississauga, Canada), 2 mM NaCl, 2 mM NaH2PO4), fluorescence (360 nm excitation, 460 nm emission) was then measured and DNA content was determined via standard curve.
BeWo cell glycolytic function
The optimized Glycolysis Stress Test was utilized to quantity the glycolytic function of NEFA-treated (100 μM) BeWo CTs and SCTs. At 72H culture media was exchanged with Glycolysis Stress Test assay media (Seahorse XF base media supplemented with 2 mM L-glutamine, pH 7.4), and cells were incubated at 37°C in 100% atmospheric air for 45 mins. Following basal Extracellular Acidification Rate (ECAR) readings, injections of D-glucose; oligomycin; and 2-Deoxy-D-glucuose were utilized to measure parameters of glycolysis function (Supplementary Fig. 1C/D). ECAR readings were normalized to total DNA content as described above.
Immunoblot analysis
BeWo cells were plated at 1×106 in 100mm cell culture plates and cultured with NEFAs (100 μM) as described. Total protein was collected with radioimmunopreciptation assay (RIPA) buffer containing protease inhibitors (leupeptin and aprotinin (1 μg/mL each; Bioshop, Burlington, Canada); and phenylmethylsulfonyl fluoride (PMSF, 1mM, Bioshop)) and phosphatase inhibitors (sodium orthovanadate (1 mM), New England Biolabs; and sodium fluoride (25 mM), Bioshop). The lysates were sonicated with 5 bursts at 30% output (MISONIX: Ultrasonic liquid processor) and centrifuged at 12,000g at 4°C for 15 mins. The supernatant was collected, and proteins quantified via Bicinchoninic Acid (BCA) assay (Pierce, ThermoFisher Scientific).
Protein abundance was then determined via immunoblotting. Protein were separated by molecular weight on sodium dodecyl sulfate polyacrylamide gels (10–15% acrylamide) and transferred to Immobilon polyvinylidene fluoride (PVDF) membrane (EMD Millipore, Fisher Scientific). Transfer of protein was confirmed with Ponceau-S stain (0.1% Ponceau-S in 0.5% acetic acid), and ponceau band density imaged with a ChemiDoc Imager (BioRad) for total lane protein normalization (Supplementary Fig. 3). Respective primary antibodies and horseradish peroxidase-labelled secondary antibodies were used to visualize proteins of interest (Supplementary Table 2). Protein bands were imaged with Clarity western ECL substrate (BioRad) on a ChemiDoc. Protein band density was analyzed with ImageLab software (BioRad) and protein abundance was normalized to total lane protein [53].
ETC complex I and II activity
ETC complex I and II was assessed as previously described, and adapted for use with cell culture lysates [54]. BeWo cells were plated at a density of 4×105 cells/well in 60mm plates and collected by scraping following the T72 hr NEFA-treatment (100 μM). Cells were lysed in ETC lysis buffer (250 mM sucrose, 5 mM HEPES, 100 μg/mL saponin) containing protease and phosphatase inhibitors (as described above), centrifuged at 5000 g for 5 mins at 4°C and resuspended in Phosphate buffer (25 mM K2HPO4). Enzyme assays were performed at 37°C using a Spectramax plate spectrophotometer (Molecular Devices) in a 96-well plate. Enzyme activities were expressed relative to total protein concentration for each cell lysate (determined by BCA assay).
Complex I activity was assessed as rate of NADH oxidation (measured at 340 nm) following the addition of cell lysate (approximately 15 μg total protein) to assay buffer (25 mM KH2PO4, 2.5 mg/mL BSA, 0.13 mM NADH, 2 mM KCN, 2 μg/mL antimycin A, 65 μM CoQ1). Complex I activity was calculated from the difference between rates with and without rotenone (10 μM) to account for non-specific NADH oxidation.
Complex II activity was assessed as rate of DCPIP oxidation (measured at 600 nm) following addition of CoQ1 (100 μM) to assay mix containing cell lysate (approximately 10 μg total protein) in assay buffer (25 mM KH2PO4, 20 mM monosodium succinate, 50 μM DCPIP, 2 mM KCN, 2 μg/mL antimycin A, 2 μg/mL rotenone).
Lactate Dehydrogenase and Citrate Synthase activity
Lactate dehydrogenase (LDH) and citrate synthase activity were assessed as previously described and adapted for use with cell culture lysates [55,56]. BeWo cells were plated at a density of 4×105 cells/well in 60 mm cell culture dishes collected by scraping at 72H. Cells were subsequently lysed in glycerol lysis buffer (20 mM Na2HPO4, 0.5 mM EDTA, 0.1% Triton X-100, 0.2% BSA, 50% glycerol) containing protease and phosphatase inhibitors (as described above) and stored at −80°C until analyzed [57]. Assays were performed at 37°C using a 96-well plate. Enzyme activity rates were normalized to total protein content of cell lysates determined via BCA assay.
LDH activity was assessed as the rate of NADH oxidation (measured at 340 nm) following addition of cell lysate (corresponding to approximately 1 μg total protein) to assay buffer (0.2 mM NADH, 1mM sodium pyruvate).
Citrate synthase activity was assessed as rate of Ellman’s reagent consumption (measured at 412 nm), following addition of oxaloacetate (0.33 mM) to assay mix containing cell lysate (approximately 30 μg total protein) in assay buffer (0.15 mM acetyl CoA, 0.15 mM DTNB).
Analysis of total Pyruvate Dehydrogenase Activity and PDHE1-subunit activity
Total PDH activity was assessed at 72H using the PDH Activity Assay Kit (Millipore Sigma). In brief, BeWo cells were plated at a density of 4*105 cells/well in 60mm cell culture plates and treated with NEFAs as previously described. At 72H cells were lysed in 400 μL of the provided lysis buffer and 50 μL of cell lysate was mixed with an equal volume of assay mix and PDH enzyme activity was assessed as per the provided kit instructions.
The activity of the PDH-E1 enzyme subunit was assessed as previously described, and modified for use with the BeWo cell line [58]. BeWo cells were plated at a density of 4×105 cells/well in 60mm plates and treated with NEFAs as previously described. At 72H cells were collected by scraping and were lysed by sonication with 10 bursts at 15% output (MISONIX: Ultrasonic liquid processor) in 50 mM K2HPO4 buffer containing protease and phosphatase inhibitors. PDH-E1 enzyme activity was assessed at 37°C using a Spectramax plate spectrophotometer (Molecular Devices) in a 96-well plate, as the rate of DCPIP oxidation (measured at 600 nm) following the addition of cell lysate (approximately 20 μg total protein) to assay buffer (50 mM KH2PO4, 1 mM MgCl2, 0.2 mM thymine pyrophosphate, 0.1 mM DCPIP, 2 mM sodium pyruvate). Enzyme activities were expressed relative to total protein concentration for each cell lysate (determined by BCA assay).
Statistical Analysis
A Randomized Block Design One-Way ANOVA and Dunnett’s Multiple Comparisons Test of raw fluorescence values was utilized to compare cell viabilities of BeWo cells treated with each NEFA to respective differentiation state BSA-alone control. BeWo percent fusion data, as well as the Mito Stress Test parameters of Spare Respiratory Capacity and Coupling Efficiency (expressed as expressed as percent basal OCR) and the Glycolysis Stress Test parameter of Reserve Capacity (expressed as percent ECAR) were log-transformed and statistically analyzed via Two-Way ANOVA (2WA) and Bonferroni’s Multiple Comparisons. A Randomized Block Design 2WA and Sidak’s Multiple Comparisons Test using raw data was utilized to analyze statistical differences in the relative CGB mRNA transcript abundance; live-cell caspase 3/7 activity; protein abundance; metabolic enzyme activities; as well as all other Seahorse Mito Stress Test and Glycolysis Stress Test parameters between the NEFA-treatment groups for the BeWo CT and SCT cells [59]. Data were then expressed as percent of untreated CT control (basal F12K media) for use in experimental figures. All statistical analyses were performed with GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA).
Results
Characterization of un-differentiated and differentiated BeWo culture treated with NEFAs
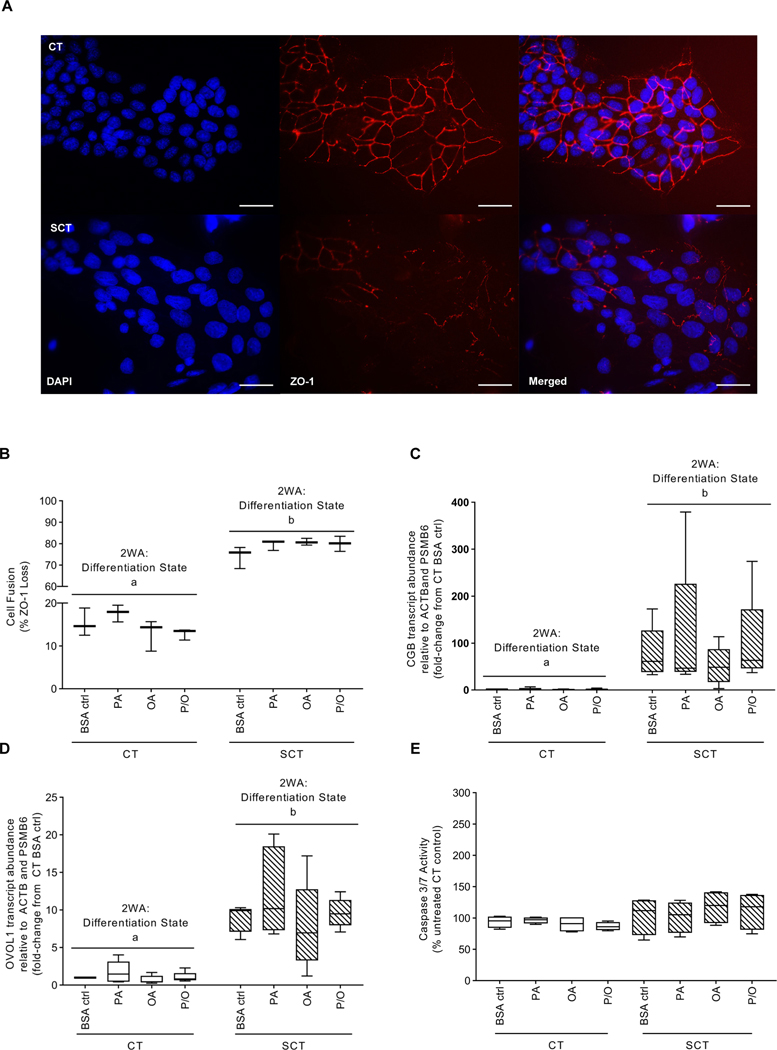
BeWo cells treated with 8-Br-cAMP did display decreased ZO-1 protein expression and increased cell fusion (mean 78% fusion) compared to CT cells (mean 15% fusion) (Fig. 2A,B; 2WA: differentiation state p<0.001). Additionally, BeWo cells treated with 8-Br-cAMP displayed elevated CGB (mean expression 86-fold higher) and OVOL1 (mean expression 9-fold higher) mRNA abundance (23–85 fold higher expression) consistent with the differentiation process (Fig. 2CD; 2WA: differentiation state p<0.01)
Figure 2: 100 μM of PA, OA or P/O does not negatively impact BeWo cell syncytialization or upregulate pro-apoptotic pathways at 72H.
(A) Representative ZO-1, DAPI and merged immunofluorescent images of BeWo CT and SCT cells (scale bar = 50 μm); (B) Percent fusion of NEFA-treated BeWo CT and SCT cells; data is expressed as percentage of nuclei lacking ZO-1 staining. Relative mRNA abundance of (C) CGB and (D) OVOL1 in NEFA-treated BeWo CT and SCT cells; RT-qPCR data is presented as the fold-change of target mRNA abundance compared to CT BSA ctrl samples relative to the geometric mean of ACTB and PMSB6 mRNA abundance. (E) Live-cell caspase activity of NEFA-treated BeWo CT and SCT cells; data is presented as percent untreated CT control luminescence values (B n=3/group; C-E n=5/group; different letters denote statistical significance p<0.05; B percent fusion data was log transformed and analyzed via two-way ANOVA (2WA); C,D relative mRNA abundance data was analyzed via randomized-block 2WA).
Cell viability of BeWo CT and SCT cells following a prolonged (72H) NEFA treatment was assessed to ensure that subsequent mitochondrial activity measurements were not confounded by a potential stress activated cell death program. A 100 μM NEFA treatment (PA, OA and P/O) did not negatively impact BeWo cell viability in either CT or SCT cells relative to BSA-alone treated controls at T72H (Table 1).
Table 1.
Cell viability of NEFA-treated BeWo CT and SCT cells. Data are mean ± SEM.
| NEFA Treatment (μM) | CT Cell Viability (% CT F12K) | SCT Cell Viability (% SCT F12K) | |
|---|---|---|---|
| BSA Control | 72.35 ± 2.61 | 46.25 ± 4.17 | |
| PA | |||
| 50 | 86.83 ± 3.56 * | 87.08 ± 9.46 * | |
| 100 | 77.34 ± 4.92 | 61.40 ± 12.14 * | |
| 200 | 55.03 ± 10.31 | 45.32 ± 16.53 | |
| OA | |||
| 50 | 88.10 ± 4.93 | 73.70 ± 59.02 * | |
| 100 | 72.97 ± 6.13 | 59.02 ± 10.18 * | |
| 200 | 48.78 ± 10.19 | 34.54 ± 11.59 | |
| P/O | |||
| 50 | 85.57 ± 4.39 | 70.27 ± 10.72 * | |
| 100 | 76.37 ± 6.09 | 57.92 ± 9.71 | |
| 200 | 57.87 ± 10.59 | 40.18 ± 14.48 | |
denotes significantly different cell viability relative to respective differentiation state BSA-control (n=4/group)
The 100 μM NEFA treatments were additionally not associated with altered BeWo fusion in either CT and SCT cells as measured with ZO-1 expression (Fig. 2A,B). Further, these 100 μM treatments did not significantly alter the mRNA abundance of CGB or OVOL1 (markers of syncytialization in BeWo cells) within the CT and SCT cells (Fig. 2C,D)..
Additionally, in each NEFA-treatment there was no significant difference in live-cell caspase 3/7 activity compared to respective differentiation state BSA-control sample (Fig. 2E; 2WA: differentiation state and 2WA: treatment p>0.05).
The impact of prolonged NEFA exposure upon respiratory activity of BeWo cells
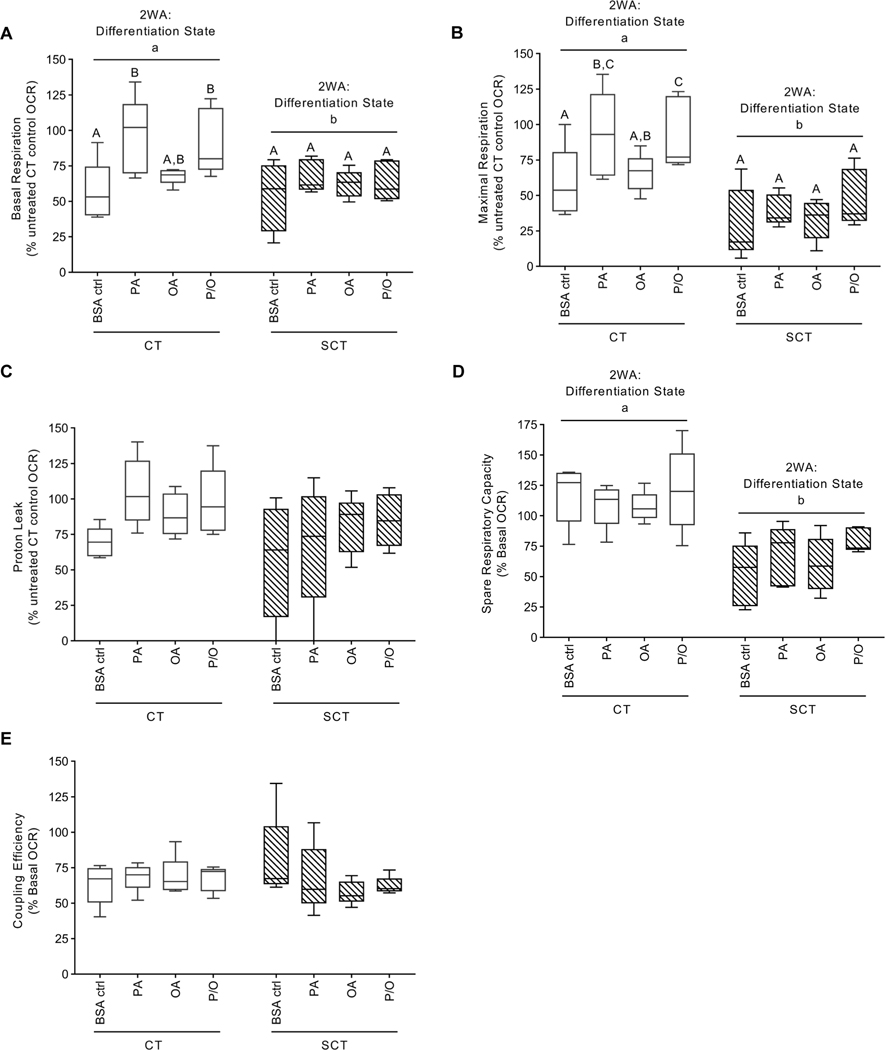
BeWo CT cells treated with PA (100 μM) and P/O (containing 50 μM PA) for 72H exhibited a significant increase in basal mitochondrial activity compared to control CT samples (Fig. 3A, 2WA: NEFA treatment p<0.01). Under DNP-stimulated conditions CT cells exposed to PA exhibited a significant increase in OCR compared to BSA-alone treated CT cells (Fig. 3B, 2WA: NEFA Treatment p<0. 05). Conversely, BeWo SCT cells displayed diminished respiratory activity and had significantly lower OCR under basal and DNP-stimulated conditions compared to BeWo CT cells (Fig. 3A–B, 2WA differentiation state p<0.05). This diminished respiratory activity observed in BeWo SCT cells was associated with a significant reduction in spare respiratory capacity (Fig 3D., 2WA differentiation state p<0.001). No differences in BeWo cell proton leak or coupling efficiency was observed with differentiation state or NEFA-treatment (Fig. 3C,E)
Figure 3. Mitochondrial respiratory activity of BeWo cells following prolonged NEFA treatment.
Oxygen Consumption Rate (OCR) of experimental media was measured under basal conditions and following subsequent injections of: Oligomycin (1.5 μg/mL); Dinitrophenol (50 μM); and Rotenone and Antimycin A (0.5 μM each) to calculate the (A) Basal Respiration, (B) Maximal Respiration, (C) Proton Leak, (D) Spare Respiratory Capacity, and (E) Coupling Efficiency in NEFA-treated BeWo CT and SCT cells (n=5/group). (A, B, C) Statistical analysis was performed on DNA-normalized OCR rates via Two-Way Randomized Block ANOVA, Sidak’s multiple comparisons Test; (D, E) percent basal OCR was log transformed and analyzed via Two-Way ANOVA). (A, B, C) Respiratory activity data is expressed as OCR normalized to Fsutotal DNA content and presented as percent CT untreated control; (D, E) data is represented as percent basal OCR for each condition. Different letters denote statistical significance (p<0.05, n = 5/group).
Glycolytic function in BeWo cells is unaltered with NEFA treatments
Basal glycolysis, maximum glycolysis, glycolytic reserve capacity, and non-glycolytic acidification as measured by the Seahorse XF Glycolysis Stress Test, were not impacted after cells were differentiated to SCT cells and were unaltered with NEFA treatment (Table 2).
Table 2:
NEFA treatments did not affect the glycolytic activities of BeWo CT or SCT cells. Data are mean ± SEM (n=5/group).
| Cell Type / Treatment | Basal glycolysis1 | Max Glycolysis2 | Reserve Glycolytic Capacity3 | Non-Glycolytic Acidification4 | |
|---|---|---|---|---|---|
| CT | |||||
| BSA Control | 86.14 ± 10.71 | 72.19 ± 8.22 | 120.80 ± 6.79 | 84.25 10.01 | |
| PA | 89.99 ± 6.20 | 81.61 ± 5.62 | 134.30 ± 6.77 | 91.31 8.53 | |
| OA | 97.59 ± 11.09 | 86.11 ± 11.81 | 129.50 ± 11.57 | 87.72 11.08 | |
| P/O | 89.26 ± 6.19 | 85.82 ± 5.47 | 142.90 ± 6.68 | 91.90 7.33 | |
| SCT | |||||
| BSA Control | 83.53 ± 7.97 | 67.04 ± 6.54 | 115.50 ± 4.48 | 117.70 9.16 | |
| PA | 81.91 ± 14.13 | 66.57 ± 11.47 | 137.00 ± 27.10 | 104.90 13.76 | |
| OA | 83.51 ± 11.90 | 67.32 ± 8.49 | 118.40 ± 13.33 | 105.00 9.36 | |
| P/O | 75.55 ± 8.19 | 65.30 ± 6.97 | 131.30 ± 7.73 | 98.36 8.68 | |
% CT F12K ECAR;
% CT F12K ECAR;
% basal glycolysis;
% CT F12K ECAR
NEFA impact upon electron transport chain complex protein abundance and activity
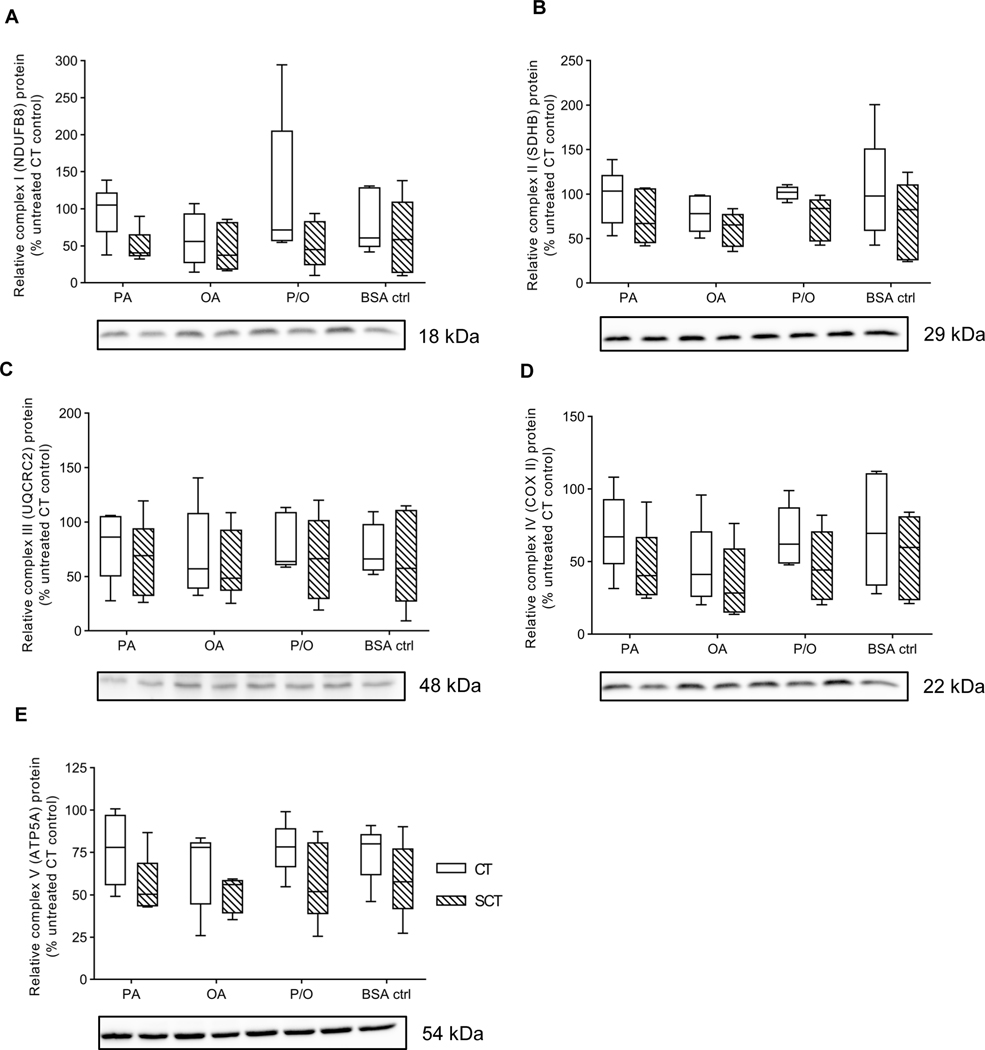
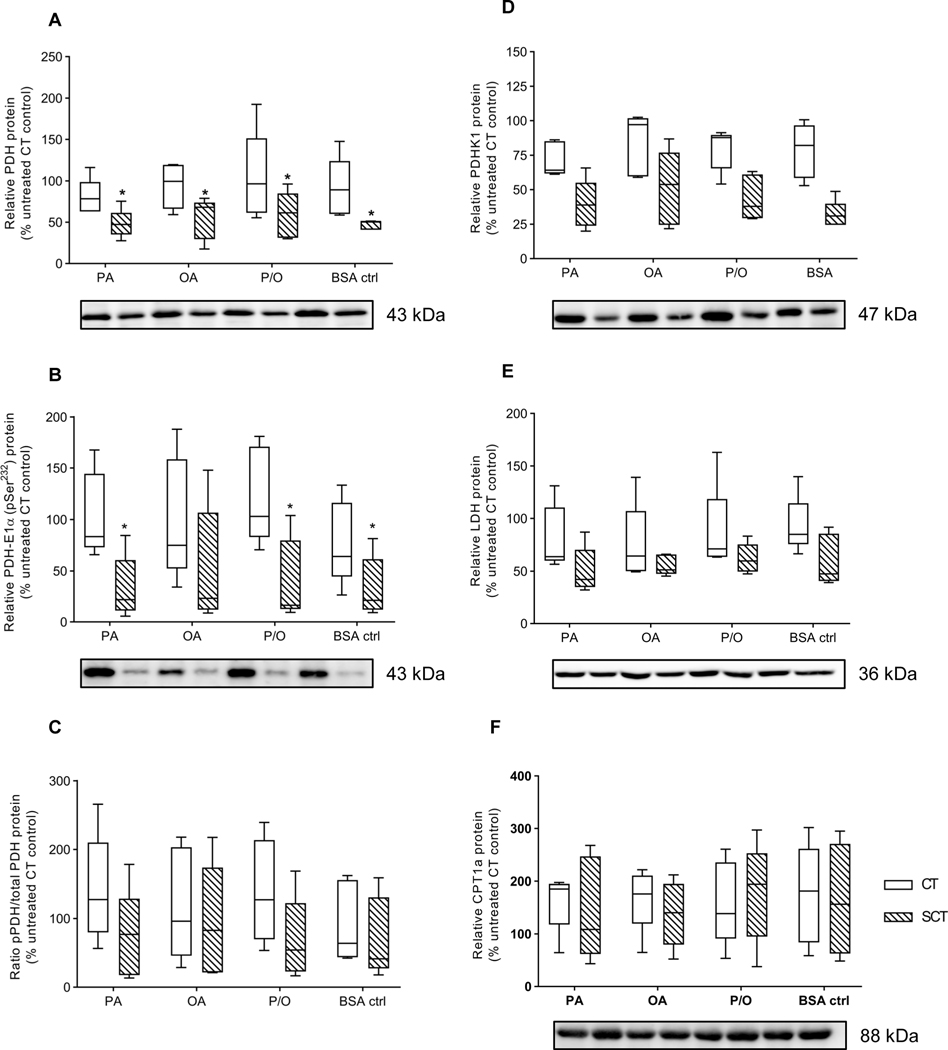
There were no significant differences in response to NEFA treatment in both CT and SCT cells for ETC complex I (NDUFB8 subunit), complex II (SDHB subunit), complex III (UQCRC2 subunit), complex IV (COX II subunit), and complex V (ATP5A subunit) protein abundance relative to total lane protein (Fig. 4). However, BeWo SCT displayed significantly increased Complex I and decreased complex II activity compared to CT cells (Table 3; 2WA: differentiation state p<0.05). Additionally, P/O-treated SCT cells exhibited a significantly reduced complex I activity relative to BSA-treated SCT controls (Table 3; 2WA treatment p<0.001).
Figure 4. Prolonged NEFA treatment did not affect BeWo Electron Transport Chain complex protein abundance.
Relative protein abundance of (A) Complex I (NDUFB8 subunit) (B) Complex II (SDHB subunit) (C) Complex III (UQCRC2 subunit) (D) Complex IV (COX II subunit), and (E) Complex V (ATP5A subunit) of the Electron Transport Chain was determined at T72H via immunoblotting. Data is presented as percent untreated CT control for each experimental replicate (n=5/group). Representative full length ponceau loading control image is available in Supplementary Figure 3.
Table 3:
Maximal activity rates of mitochondrial enzymes in NEFA-treated BeWo CT and SCT cells. Data are mean ± SEM of % untreated CT U/mg protein.
| Cell Type / Treatment | Citrate Synthase | ETC Complex I | ETC Complex II | Lactate Dehydrogenase | PDH-E1 subunit | Pyruvate Dehydrogenase | |
|---|---|---|---|---|---|---|---|
| CT | |||||||
| BSA Ctrl | 116.20 ± 22.37 | 120.7 ± 7.71 | 113.8 ± 13.71 | 163.20 ± 45.50 | 99.23 ± 6.37 | 105.04 ± 6.93a | |
| PA | 93.62 ± 15.00 | 98.00 ± 10.03 | 98.99 ± 19.80 | 109.80 ± 14.41 | 101.72 ± 3.83 | 86.78 ± 10.96b | |
| OA | 85.31 ± 14.22 | 100.00 ± 5.59 | 94.46 ± 17.77 | 104.90 ± 19.23 | 102.84 ± 16.80 | 112.01 ± 10.41a | |
| P/O | 90.34 ± 5.44 | 106.20 ± 0.32 | 93.67 ± 17.50 | 133.60 ± 31.71 | 118.96 ± 21.97 | 99.56 ± 7.37ab | |
| SCT | |||||||
| BSA Ctrl | 60.68 ± 6.01 | 191.50 ± 31.49a | 61.76 ± 13.75 | 79.25 ± 18.77 | 260.45 ± 41.81 | 84.66 ± 4.99 | |
| PA | 49.80 ± 4.60 | 183.10 ± 25.80a | 62.05 ± 15.57 | 79.54 ± 16.18 | 246.35 ± 20.16 | 88.20 ± 9.06 | |
| OA | 39.80 ± 4.74 | 129.2 ± 13.85ab | 51.88 ± 17.79 | 73.79 ± 17.63 | 222.28 ± 19.02 | 80.33 ± 3.51 | |
| P/O | 63.52 ± 4.14 | 103.60 ± 17.77b | 58.96 ± 8.20 | 78.59 ± 13.26 | 233.78 ± 57.00 | 81.68 ± 8.10 | |
| *Differentiation state difference | NS | * | * | NS | * | * | |
denotes a differentiation state dependent difference in enzyme activity between BeWo CT and SCT cells (n=5/group). Different letters denote significant differences between NEFA treatments within a differentiation state.
The impact of prolonged NEFA exposure on metabolic enzyme activity
LDH activity was not significantly altered with differentiation state or NEFA-treatment (Table 3). BeWo SCT cells exhibited a significantly increased PDH-E1 subunit activity compared to CT cells (Table 3). However, BeWo CT cells displayed a significantly increased rate of total PDH enzyme activity compared to SCT cells (Table 3; 2WA differentiation state p<0.01). Additionally, BeWo CT cells treated with PA had a significantly reduced total PDH enzyme activity compared to BSA-alone treated CT controls (Table 3; 2WA differentiation state p<0.05). There was no differentiation state or NEFA dependent alteration in citrate synthase enzyme activity in BeWo cells (Table 3).
Oxidative State in unaltered in NEFA-treated BeWo cells
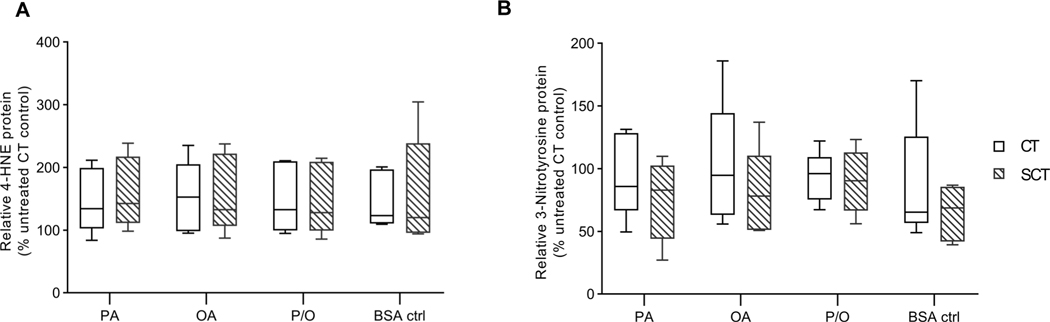
There were no differences in the protein expression of 4-Hydroxynonenal (4-HNE) and 3-Nitrotyrosine, markers of oxidative stress and nitrative stress respectively, with prolonged NEFA treatment or differentiation in BeWo cells (Fig. 5A,B).
Figure 5. Prolonged NEFA treatment does not alter oxidative state of BeWo cells.
Relative Protein abundance of (A) 4-Hydroxynonenal (4-HNE) and (B) 3-Nitrotyrosine was determined in NEFA-treated BeWo CT and SCT cells at 72H via immunoblotting. Data is presented as relative protein density normalized to total lane protein density and expressed as percent CT untreated control protein abundance (n=5/group). Representative full-length blots are available in Supplementary Figure 2. Representative full length ponceau loading control images are available in Supplementary Figure 3.
The impact of prolonged NEFA exposure on metabolic enzyme expression
There was a differentiation state dependent expression pattern in Pyruvate Dehydrogenase (PDH) in both its unphosphorylated and phosphorylated (pPDH) form. BeWo SCT cells displayed significantly reduced relative abundances of both PDH and pPDH (Fig. 6A,B; 2WA differentiation state p<0.05), however the ratio of pPDH:PDH (Fig. 6C) was not significantly altered between differentiation states. No differentiation state or NEFA-treatment specific differences were found in the relative protein abundances of either LDH or carnitine palmitoyltransferase 1a (CPT1a) in BeWo cells (Fig. 6D,E).
Figure 6. Prolonged NEFA treatments did not affect protein expression of enzymes involved in mitochondrial uptake of pyruvate or long-chain fatty acid species.
Relative protein abundance of (A) Pyruvate Dehydrogenase (PDH), (B) PDH-E1α (pSer232) (pPDH), (C) the pPDH/PDH ratio, (D) Pyruvate Dehydrogenase Kinase-1 (PDHK1) (E) Lactate Dehydrogenase (LDH), and (F) CPT1a was determined in NEFA-treated BeWo CT and SCT cells at 72H via immunoblotting. Data is presented as percent CT untreated control (n=5/group). Representative full length ponceau loading control images are available in Supplementary Figure 3.
Discussion
The current study aimed to characterize the mitochondrial function and respiratory activity in a model of the differentiating human villous trophoblast following a prolonged (72 hour) NEFA challenge. To confirm that mitochondrial respiratory activity and metabolic function was examined using a stable population of cells we analyzed cell viability, CT-to-SCT differentiation potential and apoptotic state of the treated cells. Our experiments demonstrated that NEFA treatments exceeding 200 μM produced large variances in cell viability, which is consistent with previous observations in primary human trophoblast culture systems [60]. A treatment of 100 μM of PA, OA and P/O was found to not impact BeWo cell syncytialization or upregulate pro-apoptotic pathways, yielding a stable cell culture system in which mitochondrial function could be examined. Additionally, the 100 μM of PA and OA used in this current study also aligned with published human third trimester fasting circulating lipid levels under conditions of obesity highlighting a physiological relevance of the utilized NEFA treatment [28].
Differentiation and BeWo Metabolic Function
BeWo cells displayed a significant reduction in mitochondrial respiratory activity (Mito Stress Test readouts) following 8-Br-cAMP induced differentiation to SCT cells. These reductions in basal and maximal respiration rates were consistent with previous reports in human primary trophoblast cell culture, which demonstrated that SCT cells were less metabolically active than the progenitor CT cells [61]. More strikingly, we observed these similarities despite the BeWo cells in our culture system fusing at rates around 60%, further supporting that diminished mitochondrial oxidative function is an important physiological marker of placental syncytialization.
This study provides novel insight into dynamic changes in the activity of individual ETC complexes during CT differentiation in SCT. Specifically, we observed significant differences in complex I and complex II activity between CT and SCT cell types that were independent of alterations in mitochondrial biomass as assessed by citrate synthase enzyme activity. The decreased complex II activity observed in BeWo SCT cells may allude to a biochemical mechanism that potentially explains the overall reduction in respiratory activity observed in syncytialized BeWo cells.
Additionally, the BeWo SCT cells in our study may exhibit a diminished oxidative metabolism of pyruvate substrates as evidenced by reduced protein expression and enzyme activity of total Pyruvate Dehydrogenase (PDH). These data may indicate an overall decrease in the conversion of glycolysis-derived pyruvate to acetyl-CoA in BeWo SCTs and may further highlight the mechanism by which oxidative energy production is suppressed in differentiated villous trophoblast cells. However, future studies are needed to elucidate if similar mechanisms govern the reduction in mitochondrial function in the differentiated SCT cell populations in both BeWo cells and isolated primary trophoblasts.
NEFAs and BeWo Metabolic Function
The current study demonstrated a novel NEFA-induced increase mitochondrial respiratory activity in PA and P/O-treated BeWo CT cells under basal and DNP-stimulated (maximal) conditions of the Mito Stress Test that was not associated with any specific alterations in the individual enzyme activities of ETC Complexes I and II. Previously, an overall reduction in mitochondrial respiratory activity has been reported as an indication of mitochondrial dysfunction in term placentae [19]. However, increases in mitochondrial respiratory activity have also been described as a preliminary marker of mitochondrial failure in disease states that are linked to an ultimate mitochondrial dysfunction [62,63]. In particular, mitochondrial dysfunction induced in hepatocytes under conditions of Non-alcoholic fatty liver disease (NAFLD) has been suggested to be the result of an initial increase in mitochondrial respiratory activity stimulated by saturated fat overabundance that in turn promotes increased Reactive Oxygen Species (ROS) production, and ultimately, oxidative damage to mitochondria [62,64]. We speculate that the increases in respiratory activity observed in the PA and P/O-treated BeWo CT cultures may be indicative of an early timepoint of mitochondrial dysfunction in the villous trophoblast, mirroring these other reports of activation of mitochondrial activity prior to an ultimate failure.
In many other culture systems PA treatment has been demonstrated to increase ROS production leading to cellular oxidative stress [65–67]. As oxidative stress has additionally been identified as a marker of metabolic dysfunction in the term obese placenta, we speculate that the increased respiratory activity observed in PA and P/O-treated BeWo CT cells in this study could over time increase ROS production and lead to oxidative stress [68,69]. However, at the timepoint utilized in the current study there were no indications of oxidative stress in any of the treatments, as evidenced by immunoblots for total abundance of 3-nitrotyrosine and 4-HNE. Further studies are needed to investigate if continued exposure of BeWo CT cells to the selected physiologically relevant PA treatment will lead to ROS-induced mitochondrial oxidative damage and ultimately impaired mitochondrial respiratory function.
It is important to note that OA has previously been shown to elicit anti-oxidant capabilities and rescue mitochondrial function in muscle and pancreatic beta-cells treated with high levels of saturated fats [34,70]. Thus, the co-culture of OA with PA (P/O) over longer timepoints may prevent the accumulation of PA-stimulated ROS and may potentially preserve villous trophoblast mitochondrial function as has been observed in other systems.
While this study has demonstrated the impacts of a prolonged PA exposure on placental cell metabolic function, the use of the immortalized BeWo cell line may limit the conclusions that can be drawn as they may confer properties not observed in primary culture or other ex vivo systems. Having said that, the use of a cell line model of the villous trophoblast in the current study allowed for the analysis of the effects of lipid exposure in isolation from factors of maternal body composition and gestational diet. Specifically, the BeWo choriocarcinoma cell line enabled us to analyze the metabolic function of SCTs differentiated under constant exposure to elevated dietary-NEFAs, analogous to that of a differentiating trophoblast throughout gestation in an obese pregnancy, but independent to non-dietary obesogenic environment components [71–73]. However, future studies still need to examine if prolonged NEFA challenge at physiologically relevant levels can lead to similar early markers of mitochondrial dysfunction in primary-derived trophoblast cultures. However, the use of trophoblasts from early gestational periods may be required in these future works to develop a primary culture model that, similar to this study, is independent from the influence of material body composition and gestational diet.
Conclusion
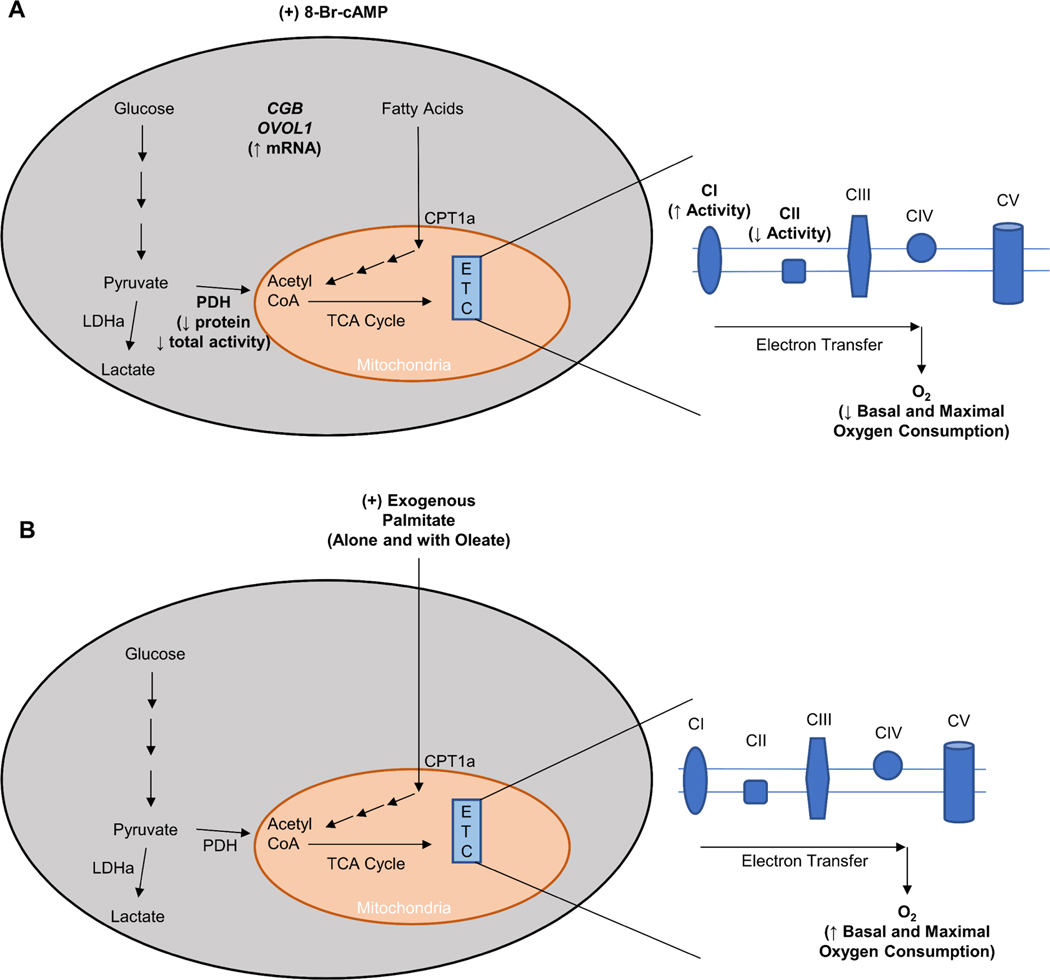
The current study aimed to characterize the mitochondrial function and respiratory activity in a model of the differentiating human villous trophoblast following a prolonged (72 hour) NEFA challenge. Differentiated BeWo SCT cells displayed a reduced basal and maximal mitochondrial respiratory activity, similar to that previously observed in primary trophoblast culture (Fig. 7A). Additionally, this study highlights that an isolated and prolonged exposure to physiological levels of dietary-NEFAs, independent from factors of maternal body composition, increases villous trophoblast cell mitochondrial respiratory function (Fig. 7B). The observed increases in basal and maximal respiratory activity in response to PA and P/O treatments in this study may be indicative of an early phenotype of placental mitochondrial impairment that may later trigger oxidative damage to the mitochondria. Overall, this study highlights that dietary fats independently modulate villous trophoblast mitochondrial function, and further demonstrates that maternal diet composition is an important regulator of the adverse placental outcomes that underlie the development of metabolic disease in the offspring.
Figure 7. Summary description of changes in BeWo cell metabolic function following (A) syncytialization (48-hour supplementation of cell culture media with 250 μM 8-Br-cAMP) and (B) 72-hour treatment with exogenous palmitate either independently (PA; 100 μM) or in conjunction with oleate (P/O; 50 μM each).
BeWo Syncytialization was associated with a suppression in basal and maximal mitochondrial function that has previously been observed following syncytialization of primary human trophoblast cells. Exogenous palmitate treatment increased basal and maximal respiratory activity in BeWo cytotrophoblast cells.
Supplementary Material
Supplementary Figure 1: Representative Seahorse Assay Tracings. Representative tracing of Oxygen Consumption Rate (OCR) throughout experimental assay for (A) BeWo CT cells and (B) BeWo SCT cells from the Mito Stress Test. Representative tracing of Extracellular Acidification Rate (ECAR) throughout experimental assay for (C) BeWo CT cells and (D) BeWo SCT cells from the Glycolysis Stress Test. Data is represented as mean OCR or ECAR of each group in technical duplicate from one experiment and presented as percent CT untreated control.
Supplementary Figure 2. Full length representative blots of oxidative stress markers. Representative images of protein bands obtained in the immunoblots for (A) 4-hydroxynonenal and (B) nitro-tyrosine. Left-to-right represents the treatments PA CT, PA SCT, OA CT, OA SCT, P/O CT, P/O SCT, BSA CT, BSA SCT.
Supplementary Figure 3. Representative images of immunoblot ponceau loading controls. Representative full length ponceau loading control bands for (A) Mitoprofile, (B) Pyruvate Dehydrogenase, (C) PDH-E1α (pSer232), (D) Pyruvate Dehydrogenase Kinase 1, (E) Lactate Dehydrogenase, (F) Carnitine Palmitoyltransferase 1a, (G) 4-Hydroxynonenal, and (H) Nitro-tyrosine immunoblots. Left-to-right represents the treatments PA CT, PA SCT, OA CT, OA SCT, P/O CT, P/O SCT, BSA CT, BSA SCT.
Acknowledgements
We wish to acknowledge Dr. Stephen Renaud for his assistance in reviewing the manuscript for submission. We additionally wish to thank Karen Nygard and Marc Courchesne for their assistance in optimizing a protocol for measuring BeWo cell fusion rate. This research was supported by funds from a National Institutes of Health (NIH) Human Placenta Project Grant (grant No. U01 HD087181-01). Zachary Easton was supported by funds from an Obstetrics & Gynaecology Graduate Scholarship (OGGS) awarded by the Obstetrics & Gynaecology department at the University of Western Ontario.
Footnotes
Declaration of Interest
The authors have no conflicts of interest to declare.
References
- 1.Chen C; Xu X; Yan Y. Estimated global overweight and obesity burden in pregnant women based on panel data model. PLoS One 2018, 13, e0202183, doi: 10.1371/journal.pone.0202183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popkin BM; Doak CM The Obesity Epidemic Is a Worldwide Phenomenon. Nutr. Rev 2009, 56, 106–114, doi: 10.1111/j.1753-4887.1998.tb01722.x. [DOI] [PubMed] [Google Scholar]
- 3.Deputy NP; Dub B; Sharma AJ Prevalence and Trends in Prepregnancy Normal Weight — 48 States, New York City, and District of Columbia, 2011–2015. MMWR. Morb. Mortal. Wkly. Rep 2018, 66, 1402–1407, doi: 10.15585/mmwr.mm665152a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Statistics Canada Women in Canada: A gender-based statistical report (89–503-X); 2012–2013 Canadian Health Measures Survey, custom tabulation 2013. [Google Scholar]
- 5.Czernichow S; Kengne A-P; Huxley RR; Batty GD; de Galan B; Grobbee D; Pillai A; Zoungas S; Marre M; Woodward M; et al. Comparison of waist-to-hip ratio and other obesity indices as predictors of cardiovascular disease risk in people with type-2 diabetes: a prospective cohort study from ADVANCE. Eur. J. Cardiovasc. Prev. Rehabil 2011, 18, 312–319, doi: 10.1097/HJR.0b013e32833c1aa3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitaker RC Predicting Preschooler Obesity at Birth: The Role of Maternal Obesity in Early Pregnancy. Pediatrics 2004, 114, e29–e36, doi: 10.1542/peds.114.1.e29. [DOI] [PubMed] [Google Scholar]
- 7.Heerwagen MJR; Miller MR; Barbour LA; Friedman JE Maternal obesity and fetal metabolic programming: a fertile epigenetic soil. AJP Regul. Integr. Comp. Physiol 2010, 299, R711–R722, doi: 10.1152/ajpregu.00310.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boney CM; Verma A; Tucker R; Vohr BR Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics 2005, 115, e290–6, doi: 10.1542/peds.2004-1808. [DOI] [PubMed] [Google Scholar]
- 9.Borengasser SJ; Kang P; Faske J; Gomez-Acevedo H; Blackburn ML; Badger TM; Shankar K. High Fat Diet and In Utero Exposure to Maternal Obesity Disrupts Circadian Rhythm and Leads to Metabolic Programming of Liver in Rat Offspring. PLoS One 2014, 9, e84209, doi: 10.1371/journal.pone.0084209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan EL; Grove KL Metabolic Imprinting in Obesity. In Frontiers in Eating and Weight Regulation; KARGER: Basel, 2009; pp. 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonnold M; Mele L; Myatt L; Hauth J; Leveno K; Reddy U; Mercer B. Waist-to-Hip Ratio versus Body Mass Index as Predictor of Obesity-Related Pregnancy Outcomes. Am. J. Perinatol 2016, 33, 618–624, doi: 10.1055/s-0035-1569986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss R; Bremer AA; Lustig RH What is metabolic syndrome, and why are children getting it? Ann. N. Y. Acad. Sci 2013, 1281, 123–140, doi: 10.1111/nyas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poyrazoglu S; Bas F; Darendeliler F. Metabolic syndrome in young people. Curr. Opin. Endocrinol. Diabetes Obes 2014, 21, 56–63, doi: 10.1097/01.med.0000436414.90240.2c. [DOI] [PubMed] [Google Scholar]
- 14.Borengasser SJ; Faske J; Kang P; Blackburn ML; Badger TM; Shankar K. In utero exposure to prepregnancy maternal obesity and postweaning high-fat diet impair regulators of mitochondrial dynamics in rat placenta and offspring. Physiol. Genomics 2014, 46, 841–850, doi: 10.1152/physiolgenomics.00059.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myatt L. Placental adaptive responses and fetal programming. J. Physiol 2006, 572, 25–30, doi: 10.1113/jphysiol.2006.104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornburg KL; O’Tierney PF; Louey S. Review: The Placenta is a Programming Agent for Cardiovascular Disease. Placenta 2010, 31, S54–S59, doi: 10.1016/j.placenta.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maloyan A; Mele J; Muralimanoharan S; Myatt L. Placental metabolic flexibility is affected by maternal obesity. Placenta 2016, 45, 69, doi: 10.1016/j.placenta.2016.06.031. [DOI] [Google Scholar]
- 18.Ireland KE; Maloyan A; Myatt L. Melatonin Improves Mitochondrial Respiration in Syncytiotrophoblasts From Placentas of Obese Women. Reprod. Sci 2018, 25, 120–130, doi: 10.1177/1933719117704908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mele J; Muralimanoharan S; Maloyan A; Myatt L. Impaired mitochondrial function in human placenta with increased maternal adiposity. Am. J. Physiol. Endocrinol. Metab 2014, 307, E419–25, doi: 10.1152/ajpendo.00025.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hastie R; Lappas M. The effect of pre-existing maternal obesity and diabetes on placental mitochondrial content and electron transport chain activity. Placenta 2014, 35, 673–83, doi: 10.1016/j.placenta.2014.06.368. [DOI] [PubMed] [Google Scholar]
- 21.Gude NM; Roberts CT; Kalionis B; King RG Growth and function of the normal human placenta. Thromb. Res 2004, 114, 397–407, doi: 10.1016/j.thromres.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 22.Osadnik K; Osadnik T; Lonnie M; Lejawa M; Reguła R; Fronczek M; Gawlita M; Wądołowska L; Gąsior M; Pawlas N. Metabolically healthy obese and metabolic syndrome of the lean: the importance of diet quality. Analysis of MAGNETIC cohort. Nutr. J 2020, 19, 19, doi: 10.1186/s12937-020-00532-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popkin BM Global nutrition dynamics: the world is shifting rapidly toward a diet linked with noncommunicable diseases. Am. J. Clin. Nutr 2006, 84, 289–298, doi: 10.1093/ajcn/84.2.289. [DOI] [PubMed] [Google Scholar]
- 24.Savard C; Lemieux S; Weisnagel S; Fontaine-Bisson B; Gagnon C; Robitaille J; Morisset A-S Trimester-Specific Dietary Intakes in a Sample of French-Canadian Pregnant Women in Comparison with National Nutritional Guidelines. Nutrients 2018, 10, 768, doi: 10.3390/nu10060768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watts V; Rockett H; Baer H; Leppert J; Colditz G. Assessing Diet Quality in a Population of Low-Income Pregnant Women: A Comparison Between Native Americans and Whites. Matern. Child Health J 2007, 11, 127–136, doi: 10.1007/s10995-006-0155-2. [DOI] [PubMed] [Google Scholar]
- 26.Denomme J; Stark KD; Holub BJ Directly quantitated dietary (n-3) fatty acid intakes of pregnant Canadian women are lower than current dietary recommendations. J. Nutr 2005, 135, 206–11, doi:135/2/206. [DOI] [PubMed] [Google Scholar]
- 27.Siega-Riz AM; Bodnar LM; Savitz DA What are pregnant women eating? Nutrient and food group differences by race. Am. J. Obstet. Gynecol 2002, 186, 480–486, doi: 10.1067/mob.2002.121078. [DOI] [PubMed] [Google Scholar]
- 28.Chen X; Scholl TO; Leskiw M; Savaille J; Stein TP Differences in maternal circulating fatty acid composition and dietary fat intake in women with gestational diabetes mellitus or mild gestational hyperglycemia. Diabetes Care 2010, 33, 2049–2054, doi: 10.2337/dc10-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon ES Non-Esterified Fatty Acids in the Blood of Obese and Lean Subjects. Am. J. Clin. Nutr 1960, 8, 740–747, doi: 10.1093/ajcn/8.5.740. [DOI] [Google Scholar]
- 30.Merzouk H; Meghelli-Bouchenak M; Loukidi B; Prost J; Belleville J. Impaired Serum Lipids and Lipoproteins in Fetal Macrosomia Related to Maternal Obesity. Neonatology 2000, 77, 17–24, doi: 10.1159/000014190. [DOI] [PubMed] [Google Scholar]
- 31.Kien CL; Bunn JY; Stevens R; Bain J; Ikayeva O; Crain K; Koves TR; Muoio DM Dietary intake of palmitate and oleate has broad impact on systemic and tissue lipid profiles in humans. Am. J. Clin. Nutr 2014, 99, 436–445, doi: 10.3945/ajcn.113.070557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kien CL; Everingham KI; Stevens RD; Fukagawa NK; Muoio DM Short-Term Effects of Dietary Fatty Acids on Muscle Lipid Composition and Serum Acylcarnitine Profile in Human Subjects. Obesity 2011, 19, 305–311, doi: 10.1038/oby.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon B; Lee H-K; Querfurth HW Oleate prevents palmitate-induced mitochondrial dysfunction, insulin resistance and inflammatory signaling in neuronal cells. Biochim. Biophys. Acta - Mol. Cell Res 2014, 1843, 1402–1413, doi: 10.1016/j.bbamcr.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Yuzefovych L; Wilson G; Rachek L. Different effects of oleate vs. palmitate on mitochondrial function, apoptosis, and insulin signaling in L6 skeletal muscle cells: role of oxidative stress. Am. J. Physiol. Metab 2010, 299, E1096–E1105, doi: 10.1152/ajpendo.00238.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itami N; Shirasuna K; Kuwayama T; Iwata H. Palmitic acid induces ceramide accumulation, mitochondrial protein hyperacetylation, and mitochondrial dysfunction in porcine oocytes†. Biol. Reprod 2018, 98, 644–653, doi: 10.1093/biolre/ioy023. [DOI] [PubMed] [Google Scholar]
- 36.McCurdy CE; Bishop JM; Williams SM; Grayson BE; Smith MS; Friedman JE; Grove KL Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J. Clin. Invest 2009, 119, 323–335, doi: 10.1172/JCI32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wesolowski SR; Mulligan CM; Janssen RC; Baker PR; Bergman BC; D’Alessandro A; Nemkov T; Maclean KN; Jiang H; Dean TA; et al. Switching obese mothers to a healthy diet improves fetal hypoxemia, hepatic metabolites, and lipotoxicity in non-human primates. Mol. Metab 2018, 18, 25–41, doi: 10.1016/j.molmet.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salati JA; Roberts VHJ; Schabel MC; Lo JO; Kroenke CD; Lewandowski KS; Lindner JR; Grove KL; Frias AE Maternal high-fat diet reversal improves placental hemodynamics in a nonhuman primate model of diet-induced obesity. Int. J. Obes 2019, 43, 906–916, doi: 10.1038/s41366-018-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pound LD; Comstock SM; Grove KL Consumption of a Western-style diet during pregnancy impairs offspring islet vascularization in a Japanese macaque model. Am. J. Physiol. Metab 2014, 307, E115–E123, doi: 10.1152/ajpendo.00131.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hulme CH; Stevens A; Dunn W; Heazell AEP; Hollywood K; Begley P; Westwood M; Myers JE Identification of the functional pathways altered by placental cell exposure to high glucose: lessons from the transcript and metabolite interactome. Sci. Rep 2018, 8, 5270, doi: 10.1038/s41598-018-22535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tobin KAR; Johnsen GM; Staff AC; Duttaroy AK Long-chain Polyunsaturated Fatty Acid Transport across Human Placental Choriocarcinoma (BeWo) Cells. Placenta 2009, 30, 41–47, doi: 10.1016/j.placenta.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Ogura K; Sakata M; Okamoto Y; Yasui Y; Tadokoro C; Yoshimoto Y; Yamaguchi M; Kurachi H; Maeda T; Murata Y. 8-bromo-cyclicAMP stimulates glucose transporter-1 expression in a human choriocarcinoma cell line. J. Endocrinol 2000, 164, 171–8. [DOI] [PubMed] [Google Scholar]
- 43.Wice B; Menton D; Geuze H; Schwartz AL Modulators of cyclic AMP metabolism induce syncytiotrophoblast formation in vitro. Exp. Cell Res 1990, 186, 306–316, doi: 10.1016/0014-4827(90)90310-7. [DOI] [PubMed] [Google Scholar]
- 44.Pang DKT; Nong Z; Sutherland BG; Sawyez CG; Robson DL; Toma J; Pickering JG; Borradaile NM Niacin promotes revascularization and recovery of limb function in diet-induced obese mice with peripheral ischemia. Pharmacol. Res. Perspect 2016, 4, e00233, doi: 10.1002/prp2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang C; Tang L; Wu X; Xiong W; Ruan H; Hussain M; Wu J; Zou C; Wu X. Glioma-associated Oncogene 2 Is Essential for Trophoblastic Fusion by Forming a Transcriptional Complex with Glial Cell Missing-a. J. Biol. Chem 2016, 291, 5611–5622, doi: 10.1074/jbc.M115.700336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malhotra SS; Suman P; Kumar Gupta S. Alpha or beta human chorionic gonadotropin knockdown decrease BeWo cell fusion by down-regulating PKA and CREB activation. Sci. Rep 2015, 5, 11210, doi: 10.1038/srep11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kusama K; Bai R; Imakawa K. Regulation of human trophoblast cell syncytialization by transcription factors STAT5B and NR4A3. J. Cell. Biochem 2018, 119, 4918–4927, doi: 10.1002/jcb.26721. [DOI] [PubMed] [Google Scholar]
- 48.Wolvetang EJ; Johnson KL; Krauer K; Ralph SJ; Linnane AW Mitochondrial respiratory chain inhibitors induce apoptosis. FEBS Lett. 1994, 339, 40–44, doi: 10.1016/0014-5793(94)80380-3. [DOI] [PubMed] [Google Scholar]
- 49.Starkov AA; Simonyan RA; Dedukhova VI; Mansurova SE; Palamarchuk LA; Skulachev VP Regulation of the energy coupling in mitochondria by some steroid and thyroid hormones. Biochim. Biophys. Acta - Bioenerg 1997, 1318, 173–183, doi: 10.1016/S0005-2728(96)00135-1. [DOI] [PubMed] [Google Scholar]
- 50.Jeschke U; Richter D-U; Möbius B-M; Briese V; Mylonas I; Friese K. Stimulation of progesterone, estradiol and cortisol in trophoblast tumor bewo cells by glycodelin A N-glycans. Anticancer Res. 2007, 27, 2101–8. [PubMed] [Google Scholar]
- 51.Downs TR; Wilfinger WW Fluorometric quantification of DNA in cells and tissue. Anal. Biochem 1983, 131, 538–47. [DOI] [PubMed] [Google Scholar]
- 52.Muschet C; Möller G; Prehn C; de Angelis MH; Adamski J; Tokarz J. Removing the bottlenecks of cell culture metabolomics: fast normalization procedure, correlation of metabolites to cell number, and impact of the cell harvesting method. Metabolomics 2016, 12, 151, doi: 10.1007/s11306-016-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thacker JS; Yeung DH; Staines WR; Mielke JG Total protein or high-abundance protein: Which offers the best loading control for Western blotting? Anal. Biochem 2016, 496, 76–78, doi: 10.1016/j.ab.2015.11.022. [DOI] [PubMed] [Google Scholar]
- 54.Kirby DM; Thorburn DR; Turnbull DM; Taylor RW Biochemical Assays of Respiratory Chain Complex Activity. In; 2007; pp. 93–119. [DOI] [PubMed] [Google Scholar]
- 55.Davies R; Mathers KE; Hume AD; Bremer K; Wang Y; Moyes CD Hybridization in Sunfish Influences the Muscle Metabolic Phenotype. Physiol. Biochem. Zool 2012, 85, 321–331, doi: 10.1086/666058. [DOI] [PubMed] [Google Scholar]
- 56.Mathers KE; Staples JF Saponin-permeabilization is not a viable alternative to isolated mitochondria for assessing oxidative metabolism in hibernation. Biol. Open 2015, 4, 858–864, doi: 10.1242/bio.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price ER; McFarlan JT; Guglielmo CG Preparing for Migration? The Effects of Photoperiod and Exercise on Muscle Oxidative Enzymes, Lipid Transporters, and Phospholipids in White-Crowned Sparrows. Physiol. Biochem. Zool 2010, 83, 252–262, doi: 10.1086/605394. [DOI] [PubMed] [Google Scholar]
- 58.Ke C-J; He Y-H; He H-W; Yang X; Li R; Yuan J. A new spectrophotometric assay for measuring pyruvate dehydrogenase complex activity: a comparative evaluation. Anal. Methods 2014, 6, 6381–6388, doi: 10.1039/C4AY00804A. [DOI] [Google Scholar]
- 59.Lew M. Good statistical practice in pharmacology Problem 2. Br. J. Pharmacol 2009, 152, 299–303, doi: 10.1038/sj.bjp.0707372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colvin BN; Longtine MS; Chen B; Costa ML; Nelson DM Oleate attenuates palmitate-induced endoplasmic reticulum stress and apoptosis in placental trophoblasts. Reproduction 2017, 153, 369–380, doi: 10.1530/REP-16-0576. [DOI] [PubMed] [Google Scholar]
- 61.Kolahi K; Valent A; Thornburg KL Cytotrophoblast, Not Syncytiotrophoblast, Dominates Glycolysis and Oxidative Phosphorylation in Human Term Placenta. Sci. Rep 2017, 1–12, doi: 10.1038/srep42941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sunny NE; Parks EJ; Browning JD; Burgess SC Excessive Hepatic Mitochondrial TCA Cycle and Gluconeogenesis in Humans with Nonalcoholic Fatty Liver Disease. Cell Metab. 2011, 14, 804–810, doi: 10.1016/j.cmet.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palmeira CM; Ferreira FML; Santos DL; Ceiça R; Suzuki K; Santos MS Higher efficiency of the liver phosphorylative system in diabetic Goto-Kakizaki (GK) rats. FEBS Lett. 1999, 458, 103–106, doi: 10.1016/S0014-5793(99)01144-8. [DOI] [PubMed] [Google Scholar]
- 64.Wei Y; Rector RS; Thyfault JP; Ibdah JA Nonalcoholic fatty liver disease and mitochondrial dysfunction. World J. Gastroenterol 2008, 14, 193, doi: 10.3748/wjg.14.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koyama T; Kume S; Koya D; Araki S; Isshiki K; Chin-Kanasaki M; Sugimoto T; Haneda M; Sugaya T; Kashiwagi A; et al. SIRT3 attenuates palmitate-induced ROS production and inflammation in proximal tubular cells. Free Radic. Biol. Med 2011, 51, 1258–1267, doi: 10.1016/j.freeradbiomed.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 66.Liu J; Chang F; Li F; Fu H; Wang J; Zhang S; Zhao J; Yin D. Palmitate promotes autophagy and apoptosis through ROS-dependent JNK and p38 MAPK. Biochem. Biophys. Res. Commun 2015, 463, 262–267, doi: 10.1016/j.bbrc.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 67.Miller TA; LeBrasseur NK; Cote GM; Trucillo MP; Pimentel DR; Ido Y; Ruderman NB; Sawyer DB Oleate prevents palmitate-induced cytotoxic stress in cardiac myocytes. Biochem. Biophys. Res. Commun 2005, 336, 309–315, doi: 10.1016/j.bbrc.2005.08.088. [DOI] [PubMed] [Google Scholar]
- 68.Malti N; Merzouk H; Merzouk SA; Loukidi B; Karaouzene N; Malti A; Narce M. Oxidative stress and maternal obesity: Feto-placental unit interaction. Placenta 2014, 35, 411–416, doi: 10.1016/j.placenta.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 69.Roberts VHJ; Smith J; McLea SA; Heizer AB; Richardson JL; Myatt L. Effect of Increasing Maternal Body Mass Index on Oxidative and Nitrative Stress in The Human Placenta. Placenta 2009, 30, 169–175, doi: 10.1016/j.placenta.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sommerweiss D; Gorski T; Richter S; Garten A; Kiess W. Oleate rescues INS-1E β-cells from palmitate-induced apoptosis by preventing activation of the unfolded protein response. Biochem. Biophys. Res. Commun 2013, 441, 770–776, doi: 10.1016/j.bbrc.2013.10.130. [DOI] [PubMed] [Google Scholar]
- 71.Abaidoo C; Warren MA; Andrews PW; Boateng KA A Quantitative Assessment of the Morphological Characteristics of BeWo Cells as an in vitro Model of Human Trophoblast Cells. Int. J. Morphol 2010, 28, 1047–1058, doi: 10.4067/S0717-95022010000400011. [DOI] [Google Scholar]
- 72.Mayhew TM Villous trophoblast of human placenta: a coherent view of its turnover, repair and contributions to villous development and maturation. Histol. Histopathol 2001, 16, 1213–24, doi: 10.14670/HH-16.1213. [DOI] [PubMed] [Google Scholar]
- 73.Huppertz B; Kingdom JCP Apoptosis in the Trophoblast—Role of Apoptosis in Placental Morphogenesis. J. Soc. Gynecol. Investig 2004, 11, 353–362, doi: 10.1016/j.jsgi.2004.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Representative Seahorse Assay Tracings. Representative tracing of Oxygen Consumption Rate (OCR) throughout experimental assay for (A) BeWo CT cells and (B) BeWo SCT cells from the Mito Stress Test. Representative tracing of Extracellular Acidification Rate (ECAR) throughout experimental assay for (C) BeWo CT cells and (D) BeWo SCT cells from the Glycolysis Stress Test. Data is represented as mean OCR or ECAR of each group in technical duplicate from one experiment and presented as percent CT untreated control.
Supplementary Figure 2. Full length representative blots of oxidative stress markers. Representative images of protein bands obtained in the immunoblots for (A) 4-hydroxynonenal and (B) nitro-tyrosine. Left-to-right represents the treatments PA CT, PA SCT, OA CT, OA SCT, P/O CT, P/O SCT, BSA CT, BSA SCT.
Supplementary Figure 3. Representative images of immunoblot ponceau loading controls. Representative full length ponceau loading control bands for (A) Mitoprofile, (B) Pyruvate Dehydrogenase, (C) PDH-E1α (pSer232), (D) Pyruvate Dehydrogenase Kinase 1, (E) Lactate Dehydrogenase, (F) Carnitine Palmitoyltransferase 1a, (G) 4-Hydroxynonenal, and (H) Nitro-tyrosine immunoblots. Left-to-right represents the treatments PA CT, PA SCT, OA CT, OA SCT, P/O CT, P/O SCT, BSA CT, BSA SCT.